Abstract
Stats1 and 3 (signal transducers and activators of transcription) can be activated simultaneously, although not necessarily to the same degree or duration, by the interaction of cells with the same polypeptide ligand (EGF, PDGF, or high concentrations of IL-6, for example). However, these two Stat proteins can mediate opposing effects on cell growth and survival. Stat1 activation slows growth and promotes apoptosis. In contrast, activated Stat3 can protect cells from apoptosis. Furthermore, a constitutively active form of Stat3, Stat3-C (bridged by S-S linkages between cysteines instead of phosphotyrosines) can induce cellular transformation of fibroblasts. We have determined that fibroblasts transformed by Stat3-C are more resistant to proapoptotic stimuli than nontransformed cells. Also, to examine the potential opposing roles in apoptosis of Stat1 and Stat3, we studied the cervical carcinoma-derived cell line, Me180, which undergoes Stat1-dependent, IFNγ-induced apoptosis. Me180 cells that express Stat3-C are protected against IFNγ-mediated apoptosis.
Signal transducers and activators of transcription (STAT) proteins are a family of latent transcription factors that are produced in many cell types and become activated by tyrosine phosphorylation and dimerization in response to a wide variety of extracellular polypeptides, many of which have effects on cell growth and survival (1, 2).
Stat1 and Stat3 are very similar (40% identical) proteins that can often be activated by the same extracellular ligand. Yet, the two proteins are also thought to have different effects on cell growth and survival. For example, fully transcriptionally active Stat1 is required for the growth restraint imposed by IFNγ on continuously cultured cells. Also, Stat1 null mouse fibroblasts do not exhibit growth arrest in response to IFNα or IFNγ (3). Furthermore, Stat1 is required for osteosarcoma-derived cells to undergo apoptosis in response to IFNγ or TNFα (4, 5). Me180 cells, a cervical carcinoma-derived cell line, are growth inhibited by low concentrations of IFNγ (6) and undergo apoptosis at higher concentrations in association with persistent Stat1 activation (7). The mechanism by which Stat1 contributes to apoptosis in these various cell lines is not known, but proteins involved in the promotion of apoptosis, such as IRF1, Caspases1, 2, and 3, have been implicated (4, 5, 7).
Stat3 is ubiquitously expressed and is transiently activated by a large number of different ligands, including EGF, PDGF, IL-6, CNTF, OSM, LIF, as well as by a number of oncogenic receptor and nonreceptor (src-like) tyrosine kinases (8). In many cancers, including lymphomas, leukemias, mycoses fungoides, multiple myeloma, brain, prostate, breast, lung, and head and neck cancers, Stat3 has been found to be persistently activated (9). Moreover, in cell culture, cellular transformation of rodent fibroblasts by the oncogene v-src results in the constitutive activation of Stat3 (10), which is required for the transformed phenotype (11, 12). Stat3 is thought to confer protection against apoptosis in many of these transformed cells. Introduction of either anti-sense Stat3 or dominant negative Stat3 in multiple myeloma-derived cell lines (13), squamous cell carcinomas (14, 15), melanoma (16), and mycosis fungoides tumor cells (17) leads to apoptosis. In addition, normal T cells that no longer express Stat3 are insensitive to the anti-apoptotic effects of IL-6 (18). We have demonstrated that a constitutively activated form of Stat3 (Stat3-C, bridged or dimerized by two cysteines instead of phosphotyrosines) can cause transformation of fibroblasts (19). In the present experiments, we demonstrated that Stat3-C transformation increases resistance to apoptotic challenge. In addition, we have shown that Stat3-C can spare Me180 cells from IFNγ (or Stat1)-dependent apoptosis. Thus, one important possible contribution made by Stat3 to cellular transformation is to protect cells against apoptosis.
Materials and Methods
Cell Culture.
NIH3T3 and Me180 cells were obtained from the American Type Culture Collection. 3Y1 cells were a generous gift of Hidesaburo Hanafusa (The Rockefeller University, New York) (20) as were NIH3T3 and 3Y1 cells transformed by v-src. NIH3T3 cells and 3Y1 cells were grown in DMEM with Pen/Strep containing 10% bovine calf serum (HyClone). NIH3T3 clones expressing Stat3-C/pBabe, v-src/pBabe or control plasmid pBabe were maintained in media containing Puromycin (1 μg/ml; Sigma), whereas 3Y1 cells expressing Stat3-C/RcCMV or control plasmid RcCMV were maintained in medium containing G418 (500 μg/ml; Geneticin GIBCO/BRL). Me180 cells were grown in RPMI with Pen/Strep containing 10% bovine calf serum. Me180 cells expressing Stat3-C/RcCMV [Me(3-C)] or control plasmid RcCMV [(Me(Rc)] were maintained in the same medium containing G418 (800 μg/ml; Geneticin GIBCO/BRL). IFNγ was a generous gift from Amgen Biologicals. IL-6 (Roche Molecular Biochemicals) was used at concentrations of 200 units/ml. TNF-α (rat) was from PeproTech (Rocky Hill, NJ).
