Abstract
We present here the characterization and optimization of a novel imaging-based positional biosensor high-content screening (HCS) assay to identify disruptors of p53-hDM2 protein–protein interactions (PPIs). The chimeric proteins of the biosensor incorporated the N-terminal PPI domains of p53 and hDM2, protein targeting sequences (nuclear localization and nuclear export sequence), and fluorescent reporters, which when expressed in cells could be used to monitor p53-hDM2 PPIs through changes in the subcellular localization of the hDM2 component of the biosensor. Coinfection with the recombinant adenovirus biosensors was used to express the NH-terminal domains of p53 and hDM2, fused to green fluorescent protein and red fluorescent protein, respectively, in U-2 OS cells. We validated the p53-hDM2 PPI biosensor (PPIB) HCS assay with Nutlin-3, a compound that occupies the hydrophobic pocket on the surface of the N-terminus of hDM2 and blocks the binding interactions with the N-terminus of p53. Nutlin-3 disrupted the p53-hDM2 PPIB in a concentration-dependent manner and provided a robust, reproducible, and stable assay signal window that was compatible with HCS. The p53-hDM2 PPIB assay was readily implemented in HCS and we identified four (4) compounds in the 1,280-compound Library of Pharmacologically Active Compounds that activated the p53 signaling pathway and elicited biosensor signals that were clearly distinct from the responses of inactive compounds. Anthracycline (topoisomerase II inhibitors such as mitoxantrone and ellipticine) and camptothecin (topoisomerase I inhibitor) derivatives including topotecan induce DNA double strand breaks, which activate the p53 pathway through the ataxia telangiectasia mutated-checkpoint kinase 2 (ATM-CHK2) DNA damage response pathway. Although mitoxantrone, ellipticine, camptothecin, and topotecan all exhibited concentration-dependent disruption of the p53-hDM2 PPIB, they were much less potent than Nutlin-3. Further, their corresponding cellular images and quantitative HCS data did not completely match the Nutlin-3 phenotypic profile.
Introduction
Protein–protein interactions (PPIs) are essential for all cellular functions including the assembly and maintenance of morphological structures, DNA replication, mRNA transcription, protein translation, protein folding, and the regulation of cellular metabolism and signaling pathways.1,2 Protein interactions between identical (homotypic) and nonidentical (heterotypic) polypeptides may be characterized by kinetic and thermodynamic parameters and span a continuum from obligate high-affinity stable contacts to nonobligate low-affinity transient interactions.1,2 Therefore, PPIs represent a large number of potential therapeutic targets distinct from the ligand-binding or active sites that are most commonly exploited for drug discovery.1–3 For many years, however, the evident lack of success in finding PPI inhibitors contributed to the widely held perception that protein–protein interaction surfaces were large, flat, and essentially not druggable.1–3 However, recent advances in PPI high throughput screening (HTS)/high-content screening (HCS) assay technologies combined with structure-based drug design approaches are accelerating the discovery of small-molecule PPI disruptors.1–3 The primary amino acid sequence of proteins together with the secondary and tertiary structures of the interacting polypeptides determines the 3D shape and chemical environment of protein surfaces at the point(s) of contact.1,2 The structural elucidation of a number of protein–protein complexes has revealed that protein-binding interfaces can be dissected into discrete patches defined by geometric clusters of atoms, and that some of these residues, referred to as “hot spots,” contribute more to the binding interactions than others.1,2 The ability to screen large chemical libraries against HTS assay formats that have been guided by structural information on the critical domains and residues involved in protein-binding interfaces bodes well for improving the success rate for finding small-molecule PPI disruptors.1–3
The pursuit of small-molecule PPI inhibitors/disruptors has prompted the development and implementation of a large number of biochemical and cell-based assay formats compatible with HTS and/or HCS.1–4 Low-throughput biochemical PPI assay formats include methods such as coprecipitation, copurification, affinity chromatography, ultracentrifugation, nuclear magnetic resonance, surface plasmon resonance (SPR), mass spectrometry, and isothermal titration calorimetry.1,2,4 Higher-throughput biochemical PPI assay formats that have been adapted to screening include capture ELISAs, cell surface binding, fluorescence polarization (FP), time-resolved fluorescence, ligand-induced changes in thermal stability (Thermofluor®), bead-based technologies (SPA, AlphaScreen, and Luminex), and resonance energy transfer (RET) assays (fluorescence FRET, luminescence LRET, and bioluminescence BRET).1,2,4 Cell-based PPIs include yeast genetic approaches where the interactions of the protein partners either enables or inhibits growth on selective media, or at a restrictive temperature, and a number of yeast 2-hybrid transcriptional reporter PPI assays including the forward, reverse, and repressed transactivation formats.1,2 Several variants of the 2-hybrid transcriptional reporter PPI assays have also been developed in mammalian cells.1,2,5 BRET assays have been extensively employed to dissect the homo- and heterodimerization of GPCRs and ion channel subunits.6 Several imaging-based HCS assay formats have also been employed to study PPIs such as the colocalization of fluorescently labeled protein (FP) partners, FRET measurements between PPI partners bearing donor and acceptor FPs, protein fragment complementation assays (PCAs), and positional biosensors that measure the PPI-induced redistribution of FPs.7–14 We describe here the characterization and optimization of a novel positional PPI biosensor (PPIB) HCS assay to identify disruptors of the interactions between p53 and hDM2.
The p53 tumor suppressor is a transcriptional activator that regulates the expression of target genes involved in processes that serve to limit the initiation, progression, or survival of cancer cells such as cell cycle arrest, DNA damage repair, apoptosis, senescence, metastasis, and angiogenesis.15–21 In 50% of human cancers, p53 is inactivated by single-point missense mutations in the DNA-binding domain, resulting in deficient regulation of p53 target genes.16,17,19–22 In a significant proportion of the remaining cancers where wild-type p53 is functional, however, hDM2 is overexpressed and blocks p53's tumor suppressor activity.23–28 hDM2 binds to the N-terminal transactivation domain of p53, thereby blocking the activation of p53 target genes. By virtue of its E3-ubiquitin-ligase activity, hDM2 also directs p53 for degradation by the proteasome.23–28 The structure of the protein–protein binding interfaces between p53 and hDM2 have been well mapped and characterized, and 3 amino acids in the N-terminus of p53 (Phe19, Trp23, and Leu26) were shown to bind to a small hydrophobic pocket on the surface of hDM2.25,29 The cis-imidazoline Nutlin analogs are the prototype disruptors of p53-hDM2 PPIs that were identified using a combination of structure-based drug design and an SPR assay that measured the disruption of the p53-hDM2 binding.30,31 Nutlin derivatives occupy the hydrophobic pocket on the surface of hDM2 by disrupting the p53-hDM2 PPI and lead to in vivo stabilization of p53 and suppression of tumor growth in xenograft models.30,31 Several strategies to reactivate p53 tumor suppressor activity are being pursued as potential cancer therapies, such as compounds that disrupt p53-hDM2 PPIs, inhibit hDM2 ubiquitin E3 ligase activity, or restore the thermal stability and DNA-binding activity of p53 DNA-binding mutants.22,30–37
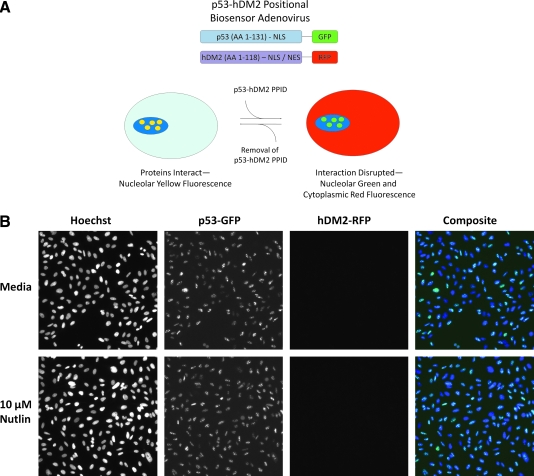
The p53-hDM2 PPIB design incorporates the N-terminal domains of the 2 interacting partners fused to monomeric fluorescent proteins (green fluorescent protein [GFP] or red fluorescent protein [RFP]) via their carboxy-terminus. The N-terminal residues 1–131 of p53 include the p53 transactivating domain that contains the binding site for hDM2, and this protein fragment is expressed as a GFP fusion protein that is targeted and anchored in the nucleolus of infected cells by the inclusion of a nuclear localization sequence (NLS) (Fig. 1A). The N-terminal residues 1–118 of hDM2 encode the domain for binding to the N-terminal transactivating domain of p53, and this fragment is expressed as a RFP fusion protein that includes both an NLS and an NES (Fig. 1A). In U-2 OS cells that are coinfected with both adenovirus constructs, the binding interactions between the hDM2 and p53 components of the biosensor resulted in both proteins becoming localized to the nucleolus (Fig. 1A). Upon disruption of the p53-hDM2 protein–protein interaction with a compound such as Nutlin-3, the p53-GFP interaction partner remained nucleolar, while the shuttling hDM2-RFP interaction partner redistributed into the cytoplasm (Fig. 1A). The disruption of the p53-HDM2 interaction biosensor was measured by acquiring images on an automated HCS imaging platform and using an image analysis algorithm to quantify the relative distribution of the hDM2-RFP shuttling component of the biosensor between the cytoplasm and nuclear regions. We describe here the characterization of the p53-hDM2 PPIB adenovirus components, the development and optimization of an imaging-based p53-hDM2 PPIB HCS assay on the ArrayScan VTI platform, the performance of the assay when we screened the Library of Pharmacologically Active Compounds (LOPAC), and the characterization of active compounds identified in the screen.
Fig. 1.
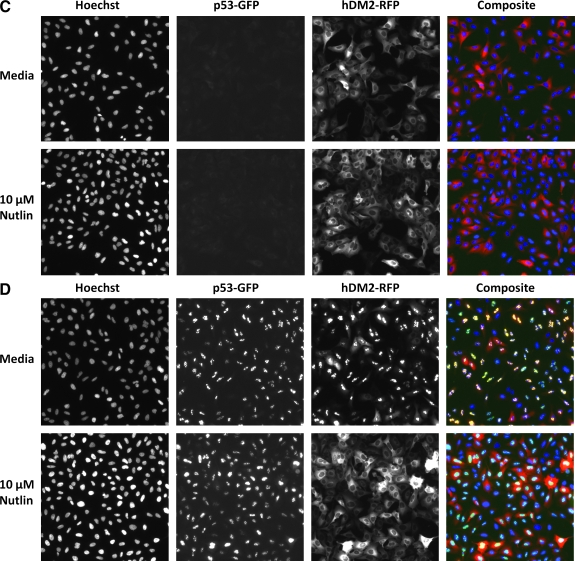
p53-hDM2 protein–protein interaction biosensor components, principle, and characterization. (A) Cartoon schematic of the protein–protein interaction partner components and the 2 interaction states of the p53-hDM2 positional biosensor. The p53-hDM2 protein–protein interaction biosensor design incorporates 2 recombinant adenovirus constructs driving expression of the N-terminal domains of the p53 and hDM2 interacting partners fused to fluorescent proteins (green fluorescent protein [GFP] or red fluorescent protein [RFP]) at their carboxy-terminus. The N-terminal residues 1–131 of p53 encompass the p53 transactivating domain that contains the binding site for hDM2 and is expressed as a GFP fusion protein that is targeted and anchored in the nucleolus of infected cells by the inclusion of a nuclear localization sequence (NLS). The N-terminal residues 1–118 of hDM2 encompass the binding site for the N-terminal transactivating domain of p53 and is expressed as an RFP fusion protein that includes both an NLS and a nuclear export sequence (NES). In U-2 OS cells that are coinfected with both adenovirus constructs the binding interactions between the hDM2 and p53 components of the biosensor resulted in both proteins becoming localized to the nucleolus, producing a yellow signal in composite images. Upon disruption of the p53-hDM2 protein–protein interaction with a compound such as Nutlin-3, the p53-GFP interaction partner remained nucleolar, while the shuttling hDM2-RFP interaction partner redistributed into the cytoplasm, and in the composite images of these cells, the nucleolus was predominantly light green/blue and the cytoplasm was predominantly red. (B–D) Individual gray-scale and 3-color composite images of U-2 OS cells from 3 fluorescent channels (Hoechst Ch1, GFP Ch2, and RFP Ch3) were sequentially acquired on the ArrayScan VTI platform using a 20 × 0.4 NA objective with the XF93 excitation and emission filter set (Hoechst, blue; FITC, green; and TRITC, red). U-2 OS cells were infected with (B) only the p53-GFP biosensor adenovirus, (C) only the hDM2-RFP biosensor adenovirus, or (D) both the p53-GFP and hDM2-RFP biosensor adenoviruses. Adenovirus-infected cells were seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, cultured overnight at 37°C, 5% CO2, and 95% humidity, and were then treated for 90 min with 0.5% dimethyl sulfoxide (DMSO) or 10 μM Nutlin-3 in 0.5% DMSO prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images from a single representative experiment of numerous experiments are presented.
