Abstract
The germline originates from primordial embryonic germ cells which give rise to sperm and egg cells and consequently, to the next generation. Germ cells of many organisms contain electron-dense granules that comprise RNA and proteins indispensable for germline development. Here we review recent reports that provide important insights into the structure and function of crucial RNA and protein components of the granules, including DEAD-box helicases, Tudor domain proteins, Piwi/Argonaute proteins and piRNA. Collectively, these components function in translational control, remodeling of ribonucleoprotein complexes and transposon silencing. Furthermore, they interact with each other by means of conserved structural modules and post-translationally modified amino acids. These data suggest a widespread use of several protein motifs in germline development and further our understanding of other ribonucleoprotein structures, for example, processing bodies and neuronal granules.
Germline cells and their granules: their components and functions
In many organisms germline (germ)cells form early during embryo development and eventually are specified to become sperm or egg. In this sense, germ cells are highly specialized. However, their union by fertilization results in a zygotic cell from which all other cell lineages are derived. As germ cells have the potential to generate an entire organism, they have been referred to as the ultimate totipotent stem cells [1].
A distinct feature of the germ cells is the presence of electron-dense ribonucleoprotein (RNP)organelles – the germ granules – that contain a number of identified proteins and RNAs that are required for germline development [2]. Germ granules have been described more than 100 years ago and are cytoplasmic non-membranous organelles with fibrous morphology (Box 1). Historically, these granules have been variously named in different organisms – for example, polar granules in flies [3], P granules in nematodes [4], germinal granules in frogs [5] and chromatoid bodies in mammals [5,6]. In addition, the term nuage (meaning cloud in French)is frequently used to describe germ granules [2,3,5,7,8]. Here, we use the term germ granules. Frequently, but not always, germ granules exhibit perinuclear localization or associate with mitochondria. Interestingly, at least some of the components of germ granules are present in electron-dense organelles of planarian somatic stem cells – neoblasts, [9] – thereby suggesting that the granules serve a functional role in maintenance of both germline and stem cells and highlighting the common features of both cell types [2].
Box 1. Germ granules: historical perspective
In the end of 19th century, Weismann proposed a theory of ‘The Continuity of the Germ plasm’ based on the idea that a special substance (germ plasm)is transferred from the parent germ cells to the germ cells of the next generation individual [92]. Subsequently, studying development of chrysomelid beetles, Hegner noticed dark granules at the posterior cytoplasm of the egg and when he destroyed the region containing these granules with a hot needle, the embryo failed to form germ cells [93,94]. Hegner called these granules ‘germ cell determinants’ and proposed that they are responsible for germline development. Later, Bounoure identified similar substance in the vegetal pole of amphibian eggs and also concluded that this substance is connected to specification of germ cells [95].
In the fruit fly Drosophila, ultraviolet (UV)irradiation of the egg posterior prevented germ cell formation [96]. However, when germ plasm from non-irradiated eggs was transplanted to the posterior pole of irradiated eggs, germ cells were able to form [97]. Germ plasm transplantation experiments [97,98] clearly demonstrated the instructive role of germ plasm in germ cell formation and suggested that germ granules at the posterior (polar granules) may be crucial for this process.
Electron microscopy of polar granules in Drosophila oocytes and early embryos revealed non-membranous organelles composed of 150–200 Å granules and a smaller fibrillar component [99]. These organelles were shown to be highly dynamic during germline development and frequently change their size, morphology, association with other organelles and composition. Based on their staining properties, polar granules were proposed to contain RNA and protein. In addition, ribosome clusters (polysomes)were found on polar granules of early embryos before and during germ cell formation and polysome-polar granule complexes were either absent or rarely observed after germ cells have been formed. Therefore, based on the dynamics of polar granule-ribosome association and on the presence of RNA in polar granules, Mahowald proposed that polar granules participate in translational control of mRNAs which leads to production of specific proteins required for germ cell formation [100].
Over 80 animals from rotifer to human in eight phyla have been reported to have germ granules in germ cells at various stages of development including primordial germ cells, oogonia and spermatogonia, oocytes and spermatocytes (reviewed in [101]). In virtually all these species, germ granules are cytoplasmic non-membranous and fibrous particles often associated with nuclear envelope or mitochondria.
Among the identified protein components of germ granules, three classes of proteins are particularly abundant: DEAD-box RNA helicases, Tudor (Tud)domain-containing proteins and Piwi proteins. These proteins interact with each other and also with RNA and these interactions are important for the formation of functional RNP assemblies. Here, we review the structural and functional features of the protein families and RNA molecules that constitute germ granules and highlight how their structural design contributes to a specific function required for germline development. In addition, we discuss studies that demonstrate the involvement of Tud domain proteins in the recognition of methylated arginines of Piwi proteins across species and the importance of these associated proteins and small Piwi-interacting RNAs (piRNAs)for the protection of germline DNA from mutations caused by mobilization of transposons. The available data suggest that functional germ granule assembly use a defined repertoire of protein-protein and protein-RNA interaction modules designed for highly dynamic particle remodeling during translational control, regulation of RNA stability and localization, all of which determine germline development.
DEAD-box RNA helicases
Structure and interactions - general
DEAD-box RNA helicases are a large family of proteins important for most steps of mRNA metabolism and transport [10,11] as well as acting in transcriptional regulation [12]. Members of this class of helicases share a central core that includes two tandem RecA-like domains with nine conserved motifs (Figure 1; [10,11]). This core is flanked by N- and C-terminal regions that are variable and, in most cases, only partially structured [13,14]. DEAD-box helicases bind cooperatively to ATP and RNA, and simultaneous binding of the two ligands results in the rearrangement of the two RecA- like domains from an ‘open’ conformation – or ensemble of conformations [14,15] – to a ‘closed’ one, with the RNA being trapped in the bound conformation. RNA and ATP binding involve residues in both domains and can be regulated by other proteins that either directly bind to the RNA interaction surface – as in the case of the RNA helicases DBP5 and eIF4A [14,16] – or stimulate the ATP binding activity [16]. In the DEAD-box helicase–RNA complexes, the backbone of the single-stranded RNA target is kinked, and this kink is generally accepted as being a major determinant of double-stranded (ds)RNA unwinding [17,18]. Additional determinants that have been reported to be important in dsRNA unwinding by DEAD-box helicases include the interaction of the RNA strand contacting the RecA-like domains with further domains/protein partners and a conformational rearrangement linked to ATP hydrolysis – whose role in dsRNA unwinding is still debated [13,19]. What are the consequences of the unwinding of RNA helices in RNP particles? Contrary to what is observed for genomic DNA, dsRNA helices are normally short – and, consistently, DEAD-box RNA helicases have been generally reported to be nonprocessive. dsRNA helices provide the majority of the basic building blocks of most known RNA structures and, in many cases alternative base pairing is possible within large RNAs [20]. Therefore, unwinding of RNA helices has the potential for an important remodeling effect on the 3D fold of RNA structures. The significant number of DEAD-box helicases important to the assembly and dissociation of large RNA–protein particles (for example, the ribosome, the exon junction complex and the germ granules)together with the structural consideration above indicate that these helicases are likely to perform a general role as RNA chaperones or architectural factors that, upon undergoing a change in their protein partners, remodel the RNP complexes. The importance and generality of DEAD-box helicases in building and reshaping large RNA–protein complexes is justified by the powerful combination of a simple and robust conformational switch and the capability to use different surface areas to interact with a large number of protein partners that regulate their activity (Figure 1; [14,16,21–23]).
