Abstract
Rambutan (Nephelium lappaceum) and lychee (Litchi chinensis) are tropical trees in the Sapindaceae that produce delicious edible fruits and are increasingly cultivated in tropical regions. These trees are afflicted with a stem canker disease associated with the ascomycete Dolabra nepheliae. Previously known from Asia and Australia, this fungus was recently reported from Hawaii and Puerto Rico. The sexual and asexual states of Dolabra nepheliae are redescribed and illustrated. In addition, the ITS and large subunit of the nuclear ribosomal DNA plus fragments from the genes RPB2, TEF1, and the mitochondrial small ribosomal subunit were sequenced for three isolates of D. nepheliae and compared with other sequences of ascomycetes. It was determined that D. nepheliae represents a new lineage within the Eurotiomycetes allied with Phaeomoniella chlamydospora, the causal agent of Petri grapevine decline.
Keywords: Canker, Litchi, Nephelium, Phaeomoniella chlamydospora, Sapindaceae, Systematics
Introduction
Rambutan (Nephelium lappaceum L.) and lychee (Litchi chinensis Sonn.) are tropical trees in the Sapindaceae that produce delicious edible fruits. Although these plants originated in Asia, they have been widely cultivated as garden fruit trees throughout the tropics and are now being developed as crop plants for commercial exploitation. Rambutan flourishes from sea level to 600 m in tropical, humid regions having evenly distributed rainfall (Morton 1987; Tindall 1994). Depending on the location, rambutan can produce up to two crops a year (Laksmi et al. 1987). Recently, production of rambutan and lychee has increased in Australia as well as in Hawaii and Puerto Rico where improved plant germplasm for disease resistance and increased fruit production is being investigated. In Hawaii, about 1 million kg of these tropical specialty fruits valued at $4.5 million were grown in 2007, with longan, lychee, and rambutan garnering higher farm prices compared to other fruit crops in this niche market of exotic fruit (NASS-USDA 2008).
A number of fungal diseases attack both Nephelium lappaceum and Litchi chinensis. One of these diseases is a stem canker disease associated with the ascomycete Dolabra nepheliae C. Booth & W.P. Ting (Booth and Ting 1964). Dolabra nepheliae was originally described from Malaysia, has been reported from Australia (Janick and Paul 2008), and was recently reported from Hawaii and Puerto Rico (Rossman et al. 2007). An asexual state was described by Zalasky et al. (1971). With the increased planting of rambutan as a specialty crop, it is likely that this fungus will spread. Using DNA sequence data, the phylogenetic placement of this species was determined. In addition, a redescription and illustrations of the sexual and asexual state of Dolabra nepheliae are provided.
Materials and methods
Morphological methods
Fresh specimens of cankers with ascomata from Hawaii and Puerto Rico were obtained as air-dried collections. Three isolates were grown from single ascospores plated on Difco corn meal agar (CM) supplemented with 0.2% dextrose and antibiotics (2 mg/ml neomycin and streptomycin). Germinated spores were transferred to Difco potato dextrose agar (PDA) and CM plates. All isolates were maintained on CM slants at 4°C. Living cultures were deposited in the Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands. The original specimens from which isolates were obtained were deposited in the U.S. National Fungus Collections (BPI). For microscopic examination, material was rehydrated and mounted in 3% KOH. Fruiting bodies were sectioned at ∼ 10 μm thick using a freezing microtome. Sections were mounted in lactic acid with cotton blue. Observations of microscopic features were made using a Zeiss Axioplan 2 microscope with bright-field illumination. Photographs and measurements of microscopic features were taken using an Olympus Q Color 5 digital camera (Olympus America, Center Valley, PA, USA) and ImagePro software (Media Cybernetics, Silver Spring, MD, USA). Specimens examined are listed following the descriptions. To determine cultural characteristics each isolate was inoculated onto two 90-mm-diameter plastic Petri plates of PDA and allowed to grow for 1 week at 25°C under alternating fluorescent (12 h) and near-ultraviolet (12 h) light. Colony diameters were averaged. Color names are based on Rayner (1970).
Nucleic acid extraction and polymerase chain reaction (PCR) amplification
Fungal genomic DNA was isolated by scraping mycelium from PDA plates. This mycelium were subsequently ground up, and the DNA was extracted using the FastDNA kit and the FastPrep instrument from MPI Biochemicals (Irvine, CA, USA). We initially evaluated the following regions: the internal transcribed spacer regions plus the 5.8S nuclear ribosomal RNA (ITS) and the large subunit (28S) for the nuclear ribosome (LSU). DNA amplifications were completed using Taq polymerase (GenScript, Piscataway, NJ, USA), with FailSafe PCR 2× PreMix E (Epicentre, San Diego, CA, USA) under the following PCR conditions: 94°C for 2 min; five cycles of 94°C for 40 s, 55°C for 45 s lowering by 0.8°C per cycle and 72°C for 90 s; 30 cycles of 94°C for 30 s, 52°C for 45 s, and 72°C for 120 s, and a final cycle for 10 min at 72°C. The ITS region was amplified with primers ITS4 and ITS5 (White et al. 1990) and LSU amplified with primers LR0R and LR5 (Vilgalys and Hester 1990). To confirm that the cultures were actually D. nepheliae and sequences obtained were not the result of contamination, DNA was reisolated twice from these cultures. To provide a confident placement of Dolabra within the class Eurotiomycetes, we also amplified a number of gene fragments used by the Assembling the Fungal Tree of Life project (AFTOL) (Schoch et al. 2009). This procedure resulted in DNA sequence data obtained from two protein-coding genes, the translation elongation factor-1 alpha (TEF1) and the RNA polymerase II largest subunit (RPB1), as well as the mitochondrial small ribosomal subunit (MSSU). Primer sets used for these genes were as follows: TEF1–983, 2218R (initially obtained from S. Rehner: http://ocid.nacse.org/research/deephyphae/EF1primer.pdf), RPB1–RPB1-Ac, RPB1-Cr (initially obtained from V. Hofstetter), and MSSU–mrSSU1, mrSSU3R (Zoller et al. 1999). Primer sequences are available at the AFTOL website (aftol.org). PCRs for these three genes were performed under conditions described previously (Lutzoni et al. 2004; Schoch et al. 2009).
