Abstract
The heat shock (HS) response is the major cellular defense mechanism against acute exposure to environmental stresses. The hallmark of the HS response, which is conserved in all eukaryotes, is the rapid and massive induction of expression of a set of cytoprotective genes. Most of the induction occurs at the level of transcription. The master regulator, heat shock transcription factor (HSF, or HSF1 in vertebrates), is responsible for the induction of HS gene transcription in response to elevated temperature. Under normal conditions HSF is present in the cell as an inactive monomer. During HS, HSF trimerizes and binds to a consensus sequence in the promoter of HS genes, stimulating their transcription by up to 200-fold. We have shown that a large, non-coding RNA, HSR1, and the translation elongation factor eEF1A form a complex with HSF during HS and are required for its activation.
Keywords: heat shock, HSF, HSR1, eEF-1A, activation
1. Introduction
The HS response is an evolutionarily ancient, conserved reaction of cells to unfavorable environmental conditions. During HS major changes in the pattern of gene expression occur, including the rapid induction of a specific set of genes. Most of these genes encode heat shock proteins (HSPs), a large family of molecular chaperones, which includes several functional and molecular weight sub-families. HSPs are essential for cell survival under normal conditions and are critical for cell survival during stress. Elevated expression of HSPs is a major cause of cancer cell resistance to therapies (1, 2). In some instances, HSPs were found to be critical for malignant transformation due to their ability to stabilize mutant, metastable forms of oncoproteins, such as ras or p53 (3). Expression of HSPs is regulated predominantly at the level of transcription by heat shock transcription factor (HSF) (4, 5). HSF is ubiquitously expressed as an inactive monomer. The activation of HSF during HS requires its trimerization, nuclear translocation, binding to a cognate DNA sequence in the promoter of HS genes, phosphorylation, and, finally, activation of transcription by rescue of transcriptionally engaged, arrested RNA polymerase II molecules in the 5' coding region of most of HSP genes (6, 7). We have shown that the first steps in the activation of HSF in response to HS require two positive regulators: a large, non-coding RNA, HSR1, which provides thermosensing capabilities, and translation elongation factor eEF1A, which promotes trimerization of HSF (8).
HSR1 is isolated by the affinity fractionation of a lysate of heat shocked cells on a column of HSF covalently attached to Sepharose . The functional characterization of HSR1 is performed in a reconstituted, in vitro system using recombinant HSF1 and eEF1A isolated from HeLa cells. Since HSF spontaneously trimerizes at high concentrations, it is expressed and purified by procedures that keep its concentration low. Activation of HSF is determined by electrphoretic mobility shift assays (EMSA).
2. Materials
2.1. Cell Culture and Lysis
Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen), supplemented with 10% fetal bovine serum (Gemini Bio-Tech), 2 mM glutamine, and antibiotic/antimycotic cocktail (Invitrogen).
Solution of trypsin (0.25%) and ethylenediamine tetraacetic acid (EDTA, 1 mM, Cellgro).
Water bath large enough to accommodate 8–9 T-75 tissue culture flasks, set to 43°C.
Teflon cell scrapers (Fisher Scientific).
HEDGM buffer: 20 mM HEPES-NaOH, pH 7.9, 0.5 mM EDTA, 0.5 mM DTT, 1.5 mM MgCl2, 10% (v/v) glycerol, 0.0025% NP-40 (Igepal CA-630, Sigma).
Roche Complete Mini Protease Inhibitors cocktail (Roche).
Dry ice/ethanol or liquid nitrogen bath for flash-freezing.
Refrigerated centrifuge accepting Eppendorf tubes and capable of generating forces of at least 20,000×G.
2.2. SDS-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Protein-denaturing SDS polyacrylamide gel electrphoresis (SDS-PAGE) is performed according to Laemmli, with minor modifications (9).
Fourty percent acrylamide/N,N'-methylene-bis-acrylamide (37.5:1) solution, filtered through a 20 micron filter. Store at 4°C.
N,N,N',N'-Tetramethyl-ethylenediamine (TEMED) from Sigma.