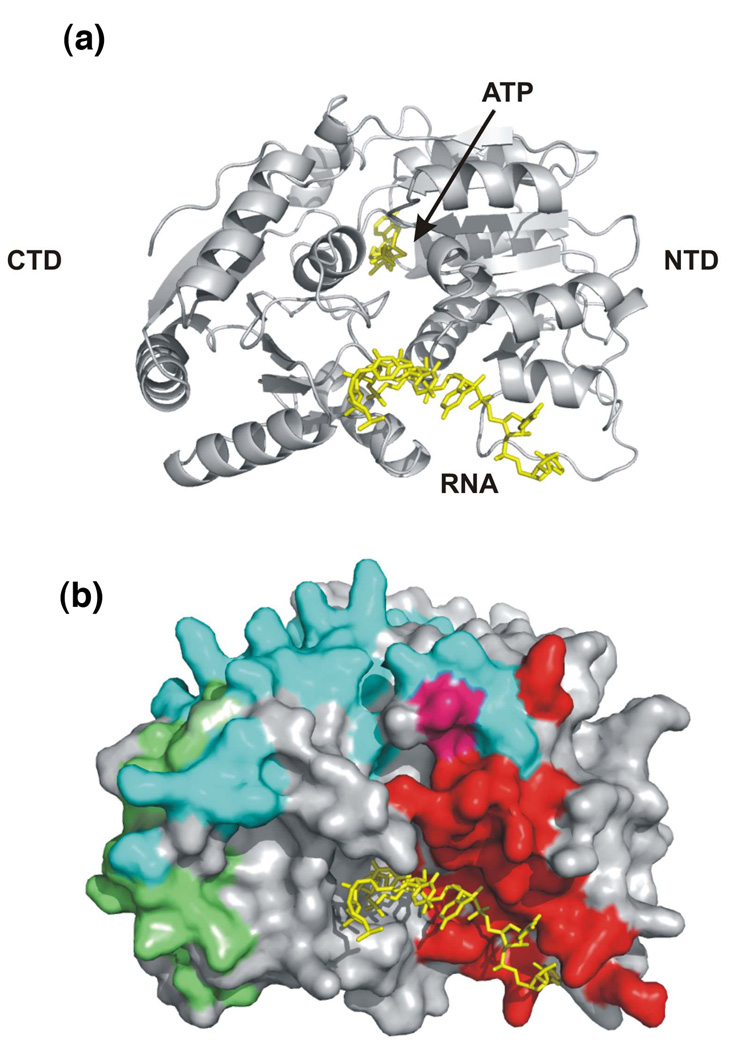
Figure 1.
Binding of different protein partners regulates the activity of DEAD-box RNA helicases. (a) Cartoon representation of the DEAD-BOX domains of DBP5 protein (grey): NTD and CTD are N-terminal and C-terminal RecA-like domains, respectively. The bound RNA and ATP are represented using yellow sticks (PDB ID: 3FHT; [14]). (b) Surface representation of the same molecule in the same orientation. The DBP5–NUP214 [14], Me31B–EDC3 [23] and eIF4AIII–MAGOH [21] (PDB IDs: 3FHC, 2WAX, 2HYI respectively)interaction surfaces are mapped on corresponding residues of DBP5 in red, green and cyan, respectively, with potentially overlapping residues mapped in a combination of the relevant colors. The figure highlights the existence of numerous, partially overlapping protein–protein (and protein–RNA)interaction surfaces of the DEAD-box helicase motif and the potential for a large number of alternative regulatory binding events. The inter-molecular surface was defined using the program InsightII (Accelerys), structural alignment of the different DEAD-box helicases was performed using the EBI DALI server [113], and the figures were created using the program Pymol (http://www.pymol.org).
Role of DEAD-box helicases in germline development
Several DEAD-box RNA helicases have been found in germ granules and implicated in granule assembly, primordial germ cell formation, oogenesis and spermatogenesis as specified below.
Vasa (Vas)helicase and its homologs are present in germ granules of various organisms, including nematode, fly, mouse and human [ 24]. In the fly, Vas is recruited by Oskar (Osk)protein during the formation of granules [25,26] and is necessary for their assembly [27]. Furthermore, Vas is required for oogenesis and primordial germ cell formation [17]. In addition to Osk, Vas has been shown to associate with other proteins [28–30], including the general translation initiation factor eIF5B [28]. This interaction is required for the development of the oocyte and for the accumulation of a transforming growth factor α (TGFα)homolog Gurken (Grk)protein in the oocyte cytoplasm – likely by activating grk mRNA translation – as well as for the formation of embryonic germ cells [28].
An important question is how germline helicases recognize their specific RNA targets. It is possible that RNA-binding protein partners or non-conserved helicase regions direct a helicase central core to its target. For example, a recent study has shown that Vas interacts specifically with a (U)-rich sequence in the 3´-untranslated region of mei-P26 mRNA and that a domain located outside of the enzymatic central core is important for target recognition [31]. Similar to Vas activation of grk mRNA translation, a Vas–eIF5B interaction has been implicated in the activation of translation of mei-P26 mRNA in the ovary, which leads to germline stem cell differentiation [31].
The elucidation of the structure of Vas in complex with an RNA and an ATP analog [18] was an important step in characterizing the basic principles of double-stranded RNA unwinding by DEAD-box helicases, but, contrary to other DEAD-box helicases from the germ granules (for example, Me31B and eIF4A – see below), no structural information exists on Vas interaction with its protein partners [17]. The structural characterization of Vas associated with its partners will be an important goal toward a better understanding of how helicases function and how their activity is regulated in germ granules.
Belle (Bel)helicase (Ded1p, An3, PL10, DBX, DBY)is closely related to Vas [32,33], and mutations in a human member of the Bel subfamily of helicases, DBY, have been associated with male infertility caused by substantial reduction or absence of germ cells [34]. Similarly, fly bel mutants are male-sterile and show defects in meiotic progression during spermatogenesis [32]. In addition, Bel is required for female fertility in flies and in bel mutants the majority of egg chambers stop development at mid-oogenesis stages and degenerate [32]. Bel colocalizes with Vas in germ granules of Drosophila female germline cells [32].
Similar to Vas and Bel, Me31B [DDX6, p54, Dhh1p, Cgh-1, RCK] helicase has been identified as a bona fide component of germ granules and me31b mutation causes reduction in germ cell number in embryos from females heterozygous for mutations in vas or tud genes [2,35]. Me31B is also present in Processing bodies (P-bodies)– cytoplasmic granules that are sites of translational repression and mRNA decay [36,37] – where it has been implicated in translational repression, mRNA decapping and the assembly of the P-bodies themselves [23,38,39]. In Drosophila egg chambers, Me31B functions to repress premature translation of osk and Bicaudal-D (BicD)mRNAs [40]. Recent structural and molecular work has shown that the decapping activator EDC3 and the translational repressor Tral bind to the C-terminal RecA-like domain of Me31B in a mutually exclusive manner, thereby explaining the Me31B-mediated switch between mRNA decapping and translational repression [23,38,39].
eIF4A helicase has been studied extensively owing to its role as a general translation initiation factor. During regular translation, it associates with eIF4E and eIF4G to form the eIF4F complex, which recognizes the 5´ cap of mRNAs [16]. Free eIF4A is a very inefficient helicase. However, it is activated by accessory binding proteins, including eIF4B, eIF4E, eIF4G and eIF4H [16]. Both eIF4A RecA-like domains have been shown to interact with regulatory proteins [16]. In Drosophila, eIF4A has been isolated as a component of germline Vas and Tud protein complexes and is localized to germ granules [35]. In addition, in genetic interaction experiments, embryos from females doubly heterozygous for mutations in eIF4a and tud or in eIF4a and vas genes were more defective in germ cell formation than embryos from any of these single heterozygotes [35].
