Abstract
The regulatory domain of phenylalanine hydroxylase (PAH, EC 1.14.16.1) consists of more than 100 amino acids at the N terminus, the removal of which significantly activates the enzyme. To study the regulatory properties controlled by the N terminus, a series of truncations and site-specific mutations were made in this region of rat PAH. These enzymes were expressed highly in Escherichia coli and purified through a pterin-conjugated Sepharose affinity column. The removal of the first 26 amino acids of the N terminus increased the activity by about 20-fold, but removal of the first 15 amino acids increased the activity by only 2-fold. Replacing serine-29 of rat PAH with cysteine from the same site of human PAH increased the activity by more than 4-fold. Mutation of serine to other amino acids with varying side chains: alanine, methionine, leucine, aspartic acid, asparagine, and arginine also resulted in significant activation, indicating a serine-specific inhibitory effect. But these site-specific mutants showed 30–40% lower activity when assayed with 6-methyl-5,6,7,8-tetrahydropterin. Stimulation of hydroxylase activity by preincubation of the enzyme with phenylalanine was inversely proportional to the activation state of all these mutants. Combined with recent crystal structures of PAH [Kobe, B. et al. (1999) Nat. Struct. Biol. 6, 442–448; and Erlandsen, H., Bjorgo, E., Flatmark, T. & Stevens, R. C. (2000) Biochemistry 39, 2208–2217], these data suggest that residues 16–26 have a controlling regulatory effect on the activity by interaction with the dihydroxypropyl side chain of (6R)-5,6,7,8-tetrahydrobiopterin. The serine/cysteine switch explains the difference in regulatory properties between human and rat PAH. The N terminus as a whole is important for maintaining rat PAH in an optimum catalytic conformation.
Phenylalanine hydroxylase (PAH, phenylalanine 4-monooxygenase, EC 1.14.16.1) catalyzes the pterin-dependent hydroxylation of L-phenylalanine to L-tyrosine (1). The reaction is the rate-limiting step in the complete catabolism of the amino acid. Deficiency of PAH results in marked elevation of phenylalanine levels, causing phenylketonuria, a disease that if untreated, ultimately leads to severe mental retardation (2).
PAH is structurally and functionally homologous to the other two tetrahydropterin-dependent aromatic amino acid hydroxylases, tyrosine hydroxylase and tryptophan hydroxylase. These enzymes share very similar catalytic mechanisms and have highly conserved catalytic domains. They are significantly different, however, in their regulatory domains, which comprise about one-third of the length of the whole enzyme. Their structural differences probably account for differences in their regulatory properties.
Partial proteolysis of rat PAH by α-chymotrypsin gives rise to a species of the hydroxylase, the catalytic activity of which matches the activity of wild type when assayed with 6-methyl-5,6,7,8-tetrahydropterin (6MPH4) or after treatment with lysolecithin (3). This limited digestion yields a dimer composed of a 35- to 36-kDa subunit, which lacks ≈11 kDa from the N terminus and ≈4–5 kDa from the C terminus (4–6). These results, combined with results of other studies, led to a picture of the structure of PAH in which the enzyme is divided into three domains: N-terminal regulatory domain; middle catalytic domain; and C-terminal tetramerization domain (1, 7). Recent x-ray crystallographic information has confirmed this picture (8, 9).
Besides protease treatment, the activity of PAH also can be increased highly by various treatments such as preincubation with substrate phenylalanine, treatment with lysolecithin, phosphorylation by cAMP-dependent protein kinase, exposure to alkaline pH, and alkylation of a sulfhydryl residue by N-methylmaleimide. Generally, these treatments are believed to involve the regulatory domain (1). It should be noted that the activation of PAH can only be observed when the assay is conducted with the natural cofactor (6R)-5,6,7,8-tetrahydrobiopterin (BH4). With synthetic cofactors like 6MPH4 or 6,7-dimethyl-5,6,7,8-tetrahydropterin (DMPH4), PAH shows the maximal activity. Furthermore, rat PAH is activated much more significantly than human PAH, despite the fact that their regulatory domains have nearly 90% identity in primary sequence, and their catalytic domains are nearly identical (99%).
It has been proposed that the regulatory domain, like a mobile structure, mediates the access to the active site of the catalytic domain. The modifications mentioned above would interfere with the interaction between the two domains and facilitate access to the active site (7). To decipher this interaction structurally, we carried out a series of truncations of the N terminus of rat PAH. Moreover, to understand the different regulatory effects between rat and human PAH more fully, we replaced the rat N terminus with a human sequence. We found that the removal of a fragment in the N terminus, residues 16–26, gave rise to a rat PAH of near-maximal activity. We also found a key residue that could explain the differences in regulatory properties between rat and human PAH.
Materials and Methods
Materials.
DNA primers for PCR and sequencing were purchased from Lofstrand Laboratories (Gaithersburg, MD). Pfu DNA polymerase was from Stratagene. DNA sequencing was carried out by using the T7 DNA polymerase kit (Amersham Pharmacia) on an automatic DNA sequencer (ALF DNA analysis system, Amersham Pharmacia). Plasmids pLNC209 (10) and pLNCB111 (11) expressing rat and human PAH, respectively, were constructed in this laboratory. Restriction enzymes, T4 DNA ligase, and DNA polymerase large fragment (Klenow) were obtained from New England Biolabs. Escherichia coli strains DH5α and BL21(DE3) competent cells were purchased from Life Technologies (Rockville, MD) and Novagen, respectively.
L-Phenylalanine, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide, and dihydropteridine reductase were obtained from Sigma. Catalase was purchased from Roche Molecular Biochemicals. BH4, 6MPH4, and DMPH4 were obtained from B. Shircks' Laboratory (Jona, Switzerland).
