Abstract
Serine proteases have been implicated in many stages of cancer development, facilitating tumor cell growth, invasion, and metastasis, and naturally occurring serine protease inhibitors have shown promise as potential anticancer therapeutics. Optimal design of inhibitors as potential therapeutics requires the identification of the specific serine proteases involved in disease progression and the functional targets responsible for the tumor-promoting properties. Here, we use the HMT-3522 breast cancer progression series grown in 3D organotypic culture conditions to find that serine protease inhibitors cause morphological reversion of the malignant T4-2 cells, assessed by inhibition of proliferation and formation of acinar structures with polarization of basal markers, implicating serine protease activity in their malignant growth behavior. We identify PRSS3/mesotrypsin upregulation in T4-2 cells as compared to their nonmalignant progenitors, and show that knockdown of PRSS3 attenuates, and treatment with recombinant purified mesotrypsin enhances, the malignant growth phenotype. Using proteomic methods, we identify CD109 as the functional proteolytic target of mesotrypsin. Our study identifies a new mediator and effector of breast cancer growth and progression.
Keywords: Proteases, Protease inhibitors, Tumor microenvironment, Three-dimensional culture models, Tumor reversion
Introduction
The serine proteases of the chymotrypsin superfamily are highly represented in human biology, with over 100 functional enzymes encoded in the human genome (MEROPS database; http://merops.sanger.ac.uk/) [1]. Many of these enzymes carry out essential physiological tasks, including key roles in digestion, blood coagulation, fibrinolysis, reproduction, the immune response, and signal transduction from the extracellular environment to the cell, while the functions of others are still being elucidated. The serine proteases share a common chemical mechanism featuring a reactive serine residue at the catalytic site, but vary tremendously in the nature and degree of substrate selectivity, ranging from promiscuous enzymes that carry out general protein digestion, to exquisitely precise enzymes that cleave only a single peptide bond within a single protein substrate.
Serine proteases are also important players in tumor growth and progression [2–6]. A broad array of extracellular membrane-bound and secreted serine proteases activate chemokines, growth factors, growth factor receptors, and other signaling receptors, contributing in a signaling capacity to tumor initiation, proliferation, and metastasis [2, 3, 5–10]. Plasmin and plasminogen activators (PLAU), tumor-associated trypsins, tissue kallikreins, and neutrophil elastase break down extracellular matrix and cellular adhesion molecules, facilitating angiogenesis, invasion, and metastasis [2, 3, 5, 6]. Several natural polypeptide and phytochemical inhibitors of serine proteases have shown promise as cancer chemopreventive or therapeutic agents [11–17]. Surprisingly, for some of these inhibitors, the specific target proteases have not been identified, and this remains a critical frontier [18].
Over the past two decades, the increasing diagnosis of breast cancer at earlier stages has resulted in a growing treatment focus on tertiary cancer prevention, identifying the best adjuvant therapies to prevent relapse after surgery to remove preinvasive or localized breast lesions [19, 20]. Antiendocrine adjuvant therapies have led to important reductions in breast cancer mortality [21, 22], but identification of new molecular targets, particularly for treatment of the 30–40% of breast cancer patients with non-endocrine-responsive tumors, remains a high priority [20, 22]. Given the importance of a permissive microenvironment for tumor growth and progression, the tumor microenvironment is an increasingly important focus for locating points of therapeutic intervention.
The stroma surrounding a developing tumor has the potential to exert either normalizing or tumor-promoting influences [23, 24]. Organotypic cell culture systems in which breast epithelial cells are grown in 3D extracelluar matrix gels have been used to dissect how extracellular factors control progression to the malignant phenotype, and how modulation of these signaling pathways can cause a stable reversion of breast cancer cells [25–27]. These same model systems can be employed to identify novel molecular targets for breast cancer therapy. Here, we use 3D models to elucidate one mechanism by which serine proteases stimulate early progression of breast cancer, as manifested by loss of responsiveness to normalizing microenvironmental stimuli, loss of cell polarity, and deregulated proliferation. We identify the atypical trypsin isoform mesotrypsin as a specific contributor to aberrant growth, and we use transcriptional and proteomic profiling approaches to define the mechanistic basis whereby mesotrypsin promotes breast cancer progression.
Materials and methods
Cell culture and reagents
HMT-3522 T4-2 mammary epithelial cells were provided by Dr. Mina J. Bissell, Lawrence Berkeley National Laboratory, Berkeley, CA, USA. Cells were cultured in H14 medium consisting of DMEM/F12 (Invitrogen, Carlsbad, CA, USA) supplemented with 250 ng/ml insulin (Sigma, St Louis, MO, USA), 10 µg/ml transferrin (Sigma), 2.6 ng/ml sodium selenite (BD Biosciences, San Jose, CA, USA), 0.1 nM estradiol (Sigma), 1.4 µM hydrocortisone (Sigma), and 5 µg/ml prolactin (Sigma) as previously described [26, 28]. 3D cultures were established by growing cells to 70–80% confluence as monolayers on collagen-coated plates (Inamed Biomaterials, Fremont, CA, USA), followed by trypsinization with 0.25% trypsin and embedding into growth factor-reduced Matrigel (BD Biosciences) using the “on-top” protocol, in which single cells are allowed to attach to a bed of 100% Matrigel before deposition of a blanketing layer of 10% Matrigel in medium, as previously described [29]. On the second day following set up of 3D cultures, fresh medium without Matrigel was overlaid, and subsequently was aspirated and replaced every 2 days.
In some experiments, the protein protease inhibitors aprotinin (United States Biochemical, Cleveland, OH, USA), soybean trypsin inhibitor (United States Biochemical), or Bowman–Birk inhibitor (Sigma) were used to supplement the culture medium. The inhibitor was added both to the upper layer of matrix in establishing 3D cultures, as well as to the culture medium replenished every 2 days.
In other experiments, recombinant human mesotrypsin was used to supplement the culture medium. Recombinant mesotrypsinogen was expressed in Escherichia coli, refolded, purified, and activated to produce mesotrypsin as previously described [30, 31]. Active mesotrypsin concentrations were quantified by active site titration using the titrant 4-nitrophenyl 4-guanidinobenzoate (Sigma) [32]. For 3D cultures treated with mesotrypsin, active mesotrypsin was added to a final concentration of 100 nM both to the upper layer of matrix and to the culture medium replenished every 2 days; this concentration was selected for maximal phenotypic effect following preliminary experiments testing a range of concentrations.
Indirect immunofluorescence
Cells from 3D cultures were stabilized by successive incubation in PBS containing 15% and then 30% sucrose, smeared onto slides, air dried, fixed with 4% formaldehyde, permeabilized with PBS containing 0.1% Triton X-100, then blocked overnight in PBS containing 0.2% Triton X-100, 0.05% Tween 20, 0.1% BSA, and 5% goat serum (Sigma). Slides were washed with PBS, incubated with primary antibodies for 2 h and then with FITC- or Alexa Fluor 488-conjugated secondary antibodies for 1 h, then nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Sigma) or with Hoechst 33342 (Invitrogen). Control slides were stained with secondary antibodies only. Primary antibodies used were as follows: anti-E-cadherin MAb clone 36 (BD Biosciences); rat anti-integrin a6 [CD49f] MAb clone NKI-GoH3 (Chemicon International, Temecula, CA, USA); anti-Ki-67 clone MIB-1 (Dako, Carpinteria, CA, USA). Fluorescence microscopy was performed using an Olympus IX71 inverted microscope and a Quantafire X1 camera. All images were obtained in grayscale and pseudocolored using Adobe Photoshop CS. For quantification of acinar size and polarity, at least 50 cell structures were assessed in four replicates for each experimental condition; data are presented as the average and SD of the replicates.
Lentiviral shRNA knockdowns
Lentiviral short hairpin RNA constructs NM_002771.2–302s1c1 (C23) and NM_002771.2–454s1c1 (C24) targeting human PRSS3 and NM_133493.1–3107s1c1 targeting human CD109 were obtained from the MISSION TRC-Hs1.0 library (Sigma). A nontarget control (NTC) lentiviral vector containing a short hairpin that does not recognize any human genes was used as a negative control in all RNAi experiments. Conditioned media containing infective lentivirus particles were produced using HEK 293FT cells and following supplier protocols. For lentiviral transduction, T4-2 cells were seeded into six-well plates at 50–60% confluency. After 24 h, medium was replaced with 600 µl/well of complete medium, 400 µl/well of viral supernatant, and a final concentration of 6 µg/ml of polybrene (Chemicon International). Cells were incubated with virus for 24 h at 37°C, then washed and grown for another 24 h in fresh culture medium before selection with 5 µg/ml puromycin. After 24 h of selection with puromycin, cells were trypsinized and used immediately to set up 3D experimental cultures, or were split and maintained under puromycin selection for up to a week prior to setting up 3D experiments.
RNA extraction, cDNA synthesis, and quantitative real-time PCR
RNA was isolated from 2D and 3D cultures using TRIZol reagent (Invitrogen) according to manufacturer protocols. cDNA was synthesized according to kit specifications using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). Quantitative real-time PCR was performed using TaqMan gene expression assays (Applied Biosystems) on an Applied Biosystems 7900HT Fast Real-Time PCR System according to manufacturer protocols. TaqMan assays employed included: GAPDH, Hs99999905_m1; PRSS1, Hs006056 31_g1; PRSS2, Hs00828418_gH; PRSS3, Hs006056 37_m1; PLAU, Hs00170182_m1; PLAT, Hs002634 92_m1; ST14, Hs00222707_m1; TMPRSS6, Hs005421 84_m1; TPSG1, Hs00202948_m1; HGFAC, Hs001735 26_m1; PRSS8, Hs00173606_m1; TMPRSS3 Hs002251 61_m1; PRSS21, Hs00199035_m1; PRSS12, Hs00186 221_m1; EGFR, Hs00193306_m1; CD109, Hs003703 47_m1; ITGAV, Hs00233808_m1; BOP1, Hs003748 84_m1; CD74, Hs00269961_m1.
