This work examines the diversity of meiotic behavior in Brassica napus allohaploids and shows that it is related to the multiple origins of this species. The results also reflect the prevalence of a major locus determining different levels of crossover suppression at the species level.
Abstract
Allopolyploid species contain more than two sets of related chromosomes (homoeologs) that must be sorted during meiosis to ensure fertility. As polyploid species usually have multiple origins, one intriguing, yet largely underexplored, question is whether different mechanisms suppressing crossovers between homoeologs may coexist within the same polyphyletic species. We addressed this question using Brassica napus, a young polyphyletic allopolyploid species. We first analyzed the meiotic behavior of 363 allohaploids produced from 29 accessions, which represent a large part of B. napus genetic diversity. Two main clear-cut meiotic phenotypes were observed, encompassing a twofold difference in the number of univalents at metaphase I. We then sequenced two chloroplast intergenic regions to gain insight into the maternal origins of the same 29 accessions; only two plastid haplotypes were found, and these correlated with the dichotomy of meiotic phenotypes. Finally, we analyzed genetic diversity at the PrBn locus, which was shown to determine meiotic behavior in a segregating population of B. napus allohaploids. We observed that segregation of two alleles at PrBn could adequately explain a large part of the variation in meiotic behavior found among B. napus allohaploids. Overall, our results suggest that repeated polyploidy resulted in different levels of crossover suppression between homoeologs in B. napus allohaploids.
INTRODUCTION
Meiosis is an obligatory process for all sexually reproducing organisms. This specialized type of cell division is essential to produce gametes, ensure genome stability throughout sexual life cycles, and generate diversity within species by creating new chromosome/allele combinations. For all these outcomes, the exclusive formation of crossovers (COs) between homologous chromosomes is required (Hamant et al., 2006). Our knowledge of the genes and mechanisms involved in meiotic recombination and CO formation in plants has received a boost in the last 6 years with the use of Arabidopsis thaliana, rice (Oryza sativa), and maize (Zea mays) as model systems (Mezard et al., 2007; Mercier and Grelon, 2008). However, little is known about natural variation in recombination rates within species (Säll, 1990; Sanchez-Moran et al., 2002; Anderson et al., 2003; Esch et al., 2007; Bovill et al., 2008), which is an important issue for understanding how recombination is regulated in the wild.
The existence of natural variability for CO regulation is particularly relevant in polyploid species, considering that most have multiple origins and that genetic systems restricting CO to homologs are required. Recurrent polyploidy is known to be the rule rather than an exception. Most nascent polyploid species should be considered as a set of genetically variable lineages produced from distinct diploid progenitors that can subsequently produce novel genotypes through hybridization and recombination (Soltis and Soltis, 1999). As these new polyploid species are not completely isolated from their diploid progenitors, the variability present in the diploids can continue to be incorporated into the polyploids. This may subsequently lead to different genetic/epigenetic changes, which can further expand the range of phenotypes. These eventualities are yet to be addressed experimentally (e.g., Dubcovsky and Dvorak, 2007; Koh et al., 2010), and little is known about the consequences of recurrent polyploidy on the establishment of polyploid species, in particular for processes that are directly relevant to natural selection, such as meiosis.
Proper chromosome segregation is a demanding process in polyploid species, including those of hybrid origin (i.e., allopolyploids). These species have more than two complete sets of chromosomes related by ancestral homology (so-called homoeologs). Homoeologs are usually still able to form COs during meiosis but must be sorted to produce viable and balanced gametes. Suppression of COs between homoeologous chromosomes is thus required to ensure fertility. Current understanding is that this process is genetically determined in many allopolyploids and usually subject to polygenic regulation (Jenczewski et al., 2003; Cifuentes et al., 2010). However, it is not known whether recurrent polyploidy drives variation in the determinants of CO between homoeologous chromosomes in allopolyploid species.
Wheat is the main model for which some data are available. Common bread wheat (Triticum aestivum; 2n = 6x = 42; genome formula AABBDD) is the product of two successive polyploidization events (2x→4x then 4x→6x); it arose from at least two maternal lineages (Hirosawa et al., 2004), had two genetically distinct D-genome progenitors (Caldwell et al., 2004), and captured a large portion of the natural genetic diversity present in its tetraploid ancestor (Dubcovsky and Dvorak, 2007). In both tetraploid and hexaploid wheat, the main locus responsible for exclusive homologous CO formation is Ph1 (Riley and Chapman, 1958; Feldman, 1966; Sears, 1976; Giorgi, 1978), which was recently defined to a region containing a cluster of cyclin-dependent kinase-related genes interrupted by a heterochromatin segment (Griffiths et al., 2006; Al-Kaff et al., 2008). This idiosyncratic structure is apparently conserved among polyploid wheat species (Griffiths et al., 2006), which is in contrast with the slight variability observed for CO suppression between homoeologous chromosomes in both tetraploid (Ozkan and Feldman, 2001) and hexaploid (Martinez et al., 2005) wheat species.
The limited data available on Brassica napus suggest that the situation could be different in this species, although it evolved roughly in the same way as wheat. B. napus (AACC; 2n = 38) is a young allopolyploid species with multiple origins (i.e., a polyphyletic species) that formed by repeated interspecific hybridization between ancestors of Brassica oleracea (CC, 2n = 18) and Brassica rapa (AA, 2n = 20) (U, 1935; Palmer et al., 1983; Song and Osborn, 1992; Allender and King, 2010). Natural euploid B. napus displays predominantly 19 bivalents at Metaphase I (MI) and an almost strict disomic inheritance; this shows that the vast majority of COs are formed between homologous chromosomes. Evidence for rare homoeologous exchanges was obtained in several B. napus cultivars (Parkin et al., 1995; Sharpe et al., 1995; Lombard and Delourme, 2001; Osborn et al., 2003; Piquemal et al., 2005; Udall et al., 2005; Howell et al., 2008), but their frequency remains very low compared with the rate in resynthesized B. napus (Parkin et al., 1995; Sharpe et al., 1995; Udall et al., 2005; Lukens et al., 2006; Gaeta et al., 2007; Szadkowski et al., 2010). Contrary to wheat, comparisons of meiosis among B. napus allohaploid plants, which carry one copy of each of the 10 A and 9 C B. napus chromosomes (AC), showed that allohaploids produced from some varieties displayed only a few univalents (i.e., chromosomes that fail to form a CO), whereas those produced from other varieties displayed mostly univalents (Olsson and Hagberg, 1955; Renard and Dosba, 1980; Attia and Röbbelen, 1986; Jenczewski et al., 2003). The quantitative trait loci (QTL) determining these phenotypes were mapped in a single population. A major locus (PrBn; Jenczewski et al., 2003) localized to linkage group C9, and four to six other additive or epistatic loci (Liu et al., 2006) were identified. Due to the polyphyletic origin of B. napus, it was logical to ask if PrBn is still the main determinant for CO suppression between homoeologs at the entire species level.
Our goal in this study was to decipher whether the diversity of meiotic behavior found among a wide range of B. napus allohaploid accessions is related to the polyphyletic origin of this species and diversity at the PrBn locus. We first characterized the meiotic behavior at MI in allohaploids produced from 29 B. napus varieties representing a range of genetic and geographic origins. We then analyzed chloroplast diversity in all these varieties to assess their maternal origins. Finally, we assayed molecular markers surrounding PrBn to reconstruct different multilocus genotypes for this region. Altogether, our results indicate that variation in CO frequency among allohaploid genotypes roughly correlates with the multiple origins of B. napus and PrBn diversity. More complex patterns of meiotic and genetic diversity were also observed. Our findings highlight the diverse nature of homoeologous recombination regulation in the wild.