Phylogenetic analysis
Initial alignments were prepared using the M-Coffee web server (Moretti et al. 2007) and, for the ITS alignment, variable regions were excluded using GBlocks with the following settings: minimum size of blocks = 10, allow gapped positions, and allow 55% flanking sequences. Outgroups were selected according to Geiser et al. (2006). For the ITS data set, phylogenetic trees were obtained by maximum-parsimony analysis with the branch-and-bound search option in PAUP* (Swofford 2002). The LSU data set was analyzed with a heuristic search with 100 random additions in the same program (Fig. 1). In both data sets, alignment gaps were treated as missing data and characters were unordered and of equal weight in all cases. Branches of zero length were collapsed, and all multiple, equally parsimonious trees were saved. Group support was evaluated by 1,000 bootstrap pseudoreplicates. Tree length was calculated and the resulting trees were printed with Tree-View v. 1.6.6 (Page 1996) and Treedyn (Chevenet et al. 2006). The complete set of sequences and the related taxa used for comparison are listed in Table 1 with their GenBank Accession numbers.
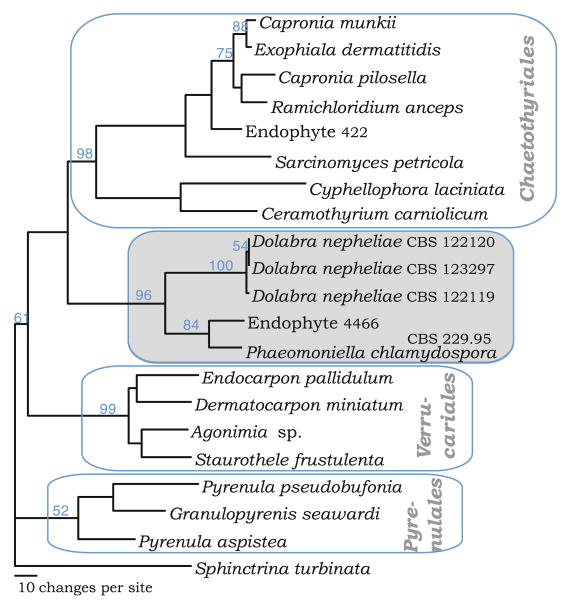
Fig. 1.
Phylogenetic placement of Dolabra nepheliae based on the large subunit (LSU) nrDNA. One of six most parsimonious trees (742 steps) with LSU sequences exemplifying the major related groups in the Eurotiomycetes. Bootstrap values from 1,000 pseudoreplicates are shown on the nodes
Table 1.
Taxa, isolate numbers, and GenBank sequence numbers used in the phylogenetic analyses
| Taxon | Isolate number | ITS | LSU | MSSU | RPB1 | TEF1 |
|---|---|---|---|---|---|---|
| Chaetothyriomycetidae | ||||||
| Chaetothyriales | ||||||
| Capronia pilosella | AFTOL 657 | DQ823099 | ||||
| Ceramothyrium carniolicum | AFTOL 1063 | EF413628 | EF413629 | EF413629 | ||
| Cyphellophora laciniata | AFTOL 1033 | EF413619 | ||||
| Exophiala dermatitidis | AFTOL 668 | DQ823100 | DQ840555 | DQ840566 | ||
| Exophiala pisciphila | AFTOL 669 | DQ823101 | DQ840556 | DQ840567 | ||
| Exophiala salmonis | AFTOL 671 | EF413609 | FJ225745 | EF413610 | EF413612 | |
| Ramichloridium anceps | AFTOL 659 | DQ823102 | FJ225752 | DQ840557 | DQ840568 | |
| Sarcinomyces petricola | AFTOL 2195 | FJ176893 | ||||
| Coryneliales | ||||||
| Caliciopsis orientalis | AFTOL 1911 | DQ470987 | FJ190654 | DQ471185 | DQ471111 | |
| Caliciopsis pinea | AFTOL 1869 | DQ678097 | FJ190653 | DQ677937 | ||
| Pyrenulales | ||||||
| Anthracothecium nanum | AFTOL 1649 | FJ358271 | FJ358403 | |||
| Granulopyrenis seawardi | AFTOL 2013 | EF411062 | ||||
| Lithothelium septemseptatum | AFTOL 12 | AY584638 | AY584620 | |||
| Pyrenula aspistea | AFTOL 2012 | EF411063 | EF411069 | |||
| Pyrenula pseudobufonia | AFTOL 387 | AY640962 | AY584720 | DQ840558 | ||
| Pyrgillus javanicus | AFTOL 342 | DQ823103 | FJ225774 | DQ842010 | ||
| Verrucariales | ||||||
| Agonimia sp. | AFTOL 684 | DQ782913 | DQ782853 | DQ782917 | ||
| Dermatocarpon miniatum | AFTOL 91 | AY584644 | AY584616 | DQ782821 | DQ782893 | |
| Endocarpon pallidulum | AFTOL 661 | DQ823097 | FJ225674 | DQ840552 | ||
| Polyblastia melaspora | AFTOL 1356 | EF413601 | EF413602 | |||
| Staurothele frustulenta | AFTOL 697 | DQ823098 | FJ225702 | DQ840553 | ||
| Wahlenbergiella mucosa | AFTOL 2264 | FJ225720 | EF689804 | |||
| Eurotiomycetidae | ||||||
| Eurotiales | ||||||
| Aspergillus fumigatus | AFTOL 1079 | Genome | Genome | |||
| Aspergillus nidulans | AFTOL 1080 | Genome | Genome | |||
