SUMMARY
Mammalian telomeres contain a single-stranded 3′ overhang that is thought to mediate telomere protection. Here we identify the TRF2-interacting factor Apollo as a nuclease that contributes to the generation/maintenance of this overhang. The function of mouse Apollo was determined using Cre-mediated gene deletion, complementation with Apollo mutants, and the TRF2-F120A mutant that cannot bind Apollo. Cells lacking Apollo activated the ATM kinase at their telomeres in S phase and showed leading-end telomere fusions. These telomere dysfunction phenotypes were accompanied by a reduction in the telomeric overhang signal. The telomeric functions of Apollo required its TRF2-interaction and nuclease motifs. Thus, TRF2 recruits the Apollo nuclease to process telomere ends synthesized by leading-strand DNA synthesis, thereby creating a terminal structure that avoids ATM activation and resists end-joining. These data establish that the telomeric overhang is required for the protection of telomeres from the DNA damage response.
INTRODUCTION
Mammalian telomeres terminate in a 50 to 400 nt single-stranded 3′ overhang that is assumed to have a crucial role in end protection (Makarov et al., 1997; McElligott and Wellinger, 1997; Wright et al., 1997). This G-rich overhang can serve as the primer for telomerase, which synthesizes the telomeric TTAGGG repeats and maintains telomere length homeostasis in S phase (Greider and Blackburn, 1987). Furthermore, strand invasion of the G-rich overhang into the duplex region of the telomere has been proposed to prevent telomere fusions (Griffith et al., 1999). This structure, known as the t-loop, would presumably prevent loading of the Ku70/80 heterodimer and the MRN (Mre11, Rad50, Nbs1) complex, thereby blocking NHEJ and ATM signaling, respectively (reviewed in (de Lange, 2009)). The single-stranded G-rich repeats also function as binding sites for the POT1 proteins, which prevent the activation of ATR, protect against post-replicative fusions of sister chromatids, and repress homologous recombination between sister telomeres (Hockemeyer et al., 2006; Lazzerini Denchi and de Lange, 2007; Wu et al., 2006; Palm et al., 2009).
The telomeric overhang is generated during replication independently of telomerase (Hemann and Greider, 1999) and its length correlates with the rate of telomere shortening in human cells lacking telomerase (Huffman et al., 2000). However, the factors involved in the generation of the telomeric overhangs have not been identified. The degradation of the terminal RNA primer used in lagging-strand DNA synthesis has been proposed to generate a 3′ overhang at lagging-end telomeres, resulting in the end-replication problem (Watson, 1972). However, evidence of telomeric overhangs at ends replicated by both leading- and lagging-strand DNA synthesis in human cells suggests additional mechanism(s) of overhang generation (Makarov et al., 1997). Specifically, the 5′ to 3′ progression of leading-strand DNA synthesis necessitates resection of the parental 5′ ends to generate 3′ overhangs at leading-end telomeres.
Recent studies have suggested a role for the shelterin component TRF2 in overhang generation at leading-end telomeres. Telomeres with compromised TRF2 activate the MRN-dependent ATM kinase pathway and undergo non-homologous end-joining (NHEJ), generating chromosome end fusions that are promoted by ATM signaling (van Steensel et al., 1998; Celli and de Lange, 2005; Lazzerini Denchi and de Lange, 2007; Dimitrova and de Lange, 2009; Deng et al., 2009; Attwooll et al., 2009). When TRF2 is deleted, the telomeric overhang signal is rapidly lost, presumably as a consequence of the frequent NHEJ events (Celli and de Lange, 2005). However, in the absence of Nbs1, TRF2 deletion still induces significant overhang loss, even though telomeric NHEJ events are infrequent and limited to leading-end telomeres (Dimitrova and de Lange, 2009). These observations led to the proposal that TRF2 recruits or activates a nuclease that generates the 3′ overhang at leading end telomeres (Dimitrova and de Lange, 2009). In the absence of TRF2, the MRN/ATM pathway was proposed to induce resection at the unprotected ends, thereby providing an alternative means of generating overhangs at the leading-end telomeres. Consistent with this proposal, the fusion of leading-end telomeres is a highly specific phenotype associated with TRF2 deletion from MRN- or ATM-deficient cells (Dimitrova and de Lange, 2009; Attwooll et al., 2009).
Among the factors recruited to telomeres by TRF2, one candidate nuclease is Apollo/SNM1B. Apollo belongs to the mammalian SNM1/Pso2 family of nucleases, which also includes SNM1A and Artemis/SNM1C. SNM1A contributes to the repair of DNA interstrand cross-links (ICLs) (Demuth et al., 2004; Dronkert et al., 2000), lesions that block DNA replication and transcription (reviewed in (Dominski, 2007)). Similarly, knockdown of Apollo/SNM1B in human cells results in hypersensitivity to ICL-inducing agents (Demuth et al., 2004; Bae et al., 2008). In contrast, Artemis/SNM1C functions as an endonuclease to remove hairpins from coding ends during V(D)J recombination (reviewed in (de Villartay, 2009) and has been suggested to contribute to homology-directed repair (HDR) and NHEJ of a subset of DSBs by removing structures that block repair reactions (Kurosawa et al., 2008; Beucher et al., 2009).