Ammonium persulfate: prepare a 20% solution in water fresh daily (see Note 1).
Separating gel buffer (4×): 1.5 M tris-HCl, pH 8.8. Store at room temperature.
Concentrating gel buffer (4×): 0.5 M tris-HCl, pH 6.8. Store at room temperature.
Ten percent (w/v) solution of sodium dodecylsulfate (SDS).
Water-saturated isobutanol. Shake equal volumes of isobutanol and water in a glass bottle and allow phases to separate. Use the top layer. Store at room temperature.
Running buffer (10×): 250 mM tris, 1.9 M glycine, pH 8.3, 0.5% (w/v) SDS. Store at room temperature.
Prestained molecular weight markers (Fermentas).
Coomassie R-250 staining solution: 50% methanol, 10% acetic acid, 0.2% Coomassie R-250 (see Note 2).
Destaining solution: 10% methanol, 10% acetic acid.
2.3. Electromobility Shift Assay (EMSA)
Forty percent (w/v) acrylamide/N,N'-methylene-bis-acrylamide (19:1) solution, filtered through a 20 micron filter. Store at 4°C.
TG buffer (10×): 250 mM tris, 1.9 M glycine, 0.5 mM EDTA, pH 8.3. Store at room temperature.
Poly(dI-dC) solution (Sigma): dissolve the content of the vial in TE buffer to a concentration of 1.25 mg/ml. Heat at 42°C for 15 min, aliquot and store at −20°C.
EMSA binding buffer (10×): 200 mM HEPES-NaOH, pH 7.9, 1 M NaCl, 40 mM MgCl2, 2 mM DTT. Store in aliquots at −20°C.
Solutions of 50% (v/v) glycerol and 10 mg/ml BSA (fraction V, Sigma
HSE oligonucleotides (IDT DNA Technologies, Iowa).
T4 polynucleotide kinase (PNK, New England Biolabs).
gamma-32P-ATP (specific activity > 6,000 Ci/mmole, MP Biomedicals, formerly ICN).
Micro Bio-Spin disposable columns (Bio-Rad).
PCR cycler (e.g., Eppendorf MasterCycler).
Loading dye solution: 0.02% bromophenol blue in 25% glycerol.
2.4. RNA Denaturing Polyacrylamide Gel Electrophoresis
TBE buffer (10×): 0.89 M tris-boric acid, 5 mM EDTA.
Four percent acrylamid/N,N'-bis-acrylamide (19:1) in 1× TBE/8 M urea. Store at 4°C, heat at 37°C to dissolve the urea (see Note 3).
RNA loading buffer: 80% deionized formamide, 0.02% bromophenol blue, 0.02% xylene cyanol, 0.1% SDS in 1× TBE.
-
RNA silver stain.
Fixation solution: 7.5% acetic acid.
Silver impregnation solution: 0.15% (w/v) AgNO3, 0.056% (v/v) formaldehyde.
Developer solution: 0.056% formaldehyde, 400 mg/L sodium thiosulfate in 3% Na2CO3.
2.5. Purification of recombinant HSF
LB medium, bacterial shaking incubator, Beckman Coulter Avanti J-25 centrifuge with JA-10 and JA-17 rotors or equivalent, sonicator (Branson 250 with microtip), 2 l Erlenmeyer flasks.
Lysis buffer: 20 mM HEPES-NaOH, pH 7.9, 0.42 M NaCl, 0.5 mM EDTA, 0.5 mM DTT (add just before use from 1 M stock), 10% (v/v) glycerol.
ATP washing buffer: Lysis buffer containing 5 mM ATP and 25 mM MgCl2.
Thrombin cleavage buffer: Lysis buffer containing 0.0025% NP-40.
Glutathione-Sepharose 4B and thrombin (GE Healthcare).
Roche Complete Mini protease inhibitors cocktail (Roche).
0.1 M solution of PMSF in ethanol or isopropanol.
2.6. Purification of eEF1A from S-100 lysate
Buffer A: 25 mM tris-HCl, pH 7.5, 1 mM DTT, 0.1 mM EDTA, 25% (v/v) glycerol.