DNA Manipulation.
PAH expression plasmids used in this paper were constructed by PCR-based techniques. All primers are listed in Table 1. A brief explanation of these constructions is illustrated in Fig. 1. All clones of PCR products were sequenced before they were subjected to expression induction. Rat PAH coding sequence is from pLNC209 and the human counterpart is from pLNC111.
Table 1.
Oligonucleotide primers used in this paper
| Name | Sequence | Size | Notes |
|---|---|---|---|
| o464 | TGGCATATGGCAGCTGTTGTATTG | 24mer | 5′ primer for rat PAH |
| o465 | TGGCATATGGAATCCAGACCTTCTC | 25mer | 5′ primer for human PAH |
| N-rev | TTAGACAGTGGCACCAATGTCAT | 23mer | Antisense primer down-stream from Bsu36I site |
| N-15 | TGGCATATGTCAGACTTTGGGCAGGAAA | 28mer | Introduction of initiator ATG upstream of residue 16 |
| N-26 | TGGCATATGGACAACTCCAATCAAAATG | 28mer | Introduction of initiator ATG upstream of residue 27 |
| N-40 | TGGCATATGCTCAAAGAGGAAGTTGGTG | 28mer | Introduction of initiator ATG upstream of residue 41 |
| N-59 | TGGCATATGATCAACCTGACACACATTG | 28mer | Introduction of initiator ATG upstream of residue 60 |
| o265 | TAATACGACTCACTATAGGG | 20mer | Sense primer upstream from XbaI and NdeI sites |
| o560 | GACAACTGCAATCAAAATGGTGC | 23mer | 29Cys(+) |
| o561 | TTGATTGCAGTTGTCTTCAATATAGC | 26mer | 29Cys(−) |
| o562 | GACAACTCCAATCAAAATGGTGC | 23mer | 29Ser(+) |
| o563 | TTGATTGGAGTTGTCTTCAATATAGC | 26mer | 29Ser(−) |
| o569 | TGAAGACAACGCCAATCAAAATGGTG | 26mer | 29Ala(+) |
| o570 | TGAAGACAACATGAATCAAAATGGTG | 26mer | 29Met(+) |
| o571 | TGAAGACAACCTCAATCAAAATGGTG | 26mer | 29Leu(+) |
| o572 | TGAAGACAACGACAATCAAAATGGTG | 26mer | 29Asp(+) |
| o573 | TGAAGACAACAACAATCAAAATGGTG | 26mer | 29Asn(+) |
| o574 | TGAAGACAACCGCAATCAAAATGGTG | 26mer | 29Arg(+) |
| o575 | GTTGTCTTCAATATAGCTTGT | 21mer | Antisense primer upstream from site serine-29 |
Figure 1.
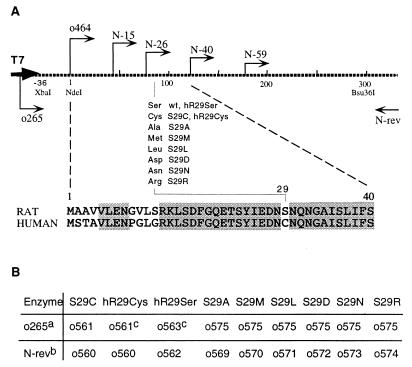
(A) A brief description of mutants made in this paper. Rat PAH coding sequence was cloned in to an expression vector pET26b(+) (Novagen), which contains a T7 promoter sequence (T7). The broken bold line represents part of the resulting plasmid pET26-rPAH, corresponding to the 5′ coding sequence. The numbers under the line indicate the position of nucleotides; the initiation codon starts with number 1. (Bottom) The first 40 amino acids from rat (RAT) and human PAH (HUMAN). Shaded areas represent identical sequences in rat and human PAH. The primers (arrows) and restriction sites used for mutagenesis are shown at relative positions. (Middle) Amino acids at site 29 of rat PAH and their corresponding enzyme names. wt, wild type. (B) Primer usage for the first round of PCR as described in Materials and Methods. a, primers in combination with primer o265; b, primers in combination with primer N-rev; c, the template was human PAH sequence.
To facilitate expression and mutation work, we first subcloned the rat PAH-coding sequence into the expression vector pET26b(+) (Novagen). PCR was conducted on plasmid pLNC209 with primers o426 and N-rev. The product was digested over night with NdeI and Bsu36I. The resulting fragment, together with a Bsu36I–SalI fragment from pLNC209, was inserted into pET26b(+), which was linearized with NdeI and SalI. This gave rise to pET26-rPAH, which has a rat PAH-coding sequence under the control of the T7 promoter. In the same way, truncated mutations (N-15, N-26, N-40, and N-59) were constructed by using the corresponding primers. The human PAH-coding sequence also was subcloned into pET26b(+), forming pET26-hPAH.
A two-step PCR procedure was used to construct rat–human N-terminal swapping and site-29-specific mutants. First, two PCRs were conducted with o265 and the antisense primer (designated with “−”) and with the sense primer (designated with “+”) and N-rev. The two products were separated on an agarose gel and used as the template for the second round of PCR. The second-round PCR was carried out with primers o265 and N-rev, initiated with a low annealing temperature such that products from the first round could anneal. The resulting product then was digested with XbaI and Bsu36I and was used to replace the counterpart in pET26-rPAH. The primer combinations and template usage for the first round of PCR are shown in Fig. 1B. E. coli DH5α was used for DNA cloning and plasmid propagation. After DNA-sequence confirmation, BL21(DE3) was transformed for protein expression.
Pterin Column Preparation.