RESULTS
Diversity of Meiotic Behavior among B. napus Allohaploids
A total of 363 allohaploid plants were isolated from 29 varieties representing different types of cultivars and a major part of B. napus genetic diversity (Table 1). We analyzed the meiotic behavior at MI of all allohaploids and one raw B. oleracea × B. rapa interspecific hybrid, which have a similar karyotype (10 A and 9 C chromosomes) (Figures 1 and 2; see Supplemental Data Set 1 online). Homologous chromosomes are absent in all these plants; thus, the chiasmata observed on bivalents and multivalents (Figure 1B) mark the sites of meiotic COs between homoeologous and/or nonhomologous chromosomes (Nicolas et al., 2007). We also observed varying numbers of univalents in all plants (Figure 1). We used the number of univalents to describe the MI meiotic behavior of B. napus allohaploids because this variable is directly correlated to the extent of CO formation between homoeologous or nonhomologous chromosomes (Nicolas et al., 2009). All the B. napus allohaploids displayed a significantly higher number of univalents than did the A×C interspecific hybrids (F-test; P < 0.001; Figure 2), suggesting that the number of COs between homoeologous/nonhomologous chromosomes was reduced in the B. napus background regardless of the accession.
Table 1.
Accessions Used to Assess and Compare Patterns of Meiotic, Plastid, and Nuclear Marker Diversity
| Species | Accession Name | Country of Origin | Plant Breedera | Pop. Type | Growth Habit | No. of Parental Plants | No. of Haploids | GenBanka | GenBank Accession No. |
| B. rapa | Z1 | Canada | AAFC | Line | AAFC | ||||
| B. oleracea | HDEM | France | INRA | Line | INRA Rennes | ||||
| B. napus spp oleifera | Akamar | Holland | VDH | Population | Winter | 4 | 25 | INRA Rennes | |
| B. napus spp oleifera | Brutor | France | Ringot | Line | Spring | 1 | 8 | INRA Rennes | |
| B. napus spp oleifera | Capricorn | UK | PBI | Population | Winter | 4 | 24 | INRA Rennes | |
| B. napus spp oleifera | Drakkar | France | INRA-Serasem | Line | Spring | 1 | 8 | INRA Rennes | |
| B. napus spp oleifera | Darmor-bzh | France | INRA-Serasem | Line | Winter | 1 | 59 | INRA Rennes | |
| B. napus spp oleifera | Eurol | France | Cargill | Line | Winter | 1 | 1 | INRA Rennes | |
| B. napus spp oleifera | Garant | Germany | Lembkes | Population | Winter | 3 | 9 | INRA Rennes | |
| B. napus spp oleifera | Hinchu | Korea | Nokpo Univ. | Line | Spring | 1 | 2 | INRA Rennes | |
| B. napus spp pabularia | asparagus kale | Population | Spring | 3 | 12 | WGB | 6224 | ||
| B. napus spp oleifera | Jet Neuf | France | Ringot | Line | Winter | 1 | 5 | INRA Rennes | |
| B. napus spp oleifera | Loras | Germany | Petkus | Population | Spring | 4 | 12 | GGB | 734 |
| B. napus spp oleifera | Maluka | Australia | NSWDA | Line | Spring | 1 | 1 | INRA Rennes | |
| B. napus spp oleifera | Maxol | France | Cargill | Line | Winter | 1 | 7 | INRA Rennes | |
| B. napus spp oleifera | Mohican | UK | CPB | Population | Winter | 4 | 33 | INRA Rennes | |
| B. napus spp oleifera | Nachan | Korea | Nokpo Univ | Line | Spring | 1 | 8 | INRA Rennes | |
| B. napus spp oleifera | Norin 1 | Japon | Fukuoka | Line | Spring | 1 | 9 | INRA Rennes | |
| B. napus spp oleifera | Norin 6 | Japon | OOSaka | Line | Spring | 1 | 10 | INRA Rennes | |
| B. napus spp oleifera | Norin 9 | Japon | OOSaka | Line | Spring | 1 | 4 | INRA Rennes | |
| B. napus spp oleifera | Norin 10 | Japon | Fukushima | Line | Spring | 1 | 9 | INRA Rennes | |
| B. napus spp oleifera | Oro | Canada | AAFC | Population | Spring | 5 | 13 | AAFC | |
| B. napus spp oleifera | Petranova | Germany | Petkus | Line | Winter | 1 | 3 | INRA Rennes | |
| B. napus spp rapifera | Rutabaga 22 | France | – | Line | Winter | 1 | 8 | INRA Rennes | |
| B. napus spp rapifera | Rutabaga 85 | France | – | Line | Winter | 1 | 10 | INRA Rennes | |
| B. napus spp oleifera | Samouraï | France | INRA-Serasem | Line | Winter | 1 | 5 | INRA Rennes | |
| B. napus spp oleifera | Spok | Germany | Dansk Plants Foraedling | Line | Spring | 1 | 7 | INRA Rennes | |
| B. napus spp oleifera | Stellar | Canada | Univ. Manitoba | Line | Spring | 1 | 8 | INRA Rennes | |
| B. napus spp oleifera | Taichung | Korea | Nokpo Univ | Line | Spring | 1 | 6 | INRA Rennes | |
| B. napus spp oleifera | Westar | Canada | AAFC | Line | Spring | 1 | 5 | INRA Rennes | |
| B. napus spp oleifera | Yudal | Korea | Nokpo Univ | Line | Spring | 1 | 51 | INRA Rennes |
AAFC, Agriculture and AgriFood Canada; INRA, Institut National de la Recherche Agronomique; VDH, van der Have BV; PBI, Plant Breeding International Cambridge; NSWDA, New South Wales Department of Agriculture; CPB, Cambridge Plant Breeder Twyford; WGB, Wellsbourne Gene Bank, Horticulture Research International; GGB, Gatersleben GeneBank, Institure of Plant Genetics and Crop Plant Research.
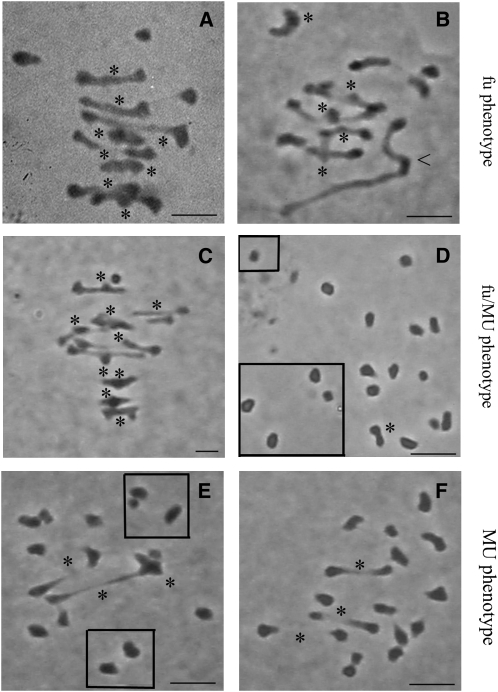
Figure 1.
Representative MI Nuclei of B. napus Allohaploids Showing Contrasting Meiotic Behaviors.
(A) Darmor-bzh: eight bivalents (II) + three univalents (I).
(B) Norin 1: one quadrivalent, 5II +4I.
(C) and (D) Norin 9: 9II + 1I – 1II + 17I,
(E) Garant: 3II + 13I.
(F) Yudal: 3II + 13I.
The univalents located peripherally (out of the frame of these high-magnification micrographs) are indicated within squares ([D] and [E]). Bivalents are indicated with an asterisk, and the quadrivalent in (B) is indicated with an arrowhead. Bars = 5 μm.
Figure 2.
Diversity of Meiotic Behavior in B. napus Allohaploids.
Symbols represent the mean number of univalents (calculated for ~20 PMCs) for every allohaploid plant isolated from the 29 B. napus accessions listed on the x axis and for five interspecific B. oleracea × B. rapa hybrids (noted A×C). Symbols with the same X-coordinate represent allohaploids isolated from the same plant. The clusters of consecutive X-coordinate samples represent three to four distinct plants sampled from the same population to account for its potential genetic heterogeneity (e.g., Mohican, Capricorn, etc.). Triangles represent allohaploids showing a high level of homoeologous recombination (fu allohaploids), diamonds represent allohaploids with an intermediate meiotic behavior, and squares represent allohaploids that showed a low level of homoeologous recombination (MU allohaploids). The two plastid haplotypes are represented by open and closed symbols, respectively.