Apoptosis Induction.
NIH3T3 and 3Y1 cells and derived cell lines were split 1–2 days before the assay such that cells were ≈50–80% confluent and growing well at the time of induction of apoptosis. NIH3T3 cells were washed with PBS once, and serum free-DMEM was added. For 3Y1 cells, the media was removed and 4 ml of PBS was added to a 100 mm dish. Cells with the lid off were treated with UV light by exposing to the germicidal lamp (peak sensitivity approximately 254 nm) in the tissue culture hood for 30 s (1.2 J/m2/s measured by J-225 Blak-Ray UV Intensity Meters, UVP, San Gabriel, CA). Following treatment, the PBS was removed and complete media prewarmed to 37°C was added. Except where indicated in the figure legends, Me180 cells were split 1–2 days before treatment such that they were 30–60% confluent. The media was replaced with complete media containing the desired ligand concentrations to initiate apoptosis.
Extranuclear DNA Fragmentation Assay.
This is a modified DNA laddering assay in which only DNA leaked into the cytoplasm because of apoptosis is detected and thus is much more sensitive than the classical DNA laddering assay (21). Following induction of apoptosis, all cells were collected (floating cells in the media as well as adherent cells) following scraping in the presence of PBS/4 mM EDTA. After washing the cells once with PBS, cell pellets were resuspended in 0.5 ml of laddering lysis buffer (25 mM Tris, pH 7.6, 0.5% Triton 100, 10 mM EDTA), incubated on ice for 20 min, followed by phenol/CHCl3 extraction and ethanol precipitation. Precipitated RNA/DNA was resuspended in TE (pH 7.6), and concentrations were determined. An aliquot of each sample was treated with RNase by adding 1/10 volume of 0.5 mg/ml RNaseA and incubated at 37°C for 1 h. Similar amounts (based on initial OD reading) of samples before and after RNaseA treatment were electrophoresed in a 1.5% agarose gel and visualized by UV fluorescence after staining by ethidium bromide. Before-RNase treated samples were used as loading controls.
SubG1 Hypodiploid FACS Analysis.
The subG1 hypodiploid apoptosis assay was performed as previously described with some modifications (22). Following induction of apoptosis or growth arrest, all cells were collected (floating cells, washed cells, and adherent cells following trypsinization). After washing with cold PBS/4 mM EDTA three times, cells were fixed in 70% ethanol O/N at −20°C. Cells were collected by centrifugation and washed once with PBS/4 mM EDTA and resuspended in PBS/4 mM EDTA containing 20 μg/ml of propidium iodide (Sigma) and 40 μg/ml RNaseA. The samples are incubated at room temperature for 30 min and then analyzed on a FACScan flow cytometer (Becton Dickenson).
Biochemical Assays.
Total cell extracts, nuclear, and cytoplasmic extracts were prepared as described with the exclusion of Nonidet P-40 in the cytoplasmic extract buffer (19). Northern and Western blots were carried out by standard methods (23). Anti-Stat3 serum (New England Biolabs) was diluted 1:1000 for Western blotting. Anti-phosphotyrosine Stat3 (Tyr705) antibody (New England Biolabs) was used at a 1:5000 dilution. Anti-FLAG monoclonal antibody (Kodak/IBI) was used at a 1:1000 dilution for Western blotting and at a 1:10 dilution for supershifting DNA-protein complexes. Anti-Bcl-2 (C-2; Santa Cruz Biotechnology), anti-Bax (N-20; Santa Cruz Biotechnology) antibodies were used at 1:1000 and 1:500, respectively, for Western blot. Survivin antibody (a gift from D. Alteri, Yale University, New Haven, CT) was used at 1 μg/ml for Western blot. Electrophoretic mobility shift assays (EMSA) were performed as previously described (24). Nuclear extracts (2–3 μg protein) from cell lines were incubated with 1 ng of 32P-labeled m67 (5′-dGATTTCCCGTAAATCAT-3′) in a 10-μl reaction volume containing DNA binding buffer [20 mM Hepes, pH 7.9, 4% Ficoll, 40 mM KCl, 10 mM MgCl2, 10 mM CaCl2, 1 mM DTT, 200 ng poly(dI:dC) (Pharmacia Biotech)]. Bcl-xL probe was kindly provided to us by Gabriel Nunez (University of Michigan Medical School, Ann Arbor, MI).