A pterin column was constructed essentially as described by Fujisawa and Nakata (12). Pterin-compound DMPH4 was conjugated to ECH Sepharose 4B (Amersham Pharmacia) with the 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) reagent. First, 6 ml of the wet gel beads was degassed and then gassed with argon. DMPH4 (60 mg) was added while the argon was bubbling. At this point, the pH was approximately 5.0. Then 300 mg of EDAC, dissolved in 0.5 ml of argon-gassed water, was dropped slowly into the suspension. The pH was maintained between 5.0 and 5.5 (if necessary, the pH was adjusted by addition of 0.2 N NaOH). The reaction was performed at room temperature for 16 h with shaking and light protection. The reaction was terminated, and the resin was washed five times with 10× volumes of water. The coupling efficiency was 10–16 μmol/gram of wet gel.
The wet resin (2 ml) was used to pack a column that can be used for 2 weeks without significant decline in binding capacity. Columns can be regenerated by alternately washing with 50 mM Tris acetate (pH 4.2)/1 M NaCl and with 50 mM Tris acetate (pH 9.8)/1 M NaCl. The column was neutralized with 20 ml of 50 mM Tris acetate, pH 7.6, before loading samples.
Enzyme Expression and Purification.
All expression was conducted at 30°C, and all purification procedures were followed at 4°C. Then, 20 ml of overnight culture was inoculated to 500 ml of 2× YT medium. When the OD at 590 nm of the culture reached 0.6, ferrous ammonium sulfate was added to 50 nM, and isopropyl β-D-thiogalactoside was added to 0.4 nM to induce expression. The cells were harvested after 4 h of induction and kept frozen at −80°C until used.
E. coli cells (≈2 g) containing wild-type or mutant PAH were suspended in 10 ml of homogenization buffer [50 mM Tris acetate, pH 7.6/150 mM KCl/50 nM EDTA/0.1 mM Fe(NH4)2(SO4)2/0.1 mM phenylalanine/0.1 mM phenylmethylsulfonyl fluoride/50 μg of leupeptin/50 μg of protinin/70 μg of pepstatin]. The mixture was then sonicated on ice for 6 min in 30-sec on/off cycles with a C2 probe (Heat Systems/Ultrasonics). Lysozyme (0.3 mg/ml) was added to the mixture, which then was incubated on ice for 20 min. Then the homogenate was centrifuged for 15 min at 25,000 × g. The supernatant was cleared further by centrifugation for 30 min at 120,000 × g before being loaded onto the activated pterin column. The column was washed sequentially with 50 ml of 50 mM Tris acetate, pH 7.6/1.2 M NaCl/5 mg of catalase/2 mM DTT; 20 ml of 50 mM Tris acetate, pH 7.6/0.5 M NaCl/2 mM DTT; and 20 ml of 10 mM NaH2PO4/Na2HPO4, pH 7.0. The enzyme was eluted with 10 mM NaOH/NaHCO3, pH 10.8. The fractions were monitored by measuring the UV absorbancy at 260 and 280 nm and were neutralized with 2 M Tris acetate (pH 6.4). KCl, Fe2+, EDTA, and glycerol were added to 50 mM, 100 nM, 50 nM, and 10%, respectively. The enzyme solution turned greenish after the ferrous ion was added, which is a good sign for the quality and concentration of the purified PAH. Aliquots were snap frozen in liquid nitrogen and stored at −80°C.
Enzyme Assay.
Initial rates were determined by the coupled-spectrophotometric assay (13, 14). The assays were monitored at 340 nm with a UV-visible spectrophotometer (Agilent 8453, Hewlett–Packard) with a temperature controller. The reaction was recorded, and the rate was calculated with Agilent CHEMSTATION.
Each assay was conducted at 25°C at atmospheric oxygen tension in a volume of 1 ml containing 0.1 M potassium phosphate (pH 6.8), 0.2 mg of catalase, 0.125 mM NADH, an excess of dihydropteridine reductase, and various amounts of BH4 or 6MPH4 and phenylalanine (25°C). The amount of NADH oxidized was calculated with an extinction coefficient of 6,220 M−1⋅cm−1. The range of phenylalanine concentration was from 0.04 to 5.0 mM in the presence of 0.030 mM BH4, and the range of BH4 was from 0.001 to 0.120 mM in the presence of 1 mM of phenylalanine. The initial rate was corrected for the contribution caused by any autooxidation of BH4. Kinetic parameters were computed by direct fitting of experimental data to the Michaelis–Menten equation with ULTRAFIT 2.1 (Biosoft, Cambridge, U.K.). Velocities were expressed as μmol of NADH oxidized/min/mg of protein, and Km values were expressed as μM.
Results
High Expression and Purification of PAH.
Recombinant rat PAH has been produced in this laboratory by inducing an E. coli host that carries pLNC209. pLNC209 is a plasmid that contains the rat PAH coding sequence under the E. coli trp/lac promoter. This expression system has certain drawbacks that precluded its use for the mutagenesis work described below. It takes a long time to express rat PAH, generally over night, which would result in serious degradation of N-terminal mutants like N-59. It is difficult to manipulate pLNC209 because of its unfavorable distribution of restriction sites. Furthermore, the expression yield is low and extra purification steps are needed.
