Leaf blades of unifacial leaves, such as those of iris, have only the abaxial identity and become flattened by a mechanism different from that in bifacial leaves. This work shows that the unifacial leaf blade is abaxialized at the gene expression level and identifies the DROOPING LEAF gene ortholog as a candidate responsible for flattened leaf blade formation in unifacial leaves.
Abstract
Angiosperm leaves generally develop as bifacial structures with distinct adaxial and abaxial identities. However, several monocot species, such as iris and leek, develop unifacial leaves, in which leaf blades have only abaxial identity. In bifacial leaves, adaxial-abaxial polarity is required for leaf blade flattening, whereas many unifacial leaves become flattened despite their leaf blades being abaxialized. Here, we investigate the mechanisms underlying the development and evolution of flattened leaf blades in unifacial leaves. We demonstrate that the unifacial leaf blade is abaxialized at the gene expression level and that an ortholog of the DROOPING LEAF (DL) gene may promote flattening of the unifacial leaf blade. In two closely related Juncus species, Juncus prismatocarpus, which has flattened unifacial leaves, and Juncus wallichianus, which has cylindrical unifacial leaves, DL expression levels and patterns correlate with the degree of laminar outgrowth. Genetic and expression studies using interspecific hybrids of the two species reveal that the DL locus from J. prismatocarpus flattens the unifacial leaf blade and expresses higher amounts of DL transcript than does that from J. wallichianus. We also show that leaf blade flattening is a trigger for central-marginal leaf polarity differentiation. We suggest that flattened unifacial leaf blades may have evolved via the recruitment of DL function, which plays a similar cellular but distinct phenotypic role in monocot bifacial leaves.
INTRODUCTION
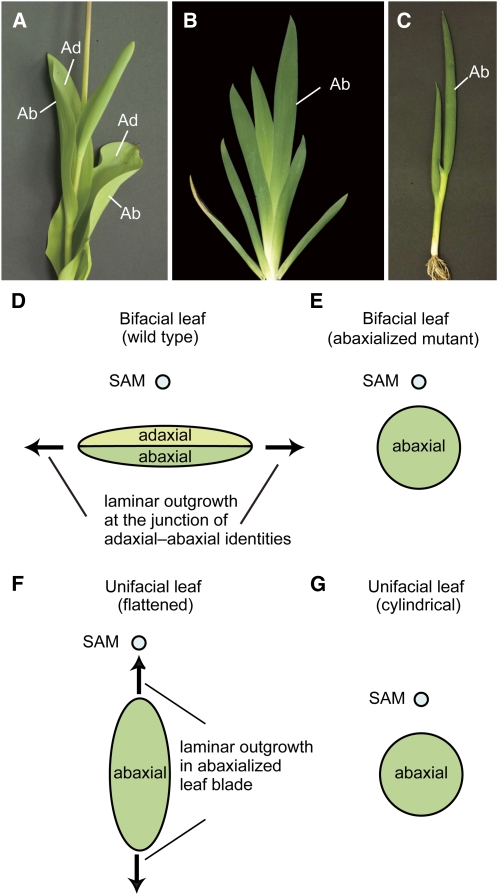
A key question in biology is how diversity in organismal morphology arises and becomes established through evolution. Leaves of angiosperms exhibit considerable morphological diversity and thus represent an attractive subject for evolutionary developmental studies (Piazza et al., 2005). The diverse leaf forms in angiosperms can be categorized as bifacial or unifacial. Bifacial leaves, such as those of Arabidopsis thaliana, snapdragon (Antirrhinum majus), and maize (Zea mays), are the more typical form of leaves that differentiate adaxial-abaxial (upper-lower) polarity with respect to the position of the shoot apical meristem (SAM) (Figures 1A and 1D). The adaxial domain of a leaf primordium is adjacent to the SAM and differentiates into the upper side of the leaf, whereas the abaxial domain is away from the SAM and differentiates into the lower side of the leaf (Steeves and Sussex, 1989). The establishment of adaxial-abaxial polarity in bifacial leaves is regulated by overlapping and antagonistic genetic interactions involving several distinct transcription factors and small regulatory RNAs (Husbands et al., 2009). In both eudicots and monocots, these include members of the Class III Homeodomain Leucine Zipper (HD-ZIPIII) gene family (McConnell et al., 2001; Juarez et al., 2004; Itoh et al., 2008b), which specify adaxial identity and are expressed in the adaxial domain of leaves, and KANADI (Eshed et al., 2001; Kerstetter et al., 2001; Candela et al., 2008; Zhang et al., 2009) and AUXIN RESPONSE FACTOR3 (ARF3)/ETTIN (ETT) genes (Pekker et al., 2005; Itoh et al., 2008a), which are expressed abaxially, where they specify abaxial identity.
Figure 1.
Leaf Blade Structures and Mechanisms of Laminar Outgrowth in Bifacial and Unifacial Leaves.
(A) Bifacial leaves in tulip (Tulipa gesneriana). Ad, adaxial; Ab, abaxial.
(B) Flattened unifacial leaves in German iris (Iris germanica).
(C) Cylindrical unifacial leaves in Welsh onion (Allium fistulosum).
(D) to (G) Schematic diagrams showing transverse sections through leaf blades and mechanisms of laminar outgrowth. Positional relationships of leaves to the SAM are indicated by circles.
(D) Bifacial leaf blade.
(E) Radialized leaf blade in abaxialized mutants.
(F) Bilaterally symmetric, flattened unifacial leaf blade.
(G) Cylindrical unifacial leaf blade.
On the other hand, unifacial leaves, which are characterized by an abaxialized leaf blade, have repeatedly evolved in a number of divergent monocot species, from early-divergent (e.g., Acoraceae) to specialized families (e.g., Iridaceae, Alliaceae, and Juncaceae) (Kaplan, 1975; Rudall and Buzgo, 2002; Yamaguchi and Tsukaya, 2010). Monocot leaves usually consist of two distinct domains along the proximal-distal axis: the proximal leaf sheath and the distal leaf blade (Kaplan, 1973). In bifacial leaves, both domains are dorsoventrally flattened and differentiate with adaxial-abaxial polarity. By contrast, the transverse shape of the unifacial leaf blade is a bilaterally symmetric, flattened structure (ensiform) (Figures 1B and 1F) or a radially symmetric structure (cylindrical/terete) (Figures 1C and 1G) with only abaxial identity, while the leaf sheath has a similar structure to that of bifacial leaves, with morphological differentiation of adaxial-abaxial polarity (Kaplan, 1975; Yamaguchi and Tsukaya, 2010). The abaxialized property of the unifacial leaf blade has been ascertained through histological analysis of morphological features. For example, in the unifacial leaf blade, epidermal and mesophyll tissues usually show only abaxial characteristics, and vascular bundles are usually arranged in a ring beneath the outer leaf surface with all of the xylem poles pointing to the center (Kaplan, 1975).
In both bifacial and unifacial leaves, flattening is an essential feature that optimizes light absorbance. In bifacial leaves, the establishment of adaxial-abaxial polarity is necessary for leaf blade flattening because laminar outgrowth is promoted at the juxtaposition of adaxial and abaxial identities (Figure 1D) (Waites and Hudson, 1995). Thus, mutants or transgenic plants that have lost adaxial-abaxial polarity develop radialized leaf blades (Figure 1E). Therefore, it is interesting that many unifacial-leafed species develop flattened leaf blades, although their leaf blades lack adaxial-abaxial polarity (Figure 1F), yet some other species develop cylindrical leaf blades, which are similar to those of abaxialized mutants in bifacial-leafed species (Figure 1G) (Rudall and Buzgo, 2002; Yamaguchi and Tsukaya, 2010). This indicates that flattened leaves have independently evolved in bifacial and unifacial leaves, and flattened leaf blade formation in unifacial leaves is regulated by unknown mechanisms that differ from those in bifacial leaves.
The development and evolution of unifacial leaves have long been subjects of debate, and considerable histological studies have been conducted (reviewed in Kaplan, 1975). However, it remains largely unknown how unifacial leaf blades become abaxialized, how and why they have repeatedly evolved in monocots, and how abaxialized leaf blades become flattened in unifacial leaves. A crucial factor hindering progress in this area has been the lack of a suitable model research system. To answer these questions, we sought to unravel the genetic basis of unifacial leaf development, focusing on the genus Juncus (Juncaceae) as a model. Juncus contains species with a wide variety of leaf forms (Cutler, 1969; Kirschner, 2002a, 2002b) and is amenable to molecular genetic studies (Yamaguchi and Tsukaya, 2010).
In this study, we investigated the mechanisms underlying development and evolution of flattened leaf blades in unifacial leaves using two Juncus species: Juncus prismatocarpus, which has flattened unifacial leaves, and Juncus wallichianus, which has cylindrical unifacial leaves. We demonstrate that the unifacial leaf blade is abaxialized at the gene expression level and identify an ortholog of the DROOPING LEAF (DL) gene (Yamaguchi et al., 2004) as a strong candidate promoting flattened leaf blade formation in unifacial leaves. Our study also provides insight into the mechanisms that regulate the specification of central-marginal leaf polarity. Based on our results, we propose a genetic framework of flattened leaf blade formation in unifacial leaves and discuss the mechanisms underlying the evolution of flattened leaf blades in unifacial leaves.