The documented presence of several DEAD-box helicases in the germ granules and their crucial role in germline development discussed above suggest that the morphology and function of the granules are dependent on active structural reshaping of RNA and RNA–protein complexes. This reshaping is likely to be controlled by other proteins that interact with the helicases and regulate both their activity and their RNA target specificity. However, the majority of these helicase partners and molecular mechanisms of this regulation are still unknown. Therefore, it will be important to identify the helicase regulatory proteins, their functional involvement and RNA targets. In addition, future studies should shed light on a specific functional outcome of a given helicase – helicase partner – RNA complex, which may include translational activation or repression and effect on RNA stability or distribution.
Tudor domain-containing proteins
Structure and interactions - general
Similar to DEAD-box RNA helicases, Tud-domain-containing proteins have been shown to localize to germ granules of several organisms, including Drosophila, zebrafish and mouse, and mutations in the genes encoding these proteins cause defects in germline development [41–49].
The Tud domain is a small (50–55 amino acids)module that engages in protein–protein interactions. The structures of Tud domains from several proteins have been solved [50–55] and have revealed a β-barrel structure that encases a pocket lined with aromatic amino acids (Figure 2). This aromatic cage has been shown to interact specifically with methylated amino acids – either symmetrically dimethylated arginines (sDMAs, arginines where each of the two NH2 moieties of the guanidinium group carries a methyl group)or methylated lysines – of target proteins. Interestingly, Tud domains have been found both in isolation and embedded in a number of different architectures, i.e. within double Tud domains, joined to non-Tudor domains [52] and sporting additional structural elements [51,56]. The architectural plasticity of the small Tud domain expands its potential for a specific recognition of target proteins.
Figure 2.
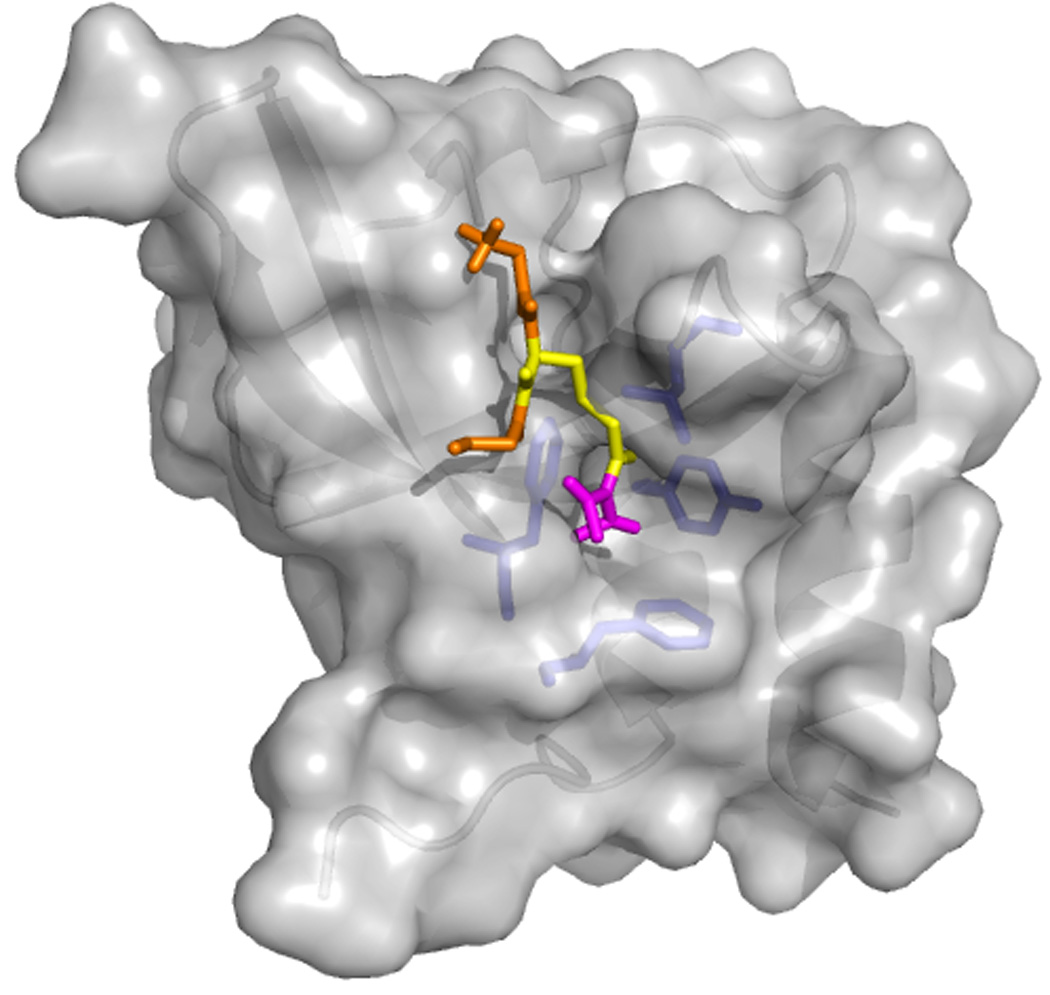
Tudor domains bind to methylated amino acids by using a conserved aromatic cage. A crystal structure of a Tudor domain – sDMA peptide is currently not available. Above is a surface representation of the crystal structure of human germline Tdrd2/Tdrkh Tudor domain with an sDMA-GRG peptide docked in the aromatic cage (adapted from [51]). The protein is in grey, with the side chains of the residues of the cage (L364, Y371, F388, F391)in blue. The peptide is in orange (Gly)and yellow (sDMA)with the Dimethyl-guanidinium group in magenta.
The majority of structural data on Tud domains have been obtained for proteins that are not germline-specific and that are responsible for the interaction with methylated amino acids during spliceosome assembly or recognition of histone tails [50,52,53,55,57]. However, a recent study on the structure of the human germline-specific Tud-domain-containing protein, Tdrd2/Tdrkh - that interacts specifically with arginine-methylated Miwi - has revealed that its Tud domain is structurally similar to non-germline Tud-domain structures and has an aromatic cage suitable for interaction with the sDMA (Figure 2; [51]; see below). Mutating the residues of the aromatic cage results in loss of Tdrkh-Miwi interaction. This example strengthens the case for a common Tud-domain – sDMA mode of recognition in non-germline and germline proteins.
The structural rationale for the choice of a specific methylated partner by Tud domain proteins is still unclear and it seems likely that multiple interactions and the larger structural context are key to specificity. Germline Tud-domain proteins contain varying number of Tud domains (Table 1). Drosophila SpnE, Ovarian tumor (Otu)and Krimp/Mtc each have one domain, whereas Tud protein contains eleven domains. In humans, nine Tud-domain-containing proteins are selectively expressed in germline cells (spermatocytes), with the number of Tud domains ranging from one (in Akap1, Tdrd5, Tdrd2/Tdrkh, Tdrd8/Stk31 and Tdrd9)to eight (in Tdrd6; [51,56]). Also, several of the Piwi targets of Tud proteins described below have potentially more than one methylation site. It has been proposed that the density of methylation sites could regulate the recruitment of the Tud-domain-containing target by allowing multiple Tud-domain-sDMA binding events within a single di-molecular interaction [51]. Alternatively the different Tud domains could interact with different methylated proteins [51], building inter-molecular bridges.