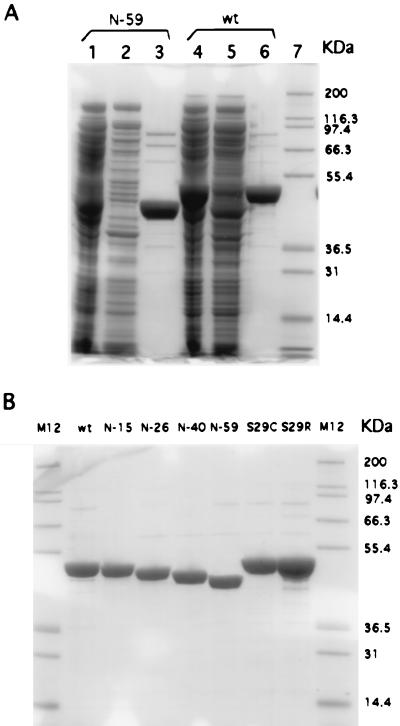
Therefore, we subcloned the full coding sequence of rat PAH from pLNC209 into a T7 expression vector pET26b(+), resulting in a plasmid named pET26-rPAH. Because we did so, rat PAH was expressed highly in E. coli in 4 h (Fig. 2, lane 4). In addition, the mutation work was facilitated, because downstream of the promoter, the vector has a polylinker that has a collection of unique restriction sites.
Figure 2.
Expression and purification of wild-type (wt) rat PAH and mutants shown on SDS/PAGE gels (10% Bis-Tris gels run with Mops running buffer, NOVEX, San Diego). Gels were stained with Colloidal blue staining kit (NOVEX). (A) Lanes 1 and 4, crude E. coli extracts; lanes 2 and 5, flow-through from pterin column after applying crude extract; lanes 3 and 6, the eluate with NaHCO3/NaOH (pH 10.8); lane 7, protein-standard markers (Mark 12, NOVEX). (B) Purified wt and mutants. Names are shown on top of the lanes. M12 is the marker.
Before conducting mutation work, we realized that it is necessary to find a method suitable for purifying N-terminal mutants. The common method for purifying rat PAH is through the use of a phenyl-Sepharose column. The crude enzyme extract is subject to activation with 25 mM phenylalanine before being applied to the column, and the enzyme is eluted after the phenylalanine is removed. However, using this method, we were unable to purify N-terminal truncation mutants such as N-59. Little enzyme eluted when phenylalanine was removed. The truncated enzyme can be eluted along with other proteins when 0.5% Tween 20 is included in the elution buffer. This phenomenon is in accord with the implication that phenylalanine activation enables the hydrophobic catalytic domain to bind to the column. In the case of N-59, half of the N terminus is removed so that its binding to the phenyl-Sepharose column will be phenylalanine independent.
For these reasons, we chose to use an affinity chromatographic approach with a pterin-conjugated Sepharose column. A pterin column was made by conjugating DMPH4 to a carboxy group on a solid phase in the presence of 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide. It was developed first by Cotton (15) for purification of rabbit PAH from partially purified materials. Nakata and Fujisawa (16) then adapted it in purification of rabbit-brain tryptophan hydroxylase. We found it very attractive, because (i) rat PAH has a fairly low dissociation constant (KD) for BH4, 10–20 nM as determined by tryptophan fluorescence quenching (17), and (ii) the catalytic domain of PAH is believed to have stronger affinity for pterin than the regulatory domain (1). A modified method for making a pterin column is described in Materials and Methods. The purification is then very simple, and pure rat PAH can be obtained at high concentration (3–6 mg/ml) in just one step from the crude extract (Fig. 2). All N-terminal mutants including N-59, which could not be purified through the phenyl-Sepharose column, were purified successfully by this method.
The Km values for BH4 and phenylalanine of purified wild-type rat PAH were: BH4, 3.7 ± 0.3 μM; phenylalanine, 320 ± 50 μM; and Vmax (BH4), 0.41 μmol of NADH oxidized/min/mg of protein. These data are consistent with those for rat PAH purified through a phenyl-Sepharose column (10). The enzyme also has a slightly greenish color after the addition of ferrous ion, which is common for recombinant rat PAH purified through a phenyl-Sepharose column after concentration by ultrafiltration.
N-Terminal Truncation.
Rat PAH is in an inhibitory state when assayed with its natural cofactor BH4. The inhibition is because of the presence of more than 100 amino acids in the N terminus. Removal of them by either limited proteolysis (4) or mutation (18, 19) gives a fully activated hydroxylase. To understand the inhibitory mechanism of the N terminus further, a series of truncations were made of the N terminus of rat PAH.
We started with N-59, which has the first 59 amino acids deleted from the recombinant construct. It contains about one-half of the regulatory domain. Because activation of rat PAH by different treatments is only expressed fully when the enzyme is assayed with BH4, the natural cofactor, but not with the synthetic cofactor 6MPH4, it was assumed that the latter case reflected the intrinsic activity of the enzyme (20). Thus, the ratio of hydroxylase activity with 6MPH4 to that with BH4 (designated as 6MPH4/BH4 ratio) provides a measure of the activation state of the enzyme (21). Therefore, we measured the catalytic activities of N-59 in the presence of BH4 or 6MPH4. In the presence of BH4, the mutant expressed an activity comparable to that of wild type in the presence of 6MPH4. The 6MPH4/BH4 ratio is 1.3 for the N-59 N-terminal deletion construct (Table 2). A comparison with the ratio of wild type, which is about 27.2, supports the conclusion that the inhibitory domain is close to the N terminus.
Table 2.
