Abstract
Transcription-coupled nucleotide excision repair (TC-NER) removes certain kinds of lesions from the transcribed strand of expressed genes. The signal for TC-NER is thought to be RNA polymerase stalled at a lesion in the DNA template. In Escherichia coli, the stalled polymerase is dissociated from the lesion by the transcription repair coupling factor (Mfd protein), which also recruits excision repair proteins to the site resulting in efficient removal of the lesion. TC-NER has been documented in cells from a variety of organisms ranging from bacteria to humans. In each case, the RNA polymerase involved has been a multimeric protein complex. To ascertain whether a gene transcribed by the monomeric RNA polymerase of bacteriophage T7 could be repaired by TC-NER, we constructed strains of Escherichia coli in which the chromosomal lacZ gene is controlled by a T7 promoter. In the absence of T7 RNA polymerase, little or no β-galactosidase is produced, indicating that the E. coli RNA polymerase does not transcribe lacZ efficiently, if at all, in these strains. By introducing a plasmid (pAR1219) carrying the T7 gene 1 under control of the E. coli lac UV5 promoter into these strains, we obtained derivatives in which the level of T7 RNA polymerase could be regulated. In cultures containing upregulated levels of the polymerase, β-galactosidase was actively produced indicating that the T7 RNA polymerase transcribes the lacZ gene efficiently. Under these conditions, we observed that UV-induced cyclobutane pyrimidine dimers were removed more rapidly from the transcribed strand of lacZ than from the nontranscribed strand, supporting the conclusion that TC-NER occurred in this gene. This response was absent in an mfd-1 mutant, indicating that the underlying mechanism may be similar to that for the bacterial RNA polymerase.
Keywords: transcription-coupled nucleotide excision repair, bacteriophage T7 RNA polymerase, lacZ, T7 promoter, pAR1219
1. Introduction
Nucleotide excision repair (NER) removes a variety of lesions from DNA. It is generally accepted that NER comprises two subpathways: transcription coupled nucleotide excision repair (TC-NER), and global genome nucleotide excision repair (GG-NER) (reviewed in [1]). The former repairs lesions in the transcribed strand of expressed genes, while the latter repairs lesions in the genome overall. When bacteriophage T7 damaged by UV light infects E. coli, the phage DNA can be repaired by the bacterial NER system, resulting in "host cell reactivation" of the phage [2]. It is not known, however, whether TC-NER can contribute to this reactivation.
In E. coli, TC-NER of damaged bacterial DNA requires RNA polymerase and Mfd, the product of the mfd gene, in addition to the UvrA, UvrB, and UvrC proteins, which recognize a DNA lesion and incise the strand containing it [1, 3, 4]. Mutants deficient in Mfd do not show TC-NER [5–7]. According to current ideas, lesions that block the RNA polymerase (e.g. cyclobutane pyrimidine dimers (CPDs)), are substrates for TC-NER. The blocked polymerase, however, occludes the lesion and prevents access by the incision complex [8–10]. The Mfd protein, also known as the transcription repair coupling factor, is required to displace the polymerase, allowing repair to occur. In addition to displacing the RNA polymerase, Mfd is thought to recruit UvrA to the site to initiate NER, thus enhancing the rate of repair [3, 4, 11, 12]. TC-NER is detected experimentally when the transcribed strand of an expressed gene is repaired more rapidly than the nontranscribed strand of the gene [13, 14].
The RNA polymerase of E. coli is a complex of several different proteins. In contrast, the RNA polymerase of T7 is a monomeric protein. The elongation complex of the T7 enzyme, like that of E. coli, can be stalled by a CPD in the transcribed strand of a gene [10]. Results obtained from studies of defined systems in vitro, however, indicate that the T7 polymerase can bypass CPDs more readily than can the multimeric polymerases [15, 16]. Furthermore, unlike the E. coli RNA polymerase, the T7 polymerase does not appear to interact with the Mfd protein in vitro [10]. Therefore, as a test of the generality of the proposed mechanism for TC-NER, it was of interest to evaluate the possibility that a gene transcribed by T7 RNA polymerase might show this type of repair.
We constructed E. coli derivatives in which a T7 promoter controls the lac operon, and the T7 RNA polymerase is supplied by a plasmid carrying the T7 gene 1. Our results show that TC-NER can occur in the lac operon when it is transcribed by the T7 RNA polymerase; furthermore, in mfd-1 derivatives, we did not observe TC-NER, indicating that even when a gene is transcribed by the T7 RNA polymerase Mfd is required for TC-NER.
Materials and Methods
2.1. Bacteria
Table 1 lists the derivatives of E. coli K-12 used.
Table 1.
