Abstract
Freshly isolated muscle stem cells (MuSCs) exhibit robust regenerative capacity in vivo that is rapidly lost in culture. Using a bioengineered substrate to recapitulate key biophysical and biochemical niche features in conjunction with a novel highly automated single cell tracking algorithm, we show that substrate elasticity is a potent regulator of MuSC fate in culture. Unlike MuSCs on rigid plastic dishes (~106kPa), MuSCs cultured on soft hydrogel substrates that mimic the elasticity of muscle (12kPa) self-renew in vitro and contribute extensively to muscle regeneration when subsequently transplanted into mice and assayed histologically and quantitatively by non-invasive bioluminescence imaging. Our studies provide novel evidence that by recapitulating physiological tissue rigidity, propagation of adult muscle stem cells is possible, enabling future cell-based therapies for muscle wasting diseases.
Many adult tissues harbor tissue-specific stem cells that have remarkable regenerative capacity when freshly isolated, a property that is rapidly lost once plated in culture. The muscle microenvironment (niche) enables freshly isolated muscle stem cells (MuSCs) to contribute extensively to skeletal muscle regeneration when transplanted in mice (1–8). In contrast, muscle stem cells grown on standard tissue culture plastic lose ‘stemness’ yielding progenitors with greatly diminished regenerative potential (3, 7, 9) and therapeutic potency (10). Indeed, the propagation of functional adult stem cells, including MuSCs, is currently not possible in culture, despite extensive research on biochemical signals. Biophysical properties such as matrix rigidity are known to alter the differentiation of cells in culture (11). Here we test the hypothesis that elastic modulus plays a crucial role in muscle stem cell self-renewal and function in muscle regeneration. We report that when MuSCs are cultured on a substrate that mimics the rigidity of muscle tissue, they self-renew to generate stem cell progeny that can potently repair damaged muscle, establishing a paradigm for overcoming a major roadblock to adult stem cell therapeutic utility.
To recapitulate muscle rigidity and uncouple biophysical from biochemical effects, we engineered a tunable polyethylene glycol (PEG) hydrogel platform. By altering the percentage of PEG polymer in the precursor solution we produced hydrogels with a range of rigidities (Figs. 1A and 1B top) including a formulation that mimics the elastic modulus of adult murine skeletal muscle (Figs. 1A and 1C) (12). Notably, polystyrene plastic traditionally used for cell culture, has an elastic modulus of ~3 GPa, more than five orders of magnitude stiffer than skeletal muscle (13). Laminin, a component of the native MuSC niche, was covalently crosslinked to the hydrogel network and used as an adhesion ligand (Fig. 1B bottom). To ensure that laminin density and surface chemistry remained consistent between hydrogel and plastic culture conditions, we generated gels that do not swell post-polymerization (Figs. 1D; S1) and cast a thin layer of PEG hydrogel (≤1 μm) on the plastic surface, allowing MuSCs to sense the stiffness of the plastic beneath (see Supplemental Methods; Fig. 1D) (12).
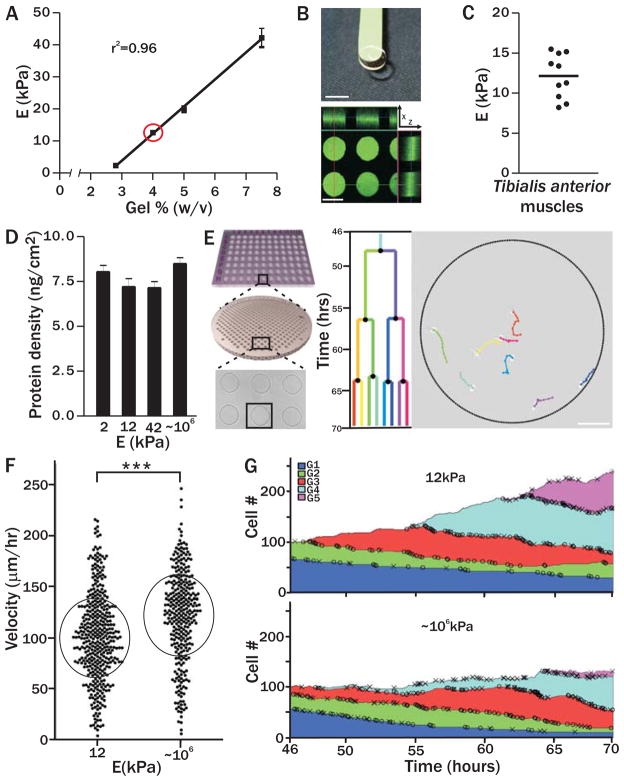
Figure 1. Pliant hydrogel promotes MuSC survival and prevents differentiation in culture.
