NENA was identified by a genetic screen for Lotus japonicus mutants impaired in arbuscular mycorrhiza and encodes a scaffold nucleoporin. nena mutants are also impaired in rhizobial root hair infection but crack entry leading to fully infected root nodules can overcome the rhizodermal nonresponsiveness of nena-1 to Nod factor.
Abstract
Legumes form symbioses with arbuscular mycorrhiza (AM) fungi and nitrogen fixing root nodule bacteria. Intracellular root infection by either endosymbiont is controlled by the activation of the calcium and calmodulin-dependent kinase (CCaMK), a central regulatory component of the plant’s common symbiosis signaling network. We performed a microscopy screen for Lotus japonicus mutants defective in AM development and isolated a mutant, nena, that aborted fungal infection in the rhizodermis. NENA encodes a WD40 repeat protein related to the nucleoporins Sec13 and Seh1. Localization of NENA to the nuclear rim and yeast two-hybrid experiments indicated a role for NENA in a conserved subcomplex of the nuclear pore scaffold. Although nena mutants were able to form pink nodules in symbiosis with Mesorhizobium loti, root hair infection was not observed. Moreover, Nod factor induction of the symbiotic genes NIN, SbtM4, and SbtS, as well as perinuclear calcium spiking, were impaired. Detailed phenotypic analyses of nena mutants revealed a rhizobial infection mode that overcame the lack of rhizodermal responsiveness and carried the hallmarks of crack entry, including a requirement for ethylene. CCaMK-dependent processes were only abolished in the rhizodermis but not in the cortex of nena mutants. These data support the concept of tissue-specific components for the activation of CCaMK.
INTRODUCTION
Arbuscular mycorrhiza (AM) is an ancient endosymbiosis between fungi of the phylum Glomeromycota (Schüssler et al., 2001) and land plants. The ubiquity of AM provides evidence for the advantage on the symbiotic partners of exchanging plant-derived carbohydrates for mineral nutrients provided by the fungus (Finlay, 2008). Root nodule symbiosis (RNS) with nitrogen-fixing bacteria, on the other hand, occurs only in a monophyletic clade within the eudicots and therefore must have evolved in a common ancestor later (Soltis et al., 1995). Both symbioses share striking similarities in the mechanisms leading to the accommodation of the respective endosymbiont.
Presymbiotic crosstalk between rhizobia and legumes leads to rhizobial production of lipochito-oligosaccharide Nod factor (NF) molecules, which induce physiological and morphological responses in root hairs, including oscillation of perinuclear calcium concentrations (Ca2+ spiking) (Ehrhardt et al., 1996), induction of symbiotic genes, and root hair curling (RHC) around entrapped rhizobia (Oldroyd and Downie, 2008). Intracellular infection threads (ITs) initiate from the center of RHC and guide the rhizobia through local cell wall decomposition and invagination of the plasma membrane toward subepidermal cells (Oldroyd and Downie, 2008). Further cortical infection is preceded by the formation of cytoplasmic bridges termed pre-ITs (van Brussel et al., 1992). By comparison, yet uncharacterized diffusible fungal factors were shown to induce the symbiotic gene ENOD11 (Kosuta et al., 2003) and Ca2+ spiking in rhizodermal cells during the presymbiotic stage of AM (Kosuta et al., 2008). After physical contact of the symbionts, passage of the hyphae through the outer cell layers to the cortex of the root is preceded by the prepenetration apparatus (PPA), a tubular rearrangement of the cytoskeleton and the endoplasmic reticulum that determines the route of intracellular infection through rhizodermal and cortical cells (Genre et al., 2005, 2008), reminiscent of (pre-)ITs.
Accordingly, genetic dissection of RNS using model legumes, including Lotus japonicus (Lotus) and Medicago truncatula, revealed host genes that turned out to be equally important in AM. This led to the concept of a shared developmental program controlled by common SYM genes, which has been adopted from AM during the evolution of RNS (Duc et al., 1989; La Rue and Weeden, 1994; Kistner and Parniske, 2002). Recognition of NFs by LysM domain receptor-like kinases NFR1 and NFR5/NFP (Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006) elicits signal transduction via SYMRK/DMI2/NORK (Endre et al., 2002; Stracke et al., 2002), a receptor kinase with extracellular Leu-rich repeats and the convergence point with AM-induced signal transduction. Unknown downstream events depend on two putative nuclear pore proteins (nucleoporins), NUP133 and NUP85 (Kanamori et al., 2006; Saito et al., 2007), and presumably lead to membrane potential alterations at the nuclear envelope involving the ion channels CASTOR and POLLUX/DMI1 (Ané et al., 2004; Imaizumi-Anraku et al., 2005; Charpentier et al., 2008) required for Ca2+ spiking. The Ca2+ and calmodulin-dependent kinase (CCaMK)/DMI3 (Lévy et al., 2004; Mitra et al., 2004) in cooperation with the nuclear protein CYCLOPS/IPD3 (Messinese et al., 2007; Yano et al., 2008) may act as decoder of Ca2+ spiking. The CCaMK-CYCLOPS complex regulates rhizobial IT development, which requires downstream activation of RNS-specific GRAS (NSP1 and NSP2) (Kaló et al., 2005; Smit et al., 2005; Heckmann et al., 2006; Murakami et al., 2006) and AP2-ERF (ERN1 to ERN3) transcription factors (Andriankaja et al., 2007; Middleton et al., 2007). These can bind to cis-regulatory elements of early nodulins, including ENOD11 and NIN, and thereby regulate target gene expression in response to NF (Andriankaja et al., 2007; Hirsch et al., 2009). NIN itself contains domains related to transcription factors as well as predicted transmembrane domains. Like NSP1 and NSP2, NIN is essential for rhizobial infection (Schauser et al., 1999).
In addition to endosymbiont accommodation, RNS involves formation of the root nodule, which provides the environment for bacterial nitrogenase activity by the expression of oxygen binding leghemoglobin (Ott et al., 2005). Organogenesis is tightly coordinated with the progression of infection and endocytosis of rhizobia into the nodule cortical cells, where they mature into nitrogen-fixing bacteroids inside organella-like symbiosomes (Van de Velde et al., 2010). The epistatic nature of genes required for early NF signaling potentially masks tissue-specific processes. Gain-of-function mutations in CCaMK (Gleason et al., 2006; Tirichine et al., 2006) and the cytokinin receptor LHK1 (Murray et al., 2007; Tirichine et al., 2007) lead to nodulation in the absence of rhizobia, thereby uncoupling infection from nodule formation. NIN and the NSP1 and NSP2 transcription factors are proposed candidates for coordinating infection with nodule formation, since they are required during NF signaling and for nodulation in autoactive CCaMK mutants (Gleason et al., 2006; Marsh et al., 2007). Natural variation of RNS also provides insights into different prerequisites for infection initiation and nodule development. Aquatic and semiaquatic legumes from tropical and subtropical regions have an intercellular infection mode that does not require intracellular entry through RHC but uses rhizodermal cracks at sites of lateral root emergence (Ndoye et al., 1994). Subsequently, rhizobia proliferate in subepidermal infection pockets that are caused by local apoptosis promoted by reactive oxygen species and ethylene (D'Haeze et al., 2003). From there on, infection proceeds inter- and intracellularly and concludes in the release of bacteroids into cortical cells of the nodule. Some legumes (e.g., Sesbania rostrata) can switch between intracellular infection during aerated conditions and crack entry during root submergence (Goormachtig et al., 2004).
AM does not involve the formation of a new plant organ. After intracellular passage through the outer cell layers, fungal hyphae enter the apoplast and extensively colonize the root cortex. Branches of apoplastic hyphae enter into cells of the inner root cortex, where highly ramified tree-like structures, the so-called arbuscules, that create a large surface area for nutrient exchange, are formed (Harrison, 2005). The phenotype of loss-of-function TILLING mutants of the AM-induced PT4 gene from M. truncatula (Javot et al., 2007) showed that phosphate transport via the arbuscule is indispensable for full arbuscule development and maintenance of the root-fungal association. Disruption of common SYM genes typically results in the abortion of fungal infection in the epidermal root layer (Kistner et al., 2005). In addition, dissection of AM development by forward genetic screens in tomato (Solanum lycopersicum), maize (Zea mays), or petunia (Petunia hybrida) discerned two additional stages (before fungal infection and during root colonization) that are controlled by the plant (Barker et al., 1998; David-Schwartz et al., 2001; Paszkowski et al., 2006; Reddy et al., 2007). We initiated a genetic screen for AM mutants in Lotus and identified the gene NENA, encoding a WD40 repeat nucleoporin required for AM fungal infection and RNS. Detailed phenotypic analysis of nena mutants revealed that nodule organogenesis is unaffected and intercellular rhizobial infection can overcome the mutants’ rhizodermal symbiotic defects.
RESULTS
Identification of nena by a Genetic Screen for AM Mutants
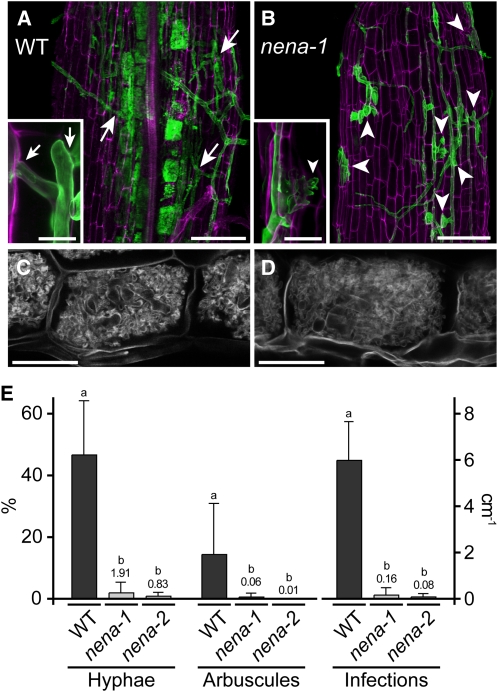
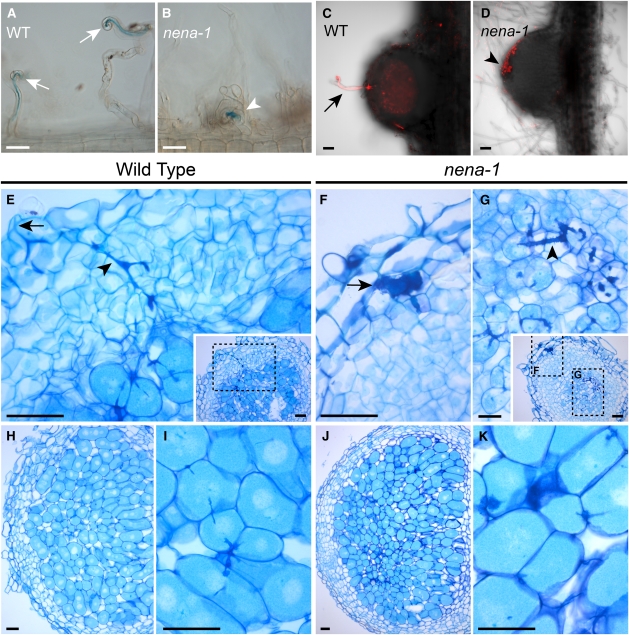
An ethyl methanesulfonate–mutagenized population of L. japonicus ecotype B-129 (Gifu) (Perry et al., 2003) was subjected to a genetic screen for mutations affecting AM symbiosis. It comprises 2204 separately propagated M3 families with distinct mutational backgrounds (SL lines) and shares common origin with the Lotus general TILLING population (Perry et al., 2009). Among the mutants isolated that exhibited heritable AM defects, we obtained one family (SL1841-N) containing individuals with a severe and early defect during AM development. In the course of this work, the causative mutation has been identified (see below), and the corresponding mutant allele was named nena-1. When cultivated together with G. intraradices-like BEG195 (Stockinger et al., 2009), nena-1 mutants displayed balloon-shaped swollen hyphal structures that had formed at sites of attempted fungal penetration of the root surface (Figure 1B). These structures were not observed on wild-type plants (Figure 1A). Confocal laser scanning microscopy (CLSM) revealed that AM fungal infection of nena-1 mutants was aborted after invasion between rhizodermal cells at the stage of intracellular passage through the outer root layers (inset in Figure 1B; see Supplemental Figure 1B online). This stage is essential for the succeeding colonization in the wild type (inset in Figure 1A; see Supplemental Figure 1A online). Consequently, roots of nena-1 mutants that were cocultivated with BEG195 at 24°C were nearly void of internal hyphae (Figure 1E). Successful infection of nena-1 was observed at low frequency. In these cases, hyphae traversed the rhizodermal layer despite obvious obstructions (see Supplemental Figure 1C online) and led to cortical colonization and formation of arbuscules that were indistinguishable from the wild type (Figures 1C and 1D).
