Abstract
Multispectral images of skin contain information on the spatial distribution of biological chromophores, such as blood and melanin. From this, parameters such as blood volume and blood oxygenation can be retrieved using reconstruction algorithms. Most such approaches use some form of pixelwise or volumetric reconstruction code. We explore the use of principal component analysis (PCA) of multispectral images to access blood volume and blood oxygenation in near real time. We present data from healthy volunteers under arterial occlusion of the forearm, experiencing ischemia and reactive hyperemia. Using a two-layered analytical skin model, we show reconstruction results of blood volume and oxygenation and compare it to the results obtained from our new spectral analysis based on PCA. We demonstrate that PCA applied to multispectral images gives near equivalent results for skin chromophore mapping and quantification with the advantage of being three orders of magnitude faster than the reconstruction algorithm.
Keywords: multispectral imaging, tissue oxygenation, blood volume, principal component analysis, modeling, biophotonics
Introduction
Assessing spatial distributions of skin chromophores such as blood and melanin can be achieved by diffuse multispectral imaging. Acquiring several wavelength images in the near-infrared spectrum together with an analytical skin model, allows fitting the data to the model, thus extracting and mapping the spatial distribution of those parameters.
Diffuse multispectral imaging of the skin and image reconstruction of skin chromophores has found its application in the clinic, successfully assessing parameters for healthy and diseased skin.1, 2, 3, 4, 5, 6, 7 Assessment of the metabolic state of skin surface lesions is often desired in clinical routines as a measure for treatment outcome. Near-infrared diffuse multispectral imaging of the skin combined with an analytical, numerical, or stochastic skin model can provide this information by producing spatial maps of skin chromophore concentrations.5, 8, 9 The main parameters of interest are blood, melanin, lipids, and water, which exhibit separable absorption coefficients in the near-infrared wavelengths range. The disadvantage of finding these parameters by fitting the data to an analytical skin model lies in the computationally expensive data postprocessing. This makes immediate conclusions difficult or even impossible if the image size is large, whereas in clinical routines it is often desired to assess the metabolic state of a tumor in real time. We are not aware of any current imaging protocol and reconstruction algorithm, which can assess quantitative blood concentrations in real time.
Principal component analysis (PCA), first introduced in 1901,10 is a statistical tool, which linearly transforms data into an orthogonal coordinate system, where the axes correspond to the inherent information within the data set. The idea is to reveal the data components (in a decreasing order) that best explain the variance in the data. PCA has found applications in fields such as face recognition,11, 12 and image compression,13 and is a common technique for finding patterns in high dimensional data.14 It is further used for analyzing and visualizing gene expression data,15, 16, 17 for dimension reduction in hyperspectral imaging,18 as well as image enhancement.19 The main advantage of PCA is the speed of computation, which is on the order of seconds per image. For the biomedical optical imaging field, PCA found its place in various applications. The usage ranges from noise reduction and image enhancement19 in multispectral data for biological cell analysis20 to pattern analysis for skin lesion classification.21 Applied to RGB images, PCA was used on relative color features for unsupervised lesion classification.22, 23, 24
When imaging the skin in the visible light range, the dominant absorbing materials are blood and melanin and should therefore explain most of the variance in multispectral data. Previous work by Tsumura et al.25 showed that skin color in digital RGB images can be described by attributing melanin and blood to the first two principal components. Fadzil et al.26 and Nugroho et al.27 used the same idea and applied PCA and independent component analysis to RGB data for blood and melanin extraction of vitiligo lesions to qualitatively evaluate the skin repigmentation progression.
In this work, we evaluate the use of PCA for retrieving quantitative near-real-time blood volume and blood oxygenation maps of skin areas of several square centimeters from a selected set of spectral images in the near-infrared range (700–1000 nm). Our experimental protocol involves imaging of healthy volunteers’ lower forearm before, during, and after arterial occlusion. Occlusion experiments were chosen, as the behavior of blood oxygenation is well known, which is ischemia during and reactive hyperemia after release of pressure.
In the first part of the paper, we describe the two layered analytical skin model used and show reconstruction results of blood volume and blood oxygenation over time. In the second part, we apply PCA to the same data set and show that the first eigenvector correlates with blood volume and the second eigenvector with blood oxygenation. Finally, we compare reconstruction results to the eigenvectors found by PCA and demonstrate the relationship between blood volume and the first eigenvector, as well as the relationship between blood oxygenation and the second eigenvector.
Materials
Instrumentation
The noninvasive, noncontact diffuse reflectance multi-spectral imaging system used in this work has been described in detail elsewhere5 and shall only be described briefly here. Polarized light from a white-light source (halogen 150 W, Techniquip, Pleasanton, California) is used for illumination of the sample. A second polarizer (Optarius, Malmesbury, UK) is placed before the detection unit, with its polarization orientation perpendicular to the incident beam polarization, thus guaranteeing diffuse reflectance measurements and removal of specular reflection.28
Images are captured by a CCD camera (Princeton Instruments CCD-612-TKB, Roper Scientific, Trenton, New Jersey) after passing consecutively one of six narrow bandpass filters (40 nm FWHM, CVI Laser, Albuquerque, New Mexico) on a filter wheel centered at 700, 750, 800, 850, 900, and 1000 nm. For calibration purposes, images from a 90% reflectance paper (Kodak, Rochester, New York) are also acquired at each image filter.