E.coli K-12 derivatives used in these experiments. Strains with HL numbers were produced for this paper.
| Designation | Genotype | Source |
|---|---|---|
| SR108 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1 | J. Cairns |
| NC397 | F-, pglΔ8, gal490, ΔCI857, Δ (cro, bioA), | D.L. Court |
| (In NC397 the cat/sacB cassette replaces the lacZ promoter (the lacI promoter, 5' end of lacI and RBS of lacZ remain), a kanamycin resistance gene and transcription terminator are inserted directly upstream of cat/sacB.) | ||
| UNC3610-45 | F- λ-, thr-1, ara-14, leuB6, Δ(gpt-proA)62, lacY1, tsx-33, supE44, galK2, rac-, hisG4(Oc), rfbD1, mgl-51, rpsL31, kdgK51, xyl-5, mtl-1, argE3, thi-1, zcf-117::Tn10, mfd-1 |
C. Selby |
| HL1132 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ | |
| HL1134 | F-, pglΔ8, gal490, λCI857, Δ(cro, bioA), kan-T7p-lacZ | |
| HL1159 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ / pAR1219 | |
| HL1160 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ / pAR1219 | |
| HL1166 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ, zcf-117::Tn10 | |
| HL1167 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ, zcf-117::Tn10, mfd-1 | |
| HL1172 | F-, pglΔ8, gal490, λCI857, Δ (cro, bioA), kan-T7p-lacZ | |
| HL1173 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ | |
| HL1176 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ / pAR1219 | |
| HL1177 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ /pAR1219 | |
| HL1179 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ, zcf-117::Tn10, mfd-1 / pAR1219 | |
| HL1180 | F- λ-, thyA36, deoC2, IN(rrnD-rrnE)1, kan-T7p-lacZ, zcf-117::Tn10, mfd-1 / pAR1219 | |
2.2 Plasmid
The plasmid used, pAR1219 [17], contains the T7 gene 1, which encodes the T7 RNA polymerase. The gene is regulated by the lac UV5 promoter. The plasmid also carries the E. coli lacI gene, which codes for the lac repressor protein. When bacteria carrying this plasmid are grown in the presence of an inducer like isopropyl-β-D-thiogalactopyranoside (IPTG), the inducer binds to the repressor, preventing it from associating with the UV5 promoter. In the absence of an inducer, the repressor can bind to the promoter and prevent synthesis of the T7 RNA polymerase.
2.3 Media
The minimal medium (MM) was Difco Minimal Broth Davis containing 0.4% glucose and 0.05% vitamin-free casamino acids. For plates, 1.5% Difco Bacto agar was added. The complex medium was LB [18]. For LB plates, 1.2% Difco Bacto agar was added. L-broth [19] containing 1 g/l of glucose was used for preparing P1vir phage.
For thymine-requiring strains, 10 µg/ml of thymine was added to all media.
When needed, kanamycin was incorporated into plates at a concentration of 25 µg/ml, chloramphenicol at a concentration of 30 µg/ml, ampicillin at a concentration of 30 µg/ml, and tetracycline at a concentration of 15 µg/ml. To select against sacB, bacteria were plated on Difco Minimal Broth Davis containing 0.2% glycerol, 5% sucrose and 0.05% vitamin-free casamino acids.
The T7 gene 1 carried on the plasmid, pAR1219, was induced by growing cultures in MM containing 1 mM IPTG.
2.4 Recombineering
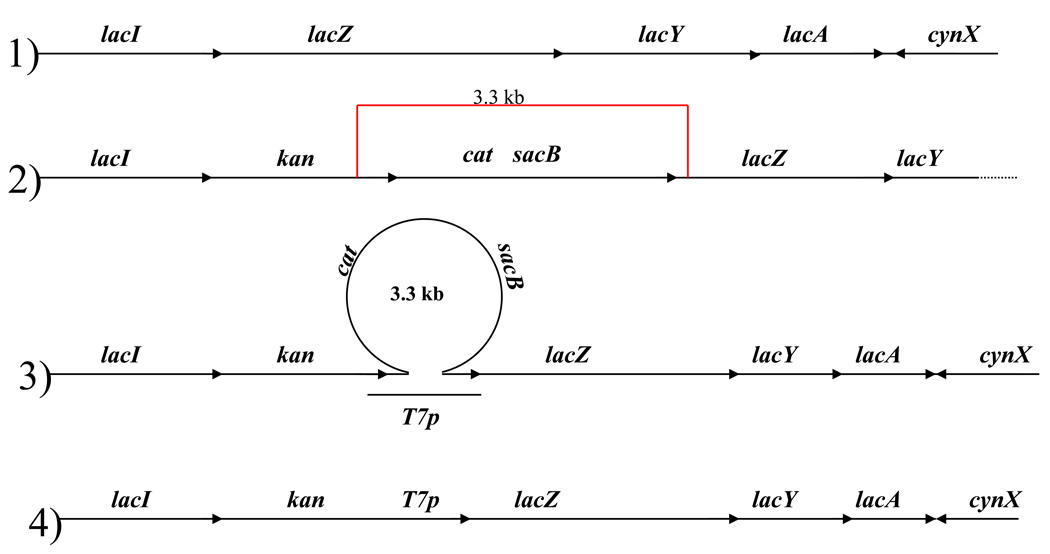
To construct E. coli derivatives with the lacZ gene under control of the T7 promoter, we used recombineering [20] with NC397 as the starting strain [21, Supporting Information]. NC397 contains the cat-sacB cassette for counterselection upstream from lacZ, and the gene conferring kanamycin resistance upstream from the cat-sacB cassette. Using a 100 base lagging strand oligonucleotide (5'GGGTGGCGGGCAGGACGCCCGCCATAAACTGCCAGGAATTtaatacgactcactataggg ACAGGAAACAGCTATGACCATGATTACGGATTCACTGGCC) containing the T7 promoter sequence (lower case letters) flanked by 40 bases of kan on one side and 40 bases of lacZ on the other, we were able to replace the cat-sacB cassette with the T7 promoter (Fig. 1). The products of recombineering were sensitive to chloramphenicol, resistant to sucrose, and resistant to kanamycin. The resulting kan-T7p-lacZ sequence was transferred to SR108 by P1 transduction. Finally, the plasmid pAR1219 [17] was introduced by electroporation.