Figure 1.
nena Is Impaired in AM Fungal Infection.
(A) to (D) Confocal micrographs of WGA-Alexa Fluor 488-stained AM fungal structures (green in [A] and [B]) associated with wild-type (WT) and nena plants. In the wild type (A), the AM fungus penetrated the outer cell layers (arrows), colonized the root cortex, and formed arbuscules. In nena-1 (B), hyphae grew on the root surface, and balloon-shaped hyphal structures (arrowheads) occurred at aborted infection sites. Insets show infection sites at higher magnification. Root cell walls were stained with propidium iodide and are shown in magenta. Arbuscules formed in nena-2 (D), did not differ from the wild type (C). AM phenotypes of nena-1 and nena-2 did not differ. Images represent observations from more than eight plants per line cocultivated with BEG195 for 3 weeks. (A) and (B) show Z-projections of GFP/RFP overlays; (C) and (D) show the GFP channel. Bars = 100 μm in (A) to (D) and 20 μm in the insets.
(E) Mean hyphal colonization (Hyphae, %), arbuscular colonization (Arbuscules, %) per root, and successful infection sites per centimeter per root (Infections) from wild-type and nena plants (n ≥ 4) after 3 weeks of cultivation at 24°C. Small values are shown by numbers above bars. Error bars show sd. Different letters above bars indicate significant differences (P ≤ 0.05, t test) between pairwise comparisons.
nena Mutants Have Temperature-Dependent Defects in AM and RNS
The AM phenotype of nena mutants was assessed at different growth temperatures. The initial AM phenotypic analyses were performed at 24°C, and the observations (Figure 1E) differed strongly to the AM phenotype obtained at 18°C growth temperature (see Supplemental Figures 6A and 6B online). The frequency of successful infections (1.36 ± 0.14 per cm root tissue) in nena-2 cultivated at 18°C approached wild-type levels (2.24 ± 0.62), although balloon-like hyphal structures were still present. Consequently, average hyphal and arbuscule colonization of nena-2 did not differ significantly from the wild type.
Aborted AM fungal infection displayed by nena was similar to AM phenotypes of common sym mutants (Kistner et al., 2005). Therefore, nena mutants were inoculated with Mesorhizobium loti strains that are compatible with wild-type L. japonicus. The nodulation assays at different temperatures revealed that nena-1, nena-2, and nena-3 formed fewer nodules than wild-type plants and that this reduction was also stronger at 24°C compared with 18 C° (see Supplemental Figure 6C online). Temperature-dependent defects in nodulation, AM colonization, and arbuscule formation have previously been described for nup133 and nup85 Lotus mutants (Kanamori et al., 2006; Saito et al., 2007).
Map-Based Cloning of NENA
To assess the phenotypic segregation and identify the causative mutation by map-based cloning, a mutant M3 individual from SL1841-N was crossed to the polymorphic mapping parent L. japonicus ecotype MG-20 (Miyakojima) (Kawaguchi et al., 2001), and F2 mapping populations were generated from self-progeny. From these, 75 out of 276 phenotyped F2 individuals were scored as AM defective, matching the segregation of a monogenic recessive trait (χ2 probability = 0.40).
After reducing the nena locus to <150 kb by map-based cloning (see Supplemental Figure 2 and Supplemental Methods online), bioinformatic analysis of the sequence within the target region annotated 32 NENA candidate genes. Due to the interaction of yeast proteins Sc Seh1 and Sc Nup85 (Brohawn et al., 2008; Debler et al., 2008) and the previous identification of the symbiosis gene NUP85 in Lotus (Saito et al., 2007), a SEH1-like annotated gene was picked out as a candidate. The candidate gene was sequenced and a C-T transition leading to a premature stop codon at amino acid 87 of the putative 326–amino acid (35.5 kD) protein was identified in the nena-1 mutant (see Supplemental Figure 3 and Supplemental Table 1 online).
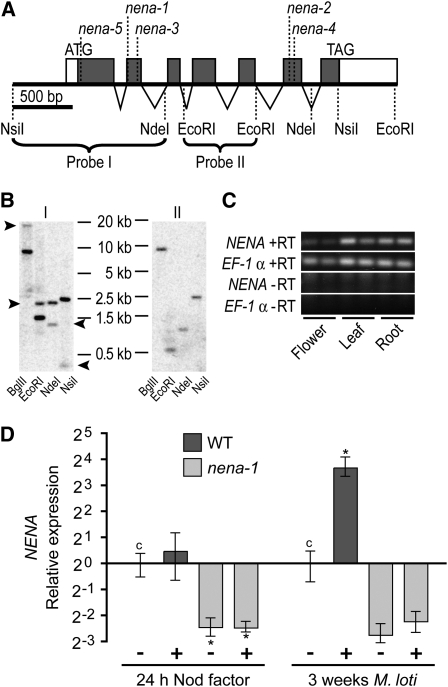
The predicted NENA gene has an open reading frame (ORF) of 2023 nucleotides that is composed of seven exons (Figure 2A). The gene structure has been verified by sequencing cDNA clones obtained from the Lotus Resource Center (Asamizu et al., 2000). The cDNA sequences carried 5′ and 3′ untranslated regions of 57 and 429 bp, respectively. The cosegregating marker SSR17 is located directly 5′ of the start codon of NENA.
Figure 2.
NENA Is a Single Copy Gene That Is Expressed in Various Tissues and Upregulated in Nodulated Roots.
(A) Gene structure of NENA. Closed boxes, open boxes, and triangles represent coding exons, untranslated regions, and introns, respectively. Positions of start and stop codons of the NENA ORF, mutations in different nena alleles, and restriction sites of selected endonucleases are indicated. Braces span the restriction fragments used as probes in DNA gel blot analyses, as referred to in (B).
(B) DNA gel blot radiographs of L. japonicus genomic DNA digested with BglII, EcoRI, NdeI, or NsiI hybridized with Probe I or II. Arrowheads mark bands that do not correspond to the genomic context of NENA but are due to partial gene duplication.
(C) Expression of NENA in leaves, flowers, and roots (two biological replicates) analyzed by RT-PCR. NENA and the reference gene EF-1α were amplified with (+RT) or without (−RT) preceding reverse transcription.
(D) Quantitative PCR analysis of NENA expression in wild-type (WT) and nena-1 roots 24 h after NF treatment or 3 weeks after M. loti inoculation (+). Expression is relative to mock (−) treated wild-type controls (c) and normalized to EF-1α levels. Mean values and se were derived from three biological replicates. Asterisks indicate significant (P < 0.05) differences to control levels.
BLAST analysis of the L. japonicus genome revealed a partial duplication of the NENA gene, ψNENA, which aligns to the first 409 nucleotides of the NENA ORF with 94% sequence identity. ψNENA has an ORF of 488 bp and has not been physically mapped yet. Interrogation of public EST and protein databases gave no indication for the expression of ψNENA. In addition, RT-PCR analyses on samples from different Lotus tissues did not show any corresponding transcripts (data not shown). Therefore, ψNENA most likely represents a pseudogene.
We confirmed the obtained in silico data by genomic DNA gel blot analysis with two probes corresponding to 1.1 kb 5′ sequence (I) and 0.5 kb from the center (II) of NENA (Figure 2B). Probe I showed two hybridization bands per genomic digest, and the patterns were as predicted by the genomic sequences surrounding NENA and ψNENA. Probe II yielded one band per digest corresponding to the predicted NENA fragments.
Using the Lotus TILLING resource (Perry et al., 2003), we obtained an allelic series for NENA that includes one additional nonsense allele (nena-2, W257Stop) and various alleles with missense mutations (nena-3 to -5; see Supplemental Figure 3 and Supplemental Table 1 online). nena-6 was identified by forward genetics using a C6+ ion beam irradiated MG-20 population (M. Kawaguchi, unpublished data). The AM and RNS phenotypes of nena-2 were the same as of nena-1 (Figure 1E), confirming the involvement of NENA in root symbiosis. The nodulation phenotype of nena-3 was comparatively weaker, but nodulation was still significantly reduced at 24°C compared with the wild type (see Supplemental Figure 6C online). No symbiotic defects have been observed in nena-4 and nena-5 (see Supplemental Table 1 online).
NENA Is Expressed in Shoots and Roots and Upregulated during Nodulation
Expression of NENA was determined by RT-PCR analysis. Transcripts were detected in all analyzed tissues, including roots, flowers, and leafs, without major variation in expression levels relative to the reference gene EF-1α (Figure 2C). Expression of NENA in wild-type roots was unaltered after NF treatment but increased 3 weeks after inoculation (WAI) with M. loti relative to mock-treated roots. Strong downregulation relative to wild-type levels indicated a posttranscriptional degradation of nena-1 mRNA possibly due to nonsense-mediated decay (Figure 2D). Therefore, nena-1 most likely is a null allele.
The NENA Gene Complements nena-1 Mutants
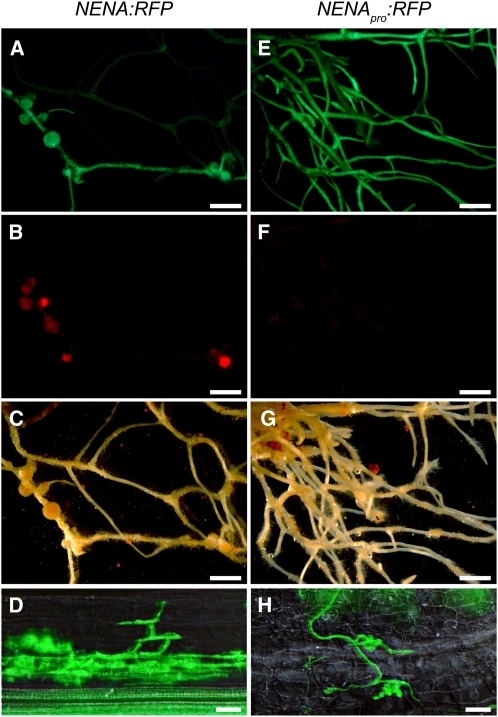
We introduced the NENA gene including 2 kb of putative promoter region lacking further predicted genes into a binary vector containing a red fluorescent protein (RFP) marker for C-terminal translational fusions (NENA:RFP). Transgenic (hairy) roots were generated by Agrobacterium rhizogenes–mediated transformation with the T-DNA construct, and complementation of the symbiotic phenotype was assayed. AM was fully restored in the nena-1 mutant background, confirming that the identified mutation is causative for the nena phenotype (Figure 3D). Introduction of NENA:RFP also restored nena-1 nodulation deficiency upon M. loti inoculation (Figures 3A to 3C, Table 1). As negative control, transformation with the same binary vector containing only the 2-kb 5′ regulatory NENA sequence (NENApro:RFP) did not complement nena-1 (Figures 3E to 3H, Table 1). Nodule colonization was visualized by rhizobial DsRed expression (Figure 3B). The binary vector contains an endoplasmic reticulum–targeted enhanced green fluorescent protein (ER-GFP) marker (Karimi et al., 2002), which was used to monitor successful transformation (Figures 3A and 3E). Furthermore, expression of NENA:RFP and NENApro:RFP was confirmed by CLSM (Figures 6B, 6C, and 6G).