Table 1.
Interactions of germline Tudor domain-containing proteins with Piwi proteins
| Tudor domain- containing proteins |
Organism | Number of Tudor domains |
Other domains present in Tudor proteinsa |
Interacting Piwi protein |
References |
|---|---|---|---|---|---|
| Tudor | Fruit fly | 11 | Not found | Aubergine, Ago3 |
[35,69,80,83] |
| SpnE | Fruit fly | 1 | DExH, HA | Not found | [111] |
| Krimp/Mtc | Fruit fly | 1 | Not found | Not found | [7,112] |
| Otu | Fruit fly | 1 | OTU | Not found | [59,60] |
| Tejas | Fruit fly | 1 | Tejas | Aubergine | [64] |
| Akap1 | Mammalsb | 1 | KH type 1 | Not found | [51,56] |
| Tdrd5 | Mammals | 1 | Tejas | Not found | [51,56,64] |
| Tdrd8/Stk31 | Mammals | 1 | Ser/Thr PK-like | Miwi | [51,56] |
| Tdrd9 | Mammals | 1 | DExH, HA | Miwi, Mili, Miwi2 |
[51,56,68] |
| Tdrd2/Tdrkh | Mammals | 1 | KH type 1 | Miwi, Mili, Miwi2 |
[51,56,68] |
| Tdrd4/Rnf17 | Mammals | 5 | RING | Miwi | [51,56,68] |
| Tdrd7 | Mammals | 3 | Tejas | Miwi | [51,56,64,68] |
| Tdrd1 | Mammals | 4 | MYND | Miwi, Mili, Miwi2 |
[46,51,56,68,80, 81,84] |
| Tdrd6 | Mammals | 8 | Not found | Miwi, Mili | [51,56,67–69] |
| Xtr | Frog | 7 | vWA | Xiwi | [58,80,82] |
DExH – RNA helicase DExH family; HA – Helicase-associated domain; OTU – Ovarian Tumor-related putative cysteine proteases; Tejas – novel ~140 amino acid domain with unknown function [64]; KH type 1 – K Homology RNA-binding domain type 1; Ser/Thr PK-like – serine/threonine protein kinase-like domain; RING – RING-type zinc finger; MYND – MYND (myeloid, Nervy, DEAF-1)-type zinc finger; vWA – von Willebrand factor type A domain (putative protein-protein interaction module).
Germline Tud domain proteins have been identified in many organisms, including planarian flatworms (Spoltud-1 and Spoltud-2; [9]), zebrafish (Tdrd7; [47])and frog (Xtr; [58])and it is likely that Tud domain proteins play an important role in establishing the network of protein–protein interactions necessary for the assembly and regulation of germ granules throughout evolution.
Tudor domains and germline development
A Drosophila gene encoding the prototypic Tud protein was named after the royal Tudor dynasty, which did not leave an heir after the death of Queen Elizabeth I in 1603. In flies, tud mutant females produce embryos that do not form primordial germ cells and develop into sterile adults. Therefore, similar to the case for vas, the strong tud mutant alleles exhibit ‘grandchildless’ phenotype [41,43,48].
The Drosophila otu transcript can be alternatively spliced to generate 98- and 104-kDa proteins [59,60]. Interestingly, the larger isoform includes a Tud domain and the expression of its mRNA is detected in the early stages of oogenesis [60]. Mutations affecting the amount of Otu104 or its activity lead to the development of tumorous egg chambers [59] and cause defects in dorsoventral polarity of the egg [60]. Three other fly Tud domain proteins, SpnE, Krimp/Mtc and Tejas are components of perinuclear nuage in the nurse cells of the fly egg chamber and function in the repression of retrotransposons in the germline [7,61–65].
In mouse, knockout mutations in tdrd1, rnf17/tdrd4and tdrd6 affect germ granule assembly and spermatogenesis [44,66,67]. In addition, similar to Drosophila SpnE, Krimp and Tejas, Tdrd1 is also involved in protection of germ cells from transposons. In particular, tdrd1 mutants exhibit derepression of L1 transposons in germ cells and show an abnormal profile of piRNAs, that have been implicated in transposon silencing [46,68]. Contrary to the tdrd1 mutant phenotype, tdrd6 mutant mice do not show significant activation of retrotransposons [67]. Similar to the tdrd6 mutant, the Drosophila tud mutant failed to show derepression of many transposons in the ovary with the exception of the Blood transposon [7,69]. These data indicate that different Tud domain proteins regulate production of different subsets of piRNAs which may result in silencing of specific transposons.
In zebrafish, the function of the Tud-domain protein Tdrd7 has been explored using antisense morpholino oligonucleotides that inhibit Tdrd7 translation [47]. Abnormal morphology of germ granules was observed in the embryos injected with Tdrd7 morpholino, although these embryos developed into fertile adults.
A Tud-domain-containing protein, named Spoltud-1, has been recently identified in the planarian species Schmidtea polychroa [9]. Interestingly, Spoltud-1 is expressed not only in germ cells but also in neoblasts, which are somatic totipotent stem cells. In neoblasts, Spoltud-1 is localized to organelles that resemble germ granules and is needed for long-term self-renewal of these stem cells. The specific localization of Tud domain proteins to germ-granule-like organelles in both germ cells and somatic stem cells supports the concept of there being functional similarity and a close relationship between stem cells and germ cells.
Tud domain proteins have been implicated in germ granule assembly, germ cell formation and protection of germline DNA from transposons. Molecular details of Tud domain-mediated interactions with methylated Piwi proteins have recently become available (see below), however, as discussed above, the rationale for the specificity of germline Tud domain proteins for their methylated targets is unclear. Defining the sets of simultaneous multiple interactions undertaken by each Tud protein is a possible key to the understanding of its target specificity.