Materials and Methods
Reagents
Nutlin-3 (mixed enantiomers), formaldehyde, and Hoechst 33342 were purchased from Sigma-Aldrich. Dimethyl sulfoxide (DMSO) (99.9% high-performance liquid chromatography-grade, under argon) was obtained from Alfa Aesar.
Cell Culture
The U-2 OS osteosarcoma cell line was acquired from the American Type Culture Collection and was maintained in McCoy's 5A medium with 2 mM l-glutamine (Invitrogen) supplemented with 10% fetal bovine serum (Gemini Bio-Products), and 100 U/mL penicillin and streptomycin (Invitrogen) in a humidified incubator at 37°C, 5% CO2, and 95% humidity.
Library of Pharmacologically Active Compounds
The 1,280-compound LOPAC was purchased from Sigma-Aldrich and was supplied at 10 mM concentration in DMSO arrayed into 96-well microtiter master plates. LOPAC compounds were assigned unique University of Pittsburgh Drug Discovery Institute (UPDDI) substance identity numbers and were handled and stored as described previously.38–41 Then, 384-well master plates containing 100 μL of 10 mM compounds in DMSO were prepared from four 96-well LOPAC master plates mapped into the quadrants of a single 384-well plate using the VPrep (Velocity11) outfitted with a 96-well transfer head. Daughter plates containing 2 μL of 10 mM compounds in DMSO were prepared and replicated from the 384-well LOPAC master plates using the VPrep (Velocity11) outfitted with a 384-well transfer head. Aluminum adhesive plate seals were applied with an Abgene Seal-IT 100 plate sealer and plates were stored at −20°C in a Matrical MatriMinistore™ automated compound storage and retrieval system. At the start of screening operations, LOPAC daughter plates were withdrawn from −20°C storage, thawed to ambient temperature, centrifuged 1–2 min at 50g, and the plate seals were removed prior to the transfer of 38 μL of McCoy's 5A media into wells using the FlexDrop liquid handler (Perkin Elmer) to generate a 500 μM intermediate stock concentration of library compounds (in 5% DMSO). The diluted compounds were mixed by repeated aspiration and dispensation using a 384-well P30 dispensing head on the evolution-P3 (EP3) liquid handling platform (Perkin Elmer) and then 5 μL of diluted compounds were transferred to the wells of assay plates. In the LOPAC primary screen, compounds were individually tested at a final concentration of 50 μM (0.5% DMSO). For the determination of the 50% inhibition concentrations (IC50), 10-point 2-fold serial dilutions of test compounds in McCoy's 5A medium-DMSO were performed using a 384-well P30 dispensing head on the EP3 liquid handling platform. The diluted compounds were mixed by repeated aspiration and dispensation using a 384-well P30 dispensing head on the EP3 and then 5 μL of diluted compounds were transferred to the wells of assay plates to provide a final concentration response ranging from 0.195 to 50 μM.
p53-hDM2 Protein–Protein Interaction Biosensor HCS Assay Protocol
The components of the p53-hDM2 protein–protein interaction biosensor have been described previously.9 Recombinant adenovirus expression constructs bearing the individual p53-GFP (TagGFP; Evrogen, Inc., Moscow, Russia) and hDM2-RFP (Tag RFP; Evrogen, Inc.) protein–protein interaction partners were utilized to infect U-2 OS cells according to the manufacturer's (Cellumen, Inc.) instructions. Typically, 1 × 107 U-2 OS cells were coinfected with the p53-GFP and hDM2-RFP adenovirus expression constructs by incubating cells with the manufacturer's (Cellumen, Inc.) recommended volume of virus in 1.5 mL culture medium for 1 h at 37°C, 5% CO2 in a humidified incubator with periodic inversion (every 10 min) to maintain cell suspension. Coinfected cells were then diluted to 5.6 × 104 cells/mL and 45 μL (2,500 cells) were seeded in each well of a 384-well collagen-coated barcoded microplate (Greiner BioOne; No. 781986,) using a Zoom liquid handler (Titertek). Plates were incubated overnight at 37°C and 5% CO2 in a humidified incubator. Compounds or plate controls (5 μL) were added to appropriate wells using a VPrep (Velocity 11) or an Evolution P3 (Perkin-Elmer) for a final screening concentration of 25–50 μM. Plates were incubated at 37°C for 90 min. Samples were fixed by the addition of 50 μL of prewarmed (37°C) 7.4% formaldehyde and 2 μg/mL Hoechst 33342 in phosphate-buffered saline (PBS), using a BioTek ELx405 (BioTek) or a MapC liquid handler (Titertek), and incubated at room temperature for 30 min, as described previously.42–44 Liquid was then aspirated and plates were then washed twice with 85 μL PBS, using the BioTek or MapC liquid handler, and sealed with adhesive aluminum plate seals (Abgene) with the last 85 μL wash of PBS in place. Fluorescent images were then acquired on an ArrayScan VTI automated imaging platform (Thermofisher Scientific).
Image Acquisition and Image Analysis
Images of 3 fluorescent channels (Hoechst, GFP, and RFP) were sequentially acquired on the ArrayScan VTI platform, using either a 10× 0.3 NA or a 20× 0.4 NA objective with the XF93 excitation and emission filter set (Hoechst, FITC, and TRITC). Excitation was provided by an X-CITE® 120-watt high-pressure metal halide arc lamp with Intelli-Lamp® technology (Photonic Solutions, Inc.). Typically with the 10× 0.3 NA objective, the ArrayScan VTI platform was set up to acquire 250 selected objects (nuclei) or 2 fields of view, whichever came first. With the 20× 0.4 NA objective, the ArrayScan VTI platform was set up to acquire 4 fields of view. The nucleic acid dye Hoechst 33342 was used to stain and identify the nucleus, and this fluorescent signal was used to focus the instrument and to define a nuclear mask for the molecular translocation (MT) image analysis algorithm. Images were analyzed using the MT image analysis algorithm as described previously.42–44
HCS Data Analysis, Observation, Statistical Analysis, and Curve Fitting
Data analysis for the LOPAC HCS assay and IC50 determinations were performed using ActivityBase™ (IDBS) and CytoMiner (UPDDI). Processed data and HCS multiparameter features were observed using Spotfire™ DecisionSite® software. An ActivityBase primary HTS template was created, which automatically calculated the percent inhibition together with plate control signal-to-background (S:B) ratios and Z′-factors. For the LOPAC screen, we utilized the mean circle ring average intensity difference in channel 3 [MCRAID-Ch3] value of the DMSO minimum plate control wells (n = 32) and the mean MCRAID-Ch3 value of the 10 μM Nutlin-3 maximum plate control wells (n = 24) to normalize the MCRAID-Ch3 compound data and to represent 0% and 100% disruption/inhibition of the p53-hDM2 interactions, respectively. The UPDDI also constructed an ActivityBase concentration–response template to calculate the percent inhibition together with plate control S:B ratios and Z′-factors for quality control purposes. IC50 values were calculated using an XLfit 4-parameter logistic model, also called the sigmoidal dose–response model: y = (A + ((B − A)/(1 + ((C/x)^D)))), where y was the percent activation and x was the corresponding compound concentration. The fitted C parameter was the IC50 and defined as the concentration giving a response half way between the fitted top (B) and bottom (A) of the curve. The A and B parameters were locked as 0 and 100, respectively. For non-HTS concentration response assays we used GraphPad Prism 5 software to plot and fit data to curves using the following sigmoidal dose–response variable slope equation: Y = Bottom + [Top − Bottom]/[1 + 10^((LogEC50 − X) × Hill slope)].
Compound Structure Classification and Clustering Analysis
Disruptors of the p53-hDM2 interaction that were identified in the LOPAC screen were confirmed in 10-point concentration-response IC50 assays and their structures were analyzed using the Leadscope Enterprise 2.4.6-1 software. The confirmed hit compounds were subjected to structure-based clustering and classification techniques based on recursive partitioning as described previously.38–41
Results
The development of an HCS assay involves the optimization of sample preparation methods, image acquisition procedures, and image analysis algorithms. The U-2 OS osteosarcoma cell line was selected as the background cell line for the p53-hDM2 PPIB characterization and optimization because it is wild type for p53, has a single copy of the hDM2 gene, and expresses normal levels of hDM2 protein.45 U-2 OS cells were also selected because their morphology is compatible with HCS assays; they are adherent, well spread, have a large well-defined cytoplasm with a large regular nucleus that lends itself to imaging and image analysis (Figs. 1 and 2).
Fig. 2.