Results
Fibroblasts Transformed by Constitutively Activated Stat3 (Stat3-C) and v-src Are Protected from Apoptosis.
Stat3-C transformed immortalized rodent fibroblasts exhibit anchorage independent growth, tumor formation in nude mice and contain increased CyclinD1 and Bcl-xL mRNA levels. The increase in the levels of these mRNAs suggests that Stat3-C-mediated transformation (19), as is the case with myc transformed cells, depends on a balance between proliferative and anti-apoptotic signals (25–27). We tested whether Stat3-C could inhibit apoptosis as part of its potential mechanism in mediating cellular transformation, by exposing Stat3-C transformed rat 3Y1 or murine NIH 3T3 cells to pro-apoptotic stimuli.
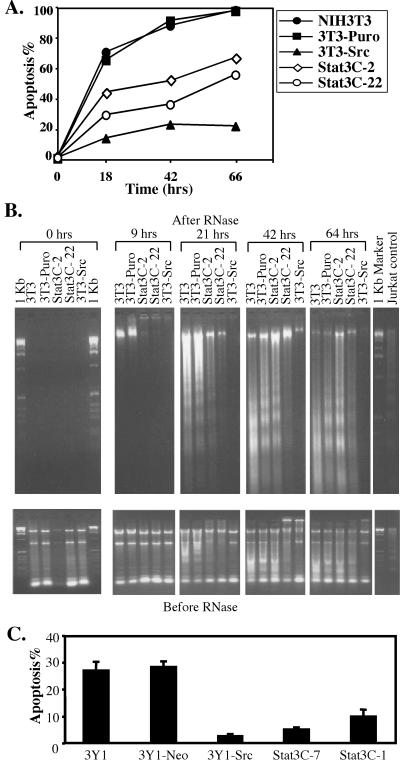
Apoptosis in NIH3T3 cells was tested by two assays (Fig. 1 A and B). Parental NIH3T3 cells (3T3), NIH3T3 cells expressing vector (3T3-Puro), Stat3-C (Stat3-C) or v-src (3T3-Src) were deprived of serum and levels of apoptosis were determined by analyzing the subG1 DNA content of the cell population at several time points (Fig. 1A). This assay quantitates hypodiploid cells generated through apoptosis secondary to DNA fragmentation. 3T3 cells and 3T3-Puro cells were sensitive to serum deprivation and, under the conditions tested, ≈70% of the cells underwent apoptosis within 18 h, ≈90% by 42 h and ≈100% after 66 h (Fig. 1A, 3T3 and 3T3-Puro lines). Two different clones expressing Stat3-C were tested. Stat3-C clone 22 (Stat3C-22) expressed somewhat more Stat3-C than clone 2 (Stat3C-2; data not shown). Both Stat3-C expressing clones showed clear protection against serum deprivation-induced apoptosis. Apoptosis in Stat3-C expressing cells after 66 h of serum starvation was comparable to the control 3T3 cells after 18 h of serum deprivation (Fig. 1A, compare Stat3C-2, Stat3C-22 vs. 3T3, 3T3-puro). The Stat3C-22 line showed more effective protection than Stat3C-2, suggesting that Stat3-C expression levels may be important. Consistent with the potential role of anti-apoptosis in transformation, the plating efficiency in soft agar or transformation potential of the cell line Stat3C-22 was ≈20% greater than clone Stat3C-2 (data not shown). It has recently been demonstrated that v-src transformed Rat-1 cells are resistant to apoptosis induced by serum deprivation in part as a function of PI3Kinase (28). We observed a similar phenomenon following serum withdrawal from v-src transformed NIH3T3 cells (Fig. 1A, compare lines of 3T3-Src and 3T3, 3T3-Puro). Consistent with its greater transforming capacity (19), v-src vs. Stat3-C transformed NIH3T3 cells were better protected from apoptosis (Fig. 1A, compare 3T3-Src 66 h vs. Stat3C-2 or Stat3C-22 18 h). Thus other molecules in addition to Stat3 probably contribute to the transforming and anti-apoptotic function of v-src.
Figure 1.