Apollo is the only member of the SNM1/Pso2 family known to function as a shelterin accessory factor. Unlike core components of shelterin, shelterin accessory factors are not abundant at telomeres; they are often not observed at all telomeres and/or show a transient telomere association; and they function at non-telomeric sites, most often in the DNA damage response (reviewed in (Palm and de Lange, 2008)). Apollo is recruited to telomeres through a C-terminal YxLxP motif that mediates its interaction with a common protein-docking site surrounding F120 of TRF2 (Chen et al., 2008). Knockdown experiments have implicated human Apollo in the protection of telomeres in S phase (van Overbeek and de Lange, 2006; Lenain et al., 2006). Here, we use conditional gene deletion and dissociation-of-function alleles to determine the role of the Apollo nuclease at mouse telomeres. We identify a function for Apollo in maintaining the telomeric overhang and preventing fusions of newly-synthesized leading-end telomeres.
RESULTS
Conditional deletion of Apollo
To generate a conditional gene deletion system in mouse embryo fibroblasts (MEFs), the Apollo gene (Dclre1b, chr. 3) was modified by gene-targeting resulting in a floxed allele (ApolloF) that contained loxP Cre recombinase target sites flanking exons 2 and 3 (Fig. 1A). Deletion of exons 2 and 3 is predicted to result in out-of-frame splicing of exon 1 to exon 4, interrupting the Apollo ORF at amino acid position 67. This strategy was favored over conditional deletion of exon 1, which might affect the neighboring Ap4b1 gene (Fig. 1A). ApolloF/F embryos (E13.5) derived from ApolloF/+ mouse intercrosses were used to establish SV40 large T antigen (SV40-LT) immortalized MEFs. Transient expression of Cre recombinase in these cells resulted in the expected deletion of the Apollo gene and concomitant loss of the full-length Apollo mRNA whereas the Ap4b1 transcript was not affected (Fig. 1B,C). Cre treatment of the ApolloF/F MEFs resulted in a slight proliferation defect that was due to the absence of Apollo since it was largely rescued by expression of the wild type protein (Fig. S1A). The cell cycle profile of SV40-LT ApolloF/F MEFs showed an elevated 4N peak due to a high basal level of tetraploid cells, which is a common phenomenon in immortalized MEFs. Cre-mediated deletion of Apollo caused a slight increase in tetraploid cells reflected in an increase in the 8N peak, but did not significantly alter the cell cycle profile or S phase index, as measured by BrdU incorporation (Fig. S1B). Thus, deletion of Apollo does not immediately block proliferation of immortalized MEFs, allowing the evaluation of Apollo function in these cells.
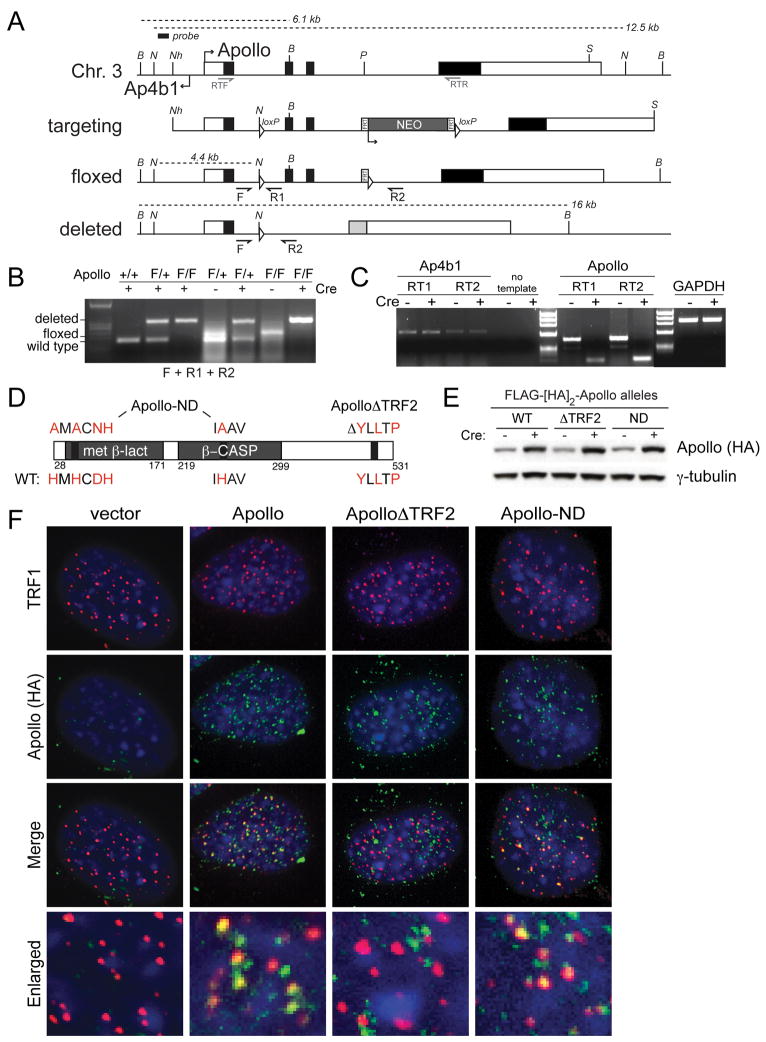
Figure 1. Deletion of mouse Apollo and complementation with mutant alleles.