Fig 1.
Strain construction. 1) represents the normal lac region (present in SR108); 2) the chromosome arrangement in NC397; 3) alignment of the recombineering oligonucleotide with the chromosome of NC397; 4) the chromosome arrangement after successful recombineering (e.g. HL1134).
Oligonucleotides for recombineering and for PCR were purchased from Midland Certified Reagent Company, Inc. (www.mcrc.com).
2.5 Transduction
Lysates of P1vir were prepared, and transductions performed, essentially as described by Miller [22].
2.6 Electroporation
Late exponential phase cultures (25 ml) were pelleted by centrifugation, the pellets were washed with 25 ml of chilled, sterile water, and pelleted again. The pellets were resuspended in 1 ml chilled water and the suspensions were transferred to 1.5 ml microfuge tubes. The cells were pelleted once more and resuspended in 100 µl chilled water. To the cell suspension, 5 µl of pAR1219 (92 mg/ml) was added, and 40 µl of the mixture was placed in an electroporation cuvette (1 mm gap). An Eppendorf Electroporator 2510, set at 1800v, was used to deliver the pulse, after which the mixture was added to 1 ml of LB medium and incubated for 2 hours at 37° C before being plated on LB+ampicillin. Ampicillin-resistant derivatives that produced substantial amounts of β-galactosidase after growth with IPTG were isolated and tested for TC-NER.
2.7 DNA Sequencing
Results of recombineering were verified using primers recommended by D.L. Court (personal communication) for amplifying and sequencing the region around, and including, the T7 promoter (Fig. 2). Sequencing was done by Sequetech Corp. (Mountain View CA). Sequencher 4.6 (Gene Codes Corp., MI) was used to compare the sequences to the wild type sequence listed in EcoCyc [23].
Fig 2.

Sequences used in this study. Base pairs 1 through 127 are from the kanamycin resistance gene. Base pairs 148 through 296 are from lacZ. Underlined sequences are those of the primers used for PCR. The bold letters denote the flanking sequences used for recombineering. Lower case letters indicate the T7 promoter sequence.
2.8 Irradiation
The UV source was an unfiltered 15 w germicidal bulb emitting primarily 254 nm. The incident dose rate was 0.6 J/m2/sec. In each experiment, the dose used was chosen to produce approximately one CPD per strand of the DNA restriction fragment in which repair was to be analyzed. Irradiated cultures were maintained in yellow light (>500 nm) to avoid photoreactivation.
2.9 Assays for β-galactosidase
Enzyme activity was assayed with o-nitrophenyl-β-D-galactoside (ONPG) essentially as described by Miller [18].
2.10 Repair assays
The frequencies of CPDs in individual strands of the lac operon were measured as previously described [24, 25]. Exponentially growing cultures of E. coli in defined medium (MM) were irradiated and reincubated to allow repair to occur. Samples (5 ml) were removed from each culture at intervals, and mixed with equal volumes of ice cold NET (100 mM NaCl, 10 mM EDTA, 10 mM Tris-HCl). The cells were pelleted by centrifugation and stored at −80° C. Subsequently, the DNA was purified on QIAGEN Genomic-tip columns, treated with restriction enzymes and T4 endonuclease V (TEV), then analyzed by electrophoresis in alkaline agarose gels. After the DNA had been transferred from the gels to Amersham Hybond-N+ membranes (GE Healthcare), each membrane was hybridized with a 32P-labeled strand-specific RNA probe for the restriction fragment of interest. Probes were made using the Promega Riboprobe System (Promega Corp., Madison, WI) with pZH-10 [14] as the template. The amount of radioactivity hybridized to the full length DNA fragment was measured using a phosphorimager (Bio-Rad GS-363 Molecular Imaging System) and associated software (Molecular Analyst, version 2.1).
CPD frequencies were calculated from the percentage of full length fragments (i.e. fragments that had no CPDs, the “0 class”), using the Poisson expression [24, 25].
At least two biological experiments were performed with each strain for each DNA fragment examined. For statistical analysis and graphing of results we used Prism 4 (GraphPad Software, Inc.). Error bars on graphs represent SEM.