Figure 3.
Transgenic Complementation of nena-1.
(A) to (D) A. rhizogenes–mediated transformation of nena-1 mutants with genomic NENA including 2 kb of 5′ regulatory sequence fused to RFP (NENA:RFP) led to restoration of RNS ([A] to [C]) and AM establishment (D).
(E) to (H) A. rhizogenes–mediated transformation of nena-1 with the 2-kb 5′ regulatory sequence fused to RFP (NENApro:RFP) did not restore RNS ([E] to [G]) and AM (H). Epifluorescence microscopy images show GFP expression in transgenic roots ([A] and [E]) and DsRed expression by M. loti in root nodules ([B] and [F]) using a GFP and a RFP filter, respectively. Corresponding white light illumination images are shown in (C) and (G). Root segments containing AM fungal structures (green) stained with WGA-Alexa Fluor 488 were visualized by DIC/epifluorescence microscopy using a GFP filter ([D] and [H]).
Bars = 2 mm in (A) to (C) and (E) to (G) and 40 μm in (D) and (H).
[See online article for color version of this figure.]
Table 1.
Complementation Analysis of nena-1 by A. rhizogenes–Mediated Transformation
| Line | Construct | Nodulation Ratioa | Nodules (sd)b |
| nena-1 | NENApro:RFP | 4/30 | 0.3 (0.7) |
| nena-1 | NENA:RFP | 35/37 | 7.4 (5.5) |
| nena-1 | NENApro:AtSeh1 | 34/45 | 5.3 (5.2) |
| nena-1 | 35Spro:NENA:GFP | 8/9 | 7.3 (6.9) |
| Wild type | NENApro:RFP | 5/5 | 6.6 (3.1) |
| Wild type | NENA:RFP | 5/5 | 10.6 (5.4) |
| Wild type | NENApro:AtSeh1 | 12/12 | 11.6 (7.4) |
Ratios indicate numbers of successfully transformed plants that formed nodules versus all plants of the indicated line that were successfully transformed with the indicated construct.
Mean nodule number per successfully transformed plant.
Figure 6.
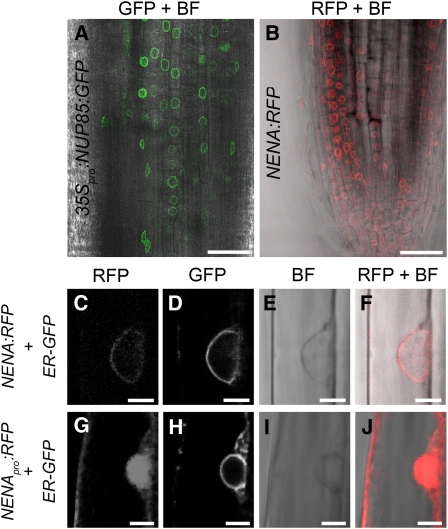
Perinuclear in Vivo Localization of NUP85 and NENA Fusion Proteins.
(A) Overlay of fluorescence and bright-field confocal micrographs showing perinuclear green fluorescence in root tip cells expressing 35Spro:NUP85:GFP.
(B) Overlay of fluorescence and bright-field confocal micrographs showing perinuclear red fluorescence in root tip cells expressing NENA:RFP and the ER-GFP marker (data not shown).
(C) to (F) Confocal micrographs of a rhizodermal cell expressing NENA:RFP ([C] and [F]) and the ER-GFP marker (D).
(G) to (J) Confocal micrographs of a rhizodermal cell expressing cytonucleoplasmic NENApro:RFP ([G] and [J]) and the ER-GFP marker (H).
Images are from wild-type (A) and nena-1 ([B] to [J]) A. rhizogenes–transformed roots and were acquired in sequential mode at excitationλ = 561 nm/detectionλ = 570 to 630 nm (RFP), excitationλ = 488 nm/detectionλ = 495 to 555nm (GFP) or bright-field (BF). Bars = 40 μm in (A) and (B) and 5 μm in (C) to (J).
Complementation of nena-1 mutants was also achieved by A. rhizogenes–mediated transformation with C-terminal NENA fusion to GFP controlled by the constitutive cauliflower mosaic virus 35S promoter (35Spro:NENA:GFP) and by the predicted NENA ortholog from Arabidopsis thaliana (At1g64350) under control of the 5′ regulatory NENA sequence from L. japonicus (Table 1).
NENA Belongs to the Sec13/Seh1 Protein Family
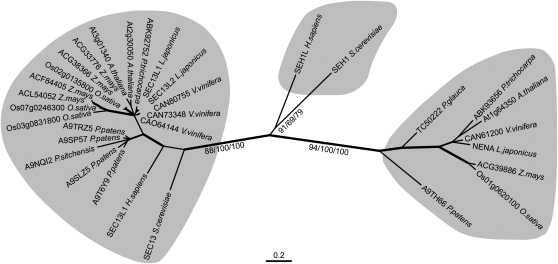
BLAST analysis of the NENA protein revealed 25% amino acid identity to Sec13 homolog 1 (Seh1) from yeast or humans. Moreover, there is 25 and 24% identity to the Sec13 protein of humans and yeast, respectively. Sec13 and Seh1 both are nucleoporins belonging to the yeast Nup84/ human Nup107-160 subcomplex (herein after referred to as the Nup84 subcomplex) (Siniossoglou et al., 1996; Belgareh et al., 2001). In addition, Sec13 together with Sec31 forms the framework of COPII vesicle coats (Fath et al., 2007). We reconstructed the phylogenetic relationships of Seh1, Sec13, and NENA-related sequences from yeast, human, and different plant species (Figure 4). The trees obtained comprise two clearly separated clusters: the first cluster contains proteins that are closely related to Sec13. The second cluster contains one member per plant species and includes NENA. Seh1 from yeast and humans form a third branch. Based on the calculated phylogenetic distances, members of the second cluster, including NENA, are more closely related to Seh1 than to Sec13. The L. japonicus genome contains in addition at least two genes, SEC13-like 1 and SEC13-like 2, which are more closely related to Sec13 than the NENA gene.
Figure 4.
Phylogenetic Relationships of Sec13/Seh1-Like Proteins.
The phylogenetic tree includes amino acid sequences from yeast, human, and representative species of bryophytes (Physcomitrella patens), gymnosperms (Picea spp), monocots (Oryza sativa and Z. mays), and dicots (Arabidopsis, L. japonicus, Populus trichocarpa, and Vitis vinifera). Three statistically supported clusters are highlighted: Sec13-related sequences on the left, yeast/human Seh1 in the middle, and plant Seh1-related sequences, including NENA, on the right. Distances correspond to the best-fitting maximum likelihood tree. Branch labels indicate bootstrap values (n = 1000) of maximum likelihood/neighbor-joining/maximum parsimony consensus trees. Thick, medium, and thin branches indicate bootstrap values ≥90, ≥80, and <80 in at least two consensus trees, respectively. Labels at the branch tips include GenBank accession numbers or gene names (accession numbers in Methods). Isoforms from splice variants were excluded from the analysis.
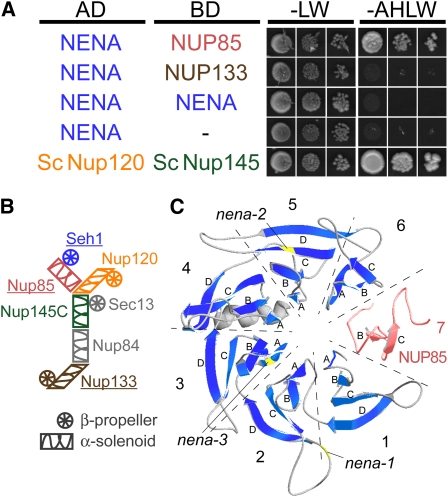
NENA and NUP85 of L. japonicus Interact in Yeast
Sc Seh1 and Sc Nup85 are bona fide constituents of nuclear pores and interact in yeast (Siniossoglou et al., 1996). In vitro reconstitution of recombinant Sc Seh1, Sc Nup85, and remaining components of the Nup84 subcomplex, including Sc Nup133, revealed their relative positions in a Y-shaped complex (Figure 5B) (Lutzmann et al., 2002). Therefore, we tested for interaction between NENA, NUP85, and NUP133 by yeast two-hybrid analysis. The results fully support homologous functions in Lotus: cotransformation of NENA fused to the Gal4 activating domain (AD) and NUP85 fused to the Gal4 binding domain (BD) allowed yeast growth on selection media, whereas no growth was observed when AD:NENA was combined with BD:NUP133, BD:NENA, or the empty BD vector (Figure 5A). This was confirmed by switching AD and BD (see Supplemental Figure 5B online). Furthermore, SEC13-like 1 and SEC13-like 2 did not show interaction with NUP85 but interacted with the corresponding partner from yeast, Sc Nup145 (see Supplemental Figure 5A online).
Figure 5.
NENA Interacts with NUP85 from L. japonicus and Adopts a β-Propeller Structure According to Homology Modeling.
(A) Gal4-based yeast two-hybrid assay for interaction between NENA as prey (AD) and NUP85, NUP133, or NENA as bait (BD). The empty bait vector (−) was used as negative control and AD:Sc NUP120 and BD:Sc NUP145 as positive control. Cotransformed yeast was grown in three dilutions on synthetic dropout medium lacking Leu and Trp (-LW) or adenine, His, Leu, and Trp (-AHLW).
(B) Schematic representation of the yeast Nup84 subcomplex and the arrangement of its components (Lutzmann et al., 2002). Putative homologs known to be required for root symbioses in L. japonicus are underlined. Color scheme refers to (A) and (C).
(C) Ribbon representation of a conceptual β-propeller formed by NENA (blue β-strands, model comprises residues 11 to 317) and the N terminus of NUP85 (red, residues 36 to 94). The model is based on crystal structures of Sc Seh1•Sc Nup85. Individual blades are delimited by dashed lines and numbered. Letters correspond to successive β-strands in each blade. Lack of β-strands in blades 6 and 7 is due to missing template data and incomplete sequence alignment. Positions of mutations in alleles nena-1, -2, and -3 are indicated.
[See online article for color version of this figure.]