Constitutively active Stat3-C and v-src inhibit apoptosis in fibroblast cells. (A) 3T3 and derived cell lines were serum starved, and apoptosis was assayed at the indicated time points by FACs quantitation of propidium iodide-stained cells in the subG1 population. (B) 3T3 and derived cell lines were serum starved, and apoptosis was assayed at the indicated time points by assaying extranuclear DNA fragmentation, as described in Materials and Methods. This method extracts both extranuclear RNA and DNA. After RNase treatment, the extranuclear DNA was electrophoresed in a 1.5% agarose gel and visualized by UV fluorescence after staining by ethidium bromide. As a loading control, the purified nucleic acids without RNase treatment were similarly analyzed by agarose gel electrophoresis and ethidium bromide staining. Jurkat cells induced into apoptosis by camptothecin are shown as controls for DNA laddering. (C) 3Y1 and derived cells were treated with UV radiation, and apoptosis was assayed 24 h after treatment by FACs quantitation of propidium iodide-stained cells in the subG1 population. A representative experiment was shown with the mean + standard deviation of the triplicate samples.
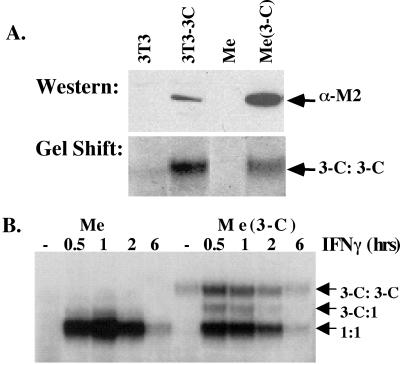
Figure 3.
Me180 cells expressing Stat3-C. (A) Stable clones of Me180 cells were isolated expressing either a control plasmid or a Stat3-C expressing plasmid. The clones were maintained in G418 containing media. Nuclear extracts were isolated from these clones and one example is shown here. Ten micrograms of protein from each cell line (3T3, 3T3–3C, Me, Me3-C) was resolved on an SDS polyacrylamide gel, transferred to nitrocellulose, and probed with anti-FLAG antisera. Two micrograms of nuclear extract from the same cell lines (see above) were incubated with radiolabeled m67 and resolved on a nondenaturing EMSA gel. (B) Parental Me180 cells (Me) and Me180 expressing Stat3-C cells (Me 3-C) were either left untreated (serum containing media) or treated with IFNγ (0.5 ng/ml) for 0.5, 1, 2, or 6 h. Nuclear extracts were isolated from these, incubated with radiolabeled m67, and resolved on an EMSA gel.
To extend the results of the subG1 assay, 3T3 and derived cell lines were analyzed by an extranuclear DNA fragmentation assay following serum deprivation (Fig. 1B). Both cytoplasmic RNAs and extranuclear DNA were extracted and analyzed directly by agarose gel electrophoresis. “Before RNase” samples (Fig. 1B, Bottom) serve as loading controls, whereas the “After RNase” samples (Fig. 1B, Top) indicate the relative levels of apoptosis. As expected, no extranuclear DNA was detected in healthy growing cells (Fig. 1B, 0 h). Extranuclear DNA, indicating early stages of apoptosis, was detected after 9 h of serum deprivation in control NIH3T3 cells (the high molecular weight band, Fig. 1B, Top, 9 h, 3T3 and 3T3-puro). At later time points, additional extranuclear DNA, some as a nucleosomal DNA ladder, was detected in the control parental 3T3 cells and 3T3-puro cells, indicating further apoptosis (Fig. 1B, 3T3 and 3T3-Puro at 21 h, 42 h and 64 h time points). Extracts of the Stat3-C clones had almost no extranuclear DNA at 9 h and significantly less extranuclear DNA at 21 h than the 3T3 controls (Fig. 1B). Protection against DNA fragmentation in the Stat3-C clones was less apparent after 42 and 64 h (Fig. 1B, 64 h). In this assay, v-src transformed 3T3 cells showed less apoptosis than the Stat3-C transformed cells at both 42 and 64 h (Fig. 1B).
The ability of either Stat3-C or v-src to prevent apoptosis was also examined in immortalized rat fibroblasts, 3Y1 cells. In contrast to NIH3T3 cells, 3Y1 cells are much less sensitive to apoptosis induced by serum deprivation, but do undergo apoptosis after UV irradiation. In response to UV irradiation, 3Y1 cells underwent apoptosis as determined by multiple criteria such as subG1 analysis, “tunnel” staining, Hoechst staining, and extranuclear DNA fragmentation assays, whereas both Stat3-C and v-src transformed 3Y1 cells were relatively resistant to UV-induced apoptosis. 3Y1 cells and 3Y1 cells expressing vector alone (3Y1-Neo) showed comparable amounts of apoptosis (≈28%) as determined by subG1 quantitation at 24 h after UV irradiation (Fig. 1C, 3Y1 and 3Y1-Neo). Two independent Stat3-C expressing clones (Stat3C-7 and Stat3C-1) as well as a v-src expressing clone (3Y1-src) were less sensitive to UV-stimulated apoptosis (5%, 10%, and 3%, respectively) (Fig. 1C).