RESULTS
Adaxial-Abaxial Identities in Unifacial Leaves
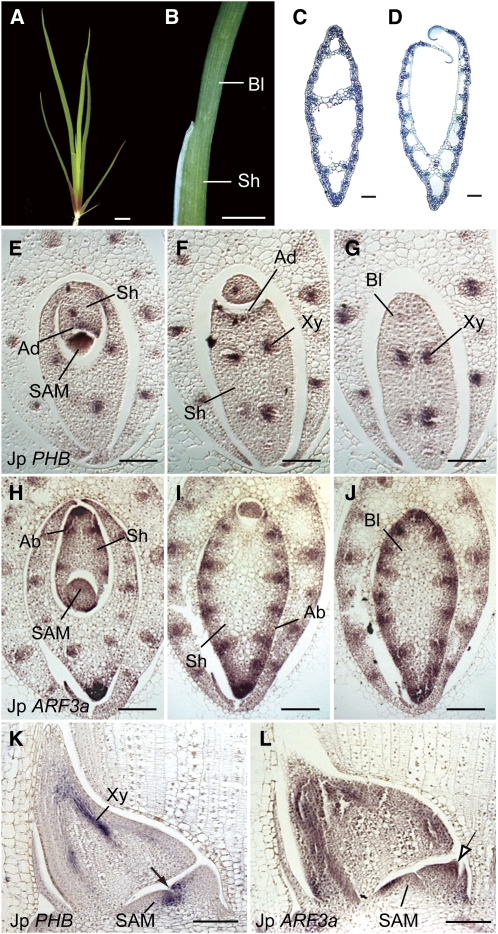
J. prismatocarpus develops typical unifacial leaves, with bilaterally symmetric flattened unifacial leaf blades and dorsoventrally flattened bifacial leaf sheaths (Figures 2A to 2D). To confirm the adaxial-abaxial identities in unifacial leaves, we first studied the expression patterns of the HD-ZIPIII (McConnell et al., 2001) and ARF3/ETT (Pekker et al., 2005) gene homologs (for their phylogenies, see Supplemental Figures 1 and 2 and Supplemental Data Sets 1 and 2 online), as they function in adaxial and abaxial domains of monocot (maize and rice [Oryza sativa]) leaves, respectively (Juarez et al., 2004; Itoh et al., 2008a, 2008b). In the leaf sheath of J. prismatocarpus, an HD-ZIPIII homolog (Jp PHB) was specifically expressed in the adaxial leaf surface and in the presumptive xylem region of procambial strands (Figures 2E and 2F), whereas an ARF3 homolog (Jp ARF3a) was specifically expressed in the abaxial domain (Figures 2H and 2I). Thus, these genes could be molecular markers of adaxial-abaxial identities in J. prismatocarpus as well as maize and rice. By contrast, in the leaf blade, the expression of Jp PHB was restricted to the presumptive xylem region (Figures 2G and 2K), whereas Jp ARF3a was expressed throughout the entire outer region of the leaf blade (Figures 2J and 2L). Thus, the leaf blade of J. prismatocarpus is indeed abaxialized at the gene expression level.
Figure 2.
Adaxial-Abaxial Identities in Unifacial Leaves of J. prismatocarpus.
(A) Seedling of J. prismatocarpus 4 weeks after germination.
(B) Lateral view of a J. prismatocarpus leaf.
(C) and (D) Transverse sections of leaf blade (C) and leaf sheath (D) of J. prismatocarpus. The top of the image is the side facing the SAM.
(E) to (G) In situ localization of Jp PHB transcripts in transverse sections of J. prismatocarpus leaf primordia.
(H) to (J) In situ localization of Jp ARF3a transcripts in transverse sections of J. prismatocarpus leaf primordia. Sections are through the SAM ([E] and [H]), the leaf sheath ([F] and [I]), and the leaf blade ([G] and [J]).
(K) and (L) In situ localization of Jp PHB (K) and Jp ARF3a (L) transcripts in longitudinal sections through the SAM. Arrow in (K) shows the adaxial expression of Jp PHB only in the basal region of the leaf primordium, which will differentiate into the leaf sheath. The outlined arrow in (L) shows expression of Jp ARF3a throughout the distal region of the leaf primordium, which will differentiate into the leaf blade.
Note that the internal region of the young leaf primordium is occupied by dividing cells (as in [G] and [J]). Air spaces in the mature leaf (as in [C]) are formed by subsequent cell death. Ad, adaxial domain; Ab, abaxial domain; Bl, leaf blade; Sh, leaf sheath; Xy, presumptive xylem region in procambial strand. Bars = 1 cm in (A) and (B) and 200 μm in (C) to (L).
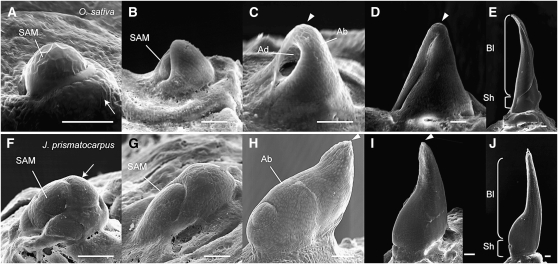
To understand the mechanisms underlying the development of unifacial leaves, we next observed the developmental patterns of unifacial leaf primordia of J. prismatocarpus under a scanning electron microscope. We also observed bifacial leaf development of rice as a comparison. In bifacial leaf development in rice, the leaf primordium arose as a small bulge on the flank of the SAM (Figure 3A). The leaf primordium then began to grow distally, enclosing the SAM (Figure 3B). During distal growth, development of adaxial and abaxial sides was coordinated, with the leaf apex being located at the junction of adaxial and abaxial domains (Figures 3C and 3D), which led to the formation of bifacial structures in both the leaf blade and the leaf sheath (Figure 3E). In the unifacial leaves of J. prismatocarpus, the leaf primordium first arose as a bulge on the flank of the SAM (Figure 3F), as in rice. However, the leaf primordium showed distinct developmental patterns soon after formation of the protrusion. During distal growth of J. prismatocarpus leaf primordia, it appeared that development of the abaxial domain was dominant (Figure 3G), and the leaf apex was located within the abaxial domain, while development of the adaxial domain was restricted to the basal region, covering the SAM (Figures 3H and 3I). As a result, the distal region of the unifacial leaf primordium, which will differentiate into the leaf blade, appeared to consist of only the abaxial identity (Figure 3J). Observations of longitudinal sections of J. prismatocarpus shoot apices also showed that the distal region of unifacial leaf primordia appeared to have only the abaxial identity, with the adaxial domain being confined to the basal region (see Supplemental Figure 3 online). To confirm these observations, we examined in situ localizations of Jp PHB and Jp ARF3a in longitudinal sections of J. prismatocarpus leaf primordia. In agreement with these observations, Jp PHB was indeed expressed adaxially only in the basal region (Figure 2K), whereas Jp ARF3a was expressed throughout the distal region and abaxially in the basal region (Figure 2L). These results indicate that the unifacial leaf blade is formed by abaxialization of the distal region of leaf primordia at a very early stage of development, rather than by postgenital fusion of adaxial leaf surfaces or leaf rotation.
Figure 3.
Development of Bifacial Leaves in Rice and Unifacial Leaves in J. prismatocarpus.
(A) to (E) Scanning electron micrographs of bifacial leaf development in rice. Leaf primordia development proceeds from (A) to (E).
(F) to (J) Scanning electron micrographs of unifacial leaf development in J. prismatocarpus. Leaf primordia development proceeds from (F) to (J).
Arrows indicate the incipient leaf primordium. Arrowheads indicate the leaf apex. Ad, adaxial domain; Ab, abaxial domain; Bl, leaf blade; Sh, leaf sheath. Bars = 50 μm.
Comparison of Unifacial Leaf Blade Development in J. prismatocarpus and J. wallichianus
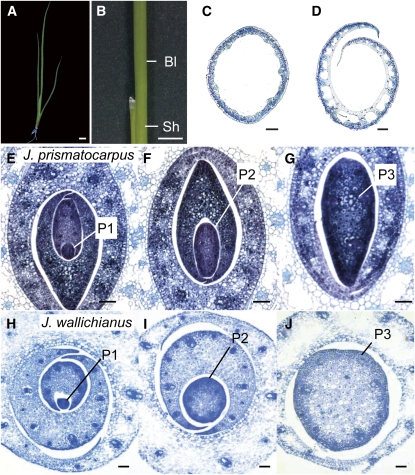
We next studied the mechanism of leaf blade flattening in unifacial leaves by comparative analysis using J. prismatocarpus and J. wallichianus, which molecular phylogenetic analysis indicated are the most closely related species of the genus (see Supplemental Figure 4 and Supplemental Data Set 3 online). The leaf blade morphologies of these species differed transversely, with J. wallichianus developing cylindrical unifacial leaves (Figures 4A to 4D) and J. prismatocarpus developing flattened unifacial leaves (Figures 2A to 2D).
Figure 4.
Differential Laminar Outgrowth in Unifacial Leaves of J. prismatocarpus and J. wallichianus.
(A) Seedling of J. wallichianus 4 weeks after germination.
(B) Lateral view of a leaf in J. wallichianus.
(C) and (D) Transverse sections of leaf blade (C) and leaf sheath (D) of J. wallichianus. Note that the internal air spaces are formed by cell death as in J. prismatocarpus, and the internal region of the young leaf primordium is occupied by dividing cells, as seen in (J).