6MPH4/BH4 ratios of rat phenylalanine hydroxylase and the activity after preincubation with phenylalanine
| Enzyme | V6MPH4* | VBH4† | Ratio‡ | Vphe§ | Ratio¶ |
|---|---|---|---|---|---|
| Wild type | 10.9 | 0.4 | 27.2 | 11.1 | 27.8 |
| N-15 | 6.5 | 0.8 | 8.1 | 6.3 | 7.9 |
| N-26 | 11.4 | 8.0 | 1.4 | 10.9 | 1.4 |
| N-40 | 12.0 | 8.6 | 1.4 | N/A‖ | N/A‖ |
| N-59 | 13.3 | 10.0 | 1.3 | N/A‖ | N/A‖ |
| S29C | 6.3 | 1.5 | 4.2 | 6.1 | 4.1 |
| hR29Cys | 6.7 | 1.9 | 3.5 | 8.9 | 4.7 |
| hR29Ser | 10.9 | 0.7 | 15.6 | 11.3 | 16.1 |
| S29A | 7.7 | 1.5 | 5.1 | 7.2 | 4.8 |
| S29M | 6.5 | 1.8 | 3.6 | 6.2 | 3.4 |
| S29L | 6.0 | 1.7 | 3.5 | 6.4 | 3.8 |
| S29D | 6.2 | 1.5 | 4.1 | 6.0 | 4.0 |
| S29N | 6.4 | 1.4 | 4.6 | 6.2 | 4.4 |
| S29R | 8.2 | 4.3 | 1.9 | 7.0 | 1.6 |
Initial rates were measured with 250 μM 6MPH4 and 5 mM phenylalanine.
Initial rates were measured with 30 μM BH4 and 1 mM phenylalanine.
6MPH4/BH4 ratio.
Initial rates for phenylalanine-activated enzymes were determined in the presence of 27 μM BH4. Enzymes were activated with 1 mM phenylalanine at concentration of 4.0 μM for 5 min at room temperature. Then 10 μl of enzyme was used to determine the initial rate in a reaction volume of 1 ml.
Vphe/VBH4, measurement of the activation by preincubation with phenylalanine.
Not available.
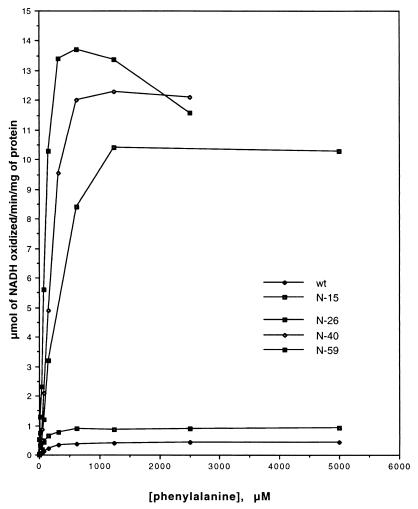
While making mutants further toward the N terminus, we noticed the similarity between rat and human PAH within the N-terminal 59 amino acids (Fig. 1, only 1–40 are shown because amino acids 41–59 are identical). The human PAH was reported to be in an activated state under the assay conditions of rat PAH (11). There is a difference in the first 12 amino acids as well as the single amino acid at site 29: serine in rat PAH and cysteine in human PAH. Considering these differences and the phosphorylation site at position 16, three truncations were made to delete the first 15, 26, and 40 residues, respectively. The catalytic activity of these truncated mutants was assayed with either BH4 or 6MPH4 (Table 2). A dramatic change of activity was observed between mutants N-15 and N-26. Compared with wild type, N-15 showed higher activity with BH4 and lower activity with 6MPH4, whereas N-26 showed very high activity with BH4 but similar activity with 6MPH4. Activities of N-40 are similar to those of N-26. This dramatic change was observed also when the phenylalanine saturation curves of these mutants were compared with the wild type (Fig. 3). The N-59 mutant was inhibited by high concentrations of phenylalanine (above 0.5 mM), which is in accord with previous reports that rat PAH activated by limited proteolysis or by treatment with lysolecithin was inhibited significantly by phenylalanine concentrations greater than 0.3 mM (4, 5). Mutants N-26 and N-40 have comparable activity to that of N-59, but they are not inhibited by high phenylalanine concentrations (Fig. 3).
Figure 3.
Phenylalanine saturation curves for wild-type rat PAH (wt) and N-terminal truncated mutants. Initial rates were recorded in the presence of 30 μM BH4 and various amounts of phenylalanine from 0.04 mM to 5 mM.
Serine/Cysteine Substitution in Rat and Human PAH.
To understand the difference in regulatory properties between rat and human PAH, the human N-terminal 29 residues were used to replace the corresponding part of rat PAH. At the same time, we changed the serine/cysteine-29 accordingly (Fig. 1). Thus, we had three mutants: hR29Cys, hR29Ser, and S29C. hR29Cys is rat PAH but has human PAH N terminus up to site 29. hR29Ser differs from hR29Cys at site 29, which has a serine in the former mutant and a cysteine in the latter. In other words, hR29Ser is a rat PAH whose first 12 amino acids are from human PAH. S29C is a site-specific point mutant of rat PAH in which the serine at 29 was changed to cysteine.
Compared with the activity of wild type, the catalytic activity of the three mutants was very informative (Table 2). hR29Ser, like wild type, has high activity with 6MPH4 but low activity with BH4. When serine-29 was replaced by cysteine, both mutants, hR29Cys and S29C, showed lower activity with 6MPH4 and significantly high activity with BH4, i.e., hR29Cys and S29C had 35 and 35% less activity compared with hRSer29 and wild type when assayed with 6MPH4, respectively. With BH4, however, they showed approximately three and four times more activity, respectively. hR29Ser has comparable activity to wild type when assayed with 6MPH4 and approximately 2-fold higher activity with BH4; thus, it has a 6MPH4/BH4 ratio of 16.0. These data suggested that the serine/cysteine substitution at site 29 could account for the difference of activity between rat and human PAH.
Site-Directed Mutagenesis of Site 29.