Me180 Cells Are Sensitive to the Growth Inhibitory Effects of IFNγ.
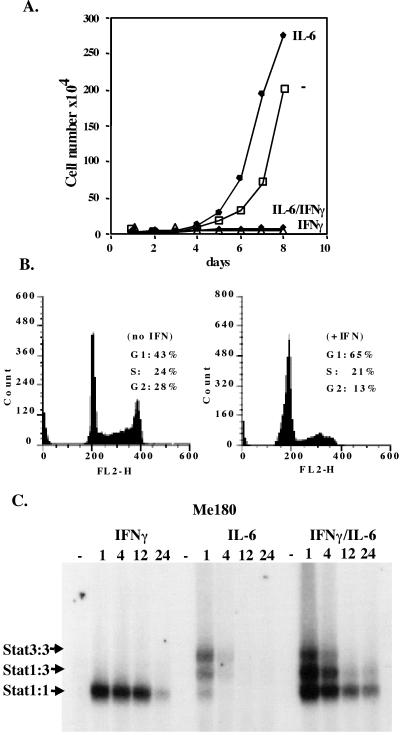
Me180 cells, derived from a cervical carcinoma, were growth arrested when treated with relatively low concentrations (0.5 ng/ml) of IFNγ (Fig. 2A). FACS analysis of Me180 cells treated with 0.5 ng/ml of IFNγ showed an arrest of the cell cycle in G1 (Fig. 2B). [At higher concentrations of IFNγ these cells undergo apoptosis as will be described later (see Fig. 4).] This growth inhibitory concentration of IFNγ led to Stat1 activation [that lasted for approximately 6–12 h] as tested by EMSA. In contrast, IL-6 treatment of Me180 cells caused a relatively transient (1 h) activation of Stat3 (Fig. 2C), and, if anything, gave a possible growth advantage (Fig. 2A). The simultaneous addition of IFNγ and IL-6 gave rise to activation of both Stat3 and Stat1 with sustained Stat1 activation lasting much longer than Stat3 activation (Fig. 2C). Addition of both of these ligands led to growth inhibition (Fig. 2A) in association with and perhaps dependent on the more extensive Stat1 activation (Fig. 2C).
Figure 2.
Me180 cells are growth inhibited by low concentrations of IFNγ. (A) 1 × 104 Me180 cells were plated onto 3-cm dishes. Once the cells had adhered (≈12 h later), IFNγ (0.5 ng/ml), IL-6 (200 units/ml), both, or neither were added to the adherent cells grown in complete media. On a daily basis, the cells were counted by trypan blue exclusion (the mean of triplicate samples). (B) 1 × 106 cells were plated onto 6-cm dishes and treated with or without IFNγ (0.5 ng/ml) for 24 h. The cells were harvested, stained with propidium iodide (PI), and analyzed by FACS. The number of cells in G1, S, and G2 was determined. (C) Me180 cells were grown and treated with either IFNγ (0.5 ng/ml), IL-6 (200 units/ml), or both for 1, 4, 12, or 24 h. Cells were harvested and nuclear extracts were isolated from the cell samples. Equal amounts of protein were incubated with radiolabeled double stranded m67(binding site for Stat1 and or Stat3) and resolved on a nondenaturing acrylamide gel.
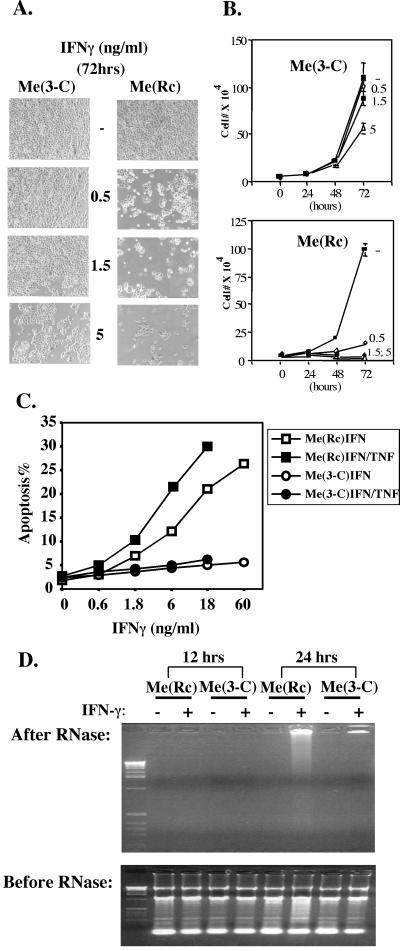
Figure 4.