[See online article for color version of this figure.]
We identified two clear-cut meiotic phenotypes among all B. napus allohaploids (Figures 1 and 2). Allohaploids isolated from a first group of 19 accessions, including Darmor-bzh, showed a high level of recombination between homoeologous/nonhomologous chromosomes; they had from 3.57 to 5.14 univalents per accession on average, mean numbers of univalents per allohaploid plant not higher than 6 (Figures 1A, 1B, and 2), and 9 to 12 chiasmata per pollen mother cell (PMC). We named this group fu, for few univalents. By contrast, allohaploids produced from a second group of seven accessions, including Yudal, showed a low level of recombination between homoeologous /nonhomologous chromosomes; they displayed from 8 to 11.4 univalents per accession on average, mean numbers of univalents per allohaploid plant not lower than 8 (Figures 1E, 1F, and 2), and two to six chiasmata per PMC. We called this group MU, for many univalents. Similar differences in the number of multivalents were observed between the fu and MU groups: 128 trivalents (III) and 281 quadrivalents (IV) but only 31 III and 16 IV were scored for the fu and MU allohaploids, respectively. The remaining three accessions (Loras, Oro, and Norin9) produced a mixture of allohaploids with either the fu or MU meiotic phenotype (Figures 1C, 1D, and 2).
Only eight allohaploids showed an intermediate meiotic phenotype (with mean numbers of univalents between 6 and 8), six of which originated from Loras, Oro, and Norin9 (Figure 2). A detailed analysis of the frequency of univalents per PMC revealed that four of them remained very similar to the MU or fu allohaploids, differing only in one additional/missing pair of univalents (e.g., Norin9-02 and Loras-07 in Supplemental Figure 1 online). The distribution of the number of univalents per PMC was clearly different for the remaining four intermediate allohaploids. Many of their MI cells had univalent scores that were rarely observed in MU/fu allohaploids (e.g., please compare Oro-09 and Loras-09 to Darmor-bzh and Yudal in Supplemental Figure 1 online). Thus, the meiotic phenotypes of only these four allohaploids (out of 363) did not noticeably match the clear-cut fu/MU dichotomy.
Slight differences were sometimes observed between allohaploids produced from a given accession (Figure 2). In general, statistically significant differences were found between allohaploids produced from every MU accession (P = 0.007; Wald Z-tests), whereas no significant difference was found between allohaploids isolated from fu accessions (P = 0.0936; Wald Z-tests). In some cases, the number of univalents determined for different allohaploids isolated from the same accession differed depending on their spatial locations in the greenhouse, suggesting a slight environmental effect across the experimental area (see Supplemental Figure 2 online). The response surfaces appeared to be the same between the different mother plants representing an open-pollinated population and between some accessions, but they were different between some others (see Supplemental Figure 2 online). This meant that spatial autocorrelation could not be introduced into the statistical models. It is also apparent in Figure 2 that variance among allohaploids isolated from MU accessions increased when the mean increased, a relationship that was taken into account in subsequent statistical analyses.
Analyses of variance, performed separately on the fu and MU groups, showed that significant differences existed between accessions within each group (P < 0.001 for both analyses). As no significant difference was observed between the different mother plants representing an open-pollinated population (Figure 2), these data were pooled before we analyzed the extent to which the number of univalents varied between accessions. Significant block effects were also found for fu allohaploids (P = 0.04; Wald Z-tests), demonstrating that a small but significant fraction of spatial and temporal heterogeneity was captured while testing for differences between accessions.
Variation between accessions within the fu or MU groups was continuous, with pairwise differences divided into a series of overlapping subgroups. No clear-cut subgroup could thus be recognized. However, it was possible to identify accessions that were significantly different from Darmor-bzh and Yudal, the two varieties we used in previous analyses (Jenczewski et al., 2003; Liu et al., 2006; Nicolas et al., 2009). We found that the number of univalents in allohaploids originating from six accessions (Akamar, Brutor, JetNeuf, Nachan, Norin10, and Taïchung), plus the fu allohaploids produced from Loras and Oro, was slightly but significantly higher than in Darmor-bzh allohaploids (see Supplemental Table 1 online). Likewise, the single fu allohaploids isolated from Maluka and Norin9 differed from Darmor-bzh allohaploids, although no statistical comparison was possible. By contrast, no B. napus allohaploid produced significantly fewer univalents than did Darmor-bzh. In the MU group, when variance stabilizing corrections were applied, only the allohaploids originating from Hinchu plus the MU allohaploids produced from Loras and Oro displayed significantly fewer univalents than did Yudal allohaploids (see Supplemental Table 1 online); only Asparagus kale produced significantly more univalents at the allohaploid stage than did Yudal.
Finally, we estimated that the variance for the number of univalents attributable to differences between the fu and MU groups (estimated at 18.54) was considerably larger than any other source of variation (for example, the variance attributable to differences between accessions from the same group was estimated at 0.3231). Due to the polyphyletic origin of B. napus, we addressed the hypothesis that these two main meiotic phenotypes could have originated from independent polyploidization events.
Diversity of Plastid Haplotypes among B. napus Accessions and Their Distribution with Respect to Allohaploid Meiotic Behavior
We first analyzed the genetic diversity of the chloroplast genome to gain insight into the maternal origins of the 29 B. napus accessions.
Sequencing both the ndhC-trnV and rbcL-accD chloroplast intergenic regions showed exactly the same extent of chloroplast diversity. Only two plastid genome (ptDNA) haplotypes were found; they differed by a total of nine single nucleotide polymorphisms and three indels (insertion/ deletion), including a 10-bp deletion (see Supplemental Data Set 2 online). Twenty four accessions, including the two rutabagas (B. napus ssp rapifera), had the same ptDNA haplotype, which was different from the ptDNA haplotypes found in the B. rapa and B. oleracea genotypes examined in this study (ptDNA haplotype 1; Figure 2; see Supplemental Data Set 2 online). The remaining five B. napus accessions, including asparagus kale (B.napus ssp pabularia), shared a second ptDNA haplotype that was more closely related to that of B. rapa Z1 and B. oleracea HDEM genotypes (ptDNA haplotype 2; Figure 2; see Supplemental Data Set 2 online). The plastid haplotypes of B. napus accessions did not appear to be completely randomly distributed with regards to the geographic origin; in particular, ptDNA haplotype 2 is found in German and Korean (Figure 2, Table 1) oilseed accessions as well as in B.napus ssp pabularia, which is thought to come out of Northern Europe/Asia.
We then compared the patterns of meiotic and ptDNA diversity and observed that the two plastid haplotypes were not randomly distributed with respect to the two main meiotic behaviors (Fisher’s exact test, P = 0.0123). Indeed, almost all the fu accessions (18 out of 19) displayed ptDNA haplotype 1, whereas a more balanced mix of ptDNA haplotypes was found among MU accessions (Figure 2). ptDNA haplotype 1 was present in B. napus accessions that produced a mixture of allohaploids with either the MU or fu meiotic phenotype (Loras, Oro, and Norin9) (Figure 2). Of note, the distribution of the two meiotic phenotypes appeared to be independent from any other source of clustering, such as the winter and spring oilseed types (Fisher’s exact test, P > 0.05).
Overall, our results confirmed that B. napus arose at least twice and suggest that the dichotomy of meiotic behaviors among B. napus allohaploids could be related to these multiple origins. To gain further insight into the origin of the natural variability for homoeologous recombination in B. napus, we analyzed genetic diversity at the PrBn locus (Jenczewski et al., 2003; Liu et al., 2006).