3. Results
3.1 Strain construction
To obtain derivatives of E. coli K-12 containing a gene located on the chromosome and transcribed by the T7 RNA polymerase, we replaced the cat-sacB cassette of NC397 with the T7 promoter using "recombineering" [20] (see Materials and Methods). This resulted in kanamycin-resistant derivatives in which the lacZ gene was controlled by the T7 promoter (Fig. 1). The kan-T7p-lac region from two of these (HL1134 and HL1172, respectively) was transferred to SR108 by P1 transduction using kanamycin to select the transductants. The plasmid, pAR1219 [17], was then electroporated into two of the transductants (HL1132 and HL1173, respectively). Among the ampicillin-resistant electroporation products, four (HL1159, HL1160, HL1176, and HL1177) that produced substantial amounts of β-galactosidase when induced with IPTG were isolated. PCR and sequencing were used to verify that the T7 promoter was intact and in the expected location. Table 2 shows the β-galactosidase levels in strains grown with (Induced) and without (Not Induced) IPTG. The low levels of β-galactosidase in induced cultures of HL1132, which does not contain pAR1219, compared to those in its parent SR108, are consistent with the idea that in the absence of the T7 RNA polymerase, the lacZ gene in this strain is not transcribed well, and that the E. coli RNA polymerase does not recognize the T7 promoter efficiently, if at all. The high levels of β-galactosidase in induced cultures of HL1159 and HL1160 indicate that the lacZ gene in these strains is efficiently transcribed by the T7 RNA polymerase produced from pAR1219. Qualitatively similar results (not shown) were obtained with HL1176 and HL1177.
Table 2.
β-galactosidase activity of strains grown with (Induced) or without (Not Induced) 1 mM IPTG. Activity is expressed as pmols of ONPG hydrolysed/min/107 colony forming units.