Buffer B: 25 mM tris-HCl, pH 7.9, 1 mM DTT, 0.1 mM EDTA, 10% (v/v) glycerol.
Q-Sepharose column (ca. 40 ml bed volume).
Phosphocellulose P11 column (ca. 15 ml bed volume, see Note 4).
FPLC system (preferably) or peristaltic pump, UV-detector connected to a recorder, and fraction collector.
2.7. Preparation of HSF-Sepharose column
Cross-linked Sepharose CL-4B (Sigma).
PD-10 disposable desalting columns (GE Healthcare).
Sodium m-periodate (Sigma).
Solution of 0.2 M sodium borate, pH 9.0.
Solution of 0.1 M sodium borate, pH 8.0.
Sodium borohydride (Sigma).
Disposable empty plastic columns (ca.12 ml total capacity, Bio-Rad).
Solution of 0.1 M ethanolamine-HCl, pH 8.0, 0.5 M NaCl.
3. Methods
Purified HSF trimerizes spontaneously at high concentration (10)(11). Therefore the conditions for HSF expression and purifications are selected to minimze the spontaneous activation of HSF by keeping its concentration relatively low. Just before coupling to periodate-activated Sepharose the HSF is exchanged into high-pH borate buffer, which also helps to prevent spontaneous trimerization by keeping HSF molecules charged. The monomeric state of HSF to be bound to the activated Sepharose is essential for the successful capture of the eEF1A-HSR1 complex from the HS cell lysate.
Once HSR has been isolated and cloned (by a combination of 3' and 5' RACE or by another strategy of choice), it can be synthesized in large amounts by in vitro transcription by phage RNA polymerase. Of all the commercially available RNA polymerases, T7 RNA polymerase provides the greatest yield. The yield tends to be higher when transcribing from a linearized plasmid template, but care should be taken to avoid the introduction of as few extra nucleotides into the resulting transcript as possible.
The active trimeric form of HSF binds to the consensus heat shock element (HSE) sequence with high affinity. HSE is defined as an array of repeats of the sequence 5'-nGAAn-3' in head-to-tail orientation. The minimum of three HSE units is required for the high affinity binding of HSF (12–14). We routinely use four-element arrays of the consensus HSE element as a probe in gel shift experiments involving HSF activation. The in vitro activation is performed by incubating the RNA at the desired temperature, followed by the addition of the proteins (HSF and eEF1A), the probe, and the degree of HSF activation is assayed by EMSA. The activation is conveniently performed in a PCR cycler, which allows for the fast and precise control of the incubation temperature. To screen antisense oligonucleotides, the oligonucleotide to be tested is pre-mixed with HSR1 at 5–10 fold molar excess.
3.1. Purification of recombinant HSF
Inoculate a single colony of TOP-10 E. coli cells transfected with GST-HSF1 plasmid (from LB/Amp plate) into 3–5 ml of LB/Amp and grow overnight in the shaker at 37°C.
Dilute the overnight starter culture 1:100–1:200 into LB/Amp and grow at 37°C until the O.D.600 reaches 0.6.
Remove the culture from the shaker and chill on ice or in the cold room for 10–15 min. In the meantime bring the temperature of the shaker to 25–28°C.
Add IPTG to a final concentration of 0.1 mM and return the culture to the 25–28°C shaker to allow GST-HSF1 synthesis to proceed for 4–6 hours.
Collect the cells by centrifugation at 5,000×G for 15 min and discard the supernatant. At this point the cells may be frozen at −80°C for later use.
Resuspend the bacteria in 20 ml of the lysis buffer per 1 liter of original culture. Add 1 tablet of Roche Complete Mini Protease Inhibitor Cocktail per 1 liter of original culture.
Sonicate for 25 min with output control set to 6 and duty cycle of 60% with constant chilling in the ice-water bath (see Note 5). The lysate should clear somewhat and become significantly darker in color.