Nuclear DNA Marker Diversity in the PrBn Region with Respect to the Allohaploid Meiotic Behavior
We analyzed the genetic diversity of eight molecular markers spanning a region of ~60 centimorgans (cM) centered on the peak of the QTL (between CB10013a and BnGMs185 markers) defined by Liu et al. (2006) as PrBn (Figure 3). Most of these markers were dominant so that our screening mainly resulted in binary band presence-absence patterns. We considered that bands showing the same electrophoretic mobility between different accessions were identical-by-descent and that all accessions that showed no band for a dominant marker had the same allele at that locus.
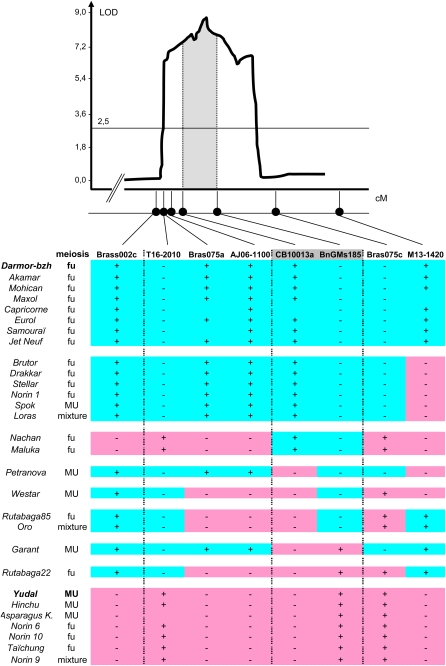
Figure 3.
Genotype of the Surveyed Accessions in the Region Surrounding PrBn, the Main Determinant for the Number of Univalents among B. napus Allohaploids.
The likelihood ratio profile at the top of the figure was modified from Liu et al. (2006); this profile was generated by composite interval mapping with a LOD score threshold of 3.3. x axis, map distances in cM; y axis, LOD score. Markers were ordered according to published genetic maps (Delourme et al., 2006, 2008; Liu et al., 2006). The contrasted genotypes of the Darmor-bzh and Yudal accessions, which were the parents of the segregating population used to map PrBn (Liu et al., 2006), were used to identify alleles for every marker: open cells represent Darmor-bzh-like alleles, while closed cells represent Yudal-like alleles. + Indicates that the band was present, and − indicates that the band was absent.
[See online article for color version of this figure.]
The most significant association with meiotic variation was observed for CB10013a (χ2 = 6.86; P = 0.008): 19 accessions (out of 25; 75%) had a CB10093a allele that matched their meiotic behavior and 37% of the overall variation for the number of univalents was accounted for by allele segregation at CB10013a. Owing to the fact that MU and fu meiotic phenotypes are both found across the most distinct genetic pools of B. napus (spring and winter oilseed types; see above), this association certainly reflects the close proximity of CB10093a to PrBn (Figure 3).
This conclusion is supported by our modeling of multilocus genotypes and their comparison to those of the Darmor-bzh and Yudal accessions, the two varieties used to identify and map PrBn (Jenczewski et al., 2003; Liu et al., 2006). We first identified a total of nine multilocus genotypes (Figure 3). The two most frequent multilocus genotypes were similar to those of Darmor-bzh and Yudal, respectively; seven accessions had the same multilocus genotype as Darmor-bzh (this number extends to 13 if marker M13-1420, which is located ~25 cM away from the peak of the QTL, is not considered), and six accessions shared the same multilocus genotype with Yudal (Figure 3). The remaining accessions had a recombinant multilocus genotype between these two archetypes, two of these showing a single marker data point that produced a double recombinant genotype. These two singletons were repeatedly found in reiterated experiments that included flanking markers.
We then compared the patterns of meiotic behavior and multilocus genotype diversity. MU accessions appeared more diverse (five multilocus genotypes out of seven accessions) than did fu accessions (six multilocus genotypes out of 19 accessions). However, the multilocus genotype in the PrBn region of most accessions matched their meiotic phenotype. Eleven fu accessions (out of 18) had the same multilocus genotype as Darmor-bzh, while two MU accessions (out of six) had the same multilocus genotype as Yudal. Focusing on the region near the peak of the PrBn QTL (between CB10013a and BnGMs185 markers) revealed that the profiles of all but one of the recombinant multilocus genotypes could match their MU/fu phenotype (see Supplemental Figure 3 online). Actually, the meiotic phenotype of only five accessions did not match their PrBn genotype; four accessions (Norin6, Norin10, Taïchung, and Rutabaga 22) had the same multilocus genotype as Yudal in the interval surrounding PrBn but behaved like Darmor-bzh allohaploids at MI; reciprocally, one accession (Spok) had the same multilocus genotype as Darmor-bzh at PrBn but behaved like Yudal allohaploids at MI (Figure 3). The three accessions that gave a mixture of fu or MU allohaploids displayed three unrelated multilocus genotypes (Figure 3): one similar to that of Darmor-bzh (Loras), one similar to that of Yudal (Norin9), and one recombinant multilocus genotype (Oro).
DISCUSSION
Natural and resynthesized B. napus are known to display very different levels of meiosis regularity and genome stability (for review, see Gaeta and Pires, 2010) that are reflected here by the reduction in CO between homoeologs in B. napus allohaploids compared with raw B. oleracea × B. rapa interspecific hybrids (Figure 2). In principle, this difference could reflect chromosome rearrangements that accentuated the divergence between B. napus homoeologous chromosomes after the inception of this species. However, although a few genetic changes were detected in several B. napus cultivars (references cited in the introduction), accumulating evidence indicates that the B. napus A/C genomes remain essentially the same as the A/C genomes of their progenitors (Bohuon et al., 1996; Parkin and Lydiate, 1997; Rana et al., 2004; Suwabe et al., 2008; Cheung et al., 2009). This suggests that natural B. napus has evolved or inherited Pairing-homoeologous loci that ensure proper chromosome recombination and segregation (such as PrBn; Jenczewski et al., 2003; Liu et al., 2006; Nicolas et al., 2009).
Our study showed that this genetic regulation occurs with varying degrees of stringency, at least among B. napus allohaploids. We effectively observed natural variation for recombination between homoeologous chromosomes that relied on two clear-cut meiotic phenotypes. Rough estimates of chiasma frequencies indicated that MU allohaploids displayed a range of two to six chiasmata per PMC, whereas the number of chiasmata per PMC varied from 9 to 12 in fu allohaploids (see also Renard and Dosba, 1980; Attia and Röbbelen, 1986). As already stated in Nicolas et al. (2009), this variation does not reflect a difference in the number of chiasmata that are formed on the recombining bivalents, which are strikingly similar between fu and MU allohaploids (~1.4 COs per bivalent on average). Conversely, it results from a difference in the number of chromosomes that form bivalents or multivalents during meiosis (Nicolas et al., 2009). In B. napus allohaploids, bivalents are not always formed between homoeologous chromosomes, as autosyndetic pairs (A-A or C-C) were also observed in the same proportion at meiosis of Darmor-bzh fu and Yudal MU allohaploids (Nicolas et al., 2007, 2009). Because these two genotypes are representative of the whole range of meiotic phenotypes (Figure 2), we would not expect more than slight variations in the distribution of COs between homoeologous and other nonhomologous chromosomes among B. napus allohaploids.
Our worldwide sample encompassed three main B. napus gene pools (ssp oleifera, ssp rapifera, and ssp pabularia), included both winter and spring oilseed types (Diers and Osborn, 1994; Lombard et al., 2000; Hasan et al., 2006), and thus represented a major part of B. napus genetic diversity. Although phenotyping other B. napus accessions could provide supplementary information, the dichotomy of meiotic behaviors found here certainly highlights the overall variation present in this species, which contains germplasm of shared recent common ancestry (Prakash and Hinata, 1980; Allender and King, 2010). This observation and the fact that no other meiotic behavior was found in a handful of other accessions (Olsson and Hagberg, 1955; Renard and Dosba, 1980; Attia and Röbbelen, 1986; Tai and Ikonen, 1988) suggest that most B. napus allohaploids display either a high (MU phenotype) or a low (fu phenotype) number of univalents, with only slight variations within these two canonical phenotypes (Figure 2). These slight variations may reflect either the segregation of genes modifying chiasma frequency or, alternatively, the occurrence of chromosomal changes that differ between accessions (discussed in Udall et al., 2005; Liu et al., 2006).