Me180 cells expressing Stat3-C are protected from IFNγ-induced apoptosis. (A) 2 × 104 Me180 cells stably transfected with the control plasmid RcCMV [Me(Rc)] and Me180 expressing Stat3-C cells (Me3-C) were plated onto 3-cm dishes. Once the cells had adhered (≈12 h later), IFNγ (0.5 ng/ml, 1.5 ng/ml, or 5 ng/ml) was added to the adherent cell culture. Then, 72 h later, the cells were washed with PBS and photographed by using phase microscopy. (B) As described above. Cells were counted on a daily basis by trypan blue exclusion (the mean of triplicate samples). (C) Me(Rc) and Me(3-C) cells were treated at the indicated IFNγ concentration (ng/ml) for 48 h, and apoptosis was assayed by FACs quantitation of propidium iodide-stained cells in the subG1 population. Whenever indicated, TNF-α was added to a concentration of 5 μg/ml. Shown are representative results of four independent experiments. (D) Me(Rc) and Me3-C cells were treated with IFNγ (18 ng/ml) for 12 and 24 h, extranuclear DNA was prepared and analyzed by agarose gel electrophoresis and ethidium bromide staining, as described in Materials and Methods.
Me180 Cells Expressing Stat3-C Are Resistant to IFNγ-Induced Apoptosis.
To examine the role of sustained or persistently activated Stat3 in Me180 cells, we introduced Stat3-C into ME180 cells and generated a number of cell lines expressing Stat3-C protein which binds DNA constitutively [detected in Fig. 3A, lane Me(3-C), by the epitope tag added at its COO-terminus (western) and by EMSA (gel shift)]. In response to IFNγ, we observed similar levels of Stat1 activation in both the parental line and in a Stat3-C reconstituted line (Fig. 3B).
It was recently demonstrated that Me180 cells undergo apoptosis in response to IFNγ and that cell lines that had lost Stat1 protein expression were resistant to IFNγ-induced apoptosis (7). We therefore wished to determine whether Me180 cells expressing Stat3-C (Me3-C) responded differently to IFNγ despite similar levels of Stat1 activation (Fig. 3B). After the addition of increasing concentrations of IFNγ, we observed a decrease in cell number and a corresponding increase in apoptotic cells in the control Me(Rc) cells (Fig. 4). In contrast, Me(3-C) cells were protected from IFNγ induced growth arrest and apoptosis (Fig. 4). After the addition of low concentrations (0.5 ng/ml) of IFNγ, we observed continued growth (Fig. 4 A and B) and no arrest in G1 of Me(3-C) cells (as determined by FACS, data not shown). At higher concentrations of IFNγ (5 ng/ml), we observed some growth arrest and apoptosis but significantly less than in the control cell line (Me3-C vs Me(Rc) (Fig. 4 A and B). Furthermore, Me(3-C) cells were protected from IFNγ-induced apoptosis as determined by analysis of the subG1 population of cells and by DNA laddering analysis of both cell types (Fig. 4 C and D). TNF-α is a known inducer of apoptosis in a number of different cell types and in certain cells may be dependent on the presence of Stat1 to induce apoptosis (5). We therefore tested whether Stat3-C expressing Me180 cells were protected against TNF-α induced apoptosis. Although TNF-α alone (even at high concentrations, 50 μg/ml) did not induce significant apoptosis in Me180 cells (data not shown), low concentrations of TNF-α (5 μg/ml) when added to IFNγ-treated Me180 cells enhanced the killing effect by IFNγ (Fig. 4C). This enhanced killing was blocked in the Stat3-C expressing cells, suggesting that Stat3-C may also inhibit IFNγ/TNF-α killing (Fig. 4C). The protective effect of Stat3-C against the IFNγ-TNF combination was also confirmed by the extra-nuclear DNA fragmentation assay (data not shown).
Pro- and Anti-Apoptotic Proteins in Stat3-C Cell Lines.
By Northern blot and Western blot analysis, we have determined levels of mRNAs encoding for proteins as well as proteins that are reported to exert pro- or anti-apoptotic effects in other cell systems. As we originally reported, the mRNA for Bcl-xL, an anti-apoptotic protein, is elevated in Stat3-C transformed 3Y1 and 3T3 fibroblasts (19). We also have assayed steady state mRNA levels in growing Me180 cells and Me(3-C) cells, the latter clearly result in protection against IFN-γ and TNF induced apoptosis. In these assays (Fig. 5A), we also found in Me(3-C) cells increased Bcl-xL mRNA levels. By Western blot analysis and RT-PCR analysis (data not shown), we observed no change in Bcl-2 levels, a decrease in the expression of Bax (a pro-apoptotic protein) in Me(3-C) cells and an increased expression of survivin (Fig. 5B). Survivin protein inhibits apoptosis and has been reported to be elevated in numerous cancer cells (29). However, we can not state with certainty which (if any) of the observed changes underlie the protection against apoptosis by Stat3 in fibroblasts or Me180 cells.