Piwi proteins in germ granules
Recognition by Tudor domains
Piwi proteins belong to a subclass of the Argonaute protein family, members of which bind to small piRNAs [70–73]. Argonaute proteins possess four domains: the amino terminal domain and the PAZ, MID and PIWI domains [74,75]. The PAZ domain has been implicated in the binding to small guide single-stranded nucleic acids such as piRNAs, which base-pair with complementary target RNAs. The PIWI domain resembles RNAse H enzymes and is involved in the cleavage of target RNAs. Indeed, Drosophila Piwi proteins Piwi, Aubergine (Aub)and Ago3 cleave target RNAs in vitro [76,77]. Piwi proteins are expressed in germline cells of many organisms (reviewed in [73]). Piwi proteins and piRNAs repress activation of transposons and, therefore, protect the germline DNA from harmful mutations caused by their mobilization. In particular, different Piwi proteins are engaged in a cycle of piRNA production (‘ping-pong’ cycle)and in germ granules, Piwi protein-associated antisense piRNAs bind transposon mRNAs which are subsequently cleaved and inactivated (Figure 3)
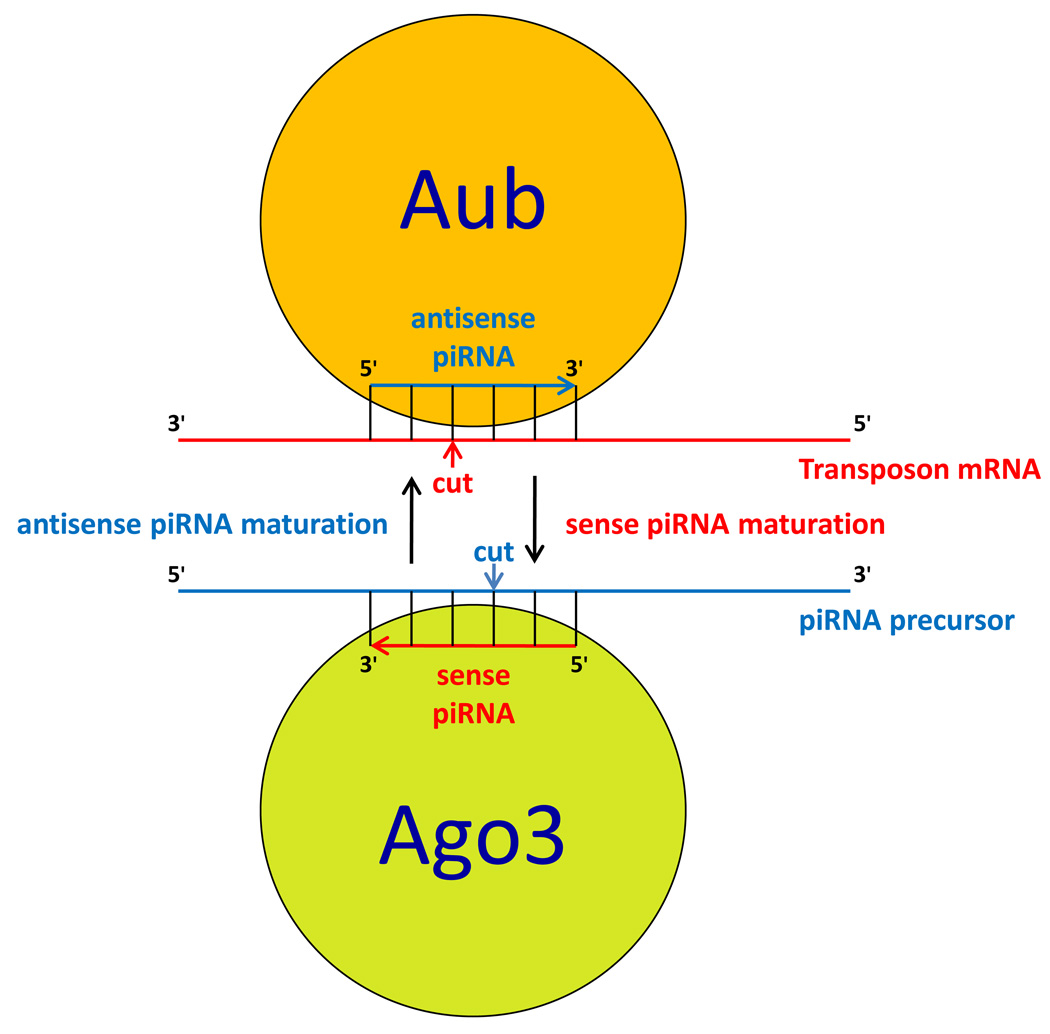
Figure 3.
Ping-pong model of transposon silencing by Piwi family proteins in germ granules. Mechanistic details for Drosophila Piwi proteins Aub and Ago3 localized in perinuclear nuage are shown, however, mammals have similar mechanism [71,73,114]. Transposon mRNA exported from the nucleus is recognized by antisense piRNA bound to Aub and then cleaved between nucleotides complementary to nucleotides 10 and 11 of the antisense piRNA. After additional processing of the cleaved RNA, it becomes a sense piRNA which, in association with Ago3, base-pairs with piRNA precursor and, similar to Aub/antisense piRNA complex, cleaves that precursor to generate another antisense piRNA. In mammals, during the ping-pong cycle, antisense and sense piRNAs are bound to Miwi2 and Mili respectively [114]. In addition to transposon mRNA degradation in cytoplasmic germ granules, some Piwi proteins have been implicated in epigenetic silencing of transposon transcription in the nucleus by promoting heterochromatin formation or DNA methylation [73,114].
Piwi proteins have been implicated in stem cell maintenance and are crucial for germline development and fertility in a variety of organisms, including Drosophila, zebrafish and mouse. Furthermore, in the planarian flatworm Schmidtea mediterranea, Piwi proteins are present in neoblasts and required for regeneration [78,79].
Recent studies in different model systems have highlighted that the different Piwi proteins are methylated by protein methyltransferases (PRMTs). In Drosophila, the methyltransferase homolog of PRMT5, dPRMT5 (Capsuléen/Dart5), methylates the Piwi proteins Piwi, Aub and Ago3, generating sDMAs [80]. As described above, Tud domains from non-germline proteins are known to interact with sDMAs [52,57], and several groups have recently provided evidence that Tud-domain-containing proteins interact with methylated Piwi proteins in the germline of mouse, frog and fly, thereby demonstrating the evolutionary conservation of germline Tud–Piwi interactions (Table 1; [35,46,51,67–69,81–84]). For example, Drosophila Aub binds to Tud protein, and sDMAs are crucial for this interaction [69,83]. Interestingly, dPRMT5 forms a complex with its protein cofactor MEP50/WD45/Valois and Tud protein [85,86], indicating that, in germ granules, Piwi proteins are available for immediate binding to Tud domains upon their methylation. Similar to the case for tud mutants, mutations in dPRMT5 and valois lead to the failure to form primordial germ cells [85–87], highlighting the functional significance of PRMT5-induced methylation for germline development.
Importance for assembly of germ granules
Piwi–Tud-domain protein interactions are important for the assembly of functional germ granules in different organisms. In the Drosophila tud1 mutant, Aub localization in the oocyte germ plasm – the specialized cytoplasm that harbours germ granules (polar granules)– is disrupted, indicating that Tud protein is required to integrate Aub into the granules [69]. Conversely, in a dPRMT5 (methyltransferase gene)mutant, Tud localization to ovarian germ granules (nuage and polar granules)is dramatically reduced [86,87]. In mouse, Tud domain protein Tdrd1 mainly localizes to the nuage organelle containing the Piwi protein Mili, and a mutation in mili causes Tdrd1 to lose nuage localization in prospermatogonia and to become evenly distributed in the cytoplasm [68]. Conversely, in tdrd1 mutants, the Piwi protein Miwi2 failed to localize to nuage during spermatogenesis [68].
The evolutionary conservation of Tud – Piwi interaction mediated by sDMAs indicates that a coordinated methylation of Piwi and perhaps other unknown proteins provides a highly efficient mechanism to quickly form a functional silencing complex to protect DNA of future generations from transposons. Biochemical characterization and structure as well as genetic analysis of this complex will be an exciting and challenging area of future research.
Concluding remarks: how to build and maintain a germ granule?