Microarrays
RNA was isolated from 3D cultures as described above. RNA integrity was confirmed by electrophoresis on an Agilent 2100 instrument, and only samples with RINs of >8 were analyzed. RNAs were labeled and hybridized to Affymetrix HU133V2 chips. Labeling, hybridization, and scanning were carried out by the Mayo Clinic Microarray Facility in Rochester, MN. Pre-processing, normalization, and background correction were carried out using the GCRMA functions of GeneSpring (Agilent Technologies, Santa Clara, CA, USA), which was used to identify genes that were significantly regulated (P < 0.01, fold change > 1.99 at false discovery rate as less than 0.05, using the standard analysis methods in the Genespring software package).
Biotinylation, cleavage, capture, and identification of shed proteins
T4-2 cells grown to 90–95% confluence in monolayer culture were washed extensively with cold PBS and labeled with a 0.5 mg/ml solution of biotin-XX, SSE (Invitrogen) in PBS/4% DMSO for 30 min at 4°C. Subsequently, cells were washed and treated with 200-nM mesotrypsin in DMEM serum-free medium for 4 h at 37°C. Conditioned medium was collected and biotinylated proteins were isolated using Streptavidin Agarose Resin (Pierce Biotechnology, Rockford, IL, USA). Recovered proteins were dissociated from streptavidin beads by boiling in SDS-PAGE loading buffer, resolved by SDS-PAGE, and stained using SilverSNAP Stain for Mass Spectrometry (Thermo Scientific/Pierce). Bands of interest were excised, subjected to in-gel tryptic digestion, extraction, and identification by nano-flow liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) at the Mayo Proteomic Research Center as previously described [33].
Western blotting
Whole cell lysates were prepared from cells grown in monolayer culture by lysis in RIPA buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate). Protein constituents of conditioned media were concentrated 20-fold by trichloroacetic acid (TCA) precipitation followed by resuspension in SDS-PAGE loading buffer. Cell extract and concentrated media samples were resolved by SDS-PAGE, blotted to Immobilon-FL PVDF membrane (Millipore, Billerica, MA, USA), and probed with appropriate primary and horseradish peroxidase-conjugated secondary antibodies according to standard protocols. Immunoblots were developed using the ECL Plus Western Blotting Detection System (GE Healthcare, Piscataway, NJ, USA) and images were captured using a ChemiDoc XRS imager (Bio-Rad Laboratories, Hercules, CA, USA); alternatively, chemiluminescent signal was recorded on Kodak BioMax MR film, and films were subsequently imaged using the ChemiDoc XRS imager.
Results
Serine protease inhibition suppresses malignant growth in a 3D culture model of human breast cancer
Defining phenotypic effects activated by secreted proteases represents a particular challenge, as their expression and activities are often modulated in response to the cellular microenvironment [34], and their proteolytic functions often target extracellular structures, many of which exist only in organized tissue architecture [2, 3, 5, 35]. Here, we used 3D epithelial culture systems in which breast epithelial cells, when cultured within basement-membrane-like matrices, organize into structures that resemble their normal tissue architecture and that recapitulate some aspects of differentiated function [36, 37]. We used the HMT3522 breast cancer cell progression series [24]; when cultured in a 3D laminin-rich reconstituted basement-membrane gel (lrBM), early passage normal breast epithelial S1 cells develop into hollow, polarized spherical acini and growth arrest, whereas the derivative T4-2 cells, which have undergone malignant transformation conferring tumorigenicity in mice, proliferate into tumor-like masses [24]. The 3D model also allows for evaluating the phenotypic effects of modulating key signaling pathways: activation of oncogenic pathways in nonmalignant cells stimulates the malignant phenotype [38], while inhibition of key oncogenic signaling pathways in T4-2 cells stimulates a phenotypic reversion [39], in which the cells organize into growth-arrested acini with basal polarity [24–26, 28, 39, 40].
We evaluated the role of serine proteases in the malignant growth phenotype of T4-2 cells by treating 3D cultures with several serine protease inhibitors at a range of concentrations. We found that treatment of 3D cultures with high concentrations (1 mg/ml; approximately 150 µM) of aprotinin attenuated the malignant phenotype, suppressing disorganized proliferation and restoring basal polarity and acinar morphology (Fig. 1). Soybean trypsin inhibitor (SBTI) produced similar effects at lower concentrations (20 µg/ml; approximately 1 µM), but marked cytotoxicity at higher concentrations, while the Bowman–Birk inhibitor from soybean had no comparable effect at concentrations up to 1 mg/ml (not shown).
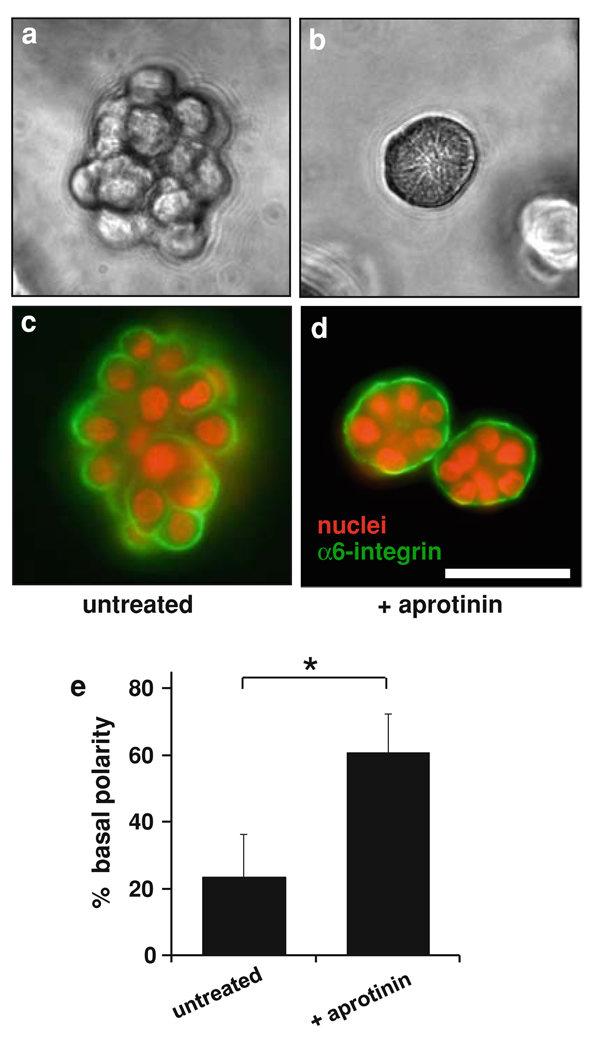
Fig. 1.
Effect of serine protease inhibitor aprotinin on growth morphology of T4-2 cells in 3D culture. Cells were cultured in Matrigel for 8 days either in the absence (panels a, c) or in the presence (panels b, d) of 1 mg/ml aprotinin. Aprotinin suppressed disorganized growth and led to formation of acini with basal polarity, as demonstrated by phase contrast microscopy (a, b), and by immunofluorescence staining for α6 integrin (c, d). Scale bar, 50 µm. e Quantitative analysis of polarity by percentage of colonies with polarized distribution of basal α6 integrin confirmed that a significantly greater proportion of colonies showed basal polarity in the aprotinin-treated culture. Data are expressed as mean ± SD. * P < 0.01 (unpaired t test)
Specific serine proteases are upregulated with progression to malignancy in the HMT3522 progression model of breast cancer
The phenotypic alteration of T4-2 cells treated with aprotinin or SBTI implicates serine proteases as important functional mediators of malignancy in these breast cancer cells. As T4-2 cells belong to the HMT3522 progression series [24, 41, 42], we hypothesized that the tumorigenic serine protease(s) of interest would show increased expression with progression to malignancy in this cell series. Accordingly, we assessed candidate proteases for increased transcription in T4-2 cells relative to nonmalignant precursor S1 cells in 3D culture. As both aprotinin and SBTI are well-known trypsin inhibitors, and as trypsins have been reported to play roles in progression of other tumor types [5, 43], we included human trypsinogen genes PRSS1, PRSS2, and PRSS3 among our candidate proteases. Further candidates, gleaned from previous reports of upregulation or correlation with malignancy in breast cancer or relevant models, included urokinase plasminogen activator (PLAU) [44], tissue plasminogen activator (PLAT) [34], matriptase-1 (ST14) [45, 46], matriptase-2 (TMPRSS6) [47], tryptase-γ1 (TPSG1) [47], and hepatocyte growth factor activator (HGFAC) [45, 46]. Additionally, our candidate list included transmembrane protease serine 3 (TMPRSS3), prostasin (PRSS8), testisin (PRSS21), and neurotrypsin (PRSS12), serine proteases identified as upregulated in T4-2 cells relative to S1 cells in 3D culture according to a recent microarray analysis [48]. Testing expression by quantitative RT/PCR, we found significant upregulation of five candidate transcripts in T4-2 cells relative to S1 cells: PRSS3 (14-fold increase), PRSS8 (14-fold increase), TMPRSS3 (4.6-fold increase), PRSS12 (2.1-fold increase), and ST14 (1.7-fold increase) (Supplemental Fig. 1).