Figure 5.
Me180 cells expressing Stat3-C contain elevated expression of Bcl-xL and survivin, and decreased expression of Bax. (A) Northern blot analysis of Bcl-xL on total RNA prepared from Me180 cells (Me) or Me180 cells stably expressing Stat3C (Me 3-C) treated with IFN-γ for 4 h (IFN-γ) or untreated (−). Blot was reprobed with GAPDH to control equal loading. (B) Western blot analysis of Bcl-2, Bax, and survivin. Fifty micrograms of whole cell extract from Me180 cells stably transfected with the control plasmid [Me(Rc)] or Me180 cells expressing Stat3C (Me 3-C) treated with IFN-γ (18 ng/ml) for 24 h (IFN-γ) or untreated (−) were blotted with the indicated antibody.
Discussion
The use of cultured mammalian cells to explore growth regulation by extracellular signaling proteins and their cognate receptors has a long and successful history. Many aberrations in signaling originally recognized in culture have led to discovery of mutant human proteins in a variety of signaling pathways that are apparently involved in human cancer. The Stat proteins are critical mediators of many of external signaling factors and therefore it is perhaps not surprising that persistent Stat activation is very commonly found in association with clinical cancers and with cultured transformed cells. That the constitutively activated Stat3 molecule, Stat3-C, also transforms immortalized fibroblasts independently of any extracellular signals (19) adds to the conviction that persistently active Stat3 in the clinical setting is important. Because cellular transformation by oncogenes is thought to depend on a balance between growth promotion, progression through the cell cycle and cell survival, i.e., prevention of apoptotsis (30–32), we turned our attention to apoptosis in Stat3-C transformed cells.
Stat3-C clearly conferred protection from apoptosis, induced by either UV irradiation or serum deprivation in fibroblasts. Stat3-C could also protect rat 3Y1 cells from other stimulators of apoptosis including cycloheximide, staurosporin and actinomycin. Furthermore, v-src transformed fibroblasts were better protected (than Stat3-C containing cells) from UV and serum deprivation induced-apoptosis, suggesting that proteins in addition to Stat3 are probably involved in the protection against apoptosis induced by v-src. However, neither Stat3-C nor v-src could protect NIH3T3 cells from Fas-mediated apoptosis (data not shown). In contrast to v-src, oncogenic ras does inhibit Fas-mediated apoptosis of 3T3 cells perhaps through inhibition of Fas expression (33, 34). Thus, different oncogenes may support cell survival through different mechanisms although the precise mechanism(s) in any case is unclear.
In addition to the experiments described here, a number of previous experiments show that activated Stat3 plays an important role in protecting transformed cancerous cells from apoptosis. The introduction of dominant negative Stat3 molecules or anti-sense molecules into multiple myeloma or head and neck cancer cells, both of which contain persistently activated Stat3, leads to their apoptosis (13–17).
In the present experiments, we have also explored the effects on apoptosis of simultaneous Stat3 and Stat1 activation. These two proteins share extensive sequence homology and their crystal structures are virtually superimposable (35, 36). Furthermore, in a DNA binding sequence selection assay, they select the same optimal DNA sequence (37). In many cases, Stat1 and Stat3 are activated simultaneously by the same cytokines and in some clinical cancers both are persistently activated. However, the biological effects of these two Stat proteins are frequently opposing. Stat3 is a proto-oncogene and inhibits apoptosis, whereas Stat1 inhibits cell growth and is required for induction of apoptosis. In the present experiments, we show that persistent activation through Stat3-C can overcome IFN-γ-induced or IFNγ/TNFα-induced apoptosis, which is likely to depend on Stat1 activation. Thus, the balance of activated Stat1 and Stat3 could be critical in deciding the fate of cells, once again adding to the conclusion that persistently active Stat3 in human tumors may be generally protective of apoptosis in cancer cells.
Acknowledgments
We thank Drs. Ray Birge, Maria Georgescu, and Young Bae Kim for helpful discussions, and Dr. Dario Alteri for the survivin antibody. We are grateful to Lois Cousseau for preparing the manuscript. J.F.B was supported by a McDonnell Foundation Award and a Culpepper Award. This work was supported by National Institutes of Health Grant AI32489 (to J.E.D.).