-
Transfer the lysate to the centrifuge tube (which fits Beckman JA-17 rotor) and centrifuge for 30 min at 35,000×G at 4°C. During the centrifugation prepare GSH-Sepharose as follows:
Resuspend the GSH-Sepharose by vigorous shaking of the bottle and transfer 1.33 ml of the suspension for every liter of original bacterial culture into a 15 ml screw-cap tube (Falcon).
Wash twice with water and twice with the lysis buffer by resuspension/centrifugation at 500×G.
Add 1 ml of the lysis buffer for every 1.33 ml of the initial GSH-Sepharose slurry.
Carefully remove the supernatant and place it in the new 50 ml screw-cap tube (Falcon). Add washed GSH-Sepharose and incubate on the rotator for 30 min in the cold room.
Collect the beads by centrifugation at 500×G, carefully remove and discard the supernatant, add 10 ml of the lysis buffer, mix and transfer the suspension to the new 15 ml screw-cap tube (Falcon).
Wash the beads successively 3 times with 10 vol of the lysis buffer by resuspension/centrifugation at 500×G.
Resuspend the beads in 10 vol of ATP washing buffer and incubate 15 min at room temperature. Repeat the wash two more times.
Wash the beads with 10 vol of the thrombin cleavage buffer. Resuspend in 1 vol of the same buffer.
Add 50 units thrombin (50 ml of the 1 u/ml stock) per each 1 ml of settled bead volume and incubate on the rotator overnight at room temperature.
Check the cleavage efficiency by measuring the protein concentration in the supernatant and/or running both supernatant and beads suspension on SDS-PAGE.
Stop the cleavage by adding 1/100 vol of PMSF. Centrifuge the beads, remove and save the supernatant and wash the beads twice with 1–2 vol of the thrombin cleavage buffer (these washes should contain significant amount of the cleaved HSF1 as well).
3.2. Preparation of the HSF-Sepharose column
This procedure covalently couples HSF to the activated Sepharose via reaction of the epsilon-amino groups of lysine residues in the protein with reactive aldehyde groups of the periodate-treated Sepharose. The mild oxidation of Sepharose by periodate results in conversion of 1,2-cis-diol groups to the corresponding dialdehydes. The aldehydes then reacts with the amino groups to yield a Schiff's bases, which are stabilized by mild reduction with sodium borohydrate.
-
Prepare periodate-activated Sepharose CL-4B.
dispense ca.1 ml resin (ca.1.33 ml 75% slurry in ethanol) into a 15 ml screw-cap tube (Falcon).
wash twice with 10 ml of water by centrifugation at 500×G followed by resuspension.
incubate for 2 h with 5 ml of 20 mg/ml sodium periodate at room temperature in the dark.
wash twice with 10 ml of 0.2 M sodium borate, pH 9.0 by centrifugation followed by resuspension.
store at +4°C for up to 1 month.
equilibrate a PD10 desalting column with 0.2 M sodium borate pH 9.0 (ca.30 ml).
apply less than 2 ml of the HSF1 prepared according to 3.1.
wash the column repeatedly (10 times) with 1 ml of 0.2 M sodium borate, pH 9.0 collecting 1 ml fractions into Eppendorf tubes.
assay the protein content of the fractions by dropping 5 μl from each fraction on a piece of Parafilm and mixing it with 20 ml Bradford reagent. Intense blue color indicates the presence of the protein.
combine the fractions containing protein (usually fractions 3–5), save an ca. 50 μl aliquot and add the pooled fractions to periodate-activated Sepharose.
incubate overnight on a rotator at room temperature.
transfer the slurry into a disposable plastic column and wash with 10 vol of 0.2 M sodium borate, pH 9.0, followed by 10 vol of 0.1 M sodium borate, pH 8.0.
dissolve pre-weighed sodium borohydride to yield a final concentration of 3 mg/ml in 0.1 M sodium borate, pH 8.0 and immediately add to the column. Plug the outlet of the column, resuspend the resin with a 1 ml pipetor and incubate it for 5 min at room temperature.
open the column outlet and wash the column with 0.1 M sodium borate until the pH of the eluate reaches 8 as measured by pH paper (ca. 10 vol). If the flow stops due to the bubbles of hydrogen, resuspend the resin by pipetting it up and down. This should resume the flow by removing most bubbles from the solution.
resuspend HSF1 Sepharose in ca. 5 vol of 0.1 M ethanolamine, pH 8.0/0.5 M NaCl and store at +4°C.