(A) PEG hydrogels with tunable mechanical properties. Young’s modulus is linearly correlated with precursor polymer concentration; (E; n=4) red circle indicates muscle elasticity. (B) Image of a pliant PEG hydrogel on a green spatula. Scale bar, 7mm (top). Confocal immunofluorescence image of hydrogel microcontact printed with laminin specifically at the bottom of hydrogel microwells (i.e. from the ‘tips’ of the micropillars). Scale bar, 125 μm (bottom). (C) Dissected tibialis anterior muscles (n=5 animals, 10 muscles total) were analyzed by rheometry (horizontal line indicates the mean). (D) Gel surface protein density did not differ significantly on PEG hydrogels of different rigidities (Young’s modulus, E; p>0.05) and was 7.6 ng/cm2 ± 1.0 ng/cm2 (n≥4). (E) Scheme of Baxter Algorithm analysis of timelapse videos. Hydrogel arrays with hundreds of microwells containing single MuSCs were followed by timelapse microscopy for 3 days. Videos were automatically processed and analyzed (Supplemental Methods). Scale bar, 100 μm. (F) Single MuSC (black data points) velocity on pliant or rigid culture substrates. Circles denote mean velocity ± standard deviation (p<0.0001). (G) Change in total MuSC number on soft (top plot) or stiff (bottom plot) substrates during timelapse acquisition. Deaths (X) and divisions (O) are shown and colors designate five cell generations (G1–G5). The proportion of cells in each generation at all timepoints is shown. Cell number is normalized to a starting population of 100 single MuSCs.
MuSCs were prospectively isolated (7) and analyzed at the single cell level on pliant or stiff culture surfaces patterned with arrays of microwells (Fig. 1E) (14), because even when FACS enriched, stem cell populations are inherently heterogeneous (4, 6–8, 14, 15). Timelapse acquisition of hundreds of single stem cells is possible using microwell technology (14); however, analysis of the resulting immense data sets remains a major challenge. To enable rapid analysis, we developed a highly automated algorithm termed the ‘Baxter Algorithm’ (Supplemental Methods), which in contrast to most commercially available software, is able to track lineage relationships over multiple cell divisions. This algorithm reduced data analysis time by ~90% with a mere 1% error rate.
The genealogic history of clones derived from a single cell was established by timelapse acquisition and automated tracking (Fig. 1E, Supplemental Video). MuSC velocity, increased on stiff (120μm/hr) compared to pliant (99μm/hr) culture substrates, (Fig. 1F, p<0.0001), in agreement with previous reports investigating cell lines (16). In addition, we observe that cell area increases as cells duplicate their content, returning to initial cell area after mitosis, further validating our segmentation parameters (Fig. S2).
MuSCs propagated on pliant hydrogel substrates do not undergo the ‘crisis’, or massive cell death previously reported in culture (9, 17). After one week of culture in soft microwells, twice as many cells give rise to clones as compared to cells cultured in rigid microwells (Fig. S3). Using the Baxter Algorithm we characterized this phenomenon at the clonal level. On rigid substrates, the overall cell number does not change over time because division is offset by death; however, on pliant substrates death is reduced and the total number of cells increases over time (Figs. 1G, S4 and S5). In both conditions, death is not sudden; indeed it is independent of time and cell division number (Figs. 1G and S4). This data demonstrates that culture on soft substrates augments MuSC survival.
Substrate rigidity also impacts gene expression, suggesting that MuSC stemness is retained on soft surfaces. MuSCs cultured for one week on soft substrates give rise to 3-fold fewer cells that express Myogenin, a myogenic transcription factor expressed by differentiated MuSCs, than MuSCs cultured on rigid substrates (Fig. S6). Timelapse analysis excludes the possibility that gene expression differences are due to differences in cell division, as we observe no statistically significant difference in MuSC time to first division (Fig. S7) or time between divisions (Fig. S8). In addition, division rate is not different on pliant compared to rigid substrates (Fig. S9 and Supplemental Methods). These in vitro studies demonstrate that substrate rigidity has no effect on cell division rate in culture but prevents differentiation and leads to increased cell numbers by enhancing viability.