Occlusion Experiments on Healthy Volunteers
A pressure cuff was used to occlude the upper right arm of five right-handed healthy volunteers with 180 mm Hg pressure. This amount of pressure was chosen to achieve arterial occlusion, and the pressure lasted for 5 min. Multispectral images were taken every 30 s before occlusion, during occlusion and for 5 min afterward, resulting in 21 time points in total. Occlusion experiments were chosen as the behavior of blood volume and blood oxygenation over time is well known, which is ischemia during, and reactive hyperemia after occlusion.4, 8, 29, 30
All volunteers signed a consent form approved by the Institutional Review Board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development under the Protocol No. 08-CH-0001.
Methods
Reconstruction of Skin Chromophores
Diffuse multispectral images obtained during the occlusion experiment were used for reconstruction of blood volume and blood oxygenation. Preprocessing of the data included spectral and spatial illumination artifact removal as described in Ref. 5. The next step in preprocessing was rigid-body registration for motion artifact removal and was performed by MultiStackReg in Image J (Bethesda, Maryland). In a last step, curvature correction was performed to remove shape based intensity bias.31
Four wavelength (λ) images were used (700, 750, 800, and 850 nm) and reconstruction was performed using MATLAB (Math Works, Natick, Massachusetts) by a least-squares nonlinear fitting of the data to the analytical skin model used.5 The analytical skin model is based on a two-layered structure, the first one being the melanin containing epidermis, the second one being the blood containing dermis, with optical properties of the skin taken from literature values,32, 33, 34 and can be written as
| (1) |
where Ie is the wavelength dependent intensity measured in the CCD camera. The attenuation by the epidermis, Ae, is based on Lambert’s law and can be written as
| (2) |
with vm the concentration of melanin, de the thickness of the epidermis and μe the absorption coefficient of the epidermis, which is based on the absorption of melanin. The attenuation by the dermis, Ad, which includes the absorption due to blood volume, vdb, and oxygenation, vboxy, is based on the analytical solution of photon migration in turbid media35 based on random walk theory and can be written as
| (3) |
| (4) |
where is the reduced scattering coefficient and μd is the absorption coefficient of the dermis, with μdeoxy and μoxy being the absorption coefficients of deoxygenated and oxygenated blood, respectively. The scaling factor S, blood volume, vdb, and blood oxygenation, vboxy, are unknown a priori and were solved for.
The scaling factor was calculated for each subject and time point and averaged over time. This so obtained subject specific scaling factor was then used to compute the 2-D maps of blood volume and oxygenation over the course of the experiment. Computational time for each 2-D image, which is in the order of 200×200 pixels, was 45 min on a 2.6-GHz, 3-GB RAM personal computer (PC).
PCA
PCA10 linearly transforms the data into an orthogonal coordinate system whose axes correspond to the principal components in the data, i.e., the first principal component accounts for as much variance in the data as possible and, successively, further components capture the remaining variance. Through an eigenanalysis, the principal components are determined as eigenvectors of the data set’s covariance matrix and the corresponding eigenvalues refer to the variance that is captured within each eigenvector.
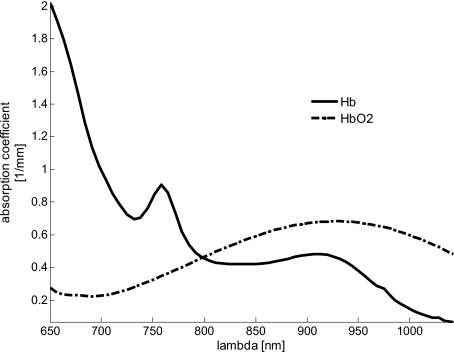
We use three wavelengths (750, 800, and 850 nm) from the occlusion experiment, which provides three 2-D images. These wavelengths were chosen because they are centered around 800 nm, which is the isosbestic point of blood, where the absorption coefficient of deoxygenated (Hb) and oxygenated (HbO2) blood are equal. Figure 1 shows the absorption spectra for Hb and HbO2, illustrating that the absorption of Hb is dominant at 750 nm and for HbO2 at 850 nm. In this wavelength range, the dominant chromophore is blood and should therefore explain most of the variance of the data.
Figure 1.
Absorption spectra of deoxygenated (Hb) and oxygenated (HbO2) blood with the isosbestic point of blood at 800 nm. For 750 and 850 nm, the absorption coefficients for oxygenated and deoxygenated blood are well separable.
After subtracting the mean of the data, PCA was performed on the collection of 3-D pixel vectors {x1,…,xn} of the zero mean data. We first diagonalize the covariance matrix
| (5) |
where X=(x1,…,xn) is the zero mean data matrix. The three eigenvectors p1,p2,p3—the principal components ordered according to the magnitude of their eigenvalues—provide the transformed data
| (6) |
where W=(p1p2p3). Rearranging the vectors in Y into matrices yields again three 2-D images. The first image represents the projected data along the first eigenvector. As mentioned above, blood is the dominant chromophore and it will turn out that the first layer correlates with blood volume. The second image is each data point’s projection along the second eigenvector, and we will show that it correlates with blood oxygenation. The computational time to process one set of data (three images at 21 timepoints) on a 2.6-GHz, 3-GB RAM PC was in the order of 1 s.
Results
In order to evaluate the potential of PCA to retrieve the spatial distribution of chromophores, we first generate a conventional pixelwise reconstruction of blood volume and oxygenation over an imaging area of several centimeters. We then compare results after applying PCA to the contributions of the principal components. The objective was to demonstrate the anticipated response (ischemia and reactive hyperemia) for all subjects and a qualitative and quantitative assessment of the consistency between reconstruction results and the eigenvectors.