Only four allohaploids (out of 363) showed a meiotic behavior between the MU and fu phenotypes (Figure 2). Three of these originated from accessions that also produced a mixture of fu and MU allohaploids (Oro and Loras). These results are reminiscent of those obtained in the segregating population of allohaploids isolated from Darmor-bzh × Yudal F1 hybrids (Jenczewski et al., 2003). Thus, it is possible that Oro and Loras are heterozygous for some determinants of CO formation between homoeologous chromosomes. Loras and Oro were obtained by open pollination and, therefore, potentially contain a bulk of more or less related genotypes. However, the explanation is not straightforward for Norin9, which was thought to be a homogeneous line but produced a mixture of fu and MU allohaploids.
The clear-cut dichotomy of meiotic phenotypes found among B. napus allohaploids is in contrast with the slight differences observed between wheat allohaploids (T. aestivum; ABD n = 21; Martinez et al., 2005 and references therein) and tall fescue (Festuca arundinacea; ABC n = 21; Eizenga and Kasperbauer, 1985) in which the number of univalents varied only from 16 to 20. Although this narrow range in wheat could be due to the small number of genotypes analyzed (three cultivars), whether it is a consequence of the polyploid history of wheat is yet to be determined. Ph1 is the main determinant of CO suppression between homoeologs (Riley and Chapman, 1958). All polyploid wheat species, including Triticum timopheevi and Triticum araraticum (AAGG), show Ph1 activity and ostensibly the same structure at the Ph1 locus (Griffiths et al., 2006). Thus, the variation in CO suppression between homoeologs observed either between T. aestivum allohaploids (Martinez et al., 2005) or between tetraploid wheats × Aegilops peregrina hybrids (Ozkan and Feldman, 2001) remains unexplained.
Compared with wheat, our study provided more straightforward support for the hypothesis that repeated polyploidy has driven extensive variation in the determinants of CO suppression between homoeologous chromosomes in B. napus. Indeed, our results suggest that MU and fu allohaploids could originate from different maternal origins. All but one fu accession showed haplotype 1 ptDNA, and the almost perfect correlation between ptDNA haplotype and MU/fu phenotype (Figures 2) was disrupted only for four accessions (out of the 26 that did not produce a mixture of MU/fu allohaploids). These exceptions were expected due to the history of oilseed rape breeding, which involved a series of crosses between established cultivars (Salisbury and Wratten, 1999; Handa, 2007). For example, only two founder spring varieties were used to improve oilseed quality in the overwhelming majority of modern spring and winter oilseed genotypes (Hasan et al., 2008). Although the pedigrees of most accessions analyzed in this study are largely unknown, there is no reason to believe that these varieties were not the products of (extensive) intercrossing (see Handa, 2007 for the genealogy of Japanese varieties), which provided many opportunities for cytoplasm exchanges. Indeed, we observed that the high diversity of ptDNA found in the MU group (Figure 2) correlated with a high diversity of multilocus genotypes around PrBn (five out of seven accessions). These results are in close agreement with the findings of Sharpe and Lydiate (2003), who showed that the genome of oilseed rape cultivars is a mosaic of blocks from ancestral genotypes. We thus speculate that the MU and fu groups originated as genetically distinct lineages produced from at least two maternal diploid progenitors. At present, we do not know if the determinant conferring variation in meiotic behavior was directly inherited from alleles segregating in diploid progenitors or if it arose through subsequent introgressions or other genetic/epigenetic changes that independently accumulated in the different nascent lineages. Regardless of the original source of variation, subsequent crossing between accessions produced new combinations of meiotic and plastid types and the original evolutionary signal has faded.
B. napus ptDNA diversity definitely appears to have originated from independent polyploidization events. As B. napus is of very recent origin, it is very unlikely that the chloroplast of this species has accumulated such a high level of polymorphisms (nine single nucleotide polymorphisms and three indels; see Supplemental Data Set 2 online) since its inception. B. napus ptDNA diversity is also unlikely to have originated from a cytoplasm introgression from related species, which could have potentially occurred from B.rapa into some Asian B. napus accessions (Handa, 2007). All Norin accessions shared ptDNA haplotype 1, which was common to most other accessions but different from the ptDNA haplotype of the B. rapa genotype here. As proposed by Palmer et al. (1983), Erickson et al. (1983), and Song and Osborn (1992), ptDNA diversity provides evidence for at least two separate origins of B. napus involving two different maternal parents. However, Song and Osborn (1992) identified four maternal lineages instead of two, with most accessions showing the same plastid DNA as Brassica montana. Contrary to Song and Osborn (1992), we found that Brutor had the same ptDNA haplotype as almost all other accessions (Figure 2; see Supplemental Data Set 2 online). We analyzed different plastid regions than did Song and Osborn (1992), so this difference could reflect the idiosyncrasy of the B. montana chloroplast genome, which has local similarities with the other B. napus ptDNAs (Song and Osborn, 1992). Second, none of the accessions in our study had a B. oleracea ptDNA haplotype, which was represented only by the New Zealand Rawara accession in the Song and Osborn study; however, we found that this accession had only 18 chromosomes, suggesting that it belongs to the B. oleracea and not the B. napus germplasm (confirmed in Allender and King, 2010). Apart from these two points, our results confirmed the conclusions of Song and Osborn (1992). The two B. napus ptDNA haplotypes occurred in very different frequencies. Most accessions had ptDNA haplotype 1, and only five accessions had ptDNA haplotype 2 (Figure 2). We also confirmed that rutabagas had the main ptDNA haplotype (ptDNA haplotype 1), whereas asparagus kale had ptDNA haplotype 2. These results are also in agreement with the recent work of Allender and King (2010).
Finally, analysis of nuclear marker diversity in the PrBn region gave further support to the hypothesis that the dichotomy of meiotic phenotypes originated from (at least) two polyploidization events. The meiotic phenotype of 22 accessions (out of 29) was consistent with their multilocus genotype in the region surrounding PrBn (Figure 3). Half of these displayed the same multilocus genotype as did Darmor-bzh or Yudal, while the other half was recombinant in the interval with alleles at the peak of the QTL being in agreement with the meiotic phenotype (see Supplemental Figure 3 online). These results confirm the most likely position for PrBn (between markers CB10013a and BnGMs185; Figure 3); owing to the rapid decay of linkage disequilibrium with distance in B. napus (Ecke et al., 2010), the statistical associations we found certainly reflects the close proximity of PrBn to the markers we used. These results also demonstrate that natural variation in meiotic behavior among B. napus allohaploids is consistent with the segregation of two alleles at the PrBn locus. This is the expected allelic composition if B. napus resulted from a double origin.
The meiotic phenotype of only eight accessions did not match their genotype in the region surrounding PrBn (Figure 3). Three of these were a mixture of MU/fu allohaploids, while they had the multilocus genotype fully or partly similar to that of Darmor-bzh or Yudal. These results echo the existence of similar allohaploids in the segregating population analyzed by Liu et al. (2006). Our study thus confirms that some accessions may have or segregate for different loci other than PrBn, which would modify the chance for homoeologous chromosomes to recombine. Although the origin, location, and mode of action of these loci remains to be determined (see Liu et al., (2006), our study provides a way of deliberately choosing genotypes to advance our understanding of the overall genetic architecture of the regulation of CO formation between homoeologous chromosomes in B. napus.