| Strain | Induced | Not Induced |
|---|---|---|
| SR108 | 6060 | 8 |
| HL1132 | 1 | 2 |
| HL1159 | 15561 | 77 |
| HL1160 | 5119 | 40 |
| HL1179 | 8422 | 40 |
| HL1180 | 6912 | 39 |
3.2 DNA Repair
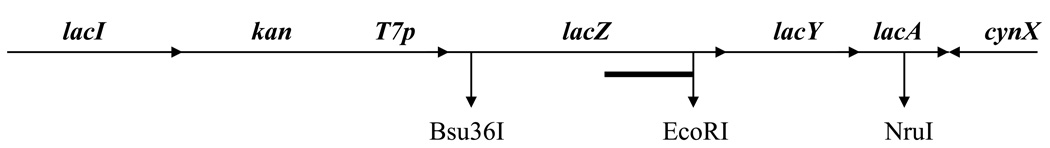
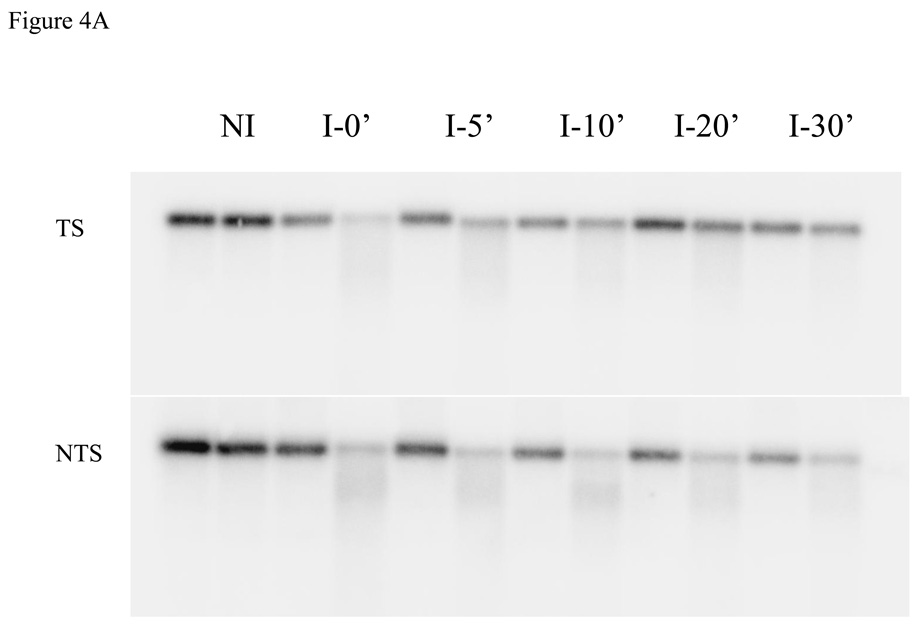
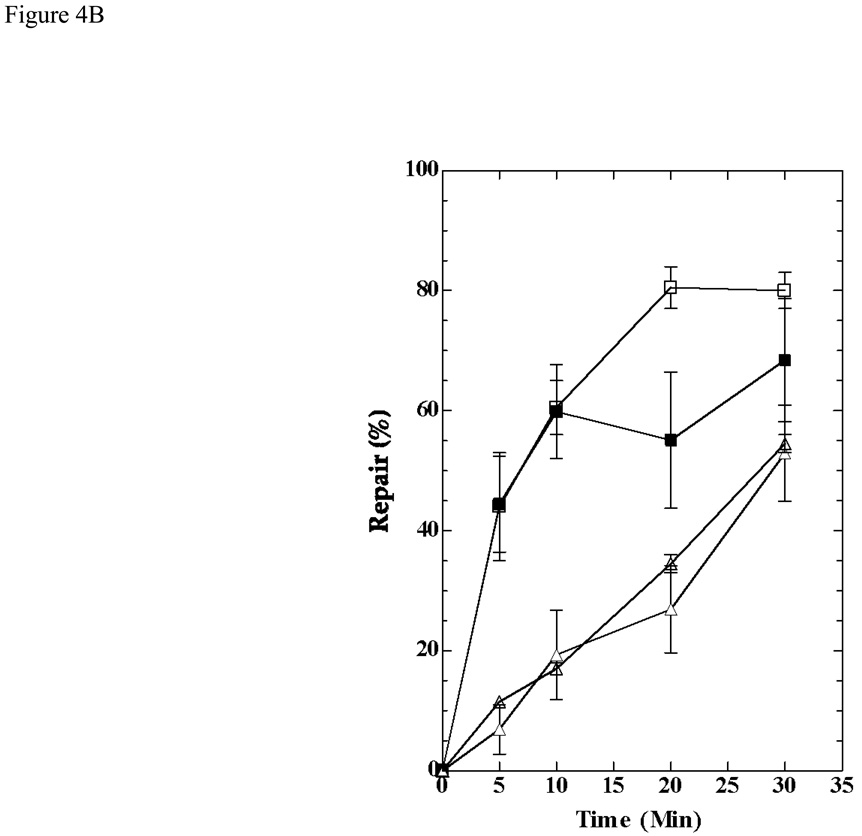
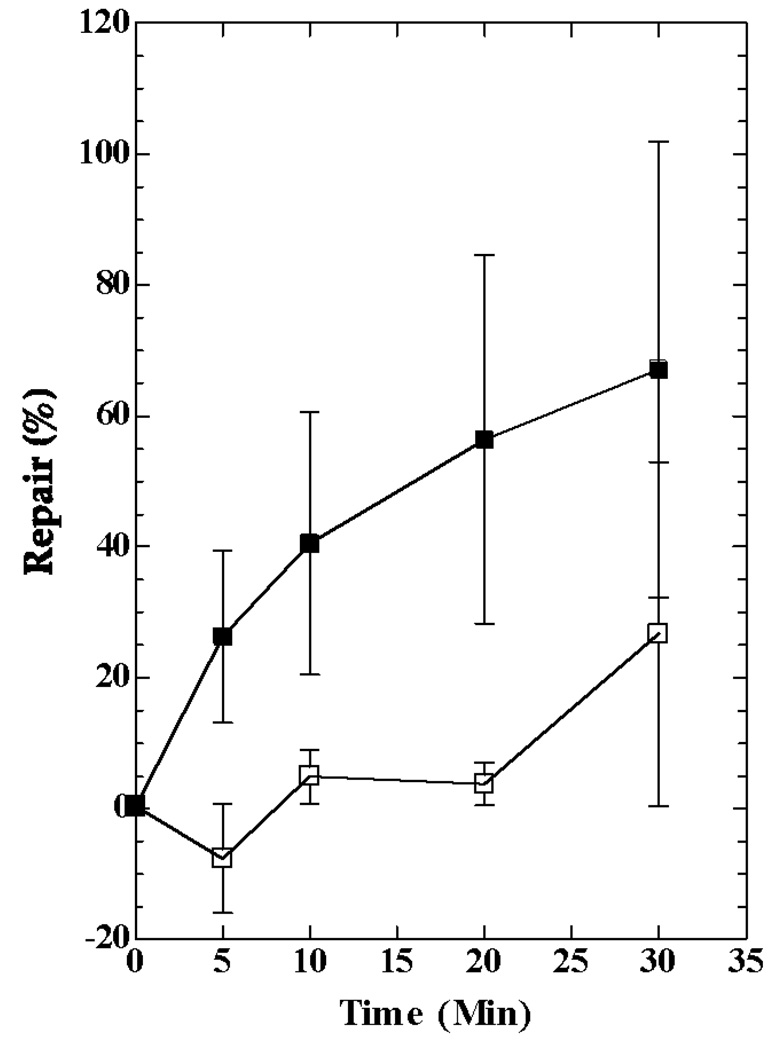
To examine DNA repair in HL1159 and HL1160, we irradiated cultures growing exponentially in MM + IPTG and measured the removal of CPDs from individual strands of a 4.8 kb Bsu36 I - NruI fragment of the lac operon (Fig. 3). In previous work [14, 25], a 6.6 kb ApaI - SacII fragment of the lac operon had been analyzed, but sequence changes in the strains we constructed precluded the use of this fragment. In both HL1159 and HL1160 the transcribed strand of the fragment was repaired more rapidly than the nontranscribed strand (Fig. 4), a response typical of TC-NER. In Fig. 4B the results obtained with HL1159 and HL1160 are combined because these strains are siblings with identical genotypes, and because they showed no significant differences in their repair responses. Similar results were obtained with HL1176 and HL1177, although fewer experiments were performed with these isolates (data not shown). Results obtained by analyzing the 6.6 kb fragment of the lac operon in the parental strain SR108, in which the lac operon is transcribed by the bacterial RNA polymerase, are shown for comparison (Fig. 4B).
Fig. 3.
Restriction fragments and hybridization probe used. The lac region of HL1159 and HL1160 is illustrated. The 4.8 kb Bsu36I - NruI fragment, and the 2.8 kb Bsu36I - EcoRI fragment, were detected using 1 kb strand specific probes produced from pZH-10 (heavy line).