(A) Targeting of the mouse Apollo locus. The structure of the genomic locus, the targeting construct, the floxed allele and the deleted allele are shown. loxP sites are represented as triangles; FRT sites surrounding the neo gene are shown as rectangles. Approximate positions of the PCR primers for genomic analysis (F and R1 and R2) and RT-PCR mRNA analysis (RTF and RTR) are shown. Restriction endonucleases and the probe used for analysis of genomic DNA: B, BamHI, N, NsiI, Nh, NheI, P, PacI, S, ScaI. (B) Genotyping PCR for Apollo using DNA from MEFs. (C) RT-PCR with two independent sets of primers for both Apollo and Ap4b1 using RNA purified from cells treated with or without Cre. GAPDH was used as a control. (D) Schematic of the mouse Apollo protein indicating regions that are altered in Apollo rescue alleles. Amino acids in red indicate important residues for nuclease activity and TRF2 interaction. (E) Immunoblotting analysis of ApolloF/F MEFs expressing the indicated Apollo alleles in the absence of Cre and at 120h after Cre. Apollo is detected with the HA.11 antibody. (F) Immunofluorescence showing the localization of the indicated Apollo alleles (detected with HA.11 Ab) in ApolloF/F MEFs at 72h after Cre infection. Telomeric loci are detected with Ab 644 to the shelterin component TRF1. DNA is stained with DAPI. See also Figure S1.
Apollo mutants
In order to assess the telomere-specific functions of Apollo, we generated an Apollo allele deficient for binding to telomeres (ApolloΔTRF2) (Fig. 1D-F and Fig. S1C). Despite a previous report documenting that Apollo is unstable when not bound to TRF2 (Freibaum and Counter, 2008), we observed that both human and mouse ApolloΔTRF2 were expressed at the same level as the wild type protein (Fig. 1E, Fig. S1D-F). Deletion of the YLLTP TRF2 binding site abolished the interaction of Apollo with TRF2 and generated a protein that was incapable of accumulating at telomeres (Fig. 1F, Fig. S1C).
To evaluate whether the function of Apollo depends on its nuclease activity, we generated a nuclease-deficient allele of Apollo, Apollo-ND, by mutating the HxHxDH motif in the metallo β-lactamase domain as well as a highly conserved histidine in the β-CASP domain (Fig. 1D-F). Both the HxHxDH motif and histidine 230 are conserved in Artemis and required for the endonucleolytic activity of this closely-related SNM1 nuclease (Callebaut et al., 2002; Pannicke et al., 2004; de Villartay et al., 2009). Consistent with the preservation of the TRF2-interacting site in Apollo-ND, the nuclease-deficient protein associated with TRF2 and localized to telomeres (Fig. 1F and Fig. S1C). Based on IF analysis (n>100 nuclei), both the wild type and Apollo-ND alleles were detectable at approximately half the telomeres in the cells. Since Apollo is not as abundant at telomeres as the shelterin components, it is possible that both wild type Apollo and Apollo-ND localize to all telomeres but escape detection because of their low abundance.
Apollo protects telomeres from activating the ATM kinase
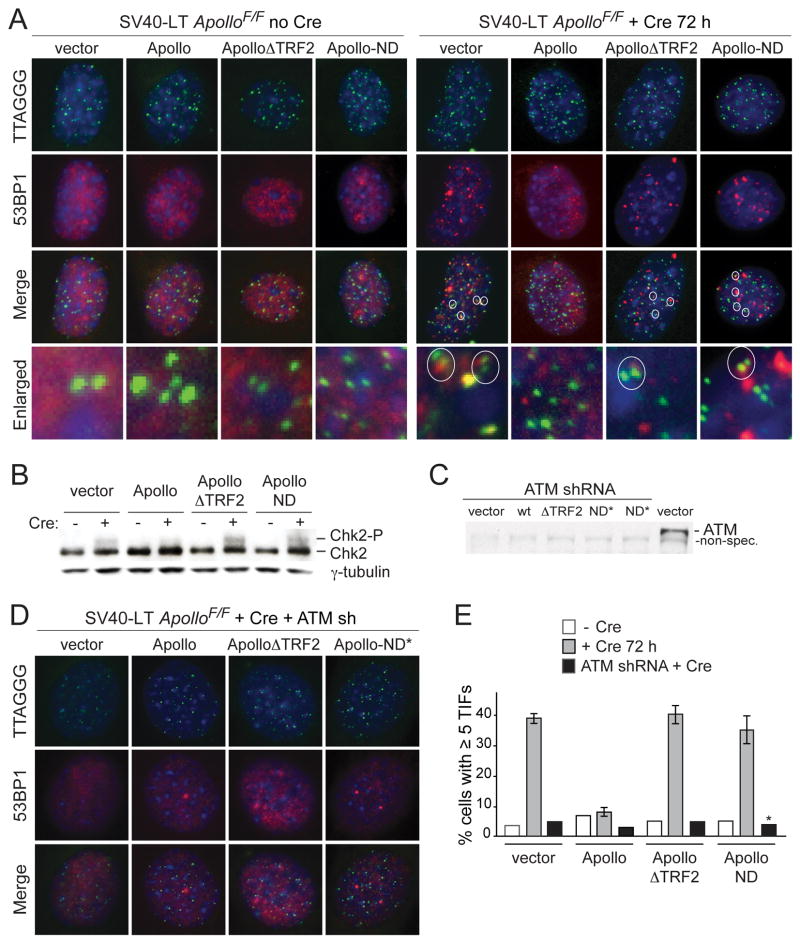
Deletion of Apollo resulted in the induction of a moderate DNA damage response. Approximately one third of the cells showed Telomere dysfunction-Induced Foci (TIFs; (Takai et al., 2003)) at a subset (~10%) of the telomeres (Fig. 2). The TIF response was accompanied by phosphorylation of Chk2, a target of the ATM kinase (Fig. 2B). Consistent with the Chk2 phosphorylation, the TIF response was ablated when cells were treated with an shRNA to the ATM kinase but unaffected by knockdown of ATR (Fig. 2C-E and Fig. S2). Thus, deletion of Apollo elicits ATM kinase signaling at a subset of the telomeres in a fraction of the cells.