(E) to (G) Transverse sections of shoot apices of J. prismatocarpus through P1 (E), P2 (F), and P3 (G) leaf blades showing directional laminar outgrowth along the median plane.
(H) to (J) Transverse sections of shoot apices of J. wallichianus through P1 (H), P2 (I), and P3 (J) leaf blades showing no directional laminar outgrowth.
Plastochron numbers of leaf blades are indicated (P1, P2, and P3). Bar in (A) and (B) = 1 cm; bar in (C) to (J) = 200 μm.
To understand the developmental mechanisms underlying flattened leaf blade formation in unifacial leaves, we first compared the development of leaf blades in the two species by making transverse sections of shoot apices. To classify stages of leaf development, we used the plastochron numbering system: plastochron1 (P1) represents the youngest primordium, P2 the next youngest, etc. (Itoh et al., 2005). In J. prismatocarpus, the leaf blade at the P1 stage was not so obviously flattened (Figure 4E) but was flattened at the P2 and P3 stages by directional outgrowth along the median plane (Figures 4F and 4G). By contrast, the leaf blade of J. wallichianus did not show such directional outgrowth and remained cylindrical throughout leaf development (Figures 4H to 4J). Thus, leaf blade flattening in unifacial leaves is regulated by mechanisms that promote directional laminar outgrowth along the median plane.
To further clarify the cell proliferation patterns during unifacial leaf development, we compared cell cycle activity during leaf development between J. prismatocarpus and J. wallichianus by examining in situ localization of HistoneH4 mRNA, which is specifically expressed in the S phase of the cell cycle (Gaudin et al., 2000). In J. prismatocarpus, we observed a concentration of HistoneH4-expressing cells on the SAM side of leaf blade primordia during P1 and P2 stages, when leaf primordia began and continued directional outgrowth toward the SAM side (Figures 5A, 5B, 5D, and 5E). After the P3 stage, HistoneH4-expressing cells were distributed uniformly throughout leaf primordia (Figures 5C and 5F). By contrast, we observed no obvious concentration of HistoneH4-expressing cells in cylindrical leaf primordia of J. wallichianus throughout leaf development (Figures 5G to 5L). These observations suggest that laminar outgrowth in J. prismatocarpus appears to be triggered by factors that promote cell proliferation of leaf primordia toward the SAM side at an early stage of development (P1 and P2 stages). Subsequently, directional laminar outgrowth in J. prismatocarpus may be maintained bidirectionally by more diffuse cell proliferation activity after the P3 stage.
Figure 5.
Cell Cycle Activity during Leaf Development in J. prismatocarpus and J. wallichianus.
(A) to (C) In situ localization of HistoneH4 transcripts in median longitudinal sections of J. prismatocarpus shoot apices. Leaf primordia development proceeds from (A) to (C).
(D) to (F) In situ localization of HistoneH4 transcripts in transverse sections through P1 (D), P2 (E), and P3 (F) leaf blades in J. prismatocarpus.
(G) to (I) In situ localization of HistoneH4 transcripts in median longitudinal sections of J. wallichianus shoot apices. Leaf primordia development proceeds from (G) to (I).
(J) to (L) In situ localization of HistoneH4 transcripts in transverse sections through P1 (J), P2 (K), and P3 (L) leaf blades in J. wallichianus.
Arrows in (A), (B), (D), and (E) indicate a concentration of HistoneH4-expressing cells at the SAM side of J. prismatocarpus leaf blades. Plastochron numbers of leaf blades are indicated (P1, P2, and P3). Bars = 200 μm.
DL Is Strongly Expressed in Flattened Unifacial Leaf Primordia of J. prismatocarpus
To identify candidate genes responsible for the laminar outgrowth in unifacial leaves, we first attempted to identify differentially expressed genes in leaf primordia between J. prismatocarpus and J. wallichianus since genome information is not currently available for Juncus. We isolated homologs of known leaf developmental genes, such as YABBY (Bowman and Smyth, 1999; Sawa et al., 1999; Siegfried et al., 1999), KANADI (Eshed et al., 2001; Kerstetter et al., 2001), HD-ZIPIII (McConnell et al., 2001), ARF3/ETT (Pekker et al., 2005), PRESSED FLOWER (PRS) (Matsumoto and Okada, 2001), and ASYMMETRIC LEAVES1/ROUGH SHEATH2/PHANTASTICA (Waites et al., 1998; Timmermans et al., 1999; Tsiantis et al., 1999; Byrne et al., 2000), and studied their expression patterns. We identified two genes that had expression patterns that significantly differed between J. prismatocarpus and J. wallichianus. One is an ortholog of the DL gene, and the other is a homolog of the PRS gene.
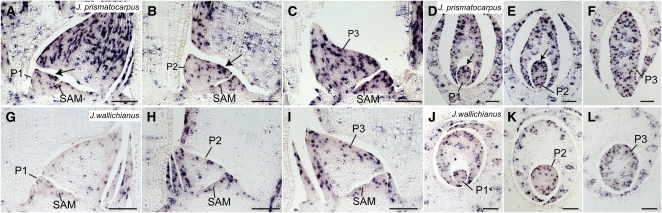
DL is a member of the YABBY gene family (see Supplemental Figure 5A and Supplemental Data Set 4 online) and has a unique function in monocot bifacial leaves, such as those of rice (Yamaguchi et al., 2004; Ishikawa et al., 2009). In rice, DL is expressed at the center of leaves, where it regulates the formation of the leaf midrib, a rigid and thickened structure at the center of the leaf, through a function to promote cell proliferation of leaf primordia toward the SAM side (Yamaguchi et al., 2004). In J. prismatocarpus, a DL ortholog (Jp DL) was strongly expressed in the central domain of leaf primordia, extending from the central large vascular bundle to the leaf surface at the SAM side during the P1 to P2 stages (Figures 6A to 6C and 6E). This is similar to the expression pattern of DL in rice leaves. The temporal pattern of Jp DL expression was correlated with the stage during which laminar outgrowth occurs. After the P3 stage, DL ceased to be expressed in mesophyll tissues and exhibited residual expression around the central large vascular bundle. At this stage, we found that DL expression also became detectable around the large vascular bundles located nearest to the secondary central domain of the flattened leaf blade (Figure 6D; discussed later). By contrast, in J. wallichianus, a DL ortholog (Jw DL) was only weakly expressed around the central large vascular bundle, and no expression was observed in proliferating mesophyll tissue throughout leaf development (Figures 6F to 6J). Therefore, expression patterns and levels of these DL orthologs correlated with the degree of laminar outgrowth. Given that rice DL plays a role in promoting cell proliferation of leaf primordia toward the SAM side, it is possible that leaf blade flattening in J. prismatocarpus is regulated by a similar DL function.
Figure 6.
Expression Pattern of DL in Leaf Primordia of J. prismatocarpus and J. wallichianus.
(A) to (E) In situ localization of Jp DL transcripts in J. prismatocarpus shoot apices.
(A) to (C) Transverse sections of shoot apices through the SAM (A), a P1 leaf blade (B), and a P2 leaf blade (C), showing strong Jp DL expression in the central domain of leaf primordia (arrows).
(D) Transverse section through a P3 leaf blade, showing Jp DL expression in the secondary central domain (arrowheads).
(E) Longitudinal section through the SAM showing strong Jp DL expression (arrow).
(F) to (J) In situ localization of Jw DL transcripts in J. wallichianus.
(F) to (I) Transverse sections of shoot apices through the SAM (F), a P1 leaf blade (G), a P2 leaf blade (H), and a P3 leaf blade (I), showing no Jw DL expression in the mesophyll tissues and weak expression around the central vascular bundle. White arrowheads indicate loss of Jw DL expression in the secondary central domain.
(J) Longitudinal section through the SAM showing weak Jw DL expression.
Bl, leaf blade; Sh, leaf sheath; Cv, central large vascular bundle. Plastochron numbers of leaf blades are indicated (P1, P2, and P3). Note that the central large vascular bundle differentiates in a slightly off-center position in Juncus leaves. Bars = 200 μm.
PRSb Is Expressed Only in the Flattened Leaf Primordia
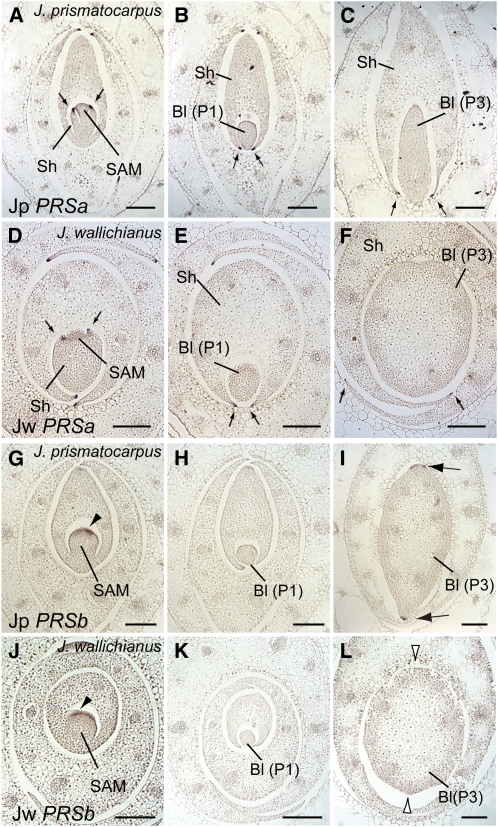
PRS is a member of the WOX (for WUSCHEL related homeobox) gene family (Haecker et al., 2004) and is necessary for the establishment of marginal domains in bifacial leaves via specific expression in leaf margins (Matsumoto and Okada, 2001; Vandenbussche et al., 2009), with loss of function in maize resulting in a narrow leaf phenotype (Nardmann et al., 2004). Phylogenetic analysis revealed that, in Juncaceae and Poaceae, the PRS genes consist of two subclasses, which we have designated PRSa and PRSb (see Supplemental Figure 6 and Supplemental Data Set 5 online). Expression patterns of PRSa were similar between J. prismatocarpus and J. wallichianus. In both species, PRSa was specifically expressed in the leaf margins of developing leaf sheaths but not in the unifacial leaf blade (Figures 7A to 7F). These results demonstrate that unifacial leaf blades do not differentiate a normal leaf margin identity, which further supports the abaxialization of unifacial leaf blades and indicates that PRSa is not involved in leaf blade flattening in unifacial leaves.