In the past few years, a number of molecular studies have established the role of DEAD-box helicases as a general architectural factor that can be regulated by diverse protein–protein interactions during RNA metabolism. Tud-domain-containing proteins are also general players in RNA metabolism, and recent data from a number of groups have demonstrated that their interactions with methylated Piwi proteins in the germ granules are essential for the assembly of these RNP particles. Furthermore, recent studies have shown that in Drosophila germ granules Tud protein and the DEAD-box helicase Vas form a complex with each other and additional DEAD-box RNA helicases, namely eIF4A and Me31B [35,88]. Also, Vas has been shown to associate with Piwi and Aub [35,89], and a mouse Vas homolog Mvh interacts with Piwi proteins [68,88,90] and Tud-domain-containing proteins Tdrd1 and Tdrd6 [67,88]. Together, these observations indicate that three classes of proteins – DEAD-box helicases, Tud domain proteins and Piwi proteins – associate within germ granules from different organisms to build the network of interactions necessary for the granules’ assembly and reshaping (Figure 4). We currently lack the molecular details of the interactions between germline DEAD-box helicases and Piwi and Tud domain proteins, but our general molecular understanding of DEAD-box helicase–RNA and Tud-domain–Piwi interactions, that is summarized above, indicates that molecular recognition in germ granules is mediated by multiple interactions, each contributing a fraction of the total affinity and specificity. This recognition model is common to many multi-domain proteins involved in different steps of RNA metabolism, including KH and RRM containing proteins, and is thought to favour reversible interactions [91].
Figure 4.
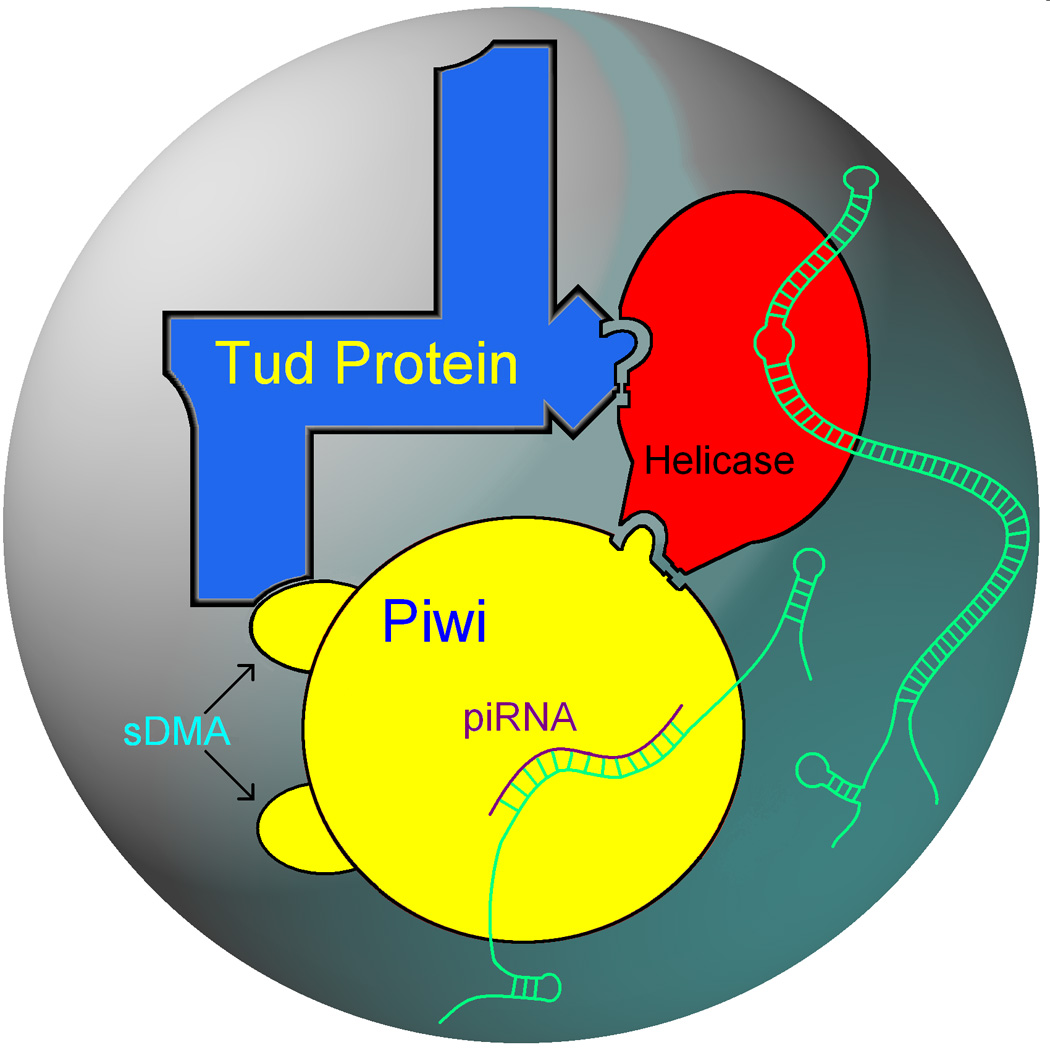
Building a germ granule. DEAD-box RNA helicases associate with Tud-domain proteins and Piwi proteins. Also, symmetrically dimethylated arginines (sDMAs)of Piwi proteins are recognized by Tud domains. These intermolecular interactions are crucial for the assembly of the functional germ granule structure. While associations between helicases and Piwi proteins and between helicases and Tud-domain proteins have been established, molecular and structural details of these interactions are not known. Helicases reshape RNA–protein complexes (RNA is indicated in green), facilitating post-transcriptional control of expression of germline genes at the level of RNA stability and translation. In addition, helicases may restructure RNAs so that they can provide a scaffold for additional germ granule components, either RNA or proteins. Piwi proteins and piRNAs guard germ cells against transposons which involves recognition of transposon mRNA by antisense piRNA as well as cleavage and eventual degradation of transposon mRNA (Figure 3). Indicated interactions and transposon silencing by Piwi-piRNA complexes have been shown in flies and mammals, demonstrating a remarkable conservation of structural principles of germ granule assembly and function throughout evolution.
Importantly, architectural factors are major components of germ granules, and are likely to play a crucial role in their regulation. RNPs are highly dynamic bodies: in germ granules this dynamic behaviour manifests itself not only in the assembly and disassembly of the particle but also in the regulation of the translation of its mRNA components (see Box 2). Future research will face the difficult but important tasks to define the precise composition of germ granules and their variations, to analyse the biophysical/structural features of the protein-protein and protein-RNA recognition events described above (including the different protein-protein interactions by DEAD-box helicases), to image the different components within germ granules during the evolution of these particles and finally to link this molecular information to the functional framework. This will require the concerted use of very diverse tools of investigation – from structural techniques to genetics. Furthermore, it will require a very challenging biochemical characterization of functionally defined but dynamic protein-RNA particles. The results of these studies will clarify rules and commonalities in the mechanisms of RNP generation and restructuring.
Box 2. RNA component of the germ granules
Drosophila germ granules and their equivalent organelles in other species comprise a heterogeneous population of RNA molecules, including short unstructured regulatory RNAs (e.g. piRNAs), mRNAs and mitochondrial ribosomal RNAs [70–73,102–104].
piRNAs are important to regulate genome stability [70] and germ granules have been proposed to both act as a vehicle to carry piRNAs from one generation to the next, which is necessary to maintain the piRNA population prior to transcription of their precursors [73], and be sites of piRNA biogenesis [71] (Figure 3). Germ granules also contain a large pool of mRNAs with important functions in development (e.g. nanos, pgc and gcl)[103]. These RNAs are differentially regulated, and it has been proposed that their translational state varies with time without the mRNA(s)dissociating from the germ granule assembly [103]. This is consistent with the observation that ribosomal structures are present on the germ granules [102]. Further, it has recently been shown that the time regulation of the translational state of the above mRNAs is determined by information encoded in the partially structured mRNA 3´-untranslated region (3´UTR)[103].