Figure 2. Apollo is required to repress telomeric ATM signaling in S phase.
(A) TIF assay on ApolloF/F MEFs expressing the indicated Apollo alleles to detect telomeric DNA damage signaling before (left) and after (right) deletion of the endogenous Apollo with Cre. Telomeres are detected using a FISH probe (green). DNA damage sites are marked with 53BP1 (red). DNA is counterstained with DAPI (blue). Circled TIFs in the enlarged images highlight the prevalence of TIF occurrence at two closely positioned telomeres or in cells with paired telomeres, indicative of DNA damage signaling during or after telomere replication. (B) Immunoblotting for the phosphorylation state of Chk2 at 6 days after Cre treatment. (C) Immunoblot showing depletion of ATM (Mat3-Sigma) 6 days after shRNA treatment and 3 days after the start of puromycin selection in ApolloF/F MEFs expressing the indicated Apollo alleles. Apollo-ND* is identical to Apollo-ND, except without the mutation of H230. (D) Effect of ATM kinase knockdown on the TIF response in Apollo deficient cells. TIF analysis as in (A) but with cells expressing an shRNA to the ATM kinase. (E) Quantification of TIF responses as assayed in (A) and (D). TIFs were scored on the basis of co-localization of 53BP1 foci with 5 or more telomeres per cell. Values for alleles +Cre indicate the mean of 3 independent experiments (> 100 nuclei per experiment), and SDs. * indicates the use of the Apollo-ND* allele in the ATM shRNA experiment. See also Figure S2.
Whereas wild type Apollo effectively repressed TIF formation and Chk2 phosphorylation in the Cre-treated ApolloF/F MEFs, ApolloΔTRF2 was unable to prevent the DNA damage response associated with Apollo loss in mouse and in human cells (Fig. 2A,B,D; Fig. S1D-F). In addition, Apollo-ND failed to prevent activation of ATM signaling at telomeres. Both mutant forms of Apollo induced a level of Chk2 phosphorylation similar to that in the absence of Apollo (Fig. 2B and data not shown). Therefore, repression of ATM signaling at telomeres appears to require an Apollo that is both nuclease-proficient and localizes to telomeres.
ATM activation in the absence of Apollo occurs in early/mid S phase
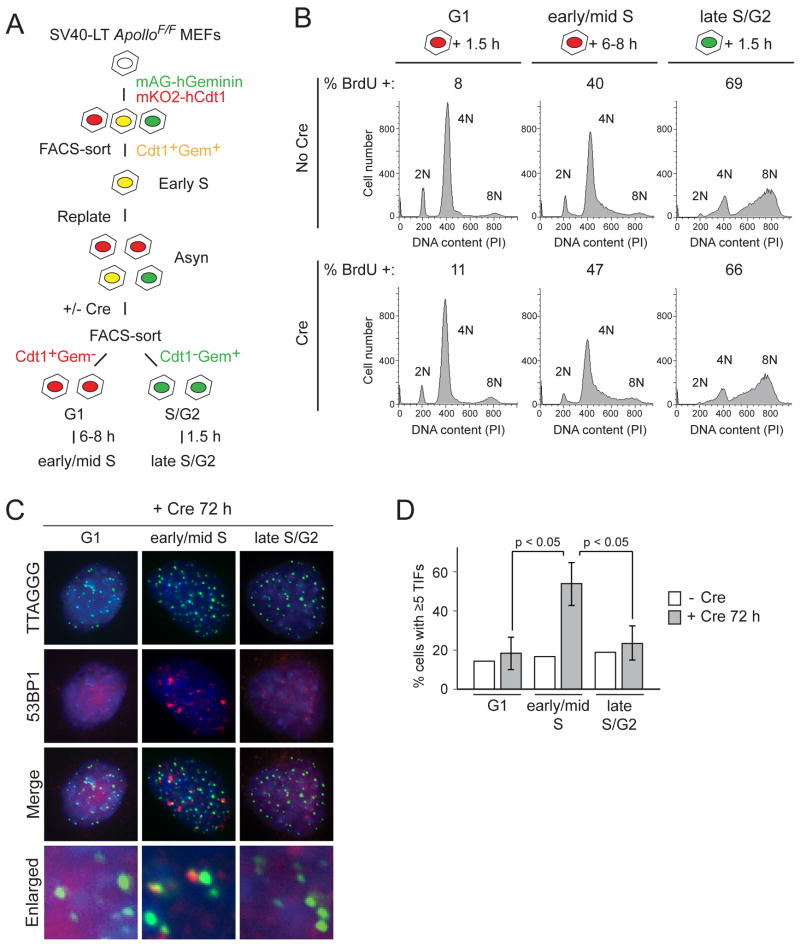
Since the TIF response was observed in approximately one third of the Apollo-deficient cells, we investigated the cell cycle dependence of this DNA damage response. We developed a new method for isolating G1, S, and late S/G2 cells based on the FUCCI (fluorescence ubiquitination-based cell-cycle indicator) system (Sakaue-Sawano et al., 2008) (Fig. 3A, Fig. S3A). FUCCI uses fluorescently-tagged Cdt1 (expressed in G1 and degraded in early S phase) and Geminin (expressed in S/G2 and degraded in mitosis) to mark cells in different phases of the cell cycle. With this approach, G1, S, and S/G2 populations are isolated by FACS sorting, avoiding the disadvantages of synchronizing cells with drug treatments. For the FUCCI-sorting method, we introduced red/orange-fluorescent Cdt1 and green-fluorescent Geminin into immortalized ApolloF/F MEFs and used FACS for both Cdt1 and Geminin to select cells that had incorporated both constructs into their genomes. The cells, which were in early S phase at the time of the FACS-sorting, were re-plated and infected with Hit&Run Cre to delete Apollo. At 72 h after Cre, wild type and Apollo-deficient cells were harvested and sorted again by FACS to isolate Cdt1+Gem− G1 and Cdt1− Gem+ late S/G2 populations (Fig. S3B,C).