| Eupenicillium javanicum | AFTOL 429 | EF413621 | FJ225778 | |||
| Eupenicillium limosum | AFTOL 2014 | EF411064 | EF411070 | |||
| Monascus purpureus | AFTOL 426 | DQ782908 | FJ225780 | DQ842012 | ||
| Penicillium freii | AFTOL 378 | AY640958 | AY584712 | |||
| Onygenales | ||||||
| Coccidioides immitis | AFTOL 1084 | Genome | Genome | Genome | ||
| Ajellomyces capsulatum (anamorph Histoplasma capsulatum) | AFTOL 1083 | Genome | Genome | Genome | ||
| Spiromastix warcupii | DQ782909 | FJ225794 | EF413613 | DQ782900 | ||
| Mycocaliciomycetidae | ||||||
| Mycocaliciales | ||||||
| Sphinctrina turbinata | AFTOL 1721 | EF413632 | FJ713611 | |||
| Incertae sedis | ||||||
| Dactylospora haliotrepha | AFTOL 758 | FJ176855 | ||||
| Dactylospora mangrovei | AFTOL 2108 | FJ176890 | FJ238411 | |||
| Dolabra nepheliae | CBS 123297 | GU345749 | GU332515 | |||
| Dolabra nepheliae | CBS 122119 | GU332516 | GU332518 | GU332520 | GU332522 | |
| Dolabra nepheliae | CBS 122120 | GU332517 | GU332519 | GU332521 | GU332523 | |
| Fungal endophyte | 4466 | DQ979444 | ||||
| Fungal endophyte | 422 | EF420091 | ||||
| Phaeomoniella chlamydospora (as Phaeoacremonium chlamydosporum) | CBS 229.95 | AF353609 | ||||
| Rock isolates (taken from Gueidan et al. 2008) | ||||||
| TRN488 | FJ358262 | FJ225766 | FJ358394 | |||
| TRN475 | FJ358260 | FJ225764 | FJ358392 | |||
| TRN107 | FJ358253 | FJ225758 | FJ358386 | |||
| TRN214 | FJ358256 | FJ225761 | FJ358389 | |||
| TRN508 | FJ358265 | FJ225770 | FJ358398 | |||
| A14 | FJ358268 | |||||
| TRN242 | FJ358257 | FJ225762 | FJ358390 | |||
| A95 | FJ358269 | FJ358401 | ||||
| Outgroup (Geoglossomycetes) | ||||||
| Geoglossum nigritum | AFTOL 56 | AY544650 | AY544740 | DQ471115 | DQ471044 |
ITS internal transcribed spacer, LSU large subunit, MSSU mitochondrial small ribosomal subunit
Additional alignments were completed by adding the newly generated sequence to initial alignment files generated by WASABI (Kauff et al. 2007) by using the ‘consensus align’ option using MUSCLE (Edgar 2004) in Geneious Pro version 4.6.5 (Biomatters, Auckland, New Zealand). A supermatrix was obtained from four genes (LSU, TEF1, RPB1, MSSU) consisting of 40% missing data. Geoglossum nigritum was used as an outgroup following Gueidan et al. (2008); this was analyzed with maximum likelihood in RAxML v 7.0.4 (Stamatakis 2006) applying unique model parameters for each gene. The dataset was thus partitioned according to each gene and separate codons (eight partitions) as in Schoch et al. (2009). The result was a general time-reversible model (GTR) applied for DNA sequences with a discrete gamma distribution and four rate classes. A tree was obtained by simultaneously running a fast bootstrap search of 1,000 pseudoreplicates followed by a search for the most likely tree under functional setting ‘a’. We also did 100 successive searches in RAxML under the GTR model with gamma rate distribution and starting each search from a randomized tree. The resultant most likely tree (log likelihood, −42731.850035) is shown in Fig. 2 with bootstrap values plotted onto the nodes. This data set was also analyzed in PAUP* by doing 1,000 bootstrap pseudoreplicates and 10 heuristic random additions per replicate. Parsimony bootstrap values for clades shown in the RAxML tree were added to the nodes. A maximum parsimony ran with 1,000 random additions in 5 trees with a length of 9,545 steps (not shown). Alignments have been deposited in Tree-BASE as S2590.
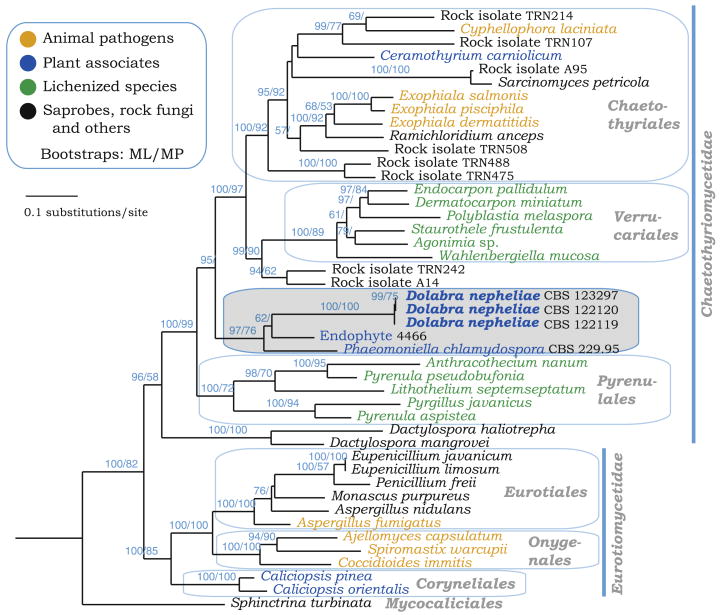
Fig. 2.