NENA Is a WD40 Repeat Protein
Structural prediction of the NENA protein indicated that the distribution of identified WD40 repeats matches well to the architecture of Sc Seh1 (see Supplemental Figure 3 online) and places NENA into the large family of β-transducin-like/WD40 repeat proteins (Neer et al., 1994). Members of this family exhibit a highly conserved three-dimensional β-propeller structure, typically composed of seven circularly arranged blades. Each blade is formed by four anti-parallel β-strands, of which the last and the first three β-strands of two adjacent blades correspond to one WD40 unit (Smith et al., 1999). Based on crystal structures of the Sc Seh1•Sc Nup85 pair (Brohawn et al., 2008; Debler et al., 2008) and the alignment of Sc Seh1 and NENA (see Supplemental Figure 3 online), we created a three-dimensional model of the NENA protein (Figure 5C). The arrangement of predicted secondary structures is largely consistent with the six-bladed β-propeller fold of Seh1, except for a helix between strands 3D and 4A and the C-terminal loop, which are not supported due to alignment gaps and the lack of template data in disordered regions. The Seh1•Nup85 β-propeller is completed and stabilized by the domain invasion motif of Sc Nup85 (Brohawn et al., 2008; Debler et al., 2008). Therefore, we created a homology model of NUP85 comprising residues 36 to 94 and fitted it to the domain invasion motif of yeast Nup85. Although complete modeling of the seventh blade, including the strands required for the Velcro closures (Smith et al., 1999) between Sc Seh1 and Sc Nup85, was not possible due to sequence divergence at the beginning of the aligned sequences, β-strands 7B and C matched well to the template structure.
NENA and NUP85 Are Located at the Nuclear Rim
To determine the subcellular localization of the NENA protein in Lotus roots, the NENA:RFP fusion construct, which complemented the symbiotic defects of nena-1 (Figures 3A to 3D, Table 2), was introduced into nena mutants by A. rhizogenes–mediated transformation. RFP signal detected by CLSM of transgenic roots was located at the nuclear rim, as shown in optical sections from rhizodermal cells (Figures 6B and 6C). The identity of the subcellular compartment was verified by perinuclear ER-GFP expression of the transformation marker and by comparison with bright-field micrographs (Figures 6E and 6F). By contrast, hairy roots that were transformed with NENApro:RFP showed red fluorescence in the cytoplasm and inside the nucleus (Figures 6G to 6J), as expected for freely diffusible RFP. Moreover, 35Spro:NUP85:GFP that has been introduced into wild-type roots by A. rhizogenes–mediated transformation was also localized at the nuclear rim (Figure 6A).
Table 2.
Analysis of NF-Induced Calcium Spiking by Microinjection of Oregon Green 488 BAPTA-1 Dextran MW10,000
| Temperaturea | Root Hairsb | Plantsb | |
| Wild type | 18°C | 53/72 | 32/40 |
| 24°C | 22/28 | 12/13 | |
| nena-1 | 18°C | 0/48 | 0/30 |
| 24°C | 1/42 | 1/21 |
Ambient temperature at which seedlings were grown and measurements were performed.
Ratios indicate positive versus total measurements.
NENA Is Required for Rhizodermal Nod Factor Response
NUP133 and NUP85 are both required for NF-induced Ca2+ spiking (Kanamori et al., 2006; Saito et al., 2007). Therefore, we measured intracellular Ca2+ concentrations by microinjection of a reporter dye into root hairs of young seedlings. Seventy-five percent of wild-type root hairs showed Ca2+ spiking after NF application. By contrast, there was only one positively scored response in 90 analyzed nena-1 root hairs (Table 2; see Supplemental Figure 7A online). This single Ca2+ spiking response was weaker and observed in a young root hair, where the nucleus was located at the base of the trichoblast. Usually, older root hairs, with more apical nuclei, are used for Ca2+ spiking measurements by microinjection (see Supplemental Figure 7B online) (Miwa et al., 2006).
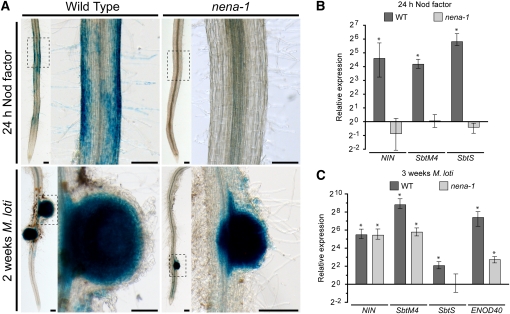
Since residual nodulation in nena might be connected to rare Ca2+ spiking events, we further investigated the expression of genes that are induced by NF or rhizobia. Among these, NIN expression provides temporal and spatial information because it is rapidly induced in rhizodermal cells, including root hairs, of the susceptible zone (Radutoiu et al., 2003) and later on in cortical cells of developing nodules (Schauser et al., 1999). Using the β-glucuronidase (GUS) reporter gene fused to the NIN promoter region, NIN induction was shown to be absent in nfr1 mutants (Radutoiu et al., 2003). Hence, we used this reporter construct to analyze NIN expression patterns in nena-1. In contrast with the wild type, none of the transformed root systems showed GUS activity in rhizodermal cells of the susceptible zone in response to NF or M. loti up to 3 days after inoculation (DAI). However, 7 and 16 DAI, transformed nena-1 mutants showed strong GUS activity in the cortex and single cells of the outer layers of developing nodules (Table 3, Figure 7A; see Supplemental Figure 8 online).
Table 3.
NINpro:GUS Expression Analysis in Transgenic Roots of Wild Type and nena-1 Genetic Background
| Treatment | Line | Blue Staininga |
| 24 h Nod factor | Wild type | 8/10 |
| nena-1 | 0/12 | |
| 1 d M. loti | Wild type | 5/6 |
| nena-1 | 0/8 | |
| 3 d M. loti | Wild type | 8/10 |
| nena-1 | 0/11 | |
| 7 d M. loti | Wild type | 8/11 |
| nena-1 | 1/12 | |
| 16 d M. loti | Wild type | 11/11 |
| nena-1 | 10/11 |
Ratios indicate numbers of plants that showed GUS expression in rhizodermal or nodule cortical cells divided by the total number of analyzed root systems per indicated treatment.
Figure 7.
Rhizodermal Nod Factor Response Is Impaired, Whereas Induction of Symbiosis Genes at Nodule Primordia Is Not Affected, in nena.
(A) Bright-field images of X-Gluc incubated roots transformed with NINpro:GUS after NF treatment or inoculation with M. loti. No blue rhizodermal staining was observed in nena-1 roots after NF treatment. Images correspond to Table 3. Boxed regions are shown at higher magnification. Bars = 0.2 mm.
(B) and (C) Quantitative PCR analysis of symbiosis gene expression in wild-type and nena-1 roots 24 h after NF treatment (B) or 3 weeks after M. loti inoculation (C). Expression is relative to mock-treated samples and normalized to EF-1α levels. Mean and se were derived from three biological replicates. Asterisks indicate significant (P < 0.05) differences in gene expression between NF or M. loti and mock treatments.
The absence of early NF responsiveness was further corroborated by quantitative RT-PCR analysis of NIN and additional early-induced symbiosis genes (Figure 7B). SbtM4 and SbtS encode subtilisin-like Ser proteases that are specifically expressed during AM and RNS. Rhizodermis-specific expression of SbtS is induced by NF and ceases after ~2 weeks of nodule development, whereas SbtM4 remains upregulated in nodules (Kistner et al., 2005; Takeda et al., 2009). While none of these genes was upregulated in nena-1 roots treated with NF, NIN, SbtM4, and the early nodulin ENOD40-1, which is strongly expressed in nodules (Takeda et al., 2005), were significantly induced in nodulated nena-1 and wild-type roots at 3 WAI with M. loti. SbtS was not induced in nena-1 but upregulated in the wild type after NF treatment and 3 weeks after M. loti inoculation (Figure 7C).
Rhizobial Infection of nena Resembles Crack Entry
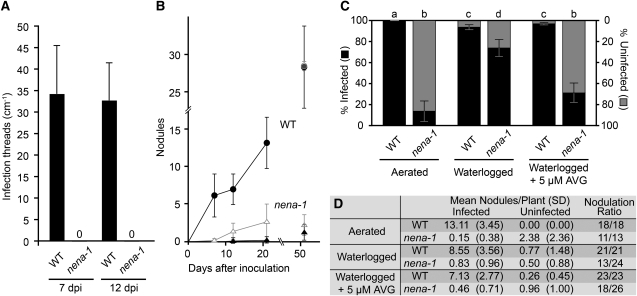
The lack of early responses to rhizobia displayed by nena-1 conflicted with our prior observation of infected nodules on nena mutants, as visualized by inoculation with DsRed expressing M. loti. Therefore, we quantified the ITs that lead to rhizobial invasion of nodules via RHC to check for rare infection events. However, no root hair ITs were observed by fluorescence microscopy of nena-1 mutants at 7 DAI (n = 19) and 12 DAI (n = 21) (Figure 8A). In parallel, plants were inoculated with M. loti strain R7A expressing the lacZ reporter and microscopically analyzed after 7 d of growth at 18°C. Wild-type plants showed normal symbiotic development, including RHC, rhizobial microcolonies, and root hair ITs (Figure 9A). By contrast, nena-1 mutants exhibited abnormal deformation of root hairs in responsive parts of the root. Occasionally, rhizobial microcolonies were observed at the base of deformed root hairs (Figure 9B). RHC or root hair ITs were not observed in any of the inspected nena-1 mutants.
Figure 8.
Rhizobial Infection of nena Does Not Occur via Root Hairs and Is Promoted by Ethylene.
(A) Quantification of root hair ITs 7 and 12 DAI with M. loti expressing DsRed and growth under aerated conditions; no ITs were observed in nena-1. Mean and sd were calculated from ≥19 (nena-1) and ≥14 wild-type (WT) root systems per time point.
(B) Nodulation time course during aerated growth conditions after inoculation with M. loti expressing DsRed. Mean and sd were calculated from 13 to 21 nena-1 (triangles) and 12 to 18 wild-type (squares) root systems per time point. Open/gray and closed/black symbols represent total and infected nodules, respectively. If all nodules were infected, only infected nodules are indicated. If all nodules were uninfected, only total nodules are indicated.
(C) and (D) Quantification of nodules from wild-type and nena-1 plants cultivated under different conditions 21 DAI with M. loti expressing DsRed.
(C) Bars indicate mean percentages of uninfected (gray) and infected (black) nodules per nodulated individual. Error bars indicate se. Different letters above bars indicate significant differences (P ≤ 0.05, t test) between pairwise comparisons.
(D) Mean per plant, sd, and number of nodulated plants versus total number of plants per line and treatment (nodulation ratio) are indicated.
Figure 9.
Rhizobial Microcolonies at the Root Surface of nena-1 Lead to Nodule Formation and Intercellular Entry.
(A) and (B) Bright-field DIC images from roots hairs 7 DAI with lacZ-expressing M. loti and 18°C growth temperature. Wild-type (WT) plants show root hair curling (arrows) and ITs, whereas nena-1 mutants display abnormal root hair deformation and occasional colony formation by rhizobia (arrowhead). Images represent observations from more than eight plants per line. Bars = 50 μm.
(C) and (D) Confocal z-projections of longitudinal 80-μm tissue sections of a young infected wild-type (C) and an uninfected nena-1 (D) nodule. Images represent samples from 16 DAI/aerated (C) and 21 DAI/waterlogged + 5 μM AVG (D) treatments, corresponding to Figure 8D.
(C) DsRed expressing rhizobia (red) have colonized the nodule via an intracellular root hair IT (arrow).
(D) An uninfected nodule developed coinciding with accumulation of rhizobia at the root surface (arrowhead).
(E) to (K) Thin sections of nodule tissue stained with toluidine blue.
(E) Young wild-type nodule with intracellular IT (arrowhead) spanning from the infection site (arrow) into the cortex.
(F) and (G) Young nena-1 nodule with a subepidermal infection pocket (arrow) and cortical ITs (arrowhead).
Insets in (E) and (G) show respective sections at lower magnification; dashed boxes indicate magnified areas. Longitudinal sections of mature nodules from the wild type (H) or nena-1 (J) and corresponding magnifications ([I] and [K]) showing colonized host cells. Plants were grown under waterlogging conditions and sampled 3 WAI with M. loti R7A. Bars = 50 μm, except (G), where bar = 20 μm.