Abbreviations
- STAT
signal transducers and activators of transcription
- EMSA
electrophoretic mobility shift assay
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041588198.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041588198
References
- 1.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin Y E, Kitagawa M, Kuida K, Flavell R A, Fu X Y. Mol Cell Biol. 1997;17:5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar A, Commane M, Flickinger T W, Horvath C M, Stark G R. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg J F, Fan Z, Brown C, Mendelsohn J, Darnell J E., Jr Cell Growth Differ. 1998;9:505–12. [PubMed] [Google Scholar]
- 7.Lee K Y, Anderson E, Madani K, Rosen G D. FEBS Lett. 1999;459:323–326. doi: 10.1016/s0014-5793(99)01283-1. [DOI] [PubMed] [Google Scholar]
- 8.Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–51. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 9.Bowman T, Garcia R, Turkson J, Jove R. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 10.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 11.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Mol Cell Biol. 1998;5:2553–8. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot R P, Jove R. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Catlett-Falcone R, Landowski T H, Oshiro M M, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna J L, Nunez G, et al. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 14.Grandis J R, Drenning S D, Chakraborty A, Zhou M Y, Zeng Q, Pitt A S, Tweardy D J. J Clin Invest. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grandis J R, Drenning S D, Zeng Q, Watkins S C, Melhem M F, Endo S, Johnson D E, Huang L, He Y, Kim J D. Proc Natl Acad Sci USA. 2000;97:4227–4232. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, Jove R, Yu H. Cancer Res. 1999;59:5059–5063. [PubMed] [Google Scholar]
- 17.Nielsen M, Kaestel C G, Eriksen K W, Woetmann A, Stokkedal T, Kaltoft K, Geisler C, Ropke C, Odum N. Leukemia. 1999;13:735–738. doi: 10.1038/sj.leu.2401415. [DOI] [PubMed] [Google Scholar]
- 18.Takeda K, Kaisho T, Yoshida N, Takeda J, Kishimoto T, Akira S. J Immunol. 1998;161:4652–4660. [PubMed] [Google Scholar]
- 19.Bromberg J, Wrzeszczynska M, Devgan G, Zhao Y, Albanese C, Pestell R, Darnell J E., Jr Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 20.Kimura G, Itagaki A, Summers J. Int J Cancer. 1975;15:694–706. doi: 10.1002/ijc.2910150419. [DOI] [PubMed] [Google Scholar]
- 21.Navarro P, Valverde A M, Benito M, Lorenzo M. Exp Cell Res. 1998;243:213–221. doi: 10.1006/excr.1998.4168. [DOI] [PubMed] [Google Scholar]
- 22.Taylor J K, Zhang Q Q, Monia B P, Marcusson E G, Dean N M. Oncogene. 1999;18:4495–4504. doi: 10.1038/sj.onc.1202836. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 24.Vinkemeier U, Cohen S L, Moarefi I, Chait B T, Kuriyan J, Darnell J E., Jr EMBO J. 1996;15:5616–5626. [PMC free article] [PubMed] [Google Scholar]
- 25.Evan G, Littlewood T. Science. 1998;281:1317–1322. doi: 10.1126/science.281.5381.1317. [DOI] [PubMed] [Google Scholar]
- 26.Juin P, Hueber A O, Littlewood T, Evan G. Genes Dev. 1999;13:1367–1381. doi: 10.1101/gad.13.11.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 28.Johnson D, Agochiya M, Samejima K, Earnshaw W, Frame M, Wyke J. Cell Death Differ. 2000;7:685–696. doi: 10.1038/sj.cdd.4400700. [DOI] [PubMed] [Google Scholar]
- 29.Altieri D C, Marchisio P C, Marchisio C. Lab Invest. 1999;79:1327–1333. [PubMed] [Google Scholar]
- 30.Weinberg R A. Cytokines Mol Ther. 1996;2:105–110. [PubMed] [Google Scholar]
- 31.Sherr C J, Weber J D. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 32.Hahn W C, Counter C M, Lundberg A S, Beijersbergen R L, Brooks M W, Weinberg R A. Nature (London) 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 33.Peli J, Schroter M, Rudaz C, Hahne M, Meyer C, Reichmann E, Tschopp J. EMBO J. 1999;18:1824–1831. doi: 10.1093/emboj/18.7.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fenton R G, Hixon J A, Wright P W, Brooks A D, Sayers T J. Cancer Res. 1998;58:3391–3400. [PubMed] [Google Scholar]
- 35.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 36.Becker S, Groner B, Muller C W. Nature (London) 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 37.Horvath C M, Wen Z, Darnell J E., Jr Genes Dev. 1995;9:984–994. doi: 10.1101/gad.9.8.984. [DOI] [PubMed] [Google Scholar]