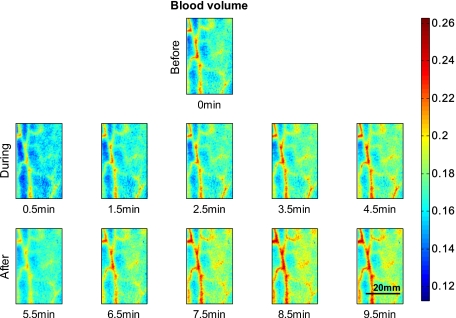
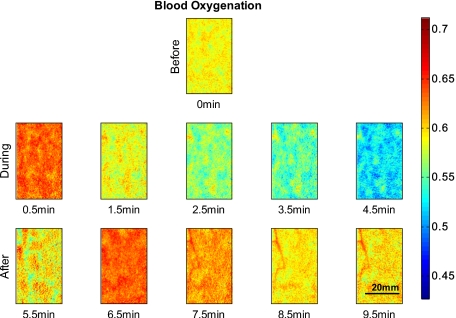
Figure 2 shows 2-D reconstruction maps of fractional blood volume concentrations over time for one representative healthy volunteer’s lower forearm. Only every second time point is shown for conciseness, with the first row showing the baseline before occlusion, rows 2 and 3 showing results during and after occlusion, respectively. Veins contain more blood than the surrounding tissue and are clearly separable in the reconstruction maps by increased blood volume. Figure 3 shows the corresponding blood oxygenation result over time, before, during, and after occlusion. Veins do not show a significant difference in blood oxygenation compared to surrounding tissue and cannot be easily separated. The overall tissue oxygenation follows the typical expected trend of ischemia during occlusion (drop of oxygenation compared to baseline) and reactive hyperemia after occlusion (over shoot compared to baseline).
Figure 2.
Fractional blood volume concentrations over time. The first row shows the reconstructed blood volume map over several centimeters of a lower forearm before occlusion. The second row shows blood volume of the same area of the arm during occlusion, and the third row shows results after release of pressure. Veins are clearly distinguishable due to the increase in blood compared to surrounding tissue.
Figure 3.
Fractional blood oxygenation over time of the same area as shown in Fig. 1. The second row shows blood oxygenation during occlusion and the expected ischemic behavior, which is a decrease in oxygenated blood. The third row shows the expected hyperemic behavior, which is an overshoot of oxygenation.
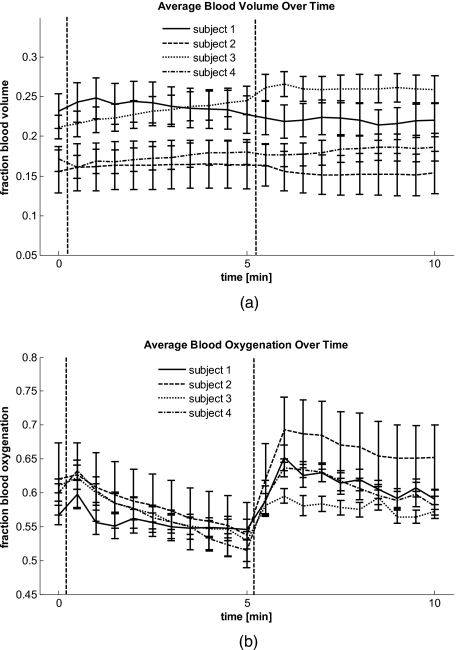
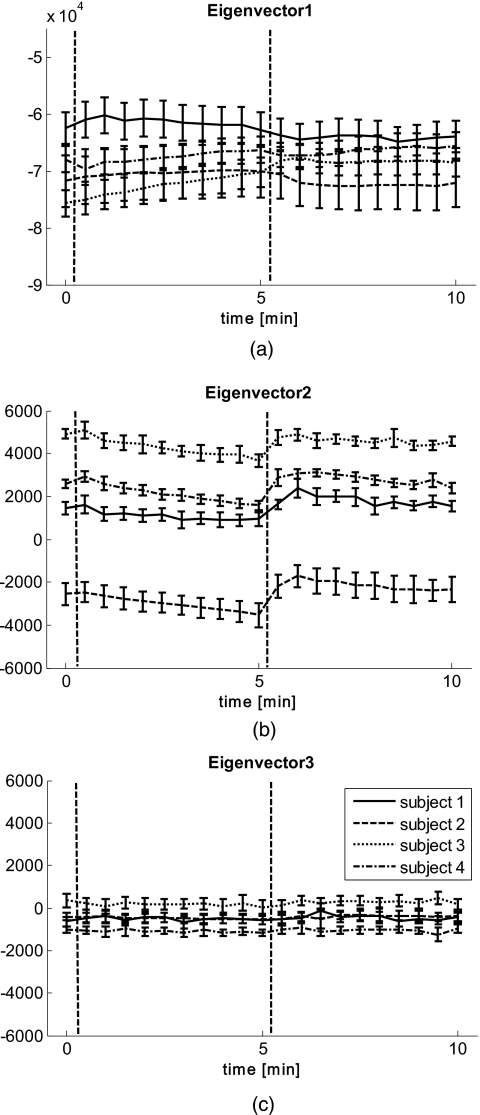
To evaluate the ischemic and hyperemic behavior even further, average concentrations of blood volume and oxygenation were calculated over the entire 2-D maps for each time point. The results for four subjects can be seen in Fig. 4a for blood volume and Fig. 4b for blood oxygenation, with error bars given by the standard deviation over all pixels per time point. Only four of the five subjects were included in the analysis, as the analysis of the fifth subject was hindered by motion artifacts greater than one-third of the image size. The dashed lines indicate the time points of beginning and ending of applied pressure. Figure 4a shows that blood volume stays over time within 5% variation, while Fig. 4b shows changes in blood oxygenation over time, corresponding to ischemia during and reactive hyperemia after occlusion.