To test for delayed nodule formation and rhizobial infection in nena, plants were analyzed at additional time points after inoculation with M. loti expressing DsRed. In the wild type, the number of nodules increased throughout the time course. All nodules were infected and colonized (Figure 9C), except at 51 DAI, where in two cases a small uninfected nodule was observed (Figures 8B and 8D). By contrast, initiation of nodulation was delayed in nena-1, followed by a moderate increase in nodulation until 21 DAI. More than 90% of these nodules were uninfected. At 51 DAI, the average number of infected nodules increased to 1.11, while uninfected nodules decreased to 1.00, suggesting a proportional infection of initially uninfected nodules between 21 and 51 DAI (Figure 8B). Nodulation in nena-1 was strongly reduced compared with the wild type, and no root hair ITs were observed in nena-1 throughout the time course. However, we observed small patches of rhizobia on the root surface that coincided with uninfected nodules. Sectioning and CLSM of such empty nodules confirmed that rhizobia were confined to the surface and did not enter the cortex (Figure 9D). By contrast, analysis of nena-1 nodules that showed rhizobial colonization at 21 DAI revealed intercellular intrusions of rhizobia spanning several cell layers from the apex into the nodule cortex. Hence, this route was the most probable entry site for further nodule colonization (see Supplemental Figure 9 online).
The infection structures observed in nena-1 were reminiscent of crack entry. Some legumes (e.g., S. rostrata) can switch between intracellular infection during aerated conditions and crack entry during root submergence (Goormachtig et al., 2004). Therefore, we tested whether the water regime had an effect on RNS in wild-type Gifu and nena-1 plants. Indeed, the proportion of infected nodules per nena-1 plant was significantly increased under waterlogging conditions compared with aerated conditions (Figures 8C and 8D). Since ethylene accumulation in the water saturated root environment is the chief cause for intercellular infection, we further tested if inhibition of endogenous ethylene production by aminoethoxyvinylglycine (AVG) treatment can suppress infection during waterlogged conditions. The proportion of infected nodules was significantly reduced by the addition of 5 μM AVG prior to rhizobial inoculation (Figures 8C and 8D). The data confirm that nodule infection under waterlogging conditions depended on ethylene and thus provide strong evidence for intercellular infection of nena-1 nodules. Infection rates did not significantly change in wild-type plants, although the average nodulation rate was reduced under waterlogging conditions with or without AVG (Figures 8C and 8D).
Further evidence for an intercellular infection mode was obtained by bright-field microscopy of 4-μm sections from young nena-1 nodules that contained subepidermal infection foci (Figure 9F). In addition, ITs were observed in the root cortex (Figure 9G). No subepidermal infection foci were observed in any of the sections from young wild-type nodules (Figure 9E). Despite the apparent defects during the early infection process, sections of mature nodules did not indicate any structural alterations in cortical infection and nodule development in nena-1 compared with the wild type (Figures 9H to 9K).
DISCUSSION
Symbiotic Infection of Rhizodermal Cells Is Blocked in nena
We found that the nena-1 mutation impairs symbiotic responses of the rhizodermis. AM development at nonpermissive temperature was mostly blocked in the outer root cell layers. The balloon-like hyphal structures at the infection sites resembled the phenotypes of other Lotus common sym mutants lacking Ca2+ spiking (Kistner et al., 2005). Ultrastructural analyses of AM infection sites of castor-2 (sym4-2) mutants indicate that abortion of infection is accompanied by the death of cells containing balloon-like hyphal swellings (Bonfante et al., 2000). Corresponding to the loss of PPA formation in M. truncatula dmi2 and dmi3 roots (Genre et al., 2005), PPA formation might also be deficient or absent in nena.
Likewise, the establishment of RNS in nena-1 was blocked at the rhizodermis. Despite scrutinizing >98 root systems and using two different and sensitive methods that both detected ITs in the wild type, we could not detect a single infection thread in root hairs of nena-1 mutants grown at 24°C, indicating that NENA is required at this stage of the symbiosis. Expression analysis of marker genes corroborated the specific lack of symbiotic responses in the rhizodermis. It has previously been shown that the expression of SbtS in response to M. loti is confined to the rhizodermis (Takeda et al., 2009). While we observed a consistent induction of SbtS in wild-type roots, SbtS was not upregulated in nena-1 after 24 h of NF treatment or 3weeks after rhizobial inoculation. Moreover, the rhizodermal NINpro:GUS induction observed in the wild type was absent in nena-1. The lack of rhizodermal responsiveness in the nena-1 mutant was further manifested in defective NF-induced Ca2+ spiking. Residual Ca2+ spiking was detected in a single nena-1 root hair; however, this low frequency occurrence of spiking cells does not support detectable infection thread formation or symbiotic gene activation in the rhizodermis. In accordance with these data, nup133 and nup85 mutants were previously shown to be impaired in NF-induced rhizodermal responses, including Ca2+ spiking and RHC and IT formation at nonpermissive temperatures (Kanamori et al., 2006; Saito et al., 2007). Residual nodulation was observed in various nup133 mutants and in nup85-2, raising the possibility that these mutants are infected via a mechanism similar to the one described here for nena mutants.
Symbiotic Development of Cortical Cells Does Not Require NENA
In striking contrast to the nonresponsiveness of the rhizodermal cell layer, nodules developed regularly on nena roots, albeit at reduced frequency. The appearance of the infected cortical region in nena-1 nodules was indistinguishable by light microscopy from wild-type nodules, indicating that rhizobial accommodation in the cortex was largely unaffected. Consistent with an intact symbiotic response of nena-1 cortical cells was the observed NINpro:GUS expression in nodule primordia and the induction of ENOD40-1 and SbtM4, which are also expressed in the nodule cortex (Takeda et al., 2005, 2009). The somewhat lower transcript abundance of these genes in nena-1 compared with wild-type roots is likely due to the lower nodule number on nena roots.
It is unclear at present why the cortical programs for nodule organogenesis and infection do not require NENA. It is unlikely that nodulation is caused by residual functional capability provided by the mutant allele because nena-1 seems to be a null allele. The formation of empty nodules on nena-1 roots coincided with the presence of superficial rhizobial microcolonies. Spot inoculation of Lotus roots with NF is sufficient to induce nodule primordia (Niwa et al., 2001). We therefore think that microcolonies on the surface of nena roots, which have not been described in other common sym mutants, produce local NF concentrations sufficiently high to trigger the formation of nodule primordia. NF molecules are bound and immobilized by cell wall material and probably do not penetrate into the root cortex (Goedhart et al., 2000). This would imply NENA-independent signaling through the rhizodermal cell layer for NF-induced activation of cortical cell division.
The apparent dispensability of NENA for cortical CCaMK activation reveals tissue-specific differences of common SYM signaling. Accumulating evidence suggests that the sole function of common SYM genes upstream of Ca2+ spiking is the efficient and context-dependent activation of CCaMK for AM fungal or rhizobial infection and nodule organogenesis. Gain-of-function versions of CCaMK introduced into the genetic background of common sym mutants, which lack NF-induced Ca2+ spiking, not only activated nodule organogenesis in the absence of rhizobia but also restored rhizobial infection via RHC and IT formation, as well as AM fungal infection (Hayashi et al., 2010; Madsen et al., 2010). These mutants also supported rhizobial colonization of the nodule inner tissue by transcellular ITs and release of bacteria from ITs into nodule cortical cells. NFR1 and NFR5, by contrast, were indispensable for infection via root hair ITs but not required for NF-dependent IT formation inside nodules induced by autoactive CCaMK, suggesting alternative NF receptors operate in the cortex (Madsen et al., 2010). These data suggest that not only NF recognition but also downstream signaling via components of the common SYM network required for Ca2+ spiking and CCaMK activation differs between rhizodermal and nodule cortical tissue.
In contrast with rhizobial infection of Lotus, AM fungi have a propensity to overcome a genetic block for rhizodermal infection (Wegel et al., 1998). This revealed that mutants defective in SYMRK, NENA, NUP85, NUP133, CASTOR, and POLLUX all support arbuscule development in the cortex. In the case of symrk, it has been shown that AM fungal hyphae enter the root via an extracellular route, which is consistent with the mutant’s inability to provide intracellular access to rhizodermal and subjacent cell layers (Demchenko et al., 2004). Successful hyphal penetration of the outer root layer in nena mutants, leading to arbuscule formation, was clearly different from the wild type and might occur similarly to symrk mutants. Only mutants defective in the common SYM genes CCaMK and CYCLOPS, which are positioned downstream of Ca2+ spiking, are blocked in arbuscule development (Demchenko et al., 2004; Kistner et al., 2005). These observations together with the intact rhizobial infection of nena cortical cells suggests that not only in AM but also during nodule development the common SYM genes upstream of Ca2+ spiking are more stringently required in the rhizodermal cell layer than in the cortex. This opens the possibility that common SYM-mediated Ca2+ spiking may be dispensable for cortical responses in RNS and, in turn, implies that an alternative regulation of CCaMK may exist in the cortex.
nena Reveals an Intercellular Rhizobial Entry Mode in L. japonicus
By employing an intercellular infection mode, which carries the hallmarks of crack entry, nena overcomes the requirement for symbiotic responsiveness of the rhizodermis. Ethylene is a potent inhibitor of rhizodermal Ca2+ spiking (Oldroyd et al., 2001), and its negative regulatory role in RNS has been confirmed genetically (Penmetsa and Cook, 1997; Penmetsa et al., 2003). Water-tolerant legumes evade the inhibitory effect of ethylene and even take advantage of increased ethylene concentrations during root submergence (Goormachtig et al., 2004). The subepidermal infection pockets observed in nena-1 resemble those seen during typical crack entry (Ndoye et al., 1994). As in S. rostrata aerated roots, the root hair is the primary route for nodule infection during aerated conditions in Lotus, but this is blocked in nena and hence leads to the formation of mostly uninfected nodules. During waterlogging, nodule infection is significantly promoted in nena-1. Importantly, as in S. rostrata, rhizobial infection of nena-1 nodules during waterlogging is suppressed by the ethylene biosynthesis inhibitor AVG, providing compelling evidence that infection occurs via crack entry. Because of the promoting effect of ethylene on nena-1 infection, residual Ca2+ spiking in rhizodermal cells is unlikely to play a role in initiating infection in nena-1. At the permissive temperature, single infection events via root hair ITs were found in nup133-1 and nup133-4 mutants (Kanamori et al., 2006). Although it is possible that rare root hair infection also occurs in nena mutants grown at permissive temperatures, the lack of Ca2+ spiking in seedlings grown and examined at 18°C indicates that, independent of the temperature, crack entry is the predominant infection route on nena mutants under waterlogging conditions.