PRSS3 knockdown suppresses malignant growth of T4-2 cells in 3D culture, while active mesotrypsin enhances malignant growth
To assess the individual roles in promoting malignant growth of the five proteases found to be upregulated in T4-2 cells, we evaluated the effect of RNAi knockdown via stable transduction with lentiviral shRNA constructs on inhibition of the malignant phenotype in 3D culture. The strongest effects were found with knockdown of PRSS3, where variable efficiencies of transcriptional repression of PRSS3 by two different viruses correlated closely with suppression of malignant growth in 3D cultures (Fig. 2); virus C23 reduced both PRSS3 transcript levels and colony size modestly, while virus C24 very effectively reduced PRSS3 transcript levels and more substantially reduced colony size.
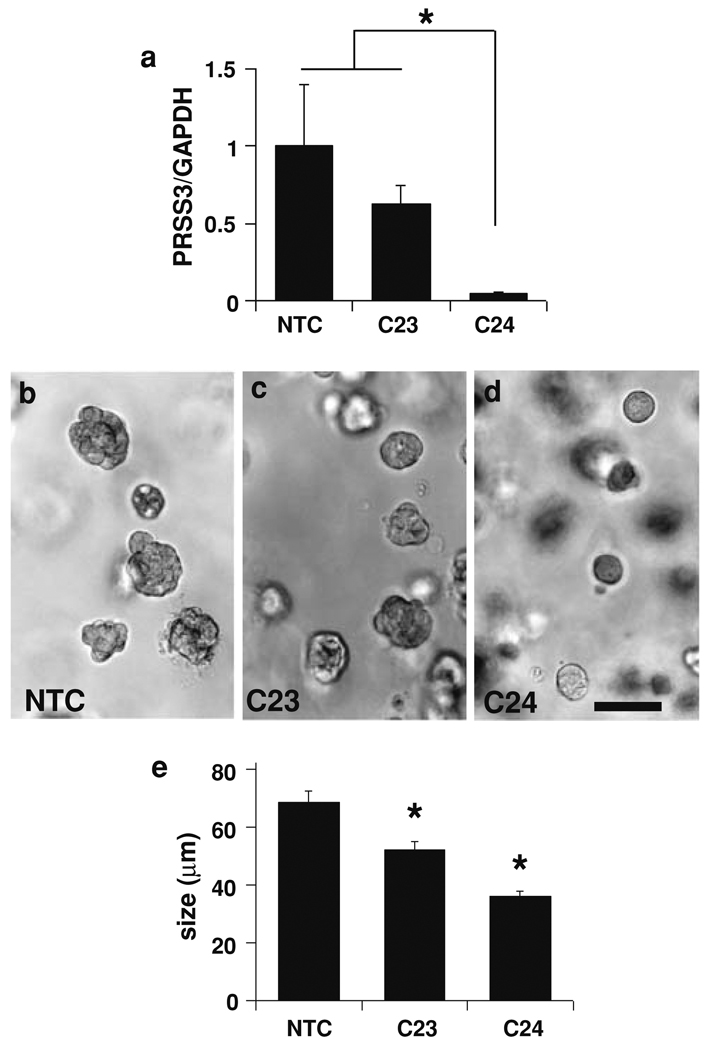
Fig. 2.
Effect of PRSS3 knockdown on malignant growth of T4-2 cells in 3D culture. a Cells transduced with lentiviral shRNA vectors C23 or C24 targeting PRSS3 revealed lower transcript levels in 2D cultures compared with cells transduced with a non-target control vector (NTC), as assessed by real-time qRT/PCR. Values represent normalized PRSS3/GAPDH transcript levels ± SEM. * P < 0.05 (unpaired t test). b–e Cells transduced with control, C23, or C24 vectors were grown in Matrigel for 7 days, then photographed and assessed for colony size. Photographs of representative fields show a reduction of colony size in cultures transduced with C23 (c) and C24 viruses (d) relative to control cultures (b), in a pattern that correlates with PRSS3 transcript levels. Scale bar, 100 µm. e Colony size was significantly reduced for both C23 and C24 transduced cultures relative to control cultures; data represented as mean ± SEM. * P < 0.001 (unpaired t test)
PRSS3 encodes mesotrypsinogen/trypsinogen IV, proteolytic activation of which is expected to produce active mesotrypsin. We tested the effect of active recombinant mesotrypsin on growth of T4-2 cells in 3D culture, and found that mesotrypsin enhanced malignant growth, significantly increasing the average colony size (Fig. 3).
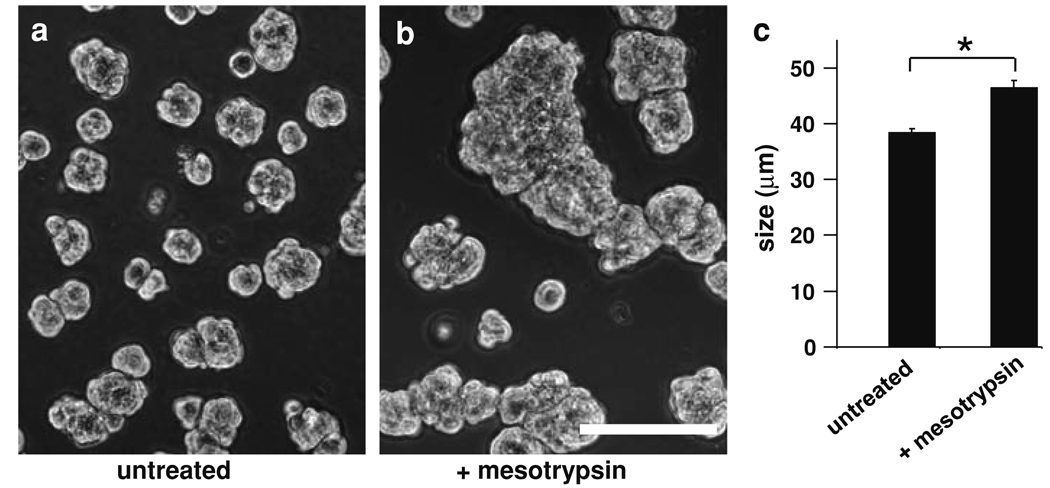
Fig. 3.
Effect of recombinant mesotrypsin treatment on malignant growth of T4-2 cells in 3D culture. Cells were grown in Matrigel for 4 days in the absence (a) or presence (b) of 100-nM mesotrypsin. Scale bar, 100 µm. c Average colony size was significantly increased for mesotrypsin-treated versus untreated cells; data represented as mean ± SEM. * P < 0.0001 (unpaired t test)
Mesotrypsin exposure modulates gene transcription in T4-2 cells in 3D culture
To gain insight into potential mechanisms involved in the promotion of malignant growth in breast cancer cells by mesotrypsin, we performed transcriptional profiling on T4-2 cells grown in 3D culture using Affymetrix GeneChip Human Genome U133 Plus 2.0 microarrays detecting 54,695 transcripts. Comparing control cultures to (a) cultures in which PRSS3 transcription was inhibited by shRNA knockdown using virus C24 and (b) PRSS3 knockdown cultures treated with recombinant mesotrypsin, we found 68 distinct transcripts that were downregulated by PRSS3 knockdown but upregulated by recombinant mesotrypsin; 42 distinct transcripts showed the opposite effect, being upregulated by PRSS3 knockdown and downregulated by mesotrypsin treatment (Fig. 4a, b). Several candidate transcripts of particular relevance to breast cancer malignant growth were selected for validation by qRT/PCR, which confirmed transcriptional regulation by mesotrypsin (Fig. 4c).
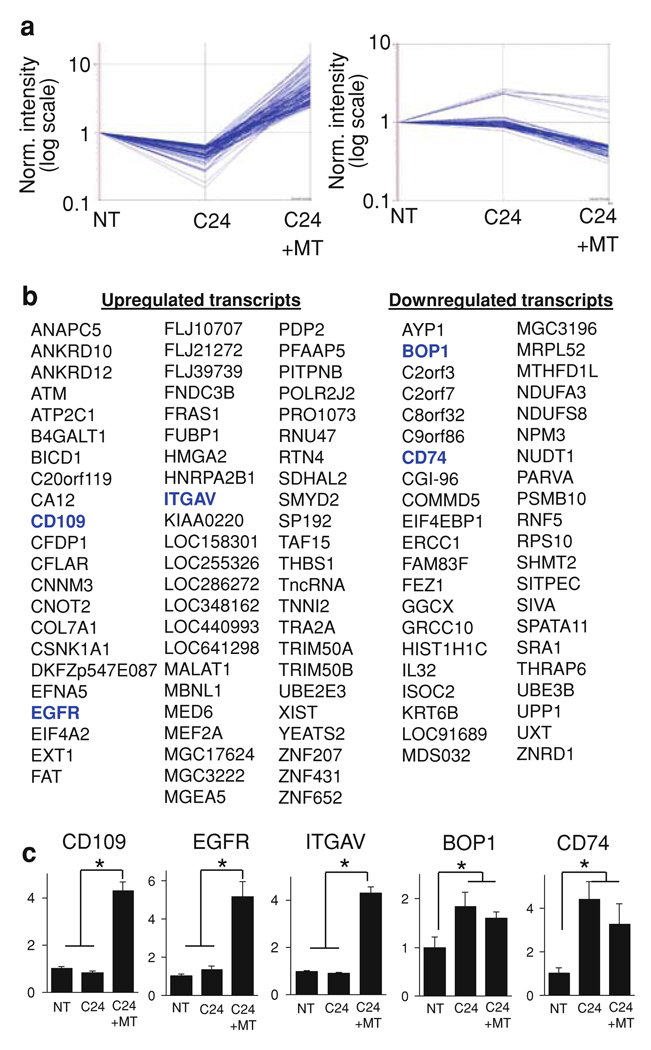
Fig. 4.