3.3. Purification of eEF-1A from S-100 lysate
The S-100 lysate, which is the starting material for this procedure, can be obtained commercially or prepared according to the published method (15). This protocol is an adaptation of the original procedure described by Merrick (16). With minimal modifications the procedure can be easily adapted to use tissue, for example rat liver, as the source of the protein. This protocol assumes that 50 ml of the S-100 lysate is used; the column and gradient volumes can be scaled up or down if greater or lesser amounts are used.
Apply 50 ml of the S-100 lysate to a 40 ml DEAE-Sepharose column equilibrated with buffer A containing 50 mM KCl. Collect the flow-through, which contains eEF-1A.
Apply the flow-through to a 15 ml phosphocellulose P11 column equilibrated with buffer B containing 50 mM KCl and wash the column with at least 3 volumes of the same buffer.
Apply a linear gradient of 50–650 mM KCl (10 column volumes) in buffer B and collect 3 ml fractions. Assay a 20 ml aliquot from each fraction by electrophoresis in a 10% SDS-PAGE and stain the gel with Coomassie. eEF-1A should be visible as a prominent band at ca. 50 kDa M.W..
Pool the fractions containing eEF-1A, concentrate them using a Centricon or similar ultrafiltration device with a 10 kDa MWCO membrane to ca. 2 ml and exchange into the storage buffer. The protein is stable at 4°C for at least a month or indefinitely at −80°C.
3.4. Isolation of HSR from heat shocked cells
This procedure has been used extensively with cells grown in monolayer, such as HeLa, BHK, or 3T3. However, it should be easily adaptable to the cells grown in suspension, if the ratio of cells to HSF-Sepharose is kept constant. Monolayer cells are grown in T-75 phenolic cap flasks (Corning) to ca. 75–90% confluency.
Tighten the caps of flasks to be heat shocked and submerge them to a water bath set to 43°C. Sealing the caps with Parafilm is not required when using phenolic that have been tightened properly. Allow heat shock to proceed for 45 min to 1 h.
Remove flasks from the water bath and pat them dry with paper towels to prevent water from entering the flasks. Aspirate the medium using a Pasteur pipet connected to a vacuum source. Rinse the cells once with ice-cold PBS (3 ml per T-75 flask) taking care not to dislodge any cells and use a Teflon cell scraper (rubber policeman, Corning) to scrape the cells into 3 ml of fresh ice-cold PBS.
Collect the cells by centrifugation for 5 min at 500×G. Remove the PBS by aspiration.
Resuspend the cell pellet in 100 ml per T-75 flask of Lysis Buffer. Incubate on ice for 10 min.
Lyse the cells by three cycles of flash freezing in ethanol/dry ice or liquid nitrogen / thawing at room temperature. Thawing can be performed at 37°C if the incubation time does not exceed the minimum required to thaw the cells completely.
Centrifuge the lysate for 15 min at 25,000×G at 4°C. Discard the pellet. In the meantime prepare the HSF-Sepharose beads (15 ml per T-75 flask) by washing them once in lysis buffer containing 0.21 M NaCl.
Dilute the cell lysate two-fold with HEDGM buffer (to give the final salt concentration of 0.21 M) and add it to the washed HSF-Sepharose. Incubate for a minimum of 4 h (or overnight) on a rotator at 4°C.
Wash the beads 3 × 15 min with 10 bed volumes of HEDGM buffer containing 0.21 M NaCl.
Resuspend the beads in an equal volume of HEDGM/0.21 M buffer and incubate 45 min at 43°C with occasional mixing. Centrifuge 1 min at maximum speed in a tabletop centrifuge, remove and save the supernatant and add one bed volume of HEDGM/0.21 M buffer pre-heated to 43°C. Repeat two more times.