A maximum-likelihood tree obtained with RAxML from four genes—LSU nrDNA, RPB2, TEF1, mitochondrial small ribosomal subunit (MSSU)—data from representative lineage in Eurotiomycetes. Orders and subclasses are indicated, and bootstrap values (ML, maximum likelihood; MP, maximum parsimony) are plotted on the nodes. Bootstrap values below 50% are not shown. The outgroup branch with Geoglossum nigritum is not shown in the figure
Results
Sequencing and analyses
ITS sequences for the three isolates of Dolabra nepheliae from Hawaii and Puerto Rico and from the two related host genera, Litchi and Nephelium, were compared and showed that these were the same species (data not shown). The ITS sequences were compared with sequences in the GenBank database using BLAST analyses. The results suggested that these isolates were most closely related to species in the Chaetothyriales, Verrucariales, the grape pathogen Phaeomoniella chlamydosporia, and several unnamed environmental isolates in the Eurotiomycetes. The closest relationship was with Phaeomoniella chlamydospora, the causal agent of Petri grapevine decline (Crous and Gams 2000; Dupont et al. 2000; Groenewald et al. 2001) and fungal Endophyte 4466 obtained from an environmental study of Picea mariana (Higgins et al. 2007). An ITS data set moderately supported shared ancestry with these sequences (data not shown).
To determine more accurately the phylogenetic placement of Dolabra nepheliae, we compared three LSU sequences of D. nepheliae with a range of taxa and environmental isolates using maximum parsimony as shown in Fig. 1. Although these results indicate that D. nepheliae falls within the Eurotiomycetes, this species does not group within any of the major orders Chaetothyriales, Pyrenulales, or Verrucariales. Rather, D. nepheliae forms a separate clade with Phaeomoniella chlamydospora and Endophyte 4466 mentioned above (Hoffman and Arnold 2008).
To further test the placement of D. nepheliae with gene targets used by the AFTOL project, we amplified three additional gene fragments (TEF1, RPB1, MSSU) and combined these in a supermatrix with LSU and similar gene sequences produced by AFTOL. We also included a subset of rock-inhabiting fungi in the Eurotiomycetes from Gueidan et al. (2008). Following Wiens (2006), we included taxa in our supermatrix even though considerable data were missing (40%). The resultant phylogeny is shown in Fig. 2. It is congruent with two recent studies in the Ascomycota and Eurotiomycetes, specifically those by Schoch et al. (2009) and Gueidan et al. (2008). This phylogeny groups together D. nepheliae with Phaeomoniella chlamydospora and Endophyte 4466 with strong support outside the known orders of the Eurotiomycetes. The exact placement of this group was in conflict between the maximum-parsimony and maximum-likelihood analyses. Dolabra nepheliae was sister to the Pyrenulales with poor bootstrap support in the parsimony analyses (55% bootstrap, not shown), possibly the result of long branch attraction, and sister to both the Chaetothyriales and Verrucariales using maximum likelihood (95% bootstrap).
Taxonomy
Associated with corky bark symptoms, small to large irregular patches of raised bark on main trunk and lateral branches. Symptoms similar on root stock. Cankers on woody branches, slightly roughened to irregularly globose, extending from surface up to 1 cm, with deep fissures in which ascomata develop. Ascomata crowded, aggregated, lining fissures, superficial, dark brown to black, ovoid to elongate with irregularly schizogenous, ostiolar opening, collapsed laterally with apex sunken when dried, surface smooth to slightly roughened, 350–750 μm high × 225–375 wide, with long stalked base. Ascomatal wall gelatinous, of two regions: outer region black, of thick-walled cells; inner region of hyaline, thin-walled cells. Pseudoparaphyses abundant, hyaline, unbranched, septate, extending beyond asci to upper region of ascomata. Asci bitunicate, narrowly clavate, 100–130 × 10–14 μm. Ascospores long cylindrical, curved, with rounded ends, 96–136 × 2.5– 3.5 μm, hyaline.
Pycnidia dark brown to black, ovoid, scattered to clustered, 110–150 × 110–125 μm diameter, smooth. Pycnidial wall of two regions: outer region of brown to black, thick-walled cells; inner region of hyaline cells, 2–8 rows; opening by an irregular slit, occasionally opening completely to appear discoid. Conidiophores hyaline, septate, short, formed from inner cells of the pycnidial wall. Conidiogenous cells holoblastic, hyaline, smooth, cylindrical 2.0–3.4 × 8.3–10.7 μm. Conidia long fusiform, slightly curved to lunate, 35–76 × 19–4.2 μm, (2–4) 5-septate, smooth, hyaline, base slightly truncate, apex rounded. Pycnidia developing toward center of colony after 1 week on PDA, appearing same as pycnidia on substratum.
Growth on PDA after 1 week in 25°C: colony 0.8–2.0 cm diameter, with heaped mycelium, white at margin, becoming slightly darker or dark green (dull green) toward center when pycnidia produced; reverse olivaceous buff, becoming olivaceous toward center. After 2 weeks on PDA, 1.8–2.6 cm diameter, appearing similar to after 1 week, white at margin, reverse olivaceous buff.
Hosts
Litchi chinensis Sonn. (lychee) and Nephelium lappaceum L. (rambutan), Sapindaceae. Also reported on Nephelium mutabile Blume (pulasan) (Zalasky et al. 1971).
Distribution
Australia (Janick and Paul 2008), Malaysia (type locality, Booth and Ting 1964), USA: Hawaii, Puerto Rico (Rossman et al. 2007).
Type specimens examined
MALAYSIA: Selangor, Petaling Jaya, on bark of Nephelium lappaceum, 18 October 1962, W.P. Ting (IMI 96355 Holotype); ibid., 25 September 1962 (IMI 95969 Topotype).