Figure 4.
(a) Average blood volume and (b) Blood oxygenation over time for four subjects. The dashed lines indicate start and end of occlusion.
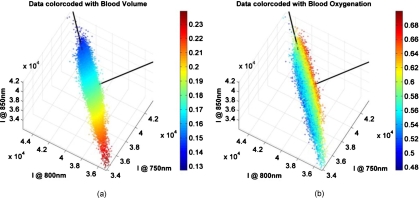
Figure 5 shows a set of three wavelength images (750, 800, and 850 nm), represented in a 3-D scatter plot in the wavelength space, with each pixel being color coded by the reconstruction results of blood volume [Fig. 5a] and blood oxygenation [Fig. 5b]. Data shown in Fig. 5 are from the same subject as in Figs. 23 at the twelveth time point, which is the first point after occlusion. All other subjects and time points show a similar behavior, which can be summarized as follows: (i) the data are of elliptical distribution, indicating that they can be described by a linear transformation. (ii) The reconstruction values of blood volume lay along the main axis of the ellipse, which will be described as eigenvector 1 [vertical black line in Figs. 5a, 5b], and are well separable. (iii) The reconstruction results of blood oxygenation show a separation perpendicular to the main axis of the ellipse, which will be described as eigenvector 2 [horizontal black line in Figs. 5a, 5b]. All three points combined indicate that a linear transform of the data by PCA is not only valid, but that it should be possible to separate blood volume and oxygenation. The separation between blood volume and oxygenation with PCA can only be possible if alignment perpendicular to each other is given. This is an inherent necessity for PCA because data are being described with eigenvectors, which are perpendicular to each other.
Figure 5.
3-D scatterplots of wavelength specific intensity data color coded with (a) blood volume and (b) blood oxygenation. The black lines indicate the eigenaxes found by PCA. Eigenvector 1 is aligned with blood volume, and eigenvector 2 with blood oxygenation. (Color online only.)
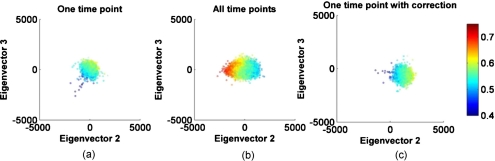
We subsequently performed PCA for each set of wavelength images and time points for all subjects. Doing so, we obtained three eigenvectors per time point and subject. Eigenvector 1 was found to be the same for each subject and over all 21 time points, except for small angular shifts (data not shown), and being well aligned with blood volume [see Fig. 5a]. Stability over time and subjects in eigenvector 1 indicates reliability in calculation. Eigenvector 2 and 3 showed large deviations within one subject and no consistent pattern was found over time, indicating that the separation between eigenvectors 2 and 3 was ambiguous. An ambiguity in eigenvectors 2 and 3 indicates that blood volume and oxygenation cannot be reliably separated. Neither eigenvector 2 nor 3 showed any temporal change similar to blood oxygenation over time. Figure 6a demonstrates this nonspecific behavior, showing a transformed data set along the second and third eigenvector, color coded by blood oxygenation. Data shown here are from the same subject as in Fig. 5, but at different time points. Because the apparent structure of the oxygenation results is not aligned with either eigenvector, PCA failed to attribute one specific direction to blood oxygenation when performed on one single data set of three wavelength images.
Figure 6.
Transformed data after PCA in the eigenvectors 2 and 3 plane, color coded with the corresponding blood oxygenation results. (a) shows PCA performed on one time point, (b) over the data including all time points, and (c) shows the transformed data of one time point using the set of eigenvectors obtained in (b). (Color online only.)
The blood oxygenation color-coded data appeared aligned and perpendicular to the primary axis of the ellipse when looking at each time point separately. The next step in the analysis was therefore to create one large data set per subject, which included all time points (21×3 wavelength images) and resulted into one set of eigenvectors per subject. Figure 6b shows the result of the same subject for the large data set color coded by blood oxygenation. The data are well aligned along the second eigenvector with blood oxygenation values being aligned in the same direction, therefore showing a dependence of the second eigenvector on oxygenation. Figure 6c shows the same data as in Fig. 6a (one time point) but transformed with the eigenvectors found by the data set including all time points. When applying the subject specific eigenvector set, the data and their oxygenation become aligned to the second eigenvector. This phenomenon is consistent for all subjects and time points, making it possible to separate blood volume and oxygenation.