Figure 7.
Expression Patterns of PRSa and PRSb in Leaf Primordia of J. prismatocarpus and J. wallichianus.
(A) to (C) In situ localization of Jp PRSa transcripts in transverse sections of J. prismatocarpus shoot apices.
(D) to (F) In situ localization of Jw PRSa transcripts in transverse sections of J. wallichianus shoot apices.
(A) and (D) Transverse sections through the SAM.
(B) and (E) Transverse sections through P1 leaf blades.
(C) and (F) Transverse sections through P3 leaf blades. Arrows in (A) to (F) indicate PRSa expression in leaf margins of the leaf sheath.
(G) to (I) In situ localization of Jp PRSb transcripts in transverse sections of J. prismatocarpus shoot apices.
(J) to (L) In situ localization of Jw PRSb transcripts in transverse sections of J. wallichianus shoot apices.
(G) and (J) Initial PRSb expression in presumptive leaf marginal domains (arrowheads).
(H) and (K) Loss of PRSb expression in leaf blades at the P1 stage.
(I) and (L) PRSb expression in P3 stage leaf blades in margin-like domains only in J. prismatocarpus ([I], arrows), but not in J. wallichianus ([L], white arrowheads).
Bl, leaf blade; Sh, leaf sheath. Plastochron numbers of leaf blades are indicated in parenthesis. Bars = 200 μm.
On the other hand, expression patterns of PRSb differed between the two species. In both species, PRSb was expressed in the presumptive leaf marginal domains before the initiation of leaf primordia (P0 stage; Figures 7G and 7J). In J. prismatocarpus, PRSb (Jp PRSb) expression was not initially observed in leaf primordia during the P1 to P2 stages (Figure 7H) but became detectable in the margin-like regions of flattened leaf blades at the P3 stage (Figure 7I). By contrast, expression of J. wallichianus PRSb (Jw PRSb) was not observed in the cylindrical leaf blades throughout leaf development (Figures 7K and 7L). Thus, PRSb was expressed in margin-like regions in flattened leaf blades of J. prismatocarpus but not in cylindrical leaf blades of J. wallichianus. These results suggest that PRSb may also regulate the flattening of unifacial leaf blades by promoting marginal growth.
Genetic Analysis of Leaf Blade Flatness Using Interspecific Hybrids
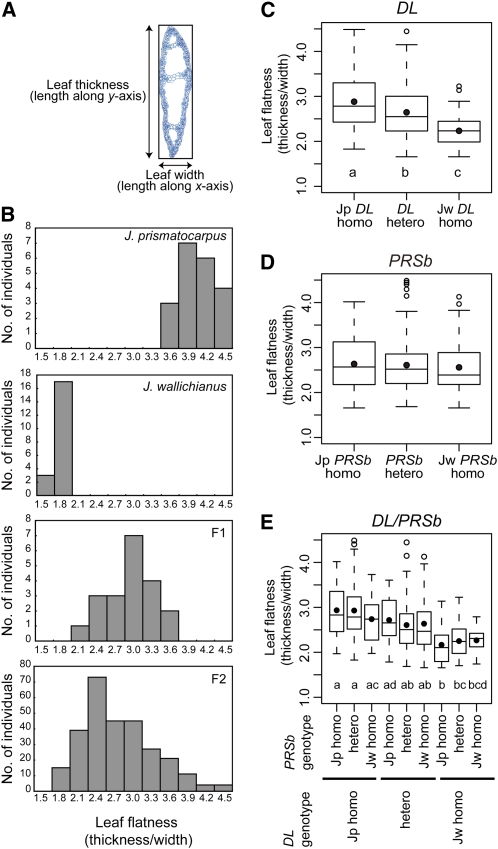
To reveal whether DL, PRSb, or both are responsible for the differences in laminar outgrowth between J. prismatocarpus and J. wallichianus, we performed genetic analysis by generating the interspecific hybrids between the two species. We found that the two species could be hybridized and the F1 hybrids produced fertile seeds. We evaluated leaf flatness by calculating the ratio of leaf thickness to leaf width in transverse leaf sections (Figure 8A).
Figure 8.
Genetic Analysis of Leaf Flatness Using Interspecific Hybrids between J. prismatocarpus and J. wallichianus.
(A) Schematic of leaf flatness analysis. The ratio of leaf thickness to leaf width was calculated to evaluate leaf flatness, with a larger value indicating a more flattened leaf.
(B) Histograms showing leaf flatness distribution in each generation. Generations are indicated at the top right.
(C) to (E) Box plots showing differences in leaf flatness in 284 siblings of the F2 generation, depending on DL (C) or PRSb (D) genotypes, and in all combinations of the DL and PRSb genotypes (E). Each box encloses 50% of the distribution, with the horizontal line marking the median and the dot marking the mean. The lines extending from each box indicate the minimum and maximum values that fall within 1.5 times the height of the box. Open circles indicate outliers. Genotypes are indicated beneath the plots. Sample numbers are shown in Supplemental Table 1 online. Different letters (i.e., a to d) below the columns in (C) and (E) indicate significant differences between genotypes. One-way ANOVA and Tukey’s HSD test (α = 0.05, with Bonferroni correction) were used for multiple comparisons.
[See online article for color version of this figure.]
We first analyzed leaf flatness in the F1 and F2 generations of interspecific hybrids between J. prismatocarpus and J. wallichianus to understand the inheritance pattern of leaf flatness. In the F1 generation, leaf blades were somewhat flattened (Figure 8B; see Supplemental Table 1 online). In the F2 generation, the distribution of leaf flatness was broader and more continuous (Figure 8B; see Supplemental Table 1 online). These results indicate that the difference in leaf flatness between J. prismatocarpus and J. wallichianus is a polygenic trait and is regulated by at least two loci, including dominant or semidominant factors, which promote laminar outgrowth in J. prismatocarpus.
Next, we analyzed the genetic linkage between leaf flatness and DL or PRSb genotypes in 284 siblings of the F2 generation. We found that differences in the DL genotype corresponded with significant differences in leaf flatness (Figure 8C; see Supplemental Table 1 online). Leaves were more flattened in homozygous Jp DL plants than in homozygous Jw DL plants. Leaf flatness was intermediate between these phenotypes when DL was heterozygous. On the other hand, leaf flatness in the F2 generation was not affected by the PRSb genotype (Figure 8D; see Supplemental Table 1 online). Combinations of each DL and PRSb genotype did not have synergistic effects on leaf flatness because differences in leaf flatness depended only on the DL genotypes (Figure 8E). These results indicate that the DL locus or a locus tightly linked to the DL locus is one of the loci responsible for the laminar outgrowth difference between the two species and that the allele at this locus of J. prismatocarpus works as a semidominant factor that promotes laminar outgrowth. On the other hand, PRSb is not directly involved in the difference in leaf flatness between J. prismatocarpus and J. wallichianus, with Jp PRSb and Jw PRSb possessing similar functions.
Expression Analysis of the DL Locus
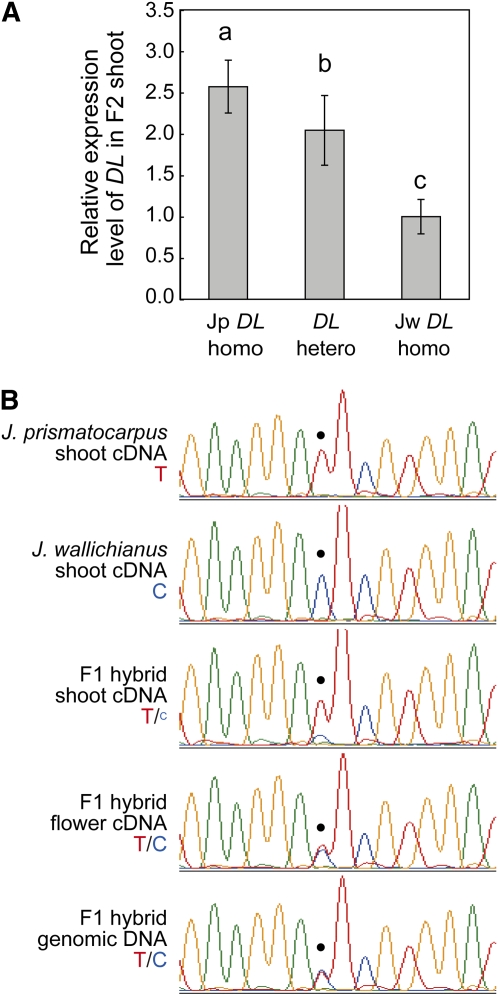
Genetic analysis indicated that the DL locus was a particularly intriguing candidate for flattened unifacial leaf blade formation and suggested that the activity of DL differed between the two species. As the putative DL protein amino acid sequences of the two species were identical (see Supplemental Figure 5B online), the differential DL activity was possibly the result of differential DL expression between the two species, as suggested by in situ expression analysis (Figure 6). To confirm this speculation, we used real-time RT-PCR analysis to study the relationship between DL expression level and DL genotype in the F2 generation. We found that the total amounts of DL transcripts increased as the copy number of Jp DL increased (Figure 9A). These results indicate that the Jp DL locus expresses higher amounts of DL transcripts than the Jw DL locus in the F2 generation.