Based on surveys of infection strategies in different legume lineages, it has been proposed that root hair infection is a more recent trait than the ancestral intercellular infection involving cortical ITs (Sprent, 2007). Crack entry might have been maintained in legumes that are challenged to engage in RNS under submerged conditions. The observation of crack entry in nena provides genetic support for an ancient nature of this trait in legumes and might be a relic of the common ancestor of Lotus spp and Sesbania spp, which both belong to the same subclade within the Robinioids (Wojciechowski et al., 2004). Intercellular infection occurring in Lotus uliginosus, a temperate legume adapted to wetland conditions, further substantiates the conservation of crack entry by other members of this genus (James and Sprent, 1999). Rhizobial infection structures indicating intercellular infection of nodule primordia were also observed in the Lotus root hairless1 (rhl1) mutant, but further evidence for crack entry as defined by its dependence on ethylene was not provided (Karas et al., 2005). In addition to rare rhizobial entry between rhizodermal cells, intracellular infection of NF-induced cortical root hairs was shown, and this was proposed as the main route to sustain RNS in the absence of epidermal root hairs. It is of note that the symbiotic signaling program was most likely not perturbed by rhl1. The recent demonstration of rare rhizobial infection of synthetic mutants carrying the gain-of-function CCaMK allele snf1 together with nfr1 and/or nfr5 loss-of-function alleles is in accordance with crack entry of rhizobia overcoming a genetic block of rhizodermal infection (Madsen et al., 2010). This supports the idea that crack entry is evolutionary older and in part genetically independent of rhizobial root hair infection.
NENA Is a Scaffold Nucleoporin Required for Calcium Spiking
Together with NUP133 and NUP85, NENA represents the third common SYM protein that shows sequence similarity to a nucleoporin from the Nup84 subcomplex of the nuclear pore. For all three proteins, conservation compared with their counterparts in yeast or humans does not exceed 25% identity. Irrespective of high sequence divergence among homologous nucleoporins from various organisms, nuclear pore components are structurally conserved, and certain protein domains therein, such as WD40 repeats, are found across kingdoms (Bapteste et al., 2005). Based on our yeast two-hybrid data, in silico structural and phylogenetic analyses, and in vivo protein localization, we conclude that NENA represents the Lotus version of nucleoporin Seh1. These data further suggest that NENA and NUP85 function together as scaffold proteins within the nuclear pore complex (NPC).
The specific involvement of NENA in symbiotic signaling is curious. The Nup84 subcomplex is part of the NPC, a macromolecular assembly of ~30 different proteins in multiple copies (Alber et al., 2007). Disruption of the Nup84 subcomplex by deletion of individual components typically leads to severe developmental defects in yeast and mammalian cells due to impaired NPC assembly (Siniossoglou et al., 1996; Harel et al., 2003; Walther et al., 2003). The absence of obvious pleiotropic defects in the different nena mutant backgrounds may be due to partial and temperature-dependent redundancy with other structurally related nucleoporins. Different degrees of redundancy among the components of the Lotus Nup84-like subcomplex might further account for the phenotypic differences between nup133, nup85, and nena mutants, for example, the extent of residual nodulation (Kanamori et al., 2006; Saito et al., 2007). (Further details are provided in the Supplemental Discussion online.)
Detailed microscopy analysis using nuclear-targeted cameleon for Förster resonance energy transfer-mediated Ca2+ measurements indicated that Ca2+ spiking originates at the nuclear periphery and spreads to the center of the nucleus (Sieberer et al., 2009). By analogy to animal cells, the lumen of the nuclear envelope is a likely Ca2+ source (Gerasimenko et al., 1995). In this context, we propose two models for the symbiotic function of the NPC. First, scaffold nucleoporins, including NENA, might be involved in the selective nuclear import of proteins required for NF-induced Ca2+ spiking. The import of protein in general does not seem to be affected, as no difference in GFP:CYCLOPS localization was detected between transgenic roots from the wild type and the nena-1 background (see Supplemental Figure 10 online). In Arabidopsis, for example, a screen for suppressors of the constitutively active TIR-NB-LRR–type R gene, snc1, resulted in the identification of three mutants that are functionally linked to nucleocytoplasmic transport (nup96/mos3, importin α/mos6, and nup88/mos7) (Zhang and Li, 2005; Palma et al., 2005; Cheng et al., 2009). Interestingly, MOS7 turned out to be specifically required for the nuclear import of SNC1 and other defense-related proteins, while nuclear and cytoplasmic pools of control proteins remained unaffected in the mos7 mutant (Cheng et al., 2009).
Second, the NPC might be involved in symbiotic Ca2+ signaling by regulating nuclear pools of second messengers. It is currently believed that Ca2+ spiking involves a synchronized flux of second messengers or effector enzymes from the cytoplasm into the nucleus after NF triggering and throughout the period of Ca2+ oscillations (Oldroyd and Downie, 2008). A reduced permeability of the nuclear envelope caused by a structural defect or a general reduction in abundance of nuclear pores might be detrimental, therefore, for NF-induced Ca2+ spiking in root hairs, while not affecting vital nucleocytoplasmic transport processes.
METHODS
Plant Growth and AM Assay
Seeds of Lotus japonicus ecotypes Miyakojima MG-20, Gifu B-129 wild type, and nena-1 to 6 (see Supplemental Table 1 online) were scarified and surface sterilized with 1% NaClO. Imbibed seeds were germinated on 1% Bacto Agar (Difco) at 18 or 24°C for 5 to 6 d. Seedlings were cultivated in chive (Allium schoenoprasum) nurse pots containing G. intraradices-like BEG195 (Stockinger et al., 2009) as described (Kistner et al., 2005), except that sand/vermiculite (1/1 volume) was used as substrate. After 3 weeks of growth in Sunbags (Sigma-Aldrich) at 18 or 24°C constant, 16-h-light/ 8-h-dark cycles, roots were harvested and cleared with 10% KOH at 90°C for 15 min. AM fungal structures were stained with 5 μg/mL WGA-Alexa Fluor 488 conjugate (Molecular Probes) and quantified under the epifluorescence microscope using the magnified intersections method (McGonigle et al., 1990). Data were obtained by two independent experiments with at least four plants per line and temperature. Roots were counterstained with 1 μg/mL propidium iodide. For detailed AM phenotype analysis, stacked micrographs were acquired by CLSM.
AM Mutant Screen
M3 individuals of the bulked TILLING population (Perry et al., 2003) were greenhouse cultivated in chive nurse pots for 4 weeks. AM fungal structures were stained with ink and vinegar (Vierheilig et al., 1998), individual root samples were mounted on slides, and colonization patterns were scored using a stereomicroscope at ×30 to ×200 magnification. M4 self-progeny of scored mutants was rescreened for confirmation. AM mutants were checked for nodulation capacity by examining roots 1 month after inoculation with M. loti strain R7A applied at an optical density of 0.01 at 600 nm (OD600). Plants were grown in white peat/bark humus soil (Fruhstorfer Typ P; Hawita) under greenhouse conditions.
Infection Thread and Nodulation Assays
Germinated seedlings (see above) were inoculated as described with Mesorhizobium loti strains R7A carrying pXLGD4 for lacZ expression (Stracke et al., 2002) or MAFF303099 expressing DsRed (Markmann et al., 2008), with the following modifications: bacterial cultures were diluted to OD600 0.005 in 80 mL half-strength BandD medium (Broughton and Dilworth, 1971) and added to 300 mL autoclaved growth substrate.
For waterlogging experiments, seedlings were grown in closed Weck jars containing expanded clay granules (Seramis; Mars). AVG solution at 5 μM final concentration was added immediately before transfer of the seedlings to Weck jars. For aerated growth conditions, seedlings were transferred to polypropylene plant pots containing sand/vermiculite (1/1 volume) and watered to field capacity at 3-d intervals. All plants were cultivated under 16/8-h light/dark cycle at a constant 24°C temperature, unless stated otherwise. ITs and nodules that contained rhizobia were visualized by DsRed fluorescence or stained for β-galactosidase activity (Lombardo et al., 2006) and scored by fluorescence and bright-field microscopy. Eighty-micrometer tissue sections for CLSM analysis were prepared with a Vibratome microtome (Leica VT1000S) after embedding nodules in 6% low melting agarose. For bright-field microscopy of nodule colonization, root sections were fixed in 1.5% glutaraldehyde (Sigma-Aldrich), dehydrated, and embedded in Technovit 7100 (Kulzer). Four-micrometer histological sections were prepared with a microtome (Leica RM2125RT) and stained with 0.1% toluidine blue in benzoate buffer, pH 4.4.
Calcium Spiking Analysis
Ca2+ imaging was performed by microinjection of the fluorescent ratiometric Ca2+ indicator Oregon Green 488 BAPTA-1 dextran MW10,000 (Invitrogen) and reference dye Texas Red dextran MW10,000 (Invitrogen) as described previously (Charpentier et al., 2008). Measurements were performed at 18 or 24°C ambient temperatures on growing root hairs of L. japonicus Gifu B-129 and nena-1 seedlings that were grown for 2 d in the dark.
Transgenic Complementation and Subcellular Localization
The NENA sequence from 1911 bp upstream of the start codon to the last base pair before the stop codon was amplified by nested PCR with primer pairs N-172/157 and N-171/168 (see Supplemental Table 2 online) from genomic Gifu wild-type DNA and cloned into pENTR/D-TOPO (Invitrogen), giving rise to pENTR-NENA. From that construct, just the putative promoter region, NENApro, was PCR amplified with primers N-171/173 and cloned into pENTR/D-TOPO. To test complementation of nena by the Arabidopsis thaliana ortholog of NENA, At SEH1 genomic CDS was amplified by nested PCR with primers S-176/175 and 5′-phosphorylated primers S-177/178 and ligated with PCR-amplified pENTR-NENA fragment lacking the NENA CDS using primer pair N-173/179. All three entry clones were recombined during Gateway LR reactions (Invitrogen) with the destination vector pK7RWG2 (Karimi et al., 2002) modified by having KanR replaced by ER-GFP and lacking the 35S promoter (kindly provided by M. Antolin-Llovera, Biocenter LMU Munich). To confirm subcellular localization, NENA genomic coding sequence, amplified with primers N-158/168, was cloned into pENTR/D-TOPO (Invitrogen) and subsequently Gateway transferred into pK7FWG2 (Karimi et al., 2002). For subcellular localization of NUP85 in hairy roots, the CDS without the stop codon was PCR amplified from the cDNA clone MFB015g09 using primers 85-162/183 and cloned into pENTR/D-TOPO. The resulting entry clone was Gateway transferred into pK7FWG2. The fidelity of all entry clones was confirmed by sequencing. A T-DNA construct with GFP fused to the N terminus of CYCLOPS (Yano et al., 2008) was also used for subcellular localization in the wild type and nena-1. T-DNA constructs were transformed into Gifu wild type and nena-1 via Agrobacterium rhizogenes strain AR1193 as described (Charpentier et al., 2008). Subcellular localization of translational fusion proteins in young hairy roots was assessed by CLSM. Nodulation and AM colonization was assayed as described above.
DNA Gel Blotting
Probes I (1148 bp) and II (546 bp) were labeled with [α32P]dCTP using the NEBlot kit (New England Biolabs) after excision of the respective restriction fragments of pENTR-NENA triple-digested with EcoRI, NdeI, and NsiI. Twenty micrograms of MG-20 genomic DNA were digested with BglII, EcoRI, NdeI, or NsiI, size separated by agarose gel electrophoresis, and blotted on Hybond-N+ (GE Healthcare). Nylon membranes were hybridized with Probe I or II in roller bottles at 67°C overnight, washed with increasing stringency (final wash: 0.1× SSPE and 0.1% SDS, 63°C for 1 h), and visualized on a Typhoon scanner (GE Healthcare) after exposure to a phosphor screen.