Figure 3. Cell cycle dependence of ATM signaling.
(A) Schematic of the FUCCI system to sort cells in G1 and S phase. SV40-LT ApolloF/F MEFs were transduced with mKO2-hCdt1 (red) and mAG-hGeminin (green). Cells were selected by FACS for integration of both plasmids, replated, and infected with Hit&Run Cre. Cells were harvest at the desired time, sorted for Cdt1+Gem− and Cdt1−Gem+ populations, and embedded in agarose for overhang analysis or plated on coverslips for immunofluorescence at desired timepoints. (B) Cell cycle profile and S phase index for different sorted populations at 1.5h or 6–8h after plating. Cells were pulsed for 30 min with BrdU before harvesting and fixing for cell cycle analysis. Cells were stained with FITC-anti-BrdU and propidium iodide (PI) for DNA content, and analyzed by flow cytometry. (C) TIF assay of G1, early/mid S, and late S/G2 ApolloF/F MEFs at 72h post Cre. G1 and late S/G2 cells were obtained by sorting Cdt1+Gem− and Cdt1−Gem+ populations and plating cells for 1.5h prior to fixation. Early/mid S phase cells were obtained by plating Cdt1+Gem− sorted (G1) cells on coverslips for 6–8h prior to fixation. Telomeres are detected using a FISH probe (green). DNA damage sites are marked with 53BP1 (red). DNA is counterstained with DAPI (blue). (D) Quantification of the TIF response in G1, early/mid S, and late S phase as assayed in (C). Values are the mean of three independent experiments (>60 nuclei per experiment) and SDs. p-values were determined using paired student’s t-test. See also Figure S3.
The FACS-sorted cells were plated on coverslips and fixed at different time points after a 30-min BrdU pulse to evaluate their S phase index. The cell cycle profile of the Cdt1+Gem− cells, either immediately after sorting or 1.5 h after plating, showed distinct 2N and 4N peaks with few cells containing intermediate DNA content (Fig. 3B and Fig. S3C). The low percentage of BrdU-positive cells confirmed that most cells were not in S phase. Given the high incidence of tetraploid cells in the asynchronous ApolloF/F MEFs (Fig. S1B), the 4N peak of the Cdt1+Gem− G1 cells likely reflects tetraploid cells in G1 rather than diploid G2 cells. At 6–8 hours after the sorted Cdt1+Gem− G1 cells were plated, the cell cycle profile showed a large percentage of cells progressing into S phase, exhibiting ~40% BrdU incorporation and increased DNA content (Fig. 3B). The cell cycle profiles of Cdt1− Gem+ late S/G2 cells, both upon sorting and at 1.5 h after plating, contained a large number of cells nearing either a 4N or an 8N DNA content, and ~70% of the cells incorporated BrdU (Fig. 3B, Fig. S3C). The different populations had similar cell cycle profiles in the presence and absence of Apollo.
Using the FUCCI-sorting method, we found that the TIFs seen in the absence of Apollo occured in early/mid S phase (Fig. 3C,D). Whereas the percentage of TIF positive G1 or late S/G2 cells did not increase, Apollo-deficient cells in mid-S phase had a strikingly higher TIF response than the controls (Fig. 3D). Thus, the absence of Apollo results in a telomeric DNA damage signal in early/mid S phase. Consistent with this conclusion, the TIFs induced by Apollo deletion were primarily found in cells in which many of the TTAGGG repeat signals were paired (Fig. 2A), suggestive of recent telomere replication.