The 3´UTR contains the targets of most proteins regulating mRNA transport and metabolism, as well as translation, and can fulfill a regulatory role not only in cis but also in trans [105,106]. The length of the 3´UTR varies from a few hundreds to many thousands of nucleotides and regulatory proteins recognize both sequence and structural features [107]. However, although secondary structure prediction and structural probing have confirmed the existence of structured regions within a number of 3´UTRs and defined the domain structure of a few 3´UTRs (e.g. oskar and bicoid)[108–110], we have no definitive information on the global fold of these large regulatory sections of the mRNAs. Indeed, it is unclear if a definitive global fold for a 3´UTR can be obtained in the absence of its protein partners, as their association with the mRNA could change the mRNA fold. DEAD-box helicases have been shown to change the local RNA structure, and to be associated with the 3´UTR in germ granules, and represent a potential tool in the regulation of the 3´UTR conformation. Understanding whether and how the local architectural changes in the RNA caused by binding of DEAD-box helicases are translated in changes of the global fold of the bound 3´UTR will have to wait for structural information on 3´UTR holocomplexes. However, it seems possible that the cell takes advantage of the possibility to regulate the access and binding of the different protein effectors to the different regions/domains within a 3´UTR by changing the global fold of the RNA. In this case, the reshaping of the 3´UTR structure would represent an important additional regulatory level for the assembly of the large functional units that control the mRNA translational state and positioning.
Acknowledgements
We thank Jinrong Min for providing the coordinates of the Tdrd2/Tdrkh Tudor domain structure and the docked peptide. Also, we thank Dan Varonin for his help with generating Figures 3 and 4, T. Michael Creed and Christina A. Jackson for critical reading of the manuscript. Support for research in the A.L.A. laboratory is provided by the NIH grant award R15GM087661 from the National Institute of General Medical Sciences, and work in the A.R. laboratory is supported by the MRC (U117574558)and the Wellcome Trust (WT022088MA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cinalli RM, et al. Germ cells are forever. Cell. 2008;132:559–562. doi: 10.1016/j.cell.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127:891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 3.Mahowald AP. Assembly of the Drosophila germ plasm. Int. Rev. Cytol. 2001;203:187–213. doi: 10.1016/s0074-7696(01)03007-8. [DOI] [PubMed] [Google Scholar]
- 4.Updike D, Strome S. P granule assembly and function in Caenorhabditis elegans germ cells. J. Androl. 2010;31:53–60. doi: 10.2164/jandrol.109.008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kloc M, et al. The Balbiani body and germ cell determinants: 150 years later. Curr. Top. Dev. Biol. 2004;59:1–36. doi: 10.1016/S0070-2153(04)59001-4. [DOI] [PubMed] [Google Scholar]
- 6.Yokota S. Historical survey on chromatoid body research. Acta Histochem. Cytochem. 2008;41:65–82. doi: 10.1267/ahc.08010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snee MJ, Macdonald PM. Live imaging of nuage and polar granules: evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J. Cell Sci. 2004;117:2109–2120. doi: 10.1242/jcs.01059. [DOI] [PubMed] [Google Scholar]
- 9.Solana J, et al. Spoltud-1 is a chromatoid body component required for planarian long-term stem cell self-renewal. Dev. Biol. 2009;328:410–421. doi: 10.1016/j.ydbio.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linder P. Dead-box proteins: a family affair - active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linder P, et al. Birth of the D-E-A-D box. Nature. 1989;337:121–122. doi: 10.1038/337121a0. [DOI] [PubMed] [Google Scholar]
- 12.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Campo M, Lambowitz AM. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol. Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.von Moeller H, et al. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 15.Theissen B, et al. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. USA. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marintchev A, et al. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linder P, Lasko P. Bent out of shape: RNA unwinding by the DEAD-box helicase Vasa. Cell. 2006;125:219–221. doi: 10.1016/j.cell.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 18.Sengoku T, et al. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 19.Karow AR, Klostermeier D. A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res. 2009;37:4464–4471. doi: 10.1093/nar/gkp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Andersen CB, et al. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 22.Bono F, et al. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Tritschler F, et al. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol. Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Raz E. The function and regulation of vasa-like genes in germ-cell development. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-3-reviews1017. REVIEWS1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breitwieser W, et al. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 1996;10:2179–2188. doi: 10.1101/gad.10.17.2179. [DOI] [PubMed] [Google Scholar]
- 26.Ephrussi A, Lehmann R. Induction of germ cell formation by oskar. Nature. 1992;358:387–392. doi: 10.1038/358387a0. [DOI] [PubMed] [Google Scholar]
- 27.Schupbach T, Wieschaus E. Maternal-effect mutations altering the anterior-posterior pattern of the Drosophila embryo. Roux's Arch. Dev. Biol. 1986;195:302–317. doi: 10.1007/BF00376063. [DOI] [PubMed] [Google Scholar]
- 28.Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- 29.Liu N, et al. Fat facets interacts with vasa in the Drosophila pole plasm and protects it from degradation. Curr. Biol. 2003;13:1905–1909. doi: 10.1016/j.cub.2003.10.026. [DOI] [PubMed] [Google Scholar]
- 30.Styhler S, et al. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev. Cell. 2002;3:865–876. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- 31.Liu N, et al. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3' UTR. Genes Dev. 2009;23:2742–2752. doi: 10.1101/gad.1820709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnstone O, et al. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 2005;277:92–101. doi: 10.1016/j.ydbio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Salinas LS, et al. The DEAD box RNA helicase VBH-1 is required for germ cell function in C. elegans. Genesis. 2007;45:533–546. doi: 10.1002/dvg.20323. [DOI] [PubMed] [Google Scholar]
- 34.Foresta C, et al. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum. Mol. Genet. 2000;9:1161–1169. doi: 10.1093/hmg/9.8.1161. [DOI] [PubMed] [Google Scholar]
- 35.Thomson T, et al. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech. Dev. 2008;125:865–873. doi: 10.1016/j.mod.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 37.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minshall N, et al. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell. 2009;20:2464–2472. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tritschler F, et al. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Cell Biol. 2008;28:6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura A, et al. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 41.Arkov AL, et al. The role of Tudor domains in germline development and polar granule architecture. Development. 2006;133:4053–4062. doi: 10.1242/dev.02572. [DOI] [PubMed] [Google Scholar]
- 42.Bardsley A, et al. Distribution of tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development. 1993;119:207–219. doi: 10.1242/dev.119.1.207. [DOI] [PubMed] [Google Scholar]
- 43.Boswell RE, Mahowald AP. tudor, a gene required for assembly of the germ plasm in Drosophila melanogaster. Cell. 1985;43:97–104. doi: 10.1016/0092-8674(85)90015-7. [DOI] [PubMed] [Google Scholar]
- 44.Chuma S, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc. Natl. Acad. Sci. USA. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosokawa M, et al. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 46.Reuter M, et al. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 47.Strasser MJ, et al. Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Dev. Biol. 2008;8:58. doi: 10.1186/1471-213X-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomson T, Lasko P. Drosophila tudor is essential for polar granule assembly and pole cell specification, but not for posterior patterning. Genesis. 2004;40:164–170. doi: 10.1002/gene.20079. [DOI] [PubMed] [Google Scholar]