Figure 9.
Expression Level of DL Depends on the DL Genotype in the Interspecific Hybrid.
(A) Quantitative real-time RT-PCR analysis of DL transcripts in the interspecific hybrid F2 generation of each DL genotype. Data (mean ± sd) are presented as relative expression units after normalization to a TUBULIN gene (n = 12). Different letters (i.e., a to c) above the columns indicate significant variations between the genotypes based on one-way ANOVA and Tukey’s HSD test (α = 0.05, with Bonferroni correction).
(B) Chromatograms of sequenced RT-PCR products, showing allele-specific DL expression in the interspecific hybrid F1 generation. Templates are indicated on the left. The dot indicates the position of a T (red) or C (blue) single nucleotide polymorphism between Jp DL and Jw DL.
To further clarify if differential DL expression between the Jp DL and Jw DL loci was due to cis-regulatory changes at the DL locus, to differential trans-acting factors linked to the DL locus, or to differences in leaf shape, we next examined allele-specific DL expression levels in the F1 hybrid using a single nucleotide polymorphism located in the 3′ untranslated region of DL cDNA (Figure 9B). In the F1 shoot, the Jp DL allele expressed higher amounts of DL transcripts than the Jw DL allele (Figure 9B). As the F1 hybrid contains both Jp DL and Jw DL alleles in an identical trans-acting environment and in an identical leaf shape background, this result indicates that differential DL expression is due to cis-regulatory changes in the DL locus itself and not to trans-acting factors or differences in leaf shape. In rice, DL is also expressed in developing carpel primordia, where it regulates carpel identity (Yamaguchi et al., 2004). In situ hybridization of DL in developing flowers of the two Juncus species revealed that DL orthologs were also expressed in carpel primordia both in J. prismatocarpus and in J. wallichianus at similar levels (see Supplemental Figure 7 online). Allele-specific expression analysis in the F1 flower showed that almost the same amount of DL mRNA was expressed from both DL alleles (Figure 9B). Thus, differential cis-regulatory activity of DL between the two species was organ specific.
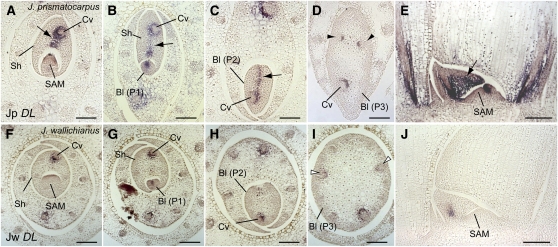
Regulation of Leaf Central-Marginal Polarity Differentiation
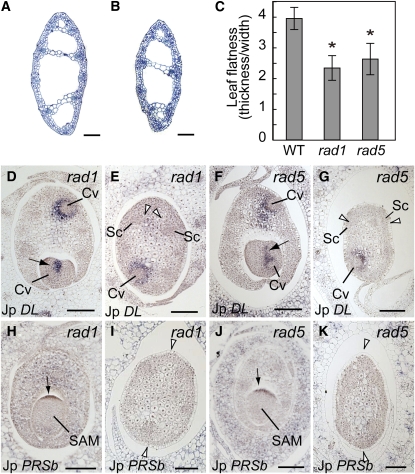
During the P3 stage of leaf blade development in J. prismatocarpus, DL expression also became detectable around the large vascular bundles located nearest to the secondary central domain of the flattened leaf blade (Figure 6D). Based on this expression of DL, together with the expression of PRSb in the margin-like domains of the flattened unifacial leaf blade of J. prismatocarpus (Figure 7I), but not in the cylindrical leaf blade of J. wallichianus (Figure 7L), we assumed that central-marginal polarity was reorganized to follow the flattened leaf shape in the late stages of leaf development. To test this possibility, we isolated mutants of J. prismatocarpus with a radialized leaf blade phenotype (radial leaf1, rad1; radial leaf5, rad5; Figures 10A to 10C) and examined expression patterns of DL and PRSb in these mutants. As in the wild type, DL and PRSb were initially expressed in these mutants in the primary central domain (Figures 10D and 10F) and in the presumptive leaf marginal domain (Figures 10H and 10J), respectively. However, unlike the wild type, we did not observe the late expression of DL or PRSb in the secondary central and marginal domains in these mutants (Figures 10E, 10G, 10I, and 10K). As we found no obvious mutation in the DL and PRSb loci of these mutants, loss of expression of these genes in the later stages of mutant leaf development is probably not caused by defects in cis-regulation or mRNA stability. Thus, these observations indicate that leaf blade flattening induces DL and PRSb expression in the secondary central and marginal domains, respectively, and suggest that central-marginal polarity can differentiate somewhat autonomously via a leaf flatness–dependent mechanism. Considering also our linkage analysis results, we further suggest that the loss of PRSb expression in the leaf blades of J. wallichianus results from the loss of blade flattening in this species and not from differential PRSb promoter activity.
Figure 10.
DL and PRSb Expression in the Radialized Leaf Blades of J. prismatocarpus rad1 and rad5 Mutants.
(A) and (B) Transverse sections of rad1 (A) and rad5 (B) leaf blades showing the radialized leaf blade phenotype.
(C) Leaf flatness in the wild type, rad1, and rad5. Data are mean ± sd. Wild type, n = 20, mean = 3.95 ± 0.36; rad1, n = 12, mean = 2.34 ± 0.40; rad5, n = 12, mean = 2.63 ± 0.5. Asterisks indicate significant difference compared with the wild type (P < 0.05, t test).
(D) to (G) In situ localization of Jp DL transcripts in transverse sections of rad1 ([D] and [E]) and rad5 ([F] and [G]) shoot apices at an early ([D] and [F]) and late ([E] and [G]) developmental stage of leaf primordia. Arrows in (D) and (F) show initial Jp DL expression in the primary central domain. Arrowheads in (E) and (G) show loss of Jp DL expression in the secondary central domain.
(H) to (K) In situ localization of Jp PRSb transcripts in transverse sections of rad1 ([H] and [I]) and rad5 ([J] and [K]) shoot apices at an early ([H] and [J]) and late ([I] and [K]) developmental stage of leaf primordia. Arrows in (H) and (J) show initial Jp PRSb expression in the presumptive leaf marginal domain. Arrowheads in (I) and (K) show loss of Jp PRSb expression at a later stage of leaf development.
Cv, central large vascular bundle; Sc, large vascular bundle in the secondary central domain. Bars = 200 μm.
DISCUSSION
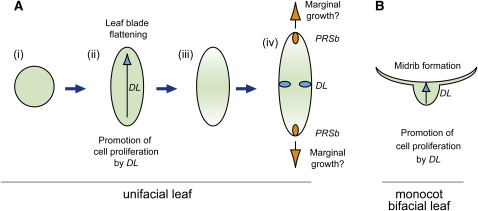
We investigated the genetic mechanisms underlying flattened leaf blade formation in unifacial leaves. Based on the results, we propose the following model of laminar outgrowth in unifacial leaves (Figure 11). Developmentally, the default shape of the unifacial leaf blade is cylindrical, as in J. wallichianus, because of abaxialization (Figure 11A, i). However, in monocots, DL functions to thicken leaf primordia by promoting cell proliferation toward the shoot apex. Such DL function in unifacial leaves may lead to flattened leaf blade formation, as in J. prismatocarpus (Figure 11A, ii), while in monocot bifacial leaves it leads to leaf midrib formation (Figure 11B). Flattening of the unifacial leaf blade then triggers the differentiation of a gradient of central-marginal polarity corresponding to the flattened leaf shape (Figure 11A, iii), which induces DL and PRSb expression in the secondary central and marginal domains, respectively (Figure 11A, iv). The developmental and evolutionary mechanisms of leaf blade flattening in unifacial leaves are discussed below.
Figure 11.
Genetic Framework of Flattened Leaf Blade Formation in Unifacial Leaves.
(A) Model of laminar outgrowth and autonomous differentiation of central-marginal polarity in flattened unifacial leaves. (i) Cylindrical leaf blade as a result of abaxialization. (ii) Leaf blade flattening through DL function. (iii) Differentiation of gradient of central–marginal polarity to follow the flattened leaf shape. (iv) Induction of DL and PRSb expression in the secondary central and marginal domains, respectively.
(B) DL function in monocot bifacial leaves.
Blue arrows indicate the DL function to promote cell proliferation of leaf primordia toward the SAM side. Orange arrows indicate a putative PRSb function to promote marginal growth in flattened unifacial leaves.