Fig. 4.
Repair of CPDs in the lac operon of cultures induced with IPTG. A) Image showing repair in the 4.8 kb band of the lac operon obtained by analyzing DNA from HL1159 irradiated with 54 J/m2. B) HL1159 and HL1160 were irradiated with 54 J/m2, and the 4.8 kb fragment of the lac operon was analyzed. Results from the two strains were combined. HL108 was irradiated with 40 J/m2 and the 6.6 kb fragment of the lac operon analyzed. Symbols: squares, transcribed strands; triangles, nontranscribed strands; solid symbols, HL1159 and HL1160 (combined data); open symbols, SR108
In addition to analyzing the 4.8 kb fragment, we examined a smaller fragment (2.8 kb), produced by digestion with Bsu36 I and EcoRI. Unlike the 4.8 kb fragment, this fragment is entirely within the lacZ gene (Fig. 3). When DNA from cultures induced with IPTG was analyzed, we found, as before, that the transcribed strand of lacZ was repaired rapidly; however, when DNA from uninduced cultures was examined, we found that the transcribed strand was repaired slowly (Fig. 5). Thus, rapid repair of the transcribed strand of lacZ depends upon the gene being actively transcribed, as expected for TC-NER.
Fig. 5.
Repair of CPDs in the transcribed strand of the 2.8 kb DNA fragment of the lac operon in cultures of HL1159 irradiated with UV (72 J/m2). Symbols: solid squares, induced cultures; open squares, uninduced cultures
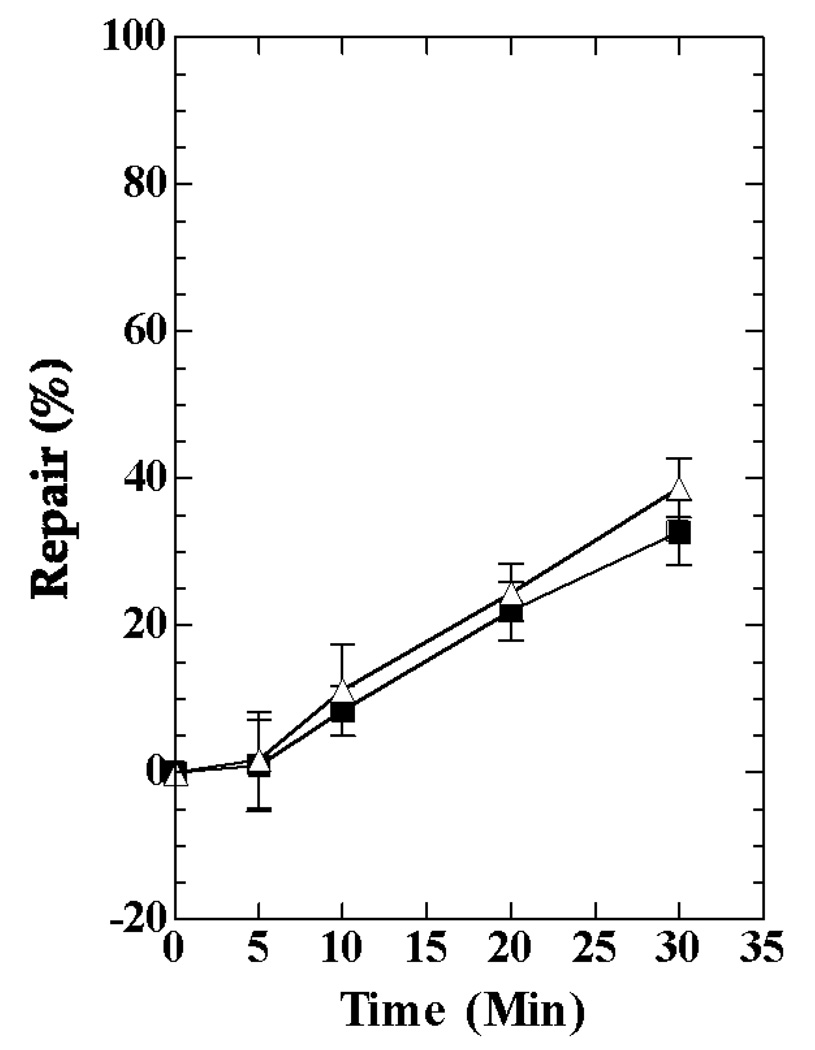
Having ascertained that TC-NER could be observed in the lacZ gene transcribed by the T7 RNA polymerase, we examined mfd-1 derivatives. Using UNC361045 [10] as the donor, the mfd-1 mutation linked to zcf-117::Tn10 was introduced into HL1132 by P1 transduction. Tetracycline-resistant transductants were isolated and tested for mfd-1 using UV sensitivity as the initial criterion. A UV-sensitive transductant (HL1167) was isolated. The plasmid p1219 was introduced into it by electroporation. Production of β-galactosidase was measured in the resulting strains (HL1179, HL1180, respectively). Both strains showed higher levels of β-galactosidase when induced with IPTG than when not induced (Table 2), consistent with transcription of lacZ by the T7 RNA polymerase. In these strains, no TC-NER of the lacZ gene was observed (Fig. 6). The repair results obtained for HL1179 and HL1180 were combined for presentation in Fig. 6 because they were siblings with identical genotypes and showed no significant differences in their repair responses.