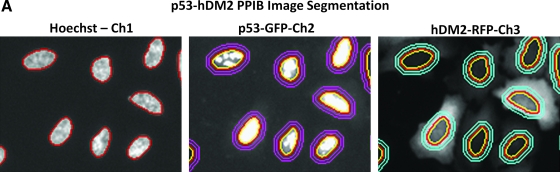
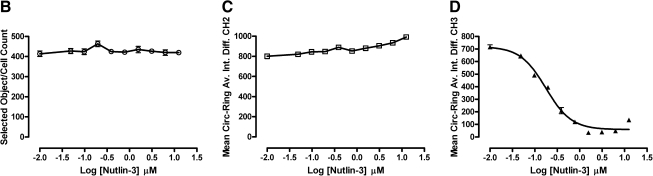
Molecular translocation image analysis algorithm. p53-hDM2 protein–protein interaction biosensor (PPIB) adenovirus-infected cells were seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, cultured overnight at 37°C, 5% CO2, and 95% humidity, and were then treated for 90 min with the indicated concentrations of Nutlin-3, mixed enantiomers, in 0.5% dimethyl sulfoxide prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images from 3 fluorescent channels were sequentially acquired on the ArrayScan VTI platform using a 20× 0.4 NA objective with the XF93 excitation and emission filter set and were analyzed with the molecular translocation (MT) image analysis algorithm. (A) Image segmentation. Hoechst-stained objects in Ch1 that exhibited the appropriate fluorescent intensities above background and size characteristics (width, length, and area) were identified and classified by the image segmentation as nuclei. The nuclear mask derived from Ch1 (red circle) was then used to segment the images from Ch2 and Ch3 into nuclear (Circ) and cytoplasmic (Ring) regions. The nuclear mask was eroded to reduce cytoplasm contamination within the nuclear area, and the reduced mask (yellow circle) was used to quantify the amount of target channel, p53-green fluorescent protein (GFP) in Ch2 and hDM2-red fluorescent protein (RFP) in Ch3, fluorescence within the nuclear region. The nuclear mask was then dilated to cover as much of the cytoplasmic region as possible without going outside the cell boundary. Removal of the original nuclear region from this dilated mask creates a ring mask that covers the cytoplasmic region outside the nuclear envelope. The number of pixels away from the nuclear mask and the number of pixels (width) between the inner and outer ring masks were selectable within the MT bioapplication software. The ring masks were then used to quantify the amount of target channel, p53-GFP (Ch2, mauve rings) or hDM2-RFP (Ch3, light blue rings), fluorescence within the cytoplasmic region. (B–D) Selected quantitative data outputs from the MT image analysis algorithm: (B) The selected object or cell counts (selected object counts per valid field of view [SCCPVF]) derived from Hoechst-stained nuclei in Ch1; (C) To quantify the relative distribution of the p53-GFP within the nucleus and the cytoplasmic regions, the MT image analysis algorithm calculates a mean average intensity difference by subtracting the average p53-GFP intensity in the Ring (cytoplasm) region from the average p53-GFP intensity in the Circ (nuclear) region of Ch2; Mean Circ–Ring Average Intensity Difference Channel 2 (MCRAID-Ch2); (D) To quantify the relative distribution of the hDM2-RFP within the nucleus and the cytoplasmic regions, the MT image analysis algorithm performs a similar calculation in Ch3 to generate a mean circle ring average intensity difference in channel 3 (MCRAID-Ch3) value. The data are presented as the mean SCCPVF (B), MCRAID-Ch2 (C), and MCRAID-Ch3 (D) values ± SD from 12 wells (n = 12). For the SCCPVF (B) and MCRAID-Ch2 (C) data, lines connecting the data were plotted using GraphPad Prism software 4.03. For the MCRAID-Ch3 (D) data, the resulting nonlinear regression curve was plotted using the following sigmoidal dose–response variable slope equation: Y = Bottom +[Top − Bottom]/[1 + 10^((LogEC50 − X) × Hill slope)], using Graphpad Prism software 4.03. Data from a single representative experiment of numerous experiments are presented.
p53-hDM2 PPIB Sample Preparation and Image Acquisition
The activities of the p53-GFP and hDM2-RFP protein interaction partners of the PPIB were previously characterized in U-2 OS cells that were transiently cotransfected with recombinant plasmid expression vectors encoding these 2 components.9 To simplify the HCS procedure and to reduce assay variability associated with inconsistent transfection efficiencies, the manufacturer (Cellumen, Inc.) subcloned the individual p53-GFP and hDM2-RFP biosensor components into recombinant adenovirus expression constructs. The single-step adenovirus coinfection process eliminates the steps to prepare the plasmid DNA–liposome complexes for both biosensor components prior to each transfection and produces much higher efficiency expression and coexpression levels. The N-terminal residues 1–131 of p53 include the binding site for hDM2 that overlaps with the p53 transactivation domain, and the p53-GFP fusion protein was targeted and anchored in the nucleolus by the inclusion of an NLS (Fig. 1A). In Figure 1B–D, images of 4 fields of view in 3 fluorescent channels (Hoechst Ch1, GFP Ch2, and RFP Ch3) were sequentially acquired on the ArrayScan VTI platform using a 20× 0.4 NA objective with the XF93 excitation and emission filter set (Hoechst, FITC, and TRITC). After overnight (16–18 h) culture, U-2 OS cells infected with the p53-GFP adenovirus alone produced a bright punctate staining pattern localized within the nucleolar region of the nucleus that was unaffected by 30 min exposure to 10 μM Nulin-3 (Fig. 1B). The lack of appreciable bleed through of the p53-GFP fluorescence captured in Ch2 into Ch3 was indicated by the lack of a signal in the hDM2-RFP images and the predominant green and blue signals apparent in the composite images (Fig. 1B). For HCS imaging assays, it is important to establish how the combination of sample preparation methods, fluorophores, and excitation/emission filters selected for the assay impacts the cross talk between the fluorescent images of each channel and the multiparameter data derived from these signals.46,47 The N-terminal residues 1–118 of hDM2 encompass the binding site for the N-terminal transactivation domain of p53, and the inclusion of both an NLS and an NES enabled the protein to shuttle between the cytoplasm and nuclear compartments (Fig. 1A). After overnight culture of U-2 OS cells infected with the hDM2-RFP adenovirus alone, the hDM2-RFP produced a diffuse staining throughout the cytoplasm that was unaffected by 30 min exposure to 10 μM Nulin-3 (Fig. 1C). Again, the lack of appreciable bleed through of the hDM2-RFP fluorescence captured in Ch3 into Ch2 was indicated by a lack of a signal in the p53-GFP images and the predominant red and blue signals evident in the composite images (Fig. 1C). After overnight culture of U-2 OS cells coinfected with both adenovirus constructs, the hDM2-RFP shuttling partner interacted with the anchored p53-GFP partner and both proteins exhibited a bright punctate staining pattern colocalized within the nucleolus (Fig. 1D). The colocalization of the p53-GFP and hDM2-RFP proteins within coinfected U-2 OS cells was indicated by the predominant yellow color in the nucleolus of the composite images (Fig. 1D). The nucleolar localization of the anchored p53-GFP component of the biosensor serves to emphasize the interaction with the hDM2-RFP shuttling component of the biosensor by producing a dramatic redistribution of the hDM2-RFP from the cytoplasm into the nucleolus that produces the bright distinctive punctate staining of the 2 fusion proteins colocalized within this compartment. However, upon exposure of coinfected U-2 OS cells to 10 μM Nulin-3 for 30 min, the p53-hDM2 protein–protein interaction was disrupted and the hDM2-RFP shuttling partner redistributed into the cytoplasm, whereas the anchored p53-GFP partner remained localized within the nucleolus of the nucleus (Fig. 1D). In the composite images of coinfected Nutlin-3-treated U-2 OS cells the nucleolus was predominantly light green/blue, and the cytoplasm was predominantly red (Fig. 1D).
p53-hDM2 PPIB Image Analysis—Molecular Translocation Algorithm
Images from the p53-hDM2 PPIB were analyzed using the MT image analysis algorithm. Hoechst-stained objects in Ch1 that exhibited the appropriate fluorescent intensities above background and morphological size characteristics (width, length, and area) were identified and classified by the image segmentation as nuclei. The nuclear mask derived from Ch1 (Fig. 2A, red circle) was then used to segment the images from Ch2 and Ch3 into nuclear (Circ) and cytoplasmic (Ring) regions. The nuclear mask was eroded to reduce cytoplasm contamination within the nuclear area, and the reduced mask (Fig. 2A, yellow circle) was used to quantify the amount of target channel, p53-GFP in Ch2 and hDM2-RFP in Ch3, fluorescence within the nuclear region. The nuclear mask was then dilated to cover as much of the cytoplasmic region as possible without going outside the cell boundary. Removal of the original nuclear region from this dilated mask created a ring mask that covered the cytoplasmic region outside the nuclear envelope. The number of pixels away from the nuclear mask and the number of pixels (width) between the inner and outer ring masks were selectable within the MT bioapplication software. The ring masks were then used to quantify the amount of target channel, p53-GFP (Ch2, mauve rings) or hDM2-RFP (Ch3, light blue rings), fluorescence within the cytoplasmic region (Fig. 2A). The MT image analysis algorithm outputs quantitative data such as the total (mean nuclear total intensity channel 1) and average (mean nuclear average intensity channel 1) fluorescent intensities of the Hoechst-stained objects in Ch1, the selected object or cell counts (selected object counts per valid field of view) from Ch1 (Fig. 2B), and the total and average fluorescent intensities of the p53-GFP Ch2 and the hDM2-RFP Ch3 signals in the nuclear (Circ) or cytoplasm (Ring) regions as an overall well average or individual cell basis. To quantify the relative distribution of the p53-GFP within the nucleus and the cytoplasmic regions, we used the MT image analysis algorithm and calculated a mean average intensity difference by subtracting the average p53-GFP intensity in the Ring (cytoplasm) region from the average p53-GFP intensity in the Circ (nuclear) region of Ch2 (mean Circ-Ring average intensity difference channel 2 [MCRAID-Ch2]) (Fig. 2C). To quantify the relative distribution of the hDM2-RFP within the nucleus and the cytoplasmic regions, we performed a similar calculation with the MT image analysis algorithm in Ch3 to generate a MCRAID-Ch3 value (Fig. 2D). High MCRAID values indicated that the hDM2-RFP biosensor was predominantly localized within the nuclear region, whereas low MCRAID values indicate a more prominent localization within the cytoplasm. Treatment of U-2 OS cells coinfected with the p53-hDM2 PPIB adenoviruses with Nutlin-3 at concentrations ranging between 0.048 and 12.5 μM had no effect on either the total cell counts (Fig. 2B) or the p53-GFP MCRAID-Ch2 values (Fig. 2C), indicating that the p53-GFP component of the biosensor remained in the nucleus. In sharp contrast, however, Nutlin-3 treatment induced a concentration-dependent decrease in the hDM2-RFP MCRAID-Ch3 signal that was consistent with the redistribution of the hDM2-RFP from the nucleus to the cytoplasm. In this experiment, Nutlin-3 exhibited an IC50 of 0.175 ± 0.028 μM for the disruption of the p53-hDM2 PPI.
p53-hDM2 PPIB HCS Assay Development and Optimization
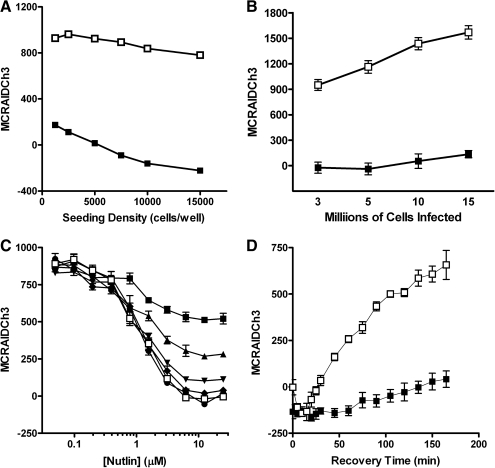
For the remaining assay development and optimization studies, images of the Hoechst, GFP, and RFP fluorescent channels were sequentially acquired on the ArrayScan VTI platform using a 10 × 0.3 NA with the XF93 excitation and emission filter set. The ArrayScan VTI was directed to acquire 250 selected objects (nuclei) or 2 fields of view, whichever came first. To identify an optimal cell seeding density for the 384-well assay plates, we evaluated the robustness and reproducibility of the assay signal window in wells seeded with U-2 OS cells coinfected with the p53-hDM2 PPIB adenoviruses at cell seeding densities ranging between 1 × 103 and 15 × 103 cells per well (Fig. 3A). To evaluate the impact of cell seeding density on the robustness and reproducibility of the assay signal window and to normalize the data, we used wells treated with 0.5% DMSO to represent maximum control wells containing cells in which the interaction between p53 and hDM2 was intact (0% inhibition) and the corresponding MCRAID-Ch3 signal was high, and wells treated with 10 μM Nutlin-3 to represent minimum control wells containing cells in which the interaction between p53 and hDM2 was maximally disrupted (100% inhibition) and the corresponding MCRAID-Ch3 signal was low. Although a robust and reproducible hDM2-RFP MCRAID-Ch3 assay signal window was maintained between the maximum and minimum control wells for all of the cell seeding densities tested (Fig. 3A), we selected a cell seeding density of 2,500 cells per well to minimize the cell culture burden for all further assay development and screening experiments. To evaluate the scalability of the p53-hDM2 PPIB adenovirus coinfection process, we incubated varying numbers of U-2 OS cells ranging between 3 × 106 and 1.5 × 107 cells in 1.5 mL of media with the manufacturer's recommended volumes of adenovirus for 60 min (Fig. 3B). A robust and reproducible hDM2-RFP MCRAID-Ch3 assay signal window was maintained between the maximum and minimum control wells for all of the cell densities coinfected with the recommended volume of adenovirus (Fig. 3B), indicating that the coinfection process could be appropriately scaled for both assay development and screening purposes. To evaluate the time dependence of the Nutlin-3-induced disruption of the p53-hDM2 interaction, we conducted independent concentration response experiments that were fixed at the indicated time points, ranging from 5 to 30 min after compound addition (Fig. 3C). Within the 1–25 μM Nutlin-3 concentration range, the disruption of the p53-hDM2 interactions was discernable as early as 5 min after compound addition, the magnitude of the Nutlin-3-induced disruption increased through 20 min of compound exposure, and thereafter, maximal disruption appeared to be maintained through 30 min (Fig. 3C). Interestingly, the Nutlin-3-induced disruption of the p53-hDM2 PPIB was reversible, as indicated by the time-dependent recovery of the hDM2-RFP MCRAID-Ch3 signal in wells that were washed 3 times with PBS after 30 min exposure to 10 μM Nutlin-3, compared with wells that were maintained in the constant presence of 10 μM Nutlin-3 (Fig. 3D). In comparison to DMSO controls, under conditions of constant exposure to 10 μM Nutlin-3, the p53-hDM2 interactions were maximally disrupted for up to 6 h with very little apparent drift in the hDM2-RFP MCRAID-Ch3 signal (Fig. 3D and data not shown). We selected 90 min as our standard compound incubation period for all further experiments.