Unifacial Leaf Blades Are Abaxialized at the Gene Expression Level
In the leaf blade of J. prismatocarpus, ARF3a is expressed throughout the outer region of the leaf blade, while PHB is only expressed in the presumptive xylem region. These results demonstrate that the unifacial leaf blade is abaxialized at the gene expression level. Loss of PRSa expression in the leaf blade also supports the abaxialization of the unifacial leaf blade because PRSa is expressed at the junction of adaxial and abaxial identities in the leaf sheath. Observations of developmental patterns of unifacial leaf primordia, together with in situ expression analysis, have shown that the distal region of the unifacial leaf primordia is abaxialized from a very early stage, which leads to the formation of abaxialized leaf blades. The abaxialization effect seems to be somewhat incomplete, as the basal sheath region acquires adaxial-abaxial polarity in unifacial leaves, indicating that the distal region of monocot leaves may be more sensitive to the abaxialization effect than the basal region. In maize, the milkweed pod1 mutant, a loss-of-function mutant of a KANADI homolog, shows adaxialization only in the leaf sheath (Candela et al., 2008), which supports the notion of differential sensitivity to adaxial-abaxial polarity defects between the leaf blade and the leaf sheath in monocots.
It has been suggested that alterations to adaxial-abaxial patterning mechanisms could be major driving forces in modifying leaf forms (Kim et al., 2003; Gleissberg et al., 2005; Johnston et al., 2010). The unifacial leaf is one of the most interesting examples in which alterations in leaf adaxial-abaxial polarity have given rise to a novel leaf form. Identification of genes responsible for unifacial leaf development is essential for a better understanding of the developmental and evolutionary mechanisms underlying unifacial leaf blade formation. The establishment of adaxial-abaxial polarity is regulated by several distinct families of transcription factors and small regulatory RNAs (Husbands et al., 2009). It is possible that a genetic change or changes in one or more of these regulators may have resulted in unifacial leaf development. Further functional studies of each regulator are expected to reveal the genetic mechanisms of leaf blade abaxialization in unifacial leaves.
DL May Promote Flattened Leaf Blade Formation in J. prismatocarpus
Observations of developmental patterns of leaf blades in J. prismatocarpus and J. wallichianus have demonstrated that leaf blade flattening in unifacial leaves is promoted by directional laminar outgrowth along the median plane of the leaf primordia. At the cellular level, the directional laminar outgrowth appears to be initially triggered by active cell proliferation on the SAM side of the leaf blade, as demonstrated by a concentration of HistoneH4-expressing cells in J. prismatocarpus leaf primordia. Subsequently, laminar outgrowth may be bidirectionally maintained by more diffuse cell proliferation activity. A similar observation has been reported in another flattened unifacial-leafed species, Acorus calamus, where unifacial leaf blade flattening is initially mediated by adaxial (i.e., SAM side) meristematic activity and then bidirectionally proceeds by more diffuse meristematic activity (Kaplan, 1970). Therefore, leaf blade flattening of unifacial leaves, which probably has a conserved mechanism among monocots, could be dissected into two developmental processes: active cell proliferation at the SAM side of leaf primordia at an early stage of development and more diffuse cell proliferation during later stages.
We identified the DL ortholog as a candidate responsible for leaf blade flattening in J. prismatocarpus. Expression levels and patterns of DL correlate with the degree of laminar outgrowth in J. prismatocarpus and J. wallichianus. Genetic analysis using interspecific hybrids demonstrates that the chromosome region containing the DL locus from J. prismatocarpus flattens the unifacial leaf blade. Although further examinations are required to finely map the locus, expression analysis demonstrates that the expression activities of DL differ between the two species, probably due to cis-regulatory changes at the DL locus itself, rather than differences in trans-acting factors of DL. These results suggest that DL is one of the genetic factors that promote laminar outgrowth in J. prismatocarpus. During unifacial leaf blade flattening, DL is probably involved in active cell proliferation on the SAM side of leaf primordia at an early stage of leaf development because strong DL expression is observed during the P1 and P2 stages and DL has a similar function in rice in promoting cell proliferation in the leaf primordia toward the SAM side. Functional studies, such as those involving the generation of near-isogenic lines carrying the Jp DL allele in a J. wallichianus background and vice versa, or the isolation of loss- and/or gain-of-function DL lines by mutation or transgenic approaches, would further reveal the exact role of DL in unifacial leaf development.
Genetic analysis indicated that factors other than DL are also involved in the differential laminar outgrowth observed between J. prismatocarpus and J. wallichianus. Such factors may interact with DL at an early stage or regulate more diffuse cell proliferation activity during the later stages of leaf development. Identifying such factors by quantitative trait locus mapping using interspecific hybrids and by isolating the causative genes for the rad mutants in J. prismatocarpus would further clarify the genetic mechanisms underlying leaf blade flattening in unifacial leaves.
Identification of cis-regulatory differences at the DNA sequence level in the DL locus of the two Juncus species would further deepen our understanding of the mechanisms that regulate DL expression. As flattened unifacial leaves are widespread in monocots from the earliest divergent extant family, Acoraceae, to specialized families, such as Iridaceae and Juncaceae (Rudall and Buzgo, 2002; Yamaguchi and Tsukaya, 2010), and expression patterns of DL are similar between bifacial leaves of rice and flattened unifacial leaves of J. prismatocarpus, it is likely that expression activity in leaf primordia is reduced in J. wallichianus. Interestingly, the differential cis-regulatory activity of DL is organ specific: Jp DL and Jw DL alleles express almost the same amount of DL transcripts in carpels of the interspecific hybrid F1 generation. It has been suggested that organ-specific cis-regulatory changes of multifunctional genes may provide a mechanism for generating morphological diversity, while preserving their roles in other developmental processes (Carroll, 2000; Shapiro et al., 2004; Prud'homme et al., 2006). In the evolution of unifacial leaves, DL expression may have been modified multiple times to generate diversity in transverse forms of unifacial leaf blades. Thus, it will be of great interest to identify the actual sequences of the cis-regulatory elements of DL, so as to examine their roles in the evolution of flattened and radialized unifacial leaves.
Regulation of Central-Marginal Leaf Polarity Differentiation
The mechanisms that regulate the specification of central-marginal leaf polarity are largely unknown even in model species because of a lack of useful mutants. For example, it remains unknown whether specification of central-marginal polarity formation requires positional cues regarding the relationship to the SAM. It is also unclear whether specification of leaf margin identity requires positional information from the junction of leaf adaxial-abaxial domains, where leaf margins generally develop in bifacial leaves. In this study, we have shown that leaf blade flattening is a key that triggers DL and PRSb expression in the secondary central and marginal domains of the flattened leaf blade, respectively. This finding indicates that central-marginal leaf polarity can somewhat autonomously differentiate depending on the flattened leaf shape without information from a source outside of the leaf, such as the SAM. In addition, specification of leaf margin identity could be partly independent of adaxial-abaxial polarity, although positional information from the junction of adaxial-abaxial polarity is necessary for PRSa expression and probably for rigid specification of leaf margin identity. We propose that once the leaf blade is flattened, a gradient of central-marginal polarity is formed by an as yet unidentified factor, which is distributed in a symmetrical pattern in the flattened leaf blade. The plant hormone auxin is known to act as a gradient signal in multiple contexts throughout plant development (De Smet and Jurgens, 2007; Bowman and Floyd, 2008). During Arabidopsis leaf development, auxin is symmetrically distributed on either side of the midvein (Reinhardt et al., 2003; Zgurski et al., 2005) and is therefore a potential candidate for this unidentified molecule. Thus, it will be of great interest to study the distribution of auxin in unifacial leaves and to determine its relationship with central-marginal polarity differentiation.
The functions of DL and PRSb in the secondary central and marginal domains are also of interest. Morphological differentiation of the secondary central-marginal polarity is often observed in flattened unifacial leaves, although it is not clearly evident in J. prismatocarpus. Some flattened unifacial-leafed species, such as A. calamus or Iris ensata, develop a clear secondary midrib in the central domain of the flattened leaf blade, and margin-like tissues often differentiate at both tips of the flattened blade (Kaplan, 1970; Yamaguchi and Tsukaya, 2010). In these species, DL and PRSb homologs may regulate the formation of the secondary midrib and margin-like tissues, respectively. It is also possible that PRSb expression in margin-like regions promotes flattening of unifacial leaf blades by promoting marginal growth, although PRSb is not directly involved in the difference in leaf flatness between J. prismatocarpus and J. wallichianus. Loss of PRSb expression in J. wallichianus is probably a secondary effect due to a lack of leaf blade flattening in this species, and Jw PRSb would possess a cryptic function equivalent to that of Jp PRSb, as indicated by genetic analysis. Isolation of PRSb mutants from J. prismatocarpus would help to reveal the role of this gene during flattened unifacial leaf development.
Evolution of Leaf Flattening in Unifacial Leaves
Although the independent evolution of similar morphological traits is widespread, the underlying mechanisms are not fully understood (Gould, 2002; West-Eberhard, 2003; Yoon and Baum, 2004). Flattened leaves have independently evolved in bifacial and unifacial leaves, probably for efficient light capture. Our research demonstrates that flattening of leaf blades in unifacial leaves is promoted by DL function, which plays a distinct phenotypic role in bifacial leaves, namely, in leaf midrib formation. However, DL seems to play a similar function at the cellular level both in the bifacial and unifacial leaf development to promote cell proliferation of leaf primordia toward the shoot apex. Such DL function may have been easily co-opted to make abaxialized leaf blades flatten during the evolution of unifacial leaves. Thus, a preexisting gene function can be recruited to play a distinct phenotypic role in organisms with different body plans, without changing the gene’s cellular function, and such recruitment can give rise to convergence of similar morphological traits.