Fig. 6.
Repair of CPDs in the lac operon of mfd strains induced with IPTG. HL1179 and HL1180 were irradiated with UV (72 J/m2) and the 4.8 kb fragment of the lac operon was analyzed. Results from the two strains were combined. Symbols: squares, transcribed strands; triangles, nontranscribed strands.
4. Discussion
Our results show that a gene transcribed by the RNA polymerase of bacteriophage T7 can be repaired by TC-NER in E. coli. When the lacZ gene was transcribed by T7 RNA polymerase, CPDs were removed more rapidly from the transcribed strand than from the nontranscribed strand of the gene (Fig. 4). Rapid repair of the transcribed strand occurred only when the gene was actively transcribed (Fig. 5) and only when the wild type allele of mfd was present (Fig. 6). These responses are the hallmarks of TC-NER in E. coli [14].
The signal for TC-NER is thought to be RNA polymerase blocked at a DNA lesion. For purified T7 RNA polymerase in vitro, many "bulky" lesions in DNA, including CPDs, are only partial blocks to transcription [15, 16, 26]; nevertheless, except for the 20 minute samples, the amount of repair of CPDs in the transcribed strand of lacZ was essentially the same whether the gene was transcribed by the T7 RNA polymerase or the E. coli RNA polymerase (Fig. 4B).
The mechanism by which the T7 RNA polymerase promotes TC-NER may be similar to the one used by the E. coli polymerase. In wild type E. coli K-12 the Mfd protein is required for TC-NER, and Mfd-deficient mutants do not show this response [5–7]. The Mfd protein interacts with the β subunit of the E. coli RNA polymerase when the polymerase is blocked at a lesion, such as a CPD, in the transcribed strand of a gene [11, 27]. Mfd dissociates the RNA polymerase from the lesion, allowing repair to occur. In addition, it enhances the rate of repair in the transcribed strand of the gene [3, 4, 12]. The mechanism by which this occurs is not well understood, but Mfd has a UvrB homology module [10] that may interact with UvrA in the UvrA2B2 complex [28], and facilitate the binding of UvrB to DNA. Studies of the purified Mfd protein in vitro indicated that, although it interacted with the E. coli RNA polymerase, it did not interact with the T7 polymerase, and it did not displace the T7 polymerase stalled at a lesion in DNA [10]. It therefore seemed unlikely that TC-NER mediated by the T7 RNA polymerase would depend upon Mfd; however, our results with the mfd-1 derivatives show that Mfd is required for T7 RNA polymerase-dependent TC-NER (Fig. 6). The common requirement for Mfd may indicate that TC-NER in E. coli involves similar mechanisms whether the bacterial or the phage RNA polymerase recognizes the lesions; however, there may be another factor (or factors) required by the T7 polymerase, but not the bacterial polymerase, that was missing from the reactions in vitro.
Acknowledgments
This work was supported by grant CA-090915 to P.C.H. from the National Cancer Institute (NIH). We thank Dr. D. L. Court (NCI) for essential bacterial strains and advice; Dr. F. W. Studier (Brookhaven National Laboratory) for pAR1219, Dr. B. Van Houten (University of Pittsburgh), and Dr. G. B. Sancar (University of North Carolina) for helpful discussions, and Dr. G. Spivak (Stanford) for critical reading of the manuscript.
Abbreviations
- NER
nucleotide excision repair
- TC-NER
transcription-coupled nucleotide excision repair
- GG-NER
global genomic nucleotide excision repair
- IPTG
isopropyl-β-D-thiogalactopyranoside
- ONPG
o-nitrophenyl-β-D-galactopyranoside
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell. Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 2.Kuemmerle NB, Masker WE. In vitro packaging of UV radiation-damaged DNA from bacteriophage T7. J. Virol. 1977;23:509–516. doi: 10.1128/jvi.23.3.509-516.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savery NJ. The molecular mechanism of transcription-coupled DNA repair. Trends. Microbiol. 2007;15:326–333. doi: 10.1016/j.tim.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Deaconescu AM, Savery N, Darst SA. The bacterial transcription repair coupling factor. Curr. Opin. Struct. Biol. 2007;17:96–102. doi: 10.1016/j.sbi.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Selby CP, Sancar A. Gene- and strand-specific repair in vitro: Partial purification of a transcription-repair coupling factor. Proc. Natl. Acad. Sciences. USA. 1991;88:8232–8236. doi: 10.1073/pnas.88.18.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selby CP, Witkin EM, Sancar A. Escherichia coli mfd mutant deficient in "mutation frequency decline" lacks strand-specific repair: in vitro complementation with purified coupling factor. Proc. Natl. Acad. Sciences. USA. 1991;88:11574–11578. doi: 10.1073/pnas.88.24.11574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellon I, Champe GN. Products of DNA mismatch repair genes mutS and mutL are required for transcription-coupled nucleotide excision repair of the lactose operon in Escherichia coli. PNAS. 1996;93:1292–1297. doi: 10.1073/pnas.93.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selby CP, Sancar A. Transcription preferentially inhibits nucleotide excision repair of the template strand in vitro. J. Biol. Chem. 1990;265:21330–21336. [PubMed] [Google Scholar]