Additional specimens examined
USA: Hawaii, Hilo, in USDA greenhouse, on Nephelium lappaceum, causing cankers, 18 June 2007, Lisa Keith, det. and isol. Amy Rossman AR 4422 = CBS 122119 (BPI 878189); Hawaii, Island of Kauai, Kilauea, on Litchi chinensis, 19 November 1984, George Wong and Chuck Hodges (BPI 626373, IMI 293693); Puerto Rico: Mayaguez, in USDA greenhouse, on small twigs of L. chinensis, May 2007, Ricardo Goenaga, det. and isol. Amy Rossman AR 4421 = CBS 123297 (BPI 878188); Mayaguez, in USDA greenhouse, on N. lappaceum, causing cankers, April 2007, Ricardo Goenaga, det. and isol. Amy Rossman AR 4426 = CBS 122120 (BPI 878190). In addition, the fungus was observed in 2001 associated with galls on twigs and branches of rambutan trees grown in a commercial orchard in the Papaikou district of the island of Hawaii.
Notes
The description presented in the original publication and the type specimens agree with observations of the more recently collected specimens from Hawaii and Puerto Rico. The specimens from Hawaii and Puerto Rico demonstrate the extensive cankers and deep fissures caused by this fungus that result in a serious disease. The type specimens showed only superficial wounding of the upper layers of tissue. The ascomatal and pycnidial wall structure is described here in detail for the first time.
Discussion
Dolabra nepheliae is distinctive in producing numerous, crowded, large, elongated fruiting bodies on well-developed cankers on the stems of the woody plant hosts, species of Litchi and Nephelium in the Sapindaceae. This fungus was originally described from Malaysia on cultivated Nephelium lappaceum and is also reported on pulasan (N. mutabile) and from Australia (Zalasky et al. 1971; Janick and Paul 2008) with specimens apparently deposited in KLA, although these could not be located. The specimen previously collected in 1984 by George Wong and Chuck Hodges on Litchi chinensis suggests that this fungus has been in Hawaii for some time, most likely on plants grown in residential gardens. This fungus is now also known from Puerto Rico (Rossman et al. 2007), possibly imported from Hawaii on plant germplasm.
The disease appears on the bark of older branches and stems causing the formation of corky tissue; new growth is not initially affected by the disease. Symptoms of the disease extend from older to younger tissue as the fungus grows out from the main trunk to the stems and eventually the twigs. In severe cases, dieback of entire branches may occur. The disease progresses slowly, and it may take years for a tree to be severely affected, even in cases in which new growth such as chupons or new branches appear to be free of disease. Trees do not die from the disease, but severe cases show reduced tree growth. A field survey at the Waiakea Agriculture Experiment Station in Hilo, Hawaii, where 55 accessions are grown, did not result in a pattern of symptom development, indicating that location and variety do not appear to have any effect (K. Nishijima, personal communication). Rambutan and litchi trees grown in a semiarid environment do not show symptoms of the disease even several years after planting.
Dolabra nepheliae is characterized by bitunicate asci and abundant, unbranched, thin-walled, septate pseudoparaphyses that suggest placement in the Pleosporales, Dothideomycetes. Booth and Ting (1964) stated that “the structure of the locules in the upper half of the stroma is typical of the Pleosporales…” and “the ascocarps suggest a member of the Nitschkiaceae…”. However, as discussed below, neither of these placements is correct. The anamorph known to be associated with the canker on rambutan trees as well as in culture was first described and illustrated by Zalasky et al. (1971) (see Figs. 8–11). Although they referred to this anamorph as Rhabdospora nepheliae, this scientific name was not validly published because of the lack of a Latin description, nor was a type specimen designated.
Fig. 8.

Anamorph of Dolabra nepheliae. Pycnidia developing in culture. Bars 100 μm
Fig. 11.
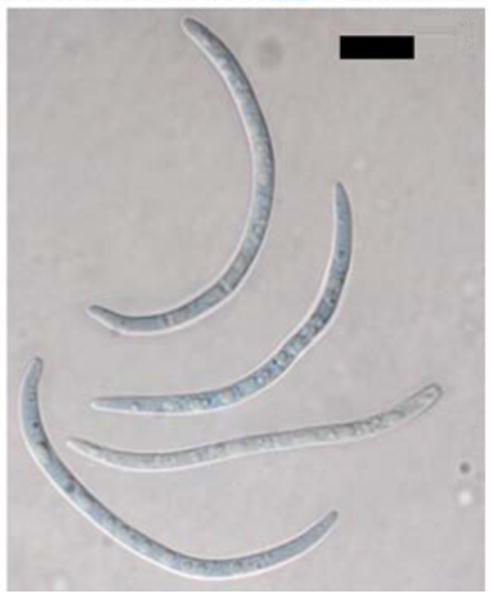
Anamorph of Dolabra nepheliae. Conidia. Bars 10 μm
Instead of confirming our initial assessment as a member of the Pleosporales, Dothideomycetes, the multigene sequence data suggest that Dolabra nepheliae represents a new lineage, placed as incertae sedis in the subclass Chaetothyriomycetidae of the Eurotiomycetes (see Figs. 1, 2). The Eurotiomycetes includes diverse orders ranging from the so-called “black yeasts” of the Chaetothyriales and lichenized Pyrenulales and Verrucariales to the “common moulds” of the Eurotiales and host-specialized Coryneliales and Onygenales (Geiser et al. 2006). The majority of the “black yeasts,” animal and human opportunistic pathogens, were previously thought to be related to other loculoascomycetes in the Dothideomycetes because of similarities in morphology and development of the ascomata (Luttrell 1951; Lumbsch and Huhndorf 2007). Since the advent of phylogenies derived from molecular data, these two groups of fungi having bitunicate asci have proven to be distinct and separate (Winka et al. 1998; Spatafora et al. 2006).
Morphological characters such as the bitunicate asci are common in the subclass Chaetothyriomycetidae. The phylogenetic placement of Dolabra nepheliae in the Chaetothyriomycetidae, Eurotiomycetes, is surprising given that this species has pseudoparaphyses. Most members of the Eurotiomycetes including the Chaetothyriales lack interthecial elements, although pseudoparaphyses are present in the Verrucariales (Gueidan et al. 2009). Their presence in Dolabra suggests convergence between the Dothideomycetes and Eurotiomycetes. The characteristic feature of Dolabra nepheliae most indicative of the Eurotiomycetes is the schizogenous ascomatal apex that splits to form an irregularly shaped pore and the stalked base of the ascomata. The other two isolates in this clade represent fungi with little morphology. Phaeomoniella chlamydospora lacks a fruiting body, producing only a hyphomycetous state.