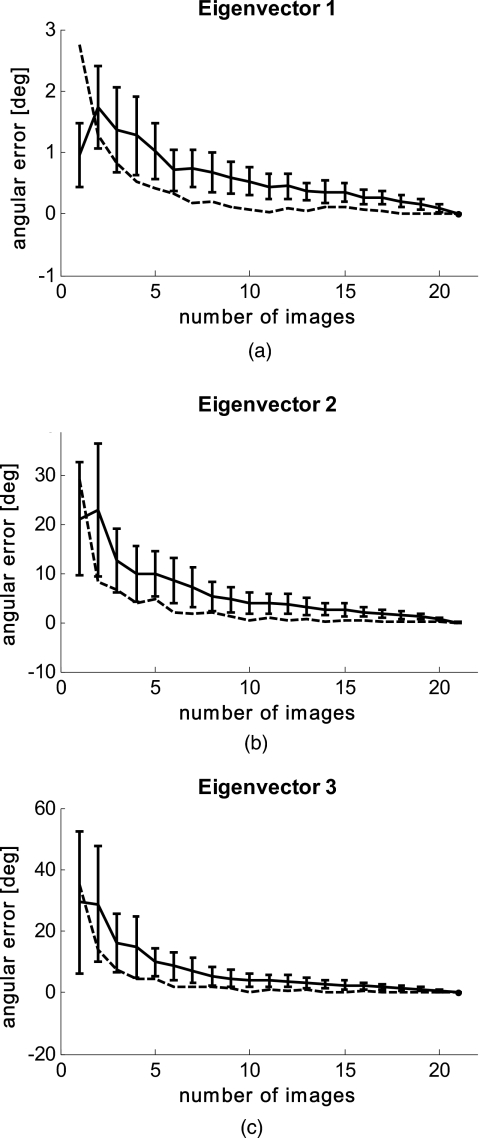
In Fig. 7, we demonstrate the validity of image summation (creating a single data set for all time points) for PCA analysis. Here we assume that the set of 21 images is sufficient to develop a patient-specific component space. We plot n against angular offset of the eigenaxes, where n is the number of images used to generate the PCA summation space. For each n we generate 100 random choices of images (note that duplication occurs only for n=1 and n=21). Figure 7a shows the plot of average angular separation of an individual summation space from the average space (21 images), with standard deviation for eigenvector 1 (data in blue). The separation of the final angular eigensystem from ground truth (given as the eigensystem of the entire image ensemble) is also shown (data in red). Figures 7b, 7c show the same data for eigenvectors 2 and 3.
Figure 7.
Validity of image summation for PCA. Solid lines are the average distance from local ensemble plus their standard deviation. Dashed lines are the distance of local ensemble average from “truth” (21 image ensemble). (a) shows data from eigenvector 1; (b) from eigenvector 2; and (c) from eigenvector 3.
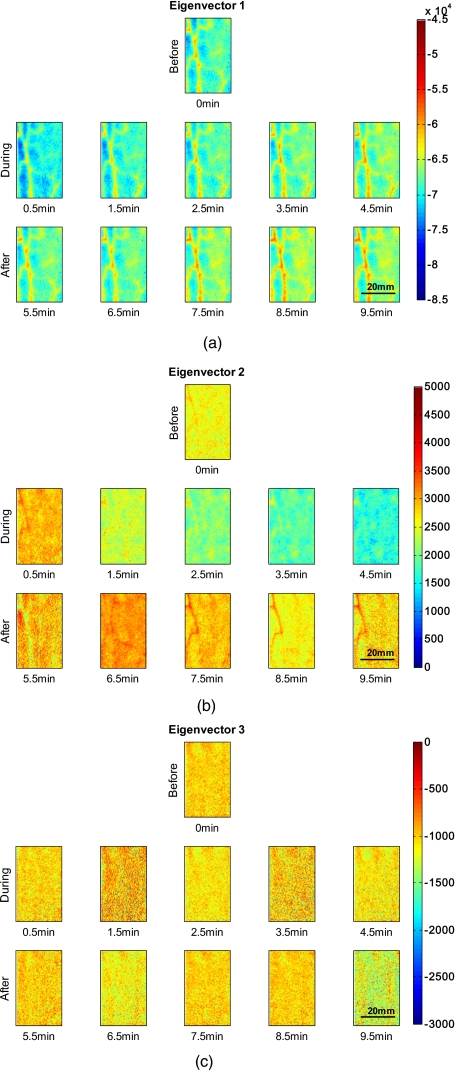
Figure 8 shows the 2-D representation of the transformed data along the three eigenvectors over time for the same subject as in Figs. 23. The eigenvectors used were obtained by taking a combined data set of all time points per subject. Figure 8a shows the result for the first eigenvector, which shows the same structures and temporal behavior as blood volume in Fig. 2. Eigenvector 2 can be seen in Fig. 8b, which shows a similar temporal and spatial appearance as blood oxygenation, as seen in Fig. 3. Figure 8c shows eigenvector 3, which does not show significant trends or structures and might be attributed to noise. It shall be mentioned that the scales in the three images are different from each other and that the first eigenvector is one magnitude larger than eigenvectors 2 and 3.
Figure 8.
Eigenvectors obtained by PCA over time for one subject. (a) shows the first eigenvector, which corresponds to blood volume; (b) shows the second eigenvector, which corresponds to blood oxygenation; and (c) shows the third eigenvector.
The results in Fig. 9 show the average distances along the eigenvectors. The average was taken as in Fig. 4 over the entire 2-D maps for each time point, with error bars given by the standard deviation over all pixels per time point. The qualitative behavior over time for eigenvector 1 [Fig. 9a] matches the behavior seen in Fig. 4a; eigenvector 2 [Fig. 9b] shows the same temporal behavior as Fig. 4b. Combined with the results from Figs. 56, the data suggest that the first eigenvector can be described by blood volume and the second one by blood oxygenation. The third eigenvector [Fig. 9c] does not show any change over time.
Figure 9.
Average eigenvectors over time for four subjects. The dashed lines indicate start and end of occlusion. (a) shows eigenvector 1, (b) eigenvector 2, and (c) eigenvector 3.