The increase in activity of rat PAH by replacement of serine at 29 with cysteine prompted us to study this single residue further. Serine is an amino acid that has a hydroxyl group that can be phosphorylated by selective protein kinases. However, there is no report indicating that this site can be phosphorylated in vivo or in vitro. Cysteine differs from serine by a sulfur substitution. The mercapto group can be oxidized to form a disulfide bond with another mercapto group. To test this possibility, we boiled the three mutants and wild type in the presence or absence of DTT. After resolution on an SDS/PAGE gel, all gave a monomer as the major form regardless of whether DTT was present, an indication that there is no intersubunit disulfide bond. The recent rat PAH crystal structure (9) also indicated that site 29 is not in proximity to other cysteines in the same subunit.
The side chain of cysteine is more hydrophobic and larger than that of serine. But the hydroxyl group of serine can form hydrogen bonds with other polar atoms. To investigate the role of serine/cysteine-29 in the regulation of the activity of PAH, we mutated this site specifically to negatively charged aspartic acid, polar uncharged asparagine, positively charged arginine, and nonpolar uncharged residues (with increasing side-chain size) alanine, leucine, and methionine. Despite different side chains, these mutants all have similar activity to that of S29C except S29R, which expressed significantly higher activity with BH4 within this series of mutants. These data suggest a serine-specific inhibitory effect at site 29.
Although they showed higher activity when assayed with BH4, these mutants, compared with that of wild type, have 30–50% lower activity with 6MPH4. N-15 and hR29Ser also gave low activity with 6MPH4. These data indicated that intact N terminus was essential for the enzyme to exhibit its maximal activity.
Kinetic Data and Phenylalanine Activation.
We documented later the kinetic values for these mutants and measured their activities after preincubation with phenylalanine (Table 3). These kinetic parameters were calculated from initial rates measured at pH 6.8, a constant concentration of BH4, and varying concentrations of phenylalanine or a constant concentration of phenylalanine and varying concentrations of BH4. From wild-type to truncation mutants N-15, N-26, N-40, and N-59, there is a continuous increase in Km values for BH4 and a continuous decrease in Km values for phenylalanine. Notably, the Km for BH4 of N-26 is more than twice the value of N-15, whereas the Vmax of N-26 is about 11 times that of N-15. The BH4 Km values for those site-29 mutations except S29R, their values of Km for BH4 are about twice that of wild type, and their Vmax values are approximately four to six times that of wild type. S29R stands out from these mutants because of its higher values of Km for BH4 and Vmax and lower Km for phenylalanine. The Km for BH4 and Vmax of hR29Ser are almost the same as those of N-15, whereas its Km for phenylalanine is like that of wild-type rat PAH.
Table 3.
Kinetic parameters for rat phenylalanine hydroxylase
| Enzyme | Km phe, μM | KmBH4, μM | Vmax |
|---|---|---|---|
| Wild type | 307 | 4.2 | 0.4 |
| N-15 | 193 | 7.5 | 0.9 |
| N-26 | 178 | 16.5 | 10.6 |
| N-40 | 152 | 18.1 | 11.7 |
| N-59 | 96* | 21.1 | 13.0 |
| S29C | 255 | 10.5 | 1.85 |
| hR29Cys | 296 | 11.7 | 2.4 |
| hR29Ser | 485 | 7.5 | 0.7 |
| S29A | 237 | 9.1 | 1.9 |
| S29M | 247 | 7.5 | 2.1 |
| S29L | 345 | 8.9 | 2.3 |
| S29D | 431 | 7.0 | 1.6 |
| S29N | 197 | 8.1 | 1.7 |
| S29R | 175 | 13.5 | 5.4 |
Initial velocities were determined as described in Materials and Methods. Km phe and Km BH4 values were determined with constant concentrations of 30 μM BH4 and 1 mM phenylalanine, respectively. Vmax values were determined in the presence of 30 μM BH4 with various amounts of phenylalanine. Enzyme concentrations varied among different species: 0.5 μM for wild type, N-15, and hR29Ser; 0.04 μM for N-26, N-40, and N-59; 0.1 μM for other mutants.
S0.5 value, i.e., the concentration of phenylalanine at which 50% of maximal PAH activity is expressed.
It was of interest to measure the activation by preincubation of those site-specific mutants with 1 mM phenylalanine. Assayed in the presence of BH4, preincubation with phenylalanine further activated these mutants (Table 2). Under our assay conditions, their activities matched those of the enzyme assayed in the presence of 6MPH4, which were considered to be the highest values. The values of Vphe/VBH4 are very close to those of 6MPH4/BH4. In other words, the higher the state of activation, the lower the extent of activation by preincubation with phenylalanine. It is noticeable that the hR29Cys mutant has relatively low activity with 6MPH4 but has exceptionally high activity after preincubation with phenylalanine.
Discussion
Here we report studies of the N terminus, the regulatory domain of rat PAH and the corresponding region of human PAH. We found that residues from 16 to 26 play an inhibitory role. Their removal gives rise to an almost fully activated rat PAH. We compared the N-terminal 59 amino acids from rat and human PAH by substituting the different residues in a rat PAH. We also found that a key amino acid at site 29 (serine in rat and cysteine in human) results in the same difference in regulatory properties between rat and human PAH. The effect of the serine at 29 seems unique because the site-directed mutation at this site also produced activation of rat PAH.
High Expression and Purification of PAH.