The Eurotiomycetes are an ecologically diverse class with well-defined clades focused on specific life strategies including lichens (most of the Pyrenulales, Verrucariales), animal pathogens (Onygenales), and plant-associated fungi (Dolabra, Phaeomoniella, Coryneliales), as highlighted in Fig. 2. Within the class, new lineages continue to emerge, such as a clade of marine fungi associated with the breakdown of wood on mangroves Dactylospora mangrovei and D. manglicola, previously placed in the lichenized order Lecanoromycetes on the basis of morphology (Kohlmeyer and Kohlmeyer 1979; Jones et al. 1999; Schoch et al. 2009). Plant-associated fungi are included in the Chaetothyriales and Coryneliales. The latter are parasites of the Podocarpaceae, have fissitunicate asci and ascolocular ascomata, and, until recently, were considered an order of uncertain position within the Pezizomycotina (Eriksson 2006). In many ways fungi in the Coryneliales are unique, producing thin-walled asci that were shown to have multiple layers (Johnston and Minter 1989). The Dolabra lineage is only distantly related to Coryneliales but within the context of the Eurotiomycetes presents another example of a plant-associated fungus. Given the relationship of the two plant pathogens, Dolabra nepheliae with Phaeomoniella chlamydospora, it seems realistic to expect to discover additional related fungi involved in plant disease in this novel lineage. Our study clearly suggests an underestimation of morphological and ecological diversity within the Eurotiomycetes.
Fig. 3.

Dolabra nepheliae. Cankers on woody trunks of Nephelium lappaceum caused by D.nepheliae.
Fig. 4.

Dolabra nepheliae. Ascomata developing within fissures of cankers on N.lappaceum. Bars 300 μm
Fig. 5.
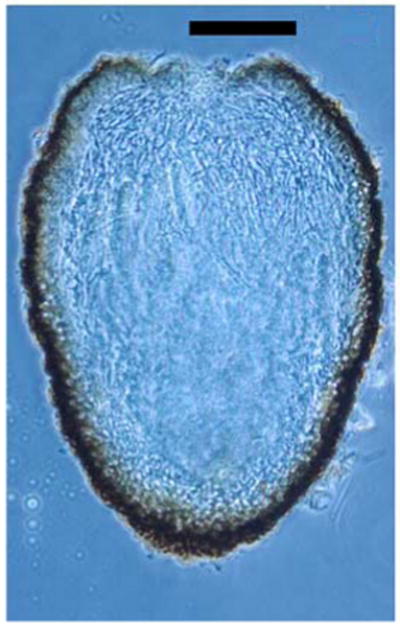
Dolabra nepheliae. Transverse section of immature ascoma showing ascomatal wall and centrum filled with pseudoparaphyses (section by G. Samuels). Bars 100 μm
Fig. 6.
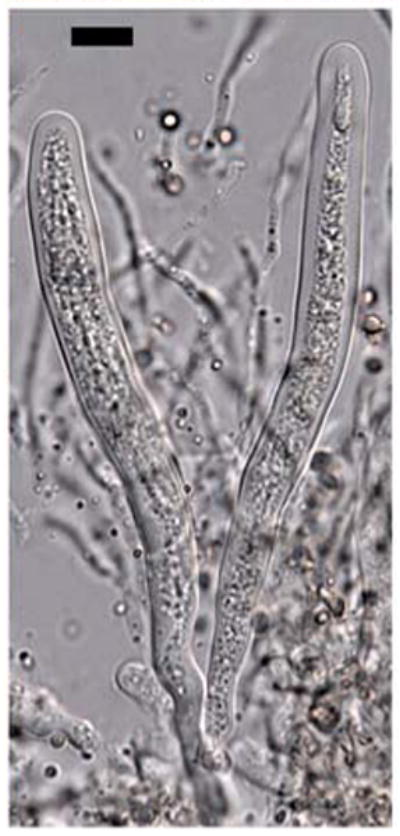
Dolabra nepheliae. Bitunicate asci and pseudoparaphyses. Bars 10 μm
Fig. 7.
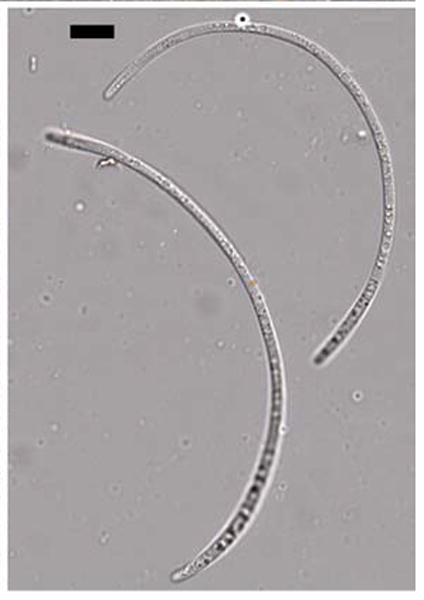
Dolabra nepheliae. Ascospores. Bars 10 μm
Fig. 9.