PRSS3 knockdown and treatment with recombinant mesotrypsin alter expression patterns of multiple genes in T4-2 cells grown in 3D culture. Cells transduced with lentiviral shRNA non-target vector (NT) or PRSS3-targeting vector C24 (C24), or cells transduced with C24 and also treated with recombinant mesotrypsin (C24 + MT), were grown in 3D for 5 days, then RNA was harvested and hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 microarrays. a Relative to the control cells (NT), 68 transcripts were downregulated by knockdown of PRSS3 (C24) and upregulated by mesotrypsin treatment (left panel), while 42 transcripts showed the opposite effect, being upregulated by PRSS3 knockdown and downregulated by mesotrypsin treatment (right panel). b List of the individual transcripts upregulated by mesotrypsin (right three columns) or downregulated by mesotrypsin (left two panels). c Transcriptional regulation in 3D cultures of several candidate mesotrypsin-responsive genes was validated by qRT/PCR, including the upregulated transcripts CD109, EGFR, and ITGAV and the downregulated transcripts BOP1 and CD74. Values represent relative transcript levels normalized to GAPDH, ± SEM. * P < 0.05 (unpaired t test)
Enhancement of malignant growth by mesotrypsin is mediated by cleavage of CD109
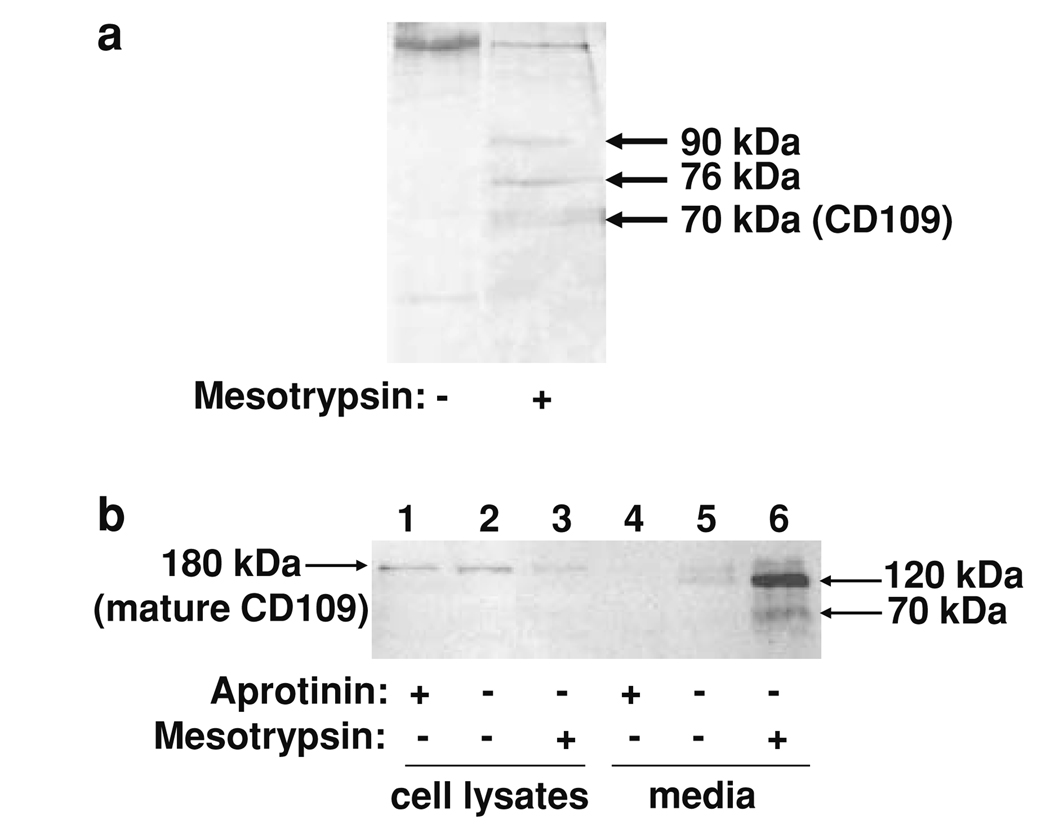
Our data showing suppression of malignant growth by treatment with aprotinin or SBTI, and enhancement of malignant growth by treatment with recombinant mesotrypsin, suggest that secreted mesotrypsin acts extracellularly to stimulate cancer growth. To identify physiological substrates of mesotrypsin through which these effects might be mediated, we designed a proteomic screen to identify cell surface proteins shed from T4-2 cells by mesotrypsin treatment. Live cells were treated with a membrane impermeant activated biotin reagent to label all cell surface proteins, and then treated with mesotrypsin to allow proteolysis of mesotrypsin targets. Biotinylated proteins shed from the cell surface into the medium were affinity purified, resolved by SDS-PAGE, and identified by nanoLC-MS/MS. As shown in Fig. 5a, we identified a 70-kDa protein band from a mesotrypsin-treated sample for which the top probability match, with 38 matching peptides and 31% sequence coverage, was CD109, a 180-kDa glycosylphosphatidylinositol-linked cell surface glycoprotein.
Fig. 5.
Identification of CD109 as a mesotrypsin proteolytic target. a Biotinylation and capture of cell surface proteins from conditioned medium of T4-2 cells yielded several silver-stained bands that were present after mesotrypsin treatment (right lane) but absent in an untreated control (left lane); the 70-kDa band was identified as CD109 by nanoLC-MS/MS. b T4-2 cells were grown in monolayer culture without treatment or in the presence of 100 µM aprotinin or 200 nM recombinant mesotrypsin, and then whole cell protein lysates and conditioned media were recovered and analyzed by western blotting using a sheep anti-human CD109 polyclonal primary antibody (R&D Systems, Minneapolis, MN, USA) at 0.15 µg/ml, and a rabbit anti-sheep HRP-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1:1,000 dilution. Immunostaining of cell lysates for CD109 was reduced when cells were treated with mesotrypsin (lane 3) relative to untreated cells (lane 2) or cells treated with aprotinin (lane 1). Conditioned medium from the mesotrypsin-treated cells contained 120 and 70 kDa CD109 fragments (lane 6). Conditioned medium from untreated cells showed evidence of limited CD109 shedding (lane 5), which was suppressed by aprotinin (lane 4)
We next confirmed by western blot with immunostaining for CD109 that this protein is shed from the surface of T4-2 cells upon treatment with recombinant mesotrypsin (Fig. 5b). We detect a relatively small amount of the shed 120 kDa CD109 ectodomain band in the media of untreated cells (Fig. 5b, lane 5); this presumably represents the low level of CD109 shedding that occurs during the 4 h course of the experiment, mediated by endogenous mesotrypsin and possibly other endogenous serine proteases. We have detected this band consistently in multiple repetitions of the experiment, and have also consistently observed an absence of the band in cultures treated with 1 mg/ml aprotinin (Fig. 5b, lane 4), confirming that the relevant sheddases are all serine proteases. By contrast with the low level of endogenous shedding seen in untreated cultures, a very large amount of the shed 120 kDa CD109 ectodomain, along with a smaller amount of the 70 kDa fragment, are present following a 4 h treatment with recombinant mesotrypsin (Fig. 5b, lane 6), showing that mesotrypsin can efficiently mediate or induce shedding of CD109.