The high expression of PAH in E. coli and the use of a simple and efficient purification method are essential to the work we presented above. Under the control of the T7 promoter, the expression of rat PAH is increased significantly in short times of incubation. We were able to express high amounts of recombinant PAH in 4 h at 30°C after induction. This high expression facilitated the purification of PAH with the use of the pterin column. The pterin compound DMPH4 has been shown to be conjugated with the Sepharose support through its 5-amino group (22). This modification stabilized the pterin compound from oxidation under ambient conditions. This modification apparently does not affect its binding to the catalytic domain, which is in accordance with the fact that the pyrimidine ring of the pterin is the essential part for catalyzing the hydroxylation (1). In fact, we benefited from this method not only because of the simple approach but also because of its strong binding to the catalytic domain as we were studying the regulatory domain.
The Interaction Between BH4 and the PAH N Terminus.
It has long been suggested that measuring the 6MPH4/BH4 ratio would indicate the activation state of rat PAH. The suggestion was based on the fact that the activation of rat PAH by different treatments is only expressed fully when the enzyme is assayed with its natural cofactor BH4. It seems likely that a critical structural difference between BH4 and 6MPH4 is the side chain at the C6 site where 6MPH4 has a simple methyl group, and BH4 has a dihydroxypropyl group. One effect of this difference is that steric hindrance would block BH4 from entering into the catalytic site. The recent crystal structure (9) showed that the active site is in a deep pocket and that residues 19–33 of the N terminus extend from the regulatory core to the open top of this pocket, and that this part of the N terminus serves as a gate to the catalytic site. This structure is consistent with the model Hufton et al. proposed (7) where the N terminus mediates the access to the active site. The other effect is that BH4 might use these two hydroxyl groups to interact with the N terminus, expressing its inhibitory function. Phillips and Kaufman (23) reported that the presence of BH4 inhibits the in vitro phosphorylation of the serine-16 of rat PAH. Adding phenylalanine to the reaction could overcome the inhibition completely, which was believed to be caused by deflection of the N terminus away from the active site, in a manner analogous to partial proteolysis and lysolecithin treatment. BH4 can also protect rat PAH to some extent from the action of a protease (4) or affect the proteolytic cleavage pattern (24). In addition, rat PAH has a very low Km for BH4 compared with that for 6MPH4, suggesting a stronger affinity for BH4. Furthermore, interaction with the N terminus may affect the transient state of BH4 during peroxidation. When it is attacked by molecular O2, the C4 atom will undergo a transition from a sp2 hybrid orbit (flat state) to a sp3 orbit (pyramid state). This transition could be favored or blocked by extra force put on the side chain. Recently, Erlandsen et al. reported a crystal structure of the BH2-bound catalytic domain of human PAH (25). They found that the pyrimidine ring in the structure is nonplanar, suggesting a transient state. Superimposing this structure onto that of ligand-free rat PAH with regulatory and catalytic domains (9), they also found that the C2′ hydroxyl group could form a hydrogen bond to serine-23 in the regulatory domain.
In the PHOH-24 structure (rat PAH without C-terminal 24 amino acids; ref. 9), the first 18 amino acids containing the serine-16, the phosphorylation site, were not visualized clearly because of a lack of electron density in this region. The deletion of the first 15 amino acids increased the activity of rat PAH about 2-fold, comparable to the effect of phosphorylation of serine-16 (26). Replacement of the first 12 amino acids with a human sequence (hR29Ser) showed less effect on activity, compared with that of N-15.
In summary, these data suggest that residues 16–26, through their interaction with the side chain of BH4, are responsible for the essential part of the inhibitory effect of the N terminus. Whether this interaction causes a delay of the biopterin entering into or releasing from the catalytic site or whether it makes a difference in the catalytic state of the pterin ring may be answered by further studies of this region.
On the basis of the high homology of rat and human PAH, it is possible that these residues would play the same role in human PAH. But a human mutation (residues 62–452) was reported not to be activated (19). Their result might represent an artifact of the large N-terminal fusion (maltose-binding protein) used in the experiments.
Serine-29.
The serine/cysteine substitution at site 29 accounts for some of the different properties of rat and human PAH. Like human enzymes, cysteine enzymes (S29C and hR29Cys) are in a highly activated state, as indicated by a low 6MPH4/BH4 ratio. Compared with serine enzymes (rat PAH and hR29Ser), they have lower Km values for phenylalanine and higher values for BH4. In addition, they could not be purified through a phenyl-Sepharose column (data not shown), reminiscent of the experience with recombinant human PAH (ref. 27, and P.G., unpublished results). One explanation could be that, as the cysteine enzymes are in a highly activated state, their hydrophobic catalytic domains are already exposed and able to bind to the column independent of the presence or absence of phenylalanine.
It is intriguing that any mutation at serine-29 caused a significant stimulation in activity. The size and polarity of the side chain of these amino acids do not seem to be critical determinants of activity. Therefore, keeping rat PAH in an inhibitory state might be due to the hydroxyl group of serine.
Indication for Studies of Human PAH.
Mutations in the human PAH gene cause the inherited disease hyperphenylalaninemia. A significant fraction of these natural mutations are located in the N terminus, among which the three most frequently observed mutations are G46S, L48S, and I65T. Their calculated frequencies of mutation are 35.35, 24.74, and 38.89%, respectively (28). Although the exact effect of serine or threonine in these mutations remains to be determined, the specific inhibitory effect of serine-29 of rat PAH might apply to these mutants. If analogous to serine-29 PAH, these mutants would have a significantly reduced catalytic activity compared with wild-type human PAH. Unfortunately, only a few studies have been carried out on N-terminal mutations that include measurements of their catalytic activity. I65T was shown to have a residual activity of approximately 26% of wild type when expressed in COS cells (29). To understand fully the mutations in the PAH gene that lead to phenylketonuria, it is necessary to characterize these natural mutants. The expression and purification system reported here may facilitate such studies.