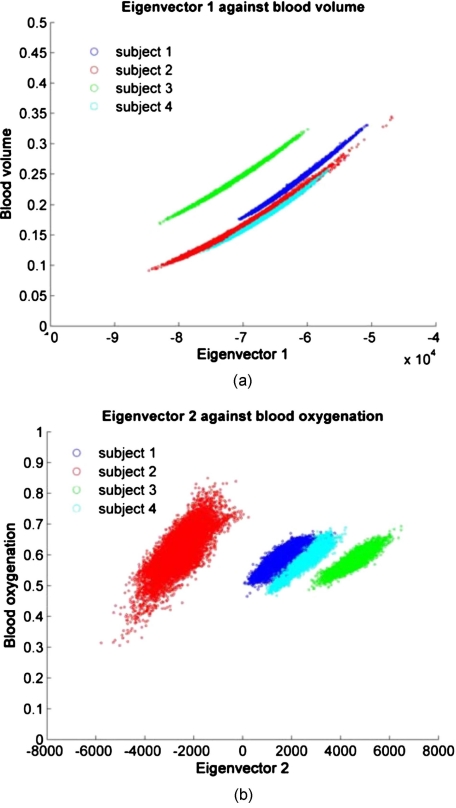
Figure 10a shows the direct comparison between eigenvector 1 and blood volume and Fig. 10b between eigenvector 2 and blood oxygenation for all pixels and time points per subject. A clear, almost linear relationship can be seen between eigenvector 1 and blood volume, indicating that these two are uniquely correlated. The same trend can be seen for all subjects, but subject 3 shows a shift toward higher blood volume values. Figure 10b shows the results for eigenvector 2 versus blood oxygenation. The relationship is not as linear but blurred compared to Fig. 10a, as well as shifted along the eigenvector axis. This indicates that blood oxygenation values can be recovered within a constant shift compared to reconstruction results.
Figure 10.
(a) Blood volume versus eigenvector 1 and (b) blood oxygenation versus eigenvector 2.
Discussion
Diffuse multispectral imaging of skin in the near-infrared wavelength range allows for quantitative chromophore assessment, which can be used to attain functional information. Blood volume and oxygenation are especially of interest for skin-lesion treatment follow-up as a parameter of the angeogenic process and metabolic state of a tumor. Previous work of assessing these parameters in healthy and diseased skin1, 2, 3, 4, 5, 6, 7, 8, 9 included fitting the acquired intensity data to an analytical model of photon migration in diffuse media. Reconstructed concentration maps are therefore based on the wavelength-dependent absorption coefficient and require an accurate model. Furthermore, model-based reconstruction is computationally intense and time consuming, therefore not applicable for real-time assessment in a clinical setting.
PCA has been used as a tool to analyze digital RGB images for extracting blood and melanin concentration and for lesion segmentation in various conditions.21, 22, 23, 24, 25 To our best knowledge, no attempt was done for extraction of blood oxygenation using PCA from RGB or multispectral images or for comparison of PCA to reconstruction results. The reconstruction results of four healthy volunteers undergoing occlusion of the upper arm and imaged at the forearm (Fig. 234) presented in this work demonstrate that the spatial distribution (Fig. 8) and temporal behavior (Fig. 9) of eigenvector 1 matches that of blood volume and eigenvector 2 matches that of blood oxygenation. In order to obtain these results, one data set, including all time points per subjects, was created, because we have seen that PCA does not otherwise pick up on blood oxygenation. Surprisingly, the subject-specific eigenvector set is the same for all four subjects with only small angular shifts of <7 deg (data not shown). This overlap in eigenvectors indicates that it might be possible to create one general set of eigenvectors describing blood volume and oxygenation. This of course raises the question if such a description is valid when applied to a different data set, i.e., from skin lesions, and if the transformation still holds. In addition, arterial blood could not be imaged because arteries lie deeper in the skin. It would be of interest to validate the PCA results with arterial oxygenation values, which are on the order of 99%.
The results also indicate an almost linear dependence of the first eigenvector with blood volume [Fig. 10a], demonstrating that values from PCA can be converted to actual concentrations of blood volume. Only subject 3 showed a deviation from the others in terms of a shift toward higher values of blood volume. This shift might be explained by the nature of our reconstruction algorithm, which assumes the same epidermal thickness for all subjects’ forearms (60 μm). We have data (unpublished), which shows that a difference of the epidermal thickness within the standard deviation of reported literature values for arms will lead to a significant overestimation of blood volume. The shift in Fig. 10a, can therefore be explained by the inaccuracy of the reconstruction rather than due to PCA errors.
Figure 10b indicates that there is a defined relationship between eigenvector 2 and blood oxygenation, but it is much more blurred and shifted along the eigenaxis. We hypothesize that the blurring might be partially explained by reconstruction errors of blood oxygenation as well as PCA itself. As mentioned above, PCA applied to one single image did not successfully align the axis of oxygenation changes to one specific eigenaxis and a data set including all time points was required to do so. The data were therefore centered on the mean of the large data set, not the individual ones. Because the second principal component is one order of magnitude smaller than the first component, even small shifts in the data over time will lead to relatively larger errors (blurring) compared to the first component. The shift along the second eigenvector, in particular for subject 2 [Fig. 10b], might well be explained by centering the large data set rather than the individual ones. When transforming the data with a given set of eigenvectors, the resulting data are not necessarily centered on the origin, leading to shifts along the axes.
The results from the direct comparison between reconstruction of blood volume and oxygenation with the PCA results of the same multispectral image set indicates that PCA may be a viable alternative tool for skin chromophore assessment. Future work will have to include phantom experiments as well as skin structure assessment for improvement of the reconstruction results and thus explaining remaining variation in the direct comparison between PCA and reconstruction results. Because PCA is considerably faster (three orders of magnitude) compared to the time-consuming reconstruction algorithms commonly used, it may provide a significant advantage for extracting metabolic information results in real time. Future work will include applying PCA to multispectral data from skin surface lesions, as well as acquiring more data from healthy volunteers to increase statistical power.