To establish definitively that MuSCs cultured on pliant substrates retain stemness, their function was assessed in vivo. MuSCs were isolated from mice constitutively expressing firefly luciferase (Fluc) (18) and green fluorescent protein (GFP) and cultured for 7 days. Cells were harvested, counted and cultured MuSC progeny were transplanted into the tibialis anterior (TA) muscles of immunodeficient mice depleted of endogenous MuSCs by 18Gy leg-irradiation (15, 19). We assayed donor cell behavior quantitatively over time by non-invasive in vivo bioluminescence imaging (BLI) of luciferase activity, which correlates with cell number (Fig. 2A) (7). The engraftment threshold that was set as the bioluminescence value corresponding with histological detection of donor derived myofibers (Fig. S10). This in vivo functional assay is critical for validating the stemness of cultured MuSCs.
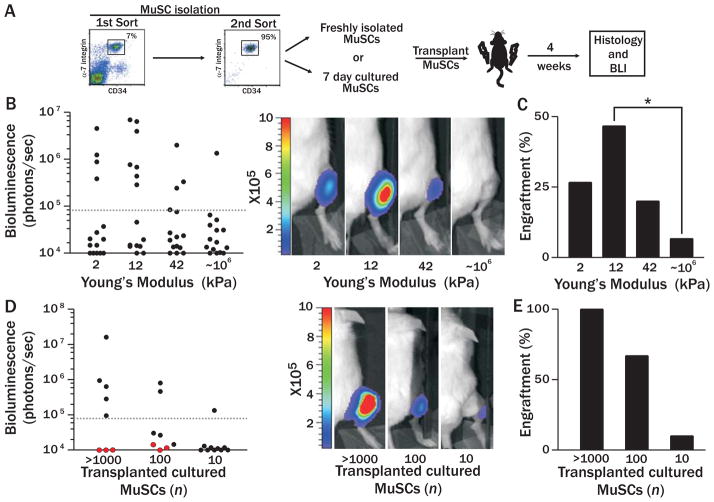
Figure 2. Cultured MuSC engraftment is modulated by substrate elasticity.
(A) Scheme of in vivo transplantation experiments. (B) Scatter graph of BLI values of recipient mice one month after transplantation with 100 GFP/Fluc MuSCs after 7 day culture on substrates of varying stiffness (left; n=15). Representative bioluminescence images of animals transplanted with each culture condition are shown (right; photons/sec/cm2/sr). (C) Percentage of animals from each experimental condition that had a BLI value above the engraftment threshold. Fisher’s exact test p<0.05. (D) Scatter graph of BLI values of recipient mice one month after transplantation with different numbers of Fluc MuSCs cultured for 7 days on either hydrogel (black) or plastic (red). Representative bioluminescence images of animals transplanted with each culture condition are shown (right; photons/sec/cm2/sr). (E) Percentage of total transplanted animals in each experimental condition exhibiting a BLI value above the engraftment threshold.
In vivo functional assays show that MuSCs cultured on substrates matching the physiological modulus of muscle tissue most potently retain stemness. Hydrogel substrates were tuned to mimic the endogenous in vivo mechanical properties of brain, muscle and cartilage (2, 12, or 42 kPa) compared to polystyrene plastic (~106 kPa). In agreement with previous reports (3, 7, 9), we observe markedly reduced engraftment from MuSCs cultured on plastic (Fig. 2B). Strikingly, the highest bioluminescent signals are obtained from mice transplanted with MuSCs cultured on the most pliant hydrogels, whereas both the extent and rate of engraftment are decreased on the stiffest culture substrates (Fig. 2B). Notably, the culture substrate that recapitulates skeletal muscle elasticity (12kPa) is the only condition that leads to a statistically significant increase in the percentage of mice with MuSC engraftment compared to tissue culture plastic (Fig. 2C).
To determine the proportion of cultured cells with engraftment potential, we cultured MuSCs for one week on either soft or rigid substrates and then performed dilution analysis. None of the mice transplanted with stem cells cultured on a rigid plastic substrate exhibit a BLI signal above the threshold of detection (Fig. 2D; red circles). By contrast, engraftment occurs with 100% frequency when ≥1000 hydrogel-cultured (12kPa) stem cells are transplanted (Fig. 2D; black circles), similar to freshly isolated cells (7). Moreover, 10% of mice transplanted with only 10 hydrogel cultured cells exhibit engraftment (Fig. 2E), on par with transplantations of 10 freshly isolated cells (16%) (7).