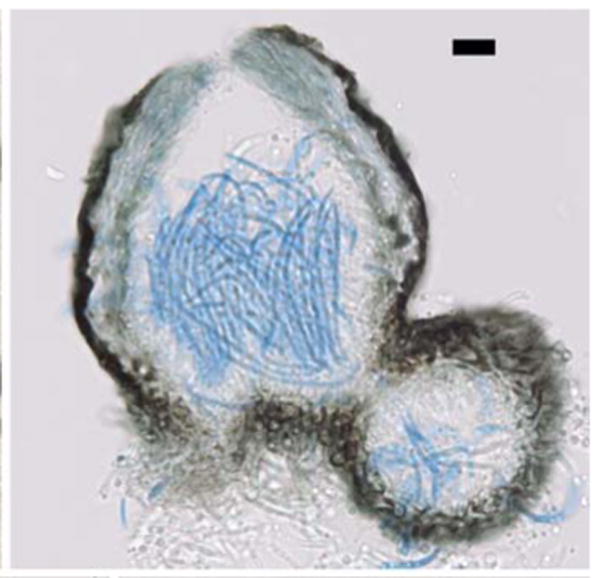
Anamorph of Dolabra nepheliae. Transverse section of pycnidia produced in culture. Bars 10 μm
Fig. 10.
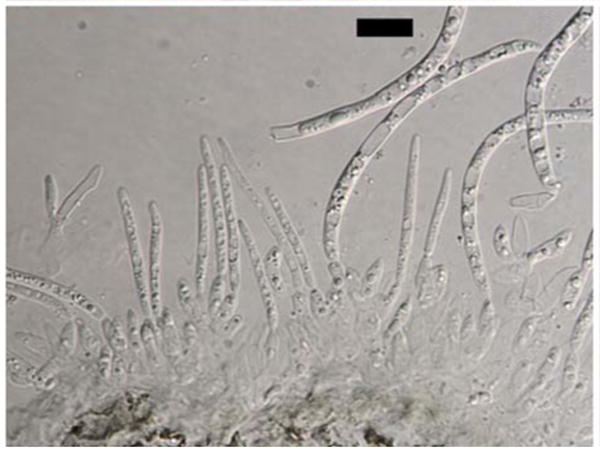
Anamorph of Dolabra nepheliae. Conidiogenous cells with developing conidia. Bars 10 μm
Acknowledgments
The authors appreciate the generous advice given by Richard P. Korf, Donald H. Pfister, and Gary J. Samuels in identifying this fungus. We thank Lisa Castlebury for her presubmission review and Steve Rehner and Valerie Hofstetter for providing primers. The able technical assistance of Audrey McVay at Oregon State University is gratefully acknowledged. The skilled support of Tunisha Phipps and Sasha Allen at the Systematic Mycology and Microbiology Laboratory was critical in taking care of the fungal cultures involved in this research. This research was supported in part by the Intramural Research Program of the NIH, National Library of Medicine.
Contributor Information
Amy Y. Rossman, Email: amy.rossman@ars.usda.gov, Systematic Mycology and Microbiology Laboratory, USDA Agricultural Research Service, Beltsville, MD 20705, USA.
Conrad L. Schoch, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, 45 Center Drive, MSC 6510, Bethesda, MD 20892, USA
David F. Farr, Systematic Mycology and Microbiology Laboratory, USDA Agricultural Research Service, Beltsville, MD 20705, USA
Kate Nishijima, Tropical Crop and Commodity Protection Research Unit, PBARC, USDA-ARS, Hilo, HI 96720, USA.
Lisa Keith, Tropical Plant Genetic Resources and Disease Research Unit, PBARC, USDA-ARS, Hilo, HI 96720, USA.
Ricardo Goenaga, Tropical Agriculture Research Station, USDA-ARS, Mayaguez, PR 00680, USA.
References
- Booth C, Ting WP. Dolabra nepheliae gen. nov., sp. nov., associated with canker of Nephelium lappaceum. Trans Br Mycol Soc. 1964;47:235–237. [Google Scholar]
- Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Gams W. Phaeomoniella chlamydospora gen. et comb. nov., a causal organism of Petri grapevine decline and esca. Phytopathol Mediterr. 2000;39:112–118. [Google Scholar]
- Dupont J, Laloui W, Roquebert MF. Partial ribosomal DNA sequences show an important divergence between Phaeoacremonium species isolated from Vitis vinifera. Mycol Res. 2000;102:631–637. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson OE. Outline of Ascomycota. Myconet. 2006;12:1–82. [Google Scholar]
- Geiser DM, Gueidan C, Miadlikowska J, Lutzoni F, Kauff F, Hofstetter V, Fraker E, Schoch CL, Tibell L, Untereiner WA, Aptroot A. Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia. 2006;98:1053–1064. doi: 10.3852/mycologia.98.6.1053. [DOI] [PubMed] [Google Scholar]
- Groenewald M, Kang JC, Crous PW, Gams W. ITS and beta tubulin phylogeny of Phaeoacremonium and Phaeomoniella species. Mycol Res. 2001;105:651–657. [Google Scholar]
- Gueidan C, Ruibal CV, de Hoog GS, Gorbushina AA, Untereiner WA, Lutzoni F. An extremotolerant rock-inhabiting ancestor for mutualistic and pathogen-rich fungal lineages. Stud Mycol. 2008;62:111–119. doi: 10.3114/sim.2008.61.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueidan C, Savic S, Thus H, Roux C, Keller C, Tibell L, Prieto M, Heiomarsson S, Breuss O, Orange A, Froberg L, Wynns AA, Navarro-Rosines P, Krzewicka B, Pykala J, Grube M, Lutzoni F. Generic classification of the Verrucariaceae (Ascomycota) based on molecular and morphological evidence: recent progress and remaining challenges. Taxon. 2009;58:184–208. [Google Scholar]
- Higgins KL, Arnold AE, Miadlikowska J, Sarvate SD, Lutzoni F. Phylogenetic relationships, host affinity, and geographic structure of boreal and arctic endophytes from three major plant lineages. Mol Phylogenet Evol. 2007;42:543–555. doi: 10.1016/j.ympev.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Hoffman MT, Arnold AE. Geographic locality and host identity shape fungal endophyte communities in cupressaceous trees. Mycol Res. 2008;112:331–344. doi: 10.1016/j.mycres.2007.10.014. [DOI] [PubMed] [Google Scholar]
- Janick J, Paul RE. The Encyclopedia of Fruit and Nuts. CABI; Oxfordshire: 2008. [Google Scholar]
- Johnston PJ, Minter DW. Structure and taxonomic significance of the ascus in the Coryneliaceae. Mycol Res. 1989;92:422–430. [Google Scholar]
- Jones EBG, Abdel-Wahab MA, Alias SA, Hsieh SY. Dactylospora mangrovei sp. nov. (Discomycetes, Ascomycota) from mangrove wood. Mycoscience. 1999;40:317–320. [Google Scholar]
- Kauff F, Cox CJ, Lutzoni F. WASABI: an automated sequence processing system for multigene phylogenies. Syst Biol. 2007;56:523–531. doi: 10.1080/10635150701395340. [DOI] [PubMed] [Google Scholar]