TRF2-F120A reproduces the ATM signaling phenotype of Apollo loss
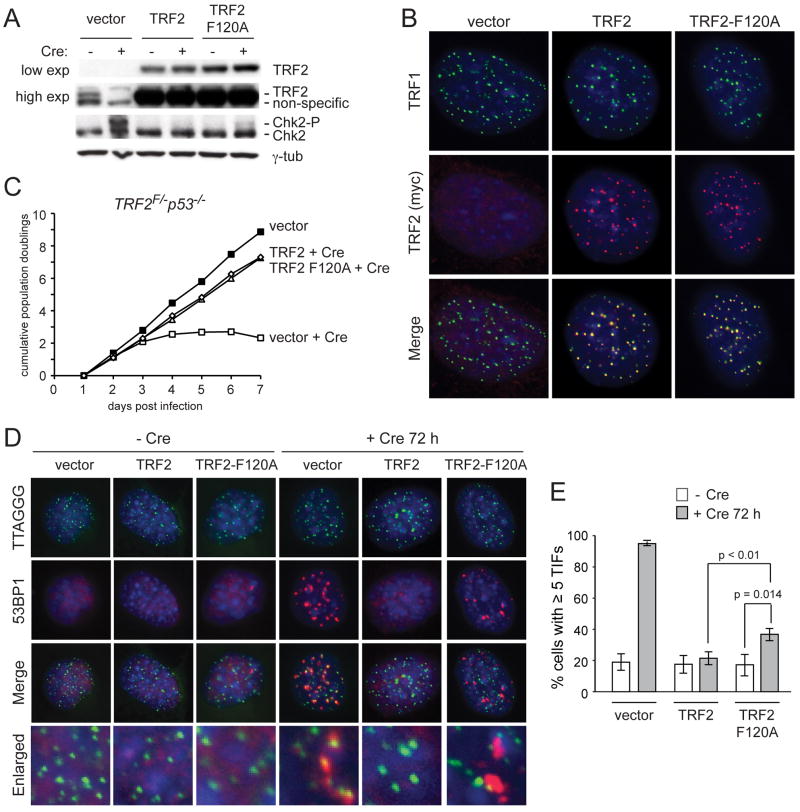
In an alternative approach to assess the role of Apollo at telomeres, we generated an allele of mouse TRF2 with a phenylalanine-to-alanine point mutation at amino acid 120, the conserved residue required for the interaction between human TRF2 and Apollo (Chen et al., 2008). Co-immunoprecipitation of co-transfected mouse Apollo and TRF2-F120A confirmed that the F120A mutation abolished the interaction between the proteins, as previously reported for the human orthologs (Fig. S1C). TRF2-F120A or wild type TRF2 were introduced into TRF2F/− p53−/− MEFs and the endogenous TRF2 was deleted with Cre. TRF2-F120A was overexpressed to the same level as the exogenous wild type TRF2 (Fig. 4A) and co-localized with TRF1 at telomeres (Fig. 4B). While cells lacking TRF2 exhibited a severe defect in proliferation, TRF2-F120A rescued this growth defect to a similar extent as wild type TRF2 (Fig. 4C). This result is consistent with the ability of TRF2-F120A to repress the frequent telomere fusions associated with TRF2 deletion (see below).
Figure 4. TRF2-F120A elicits a DNA damage response similar to Apollo deletion.
(A) Immunoblot for TRF2 and Chk2 in TRF2F/− p53−/− MEFs expressing the indicated TRF2 alleles without Cre and at 144 hours after Hit&Run Cre. (B) IF showing localization of TRF2 alleles in TRF2F/− p53−/− MEFs at 72h after Cre. TRF2 alleles are detected with the myc antibody 9B11. Telomeres are detected with the TRF1 antibody #644. (C) Growth curve showing cumulative population doublings after infection with Cre. Filled squares: vector (no Cre). Open squares: vector +Cre. Open circles: TRF2 +Cre. Open triangles: TRF2-F120A +Cre. (D) TIF assay on TRF2F/− p53−/− MEFs expressing the indicated TRF2 alleles to detect telomeric DNA damage signaling before (left) and after (right) deletion of the endogenous TRF2 with Cre. Telomeres are detected using a FISH probe (green). DNA damage sites are marked with 53BP1 (red). DNA is counterstained with DAPI (blue). (E) Quantification of TIF response as assayed in (D). TIFs were scored on the basis of co-localization of 53BP1 foci with 5 or more telomeres per cell. Values indicate the mean of 3 independent experiments (>100 nuclei per experiment) and SDs. p-values were determined based on paired student’s t-test.
We examined the ability of TRF2-F120A to suppress the ATM-mediated DNA damage response induced in the absence of TRF2. WhereasTRF2 deletion resulted in 80–90% cells with TIFs at most telomeres, expression of the TRF2-F120A mutant reduced the percentage of TIF-positive cells by approximately 2-fold and the remaining TIF-positive cells had fewer TIFs/cell (Fig. 4D,E). Expression of TRF2-F120A also diminished the level of Chk2 phosphorylation elicited by TRF2 deletion (Fig. 4A). Notably, however, the fraction of cells with ≥ 5 TIFs in the presence of TRF2-F120A remained significantly greater than when TRF2-deficient cells were complemented with wild type TRF2 (Fig. 4D,E). As was observed for Apollo deletion, 30–40% of the cells were TIF positive and these cells often contained paired TTAGGG repeat FISH signals (Fig. 4D,E and data not shown). Furthermore, like cells lacking Apollo, the TRF2-F120A cells showed a low level of Chk2 phosphorylation (Fig. 4A). Thus, disrupting the Apollo binding site of TRF2 elicits a DNA damage response that resembles the phenotype of Apollo deletion. Epistasis analysis involving TRF2F/F ApolloF/F MEFs complemented with TRF2-F120A might be able to demonstrate that the phenotypes of TRF2-F120A and Apollo loss are indeed identical.