Conclusion
We acquired multispectral images from healthy volunteers’ lower forearms during an occlusion experiment and compared reconstruction results for blood oxygenation and blood volume with results from PCA. Reconstruction was performed by fitting the data to our two layered analytical skin model and blood oxygenation results showed the expected course of ischemia during occlusion and reactive hyperemia afterwards. PCA was performed on a large data set of all time points per subject, and one subject specific set of eigenvectors was used to transform the time-dependent data. Results showed that the first principal component corresponds well to the time course and spatial distribution of blood volume, the second one to blood oxygenation, and the third one did not show any temporal or spatial change. A direct comparison between reconstruction results and principal components showed a linear dependence between blood volume and blood oxygenation with the first and second component, respectively. The correspondence between blood oxygenation and the second principal component was blurred, and a shift between subjects was observed, indicating that the dependence is more susceptible to noise and errors. The results are encouraging and demonstrate the potential of PCA for quantitative skin chromophore assessment. The biggest advantage of PCA compared to reconstruction algorithms is its computational inexpensiveness, with PCA being three orders of magnitude faster. This allows for real-time mapping of skin chromophores with PCA and would therefore find great use in clinical routines.
Acknowledgments
The research was funded by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The Graduate Partnership Program at the National Institutes of Health and the Department of Physics at the University of Vienna in Austria are also acknowledged.
References
- Jacques S. L., Ramella-Roman J. C., and Lee K., “Imaging skin pathology with polarized light,” J. Biomed. Opt. 7(3), 329–340 (2002). 10.1117/1.1484498 [DOI] [PubMed] [Google Scholar]
- Mantis G. and Zonios G., “Simple two-layer reflectance model for biological tissue applications,” Appl. Opt. 48(18), 3490–3496 (2009). 10.1364/AO.48.003490 [DOI] [PubMed] [Google Scholar]
- Miyamae Y., Yamakawa Y., Kawabata M., and Ozaki Y., “A noninvasive method for assessing interior skin damage caused by chronological aging and photoaging based on near-infrared diffuse reflection spectroscopy,” Appl. Spectrosc. 62(6), 677–681 (2008). 10.1366/000370208784658156 [DOI] [PubMed] [Google Scholar]
- Tseng S. H., Bargo P., Durkin A., and Kollias N., “Chromophore concentrations, absorption and scattering properties of human skin in-vivo,” Opt. Express 17(17), 14599–14617 (2009). 10.1364/OE.17.014599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A., Chernomordik V. V., Riley J. D., Hassan M., Amyot F., Dasgeb B., Demos S. G., Pursley R., Little R. F., Yarchoan R., Tao Y., and Gandjbakhche A. H., “Using noninvasive multispectral imaging to quantitatively assess tissue vasculature,” J. Biomed. Opt. 12(5), 051604 (2007). 10.1117/1.2801718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakian C., Pretty I., Ellwood R., and Hamlin D., “In vivo quantification of gingival inflammation using spectral imaging,” J. Biomed. Opt. 13(5), 054045 (2008). 10.1117/1.2982536 [DOI] [PubMed] [Google Scholar]
- Attas M., Hewko M., Payette J., Posthumus T., Sowa M., and Mantsch H., “Visualization of cutaneous hemoglobin oxygenation and skin hydration using near-infrared spectroscopic imaging,” Skin Res. Technol. 7(4), 238–245 (2001). 10.1034/j.1600-0846.2001.70406.x [DOI] [PubMed] [Google Scholar]
- Kainerstorfer J., Amyot F., Demos S. G., Hassan M., Chernomordik V., Hitzenberger C. K., Gandjbakhche A. H., and Riley J. D., “Quantitative assessment of ischemia and reactive hyperemia of the dermal layers using multi-spectral imaging on the human arm,” Proc. SPIE 7369, 73690P (2009). 10.1117/12.831664 [DOI] [Google Scholar]
- Vogel A., Dasgeb B., Hassan M., Amyot F., Chernomordik V., Tao Y., Demos S. G., Wyvill K., Aleman K., Little R., Yarchoan R., and Gandjbakhche A. H., “Using quantitative imaging techniques to assess vascularity in AIDS-related Kaposi’s sarcoma,” in Conf. Proc. IEEE Eng. Med. Biol. Soc., Vol. 1, pp. 232–235 (2006). [DOI] [PubMed] [Google Scholar]
- Pearson K., “On lines and planes of closest fit to systems of points in space,” Philos. Mag. 2(6), 559–572 (1901). [Google Scholar]
- Moon H. and Phillips P. J., “Computational and performance aspects of PCA-based face-recognition algorithms,” Perception 30(3), 303–320 (2001). 10.1068/p2896 [DOI] [PubMed] [Google Scholar]
- Turk M. and Pentland A., “Eigenfaces for recognition,” J. Cogn Neurosci. 3(1), 71–86 (1991). 10.1162/jocn.1991.3.1.71 [DOI] [PubMed] [Google Scholar]
- Du Q. and Fowler J. E., “Low-complexity principal component analysis for hyperspectral image compression,” Int. J. High Perform. Comput. Appl. 22, 438–448 (2008). 10.1177/1094342007088380 [DOI] [Google Scholar]