Unifacial leaves have repeatedly evolved in monocots (Rudall and Buzgo, 2002; Yamaguchi and Tsukaya, 2010), indicating the existence of backgrounds that allow unifacial leaf development in monocots. Since DL orthologs function in leaf development in monocots alone (Bowman and Smyth, 1999; Yamaguchi et al., 2004; Fourquin et al., 2005; Ishikawa et al., 2009; Wang et al., 2009), monocot leaves may possess the unique ability to become flattened by escaping from a developmental constraint to be radialized, even when they are abaxialized. Thus, the specific function of DL in leaf development in monocots may account for one genetic background that has allowed the repeated evolution of unifacial leaves in monocots.
METHODS
Plant Materials and Growth Conditions
Juncus prismatocarpus subsp leschenaultii Kirschner and Juncus wallichianus Laharpe were collected from wild populations in Okazaki, Japan. Herbarium specimens were verified by Futoshi Miyamoto at Tokyo University of Agriculture, Japan. At least six generations of each species were self-pollinated before use. Seeds of both species were cold-treated for 1 week at 4°C and germinated on agar plates containing Murashige and Skoog salts. Seedlings were grown in soil under a 16-h-light/8-h-dark cycle at 22°C. For mutagenesis, seeds of J. prismatocarpus were treated with 0.3% ethyl methanesulfonate for 14 h at room temperature. Approximately 5000 M2 plants were screened.
RNA Extractions and Degenerate PCR
Total RNA was isolated from 4-week-old seedlings of J. prismatocarpus and J. wallichianus using Plant RNA Isolation Reagent (Invitrogen), with subsequent treatment with DNaseI (Invitrogen). Total RNA (1 μg) was used for first-strand cDNA synthesis using the SuperScript III first-strand synthesis system (Invitrogen), and 0.5 μL of this reaction was used as the template for PCR amplification. Degenerate primers were designed using the CODEHOP program (Rose et al., 2003). Amplified DNA fragments were gel-extracted and cloned into pCRII-TOPO (Invitrogen), and at least 16 clones per fragment were sequenced. Primer sequences are listed in Supplemental Table 2 online.
5′ and 3′ Rapid Amplification of cDNA Ends
To determine 5′ and 3′ sequences of homologous genes, 5′ and 3′ rapid amplification of cDNA ends was performed. cDNA was generated using a GeneRacer kit (Invitrogen) according to the manufacturer’s protocol, using 2 μg of total RNA per reaction. Amplified products were cloned into pCRII-TOPO (Invitrogen), and at least eight clones per fragment were sequenced. Full-length cDNAs were reamplified by RT-PCR. Primer sequences are listed in Supplemental Table 2 online.
In Situ Hybridization
Shoot apices and inflorescences were fixed in 3% paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer for ~16 h at 4°C. Tissue was then dehydrated in a graded ethanol series, replaced with xylene, and embedded in Paraplast Plus (Oxford Labware). Hybridization was performed as previously described (Yamaguchi et al., 2004) on 9-μm paraffin sections. A portion of the cDNA sequences were amplified by RT-PCR, cloned into pCRII-TOPO (Invitrogen), and used to generate sense and antisense probes. Primer sequences are listed in Supplemental Table 2 online. In all cases, sense probes produced no signal.
Real-Time PCR
Total RNA was isolated from lateral branches that produced four visible leaves using Plant RNA Isolation Reagent, with subsequent treatment with DNaseI. Accumulation levels of DL transcripts were analyzed using a 7500 Real-Time PCR system (Applied Biosystems) by monitoring amplification with SYBR Premix Ex Taq II (Takara), as described in the manufacturer’s protocol. All data are presented as relative expression units after normalization to a TUBULIN gene. All data were reproduced in two or more additional independent experiments.
Analysis of Leaf Flatness
The middle portions of leaf blades that were formed at the final vegetative nodes were sampled after maturation, and cross sections were made by hand. Images of sections were taken and boxes enclosing the transverse planes were drawn on digital images. Leaf thickness (length along y axis) and width (length along x axis) were measured using Image J software (http://rsbweb.nih.gov/ij/). The ratio of leaf thickness to leaf width was calculated to evaluate leaf flatness. One-way analysis of variance (ANOVA) and the Tukey’s HSD test (α = 0.05, with Bonferroni correction) were used for multiple comparisons.
Genotyping Interspecific Hybrids
Genomic DNA from the DL and PRSb loci were amplified using PCR, then treated with the restriction enzymes NruI and NaeI, respectively, and separated by agarose gel electrophoresis. Primer sequences are listed in Supplemental Table 2 online.
Allele-Specific DL Expression
Total RNA was isolated from lateral branches that produced four visible leaves and young inflorescences (5 mm in length) of interspecific F1 hybrids using Plant RNA Isolation Reagent, with subsequent treatment with DNaseI. Total RNA (1 μg) was used for first-strand cDNA synthesis using the SuperScript III first-strand synthesis system, and 0.5 μL of this reaction was used as the template for PCR amplification. A portion of DL cDNA was amplified by 20 cycles of PCR and then purified and sequenced directly. The genomic DNA of the F1 hybrid was used as a control. Primer sequences are listed in Supplemental Table 2 online.
Phylogenetic Analysis
The amino acid sequences of HD-ZIPIII, ARF3/ARF4, YABBY, and WOX proteins were first aligned by ClustalW and readjusted manually. The phylogenetic trees were generated using MEGA4 (Tamura et al., 2007) by the neighbor-joining method (Saitou and Nei, 1987) with 1000 iterations of bootstrap analysis. For phylogenetic analysis of Juncus species, the nuclear ribosomal DNA internal transcribed spacer (ITS) region sequences were aligned by ClustalW and readjusted manually. Maximum likelihood and maximum parsimony analyses were performed using PAUP* version 4.0b10 (Swofford, 2002) as described by Roalson (2005). Sequences used to generate the phylogeny are presented in Supplemental Data Sets 1 to 5 online. Accession numbers used in the phylogenetic analyses are shown in Supplemental Table 3 online.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: AB539879 (Jp PHB), AB539877 (Jp ARF3a), AB539878 (Jp DL), AB539882 (Jw DL), AB539880 (Jp PRSa), AB539881 (Jp PRSb), AB539883 (Jw PRSa), AB539884 (Jw PRSb), and AB540127 (Jw ITS).
Author Contributions
T.Y. conceived the project, designed the study, performed the experiments, and wrote the article. S.Y. performed statistical analysis. H.T. designed and directed the study and wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phylogenetic Tree of HD-ZIPIII Proteins.
Supplemental Figure 2. Phylogenetic Tree of ARF3/ARF4 Proteins.
Supplemental Figure 3. Serial Longitudinal Sections of J. prismatocarpus Shoot Apices.
Supplemental Figure 4. Simplified Phylogenetic Tree of Juncus Showing the Relationship between J. prismatocarpus and J. wallichianus.
Supplemental Figure 5. Phylogeny of YABBY Family Proteins and Putative Amino Acid Sequences of J. prismatocarpus and J. wallichianus DL Proteins.
Supplemental Figure 6. Phylogenetic Tree of WOX Proteins.
Supplemental Figure 7. In Situ Localization of DL Transcripts in Carpel Primordia of J. prismatocarpus and J. wallichianus.
Supplemental Table 1. Leaf Flatness in Parental Lines and the F1 and F2 Generation of Interspecific Hybrids.
Supplemental Table 2. Primers Used in This Study.
Supplemental Table 3. Accession Numbers Used in Our Phylogenetic Analyses.
Supplemental Data Set 1. Sequences Used to Generate the Phylogenetic Tree of HD-ZIPIII Proteins Presented in Supplemental Figure 1.
Supplemental Data Set 2. Sequences Used to Generate the Phylogenetic Tree of ARF3/ARF4 Proteins Presented in Supplemental Figure 2.
Supplemental Data Set 3. Sequences Used to Generate the Phylogenetic Tree of Juncus Presented in Supplemental Figure 4.
Supplemental Data Set 4. Sequences Used to Generate the Phylogenetic Tree of YABBY Family Proteins Presented in Supplemental Figure 5A.
Supplemental Data Set 5. Sequences Used to Generate the Phylogenetic Tree of WOX Proteins Presented in Supplemental Figure 6.
Acknowledgments
We thank M. Futoshi for verifying the species used in this study and for helpful comments. We thank C. Yamaguchi, M. Nagura, T. Kadowaki, A. Irie, and A. Goto for technical assistance. This work was supported by a Grant-in-Aid for Creative Scientific Research to H.T. from the Japan Society for the Promotion of Science, a Grant-in-Aid for Scientific Research on Priority Areas to H.T., and a Grant-in-Aid for Young Scientists (B) to T.Y. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, as well as a grant from the Sumitomo Foundation to T.Y.