Combine the resulting fractions saving a 15 μl aliquot for SDS-PAGE. Add to final concentrations: SDS to 0.25%, EDTA to 12.5 mM, and proteinase K to 0.2 mg/ml. Incubate at 50°C for 30 min.
Extract twice with phenol/chloroform/isoamyl alcohol, precipitate with ethanol or isopropanol in the presence of 0.5 mg/ml glycogen. Centrigfuge and dissolve the RNA pellet in 20 ml TE buffer.
-
Analyze the RNA by electrophoresis in denaturing 4% PAAG in the presence of 8 M urea. Mix 2 μl of the RNA with 3 μl of RNA loading buffer, heat at 80°C for 5 min, load on the gel and electrophorese until the xylene cyanol (the second dye) has migrated to the end of the gel (see Note 6). Silver stain the gel as follows:
Soak the gel for 30 min in 7.5% acetic acid.
Rinse 3×5 min in water.
Soak the gel for 30 min in the silver impregnation solution.
Rinse briefly with copious amount of water.
Soak the gel in the development solution and watch for the bands to appear (usually, within 5–10 min).
Stop the development by pouring off the developer and soaking the gel in 7.5% acetic acid.
3.5. Synthesis of HSR1 by In Vitro Transcription
HSR1 is cloned in the pBluescript SK- vector (pBSKM-HSR1) in an orientation such that T7 RNA polymerase generates the sense transcript after digestion of the plasmid with SmaI and T3 RNA polymerase generates the antisense transcript after digestion of the plasmid with EcoRV. It is essential to cleave the plasmid with restriction enzymes that produce blunt ends, if possible, as this will minimize non-specific transcription initiation on protruding single stranded ends. Alternatively, a PCR product containing the T7 promoter sequence in one of the primers can be used as template to minimize the introduction of extra nucleotides in the RNA sequence.
-digest 20 mg pBSKM-HSR1 with the appropriate enzyme in a 50 μl reaction for 2–4 h.
-purify the digested plasmid or PCR fragment by agarose gel electrophoresis followed by phenol/chloroform extraction and ethanol precipitation.
-resuspend the digested plasmid or PCR fragment in ca. 20 μl TE, pH 8.0.
- -assemble the transcription reaction as follows (following the order of reagent addition):
water: to 200 μl 5× transcription buffer: 40 μl 1× 100 mM DTT: 10 μl 5 mM 25 mM rNTP (ROCHE): 30 μl 3.75 mM template: 50 mg/ml Pyrophosphatase: 1 U/ml T7 RNA polymerase: 1000–2000 U/ml -incubate 4–6 h at 37°C. After 2 h incubation another aliquot of RNA polymerase may be added.
-add RNase-free DNase I (40–50 Units) and incubate for 30 min at 37°C.
-precipitate the RNA by your method of choice (e.g. phenol/chloroform extraction followed by ethanol or isopropanol, or LiCl, etc).
3.6. In vitro HSF Activation Assay
The typical range of concentrations in which HSF is activated in this system is between 1 and 5 nM final. Higher concentrations, especially in excess of 100 nM, tend to cause spontaneous trimerization and activation of HSF. Because of the fluctuations in the quality of the recombinant HSF preparations, it may be necessary to titrate each new preparation of HSF to determine the concentration at which it is not activated spontaneously. The final concentration of eEF-1A is 100 nM. The concentration of proteins is determined by UV absorption at 280 nm in 6 M guanidin-HCl/20 mM phosphate buffer, pH 6.5 (e(HSF)=28670 l*M−2 and e(eEF-1A)=45755 l*M−2 in a 1 cm cuvette). The final concentration of HSR1 is 0.1 nM. A low concentration of HSR1 is crucial because at higher concentrations the equilibrium concentration of the HSF-activating conformation of HSR1 is high enough to cause activation to occur independent of incubation temperature. However, for the antisense oligo screen, this “constitutive” activation is beneficial as it considerably simplifies the experimental setup.