TRF2-bound Apollo prevents fusion of leading-end telomeres
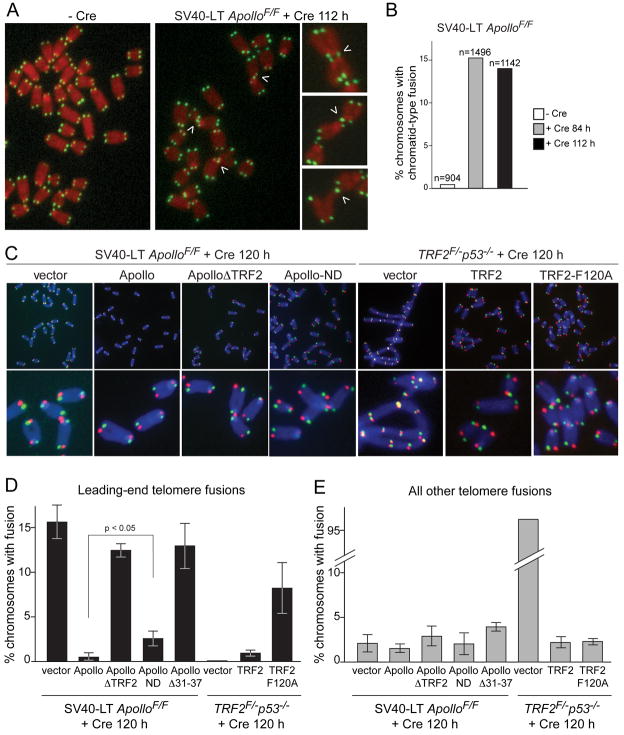
We next determined whether the telomere dysfunction induced by Apollo deletion is associated with aberrant DNA repair at telomeres. Apollo-deficient MEFs showed a distinctive telomere fusion phenotype on metaphase spreads (Fig. 5). Although the telomere fusions were 5–10 times less frequent compared to when TRF2 is deleted, the fusion phenotype of Apollo-deficient cells was highly significant. Strikingly, the telomere fusions observed between 84–120 hours after introduction of Cre were nearly all of the chromatid-type, indicating a post-replicative fusion event (Fig. 5A). Later time points included chromosome-type fusions that were most likely due to duplication of chromatid-type fusions after their segregation into daughter cells (data not shown). These secondary chromosome-type fusions were not likely to reflect the function of Apollo, and their incidence was affected by the proliferation rate of the cells. Therefore, later time-points after Apollo deletion were not analyzed.
Figure 5. TRF2-bound Apollo prevents leading-end telomere fusions.
(A) Telomere fusions in metaphase spreads from Apollo-deficient cells. Metaphase spreads were obtained from ApolloF/F MEFs before or after introduction of Cre and processed for telomeric FISH (FITC, green). DNA was stained with DAPI (false-colored in red). Arrowheads highlight chromatid-type fusion events. (B) Quantification of chromatid-type fusion events after deletion of Apollo. (C) CO-FISH analysis of leading and lagging end telomeres. Metaphases harvested from ApolloF/F MEFs or TRF2F/− p53−/− MEFs expressing the indicated rescuing alleles were incubated with BrdU/BrdC and treated with ExoIII and UV to remove the newly-synthesized DNA strand. The undigested parental telomeric DNA strands were detected with TAMRA-(TTAGGG)3 (red) and FITC-(CCCTAA)3 (green). DNA is stained with DAPI (blue). The telomere replicated by leading-strand DNA synthesis is highlighted in red, and the telomere replicated by lagging-strand DNA synthesis is highlighted in green. (D) Quantification of leading-end telomere fusions from metaphase analyses shown in (C) at 120 hours post Cre. Values represent means of three or more experiments (chromosome number >1000 per experiment) and SDs. (E) Quantification of other telomere fusions events (lagging-to-lagging, lagging-to-leading chromatid-type fusions, and chromosome-type fusions). Values represent means of three or more experiments (>1000 chromosomes per experiment) and SDs. See also Figure S4.
Leading-strand and lagging-strand DNA synthesis generates two distinct types of telomeres that could be vulnerable to post-replicative fusions. To determine whether Apollo was important for the protection of both types of telomeres, we used Chromosome Orientation Fluorescent in situ Hybridization (CO-FISH; (Bailey et al., 2001)) to distinguish between telomere ends generated by leading- and lagging-strand DNA synthesis (referred to as leading-end and lagging-end telomeres). CO-FISH analysis of the Apollo null cells showed that the fusions exclusively involved telomeres generated by leading-strand DNA synthesis (shown in red) (Fig. 5C-E). Thus, Apollo has a specific role in protecting leading-end telomeres, resulting in chromatid-type fusions when Apollo is absent. The absence of sister fusions suggests that the lagging-end telomeres remain protected in Apollo deficient cells.