References
- Bowman J.L., Floyd S.K. (2008). Patterning and polarity in seed plant shoots. Annu. Rev. Plant Biol. 59: 67–88 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126: 2387–2396 [DOI] [PubMed] [Google Scholar]
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 [DOI] [PubMed] [Google Scholar]
- Candela H., Johnston R., Gerhold A., Foster T., Hake S. (2008). The milkweed pod1 gene encodes a KANADI protein that is required for abaxial/adaxial patterning in maize leaves. Plant Cell 20: 2073–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. (2000). Endless forms: The evolution of gene regulation and morphological diversity. Cell 101: 577–580 [DOI] [PubMed] [Google Scholar]
- Cutler D. (1969). Anatomy of the Monocotyledons. IV. Juncales. (Oxford, UK: Clarendon; ). [Google Scholar]
- De Smet I., Jurgens G. (2007). Patterning the axis in plants–Auxin in control. Curr. Opin. Genet. Dev. 17: 337–343 [DOI] [PubMed] [Google Scholar]
- Eshed Y., Baum S.F., Perea J.V., Bowman J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11: 1251–1260 [DOI] [PubMed] [Google Scholar]
- Fourquin C., Vinauger-Douard M., Fogliani B., Dumas C., Scutt C.P. (2005). Evidence that CRABS CLAW and TOUSLED have conserved their roles in carpel development since the ancestor of the extant angiosperms. Proc. Natl. Acad. Sci. USA 102: 4649–4654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudin V., Lunness P.A., Fobert P.R., Towers M., Riou-Khamlichi C., Murray J.A.H., Coen E., Doonan J.H. (2000). The expression of D-Cyclin genes defines distinct developmental zones in snapdragon apical meristems and is locally regulated by the Cycloidea gene. Plant Physiol. 122: 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleissberg S., Groot E.P., Schmalz M., Eichert M., Kolsch A., Hutter S. (2005). Developmental events leading to peltate leaf structure in Tropaeolum majus (Tropaeolaceae) are associated with expression domain changes of a YABBY gene. Dev. Genes Evol. 215: 313–319 [DOI] [PubMed] [Google Scholar]
- Gould S.J. (2002). The Structure of Evolutionary Biology. (Cambridge, MA: Belknap; ). [Google Scholar]
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T. (2004). Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668 [DOI] [PubMed] [Google Scholar]
- Husbands A.Y., Chitwood D.H., Plavskin Y., Timmermans M.C. (2009). Signals and prepatterns: New insights into organ polarity in plants. Genes Dev. 23: 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa M., Ohmori Y., Tanaka W., Hirabayashi C., Murai K., Ogihara Y., Yamaguchi T., Hirano H.Y. (2009). The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses. Genes Genet. Syst. 84: 137–146 [DOI] [PubMed] [Google Scholar]
- Itoh J., Nonomura K., Ikeda K., Yamaki S., Inukai Y., Yamagishi H., Kitano H., Nagato Y. (2005). Rice plant development: From zygote to spikelet. Plant Cell Physiol. 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Itoh J.I., Hibara K.I., Sato Y., Nagato Y. (2008b). Developmental role and auxin responsiveness of class III homeodomain leucine zipper gene family members in rice. Plant Physiol. 147: 1960–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh J.I., Sato Y., Nagato Y. (2008a). The SHOOT ORGANIZATION2 gene coordinates leaf domain development along the central–marginal axis in rice. Plant Cell Physiol. 49: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Johnston R., Candela H., Hake S., Foster T. (2010). The maize milkweed pod1 mutant reveals a mechanism to modify organ morphology. Genesis, in press [DOI] [PubMed] [Google Scholar]
- Juarez M.T., Kui J.S., Thomas J., Heller B.A., Timmermans M.C. (2004). MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kaplan D.R. (1970). Comparative foliar histogenesis in Acorus calamus and its bearing on the phyllode theory of monocotyledonous leaves. Am. J. Bot. 57: 331–361 [Google Scholar]
- Kaplan D.R. (1973). The problem of leaf morphology and evolution in the monocotyledons. Q. Rev. Biol. 48: 437–457 [Google Scholar]
- Kaplan D.R. (1975). Comparative developmental evaluation of the morphology of unifacial leaves in the monocotyledons. Bot. Jahrb. Syst. Pflanzengesch. Pflanzengeogr. 95: 1–105 [Google Scholar]
- Kerstetter R.A., Bollman K., Taylor R.A., Bomblies K., Poethig R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411: 706–709 [DOI] [PubMed] [Google Scholar]
- Kim M., McCormick S., Timmermans M., Sinha N. (2003). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424: 438–443 [DOI] [PubMed] [Google Scholar]
- Kirschner J. (2002a). Juncaceae 2: Juncus subg. Juncus, Species Plantarum: Flora of the World, Part 7. (Canberra, Australia: ABRS; ). [Google Scholar]
- Kirschner J. (2002b). Juncaceae 3: Juncus subg. Agathryon, Species Plantarum: Flora of the World, Part 8. (Canberra, Australia: ABRS; ). [Google Scholar]
- Matsumoto N., Okada K. (2001). A homeobox gene, PRESSED FLOWER, regulates lateral axis-dependent development of Arabidopsis flowers. Genes Dev. 15: 3355–3364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J.R., Emery J., Eshed Y., Bao N., Bowman J., Barton M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709–713 [DOI] [PubMed] [Google Scholar]
- Nardmann J., Ji J., Werr W., Scanlon M.J. (2004). The maize duplicate genes narrow sheath1 and narrow sheath2 encode a conserved homeobox gene function in a lateral domain of shoot apical meristems. Development 131: 2827–2839 [DOI] [PubMed] [Google Scholar]
- Pekker I., Alvarez J.P., Eshed Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17: 2899–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza P., Jasinski S., Tsiantis M. (2005). Evolution of leaf developmental mechanisms. New Phytol. 167: 693–710 [DOI] [PubMed] [Google Scholar]
- Prud'homme B., Gompel N., Rokas A., Kassner V.A., Williams T.M., Yeh S.D., True J.R., Carroll S.B. (2006). Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440: 1050–1053 [DOI] [PubMed] [Google Scholar]
- Reinhardt D., Pesce E.R., Stieger P., Mandel T., Baltensperger K., Bennett M., Traas J., Friml J., Kuhlemeier C. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426: 255–260 [DOI] [PubMed] [Google Scholar]
- Roalson E.H. (2005). Phylogenetic relationships in the Juncaceae inferred from nuclear ribosomal DNA internal transcribed spacer sequence data. Int. J. Plant Sci. 166: 397–413 [Google Scholar]
- Rose T.M., Henikoff J.G., Henikoff S. (2003). CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 31: 3763–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudall P.J., Buzgo M. (2002). Evolutionary history of the monocot leaf. Developmental Genetics and Plant Evolution, Cronk Q.C.B., Bateman R.M., Hawkins J.A., (London: Taylor & Francis; ), pp. 431–458 [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sawa S., Watanabe K., Goto K., Liu Y.G., Shibata D., Kanaya E., Morita E.H., Okada K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13: 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M.D., Marks M.E., Peichel C.L., Blackman B.K., Nereng K.S., Jonsson B., Schluter D., Kingsley D.M. (2004). Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428: 717–723 [DOI] [PubMed] [Google Scholar]
- Siegfried K.R., Eshed Y., Baum S.F., Otsuga D., Drews G.N., Bowman J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126: 4117–4128 [DOI] [PubMed] [Google Scholar]
- Steeves T.A., Sussex I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Swofford D.L. (2002). PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10. (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Timmermans M.C., Hudson A., Becraft P.W., Nelson T. (1999). ROUGH SHEATH2: A Myb protein that represses knox homeobox genes in maize lateral organ primordia. Science 284: 151–153 [DOI] [PubMed] [Google Scholar]
- Tsiantis M., Schneeberger R., Golz J.F., Freeling M., Langdale J.A. (1999). The maize rough sheath2 gene and leaf development programs in monocot and dicot plants. Science 284: 154–156 [DOI] [PubMed] [Google Scholar]
- Vandenbussche M., Horstman A., Zethof J., Koes R., Rijpkema A.S., Gerats T. (2009). Differential recruitment of WOX transcription factors for lateral development and organ fusion in petunia and Arabidopsis. Plant Cell 21: 2269–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites R., Hudson A. (1995). phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121: 2143–2154 [Google Scholar]
- Waites R., Selvadurai H.R., Oliver I.R., Hudson A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93: 779–789 [DOI] [PubMed] [Google Scholar]
- Wang A., Tang J., Li D., Chen C., Zhao X., Zhu L. (2009). Isolation and functional analysis of LiYAB1, a YABBY family gene, from lily (Lilium longiflorum). J. Plant Physiol. 166: 988–995 [DOI] [PubMed] [Google Scholar]
- West-Eberhard M.J. (2003). Developmental Plasticity and Evolution. (Oxford, UK: Oxford University Press; ). [Google Scholar]
- Yamaguchi T., Nagasawa N., Kawasaki S., Matsuoka M., Nagato Y., Hirano H.Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16: 500–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Tsukaya H. (2010). Evolutionary and developmental studies of unifacial leaves in monocots: Juncus as a model system. J. Plant Res. 123: 35–41 [DOI] [PubMed] [Google Scholar]
- Yoon H.S., Baum D.A. (2004). Transgenic study of parallelism in plant morphological evolution. Proc. Natl. Acad. Sci. USA 101: 6524–6529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgurski J.M., Sharma R., Bolokoski D.A., Schultz E.A. (2005). Asymmetric auxin response precedes asymmetric growth and differentiation of asymmetric leaf1 and asymmetric leaf2 Arabidopsis leaves. Plant Cell 17: 77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.H., Xu Q., Zhu X.D., Qian Q., Xue H.W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]