Expression Analysis
Total RNA was extracted with CTAB buffer and acidic phenol as described (Kistner et al., 2005). RNA samples were TURBO DNase (Ambion) treated, and RNA integrity (RIN ≥ 7) was verified with a 2100 Bioanalyzer (Agilent). Absence of genomic DNA was confirmed by PCR. Approximately 200 ng of total RNA were used for first-strand cDNA synthesis using the SuperScript VILO kit (Invitrogen) according to the manual. For subsequent PCRs, 2 μL of cDNA template were used per 20 μL total volume. For tissue-specific analysis, samples were taken from different organs of two flowering Gifu wild-type plants. NENA and EF-1α transcript levels were visualized by ethidium bromide staining following agarose gel electrophoresis of PCR products after 28, 31, and 34 cycles with primer pairs N-174/167 or EF1-U23/L19. Quantitative expression analysis was performed by real-time PCR using Fast SYBR Green Master Mix (Applied Biosystems) and a CFX96 detection system (Bio-Rad). Samples were generated from whole roots of seven to eight pooled Gifu wild-type or nena-1 seedlings that were grown for 2 weeks on plates (half-strength BandD medium, 0.75% GELRITE [Roth], and 2 mM MgSO4) or in Weck jars and were treated with 1 μM purified NF or inoculated with MAFF303099, respectively. All plants, including the corresponding mock (half-strength BandD solution) controls, were grown at 24°C. Target transcripts were PCR amplified using primer pairs N-174/167, 40-203/204, NIN-201/202, M4-199/200, SbtS-007/008, and EF1-U23/L19 and the following cycles: 20 s at 95°C, 40× (3 s at 95°C, 20 s at 57°C, 20 s at 72°C, plate read), 10 s at 95°C, melt curve 65 to 95°C with 0.5°C/5-s increments. Amplification efficiencies and Ct values were calculated with LinRegPCR (Ruijter et al., 2009). Subsequently, relative expression normalized to the reference gene EF-1α, standard error, and statistical significance based on three biological replicates were calculated using REST 2009 software (Pfaffl et al., 2002).
Promoter GUS Analysis
A T-DNA construct with the GUS reporter gene expressed by the NIN promoter (Radutoiu et al., 2003) was transformed into Gifu wild type and nena-1 via A. rhizogenes strain AR1193. Plants with hairy roots were transferred onto plates or into Weck jars and 4 to 7 d later treated with NF or MAFF303099, respectively (see above). Growth temperature was 24°C. Treated roots were cut off and incubated in staining solution (0.5 mg mL−1 X-Gluc, 100 mM sodium phosphate, pH 7.0, 5 mM EDTA, pH 7.0, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide, and 0.1% Triton X-100) for 12 h at 37°C in the dark. A stereomicroscope was used for inspection and documentation.
Yeast Two-Hybrid Analysis
cDNA clones covering the full-length coding regions of NENA (MWM052c09), NUP85 (MFB015g09), and NUP133 (MFBL049d04) were obtained from the Lotus Resource Centre (Asamizu et al., 2000) and PCR amplified with primer pairs N-158/159, 85-162/163, and 133-160/161. Coding sequences of SEC13-like 1 and SEC13-like 2 were amplified by nested PCR from Gifu wild-type cDNA using primer pairs 13-1-195/196 and 191/194 and 13-2-197/198 and 192/193. Nup120 and Nup145 were amplified from genomic DNA of Saccharmyces cerevisiae S288c using primers 120-5′/3′ and 145-5′/3′, respectively. PCR products were cloned into pENTR/D-TOPO (Invitrogen) and subsequently inserted by LR Clonase II (Invitrogen) into Gateway-compatible bait or prey destination vectors derived from pBD-Gal4 Cam (Stratagene) or pGAG424 (Clontech) as described (Yano et al., 2008). The fidelity of all entry clones was confirmed by sequencing. Yeast two-hybrid analysis was performed with the yeast strain AH109 (Clontech) following standard procedures (Stratagene Product Manual 235702; Yeast Protocols Handbook PT3024-1, Clontech).
Microscopy
The following microscopes and conditions were used for this work: a fluorescence stereomicroscope (Leica MZ16 FA) with ×1 and ×2 objectives; inverted microscope (Leica DMI6000 B) with ×10/0.25, ×20/0.5, ×40/0.75 dry objectives, GFP and N3 filter cubes; confocal laser scanning microscope (Leica SP5) with ×20/0.5 dry, ×63/1.2 water immersion objectives, argon and DPSS lasers were used. RFP was excited at λ = 561 nm and detected at λ = 570 to 630 nm, and GFP was excited at λ = 488 nm and detected at λ = 495 to 555 nm. Images were acquired and processed with LAS AF software.
Phylogenetic Analysis
Protein sequences similar to NENA from different plant species with annotated genomes were retrieved by WU-blastp from the Uniprot database. Multiple hits corresponding to one gene, including splice variants, were discarded. Multiple sequence alignment was performed with MAFFT online and edited manually. The final alignment used for phylogenetic analysis is shown in Supplemental Figure 4 online and is available as a PHYLIP text file in Supplemental Data Set 1 online. Neighbor-joining (NJ) and parsimony analyses were performed online (http://mobyle.pasteur.fr/cgi-bin/portal.py) using PHYLIP (version 3.5c; distributed by J. Felsenstein, Department of Genome Sciences, University of Washington, Seattle) and Quartet Puzzling Maximum-Likelihood analysis using TREE-PUZZLE (Schmidt et al., 2002). The Quartet Puzzling Maximum-Likelihood tree was reconstructed from 1000 puzzling steps, exact parameter estimation using Quartet sampling + NJ, amino acid frequency, and rate heterogeneity estimations from the data set and the JTT model of substitution. Distances for NJ were obtained from 1000 bootstrap replicates using the JTT model with coefficient of variation = 0.743089312 and fraction of invariant positions = 0.000117. Consensus NJ and parsimony trees were constructed from 1000-fold bootstrapped analyses.
Homology Modeling
SMART (Schultz et al., 1998) search algorithm was used to define the domain composition of the NENA protein. A three-dimensional model of NENA was generated using DeepView and SWISS-MODEL (Guex and Peitsch, 1997; Arnold et al., 2006). For this, NENA and the N terminus of Nup85 were aligned to the PDB templates 3eweC, as shown in Supplemental Figure 1, and 3eweD, respectively.
Accession Numbers
Sequence data in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL/DDBJ databases under the following accession numbers: Lj T34D07/TM1188/CM0060 (AP007861), NENA (AB506696), SEC13-like 1 (AB506697), SEC13-like 2 (AB506698), yeast Seh1 (P53011), human Seh1-like (A8K5B1), yeast Sec13 (Q04491), human Sec13-like 1 (P55735), and Arabidopsis Seh1-like (At1g64350).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. 3D Projection of Typical AM Infection Sites in the Wild Type and nena-1.
Supplemental Figure 2. Map-Based Cloning of NENA.
Supplemental Figure 3. Alignment of Seh1 and Sec13 Related Protein Sequences from L. japonicus (Lj), Arabidopsis thaliana (At), Homo sapiens (Hs), and Saccharomyces cerevisiae (Sc).
Supplemental Figure 4. Multispecies Alignment of Seh1 and Sec13 Related Proteins.
Supplemental Figure 5. In Contrast with NENA, SEC13-like 1 and SEC13-like 2 Do Not Interact with NUP85 in the Gal4-Based Yeast Two-Hybrid Assay.
Supplemental Figure 6. AM and Nodulation Defects of nena Are Temperature Dependent.
Supplemental Figure 7. Calcium Spiking in Root Hairs of L. japonicus Gifu Wild Type and nena-1.
Supplemental Figure 8. The NIN Promoter Is Not Induced during Early Rhizodermal Response to M. loti but Active during Nodule Formation in nena-1.
Supplemental Figure 9. Intercellular Infection of Outer Nodule Cell Layers in nena-1.
Supplemental Figure 10. GFP-Cyclops Localizes to the Nucleus in nena-1.
Supplemental Table 1. nena Alleles and Symbiotic Phenotypes.
Supplemental Table 2. Primers Used in This Work.
Supplemental Data Set 1. PHYLIP Text File of the Alignment Shown in Supplemental Figure 4, Corresponding to the Phylogenetic Tree Shown in Figure 4.
Supplemental Discussion.
Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Gabi Büttner for technical assistance, Meritxell Antolin Llovera for the modified pK7RWG2 vector, and Jens Stougaard for the NINpro:GUS construct. Furthermore, we thank the Biological Resource Center in L. japonicus and Glycine max, Frontier Science Research Center, University of Miyazaki, for providing cDNA clones MWM052c09, MFB015g09, and MFBL049d04. This work was funded by a grant from the Deutsche Forschungsgemeinschaft. TILLING in L. japonicus was funded by two consecutive grants from the UK Biotechnology and Biological Sciences Research Council and one John Innes Centre joint research grant to M.P. and T.L.W.