Fig. 3.
Optimization of the p53-hDM2 protein–protein interaction biosensor (PPIB) high-content screening assay. (A) Cell seeding density. p53-hDM2 PPIB adenovirus-infected cells were seeded at the indicated cell densities in the wells of 384-well Greiner collagen-coated assay plates, cultured overnight at 37°C, 5% CO2, and 95% humidity, and were then treated for 90 min with either 0.5% dimethyl sulfoxide (DMSO) (□) or 10 μM Nutlin-3 in 0.5% DMSO (▪) prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. (B) Scalability of the p53-hDM2 PPIB adenovirus coinfection process. U-2 OS cells ranging between 3 × 106 and 1.5 × 107 cells in 1.5 mL of media were coincubated with the manufacturer's recommended volumes of the p53-hDM2 PPIB adenoviruses for 30 min. Coinfected cells were then seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, cultured overnight at 37°C, 5% CO2, and 95% humidity, and were then treated for 90 min with either 0.5% DMSO (□) or 10 μM Nutlin-3 in 0.5% DMSO (▪) prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. (C) Time course of Nutlin-3 disruption. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with the indicated concentrations of Nutlin-3 and then fixed and stained with Hoechst at 5 min intervals up to 30 min total exposure time; (▪) 5 min, (▴) 10 min, (▾) 15 min, (♦) 20 min, (●) 25 min, and (□) 30 min. (D) Reversibility and stability of the p53-hDM2 PPIB assay signal window. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with 10 μM Nutlin-3 for 90 min. Half the wells were then washed 3 times with fresh McCoy's 5A medium (□) and the remainder were left untouched (▪). Cells were incubated further at 37°C, 5% CO2, and 95% humidity for the indicated times prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images from 3 fluorescent channels were sequentially acquired on the ArrayScan VTI platform using a 10× 0.3 NA objective with the XF93 excitation and emission filter set and were analyzed with molecular translocation (MT) image analysis algorithm. The mean circle ring average intensity difference in channel 3 (MCRAID-Ch3) data derived from the MT image analysis algorithm is a measure of the relative distribution of the hDM2-red fluorescent protein within the nucleus and the cytoplasmic regions and was used as the primary indicator of the interactions between p53 with hDM2. The data are presented as the mean MCRAID-Ch3 values ± SD from (A) n = 32 wells, (B) n = 96 wells, (C) n = 4 wells, and (D) n = 6 wells. The lines connecting the data were plotted using Graphpad Prism software 4.03. Data from a single representative experiment of 3 or more experiments are presented in A–C, and from a single experiment in D.
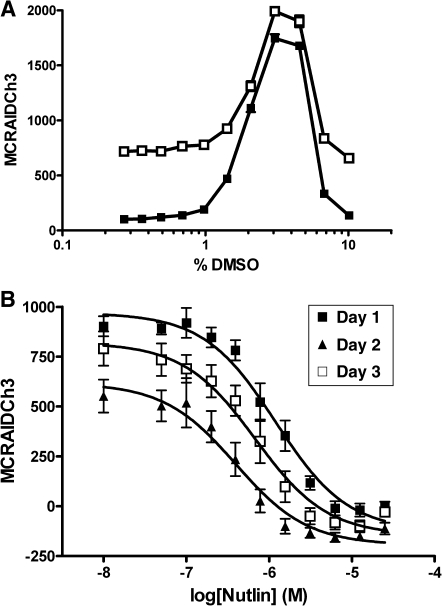
As most compound libraries are dissolved in DMSO, we evaluated the DMSO tolerance of the p53-hDM2 PPIB assay (Fig. 4A). At DMSO concentrations above 1.0%, the separation between the p53-hDM2 PPIB DMSO minimum and Nutlin-3-treated maximum controls collapsed dramatically (Fig. 4A). Consistent with our previous experience with other HCS assays,42–44 at DMSO concentrations >1%, U-2 OS cells changed from a well-spread and well-attached morphology to a more rounded loosely attached morphology that interfered with the ability of the image analysis algorithm to segment the Ch2 and Ch3 images into distinct cytoplasm and nuclear regions. Based on these observations, we elected to conduct all further p53-hDM2 PPIB assays for compound testing at ≤0.5% DMSO. Using our optimized p53-hDM2 PPIB assay conditions with respect to the 384-well cell seeding density, adenovirus coinfection conditions, the length of compound exposure, and DMSO tolerance, we conducted 3 independent concentration–response experiments to evaluate the reproducibility of the Nutlin-3 IC50 for disruption of the p53-hDM2 interaction (Fig. 4B). On average, Nutlin-3 exhibited an IC50 of 0.607 ± 0.382 μM (n = 5) in the p53-hDM2 PPIB assay (Figs. 2D, 4B, and 8D).
Fig. 4.
Dimethyl sulfoxide (DMSO) tolerance and Nutlin-3 IC50 determinations. (A) DMSO tolerance. U-2 OS cells were coinfected with the p53-hDM2 protein–protein interaction biosensor (PPIB) adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with 0.5% DMSO (□) or 10 μM Nutlin-3 (▪) at the indicated concentrations of DMSO for 90 min prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. (B) Nutlin-3 IC50 determinations from 3 independent experiments. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with the indicated concentrations of Nutlin-3 for 90 min prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images were acquired on the ArrayScan VTI platform and analyzed with molecular translocation image analysis algorithm as described for Figure 2. The mean circle ring average intensity difference in channel 3 (MCRAID-Ch3) data was used as the primary indicator of the interactions between p53 and hDM2. The data are presented as the mean MCRAID-Ch3 values ± SD from (A) n = 16 wells, (B) n = 16 wells at day 1, and n = 24 wells at days 2 and 3. For the IC50 determinations, the resulting nonlinear regression curves were plotted using the following sigmoidal dose–response variable slope equation: Y = Bottom + [Top − Bottom]/[1 + 10^((LogEC50 − X) × Hill slope)], using Graphpad Prism software 4.03. Data from a single representative experiment of 2 independent DMSO-tolerance experiments are presented in A, and the data from 3 independent IC50 experiments are presented in B. The IC50s for the disruption of the p53-hDM3 PPIB were 1.196, 0.394, and 0.680 μM on days 1, 2, and 3, respectively.
Fig. 8.
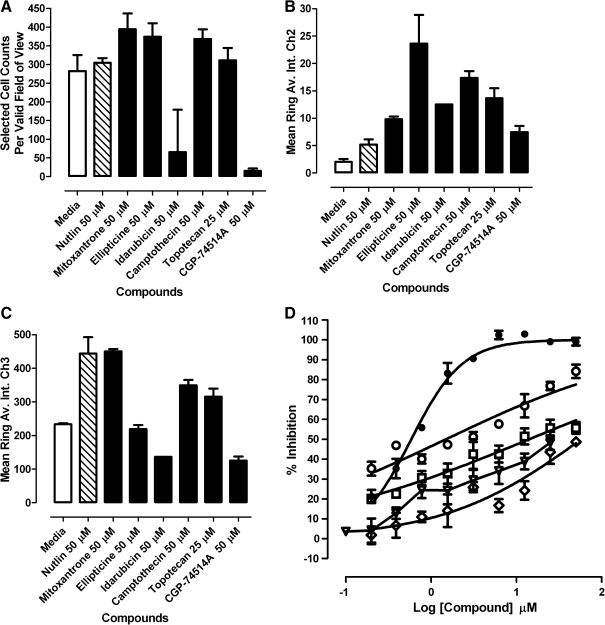
Quantitative image analysis data for the p53-hDM2 protein–protein interaction biosensor (PPIB) Library of Pharmacologically Active Compounds actives. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with the indicated concentrations of the putative active compounds for 90 min prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images were acquired on the ArrayScan VTI platform and analyzed with the molecular translocation (MT) image analysis algorithm as described for Figure 2. Selected quantitative data outputs from the MT image analysis algorithm: (A) The selected object or cell counts (selected object counts per valid field of view [SCCPVF]) derived from Hoechst-stained nuclei in Ch1; (B) the mean average fluorescent intensity in the Ch2 (p53-green fluorescent protein) cytoplasm ring region (mean ring average intensity in channel 2 [MRAI-Ch2]); (C) the mean average fluorescent intensity in the Ch3 (hDM2-red fluorescent protein [RFP]) cytoplasmic ring region (mean ring average intensity in channel 3 [MRAI-Ch3]); and (D) the mean circle ring average intensity difference in channel 3 (MCRAID-Ch3) values used to quantify the relative distribution of the hDM2-RFP between the nucleus and the cytoplasmic regions in Ch3. The data are presented as the mean SCCPVF (A), MRAI-Ch2 (B), and MRAI-Ch3 (C) values ± SD from triplicate wells (n = 3) at the top compound concentration tested 50 μM (25 μM for topotecan). Medium control (open bars), Nutlin-3 control (hashed bars), and compounds (black bars). For the MCRAID-Ch3 concentration–response data (D), the mean MCRAID-Ch3 values ± SD from triplicate wells (n = 3) at each compound concentration together with their resulting nonlinear regression curves were plotted using the following sigmoidal dose–response variable slope equation: Y = Bottom + [Top − Bottom]/[1 + 10^((LogEC50 − X) × Hill slope)], using Graphpad Prism software 4.03. Data from a single representative experiment of 3 experiments are presented.
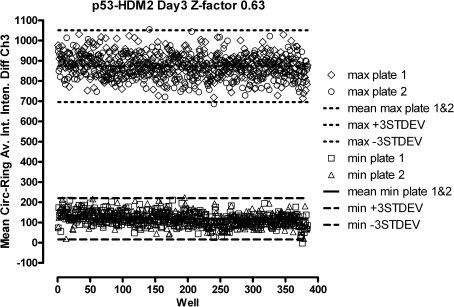
Three-Day Assay Signal Window and Z-factor Determination
The UPDDI has adopted a rigorous process to evaluate the robustness and reproducibility of a screening assay signal window and to determine Z-factors.38–41,48,49 The process consists of 3 independent experiments of 2 full plates each of the minimum (10 μM Nutlin-3) and maximum (0.5% DMSO) plate controls conducted on 3 separate days (Fig. 5, Table 1). Figure 5 is a scatter plot of the hDM2-RFP MCRAID-Ch3 signals from the 2 full 384-well plates each of maximum and minimum controls performed on day 3. The maximum and minimum plate controls performed very reproducibly and their respective hDM2-RFP MCRAID-Ch3 population responses were well separated from each other (Fig. 5). Overall, the p53-hDM2 PPIB assay exhibited an average 3-day Z-factor of 0.56 ± 0.08 and an average 3-day S:B ratio of 6.28 ± 0.98. Our only cause for concern was that the CVs of the minimum plate controls exceeded 20% (Table 1). However, variability in the lower values of the minimum controls of HTS assays can often have a larger impact and produce higher CVs,38–41 but based on the assay signal window, Z-factor, and S:B ratio performance data, we concluded that the p53-hDM2 PPIB assay was compatible with HCS (Table 1).