We showed that repeated polyploidy in B. napus probably resulted in different levels of CO suppression between homoeologs in allohaploids. We also confirmed that PrBn is still the major determinant for this trait at the species level. Interestingly Radoev et al. (2008) recently mapped a QTL that colocalizes with PrBn and is involved in determining the number of seeds per silique in a cultivar × resynthesized segregating population. As the number of seeds per silique is directly related to meiotic regularity, which is different between natural and resynthesized B. napus (for review, see Nicolas et al., 2008), this result could indicate a broader role for PrBn in regulating the diploid-like meiotic behavior of B. napus. What remains unclear then is the reason why all B. napus accessions display a diploid-like meiotic behavior, regardless of their genotype at the PrBn locus. Likewise a conventional cytological approach does not reveal any obvious difference of chiasmata between allotetraploids (see Supplemental Figure 4 online). However, this method is not sensitive enough to decipher if allohaploids with higher chiasma frequencies have mostly originated from allotetraploid accessions showing the higher values in this parameter. All these questions must be properly addressed before we can understand if “the tape of (meiosis) evolution would replay in the very same or similar way each time at the level of independently formed (B. napus) polyploid lines” (Soltis et al., 2009).
METHODS
Plant Material
Twenty-nine Brassica napus (AACC; 2n = 38) accessions were selected to represent broadly the genetic diversity present in this crop species (Table 1). Our selection encompassed the range of morphological forms that occur in this species and that are commonly used to divide it into three groups or subspecies: the oilseed crop B. napus ssp oleifera (represented by 26 accessions here), the Rutabaga B. napus ssp napobrassica or rapifera (represented by two accessions here), and B. napus ssp pabularis or pabularia (one accession: the asparagus kale). Molecular analyses confirmed that these three groups represent distinct gene pools (Song et al., 1988; Diers and Osborn, 1994). Our selection of oilseed accessions included both winter and spring types, which were shown to be differentiated gene pools (Diers and Osborn, 1994; Lombard et al., 2000; Hasan et al., 2006) and accessions from broad geographic origins, which was shown to correlate with the patterns of genetic diversity in this germplasm (Diers and Osborn, 1994; Chen et al., 2008). Most of the cultivars (22 out of 29) were represented by a single genetically homogeneous line, which was obtained after several generations of selfing (Table 1). A few cultivars (7 out of 29) were generated following open-pollination and therefore potentially contained a bulk of more or less related genotypes (so-called populations in Table 1). Finally three interspecific hybrids, which were all derived from a cross between Brassica oleracea cv HDEM and B. rapa cv Z1, were used for comparisons with B. napus allohaploids.
All allohaploids were isolated using microspore culture as described by Polsoni et al. (1988). Allohaploids were produced from a single plant when the cultivar was considered a genetically homogeneous line. When considering a population, allohaploids were produced separately from three to four distinct plants to account for potential genetic heterogeneity of the determinant influencing homoeologous recombination. B. napus accessions responded very differently to allohaploid production, so different numbers of allohaploids were generated for every genotype (Table 1). For some accessions, it even proved impossible to produce allohaploids through microspore culture. Thus, the number of accessions used in this study was smaller than an original larger selection (data not shown).
A total of seven microspore cultures were needed to isolate the 363 allohaploids analyzed here. Due to flowering time differences, eight series of allohaploids were successively grown in the greenhouse. For each series, allohaploid plants were randomly arranged on a grid in the greenhouse. Allohaploids produced from the two control accessions (Darmor-bzh and Yudal) were systematically and regularly dispersed across the entire grid. The Darmor-bzh and Yudal genotypes were the parents of the segregating allohaploid population that was used to map loci suppressing COs between homoeologous chromosomes (Jenczewski et al., 2003; Liu et al., 2006). Floral buds were sampled on the same date for all allohaploids of a series.
Meiotic Observations
Floral buds were fixed in Carnoy’s solution (ethanol-chloroform-acetic acid, 6:3:1) for 24 h and stored in 50% ethanol. Observations on the PMCs were performed at the MI stage from anthers squashed and stained in a drop of 1% acetocarmine solution. On average, 20 PMCs were examined for each allohaploid.
Statistical Analysis
Statistical analyses were performed on the number of univalents because this variable was scored most reliably and measured the extent of homoeologous recombination in a synthetic way, reflecting by subtraction the number of chromosomes associated both as bivalents and multivalents. The model employed for each group was Yglij = μg + γl + bg,i + εg,lij, where μg is the mean for group g (g is either group fu or MU), γl is the effect of accession l, bg,i is the random effect of block i, and εg,lij is a residual error term. The bg,i and εg,lij random effects were assumed to follow independent normal and centered distributions. In this design, each block included plants that were grown at the same time in two contiguous rows in the greenhouse; different blocks thus refer to either allohaploids from the same series but grown on rows separated by a walkway or allohaploids from different series that were grown at different times in the greenhouse. The random block effect, which was estimated using the plants that were dispersed throughout each experimental area and in the different experiments, was used to account roughly for uncontrolled environmental variation (either across the area of each experiment or between experiments that followed one another in time). More sophisticated methods to account for prospective spatial autocorrelations were made inappropriate by the accession-dependant response surfaces (see Supplemental Figure 2 online). When required (notably for the MU group), heteroscedasticity was accommodated when running the model. All analyses were performed using the PROC MIXED procedure of SAS (SAS Institute, 1999). The PROC VARCOMP procedure of SAS (SAS Institute, 1999) was used to quantify and compare the effect of different factors of the models on the variability observed between allohaploids.
Amplification and Sequencing of Plastid Intergenic Regions
Intergenic regions ndhC-trnV and rbcL-accD from Brassica chloroplast were directly sequenced following PCR amplification (primer details are given in Supplemental Data Set 1 online). The thermal cycling profile was 35 cycles, each with 30 s denaturation at 94°C, 30 s annealing at 53°C, and an extension of 1 min at 72°C. A final extension of 10 min at 72°C was included. PCR products were sequenced by Genoscreen (France). Multiple sequence alignments were performed using default parameters of ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Molecular Analysis of the Region Surrounding PrBn
Eight molecular markers (see details in Supplemental Data Set 1 online) were selected from published genetic maps (Delourme et al., 2006, 2008; Liu et al., 2006; Cheng et al., 2009) to span the region within which PrBn was mapped as a QTL (Liu et al., 2006). We confirmed the position of GMs185 (Cheng et al., 2009) within this interval by selective genetic mapping, as detailed in Supplemental Data Set 1 online. PCR and electrophoresis were performed using the same protocols as described by Foisset et al. (1996) and Delourme et al. (2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Description of B. napus Allohaploids Showing an Intermediate Meiotic Behavior.
Supplemental Figure 2. Spatial Variation of Meiotic Behavior Measured for Allohaploids Isolated from the Same Plants but Positioned at Different Locations in the Greenhouse.
Supplemental Figure 3. Hypothetical Position of PrBn Locus Owing to the Multilocus Genotypes of Recombinant Varieties at the PrBn Region.
Supplemental Figure 4. Representative Metaphase I Nuclei of B. napus Allotetraploid Accessions Showing No Difference in Chiasma Frequency.
Supplemental Table 1. Differences of the LS Means between Accessions within the MU and fu Groups.
Supplemental Data Set 1. Markers and Averaged Meiotic Behaviors of All B. napus Allohaploids Used in This Study.
Supplemental Data Set 2. Multiple Alignments of ndhC-trnV and rbcL-accD Chloroplast Intergenic Sequences.