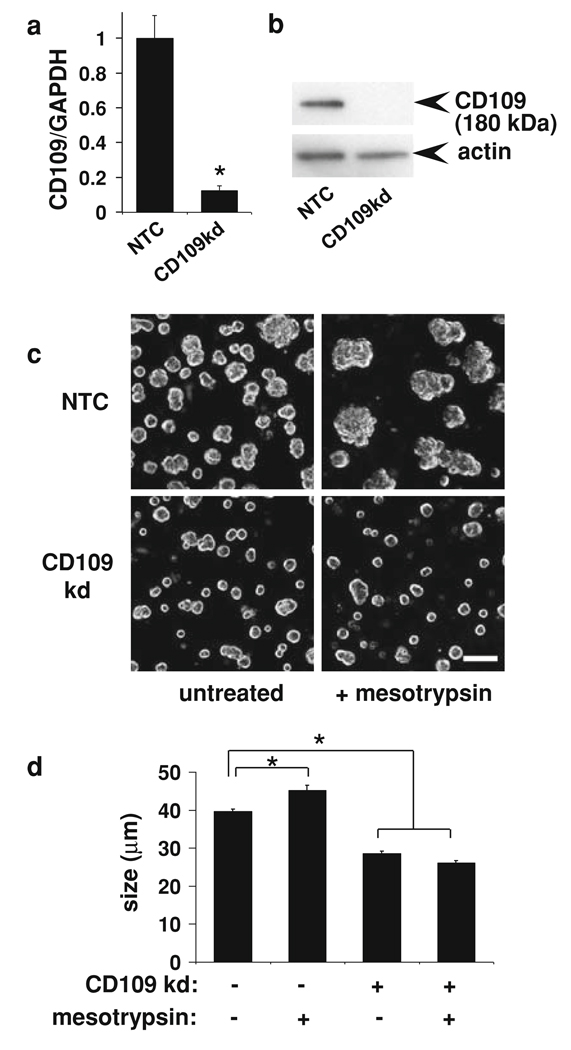
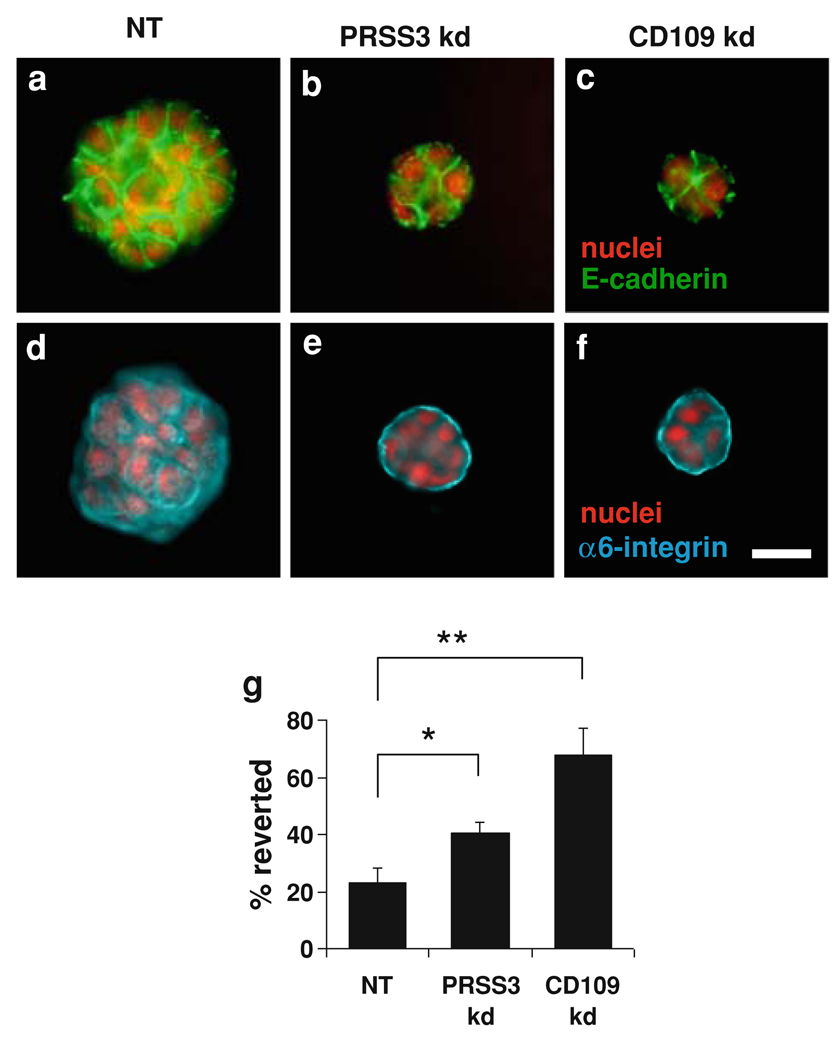
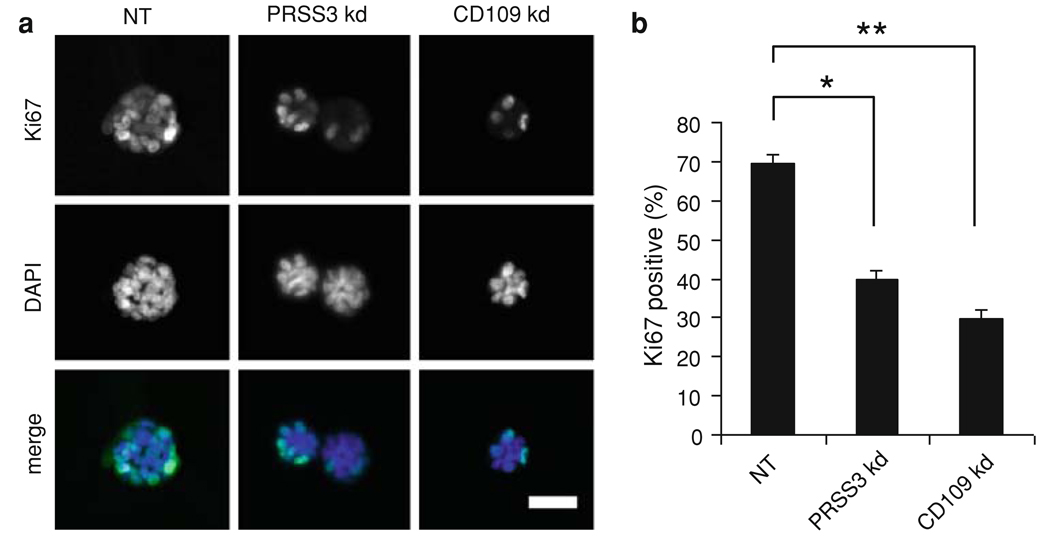
To directly assess the potential role of CD109 in the malignant growth of T4-2 cells, we inhibited CD109 expression by stable transduction with a lentiviral shRNA construct. Knockdown of CD109 in T4-2 cells grown in 3D culture was confirmed both by qRT/PCR (Fig. 6a) and by western blot (Fig. 6b). Mesotrypsin treatment of the control cells stimulated malignant growth and led to larger colonies, whereas CD109 knockdown produced small colonies unresponsive to mesotrypsin, as shown in Fig. 6c, d. Knockdown of CD109 also caused a reversion of the T4-2 cells, as assessed by basal localization of α6-integrin and lateral localization of E-cadherin (Fig. 7). These results, taken in combination with the demonstration of CD109 shedding from the cell surface by mesotrypsin (Fig. 5), suggest that CD109 is activated by cleavage to stimulate growth, and in the absence of CD109, mesotrypsin dependent malignant growth is abrogated. To directly assess the impact of PRSS3/mesotrypsin and its target CD109 on cell proliferation, we stained 3D cultures for the proliferation marker Ki-67. We found that inhibition of either PRSS3 or CD109 expression by stable transduction with lentiviral shRNA constructs resulted in a significant reduction in Ki-67 staining (Fig. 8), suggesting that stimulation of proliferation is one mechanism by which mesotrypsin promotes malignant growth.
Fig. 6.
CD109 knockdown blocks malignant growth response to mesotrypsin in T4-2 cells in 3D culture. Cells transduced with lentiviral shRNA non-target vector (NT) or a vector targeting CD109 (CD109kd) were grown in Matrigel with or without 100 nM mesotrypsin for 4 days, and then photographed and harvested for RNA. a CD109 transcript levels in 3D cultures were assessed by realtime qRT/PCR; values represent normalized CD109/GAPDH transcript levels ± SEM. * P < 0.005 (unpaired t test). b CD109 protein level was assessed by western blot of 2D culture lysates from the same knockdowns shown in a, immunostaining with anti-human CD109 monoclonal antibody clone HU17 (eBioscience, San Diego, CA, USA) at 3 lg/ml and goat anti-mouse HRP-conjugated secondary antibody (Pierce Biotechnology) at 1:10,000 dilution. The blot was subsequently stripped and reprobed for actin as a loading control, using goat polyclonal actin (I-19) (Santa Cruz Biotechnology) at 1:2,000 dilution, and a donkey anti-goat HRP-conjugated secondary antibody (Santa Cruz Biotechnology) at 1:15,000 dilution. c Photographs of representative fields of 3D cultures with and without shRNA knockdown of CD109, and with and without treatment with 100 nM recombinant mesotrypsin, are shown. Scale bar, 100 µm. d Quantitative analysis of colony size shows a significant size increase in NTC + mesotrypsin versus untreated control cultures, and a significant reduction in CD109kd cultures, both with and without mesotrypsin treatment, relative to untreated controls. Data are shown as mean ± SEM. * P < 0.0001 (unpaired t test)
Fig. 7.
Effects of PRSS3 knockdown and CD109 knockdown on the growth morphology and cell polarization of T4-2 cells in 3D culture. Cells transduced with control lentiviral shRNA vector (NT) or with PRSS3-targeting vector C24 (PRSS3 kd) or CD109-targeting vector (CD109 kd) were cultured in Matrigel for 7 days. Knockdown of PRSS3 or CD109 suppressed disorganized growth and led to formation of acini with basal polarity, as demonstrated by immunofluorescence staining for E-cadherin (a–c) and α6-integrin (d–f). Scale bar, 25 µm. g Quantitative analysis of polarity by percentage of colonies with polarized distribution of basal α6-integrin confirmed that a significantly greater proportion of colonies in cultures with PRSS3 and CD109 knockdown showed basal polarity. Data are expressed as mean ± SD. * P < 0.05; ** P < 0.01 (unpaired t test)
Fig. 8.