Fig. 5.
Three-day assay signal window and Z-factor determination. U-2 OS cells were coinfected with the p53-hDM2 protein–protein interaction biosensor adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Two maximum control plates (max plate 1 ⋄, max plate 2 ○) were then treated with 0.5% dimethyl sulfoxide and 2 minimum control plates (min plate 1 □, min plate 2 ▵) were treated with 10 μM Nutlin-3 for 90 min prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images were acquired on the ArrayScan VTI platform and analyzed with the molecular translocation image analysis algorithm as described for Figure 2. The mean circle ring average intensity difference in channel 3 (MCRAID-Ch3) data was used as the primary indicator of the interactions between p53 and hDM2. Three independent experiments of 2 full 384-well plates each of maximum and minimum plate controls were conducted on 3 separate days (Table 1). A scatter plot of the hDM2-red fluorescent protein MCRAID-Ch3 signals from the 2 full 384-well plates each of maximum (n = 768) and minimum (n = 768) controls performed on day 3 is presented.
Table 1.
Three-Day Assay Signal Window and Z-Factor Determination
| Class | Day | Plate | Max/Min | Mean | SD | CV | Z-Factor | SB |
|---|---|---|---|---|---|---|---|---|
| Intraplate |
1 |
1 |
Max |
1,436.18 |
75.02 |
5.22 |
0.56 |
5.92 |
| 2 |
Max |
1,356.41 |
94.46 |
6.96 |
0.52 |
5.48 |
||
| 3 |
Min |
242.78 |
98.59 |
40.61 |
0.48 |
5.59 |
||
| 4 |
Min |
262.21 |
111.29 |
42.44 |
0.44 |
5.17 |
||
| All plates |
|
|
|
|
0.48 |
5.53 |
||
| 2 |
1 |
Max |
1,149.79 |
64.67 |
5.62 |
0.59 |
5.82 |
|
| 2 |
Max |
1,184.73 |
64.96 |
5.48 |
0.57 |
5.83 |
||
| 3 |
Min |
197.44 |
65.81 |
33.33 |
0.60 |
6.00 |
||
| 4 |
Min |
197.29 |
71.5 |
36.24 |
0.59 |
6.01 |
||
| All plates |
|
|
|
|
0.58 |
5.91 |
||
| 3 |
1 |
Max |
879.71 |
59.28 |
6.74 |
0.64 |
7.56 |
|
| 2 |
Max |
866.21 |
58.34 |
6.74 |
0.63 |
7.34 |
||
| 3 |
Min |
116.34 |
33.39 |
28.7 |
0.63 |
7.45 |
||
| 4 |
Min |
119.91 |
34.66 |
28.91 |
0.63 |
7.22 |
||
| All plates |
|
|
|
|
0.63 |
7.39 |
||
| Interplate |
1 |
1 and 2 |
Max |
1,396.3 |
94.15 |
6.74 |
0.48 |
5.53 |
| 3 and 4 |
Min |
252.49 |
105.56 |
41.81 |
|
|
||
| 2 |
1 and 2 |
Max |
1,167.26 |
62.17 |
5.75 |
0.58 |
5.91 |
|
| 3 and 4 |
Min |
197.37 |
68.7 |
34.81 |
|
|
||
| 3 |
1 and 2 |
Max |
872.96 |
59.19 |
6.78 |
0.63 |
7.39 |
|
| 3 and 4 |
Min |
118.12 |
34.07 |
28.84 |
|
|
||
| Day to day | 1 and 2 |
All plates |
Max |
1,281.78 |
140.71 |
10.98 |
0.34 |
5.70 |
| Min |
224.93 |
93.22 |
41.44 |
|
|
|||
| 2 and 3 | All plates | Max |
1,020.11 |
160.19 |
15.7 |
0.21 |
6.47 |
|
| Min | 157.75 | 67.15 | 42.57 |
The p53-hDM2 protein–protein interaction biosensor data presented in this table were generated in 3 independent experiments of 2 full plates each of the maximum (0.5% dimethyl sulfoxide) and minimum (10 μM Nutlin-3) plate controls conducted on 3 separate days.
1,280-Compound LOPAC HCS
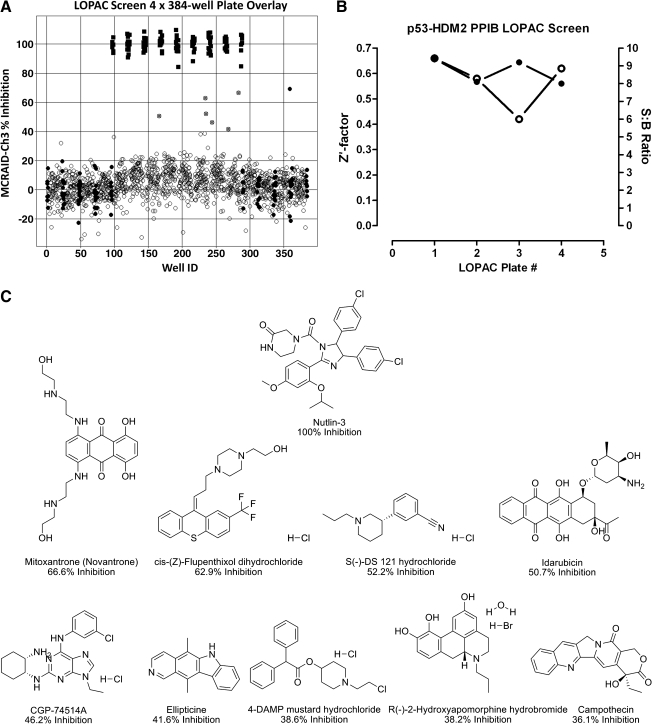
We utilized the MCRAID-Ch3 data derived from the MT image analysis algorithm as the primary output for the interactions between p53 with hDM2. For the LOPAC screen, we utilized the mean MCRAID-Ch3 value of the DMSO maximum plate control wells (n = 32) and the mean MCRAID-Ch3 value of the 10 μM Nutlin-3 minimum plate control wells (n = 24) to normalize the MCRAID-Ch3 compound data and to represent 0% and 100% disruption/inhibition of the p53-hDM2 interactions, respectively. Figure 6A shows a 4-plate overlay scatter plot of the normalized percent inhibition data from the 1,280-compound LOPAC screen. The p53-hDM2 PPIB HCS assay exhibited a robust and reproducible assay signal window between the Nutlin-3 and DMSO-treated plate controls and performed very consistently from plate to plate (Fig. 6B). For the four 384-well plates screened during the p53-hDM2 LOPAC screen, the Z′-factors were 0.66, 0.58, 0.44, and 0.62 for plates 1, 2, 3, and 4, respectively (Fig. 6B). The corresponding plate S:B ratios were 9.4, 8.1, 9.2, and 8.0 for plates 1, 2, 3, and 4, respectively (Fig. 6B). The primary HCS active criterion was set at ≥35% inhibition of MCRAID-Ch3, and the chemical structures of the 9 (0.7%) putative actives identified in the 1,280 compounds screened at 50 μM (Fig. 6A, gray circles) are presented in Figure 6C. For comparison, the chemical structure of Nutlin-3 is also presented (Fig. 6C). CGP-74514A was cited as a Cdk1 inhibitor, and 4 other compounds were cited as anticancer drugs in the Sigma-Aldrich catalog (mitoxantrone, idarubicin, ellipticine, and campothecin). Three compounds were listed as dopamine receptor ligands (cis-(Z)-flupenthixol dihydrochloride, S(−)-DS 121 hydrochloride, and R(−)-2,10,11-trihydroxyaporphine hydrobromide), and 1 was listed as a muscarinic receptor ligand (4-diphenylacetoxy-N-(2-chloroethyl)piperidine hydrochloride).
Fig. 6.
p53-hDM2 protein–protein interaction biosensor (PPIB) Library of Pharmacologically Active Compounds (LOPAC) high-content screen. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Diluted compounds and plate controls were transferred from the 4 × 384-well LOPAC daughter plates or control blocks to the p53-hDM2 PPIB assay plates to provide a final screening concentration of 50 μM and then incubated for 90 min prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Images were acquired on the ArrayScan VTI platform and analyzed with the molecular translocation image analysis algorithm as described for Figure 2. The mean circle ring average intensity difference in channel 3 (MCRAID-Ch3) data was used as the primary indicator of the interactions between p53 and hDM2. (A) Four-plate overlay of percent inhibition for the LOPAC Screen. An ActivityBase primary HTS template was created that automatically calculated the percent inhibition. The mean MCRAID-Ch3 value of the dimethyl sulfoxide (DMSO) minimum plate control wells (●, n = 32 per plate) and the mean MCRAID-Ch3 value of the 10 μM Nutlin-3 maximum plate control wells (▪, n = 24) were used to normalize the MCRAID-Ch3 compound data (○) and to represent 0% and 100% disruption/inhibition of the p53-hDM2 interactions, respectively. Potential active compounds (gray circle, ●) with greater than 40% inhibition are indicated. (B) High-content screening (HCS) performance. An ActivityBase primary HTS template was created, which automatically calculated the plate control signal-to-background (S:B) ratios and Z′-factors using the MCRAID-Ch3 values of the DMSO minimum plate control wells (n = 32 per plate) and the 10 μM Nutlin-3 maximum plate control wells (n = 24). Z′-factors (∘) and S:B ratios (●) for the four 384-well plates of the LOPAC screen. (C) Chemical structures, names, and percent inhibition of the LOPAC HCS actives. The chemical structures, names, and percent inhibition for 9 compounds that exhibited ≥35% inhibition and for Nutlin-3 are presented.