Supplementary Material
Acknowledgments
We thank Jean-Claude Letanneur for his significant contribution to the production/care of plant material, Hervé Monod (Institut National de la Recherche Agronomique [INRA], Unité Mixte de Recherche 118, Unité de Mathématique et Informatique Appliquées, Jouy-en-Josas, France) for his fruitful advice on statistical analyses, Françoise Budar (INRA Versailles) for providing chloroplast intergenic region primers, and Biogenouest for genotyping facilities. We thank K.C. Falk (Agriculture and Agri-Food, Canada) for the B. rapa Z1 line and M. Manzanares (Agrocampus-Rennes, France) for the B. oleracea HDEM line. L. Grandont, W. Crismani, M. Grelon, and C. Mézard (INRA Versailles) as well as K. Alix (Unité Mixte de Recherche de Génétique Végétale du Moulon, France) are gratefully acknowledged for their critical reading and valuable comments on the manuscript. Leigh Gebbie is acknowledged for English corrections. M.C. is supported by a Marie Curie postdoctoral fellowship (PIEF-GA-2008-219661). This work was conducted with the financial support of “BRG-Bureau des Ressources Génétiques,” project “BRG-07-038, Diversité nucléotidique de quelques gènes impliqués dans la formation des crossing-over à l’échelle d’une collection de variétés de colza (Brassica napus)” and “ANR-Agence Nationale de la Recherche–The French National Research Agency” under the “Programme Biodiversité,” project “ANR-05-BDIV-015, Effet de la polyploïdie sur la biodiversité et l’évolution du génome des plantes.”
References
- Allender C.J., King G.J. (2010). Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Kaff N., Knight E., Bertin I., Foote T., Hart N., Griffiths S., Moore G. (2008). Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum: With deletion mutants and expression profiling. Ann. Bot. (Lond.) 101: 863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L.K., Doyle G.G., Brigham B., Carter J., Hooker K.D., Lai E., Mindy R., Stephen M.S. (2003). High-resolution crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165: 849–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia T., Röbbelen G. (1986). Meiotic pairing in haploids and amphihaploids of spontaneous versus synthetic origin in rape, Brassica napus L. Can. J. Genet. Cytol. 28: 330–334 [Google Scholar]
- Bohuon E.J.R., Keith D.J., Parkin I.A.P., Sharpe A.G., Lydiate D.J. (1996). Alignment of the conserved C genomes of Brassica oleracea and Brassica napus. Theor. Appl. Genet. 93: 833–839 [DOI] [PubMed] [Google Scholar]
- Bovill W.D., Deveshwar P., Kapoor S., Able J.A. (2008). Whole genome approaches to identify early meiotic gene candidates in cereals. Funct. Integr. Genomics 9: 219–229 [DOI] [PubMed] [Google Scholar]
- Caldwell K.S., Dvorak J., Lagudah E.S., Akhunov E., Luo M.C., Wolters P., Powell W. (2004). Sequence polymorphism in polyploid wheat and their d-genome diploid ancestor. Genetics 167: 941–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Nelson N.M., Ghamkhar K., Fu T., Cowling W.A. (2008). Divergent patterns of allelic diversity from similar origins: The case of oilseed rape (Brassica napus L.) in China and Australia. Genome 51: 1–10 [DOI] [PubMed] [Google Scholar]
- Cheng X., Xu J., Xia S., Gu J., Yang Y., Fu J., Qian X., Zhang S., Wu J., Liu K. (2009). Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor. Appl. Genet. 118: 1121–1131 [DOI] [PubMed] [Google Scholar]
- Cheung F., Trick M., Drou N., Lim Y.P., Park J.Y., Kwon S.J., Kim J.A., Scott R., Pires J.C., Paterson A.H., Town C., Bancroft I. (2009). Comparative analysis between homoeologous genome segments of Brassica napus and its progenitor species reveals extensive sequence-level divergence. Plant Cell 21: 1912–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M., Grandont L., Moore G., Chevre A.M., Jenczewski E. (2010). Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 186: 29–36 [DOI] [PubMed] [Google Scholar]
- Delourme R., et al. (2006). Genetic control of oil content in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 113: 1331–1345 [DOI] [PubMed] [Google Scholar]
- Delourme R., Piel N., Horvais R., Pouilly N., Domin C., Vallée P., Falentin C., Manzanares-Dauleux M.J., Renard M. (2008). Molecular and phenotypic characterization of near isogenic lines at QTL for quantitative resistance to Leptosphaeria maculans in oilseed rape (Brassica napus L.). Theor. Appl. Genet. 117: 1055–1067 [DOI] [PubMed] [Google Scholar]
- Diers B.W., Osborn T.C. (1994). Genetic diversity of oilseed Brassica napus germplasm based on restriction fragment length polymorphisms. Theor. Appl. Genet. 84: 662–668 [DOI] [PubMed] [Google Scholar]
- Dubcovsky J., Dvorak J. (2007). Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecke W., Clemens R., Honsdorf N., Becker H.C. (2010). Exent and structure of linkage disequilibrium in canola quality winter rapeseed (Brassica napus L.). Theor. Appl. Genet. 120: 921–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizenga G.C., Kasperbauer M.J. (1985). Chromosome pairing in tall fescue haploids derived by anther-panicle culture. J. Hered. 76: 99–102 [Google Scholar]
- Erickson L.R., Straus N.A., Beversdorf W.D. (1983). Restriction patterns reveal origins of chloroplast genomes in Brassica amphidiploids. Theor. Appl. Genet. 65: 201–206 [DOI] [PubMed] [Google Scholar]
- Esch E., Szymaniak J.M., Yates H., Pawlowski W.P., Buckler E.S. (2007). Using crossover breakpoints in recombinant inbred lines to identify quantitative trait loci controlling the global recombination frequency. Genetics 177: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M. (1966). The effect of chromosomes 5B, 5D and 5A on chromosomal pairing in Triticum aestivum. Proc. Natl. Acad. Sci. USA 19: 549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisset N., Delourme R., Barret P., Hubert N., Landry B.S., Renard M. (1996). Molecular mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled-haploid progeny. Theor. Appl. Genet. 93: 1017–1025 [DOI] [PubMed] [Google Scholar]
- Gaeta R.T., Pires J.C. (2010). Homoeologous recombination in allopolyploids: The polyploid ratchet. New Phytol., in press [DOI] [PubMed] [Google Scholar]
- Gaeta R.T., Pires J.C., Iniguez-Luy F., Leon E., Osborn T.C. (2007). Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell 19: 3403–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgi B. (1978). A homoeologous pairing mutant isolated in Triticum durum cv. Capelli. Mut. Breed. Newsletter 11: 4–5 [Google Scholar]
- Griffiths S., Sharp R., Foote T.N., Bertin I., Wanous M., Reader S., Colas I., Moore G. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752 [DOI] [PubMed] [Google Scholar]
- Hamant O., Ma H., Cande W.Z. (2006). Genetics of meiotic prophase I in plants. Annu. Rev. Plant Biol. 57: 267–302 [DOI] [PubMed] [Google Scholar]
- Handa H. (2007). Investigation of the origin and transmission of linear mitochondrial plasmid based on phylogenetic analysis in Japanese rapeseed varieties. Genome 50: 234–240 [DOI] [PubMed] [Google Scholar]
- Hasan M., Friedt W., Pons-Kühnemann J., Freitag N.M., Link K., Snowdon R.J. (2008). Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus). Theor. Appl. Genet. 116: 1035–1049 [DOI] [PubMed] [Google Scholar]
- Hasan M., Seyis F., Badani A.G., Pons-Kühnemann J., Friedt W., Lühs W., Snowdon R.J. (2006). Analysis of genetic diversity in the Brassica napus L. Gene pool using SSR markers. Genet. Resour. Crop Evol. 53: 793–802 [Google Scholar]
- Hirosawa S., Takumi S., Ishii T., Kawahara T., Nakamura C., Mori N. (2004). Chloroplast and nuclear DNA variation in common wheat: Insight into the origin and evolution of common wheat. Genes Genet. Syst. 79: 271–282 [DOI] [PubMed] [Google Scholar]