- 9.Donahue BA, Yin S, Taylor JS, Reines D, Hanawalt PC. Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8502–8506. doi: 10.1073/pnas.91.18.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Selby CP, Sancar A. Molecular mechanism of transcription-repair coupling. Science. 1993;260:53–58. doi: 10.1126/science.8465200. [DOI] [PubMed] [Google Scholar]
- 11.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. II. Catalytic properties. J. Biol. Chem. 1995;270:4890–4895. doi: 10.1074/jbc.270.9.4890. [DOI] [PubMed] [Google Scholar]
- 12.Assenmacher N, Wenig K, Lammens A, Hopfner KP. Structural Basis for Transcription-coupled Repair: the N Terminus of Mfd Resembles UvrB with Degenerate ATPase Motifs. J. Mol. Biol. 2006;355:675–683. doi: 10.1016/j.jmb.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 13.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 14.Mellon I, Hanawalt PC. Induction of the Escherichia coli lactose operon selectively increases repair of its transcribed DNA strand. Nature. 1989;342:95–98. doi: 10.1038/342095a0. [DOI] [PubMed] [Google Scholar]
- 15.Smith CA, Baeten J, Taylor JS. The ability of a variety of polymerases to synthesize past site-specific cis-syn, trans-syn-II, (6-4), and Dewar photoproducts of thymidylyl-(3'-->5')-thymidine. J. Biol. Chem. 1998;273:21933–21940. doi: 10.1074/jbc.273.34.21933. [DOI] [PubMed] [Google Scholar]
- 16.Kalogeraki VS, Tornaletti S, Hanawalt PC. Transcription arrest at a lesion in the transcribed DNA strand in vitro is not affected by a nearby lesion in the opposite strand. J. Biol. Chem. 2003;278:19558–19564. doi: 10.1074/jbc.M301060200. [DOI] [PubMed] [Google Scholar]
- 17.Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. PNAS. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 19.Maloy SR. Experimental Techniques in Bacterial Genetics. Jones and Bartlett; 1990. [Google Scholar]
- 20.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. Recombineering: genetic engineering in bacteria using homologous recombination. Chapter 1. Curr. Protoc. Mol. Biol. 2007 doi: 10.1002/0471142727.mb0116s78. Unit 1.16. [DOI] [PubMed] [Google Scholar]
- 21.Svenningsen SL, Costantino N, Court DL, Adhya S. On the role of Cro in lambda prophage induction. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4465–4469. doi: 10.1073/pnas.0409839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 23.Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic. Acids. Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spivak G, Hanawalt PC. Determination of damage and repair in specific DNA sequences. Methods:. A. Companion. to. Methods. in. Enzymology. 1995;7:147–161. [Google Scholar]
- 25.Ganesan AK, Smith AJ, Savery NJ, Zamos P, Hanawalt PC. Transcription coupled nucleotide excision repair in Escherichia coli can be affected by changing the arginine at position 529 of the beta subunit of RNA polymerase. DNA Repair. (Amst) 2007;6:1434–1440. doi: 10.1016/j.dnarep.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scicchitano DA. Transcription past DNA adducts derived from polycyclic aromatic hydrocarbons. Mutat. Res. 2005;577:146–154. doi: 10.1016/j.mrfmmm.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Selby CP, Sancar A. Structure and function of transcription-repair coupling factor. 1. Structural domains and binding properties. J. Biol. Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 28.Malta E, Moolenaar GF, Goosen N. Dynamics of the UvrABC nucleotide excision repair proteins analyzed by fluorescence resonance energy transfer. Biochemistry. 2007;46:9080–9088. doi: 10.1021/bi7002235. [DOI] [PubMed] [Google Scholar]
- 29.Hanawalt PC. Photophysiology: Current Topics. Academic Press; 1968. Recovery from Photochemical Damage; pp. 203–251. [Google Scholar]
- 30.Sancar A, Franklin KA, Sancar GB. Escherichia coli DNA photolyase stimulates uvrABC excision nuclease in vitro. Proc. Natl. Acad. Sci. U. S. A. 1984;81:7397–7401. doi: 10.1073/pnas.81.23.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sancar GB, Smith FW. Interactions between yeast photolyase and nucleotide excision repair proteins in Saccharomyces cerevisiae and Escherichia coli. Mol. Cell. Biol. 1989;9:4767–4776. doi: 10.1128/mcb.9.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mazon G, Philippin G, Cadet J, Gasparutto D, Fuchs RP. The alkyltransferase-like ybaZ gene product enhances nucleotide excision repair of O(6)-alkylguanine adducts in E. coli. DNA Repair. (Amst) 2009;8:697–703. doi: 10.1016/j.dnarep.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 33.Shi YB, Gamper H, Hearst JE. Interaction of T7 RNA polymerase with DNA in an elongation complex arrested at a specific psoralen adduct site. J. Biol. Chem. 1988;263:527–534. [PubMed] [Google Scholar]
- 34.Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]