LOPAC Active Confirmation and Hit Characterization
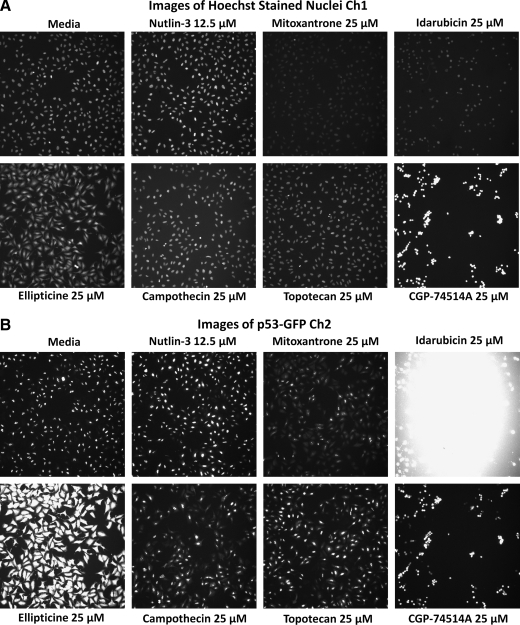
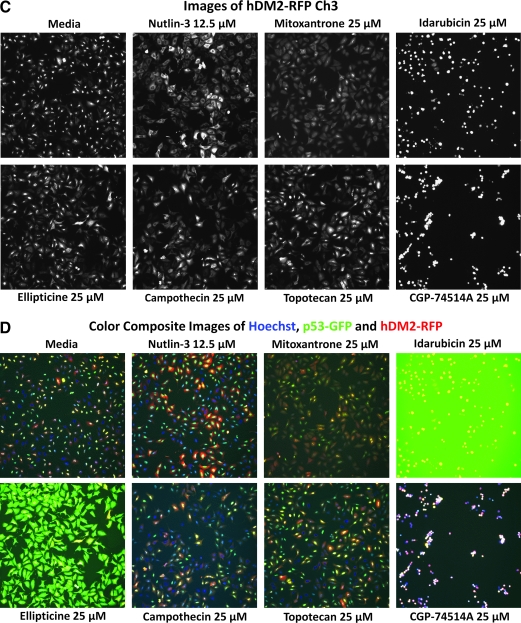
After examining the images from the active wells, the dopamine and muscarinic receptor actives were eliminated from consideration as likely focusing artifacts (data not shown). To confirm the activity of the remaining compounds in the p53-hDM2 PPIB assay and to determine their IC50s, we purchased powdered samples from Sigma-Aldrich and conducted 10-point concentration–response assays starting at a maximum of 50 μM. We also included topotecan, a water-soluble derivative of camptothecin. Representative images from the 3 fluorescent channels together with their respective composite images and the multiparameter HCS data extracted from those images are presented in Figures 7 and 8. Only mitoxantrone, ellipticine, camptothecin, and topotecan exhibited concentration-dependent inhibition of the p53-hDM2 PPIB MCRAID-Ch3 signal (Fig. 8D). However, in contrast to the Nutlin-3 data (R2 = 0.97), the inhibition data for these 4 compounds could not be reliably fit to a sigmoidal concentration–response model and we elected not to report their IC50 values. All of the LOPAC active compounds produced significantly higher average fluorescent intensities in the cytoplasmic region of Ch2 than either the DMSO or Nutlin-3 controls (Figs. 7B and 8B). CGP-74514A treatment produced a much more rounded cell morphology (Fig. 7A–D), significantly reduced the cell counts (Figs. 7A and 8A), and induced an hDM2-RFP MCRAID Ch3 signal that was >2-fold higher than the DMSO controls (data not shown). Idarubicin treatment also did not significantly reduce the hDM2-RFP MCRAID Ch3 signal compared with the DMSO controls (data not shown), but significantly reduced the cell counts (Figs. 7A and 8A), induced a rounded cell morphology (Fig. 7A–D), and produced a strong fluorescent signal throughout the well in Ch2 (Fig. 7B) and in the nucleus of cells in Ch3 (Fig. 7C). Strong fluorescent signals were observed in the images from Ch2 and Ch3 acquired from wells treated with idarubicin in the 1.56–25 μM concentration range. In addition, neither CGP-74514A nor idarubicin treatment produced an increase in the average fluorescent intensity in the cytoplasmic region of Ch3 (mean ring average intensity in channel 3 [MRAI-Ch3]), consistent with the effects of Nutlin-3 (Fig. 8C). Based on these observations, we concluded that CGP-74514A and idarubicin were not concentration-dependent inhibitors of the p53-hDM2 PPIB. Ellipticine-treated cells were strongly fluorescent in the images from Ch2 (Fig. 7C), and the fluorescent signal appeared to partially bleed through into the images from Ch1 (Fig. 7A). Although ellipticine reduced the hDM2-RFP MCRAID Ch3 signal in a concentration-dependent manner (Fig. 8D), neither was it as effective as Nutlin-3, nor did it produce an increase in the MRAI-Ch3 signal consistent with the effects of Nutlin-3 (Fig. 8C). Mitoxantrone treatment appeared to interfere with the Hoechst nuclear staining (Fig. 7A) and there were concerns that it was weakly fluorescent within cells in the images from Ch2 (Fig. 7B) and Ch3 (Fig. 7C). Mitoxantrone reduced the hDM2-RFP MCRAID Ch3 signal in a concentration-dependent manner (Fig. 8D) and also induced a significant increase in the MRAI-Ch3 signal (Fig. 8C), but it was not as potent or efficacious as Nutlin-3. Camptothecin and topotecan were also weakly fluorescent within cells in the images from Ch2 (Fig. 7B), and although they reduced the hDM2-RFP MCRAID Ch3 signal in a concentration-dependent manner, they were not as effective as Nutlin-3 at decreasing the MCRAID Ch3 signal (Fig. 8D) or at increasing the MRAI-Ch3 signal (Fig. 8C). Although mitoxantrone, ellipticine, camptothecin, and topotecan exhibited concentration-dependent disruption of the p53-hDM2 PPIB (Fig. 8D), they were much less potent than Nutlin-3 and the images and quantitative multiparameter HCS data for these compounds were not completely consistent with the Nutlin-3 response profile. Unlike Nutlin-3, which exhibited a typical sigmiodal concentration–response curve (Figs. 2D, 3C, 4B, and 8D), the curve fits for mitoxantrone, ellipticine, camptothecin, and topotecan were very shallow and failed to reach either a top or a bottom plateau in the concentration range tested. Although these compounds reduced the hDM2-RFP MCRAID Ch3 signal in a concentration-dependent manner, we elected not to report the IC50 values interpolated from these poor curve fits (Fig. 8D).
Fig. 7.
Selected images of the p53-hDM2 protein–protein interaction biosensor (PPIB) Library of Pharmacologically Active Compounds actives. U-2 OS cells were coinfected with the p53-hDM2 PPIB adenoviruses, seeded at 2,500 cells per well in 384-well Greiner collagen-coated assay plates, and cultured overnight at 37°C, 5% CO2, and 95% humidity as described earlier. Cells were then treated with the indicated concentrations of the putative active compounds for 90 min prior to fixation with 3.7% formaldehyde containing 2 μg/mL Hoechst 33342. Individual gray-scale and 3-color composite images of U-2 OS cells from 3 fluorescent channels (Hoechst Ch1, green fluorescent protein [GFP] Ch2, and red fluorescent protein (RFP) Ch3) were sequentially acquired on the ArrayScan VTI platform using a 10× 0.3 NA objective with the XF93 excitation and emission filter set (Hoechst, blue; FITC, green; TRITC, red). (A) Hoechst channel, (B) p53-GFP channel, (C) hDM2-RFP channel, and (D) composite 3-color images.
Discussion
In this report, we describe the optimization of a novel 384-well positional biosensor HCS assay to quantify the small-molecule–induced disruption of p53-hDM2 interactions. U-2 OS cells coinfected with the p53 and hDM2 PPIB adenoviruses demonstrated colocalization of the GFP and RFP fusion proteins in the nucleolus because of interactions between the NH-terminal domains of p53 and hDM2 (Fig. 1A, D). Exposure of these cells to Nutlin-3 disrupted the p53-hDM2 PPI and caused the hDM2-RFP to redistribute into the cytoplasmic region (Fig. 1A, D). The disruption of the p53-hDM2 PPIB and subsequent redistribution of the hDM2-RFP component was captured in images acquired by the ArrayScan VTI imaging platform (Fig. 1C, D). The MCRAID-Ch3 output from the MT image analysis algorithm was used to quantify the relative distribution of the hDM2-RFP between the nucleus and the cytoplasmic regions of cells (Fig. 2). To evaluate the assay signal window, to determine Z′-factors, and to normalize the HCS data, we used wells treated with 0.5% DMSO as maximum control wells in which the interaction between p53 and hDM2 was intact (high MCRAID-Ch3 signal, 0% inhibition), and wells treated with 10 μM Nutlin-3 as minimum control wells in which the interaction between p53 and hDM2 was maximally disrupted (low MCRAID-Ch3 signal, 100% inhibition) (Figs. 2–6). An optimal cell seeding density of 2,500 cells per well of 384 wells reduced the cell culture burden for assay development and screening (Fig. 3A). U-2 OS cells were efficiently and reproducibly coinfected with the p53-hDM2 PPIB adenoviruses during a simple scalable 60-min incubation step introduced between the harvesting and counting of cells and their subsequent seeding into assay plates (Fig. 3B). Disruption of the p53-hDM2 PPIB was discernable as early as 5 min after Nutlin-3 addition and increased through 20–30 min of compound exposure (Fig. 3C), and a stable assay signal window was maintained for up to 6 h (Fig. 3D and data not shown). Ninety minutes was selected as the minimum compound incubation period, and as many as one hundred 384-well plates could be processed in high throughput. DMSO of >1% concentrations altered the U-2 OS cell morphology to a rounded phenotype that invalidated the image analysis algorithm segmentation of the images into distinct cytoplasm and nuclear regions (Fig. 4A) and so all compound testings were required to be conducted at ≤0.5% DMSO. On average, Nutlin-3 exhibited an IC50 of 0.76 ± 0.41 μM (Fig. 4B), and the p53-hDM2 PPIB assay exhibited average Z-factors of 0.56 ± 0.08 and S:B ratios of 6.28 ± 0.98 (Fig. 5, Table 1), indicating that the p53-hDM2 PPIB assay signal window was suitable for HCS.
The p53-hDM2 PPIB HCS assay performed well during the 1,280-compound LOPAC screen and produced average Z′-factors of 0.57 ± 0.11 and average S:B ratios of 8.7 ± 0.7-fold (Fig. 6B). Nine (0.7%) compounds from the 1,280 LOPAC set exhibited ≥35% inhibition of the p53-hDM2 PPIB MCRAID-Ch3 signal at 50 μM (Fig. 6A, C). Four compounds, the dopamine and muscarinic receptor actives, were eliminated as focusing/imaging artifacts (data not shown). In concentration–response assays, CGP-74514A and idarubicin failed to confirm their activity in the primary HCS, significantly reduced the cell counts, and altered the morphology of the U-2 OS cells to a rounded phenotype that invalidated the segmentation of the image analysis algorithm (Figs. 7 and 8). All of the LOPAC actives exhibited significantly higher average fluorescent intensities in the cytoplasmic region of the GFP channel than either the DMSO or Nutlin-3 controls (Fig. 8B). These slightly higher MRAI-Ch2 values were the only indication that some of the compounds may have altered the nucleolar localization of the anchored p53-GFP component of the biosensor. Idarubicin was strongly fluorescent throughout the well, and ellipticine was strongly fluorescent within cells (Fig. 7B). In concentration–response assays, only mitoxantrone, ellipticine, camptothecin, and topotecan exhibited concentration-dependent inhibition of the p53-hDM2 PPIB MCRAID-Ch3 signal (Fig. 8D).