-
Prepare the labeled HSE oligo as follow:
-
In two separate tubes mix the following components:
27 ml water
5 ml sense/antisense HSE oligo
4 ml 10× PNK buffer
3 ml gamma-32P-ATP
1 ml PNK
Incubate at 37°C for 30 min.
Mix the content of the two tubes, incubate at 68°C for 10 min and let cool slowly to room temperature.
Purify the labeled HSE oligo by centrifugation through a Micro Bio-Spin 6 spin column. Store at −20°C for up to two weeks.
-
Pre-mix EMSA 10× mix, poly(dI-dC), DTT, and water according to the desired number of samples. Distribute into PCR tubes (0.2 ml) and equilibrate at 23°C in a PCR cycler.
Add RNA to the tubes and incubate at 23°C for 5 min (If performing an antisense oligo screen, add the oligo at 10 times molar excess over HSR1).
Incubate at the HS temperature (43°C, or at 23°C if performing an antisense oligo screen) for 15 min.
Bring the temperature back to 23°C and add eEF-1A and HSF1 as quickly as possible and mix well by vigorous pipetting.
Incubate at 23°C for 30 min.
Add 3 μl of 32P-HSE/loading dye mix and incubate for 20 min at 23°C
Load reactions on a 0.75 mm thick 4% PAAG/1× TG gel and electrophorese in 1× TG at no more than 20 mA until the dye migrates to bottom of the gel (ca. 3 hr for a 20×20 cm gel).
Dry the gel and expose to x-ray film or phosphoimager screen.
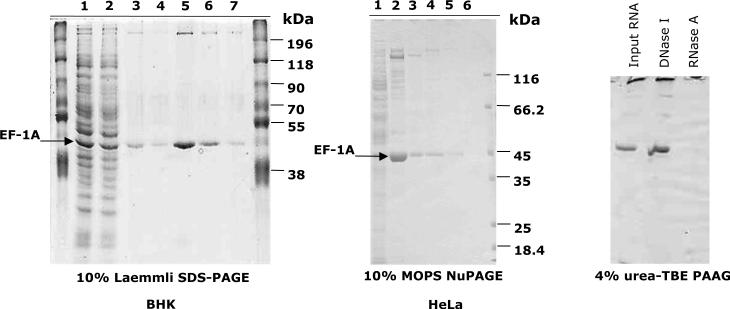
Figure 1.
Isolation of HSF1 activating fraction and HSR1 from a lysate of heat shocked HeLa (middle panel) and BHK (left panel) cells. Lane 1 (BHK and HeLa): lysate of heat shocked cells; lane 2 (BHK): supernatant after incubation of the lysate with HSF1-Sepharose beads; lane 3 (BHK) and lane 2 (HeLa): HSF1-Sepharose beads after incubation with a lysate of heat shocked cells and washes; lane 4 (BHK) and lane 3 (HeLa): HSF1-Sepharose beads after three successive elutions at 43°C; lanes 5–7 (BHK) and 4–6 (HeLa): three successive elutions with buffer at 43°C. Right panel: silver stained PAAG of RNA isolated from fractions 5–7 shown in left panel (BHK cells). Where indicated, RNA was treated with DNase I or RNase A prior to loading the gel.
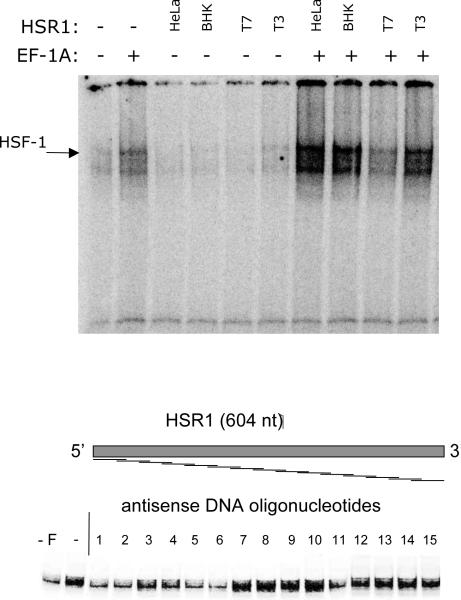
Figure 2.