References
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., Sali A., Rout M.P. (2007). The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D.G., de Carvalho-Niebel F. (2007). AP2-ERF transcription factors mediate Nod factor-dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ané J.M., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303: 1364–1367 [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Arrighi J.F., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E., Nakamura Y., Sato S., Tabata S. (2000). Generation of 7137 non-redundant expressed sequence tags from a legume, Lotus japonicus. DNA Res. 7: 127–130 [DOI] [PubMed] [Google Scholar]
- Bapteste E., Charlebois R.L., MacLeod D., Brochier C. (2005). The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol. 6: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker S.J., Stummer B., Gao L., Dispain I., Connor P.J.O., Smith S.E. (1998). A mutant in Lycopersicon esculentum Mill. with highly reduced VA mycorrhizal colonization: Isolation and preliminary characterisation. Plant J. 15: 791–797 [DOI] [PubMed] [Google Scholar]
- Belgareh N., Rabut G., Bai S.W., van Overbeek M., Beaudouin J., Daigle N., Zatsepina O.V., Pasteau F., Labas V., Fromont-Racine M., Ellenberg J., Doye V. (2001). An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P., Genre A., Faccio A., Martini I., Schauser L., Stougaard J., Webb J., Parniske M. (2000). The Lotus japonicus LjSym4 gene is required for the successful symbiotic infection of root epidermal cells. Mol. Plant Microbe Interact. 13: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Brohawn S.G., Leksa N.C., Spear E.D., Rajashankar K.R., Schwartz T.U. (2008). Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton W.J., Dilworth M.J. (1971). Control of leghaemoglobin synthesis in snake beans. Biochem. J. 125: 1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.T., Germain H., Wiermer M., Bi D., Xu F., Garcia A.V., Wirthmueller L., Despres C., Parker J.E., Zhang Y., Li X. (2009). Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haeze W., De Rycke R., Mathis R., Goormachtig S., Pagnotta S., Verplancke C., Capoen W., Holsters M. (2003). Reactive oxygen species and ethylene play a positive role in lateral root base nodulation of a semiaquatic legume. Proc. Natl. Acad. Sci. USA 100: 11789–11794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Schwartz R., Badani H., Smadar W., Levy A.A., Galili G., Kapulnik Y. (2001). Identification of a novel genetically controlled step in mycorrhizal colonization: plant resistance to infection by fungal spores but not extra-radical hyphae. Plant J. 27: 561–569 [DOI] [PubMed] [Google Scholar]
- Debler E.W., Ma Y., Seo H.S., Hsia K.C., Noriega T.R., Blobel G., Hoelz A. (2008). A fence-like coat for the nuclear pore membrane. Mol. Cell 32: 815–826 [DOI] [PubMed] [Google Scholar]
- Demchenko K., Winzer T., Stougaard J., Parniske M., Pawlowski K. (2004). Distinct roles of Lotus japonicus SYMRK and SYM15 in root colonization and arbuscule formation. New Phytol. 163: 381–392 [DOI] [PubMed] [Google Scholar]
- Duc G., Trouvelot A., Gianinazzi-Pearson V., Gianinazzi S. (1989). First report of non-mycorrhizal plant mutants (Myc-) obtained in pea (Pisum sativum L.) and fababean (Vicia faba L.). Plant Sci. 60: 215–222 [Google Scholar]
- Ehrhardt D.W., Wais R., Long S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681 [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Fath S., Mancias J.D., Bi X., Goldberg J. (2007). Structure and organization of coat proteins in the COPII cage. Cell 129: 1325–1336 [DOI] [PubMed] [Google Scholar]
- Finlay R.D. (2008). Ecological aspects of mycorrhizal symbiosis: With special emphasis on the functional diversity of interactions involving the extraradical mycelium. J. Exp. Bot. 59: 1115–1126 [DOI] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Faccio A., Barker D.G., Bonfante P. (2008). Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A., Chabaud M., Timmers T., Bonfante P., Barker D.G. (2005). Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimenko O.V., Gerasimenko J.V., Tepikin A.V., Petersen O.H. (1995). ATP-dependent accumulation and inositol trisphosphate- or cyclic ADP-ribose-mediated release of Ca2+ from the nuclear envelope. Cell 80: 439–444 [DOI] [PubMed] [Google Scholar]
- Gleason C., Chaudhuri S., Yang T., Munoz A., Poovaiah B.W., Oldroyd G.E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441: 1149–1152 [DOI] [PubMed] [Google Scholar]
- Goedhart J., Hink M.A., Visser A.J., Bisseling T., Gadella T.W., Jr. (2000). In vivo fluorescence correlation microscopy (FCM) reveals accumulation and immobilization of Nod factors in root hair cell walls. Plant J. 21: 109–119 [DOI] [PubMed] [Google Scholar]
- Goormachtig S., Capoen W., James E.K., Holsters M. (2004). Switch from intracellular to intercellular invasion during water stress-tolerant legume nodulation. Proc. Natl. Acad. Sci. USA 101: 6303–6308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D.J. (2003). Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 11: 853–864 [DOI] [PubMed] [Google Scholar]
- Harrison M.J. (2005). Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 59: 19–42 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Banba M., Shimoda Y., Kouchi H., Hayashi M., Imaizumi-Anraku H. (2010). A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Lombardo F., Miwa H., Perry J.A., Bunnewell S., Parniske M., Wang T.L., Downie J.A. (2006). Lotus japonicus nodulation requires two GRAS domain regulators, one of which is functionally conserved in a non-legume. Plant Physiol. 142: 1739–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S., Kim J., Munoz A., Heckmann A.B., Downie J.A., Oldroyd G.E. (2009). GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi-Anraku H., et al. (2005). Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 433: 527–531 [DOI] [PubMed] [Google Scholar]
- James E.K., Sprent J.I. (1999). Development of N2-fixing nodules on the wetland legume Lotus uliginosus exposed to conditions of flooding. New Phytol. 142: 219–231 [Google Scholar]
- Javot H., Penmetsa R.V., Terzaghi N., Cook D.R., Harrison M.J. (2007). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789 [DOI] [PubMed] [Google Scholar]
- Kanamori N., et al. (2006). A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 103: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B., Murray J., Gorzelak M., Smith A., Sato S., Tabata S., Szczyglowski K. (2005). Invasion of Lotus japonicus root hairless 1 by Mesorhizobium loti involves the nodulation factor-dependent induction of root hairs. Plant Physiol. 137: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inze D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawaguchi M., Motomura T., Imaizumi-Anraku H., Akao S., Kawasaki S. (2001). Providing the basis for genomics in Lotus japonicus: The accessions Miyakojima and Gifu are appropriate crossing partners for genetic analyses. Mol. Genet. Genomics 266: 157–166 [DOI] [PubMed] [Google Scholar]
- Kistner C., Winzer T., Pitzschke A., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Webb K.J., Szczyglowski K., Parniske M. (2005). Seven Lotus japonicus genes required for transcriptional reprogramming of the root during fungal and bacterial symbiosis. Plant Cell 17: 2217–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistner C., Parniske M. (2002). Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci. 7: 511–518 [DOI] [PubMed] [Google Scholar]
- Kosuta S., Chabaud M., Lougnon G., Gough C., Dénarié J., Barker D.G., Becard G. (2003). A diffusible factor from arbuscular mycorrhizal fungi induces symbiosis-specific MtENOD11 expression in roots of Medicago truncatula. Plant Physiol. 131: 952–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Hazledine S., Sun J., Miwa H., Morris R.J., Downie J.A., Oldroyd G.E. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. USA 105: 9823–9828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rue T.A., Weeden N.F. (1994). The symbiosis genes of the host. In Proceedings of the 1st European Nitrogen Fixation Conference, Kiss G.B., Endre G., (Szeged, Hungary: Officina Press; ), Pp. 147–151 [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364 [DOI] [PubMed] [Google Scholar]
- Lombardo F., Heckmann A.B., Miwa H., Perry J.A., Yano K., Hayashi M., Parniske M., Wang T.L., Downie J.A. (2006). Identification of symbiotically defective mutants of Lotus japonicus affected in infection thread growth. Mol. Plant Microbe Interact. 19: 1444–1450 [DOI] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E. (2002). Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmann K., Giczey G., Parniske M. (2008). Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 6: e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle T.P., Miller M.H., Evans D.G., Fairchild G.L., Swan J.A. (1990). A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115: 495–501 [DOI] [PubMed] [Google Scholar]
- Messinese E., Mun J.H., Yeun L.H., Jayaraman D., Rougé P., Barre A., Lougnon G., Schornack S., Bono J.J., Cook D.R., Ané J.M. (2007). A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant Microbe Interact. 20: 912–921 [DOI] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E., Long S.R. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101: 4701–4705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Sun J., Oldroyd G.E., Downie J.A. (2006). Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol. Plant Microbe Interact. 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Murakami Y., Miwa H., Imaizumi-Anraku H., Kouchi H., Downie J.A., Kawaguchi M., Kawasaki S. (2006). Positional cloning identifies Lotus japonicus NSP2, a putative transcription factor of the GRAS family, required for NIN and ENOD40 gene expression in nodule initiation. DNA Res. 13: 255–265 [DOI] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104 [DOI] [PubMed] [Google Scholar]
- Ndoye I., de Billy F., Vasse J., Dreyfus B., Truchet G. (1994). Root nodulation of Sesbania rostrata. J. Bacteriol. 176: 1060–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neer E.J., Schmidt C.J., Nambudripad R., Smith T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371: 297–300 [DOI] [PubMed] [Google Scholar]
- Niwa S., Kawaguchi M., Imazumi-Anraku H., Chechetka S.A., Ishizaka M., Ikuta A., Kouchi H. (2001). Responses of a model legume Lotus japonicus to lipochitin oligosaccharide nodulation factors purified from Mesorhizobium loti JRL501. Mol. Plant Microbe Interact. 14: 848–856 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Engstrom E.M., Long S.R. (2001). Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13: 1835–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott T., van Dongen J.T., Gunther C., Krusell L., Desbrosses G., Vigeolas H., Bock V., Czechowski T., Geigenberger P., Udvardi M.K. (2005). Symbiotic leghemoglobins are crucial for nitrogen fixation in legume root nodules but not for general plant growth and development. Curr. Biol. 15: 531–535 [DOI] [PubMed] [Google Scholar]
- Palma K., Zhang Y., Li X. (2005). An importin α homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Paszkowski U., Jakovleva L., Boller T. (2006). Maize mutants affected at distinct stages of the arbuscular mycorrhizal symbiosis. Plant J. 47: 165–173 [DOI] [PubMed] [Google Scholar]
- Penmetsa R.V., Cook D.R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275: 527–530 [DOI] [PubMed] [Google Scholar]
- Penmetsa R.V., Frugoli J.A., Smith L.S., Long S.R., Cook D.R. (2003). Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 131: 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J., Brachmann A., Welham T., Binder A., Charpentier M., Groth M., Haage K., Markmann K., Wang T.L., Parniske M. (2009). TILLING in Lotus japonicus identified large allelic series for symbiosis genes and revealed a bias in functionally defective ethyl methanesulfonate alleles toward glycine replacements. Plant Physiol. 151: 1281–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.A., Wang T.L., Welham T.J., Gardner S., Pike J.M., Yoshida S., Parniske M. (2003). A TILLING reverse genetics tool and a web-accessible collection of mutants of the legume Lotus japonicus. Plant Physiol. 131: 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Gronlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Reddy D.M.R.S., Schorderet M., Feller U., Reinhardt D. (2007). A petunia mutant affected in intracellular accommodation and morphogenesis of arbuscular mycorrhizal fungi. Plant J. 51: 739–750 [DOI] [PubMed] [Google Scholar]
- Ruijter J.M., Ramakers C., Hoogaars W.M., Karlen Y., Bakker O., van den Hoff M.J., Moorman A.F. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 19: 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195 [DOI] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A. (2002). TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18: 502–504 [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. (1998). SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüssler A., Schwarzott D., Walker C. (2001). A new fungal phylum, the Glomeromycota: Phylogeny and evolution. Mycol. Res. 105: 1413–1421 [Google Scholar]
- Sieberer B.J., Chabaud M., Timmers A.C., Monin A., Fournier J., Barker D.G. (2009). A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151: 1197–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E.C. (1996). A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791 [DOI] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. (1999). The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 24: 181–185 [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S., Morgan D.R., Swensen S.M., Mullin B.C., Dowd J.M., Martin P.G. (1995). Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc. Natl. Acad. Sci. USA 92: 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent J.I. (2007). Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytol. 174: 11–25 [DOI] [PubMed] [Google Scholar]
- Stockinger H., Walker C., Schüssler A. (2009). 'Glomus intraradices DAOM197198', a model fungus in arbuscular mycorrhiza research, is not Glomus intraradices. New Phytol. 183: 1176–1187 [DOI] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962 [DOI] [PubMed] [Google Scholar]
- Takeda N., Okamoto S., Hayashi M., Murooka Y. (2005). Expression of LjENOD40 genes in response to symbiotic and non-symbiotic signals: LjENOD40-1 and LjENOD40-2 are differentially regulated in Lotus japonicus. Plant Cell Physiol. 46: 1291–1298 [DOI] [PubMed] [Google Scholar]
- Takeda N., Sato S., Asamizu E., Tabata S., Parniske M. (2009). Apoplastic plant subtilases support arbuscular mycorrhiza development in Lotus japonicus. Plant J. 58: 766–777 [DOI] [PubMed] [Google Scholar]
- Tirichine L., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441: 1153–1156 [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107 [DOI] [PubMed] [Google Scholar]
- van Brussel A.A., Bakhuizen R., van Spronsen P.C., Spaink H.P., Tak T., Lugtenberg B.J., Kijne J.W. (1992). Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257: 70–72 [DOI] [PubMed] [Google Scholar]
- Van de Velde W., Zehirov G., Szatmari A., Debreczeny M., Ishihara H., Kevei Z., Farkas A., Mikulass K., Nagy A., Tiricz H., Satiat-Jeunemaître B., Alunni B., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Vierheilig H., Coughlan A.P., Wyss U., Piché Y. (1998). Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 64: 5004–5007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T., Mattaj I.W., Doye V. (2003). The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113: 195–206 [DOI] [PubMed] [Google Scholar]
- Wegel E., Schauser L., Sandal N., Stougaard J., Parniske M. (1998). Mycorrhiza mutants of Lotus japonicus define genetically independent steps during symbiotic infection. Mol. Plant Microbe Interact. 11: 933–936 [Google Scholar]
- Wojciechowski M.F., Lavin M., Sanderson M.J. (2004). A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am. J. Bot. 91: 1846–1862 [DOI] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li X. (2005). A putative nucleoporin 96 Is required for both basal defense and constitutive resistance responses mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17: 1306–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.