Acknowledgments
We thank Cynthia Falke and Kun Park for technical assistance.
Abbreviations
- PAH
phenylalanine hydroxylase
- 6MPH4
6-methyl-5,6,7,8-tetrahydropterin
- BH4
(6R)-5,6,7,8-tetrahydrobiopterin
- DMPH4
6,7-dimethyl-5,6,7,8-tetrahydropterin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031561698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031561698
References
- 1.Kaufman S. In: Advances in Enzymology. Meister A, editor. Vol. 67. New York: Wiley; 1993. pp. 77–264. [DOI] [PubMed] [Google Scholar]
- 2.Scriver C R, Kaufman S, Woo S L C. In: The Metabolic Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1989. pp. 495–546. [Google Scholar]
- 3.Fisher D B, Kaufman S. J Biol Chem. 1972;247:2250–2252. [PubMed] [Google Scholar]
- 4.Fisher D B, Kaufman S. J Biol Chem. 1973;248:4345–4353. [PubMed] [Google Scholar]
- 5.Abita J P, Parniak M, Kaufman S. J Biol Chem. 1984;259:14560–14566. [PubMed] [Google Scholar]
- 6.Iwaki M, Phillips R S, Kaufman S. J Biol Chem. 1986;261:2051–2056. [PubMed] [Google Scholar]
- 7.Hufton S E, Jennings I G, Cotton R G. Biochem J. 1995;311:353–366. doi: 10.1042/bj3110353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erlandsen H, Fusetti F, Martinez A, Hough E, Flatmark T, Stevens R C. Nat Struct Biol. 1997;4:995–1000. doi: 10.1038/nsb1297-995. [DOI] [PubMed] [Google Scholar]
- 9.Kobe B, Jennings I G, House C M, Michell B J, Goodwill K E, Santarsiero B D, Stevens R C, Cotton R G, Kemp B E. Nat Struct Biol. 1999;6:442–448. doi: 10.1038/8247. [DOI] [PubMed] [Google Scholar]
- 10.Citron B A, Davis M D, Kaufman S. Protein Expression Purif. 1992;3:93–100. [PubMed] [Google Scholar]
- 11.Kowlessur D, Citron B A, Kaufman S. Arch Biochem Biophys. 1996;333:85–95. doi: 10.1006/abbi.1996.0367. [DOI] [PubMed] [Google Scholar]
- 12.Fujisawa H, Nakata H. In: Methods in Enzymology. Kaufman S, editor. Vol. 142. Orlando, FL: Academic; 1987. pp. 83–87. [DOI] [PubMed] [Google Scholar]
- 13.Kaufman S. J Biol Chem. 1957;226:511–524. [PubMed] [Google Scholar]
- 14.Kaufman S. In: Methods in Enzymology. Kaufman S, editor. Vol. 142. Orlando, FL: Academic; 1987. pp. 3–17. [DOI] [PubMed] [Google Scholar]
- 15.Cotton R G. FEBS Lett. 1974;44:290–292. [PubMed] [Google Scholar]
- 16.Nakata H, Fujisawa H. J Biochem (Tokyo) 1981;90:567–569. doi: 10.1093/oxfordjournals.jbchem.a133508. [DOI] [PubMed] [Google Scholar]
- 17.Phillips R S, Parniak M A, Kaufman S. Biochemistry. 1984;23:3836–3842. doi: 10.1021/bi00312a007. [DOI] [PubMed] [Google Scholar]
- 18.Dickson P W, Jennings I G, Cotton R G. J Biol Chem. 1994;269:20369–20375. [PubMed] [Google Scholar]
- 19.Knappskog P M, Flatmark T, Aarden J M, Haavik J, Martinez A. Eur J Biochem. 1996;242:813–821. doi: 10.1111/j.1432-1033.1996.0813r.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaufman S. Adv Enzyme Regul. 1986;25:37–64. doi: 10.1016/0065-2571(86)90007-5. [DOI] [PubMed] [Google Scholar]
- 21.Hasegawa H, Kaufman S. J Biol Chem. 1982;257:3084–3089. [PubMed] [Google Scholar]
- 22.Cotton R G, Jennings I G. Eur J Biochem. 1978;85:357–363. doi: 10.1111/j.1432-1033.1978.tb12247.x. [DOI] [PubMed] [Google Scholar]
- 23.Phillips R S, Kaufman S. J Biol Chem. 1984;259:2474–2479. [PubMed] [Google Scholar]
- 24.Gibbs B S, Benkovic S J. Biochemistry. 1991;30:6795–6802. doi: 10.1021/bi00241a024. [DOI] [PubMed] [Google Scholar]
- 25.Erlandsen H, Bjorgo E, Flatmark T, Stevens R C. Biochemistry. 2000;39:2208–2217. doi: 10.1021/bi992531+. [DOI] [PubMed] [Google Scholar]
- 26.Kowlessur D, Yang X J, Kaufman S. Proc Natl Acad Sci USA. 1995;92:4743–4747. doi: 10.1073/pnas.92.11.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez A, Knappskog P M, Olafsdottir S, Doskeland A P, Eiken H G, Svebak R M, Bozzini M, Apold J, Flatmark T. Biochem J. 1995;306:589–597. doi: 10.1042/bj3060589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erlandsen H, Stevens R C. Mol Genet Metab. 1999;68:103–125. doi: 10.1006/mgme.1999.2922. [DOI] [PubMed] [Google Scholar]
- 29.John S W, Scriver C R, Laframboise R, Rozen R. Hum Mutat. 1992;1:147–153. doi: 10.1002/humu.1380010210. [DOI] [PubMed] [Google Scholar]