MuSCs cultured on a substrate that mimics muscle tissue exhibit dynamic proliferative behavior similar to freshly isolated MuSCs when transplanted in vivo. Both cell populations undergo a period of extensive proliferation that ultimately plateaus and stabilizes when homeostasis is achieved (Fig. 3A). Histology identifies GFP+ myofibers resulting from regeneration in animals transplanted with MuSCs that were freshly isolated or cultured on soft substrates (Fig. 3B). While the engraftment rate of freshly isolated MuSCs (7) and those grown on soft substrates is comparable (Fig. 2C), the extent of engraftment from cultured MuSCs is not as robust as that of freshly isolated cells (Fig. 3A), suggesting that exposure to additional biochemical cues in vitro may be required to recapitulate other key niche components necessary for maximal function in vivo.
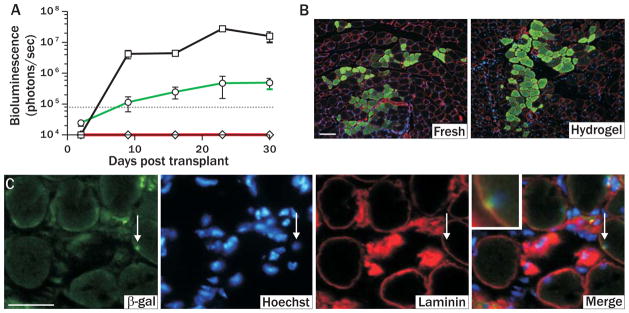
Figure 3. Culture on pliant hydrogel promotes muscle stem cell engraftment and niche repopulation in vivo.
(A) Engraftment of freshly isolated (black line, squares; 500 cells) and pliant (green line, circles; 1500 cells) or rigid (red line, diamonds; 1500 cells) substrate cultured MuSCs monitored by BLI for a period of 30 days post transplantation (p<0.05). (B) Immunofluorescence of GFP expression in transverse sections of muscles one month after transplantation with freshly isolated (left; 500 transplanted cells) or one week pliant hydrogel cultured MuSCs (right; 2500 transplanted cells). GFP=green, Laminin=red, and Hoechst=blue. Scale bar, 100μm. (C) Immunohistochemical analysis of transverse muscle tissues one month after transplant with pliant substrate cultured MuSCs. Arrow points to a donor derived cell in satellite cell position. β-galactosidase=green, Laminin=red, and Hoechst=blue. Scale bar, 50μm.
MuSCs cultured on soft substrates can home to their native satellite cell niche upon transplantation into muscle, a defining characteristic of freshly isolated MuSCs (3, 7). MuSCs isolated from mice expressing LacZ under the regulation of the Myf5 promoter (20) were cultured on soft hydrogel and subsequently transplanted into mice. Prior to harvesting tissues, recipient muscles were damaged with notexin (21), which activates MuSCs and upregulates expression of the myogenic transcription factor Myf5 (7, 20). Histological analysis of β-galactosidase (β-gal) staining reveals that, like freshly isolated cells (7), transplanted hydrogel cultured cells expressing Myf5 are found in the satellite cell niche, beneath the basal lamina and atop myofibers (Fig. 3C).
One of the defining characteristics of stem cells is their ability to make more copies of themselves upon division, or self-renew. Several elegant in vitro approaches have provided strong evidence that MuSC asymmetric and symmetric divisions occur in culture, consistent with self-renewal (4, 22–27). Our in vitro analysis of MuSC gene expression suggests that a pliant substrate supports self-renewal. We observe that 32% of doublets (two cells that arose from a single cell division) in pliant microwells are positive for the MuSC marker Pax7 (Figs. 4A, 4B and S11). In contrast, only 6% of doublets in plastic microwells have this gene expression pattern, suggesting that a pliant substrate enables MuSC expansion. Although gene expression data are suggestive, an in vivo functional assay is necessary to conclude definitively that a self-renewal division event occurred in culture.
Figure 4. Culture on pliant hydrogel promotes muscle stem cell self-renewal.