PRSS3 knockdown inhibits cell proliferation in 3D culture. Cells transduced with control lentiviral shRNA vector (NT) or with PRSS3-targeting vector C24 (PRSS3 kd) or CD109-targeting vector (CD109 kd) were cultured in Matrigel for 7 days, and then cell proliferation was assessed by expression of Ki-67. a Knockdown of PRSS3 or CD109 reduced the number of cells staining positive for Ki-67. Scale bar, 50 µm. b Quantification revealed a statistically significant reduction in number of positively staining cells. Data are expressed as mean ± SD. * P < 0.05; ** P < 0.01 (unpaired t test)
Discussion
We found that serine proteases are required for the malignant growth of HMT3522 T4-2 breast cancer cells in 3D culture, since in the presence of the serine protease inhibitors aprotinin or SBTI, these cells form acini with basal polarity and undergo growth arrest. We found that PRSS3/mesotrypsin is upregulated with progression in the HMT3522 cell series, and that knockdown of the PRSS3 gene suppressed malignant growth of T4-2 cells in 3D culture, while treatment with active recombinant mesotrypsin enhanced the malignant phenotype. All of these findings implicate mesotrypsin as a serine protease upon which malignant growth of T4-2 cells depends. Furthermore, the concentrations at which different serine protease inhibitors effected reversion of T4-2 cells were consistent with the relative affinities of these inhibitors toward mesotrypsin. BPTI reverted T4-2 cells at a concentration around 150 µM, consistent with the Ki of BPTI for mesotrypsin of 14 µM [30], while SBTI blocked the T4-2 malignant phenotype at a concentration around 1 µM, consistent with the Ki of SBTI for mesotrypsin of 0.4 µM [31, 49].
A few previous studies have examined a potential role for PRSS3 expression in cancer progression. Tumor-derived epithelial cell lines from prostate, colon, and lung have been found to express PRSS3 transcripts [50–52]. Tumor growth of PC-3 prostate cancer xenografts was inhibited by the broad-spectrum serine protease inhibitor ecotin, and an effort to identify the serine proteases expressed in this system found PRSS3 transcripts among several other candidates [53]. Additionally, comparative microarray studies of non-small cell lung cancer (NSCLC) patients found PRSS3 expression to be associated with metastasis and predictive of poor survival [43]. Furthermore, overexpression of PRSS3 in NSCLC cultures led to increased transendothelial migration, highlighting a potential functional role for mesotrypsin in metastasis [43]. By contrast, a study of esophageal squamous cell carcinoma found that promoter hypermethylation frequently silenced PRSS3 expression in this cancer, while a similar phenomenon in gastric adenocarcinoma correlated with increased invasion and tumor stage, suggesting a potential tumor suppressive role for mesotrypsin [54]. Epigenetic silencing of PRSS3 has subsequently been reported in NSCLC [55] and in bladder cancer [56], although only in the latter was the silencing associated with advancing tumor stage. It may be that the malignancy promoting function of mesotrypsin depends upon the tumor type.
Using microarray analysis and qRT-PCR validation, we found that a considerable number of genes were regulated by mesotrypsin. Among these, EGFR, ITGAV, and CD109 were upregulated by mesotrypsin, while BOP1 and CD74 were downregulated by mesotrypsin. While unlikely to be exclusively responsible, these alterations are suggestive of the signaling pathways that may underlie promotion of T4-2 malignant growth by mesotrypsin. EGFR and other members of the EGF-responsive family of growth factors have been found to play a central role in the progression of cancers of the breast and other organs [57], inhibition of EGFR is a critical area of breast cancer research [58], and inhibition of EGFR has been previously found to be sufficient for reversion of T4-2 cells [28]. Integrin-av is a component of the integrin αvβ3 heterodimer, which is expressed on aggressive tumor cells of breast and other cancers, and which promotes tumor progression [59] and metastasis of breast cancer to bone [60]. Among genes downregulated by mesotrypsin, block of proliferation-1 (BOP1) was first identified in a screen for inhibitors of proliferation [61] and has been implicated in colorectal cancer progression [62], while CD74, the receptor for the tumor growth factor macrophage migration inhibitory factor [63, 64], plays a critical role in breast cancer cell proliferation [65]. Identification of how these genes are regulated and how they contribute to the effects of mesotrypsin will be an important area for future study.
Our studies with recombinant mesotrypsin indicate that mesotrypsin acts extracellularly to promote malignant growth, presumably through proteolysis of extracellular or cell surface targets. A previous study suggested that mesotrypsin may activate protease-activated receptors (PARs) on the surface of epithelial cells[50], potentially triggering pathways that have been linked to breast cancer development and progression [66–69]; however, contrasting studies have questioned the ability of mesotrypsin to signal through epithelial cell PARs [70]. Here, we identify CD109 as an alternative potential mesotrypsin substrate involved in malignant growth. We found that mesotrypsin treatment of cells both upregulated CD109 at the transcriptional level, and led to shedding of 120 and 70 kDa CD109 ectodomain fragments; shedding was blocked by the serine protease inhibitor aprotinin. The simplest hypothesis consistent with our data is that mesotrypsin cleavage releases CD109 from the cell surface. However, as a very high concentration of aprotinin is required to inhibit mesotrypsin, and at this concentration, aprotinin inhibits a broad-spectrum of serine proteases, an alternative interpretation also consistent with our data is that mesotrypsin may stimulate increased activity of another serine protease responsible for CD109 shedding. CD109 is highly expressed in many lung, esophageal, and cervical squamous cell carcinomas [71–73], and is also diagnostic for the basal-like subtype of breast carcinoma [74]. We found that RNAi knockdown of CD109 both suppressed malignant growth and blocked responsiveness to mesotrypsin treatment, consistent with a model in which CD109 shed from the cell surface by mesotrypsin participates in a pro-growth signaling function. A member of the α2-macroglobulin/complement family of proteins involved in innate immunity [75, 76], CD109 contains an internal thioester bond that may become activated upon proteolytic cleavage, triggering covalent attachment to adjacent molecules or cells [75]. While this could provide a plausible chemical basis for a growth signaling activity, further studies must focus on elucidating the mechanism, identifying CD109 attachment sites or binding partners, and understanding how these events stimulate breast cancer malignant growth. It should also be noted that knockdown of CD109 was more effective for phenotypic reversion than knockdown of PRSS3 (Fig. 7), which suggests that other serine proteases upregulated in T4-2 cells (Supplemental Fig. 1) may also be involved in CD109-mediated pathways.
As a protease that acts to stimulate malignant growth of breast cancer cells, mesotrypsin offers a potential target for breast cancer therapy. If additional studies in other systems validate the role we have identified for mesotrypsin in breast cancer progression, the T4-2 cells and 3D culture assay could be used for the development and testing of mesotrypsin-targeted therapeutics. Other proteases have been pursued as targets for breast cancer therapy, most notably the matrix metalloproteases (MMPs). Clinical trials with MMP inhibitors in both early and late stage breast cancer gave disappointing results; trials were complicated by drug nonspecificity, dose-limiting side effects, and failure to reach therapeutic plasma levels [77–81]. Given that closely related proteases can possess contrasting protumorigenic and antitumorigenic functions [78], and thus nonspecific targeting of a class of proteases can have a negative impact on survival [79], it will likely be critical for a protease-targeted therapeutic to inhibit a unique tumor-promoting enzyme in a highly selective fashion. As a potential drug target, mesotrypsin possesses the advantage of having unique sequence, steric, and electrostatic features that differentiate it from closely related proteases and may offer an opportunity to design highly selective inhibitors [30, 49]. However, these features also render mesotrypsin unusually resistant to inhibition by several classes of polypeptide inhibitors, as it is instead remarkably proficient at metabolizing these inhibitors [30, 31]. The challenge of developing inhibitors capable of binding tightly, stably, and selectively to mesotrypsin will depend upon further structural and mechanistic studies of this intriguing enzyme.
Supplementary Material
Acknowledgments
This work was supported by United States Department of Defense Breast Cancer Research Program concept grant W81XWH-06-1-0605 (ESR), Florida Department of Health Bankhead-Coley New Investigator Research Grant 07BN-07 (ESR), and the National Cancer Institute CA122086 (DCR)
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-009-0699-0) contains supplementary material, which is available to authorized users.