In vitro reconstituted system for the activation of HSF1. Upper panel: recombinant mouse HSF1 (10 nM final) and eEF-1A (100 nM final, omitted as shown) were incubated in the absence of RNA, in the presence of HSR1 isolated from either BHK or HeLa cells, as indicated, in the presence of (25 nM final) in vitro transcribed sense (T3) or antisense (T7) mammalian HSR. Reactions were then incubated with the radiolabeled HSE oligo and separated on a native PAAG, as described in the text. The gel was dried and radioautographed. Lower panel: HSR1 antisense oligos screening. EMSA reactions were assembled as described above with the exception of the inclusion of antisense oligos at a 10-fold molar excess over HSR1.
Acknowledgement
This work was supported by the NIH grant R01 GM069800 (E.N.)
Footnotes
Ammonium persulfate solution is stable for up to two weeks if stored at −20°C and kept on ice when thawed.
Prepare Coomassie staining solution by first dissolving the dye powder in methanol (stir on a magnetic stirrer for at least 1 h), then add acetic acid and water.
Avoid heating urea-containing solutions to high temperature, as urea will decompose.
Phosphocellulose P11 column is prepared as described at: http://mitchison.med.harvard.edu/protocols/tubprep.html
The efficient cooling of the sample during sonication is essential. It is best achieved if the sonication is done in a metal vial (we routinely use tube adapters from an older Beckman ultracentrifuge) to ensure rapid and efficient dissipation of heat.
The volumes and incubation times given are for a 4% gel in a Bio-Rad mini electrophoresis apparatus. They should be adjusted accordingly if using larger gels and/or higher concentration of acrylamide. Larger 4% gels may be difficult to handle.
References
- 1.Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10:86–103. doi: 10.1379/CSC-99r.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31:164–172. doi: 10.1016/j.tibs.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Whitesell L, Lindquist SL. Hsp90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 4.Voellmy R. Sensing stress and responding to stress. EXS. 1996;77:121–137. doi: 10.1007/978-3-0348-9088-5_9. [DOI] [PubMed] [Google Scholar]
- 5.Morimoto RI, Kroeger PE, Cotto JJ. The transcriptional regulation of heat shock genes: a plethora of heat shock factors and regulatory conditions. EXS. 1996;77:139–163. doi: 10.1007/978-3-0348-9088-5_10. [DOI] [PubMed] [Google Scholar]
- 6.Westwood JT, Wu C. Activation of drosophila heat shock factor: conformational change associated with a monomer-to-trimer transition. Mol Cell Biol. 1993;13:3481–3486. doi: 10.1128/mcb.13.6.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. Rna-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 9.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 10.Zhong M, Orosz A, Wu C. Direct sensing of heat and oxidation by drosophila heat shock transcription factor. Mol Cell. 1998;2:101–108. doi: 10.1016/s1097-2765(00)80118-5. [DOI] [PubMed] [Google Scholar]
- 11.Goodson ML, Sarge KD. Heat-inducible dna binding of purified heat shock transcription factor 1. J Biol Chem. 1995;270:2447–2450. doi: 10.1074/jbc.270.6.2447. [DOI] [PubMed] [Google Scholar]
- 12.Orosz A, Wisniewski J, Wu C. Regulation of drosophila heat shock factor trimerization: global sequence requirements and independence of nuclear localization. Mol Cell Biol. 1996;16:7018–7030. doi: 10.1128/mcb.16.12.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 14.Abravaya K, Phillips B, Morimoto RI. Heat shock-induced interactions of heat shock transcription factor and the human hsp70 promoter examined by in vivo footprinting. Mol Cell Biol. 1991;11:586–592. doi: 10.1128/mcb.11.1.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by rna polymerase ii in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho JF, Carvalho MD, Merrick WC. Purification of various forms of elongation factor 1 from rabbit reticulocytes. Arch Biochem Biophys. 1984;234:591–602. doi: 10.1016/0003-9861(84)90309-6. [DOI] [PubMed] [Google Scholar]