Although none of these LOPAC actives proved to be as potent or effective as Nutlin-3 at disrupting the p53-hDM2 PPIB, they all interact with the p53 pathway. The anthracyclines mitoxantrone and idarubicin are clinically approved anticancer drugs that inhibit topoisomerase II and induce increased serine-15 phosphorylation of p53, stabilization of p53, and transactivation of p53 target genes, ensuing cell cycle arrest.50–52 Ellipticine also inhibits topoisomerase II activity, and several ellipticine derivatives have been shown to rescue a variety of p53 point mutants, restore transactivation of p53 target genes, and induce cell death in tumor cell lines and mouse xenograft tumor models.22,35,53,54 In the NCI 60 tumor cell line panel, ellipticine derivatives are more effective against cells with mutant p53,54 and ellipticine treatment enhances the nuclear localization of p53 in cell lines bearing mutant p53.55 Recently, an HCS assay was developed to identify compounds that increased the nuclear localization of mutant p53-GFP stably expressed in Saos-2 cells.55 A mutant p53-GFP fusion protein was utilized for the HCS assay because overexpression of wild-type p53-GFP induced apoptosis. After a 3.5-h exposure to compounds, ellipticine was the only active compound identified in the 1,200-compound Preswick library screened at 5 μM (0.2% DMSO).55 Ellipticine treatment increased total p53 protein levels, enhanced the nuclear localization of p53, and activated a p21 promoter-driven luciferase reporter in HCT116 cells that are wild type for p53, but not in HCT116 p53−/− cells that are null for p53.55 Using γ-H2AX foci as a marker for DNA damage, it was concluded that the ellipticine-induced nuclear localization of p53 was independent of DNA damage.55 Although ellipticine derivatives have shown moderate antitumor activity in phase I and II clinical trials, these compounds also exhibit non–p53-dependent antitumor activities and their use has been limited by toxic side-effects.22,53–55 The quinoline alkaloid topoisomerase I inhibitor camptothecin and a variety of derivatives, including topotecan and irinotecan, have been shown to induce increased serine-15 phosphorylation of p53, stabilization of p53 protein, transactivation of p53 target genes, and cell cycle arrest.50,52,56,57 The activation of the p53 pathway in response to the DNA double strand breaks induced by the anthracycline (topoisomerase II inhibitors) or camptothecin (topoisomerase I inhibitors) derivatives appears to be mediated by the ATM-CHK2 DNA damage response pathway.50,52 Although the p53-hDM2 PPIB biosensor was designed and optimized to identify compounds that disrupt the interactions between p53 and hDM2, the biosensor was also able to distinguish the activity of compounds that activate the p53 pathway, or that rescue mutant p53 function, from compounds that do not impact the p53 signaling pathway.
A GFP-assisted readout (GRIP) positional biosensor HCS assay was previously used to screen a compound library for inhibitors of p53-hDM2 PPIs.12 A stable cell line was constructed, in which hDM2 was expressed as a fusion protein with cAMP phosphodiesterase (PDE) isoform 4A4B, and p53 was expressed as a GFP fusion protein.12 Treatment of this cell line with the PDE-4A4B agonist RS25344 induces the hDM2-PDE-4A4B to relocate to compact intracellular foci, and through its interactions with the hDM2-PDE-4A4B the p53-GFP redistributes from a diffuse cytoplasm staining pattern to a granular foci pattern.12 PDE-4A4B inhibitors such as RP73401, or p53-hDM2 disruptors, such as Nutlin, disperse the RS25344-induced GFP foci phenotype.12 A library of 3,165 compounds was screened at 10 μM in the p53-hDM2 GRIP biosensor assay using the PDE-4A4B inhibitor RP73401 (10 μM) as a disruption control. Forty-one compounds produced >40% activity at 10 μM in the primary HCS, but only 30 of these were confirmed in concentration–response assays and only 20 were deemed suitable for follow-up. Fifteen (75%) of the 20 confirmed actives exhibited a PDE-4A4B inhibitor spot dispersion phenotype, and 14 of these were confirmed in a PDE-4A4B redistribution counter screen. Nutlin that had been inserted into the screening library was 1 of the other 5 compounds, and it was the only compound that activated a p53-luciferase reporter secondary assay and differentially inhibited the proliferation of tumor cell lines with wild-type p53. The authors concluded that none of the other compounds that were active in the p53-hDM2 GRIP biosensor assay were functional inhibitors of the p53-hDM2 protein–protein interaction.12
In vitro biochemical PPI assay formats typically employ interaction partners that have been purified or partially purified from cells that endogenously express the proteins, or from Escherichia coli, mammalian cell lines, or insect cells engineered to express these proteins through recombinant DNA techniques.1,2 In addition to full-length proteins, subdomains of proteins or small interacting peptides derived from one or both of the interaction partners are frequently utilized in biochemical PPI assays. Biochemical PPI assay formats are typically easier to develop and prosecute in HTS, and issues with compound permeability and/or toxicity do not limit the number of chemotypes identified. However, biochemical PPI formats can have some significant limitations. First, many biochemical PPI formats consume large amounts of the interaction partners and the expression and purification of sufficient material to screen a sizable compound library may be both difficult and costly.1,2 Second, ELISAs and cell surface-binding PPI formats involve multiple transfer and wash steps that may prove challenging to automate.1,2 However, as biochemical PPI assays are conducted under nonphysiological conditions where competing interaction partners and/or contributing proteins may have been eliminated, there are significant concerns over the biological relevance of the data.1,2
The major advantages of cell-based PPI assays are that the interacting partners are generated in situ and that the interactions occur within the native environment of the cell, where any binding cofactors or posttranslational modifications that participate in or modulate the PPIs are available.3,7–10,12–14 Imaging-based PPI formats provide an additional benefit, the subcellular localization of the protein partners can also be monitored and measured.3,7–10,12–14 However, cell-based PPI assays also have some limitations. Although there are always concerns that permeability and/or cytotoxicity may limit the chemotypes identified in cell-based screening assays, the impact of yeast cell permeability is a recurrent concern with yeast genetic and yeast 2-hybrid PPI assays.1,2 It is also important to consider that the attachment of a fluorescent fusion protein to the PPI partners may alter their behavior, or that the overexpression of PPI partners and the coexpression of proteins that normally reside in different cellular compartments may lead to the detection of interactions that do not occur in vivo.7–10,14 It is therefore desirable to express fusion proteins at levels comparable to their endogenous counterparts, and to validate assays with mutations that eliminate the PPIs or with small molecules that specifically disrupt the PPIs of interest.7–10,14 The efficiency of FRET biosensors depends upon the distance between and the orientation of the donor and acceptor molecules, and the insertion of the donor/acceptor pairs into the appropriate site(s) of the interacting polypeptides so that they can efficiently measure the interactions between the 2 proteins can often be challenging.11 With PCA methods, there are concerns that the fluorescent protein fragments may associate with each other independently of an interaction between the attached fusion partners, and that the irreversible association of the fluorescent protein fragments may lead to the trapping of transient interactions in the absence of specific interactions between the attached fusion partners.10,13,14 The p53-hDM2 PPI biosensor characterized here was designed to be both sensitive and reversible so that it reflected the dynamics of the system without disrupting it (Fig. 3D), and further, the movement of the hDM2 chimeric protein induced by different stimuli or drugs could be monitored in living cells as well as in fixed cell preparations.9
In conclusion, we have characterized and optimized a novel imaging-based positional biosensor HCS assay to identify disruptors of p53-hDM2 PPIs. The chimeric proteins of the biosensor incorporated the N-terminus PPI domains of p53 and hDM2, protein targeting sequences (NLS and NES), and fluorescent reporters, which when expressed in cells could be used to monitor p53-hDM2 PPIs through changes in the position of the hDM2 biosensor. Coinfection with the recombinant adenovirus biosensors was used to express the NH-terminal domains of p53 and hDM2, fused to GFP and RFP, respectively, in U-2 OS cells. We validated the p53-hDM2 PPIB HCS assay with Nutlin-3, a compound that occupies the hydrophobic pocket on the surface of the N-terminus of hDM2 and blocks the binding interactions with 3 N-terminal amino acids (Phe19, Trp23, and Leu26) of p53.25,29–31 Nutlin-3 is a racemic mixture of 2 enantiomers that when separated with chiral columns have significantly (150-fold) different activities.9,31 Nutlin-3 disrupted the p53-hDM2 PPIB in a concentration-dependent manner and provided a robust, reproducible, and stable assay signal window that was compatible with HCS. The p53-hDM2 PPIB assay was readily implemented in HCS, and although we did not identify any novel p53-hDM2 disruptors in the 1,280 compounds of the LOPAC library, compounds that activated the p53 signaling pathway elicited biosensor signals that were clearly distinct from the responses of inactive compounds. Mitoxantrone, ellipticine, camptothecin, and topotecan exhibited concentration-dependent disruption of the p53-hDM2 PPIB, but they were much less potent than Nutlin-3 and the cellular images and quantitative HCS data for these compounds were not completely consistent with the Nutlin-3 response profile. We have recently screened 220,017 compounds from the NIH's small-molecule library in the p53-hDM2 PPIB assay and identified 3 structurally related methylbenzo-naphthyridin-5-amine (MBNA) hits with IC50s between 30 and 50 μM. In HCT116 cells expressing wild-type p53, the MBNAs enhanced p53 protein levels, increased the expression of p53 target genes, caused a p53-dependent cell cycle arrest in G1, induced apoptosis, and inhibited cell proliferation with an IC50 of ∼4 μM. Conducting an HCS campaign with the p53-hDM2 PPIB assay enabled us to discover a novel chemotype series that disrupts p53-hDM2 protein–protein interactions in cells.