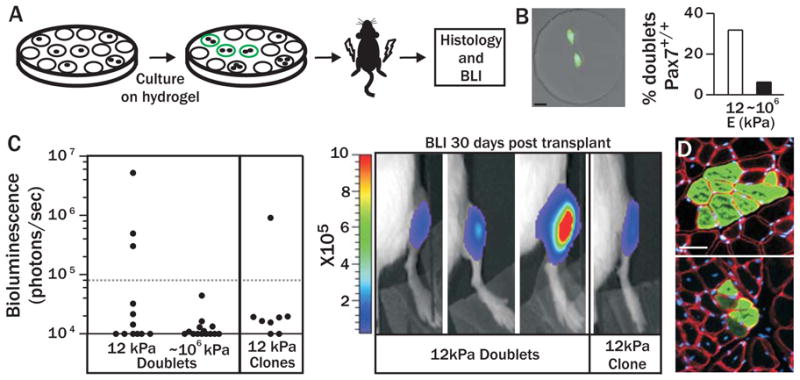
(A) Scheme of in vivo self-renewal assay. (B) Percentage of total doublets exhibiting a Pax7+/+ gene signature in pliant (n=47) or stiff (n=32) microwells (right). Representative image of a Pax7+/+ doublet (left). Native Zs-Green=green (i.e. Pax7), Hoechst=blue, scale bar, 20μm. (C) Scatter graph (left) of bioluminescent values from mice transplanted with doublets derived from pliant (n=12) or stiff (n=14) microwells or clones collected from pliant microwells (n=8). Images of animals with bioluminescent values above threshold are shown (right; photons/sec/cm2/sr). (D) Immunohistochemistry of GFP expression in transverse sections of muscles one month after transplantation with five MuSC doublets (top; GFP+ fibers persist ~13mm longitudinally) or a single clone (bottom; fibers persist ~7mm longitudinally) cultured on pliant hydrogel. GFP=green, Laminin=red, and Hoechst=blue. Scale bar, μm.
We show conclusively that stem cell self-renewal occurs using an in vivo functional assay. The transplantation of MuSCs at a population level demonstrates engraftment (Figs. 2 and 3), but does not definitively show that self-renewal divisions occurred in culture, because the population could include non-dividing cells which maintained stem cell properties. Accordingly, in this experiment, we plated MuSCs in hydrogel microwell arrays and obtained images immediately after plating and 2–3 days after culture to identify microwells that contained only one doublet. Doublets from 5 microwells were picked and pooled using a micromanipulator and 10 cells total transplanted per mouse (Fig. 4A). A detectable BLI signal indicates engraftment resulting from a self-renewal division event that must have occurred in at least one of the five transplanted doublets. Strikingly, 25% (3/12) of animals transplanted with doublets cultured on soft substrates demonstrate detectable engraftment (Fig. 4C) and contribution to regenerating myofibers (Fig. 4D, top), providing in vivo functional evidence that MuSC self-renewal division events occur in culture on pliant substrates. In contrast, doublets grown on rigid plastic microwells never exhibit engraftment following transplantation (Fig. 4C; 0/14), indicating that their regenerative potential is rapidly lost.
MuSC self-renewal on pliant hydrogel occurs even after multiple divisions. We transplanted clones that arose from a single cell which underwent 3–5 divisions. Remarkably, 12% (1/8) of animals transplanted with a single clone show engraftment, demonstrating that MuSC self-renewal capacity is retained on pliant substrates even after multiple divisions (Figs. 4C and 4D, bottom).
Here we provide novel insight into the potency of tissue rigidity, a biophysical property of the skeletal muscle microenvironment, on stem cell fate regulation. Using a novel single cell tracking algorithm to interrogate MuSC behaviors at the single cell level, we demonstrate that soft substrates enhance MuSC survival, prevent differentiation and promote stemness. Functional assays in mice demonstrate conclusively that pliant substrates permit MuSC self-renewal in culture. While the underlying mechanisms remain to be elucidated, we hypothesize that decreased rigidity preserves stemness by altering cell shape, resulting in cytoskeletal rearrangements and altered signaling as shown for cell lines (16). Despite the remarkable retention of stemness in response to a single parameter, rigidity, we anticipate further enhancement of stemness through incorporation of additional biochemical cues into our reductionist platform. Studies employing biomimetic culture platforms, such as described here for MuSCs, will broadly impact stem cell studies by facilitating in vitro propagation while maintaining stemness and the capacity to regenerate tissues, a critical step towards the development of cell-based therapies.