- Ferraz A., Esposito E., Bruns R. E., and Durán N., “The use of principal component analysis (PCA) for pattern recognition in Eucalyptus grandis wood biodegradation experiments,” World J. Microbiol. Biotechnol. 14(4), 487–490 (1998). 10.1023/A:1008875730177 [DOI] [Google Scholar]
- de Haan J. R., Wehrens R., Bauerschmidt S., Piek E., van Schaik R. C., and Buydens L. M. C., “Interpretation of ANOVA models for microarray data using PCA,” Bioinformatics 23(2), 184–190 (2007). 10.1093/bioinformatics/btl572 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Abe K., Ishikawa S., Tsutsumi S., Hirota K., Aburatani H., and Hirose M., “A PCA based method of gene expression visual analysis,” Genome Informatics 14, 346–347 (2003). [Google Scholar]
- Sghaier-Hammami B., Valledor L., Drira N., and Jorrin-Novo J. V., “Proteomic analysis of the development and germination of date palm (Phoenix dactylifera L.) zygotic embryos,” Proteomics 9(9), 2543–2554 (2009). 10.1002/pmic.200800523 [DOI] [PubMed] [Google Scholar]
- Begelman G., Zibulevsky M., Rivlin E., and Kolatt T., “Blind decomposition of transmission light microscopic hyperspectral cube using sparse representation,” IEEE Trans. Med. Imaging 28(8), 1317–1324 (2009). 10.1109/TMI.2009.2015145 [DOI] [PubMed] [Google Scholar]
- Mandelkow H., Brandeis D., and Boesiger P., “Good practices in EEG-MRI: the utility of retrospective synchronization and PCA for the removal of MRI gradient artefacts,” Neuroimage 49(3), 2287–2303 (2010). 10.1016/j.neuroimage.2009.10.050 [DOI] [PubMed] [Google Scholar]
- Nuffer L. L., Medvick P. A., Foote H. P., and Solinsky J. C., “Multispectral/hyperspectral image enhancement for biological cell analysis,” Cytometry A 69(8), 897–903 (2006). [DOI] [PubMed] [Google Scholar]
- She Z., Liu Y., and Damatoa A., “Combination of features from skin pattern and ABCD analysis for lesion classification,” Skin Res. Technol. 13(1), 25–33 (2007). 10.1111/j.1600-0846.2007.00181.x [DOI] [PubMed] [Google Scholar]
- Cheng Y., Swamisai R., Umbaugh S. E., Moss R. H., Stoecker W. V., Teegala S., and Srinivasan S. K., “Skin lesion classification using relative color features,” Skin Res. Technol. 14(1), 53–64 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hance G., Umbaugh S. E., Moss R. H., and Stoecker W. V., “Unsupervised color image segmentation,” IEEE Eng. Med. Biol. 96, 104-111 (1996). 10.1109/51.482850 [DOI] [Google Scholar]
- Umbaugh S. E., Moss R. H., Stoecker W. V., and Hance G., “Automatic color segmentation algorithms,” IEEE Eng. Med. Biol. 93, 75-82 (1993). 10.1109/51.232346 [DOI] [PubMed] [Google Scholar]
- Tsumura N., Haneishi H., and Miyake Y., “Independent-component analysis of skin color image,” J. Opt. Soc. Am. A 16(9), 2169–2176 (1999). 10.1364/JOSAA.16.002169 [DOI] [PubMed] [Google Scholar]
- Fadzil M. H., Norashikin S., Suraiya H. H., and Nugroho H., “Independent component analysis for assessing therapeutic response in vitiligo skin disorder,” J. Med. Eng. Technol. 33(2), 101–109 (2009). 10.1080/03091900802454459 [DOI] [PubMed] [Google Scholar]
- Nugroho H., Fadzil M. H., Yap V. V., Norashikin S., and Suraiya H. H., “Determination of skin repigmentation progression,” in Conf. Proc. IEEE Eng. Med. Biol. Soc. pp. 3442–3445 (2007). [DOI] [PubMed] [Google Scholar]
- Demos S. G. and Alfano R. R., “Optical polarization imaging,” Appl. Opt. 36(1), 150–155 (1997). 10.1364/AO.36.000150 [DOI] [PubMed] [Google Scholar]
- Cuccia D. J., Bevilacqua F., Durkin A. J., Ayers F. R., and Tromberg B. J., “Quantitation and mapping of tissue optical properties using modulated imaging,” J. Biomed. Opt. 14(2), 024012 (2009). 10.1117/1.3088140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merschbrock U., Hoffmann J., Caspary L., Huber J., Schmickaly U., and Lubbers D. W., “Fast wavelength scanning reflectance spectrophotometer for noninvasive determination of hemoglobin oxygenation in human skin,” Int. J. Microcirc.: Clin. Exp. 14(5), 274–281 (1994). 10.1159/000178841 [DOI] [PubMed] [Google Scholar]
- Kainerstorfer J., Amyot F., Ehler M., Hassan M., Demos S. G., Chernomordik V., Hitzenberger C. K., Gandjbakhche A. H., and Riley J. D., “Direct curvature correction for non-contact imaging modalities—applied to multi-spectral imaging,” J. Biomed. Opt. 15(4), 046013 (2010). 10.1117/1.3470094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques S. L., “Skin optics,” Oregon Med. Laser Center News, http://omlc.ogi.edu/news/jan98/skinoptics.html (1998).
- Meglinski I. V. and Matcher S. J., “Quantitative assessment of skin layers absorption and skin reflectance spectra simulation in the visible and near-infrared spectral regions,” Physiol. Meas 23(4), 741–753 (2002). 10.1088/0967-3334/23/4/312 [DOI] [PubMed] [Google Scholar]
- Prahl S., “Skin optics,” Oregon Med. Laser Center News, http://omlc.ogi.edu/spectra/hemoglobin/index.html (1998).
- Gandjbakhche A. H. and Weiss G. H., “Random walk and diffusion-like models of photon migration in turbid media,” Prog. Opt. XXXIV(V), 335–402 (1995). [Google Scholar]