References
- 1.Rawlings ND, Morton FR, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2006;34:D270–D272. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nat Rev Cancer. 2004;4:876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- 3.Duffy MJ. The urokinase plasminogen activator system: role in malignancy. Curr Pharm Des. 2004;10:39–49. doi: 10.2174/1381612043453559. [DOI] [PubMed] [Google Scholar]
- 4.Netzel-Arnett S, Hooper JD, Szabo R, Madison EL, Quigley JP, Bugge TH, Antalis TM. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–258. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- 5.Nyberg P, Ylipalosaari M, Sorsa T, Salo T. Trypsins and their role in carcinoma growth. Exp Cell Res. 2006;312:1219–1228. doi: 10.1016/j.yexcr.2005.12.024. [DOI] [PubMed] [Google Scholar]
- 6.Sun Z, Yang P. Role of imbalance between neutrophil elastase and alpha 1-antitrypsin in cancer development and progression. Lancet Oncol. 2004;5:182–190. doi: 10.1016/S1470-2045(04)01414-7. [DOI] [PubMed] [Google Scholar]
- 7.Bhatt AS, Welm A, Farady CJ, Vasquez M, Wilson K, Craik CS. Coordinate expression and functional profiling identify an extracellular proteolytic signaling pathway. Proc Natl Acad Sci USA. 2007;104:5771–5776. doi: 10.1073/pnas.0606514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen KK, Oikonomopoulou K, Baruch A, Ramachandran R, Beck P, Diamandis EP, Hollenberg MD. Proteinases as hormones: targets and mechanisms for proteolytic signaling. Biol Chem. 2008;389:971–982. doi: 10.1515/BC.2008.120. [DOI] [PubMed] [Google Scholar]
- 9.Wilson TJ, Nannuru KC, Futakuchi M, Sadanandam A, Singh RK. Cathepsin G enhances mammary tumor-induced osteolysis by generating soluble receptor activator of nuclear factorkappaB ligand. Cancer Res. 2008;68:5803–5811. doi: 10.1158/0008-5472.CAN-07-5889. [DOI] [PubMed] [Google Scholar]
- 10.Ruf W, Mueller BM. Thrombin generation and the pathogenesis of cancer. Semin Thromb Hemost. 2006;32 Suppl 1:61–68. doi: 10.1055/s-2006-939555. [DOI] [PubMed] [Google Scholar]
- 11.Armstrong WB, Kennedy AR, Wan XS, Taylor TH, Nguyen QA, Jensen J, Thompson W, Lagerberg W, Meyskens FL., Jr Clinical modulation of oral leukoplakia and protease activity by Bowman–Birk inhibitor concentrate in a phase IIa chemoprevention trial. Clin Cancer Res. 2000;6:4684–4691. [PubMed] [Google Scholar]
- 12.Kennedy AR. Chemopreventive agents: protease inhibitors. Pharmacol Ther. 1998;78:167–209. doi: 10.1016/s0163-7258(98)00010-2. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi H, Yagyu T, Inagaki K, Kondo T, Suzuki M, Kanayama N, Terao T. Therapeutic efficacy of once-daily oral administration of a Kunitz-type protease inhibitor, bikunin, in a mouse model and in human cancer. Cancer. 2004;100:869–877. doi: 10.1002/cncr.20034. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H, Suzuki M, Hirashima Y, Terao T. The protease inhibitor bikunin, a novel anti-metastatic agent. Biol Chem. 2003;384:749–754. doi: 10.1515/BC.2003.083. [DOI] [PubMed] [Google Scholar]
- 15.Malkowicz SB, McKenna WG, Vaughn DJ, Wan XS, Propert KJ, Rockwell K, Marks SH, Wein AJ, Kennedy AR. Effects of Bowman–Birk inhibitor concentrate (BBIC) in patients with benign prostatic hyperplasia. Prostate. 2001;48:16–28. doi: 10.1002/pros.1077. [DOI] [PubMed] [Google Scholar]
- 16.Putnam JB, Royston D, Chambers AF, Dunbar S, Lemmer JH, Norman P, Travis E, Vaporciyan AA, Yang S, Zacharski LR. Evaluating the role of serine protease inhibition in the management of tumor micrometastases. Oncology (Williston Park) 2003;17:9–30. quiz 31–32. [PubMed] [Google Scholar]
- 17.Nguyen HH, Aronchik I, Brar GA, Nguyen DH, Bjeldanes LF, Firestone GL. The dietary phytochemical indole-3-carbinol is a natural elastase enzymatic inhibitor that disrupts cyclin E protein processing. Proc Natl Acad Sci USA. 2008;105:19750–19755. doi: 10.1073/pnas.0806581105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lippman SM, Matrisian LM. Protease inhibitors in oral carcinogenesis and chemoprevention. Clin Cancer Res. 2000;6:4599–4603. [PubMed] [Google Scholar]
- 19.Cuzick J. Treatment of DCIS—results from clinical trials. Surg Oncol. 2003;12:213–219. doi: 10.1016/j.suronc.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Senn HJ, Morant R. Chemoprevention of breast and prostate cancers: where do we stand? Ann Oncol. 2008;19 Suppl 7:vii234–ii237. doi: 10.1093/annonc/mdn470. [DOI] [PubMed] [Google Scholar]
- 21.Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, Mandelblatt JS, Yakovlev AY, Habbema JD, Feuer EJ. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353:1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 22.Jatoi I, Chen BE, Anderson WF, Rosenberg PS. Breast cancer mortality trends in the United States according to estrogen receptor status and age at diagnosis. J Clin Oncol. 2007;25:1683–1690. doi: 10.1200/JCO.2006.09.2106. [DOI] [PubMed] [Google Scholar]
- 23.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bissell MJ, Radisky DC, Rizki A, Weaver VM, Petersen OW. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70:537–546. doi: 10.1046/j.1432-0436.2002.700907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang F, Hansen RK, Radisky D, Yoneda T, Barcellos-Hoff MH, Petersen OW, Turley EA, Bissell MJ. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. J Natl Cancer Inst. 2002;94:1494–1503. doi: 10.1093/jnci/94.19.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phe-notype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–245. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park CC, Zhang H, Pallavicini M, Gray JW, Baehner F, Park CJ, Bissell MJ. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Res. 2006;66:1526–1535. doi: 10.1158/0008-5472.CAN-05-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F, Weaver VM, Petersen OW, Larabell CA, Dedhar S, Briand P, Lupu R, Bissell MJ. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proc Natl Acad Sci USA. 1998;95:14821–14826. doi: 10.1073/pnas.95.25.14821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salameh MA, Soares AS, Hockla A, Radisky ES. Structural basis for accelerated cleavage of bovine pancreatic trypsin inhibitor (BPTI) by human mesotrypsin. J Biol Chem. 2008;283:4115–4123. doi: 10.1074/jbc.M708268200. [DOI] [PubMed] [Google Scholar]
- 31.Szmola R, Kukor Z, Sahin-Toth M. Human mesotrypsin is a unique digestive protease specialized for the degradation of trypsin inhibitors. J Biol Chem. 2003;278:48580–48589. doi: 10.1074/jbc.M310301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chase T, Jr, Shaw E. p-Nitrophenyl-p′-guanidinobenzoate HCl: a new active site titrant for trypsin. Biochem Biophys Res Commun. 1967;29:508–514. doi: 10.1016/0006-291x(67)90513-x. [DOI] [PubMed] [Google Scholar]
- 33.Dachsel JC, Taylor JP, Mok SS, Ross OA, Hinkle KM, Bailey RM, Hines JH, Szutu J, Madden B, Petrucelli L, et al. Identification of potential protein interactors of Lrrk2. Parkinsonism Relat Disord. 2007;13:382–385. doi: 10.1016/j.parkreldis.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR, III, Mueller BM, Cravatt BF. Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo. Proc Natl Acad Sci USA. 2004;101:13756–13761. doi: 10.1073/pnas.0404727101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 36.Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 37.Schmeichel KL, Bissell MJ. Modeling tissue-specific signaling and organ function in three dimensions. J Cell Sci. 2003;116:2377–2388. doi: 10.1242/jcs.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw KR, Wrobel CN, Brugge JS. Use of three-dimensional basement membrane cultures to model oncogene-induced changes in mammary epithelial morphogenesis. J Mammary Gland Biol Neoplasia. 2004;9:297–310. doi: 10.1007/s10911-004-1402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kenny PA, Bissell MJ. Tumor reversion: correction of malignant behavior by microenvironmental cues. Int J Cancer. 2003;107:688–695. doi: 10.1002/ijc.11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. J Cell Biol. 2004;164:603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muschler J, Levy D, Boudreau R, Henry M, Campbell K, Bissell MJ. A role for dystroglycan in epithelial polarization: loss of function in breast tumor cells. Cancer Res. 2002;62:7102–7109. [PubMed] [Google Scholar]
- 42.Chen HM, Schmeichel KL, Mian IS, Lelievre S, Petersen OW, Bissell MJ. AZU-1: a candidate breast tumor suppressor and biomarker for tumor progression. Mol Biol Cell. 2000;11:1357–1367. doi: 10.1091/mbc.11.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Diederichs S, Bulk E, Steffen B, Ji P, Tickenbrock L, Lang K, Zanker KS, Metzger R, Schneider PM, Gerke V, et al. S100 family members and trypsinogens are predictors of distant metastasis and survival in early-stage non-small cell lung cancer. Cancer Res. 2004;64:5564–5569. doi: 10.1158/0008-5472.CAN-04-2004. [DOI] [PubMed] [Google Scholar]
- 44.Han B, Nakamura M, Mori I, Nakamura Y, Kakudo K. Urokinase-type plasminogen activator system and breast cancer (review) Oncol Rep. 2005;14:105–112. [PubMed] [Google Scholar]