- Kohlmeyer J, Kohlmeyer E. Marine mycology: the higher fungi. Academic Press; New York: 1979. [Google Scholar]
- Laksmi LDS, Lam PF, Mondoza DB, Jr, Kosiyachinda S, Leong PC. Status of the rambutan industry in ASEAN. In: Lam PF, Kosiyachinda S, editors. Rambutan: fruit development, postharvest physiology and marketing in ASEAN. ASEAN Food Handling Bureau; Kuala Lumpur: 1987. [Google Scholar]
- Lumbsch HT, Huhndorf S. Whatever happened to the pyrenomycetes and loculoascomycetes? Mycol Res. 2007;111:1068–1078. doi: 10.1016/j.mycres.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Luttrell ES. Taxonomy of pyrenomycetes. Univ Mo Stud. 1951;24:1–120. [Google Scholar]
- Lutzoni F, Kauff F, Cox CJ, McLaughlin D, Celio G, Dentinger B, Padamsee M, Hibbett D, James TY, Baloch E, Grube M, Reeb V, Hofstetter V, Schoch C, Arnold AE, Miadlikowska J, Spatafora J, Johnson D, Hambleton S, Crockett M, Shoemaker R, Sung GH, Lucking R, Lumbsch T, O'Donnell K, Binder M, Diederich P, Ertz D, Gueidan C, Hansen K, Harris RC, Hosaka K, Lim YW, Matheny B, Nishida H, Pfister D, Rogers J, Rossman A, Schmitt I, Sipman H, Stone J, Sugiyama J, Yahr R, Vilgalys R. Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. Am J Bot. 2004;91:1446–1480. doi: 10.3732/ajb.91.10.1446. [DOI] [PubMed] [Google Scholar]
- Moretti S, Armougom F, Wallace IM, Higgins DG, Jongeneel CV, Notredame C. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 2007;35:W645–W648. doi: 10.1093/nar/gkm333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton J. Fruits of Warm Climates. Florida Flair; Miami: 1987. [Google Scholar]
- NASS-USDA (National Agricultural Statistics Service (NASS), USDA, Hawaii Field Office) NASS, USDA, and Hawaii Department of Agriculture, Agricultural Development Div.; Honolulu, HI: 2008. Hawaii Tropical Specialty Fruits. Annual Report, released August 14, 2008. http://www.nass.usda.gov/StatisticsbyState/Hawaii/Publications/FruitsandNuts/index.asp. [Google Scholar]
- Page RDM. Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Rayner RW. A Mycological Colour Chart. Commonwealth Mycological Institute and British Mycological Society; Kew: 1970. [Google Scholar]
- Rossman AY, Goenaga R, Keith L. First report of Dolabra nepheliae on rambutan and litchi in Hawaii and Puerto Rico. Plant Dis. 2007;91:1685. doi: 10.1094/PDIS-91-12-1685C. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Sung GH, Lopez-Giraldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JZ, Arzanlou M, Sybren de Hoog G, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh SO, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lucking R, Budel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, Kerry OD, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner W, Johnston PR, Stenroos S, Zuccaro A, Dyer PS, Crittenden PD, Cole MS, Hansen K, Trappe JM, Yahr R, Lutzoni F, Spatafora JW. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst Biol. 2009;58:224–239. doi: 10.1093/sysbio/syp020. [DOI] [PubMed] [Google Scholar]
- Spatafora JW, Sung GH, Johnson D, Hesse C, O'Rourke B, Serdani M, Spotts R, Lutzoni F, Hofstetter V, Miadlikowska J, Reeb V, Gueidan C, Fraker E, Lumbsch T, Lücking R, Schmitt I, Hosaka K, Aptroot A, Roux C, Miller AN, Geiser DM, Hafellner J, Hestmark G, Arnold AE, Büdel B, Rauhut A, Hewitt D, Untereiner WA, Cole MS, Scheidegger C, Schultz M, Sipman H, Schoch CL. A five gene phylogeny of Pezizomycotina. Mycologia. 2006;98:1018–1028. doi: 10.3852/mycologia.98.6.1018. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*v4.0b10 Phylogenetic analysis using parsimony (*and other methods) Sinauer Associates; Sunderland: 2002. [Google Scholar]
- Tindall HD. Rambutan cultivation FAO Plant Production and Protection. Paper 121. Food and Agriculture Organization of the United Nations; Rome: 1994. [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee SH, Taylor J. Amplification and direct sequencing of fungal ribosomal DNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. Academic Press; New York: 1990. pp. 315–322. [Google Scholar]
- Wiens JJ. Missing data and the design of phylogenetic analyses. J Biomed Inform. 2006;39:34–42. doi: 10.1016/j.jbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Winka K, Eriksson OE, Bang A. Molecular evidence for recognizing the Chaetothyriales. Mycologia. 1998;90:822–830. [Google Scholar]
- Zalasky H, Nawawi A, Ting WP, Tai LH. Dolabra nepheliae and its imperfect state associated with canker of Nephelium lappaceum and N. mutabile. Can J Bot. 1971;49:559–561. [Google Scholar]
- Zoller S, Scheidegger C, Sperisen C. PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming Ascomycetes. Lichenologist. 1999;31:511–516. [Google Scholar]