Supplementary Material
Acknowledgments
We would like to thank Kassie Koleckar, Rose Tran, IBM Almaden, the SCI3 and SSFF Stanford core facilities for technical support, Joe Sly, Bob Miller, Stefan Kobal, Samy Gobaa, Regis Doyonnas and Annelise Barron for valuable discussions, Fabio Rossi for providing us with alpha-7 integrin antibody, and the following Grants for financial support: NIH training grant CA09151 and CIRM TG2-01159 to P.M.G., NSF and NIH Training Grant 2 T32 HD007249 to K.H., John och Karin Engbloms stipendiefond to K.E.G.M., MDA 4063 to A.S., EPFL Excellence Scholarship and the Swiss Study Foundation to N.A.L., Bio X Undergraduate Research Award and HHMI 52005886 to N.K.N., Google to S.T., Leukemia and Lymphoma Society Fellow and EURYI to M.P.L., and NIH grants HL096113, AG020961, JDRF 34-2008-623, MDA Grant 4320, CIRM RB1-01292, CIRM RT1-01001 and the Baxter Foundation to H.M.B.
References
- 1.Cornelison DD, Filla MS, Stanley HM, Rapraeger AC, Olwin BB. Dev Biol. 2001 Nov 1;239:79. doi: 10.1006/dbio.2001.0416. [DOI] [PubMed] [Google Scholar]
- 2.Fukada S, et al. Exp Cell Res. 2004 Jun 10;296:245. doi: 10.1016/j.yexcr.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 3.Montarras D, et al. Science. 2005 Sep 23;309:2064. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 4.Kuang S, Kuroda K, Le Grand F, Rudnicki MA. Cell. 2007 Jun 1;129:999. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerletti M, et al. Cell. 2008 Jul 11;134:37. [Google Scholar]
- 6.Collins CA, et al. Cell. 2005 Jul 29;122:289. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Nature. 2008 Nov 27;456:502. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherwood RI, et al. Cell. 2004 Nov 12;119:543. doi: 10.1016/j.cell.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Qu-Petersen Z, et al. J Cell Biol. 2002 May 27;157:851. doi: 10.1083/jcb.200108150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gussoni E, et al. Nature. 1992 Apr 2;356:435. doi: 10.1038/356435a0. [DOI] [PubMed] [Google Scholar]
- 11.Lutolf MP, Gilbert PM, Blau HM. Nature. 2009 Nov 26;462:433. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006 Aug 25;126:677. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 13.Callister WD. Fundamentals of Materials Science and Engineering: An Interactive E-Text. 5. John Wiley & Sons; Somerset, NJ: 2000. [Google Scholar]
- 14.Lutolf MP, Doyonnas R, Havenstrite KL, Koleckar K, Blau HM. Integrative Biology. 2009;1:59. doi: 10.1039/b815718a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heslop L, Morgan JE, Partridge TA. J Cell Sci. 2000 Jun;113(Pt 12):2299. doi: 10.1242/jcs.113.12.2299. [DOI] [PubMed] [Google Scholar]
- 16.Pelham RJ, Jr, Wang Y. Proc Natl Acad Sci U S A. 1997 Dec 9;94:13661. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Partridge TA. In: Basic cell culture protocols. Polard JMJWaW., editor. Vol. 75. Humana Press, Inc; Totowa, NJ: 1997. pp. 131–144. [Google Scholar]
- 18.Cao YA, et al. Proc Natl Acad Sci U S A. 2004 Jan 6;101:221. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakeford S, Watt DJ, Partridge TA. Muscle Nerve. 1991 Jan;14:42. doi: 10.1002/mus.880140108. [DOI] [PubMed] [Google Scholar]
- 20.Tajbakhsh S, et al. Dev Dyn. 1996 Jul;206:291. doi: 10.1002/(SICI)1097-0177(199607)206:3<291::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 21.Harris JB, Johnson MA. Clin Exp Pharmacol Physiol. 1978 Nov–Dec;5:587. doi: 10.1111/j.1440-1681.1978.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 22.Abou-Khalil R, et al. Cell Stem Cell. 2009 Sep 4;5:298. doi: 10.1016/j.stem.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinin V, Gayraud-Morel B, Gomes D, Tajbakhsh S. Nat Cell Biol. 2006 Jul;8:677. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 24.Conboy MJ, Karasov AO, Rando TA. PLoS Biol. 2007 May;5:e102. doi: 10.1371/journal.pbio.0050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conboy IM, Rando TA. Dev Cell. 2002 Sep;3:397. doi: 10.1016/s1534-5807(02)00254-x. [DOI] [PubMed] [Google Scholar]
- 26.Le Grand F, Jones AE, Seale V, Scime A, Rudnicki MA. Cell Stem Cell. 2009 Jun 5;4:535. doi: 10.1016/j.stem.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zammit PS, et al. J Cell Biol. 2004 Aug 2;166:347. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.