- 45.Kang JY, Dolled-Filhart M, Ocal IT, Singh B, Lin CY, Dickson RB, Rimm DL, Camp RL. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–1105. [PubMed] [Google Scholar]
- 46.Parr C, Watkins G, Mansel RE, Jiang WG. The hepatocyte growth factor regulatory factors in human breast cancer. Clin Cancer Res. 2004;10:202–211. doi: 10.1158/1078-0432.ccr-0553-3. [DOI] [PubMed] [Google Scholar]
- 47.Overall CM, Tam EM, Kappelhoff R, Connor A, Ewart T, Morrison CJ, Puente X, Lopez-Otin C, Seth A. Protease degradomics: mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol Chem. 2004;385:493–504. doi: 10.1515/BC.2004.058. [DOI] [PubMed] [Google Scholar]
- 48.Kenny PA, Lee GY, Myers CA, Neve RM, Semeiks JR, Spellman PT, Lorenz K, Lee EH, Barcellos-Hoff MH, Petersen OW, et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol Oncol. 2007;1:84–96. doi: 10.1016/j.molonc.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katona G, Berglund GI, Hajdu J, Graf L, Szilagyi L. Crystal structure reveals basis for the inhibitor resistance of human brain trypsin. J Mol Biol. 2002;315:1209–1218. doi: 10.1006/jmbi.2001.5305. [DOI] [PubMed] [Google Scholar]
- 50.Cottrell GS, Amadesi S, Grady EF, Bunnett NW. Trypsin IV, a novel agonist of protease-activated receptors 2 and 4. J Biol Chem. 2004;279:13532–13539. doi: 10.1074/jbc.M312090200. [DOI] [PubMed] [Google Scholar]
- 51.Dozmorov MG, Hurst RE, Culkin DJ, Kropp BP, Frank MB, Osban J, Penning TM, Lin HK. Unique patterns of molecular profiling between human prostate cancer LNCaP and PC-3 cells. Prostate. 2009;69:1077–1090. doi: 10.1002/pros.20960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Zhang L, Wu Q, Boyd DD. Unbiased screening for transcriptional targets of ZKSCAN3 identifies integrin beta 4 and vascular endothelial growth factor as downstream targets. J Biol Chem. 2008;283:35295–35304. doi: 10.1074/jbc.M806965200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc Natl Acad Sci USA. 1999;96:11054–11061. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamashita K, Mimori K, Inoue H, Mori M, Sidransky D. A tumor-suppressive role for trypsin in human cancer progression. Cancer Res. 2003;63:6575–6578. [PubMed] [Google Scholar]
- 55.Marsit CJ, Okpukpara C, Danaee H, Kelsey KT. Epigenetic silencing of the PRSS3 putative tumor suppressor gene in non-small cell lung cancer. Mol Carcinog. 2005;44:146–150. doi: 10.1002/mc.20125. [DOI] [PubMed] [Google Scholar]
- 56.Marsit CJ, Karagas MR, Danaee H, Liu M, Andrew A, Schned A, Nelson HH, Kelsey KT. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–116. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 57.Normanno N, De Luca A, Bianco C, Strizzi L, Mancino M, Maiello MR, Carotenuto A, De Feo G, Caponigro F, Salomon DS. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366:2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Flynn JF, Wong C, Wu JM. Anti-EGFR therapy: mechanism and advances in clinical efficacy in breast cancer. J Oncol. 2009;2009:526963. doi: 10.1155/2009/526963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Desgrosellier JS, Barnes LA, Shields DJ, Huang M, Lau SK, Prevost N, Tarin D, Shattil SJ, Cheresh DA. An integrin alpha(v)beta(3)-c-Src oncogenic unit promotes anchorage-independence and tumor progression. Nat Med. 2009;15:1163–1169. doi: 10.1038/nm.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y, Bachelier R, Treilleux I, Pujuguet P, Peyruchaud O, Baron R, Clement-Lacroix P, Clezardin P. Tumor alp-havbeta3 integrin is a therapeutic target for breast cancer bone metastases. Cancer Res. 2007;67:5821–5830. doi: 10.1158/0008-5472.CAN-06-4499. [DOI] [PubMed] [Google Scholar]
- 61.Pestov DG, Grzeszkiewicz TM, Lau LF. Isolation of growth suppressors from a cDNA expression library. Oncogene. 1998;17:3187–3197. doi: 10.1038/sj.onc.1202260. [DOI] [PubMed] [Google Scholar]
- 62.Killian A, Sarafan-Vasseur N, Sesboue R, Le Pessot F, Blanchard F, Lamy A, Laurent M, Flaman JM, Frebourg T. Contribution of the BOP1 gene, located on 8q24, to colorectal tumorigenesis. Genes Chromosomes Cancer. 2006;45:874–881. doi: 10.1002/gcc.20351. [DOI] [PubMed] [Google Scholar]
- 63.Bach JP, Rinn B, Meyer B, Dodel R, Bacher M. Role of MIF in inflammation and tumorigenesis. Oncology. 2008;75:127–133. doi: 10.1159/000155223. [DOI] [PubMed] [Google Scholar]
- 64.Meyer-Siegler KL, Iczkowski KA, Leng L, Bucala R, Vera PL. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J Immunol. 2006;177:8730–8739. doi: 10.4049/jimmunol.177.12.8730. [DOI] [PubMed] [Google Scholar]
- 65.Verjans E, Noetzel E, Bektas N, Schutz AK, Lue H, Lennartz B, Hartmann A, Dahl E, Bernhagen J. Dual role of macro-phage migration inhibitory factor (MIF) in human breast cancer. BMC Cancer. 2009;9:230. doi: 10.1186/1471-2407-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang X, Guo YL, Bromberg ME. Formation of tissue factor-factor VIIa-factor Xa complex prevents apoptosis in human breast cancer cells. Thromb Haemost. 2006;96:196–201. [PubMed] [Google Scholar]
- 67.Jiang X, Zhu S, Panetti TS, Bromberg ME. Formation of tissue factor-factor VIIa-factor Xa complex induces activation of the mTOR pathway which regulates migration of human breast cancer cells. Thromb Haemost. 2008;100:127–133. doi: 10.1160/TH07-12-0722. [DOI] [PubMed] [Google Scholar]
- 68.Versteeg HH, Schaffner F, Kerver M, Ellies LG, Andrade-Gordon P, Mueller BM, Ruf W. Protease-activated receptor (PAR) 2, but not PAR1, signaling promotes the development of mammary adenocarcinoma in polyoma middle T mice. Cancer Res. 2008;68:7219–7227. doi: 10.1158/0008-5472.CAN-08-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arora P, Cuevas BD, Russo A, Johnson GL, Trejo J. Persistent transactivation of EGFR and ErbB2/HER2 by protease-activated receptor-1 promotes breast carcinoma cell invasion. Oncogene. 2008;27:4434–4445. doi: 10.1038/onc.2008.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grishina Z, Ostrowska E, Halangk W, Sahin-Toth M, Reiser G. Activity of recombinant trypsin isoforms on human proteinase-activated receptors (PAR): mesotrypsin cannot activate epithelial PAR-1, −2, but weakly activates brain PAR-1. Br J Pharmacol. 2005;146:990–999. doi: 10.1038/sj.bjp.0706410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hashimoto M, Ichihara M, Watanabe T, Kawai K, Koshikawa K, Yuasa N, Takahashi T, Yatabe Y, Murakumo Y, Zhang JM, et al. Expression of CD109 in human cancer. Oncogene. 2004;23:3716–3720. doi: 10.1038/sj.onc.1207418. [DOI] [PubMed] [Google Scholar]
- 72.Sato T, Murakumo Y, Hagiwara S, Jijiwa M, Suzuki C, Yatabe Y, Takahashi M. High-level expression of CD109 is frequently detected in lung squamous cell carcinomas. Pathol Int. 2007;57:719–724. doi: 10.1111/j.1440-1827.2007.02168.x. [DOI] [PubMed] [Google Scholar]
- 73.Zhang JM, Hashimoto M, Kawai K, Murakumo Y, Sato T, Ichihara M, Nakamura S, Takahashi M. CD109 expression in squamous cell carcinoma of the uterine cervix. Pathol Int. 2005;55:165–169. doi: 10.1111/j.1440-1827.2005.01807.x. [DOI] [PubMed] [Google Scholar]
- 74.Hasegawa M, Moritani S, Murakumo Y, Sato T, Hagiwara S, Suzuki C, Mii S, Jijiwa M, Enomoto A, Asai N, et al. CD109 expression in basal-like breast carcinoma. Pathol Int. 2008;58:288–294. doi: 10.1111/j.1440-1827.2008.02225.x. [DOI] [PubMed] [Google Scholar]
- 75.Lin M, Sutherland DR, Horsfall W, Totty N, Yeo E, Nayar R, Wu XF, Schuh AC. Cell surface antigen CD109 is a novel member of the alpha(2) macroglobulin/C3, C4, C5 family of thioester-containing proteins. Blood. 2002;99:1683–1691. doi: 10.1182/blood.v99.5.1683. [DOI] [PubMed] [Google Scholar]
- 76.Solomon KR, Sharma P, Chan M, Morrison PT, Finberg RW. CD109 represents a novel branch of the alpha2-macro-globulin/complement gene family. Gene. 2004;327:171–183. doi: 10.1016/j.gene.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 77.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 78.Fingleton B. MMPs as therapeutic targets—still a viable option? Semin Cell Dev Biol. 2008;19:61–68. doi: 10.1016/j.semcdb.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sparano JA, Bernardo P, Stephenson P, Gradishar WJ, Ingle JN, Zucker S, Davidson NE. Randomized phase III trial of marimastat versus placebo in patients with metastatic breast cancer who have responding or stable disease after first-line chemotherapy: Eastern Cooperative Oncology Group trial E2196. J Clin Oncol. 2004;22:4683–4690. doi: 10.1200/JCO.2004.08.054. [DOI] [PubMed] [Google Scholar]
- 80.Miller KD, Gradishar W, Schuchter L, Sparano JA, Cobleigh M, Robert N, Rasmussen H, Sledge GW. A randomized phase II pilot trial of adjuvant marimastat in patients with early-stage breast cancer. Ann Oncol. 2002;13:1220–1224. doi: 10.1093/annonc/mdf199. [DOI] [PubMed] [Google Scholar]
- 81.Miller KD, Saphner TJ, Waterhouse DM, Chen TT, Rush-Taylor A, Sparano JA, Wolff AC, Cobleigh MA, Galbraith S, Sledge GW. A randomized phase II feasibility trial of BMS-275291 in patients with early stage breast cancer. Clin Cancer Res. 2004;10:1971–1975. doi: 10.1158/1078-0432.ccr-03-0968. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.