Abbreviations
- ATM-CHK2
ataxia telangiectasia mutated-checkpoint kinase 2
- DMSO
dimethyl sulfoxide
- EP3
evolution-P3
- FITC
fluorescein iso-thiocyanate
- GFP
green fluorescent protein
- HCS
high-content screening
- hDM2
human homologue of murine double minute 2 protein
- LOPAC
Library of Pharmacologically Active Compounds
- MBNA
methylbenzo-naphthyridin-5-amine
- MCRAID-Ch3
mean circle ring average intensity difference in channel 3
- MRAI-Ch2
mean ring average intensity in channel 2
- MRAI-Ch3
mean ring average intensity in channel 3
- MT
molecular translocation
- NES
nuclear export sequence
- NLS
nuclear localization sequence
- PBS
phosphate-buffered saline
- PCA
protein fragment complementation assays
- PDE
phosphodiesterase
- PPI
protein-protein interaction
- PPIB
PPI biosensor
- RET
resonance energy transfer
- RFP
red fluorescent protein
- S:B
signal-to-background
- SCCPVF
selected object cell counts per valid field of view
- SPR
surface plasmon resonance
- TRITC
tetramethyl rhodamine iso-thiocyanate
- UPDDI
University of Pittsburgh Drug Discovery Institute
Acknowledgments
This work was supported by grants from the National Institutes of Health Molecular Library Screening Center Network (U54MH074411) (to J.S.L.), The Hartwell Foundation (to D.D.D.), and the Heinz Foundation (to J.S.L.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Colas P. High-throughput screening assays to discover small-molecule inhibitors of protein interactions. Curr Drug Discov Technol. 2008;5:190–199. doi: 10.2174/157016308785739875. [DOI] [PubMed] [Google Scholar]
- 2.Lalonde S. Ehrhardt DW. Loqué D. Chen J. Rhee SY. Frommer WB. Molecular and cellular approaches for the detection of protein-protein interactions: latest techniques and current limitations. Plant J. 2008;53:610–635. doi: 10.1111/j.1365-313X.2007.03332.x. [DOI] [PubMed] [Google Scholar]
- 3.Pagliaro L. Felding J. Audouze K. Nielsen SJ. Terry RB. Krog-Jensen C. Butcher S. Emerging classes of protein-protein interaction inhibitors and new tools for their development. Curr Opin Chem Biol. 2004;8:442–449. doi: 10.1016/j.cbpa.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Cummings M. Farnum MA. Nelen MI. Universal screening methods and applications of ThermoFluor. J Biomol Screen. 2006;11:854–863. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 5.Lievens S. Lemmens I. Tavernier J. Mammalian two-hybrids come of age. Trends Biochem Sci. 2009;34:579–588. doi: 10.1016/j.tibs.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angers S. Salahpour A. Bouvier M. Dimerization: an emerging concept for G protein-coupled receptor ontogeny and function. Annu Rev Pharmacol Toxicol. 2002;42:409–435. doi: 10.1146/annurev.pharmtox.42.091701.082314. [DOI] [PubMed] [Google Scholar]
- 7.Giuliano K. Taylor DL. Fluorescent-protein biosensors: new tools for drug discovery. Trends Biotechnol. 1998;16:135–140. doi: 10.1016/S0167-7799(97)01166-9. [DOI] [PubMed] [Google Scholar]
- 8.Giuliano K. Chen Y. Haskins JR. Fluorescent protein biosensors. Mod Drug Discov. 2003;8:33–37. [Google Scholar]
- 9.Giuliano K. Premkumar D. Taylor DL. Optimal characteristics of protein-protein interaction biosensors for cellular systems biology profiling. In: Haney S, editor. High Content Screening; Science, Techniques and Applications. John Wiley and Sons, Inc.; Hoboken, NJ: 2007. pp. 371–387. [Google Scholar]
- 10.Kerpoola T. Visualization of molecular interactions by fluorescence complementation. Nat Rev Mol Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li I. Pham E. Truong K. Protein biosensors based on the principle of fluorescence resonance energy transfer for monitoring cellular dynamics. Biotechnol Lett. 2006;28:1971–1982. doi: 10.1007/s10529-006-9193-5. [DOI] [PubMed] [Google Scholar]
- 12.Lundholt BK. Heydorn A. Bjorn SP. Praestegaard M. A simple cell-based HTS assay system to screen for inhibitors of p53-Hdm2 protein-protein interactions. Assay Drug Dev Technol. 2006;4:679–688. doi: 10.1089/adt.2006.4.679. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald M. Lamerdin J. Owens S. Keon BH. Bilter GK. Shang Z. Huang Z. Yu H. Dias J. Minami T. Michnick SW. Westwick JK. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat Chem Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 14.Michnick S. Ear PH. Manderson EN. Remy I. Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 15.Aylon YaO M. Living with p53, dying with p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Chipuk JE. Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13:994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- 17.Kastan MB. Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 18.Laureline R. Gilles G. Roux P. Control of cell migration: a tumor supressor function for p53. Biol Cell. 2006;98:141–152. doi: 10.1042/BC20050058. [DOI] [PubMed] [Google Scholar]
- 19.Sengupta S. Harris CC. p53: Traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 20.Teodoro JG. Evans SK. Green MR. Inhibition of tumor angiogenesis by p53: a new role for the guardian of the genome. J Mol Med. 2007;85:1175–1186. doi: 10.1007/s00109-007-0221-2. [DOI] [PubMed] [Google Scholar]
- 21.Voglestein B. Lane D. Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 22.Wiman KG. Strategies for therapeutic targeting of the p53 pathway in cancer. Cell Death Differ. 2006;13:921–926. doi: 10.1038/sj.cdd.4401921. [DOI] [PubMed] [Google Scholar]
- 23.Hu B. Gilkes DM. Chen J. Efficient p53 Activation and apoptosis by simultaneous disruption of binding to MDM2 and MDMX. Cancer Res. 2007;67:8810–8817. doi: 10.1158/0008-5472.CAN-07-1140. [DOI] [PubMed] [Google Scholar]
- 24.Hu BGD. Farooqi B. Sebti SM. Chen J. MDMX overexpression prevents p53 activation by the MDM2 inhibitor Nutlin. J Biol Chem. 2006;281:33030–33035. doi: 10.1074/jbc.C600147200. [DOI] [PubMed] [Google Scholar]
- 25.Schon O. Friedler A. Bycroft M. Freund SM. Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 26.Toledo F. Wahl GM. MDM2 and MDM4:p53 regulators as targets of anticancer therapy. Int J Biochem Cell Biol. 2007;39:1476–1482. doi: 10.1016/j.biocel.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wade MWE. Tang M. Stommel JM. Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281:33036–33044. doi: 10.1074/jbc.M605405200. [DOI] [PubMed] [Google Scholar]
- 28.Wu X. Bayle JH. Olson D. Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 29.Kussie PH. Gorina S. Marechal V, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 30.Vassilev LT. p53 Activation by small molecules: application in oncology. J Med Chem. 2005;48:4491–4499. doi: 10.1021/jm058174k. [DOI] [PubMed] [Google Scholar]
- 31.Vassilev LT. Vu BT. Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 32.Demma MJ. Wong S. Maxwell E. Dasmahapatra B. CP-31398 restores DNA-binding activity to mutant p53 in vitro but does not affect p53 homologs p63 and p73. J Biol Chem. 2004;279:45887–45896. doi: 10.1074/jbc.M401854200. [DOI] [PubMed] [Google Scholar]
- 33.Foster BA. Coffey HA. Morin MJ. Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507–2510. doi: 10.1126/science.286.5449.2507. [DOI] [PubMed] [Google Scholar]
- 34.Rippin TM. Bykov VJ. Freund SM. Selivanova G. Wiman KG. Fersht AR. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene. 2002;21:2119–2129. doi: 10.1038/sj.onc.1205362. [DOI] [PubMed] [Google Scholar]
- 35.Wang W. Wafik SE. Restoration of p53 to limit tumor cell growth. Curr Opin Oncol. 2008;20:90–96. doi: 10.1097/CCO.0b013e3282f31d6f. [DOI] [PubMed] [Google Scholar]
- 36.Wang W. Ho WC. Dicker DT, et al. Acridine derivatives activate p53 and induce tumor cell death through Bax. Cancer Biol Ther. 2005;4:893–898. doi: 10.4161/cbt.4.8.2134. [DOI] [PubMed] [Google Scholar]
- 37.Wang W. Takimoto R. Rastinejad F. El-Deiry WS. Stabilization of p53 by CP-31398 inhibits ubiquitination without altering phosphorylation at serine 15 or 20 or MDM2 binding. Mol Cell Biol. 2003;23:2171–2181. doi: 10.1128/MCB.23.6.2171-2181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnston P. Foster CA. Shun TY. Skoko JJ. Shinde S. Wipf P. Lazo JS. Development and implementation of a 384-well homogeneous fluorescence intensity high-throughput screening assay to identify mitogen-activated protein kinase phosphatase-1 dual-specificity protein phosphatase inhibitors. Assay Drug Dev Technol. 2007;5:319–332. doi: 10.1089/adt.2007.066. [DOI] [PubMed] [Google Scholar]
- 39.Johnston P. Soares KM. Shinde SN. Foster CA. Shun TY. Takyi HK. Wipf P. Lazo JS. Development of a 384-well colorimetric assay to quantify hydrogen peroxide generated by the redox cycling of compounds in the presence of reducing agents. Assay Drug Dev Technol. 2008;6:505–518. doi: 10.1089/adt.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston PA. Phillips J. Shun TY, et al. HTS identifies novel and specific uncompetitive inhibitors of the two-component NS2B-NS3 proteinase of West Nile Virus. Assay Drug Dev Technol. 2007;5:737–750. doi: 10.1089/adt.2007.101. [DOI] [PubMed] [Google Scholar]
- 41.Johnston PAFC. Tierno MB. Shun TY. Brummond KM. Wipf P. Lazo JS. Characterization of the Cdc25B dual specificity phosphatase inhibitor hits identified in a high throughput screen of the NIH compound library. Assay Drug Dev Technol. 2009;7:250–265. doi: 10.1089/adt.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nickischer D. Laethem C. Trask OJ, Jr., et al. Development and implementation of three mitogen-activated protein kinase (MAPK) signaling pathway imaging assays to provide MAPK module selectivity profiling for kinase inhibitors: MK2-EGFP translocation, c-Jun, and ERK activation. Methods Enzymol. 2006;414:389–418. doi: 10.1016/S0076-6879(06)14022-7. [DOI] [PubMed] [Google Scholar]
- 43.Trask OJ., Jr. Baker A. Williams RG, et al. Assay development and case history of a 32K-biased library high-content MK2-EGFP translocation screen to identify p38 mitogen-activated protein kinase inhibitors on the ArrayScan 3.1 imaging platform. Methods Enzymol. 2006;414:419–439. doi: 10.1016/S0076-6879(06)14023-9. [DOI] [PubMed] [Google Scholar]
- 44.Williams RG. Kandasamy R. Nickischer D, et al. Generation and characterization of a stable MK2-EGFP cell line and subsequent development of a high-content imaging assay on the Cellomics ArrayScan platform to screen for p38 mitogen-activated protein kinase inhibitors. Methods Enzymol. 2006;414:364–389. doi: 10.1016/S0076-6879(06)14021-5. [DOI] [PubMed] [Google Scholar]
- 45.Tovar CRJ. Filipovic Z. Higgins B. Kolinsky K. Hilton H. Zhao X. Vu BT. Qing W. Packman K. Myklebost O. Heimbrook DC. Vassilev LT. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gough AH. Johnston PA. Requirements, features and performance of high content screening platforms, in high content screening: a powerful approach to systems cell biology and drug discovery. Methods Mol Biol. 2006;356:41–61. doi: 10.1385/1-59745-217-3:41. [DOI] [PubMed] [Google Scholar]
- 47.Johnston P. Automated high content screening microscopy. In: Haney S, editor. High Content Screening. John Wiley and Sons, Inc.; Hoboken, NJ: 2008. pp. 25–42. [Google Scholar]
- 48.Tierno MBJP. Foster C. Skoko JJ. Shinde SN. Shun TY. Lazo JS. Development and optimization of high-throughput in vitro protein phosphatase screening assays. Nat Protoc. 2007;2:1134–1144. doi: 10.1038/nprot.2007.155. [DOI] [PubMed] [Google Scholar]
- 49.Zhang JH. Chung TD. Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 50.Kurose A. Tanaka T. Huang X. Halicka HD. Traganos F. Dai W. Darzynkiewicz Z. Assessment of ATM phosphorylation on Ser-1981 induced by DNA topoisomerase I and II inhibitors in relation to Ser-139-histone H2AX phosphorylation, cell cycle phase, and apoptosis. Cytometry A. 2005;68:1–9. doi: 10.1002/cyto.a.20186. [DOI] [PubMed] [Google Scholar]
- 51.Minotti G. Menna P. Salvatorelli E. Cairo G. Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 52.Zhao H. Traganos F. Darzynkiewicz Z. Phosphorylation of p53 on Ser15 during cell cycle caused by Topo I and Topo II inhibitors in relation to ATM and Chk2 activation. Cell Cycle. 2008;7:3048–3055. doi: 10.4161/cc.7.19.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peng YLC. Chen L. Sebti S. Chen J. Rescue of mutant p53 transcription function by ellipticine. Ongcogene. 2003;22:4478–4487. doi: 10.1038/sj.onc.1206777. [DOI] [PubMed] [Google Scholar]
- 54.Shi LMMT. Fan Y. O'Connor PM. Paull KD. Friend SH. Weinstein JN. Mining the National Cancer Institute Anticancer Drug Discovery Database: cluster analysis of ellipticine analogs with p53-inverse and central nervous system-selective patterns of activity. Mol Pharmacol. 1998;53:241–251. doi: 10.1124/mol.53.2.241. [DOI] [PubMed] [Google Scholar]
- 55.Xu G. Mawji IA. Macrae CJ. Koch CA. Datti A. Wrana JL. Dennis JW. Schimmer AD. A high-content chemical screen identifies ellipticine as a modulator of p53 nuclear localization. Apoptosis. 2008;13:413–422. doi: 10.1007/s10495-007-0175-4. [DOI] [PubMed] [Google Scholar]
- 56.Takeba Y. Kumai T. Matsumoto N. Nakaya S. Tsuzuki Y. Yanagida Y. Kobayashi S. Irinotecan activates p53 with its active metabolite, resulting in human hepatocellular carcinoma apoptosis. J Pharmacol Sci. 2007;104:232–242. doi: 10.1254/jphs.fp0070442. [DOI] [PubMed] [Google Scholar]
- 57.Zhang C. Zhu H. Yang X. Lou J. Zhu D. Lu W. He Q. Yang B. P53 and p38 MAPK pathways are involved in MONCPT-induced cell cycle G2/M arrest in human non-small cell lung cancer A549. J Cancer Res Clin Oncol. 2010;136:437–445. doi: 10.1007/s00432-009-0674-5. [DOI] [PubMed] [Google Scholar]