- 49.Thomson T, Lasko P. Tudor and its domains: germ cell formation from a Tudor perspective. Cell Res. 2005;15:281–291. doi: 10.1038/sj.cr.7290297. [DOI] [PubMed] [Google Scholar]
- 50.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl. Acad. Sci. USA. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friberg A, et al. Structure and ligand binding of the extended Tudor domain of D. melanogaster Tudor-SN. J. Mol. Biol. 2009;387:921–934. doi: 10.1016/j.jmb.2009.02.018. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, et al. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 54.Ramos A, et al. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction. Structure. 2006;14:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 55.Selenko P, et al. SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- 56.Jin J, et al. Eukaryotic protein domains as functional units of cellular evolution. Sci. Signal. 2009;2:ra76. doi: 10.1126/scisignal.2000546. [DOI] [PubMed] [Google Scholar]
- 57.Cote J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J. Biol. Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 58.Hiyoshi M, et al. Involvement of Xtr (Xenopus tudor repeat) in microtubule assembly around nucleus and karyokinesis during cleavage in Xenopus laevis. Dev. Growth Differ. 2005;47:109–117. doi: 10.1111/j.1440-169x.2005.00787.x. [DOI] [PubMed] [Google Scholar]
- 59.Steinhauer WR, Kalfayan LJ. A specific ovarian tumor protein isoform is required for efficient differentiation of germ cells in Drosophila oogenesis. Genes Dev. 1992;6:233–243. doi: 10.1101/gad.6.2.233. [DOI] [PubMed] [Google Scholar]
- 60.Van Buskirk C, Schupbach T. Half pint regulates alternative splice site selection in Drosophila. Dev. Cell. 2002;2:343–353. doi: 10.1016/s1534-5807(02)00128-4. [DOI] [PubMed] [Google Scholar]
- 61.Aravin AA, et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 62.Klenov MS, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35:5430–5438. doi: 10.1093/nar/gkm576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lim AK, et al. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J. Cell Biol. 2009;186:333–342. doi: 10.1083/jcb.200904063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patil VS, Kai T. Repression of Retroelements in Drosophila Germline via piRNA Pathway by the Tudor Domain Protein Tejas. Curr. Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- 65.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 66.Pan J, et al. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development. 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vasileva A, et al. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr. Biol. 2009;19:630–639. doi: 10.1016/j.cub.2009.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirino Y, et al. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA. 2010;16:70–78. doi: 10.1261/rna.1869710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- 71.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 72.Klattenhoff C, et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouasker S, Simard MJ. Structural biology: Tracing Argonaute binding. Nature. 2009;461:743–744. doi: 10.1038/461743a. [DOI] [PubMed] [Google Scholar]
- 75.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gunawardane LS, et al. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 77.Saito K, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palakodeti D, et al. The PIWI proteins SMEDWI-2 and SMEDWI-3 are required for stem cell function and piRNA expression in planarians. RNA. 2008;14:1174–1186. doi: 10.1261/rna.1085008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reddien PW, et al. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 80.Kirino Y, et al. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kojima K, et al. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells. 2009;14:1155–1165. doi: 10.1111/j.1365-2443.2009.01342.x. [DOI] [PubMed] [Google Scholar]
- 82.Lau NC, et al. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 2009;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nishida KM, et al. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang J, et al. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr. Biol. 2009;19:640–644. doi: 10.1016/j.cub.2009.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anne J, Mechler BM. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development. 2005;132:2167–2177. doi: 10.1242/dev.01809. [DOI] [PubMed] [Google Scholar]
- 86.Anne J, et al. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- 87.Gonsalvez GB, et al. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 2006;16:1077–1089. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 88.Kirino Y, et al. Arginine methylation of vasa protein is conserved across phyla. J. Biol. Chem. 2010;285:8148–8154. doi: 10.1074/jbc.M109.089821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Megosh HB, et al. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr. Biol. 2006;16:1884–1894. doi: 10.1016/j.cub.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 90.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 91.Lunde BM, et al. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weismann A. Continuity of the germ-plasm. In: Poulton EB, et al., editors. Essays upon heredity and kindred biological problems. 2nd edn. Clarendon Press; 1891. pp. 164–255. [Google Scholar]
- 93.Hegner RW. Effects of removing the germ-cell determinants from the eggs of some chrysomelid beetles. Preliminary report. Biol. Bull. 1908;16:19–26. [Google Scholar]
- 94.Hegner RW. Experiments with chrysomelid beetles. III. The effects of killing parts of the eggs of Leptinotarsa decemlineata. Biol. Bull. 1911;20:237–251. [Google Scholar]
- 95.Bounoure L. L’Origin des Cellules Reproductrices et la Probleme de la Lignee Germinale. Gauthier-Villars; 1939. [Google Scholar]
- 96.Geigy R. Action de l’ultra-violet sur le pole germinale dans l’ouef de Drosophila melanogaster. Rev. Suisse Zool. 1931;38:187–288. [Google Scholar]
- 97.Okada M, et al. Restoration of fertility in sterilized Drosophila eggs by transplantation of polar cytoplasm. Dev. Biol. 1974;37:43–54. doi: 10.1016/0012-1606(74)90168-7. [DOI] [PubMed] [Google Scholar]
- 98.Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc. Natl. Acad. Sci. USA. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mahowald AP. Fine structure of pole cells and polar granules in Drosophila melanogaster. J. Exp. Zool. 1962;151:201–215. [Google Scholar]
- 100.Mahowald AP. Polar granules of Drosophila: II. Ultrastructural changes during early embryogenesis. J. Exp. Zool. 1968;167:237–262. doi: 10.1002/jez.1401670211. [DOI] [PubMed] [Google Scholar]
- 101.Eddy EM. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cytol. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- 102.Amikura R, et al. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc. Natl. Acad. Sci. USA. 2001;98:9133–9138. doi: 10.1073/pnas.171286998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rangan P, et al. Temporal and spatial control of germ-plasm RNAs. Curr. Biol. 2009;19:72–77. doi: 10.1016/j.cub.2008.11.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rangan P, et al. Regulating gene expression in the Drosophila germ line. Cold Spring Harb. Symp. Quant. Biol. 2008;73:1–8. doi: 10.1101/sqb.2008.73.057. [DOI] [PubMed] [Google Scholar]
- 105.Jenny A, et al. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development. 2006;133:2827–2833. doi: 10.1242/dev.02456. [DOI] [PubMed] [Google Scholar]
- 106.Reveal B, et al. BREs mediate both repression and activation of oskar mRNA translation and act in trans. Dev. Cell. 2010;18:496–502. doi: 10.1016/j.devcel.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 108.Besse F, et al. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 2009;23:195–207. doi: 10.1101/gad.505709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brunel C, Ehresmann C. Secondary structure of the 3' UTR of bicoid mRNA. Biochimie. 2004;86:91–104. doi: 10.1016/j.biochi.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 110.Ferrandon D, et al. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3' UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gillespie DE, Berg CA. Homeless is required for RNA localization in Drosophila oogenesis and encodes a new member of the DE-H family of RNA-dependent ATPases. Genes Dev. 1995;9:2495–2508. doi: 10.1101/gad.9.20.2495. [DOI] [PubMed] [Google Scholar]
- 112.Barbosa V, et al. A maternal screen for genes regulating Drosophila oocyte polarity uncovers new steps in meiotic progression. Genetics. 2007;176:1967–1977. doi: 10.1534/genetics.106.069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Holm L, et al. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Aravin AA, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]