- Howell E.C., Kearsey M.J., Jones G.H., King G.J., Armstrong S.J. (2008). A and C genome distinction and chromosome identification in Brassica napus by sequential fluorescence in situ hybridization and genomic in situ hybridization. Genetics 180: 1849–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenczewski E., Eber F., Grimaud A., Huet S., Lucas M.O., Monod H., Chèvre A.M. (2003). PrBn, a major gene controlling homoeologous pairing in oilseed rape (Brassica napus) haploids. Genetics 164: 645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J., Soltis P.S., Soltis D.E. (2010). Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae). BMC Genomics 11: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Adamczyk K., Manzanares-Dauleux M., Eber F., Lucas M.O., Delourme R., Chevre A.M., Jenczewski E. (2006). Mapping PrBn and other quantitative trait loci responsible for the control of homeologous chromosome pairing in oilseed rape (Brassica napus L.) haploids. Genetics 174: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Baril C.P., Dubreuil P., Blouet F., Zhang D. (2000). Genetic relationships and fingerprinting of rapeseed cultivars using AFLP: Consequences for varietal registration. Crop Sci. 40: 1417–1425 [Google Scholar]
- Lombard V., Delourme R. (2001). A consensus linkage map for rapeseed (Brassica napus L.): Construction and integration of three individual maps from DH populations. Theor. Appl. Genet. 103: 491–507 [Google Scholar]
- Lukens L.N., Pires J.C., Leon E., Vogelzang R., Oslach L., Osborn T. (2006). Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140: 336–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M., Cuadrado C., Laurie D.A., Romero C. (2005). Synaptic behaviour of hexaploid wheat haploids with different effectiveness of the diploidizing mechanism. Cytogenet. Genome Res. 109: 210–214 [DOI] [PubMed] [Google Scholar]
- Mercier R., Grelon M. (2008). Meiosis in plants: Ten years of gene discovery. Cytogenet. Genome Res. 120: 281–290 [DOI] [PubMed] [Google Scholar]
- Mezard C., Vignard J., Drouaud J., Mercier R. (2007). The road to crossovers: Plants have their say. Trends Genet. 23: 91–99 [DOI] [PubMed] [Google Scholar]
- Nicolas S.D., Leflon M., Liu Z., Eber F., Chelysheva L., Coriton O., Chevre A.M., Jenczewski E. (2008). Chromosome speed dating during meiosis of polyploid Brassica hybrids and haploids. Cytogenet. Genome Res. 120: 331–338 [DOI] [PubMed] [Google Scholar]
- Nicolas S.D., Leflon M., Monod H., Eber F., Coriton O., Huetau V., Chevre A.M., Jenczewski E. (2009). Genetic regulation of meiotic cross-overs between related genomes in Brassica napus haploids and hybrids. Plant Cell 21: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas S.D., et al. (2007). Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175: 487–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson G., Hagberg A. (1955). Investigations on haploid rape. Hereditas 41: 227–237 [Google Scholar]
- Osborn T.C., Butrulle D.V., Sharpe A.G., Pickering K.J., Parkin I.A., Parker J.S., Lydiate D.J. (2003). Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165: 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H., Feldman M. (2001). Genotypic variation in tetraploid wheat affecting homoeologous pairing in hybrids with Aegilops peregrina. Genome 44: 1000–1006 [PubMed] [Google Scholar]
- Palmer J.D., Shields C.R., Cohen D.B., Orton T.J. (1983). Chloroplast DNA evolution and the origin of amphidiploid Brassica. Theor. Appl. Genet. 65: 181–189 [DOI] [PubMed] [Google Scholar]
- Parkin I.A.P., Lydiate D. (1997). Conserved patterns of chromosome pairing and recombination in Brassica napus crosses. Genome 40: 496–504 [DOI] [PubMed] [Google Scholar]
- Parkin I.A.P., Sharpe A.G., Keith D.J., Lydiate D.J. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131 [DOI] [PubMed] [Google Scholar]
- Piquemal J., Cinquin E., Couton F., Rondeau C., Seignoret E., Doucet E., Perret D., Villeger M.-J., Vincourt P., Blanchard P. (2005). Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor. Appl. Genet. 111: 1514–1523 [DOI] [PubMed] [Google Scholar]
- Polsoni L., Kott S.B., Beversdorf W.D. (1988). Large-scale microspore culture technique for mutation-selection studies in B. napus. Can. J. Genet. Cytol. 66: 1681–1685 [Google Scholar]
- Prakash S., Hinata K. (1980). Taxonomy, cytogenetics and origin of crop Brassica, a review. Opera Bot. 55: 1–57 [Google Scholar]
- Radoev M., Becker H.C., Ecke W. (2008). Genetic analysis of heterosis for yield and yield components in rapeseed (Brassica napus L.) by quantitative trait locus mapping. Genetics 179: 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana D., van den Boogaart T., O'Neill C.M., Hynes L., Bent E., Macpherson L., Park J.Y., Lim Y.P., Bancroft I. (2004). Conservation of the microstructure of genome segments in Brassica napus and its diploid relatives. Plant J. 40: 725–733 [DOI] [PubMed] [Google Scholar]
- Renard M., Dosba F. (1980). Etude de l'haploidie chez le colza (Brassica napus L. var oleifera Metzger). Ann. Amel. Plant. 30: 191–209 [Google Scholar]
- Riley R., Chapman V. (1958). Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 13: 713–715 [Google Scholar]
- Salisbury P.A., Wratten N. (1999). Brassica napus breeding. In Canola in Australia: The First 30 Years, P.A. Salisbury, Potter T.D., McDonald G., Green A.G., (Canberra, Australia: 10th International Rapeseed Congress Organizing Committee; ), pp. 29–35 [Google Scholar]
- Säll T. (1990). Genetic control of recombination in barley: II. Variation in linkage between marker genes. Hereditas 112: 171–178 [Google Scholar]
- Sanchez-Moran E., Armstrong S.J., Santos J.L., Franklin F.C.H., Jones G.H. (2002). Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute (1999). SAS/STAT User’s Guide, Version 8. (Cary, NC: SAS Institute; ). [Google Scholar]
- Sears E.R. (1976). Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10: 31–51 [DOI] [PubMed] [Google Scholar]
- Sharpe A.G., Lydiate D.J. (2003). Mapping the mosaic of ancestral genotypes in a cultivar of oilseed rape (Brassica napus) selected via pedigree breeding. Genome 46: 461–468 [DOI] [PubMed] [Google Scholar]
- Sharpe A.G., Parkin I.A.P., Keith D.J., Lydiate D.J. (1995). Frequent nonreciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus). Genome 38: 1112–1121 [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Buggs R.G.A., Barbazuk W.B., Schnable P.S., Soltis P.S. (2009). On the origins of species: Does evolution repeat itself in polyploid population of independent origin? Cold Spring Harb. Symp. Quant. Biol. 74: 215–223 [DOI] [PubMed] [Google Scholar]
- Soltis D.E., Soltis P.S. (1999). Polyploidy: Recurrent formation and genome evolution. Trends Ecol. Evol. 14: 348–352 [DOI] [PubMed] [Google Scholar]
- Song K., Osborn T.C. (1992). Polyphyletic origins of Brassica napus: New evidence based on organelle and nuclear RFLP analyses. Genome 35: 992–1001 [Google Scholar]
- Song K.M., Osborn T.C., Williams P.H. (1988). Brassica taxonomy based on nuclear restriction fragment length polymorphisms (RFLPs). 1. Genome evolution of diploid and amphidiploid species. Theor. Appl. Genet. 75: 784–794 [Google Scholar]
- Suwabe K., Morgan C., Bancroft I. (2008). Integration of Brassica A genome genetic linkage map between Brassica napus and B. rapa. Genome 51: 169–176 [DOI] [PubMed] [Google Scholar]
- Szadkowski E., et al. (2010). The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186: 102–112 [DOI] [PubMed] [Google Scholar]
- Tai W., Ikonen H. (1988). Incomplete bivalent pairing in dihaploids of Brassica napus L. Genome 30: 450–457 [Google Scholar]
- U N. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan J. Bot. 7: 389–452 [Google Scholar]
- Udall J.A., Quijada P.A., Osborn T.C. (2005). Detection of chromosomal rearrangements derived from homologous recombination in four mapping populations of Brassica napus L. Genetics 169: 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.