The function of Apollo in protecting leading-end telomeres required its localization at telomeres. The ApolloΔ TRF2 mutant was unable to prevent leading-end telomere fusions (Fig. 5C-E). Similarly, expression of TRF2-F120A resulted in a significant level of leading-end telomere fusions observed by CO-FISH while this mutant was fully capable of repressing the chromosome-type fusions associated with TRF2 deletion (Fig. 5C-E).
In addition, the nuclease activity of Apollo was involved in protecting the leading-end telomeres. Cells expressing Apollo-ND instead of wild type Apollo generated a statistically significant level of leading-end telomere fusions after deletion of endogenous Apollo (Fig. 5D). A more severe phenotype was observed with a second nuclease domain mutant (ApolloΔ31–37, removing the HxHxDH motif in the metallo β-lactamase domain and also containing the H230A mutation), which was expressed at similar levels as wildtype Apollo and retained its interaction with TRF2 (Fig. S4 and data not shown) but was incapable of repressing leading-end telomere fusions (Fig. 5D). The difference between the two nuclease mutants is discussed below.
Apollo contributes to maintenance of the telomeric overhang
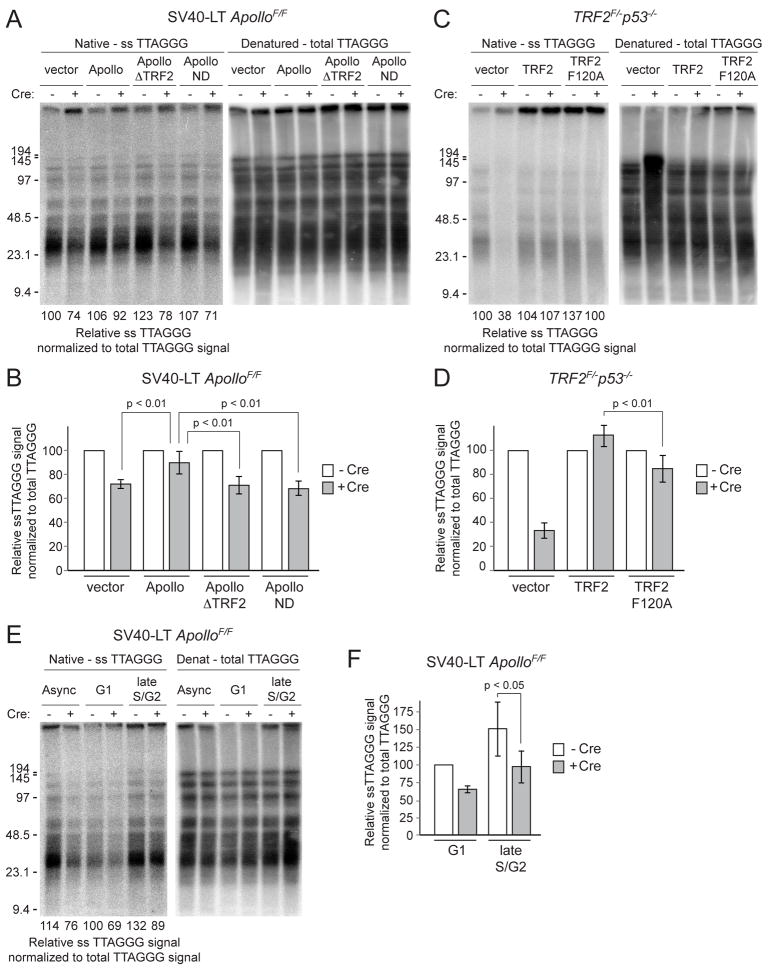
To determine whether the aberrant DNA damage response at leading-end telomeres was due to a change in the terminal telomeric structure, we assessed the telomeric overhang in the absence of Apollo. At five days after Cre-mediated deletion of Apollo, we observed a 30–40% reduction in the relative telomeric overhang signal (Fig. 6A,B). Assuming equal overhang lengths at lagging- and leading-end telomeres, complete loss of the overhang from leading-end telomeres would be expected to result in a 50% drop. Therefore, we consider the observed 30–40% reduction in overhang signal an indication of a considerable defect in overhang maintenance. The overhang phenotype was rescued by full-length wild type Apollo but not by ApolloΔTRF2 or Apollo-ND (Fig. 6A,B). Similarly, when the TRF2-F120A mutant was expressed in TRF2F/− p53−/− MEFs, the relative telomeric overhang signal was approximately 30% less than in the presence of wild type TRF2 (Fig. 6C,D). Since TRF2-F120A represses most of the telomere fusions resulting from TRF2 deletion (Fig. 5E), this overhang loss cannot be ascribed to processing by NHEJ. More likely, the diminished overhang signal in the TRF2-F120A setting is due to the lack of recruitment of Apollo to telomeres.
Figure 6. TRF2-bound Apollo maintains the 3′ telomeric overhang.
(A) Representative telomeric overhang analysis of ApolloF/F MEFs expressing the indicated rescuing alleles without Cre and at 120 h after pWzl-Cre infection. The single-stranded telomeric signal was determined by in-gel hybridization (left) of an end-labeled 32P-(AACCCT)4 telomeric oligonucleotide to native MboI-digested genomic DNA. After capture of the signal, the DNA was denatured in situ and the gel was rehybridized with the same probe to determine the total telomeric DNA signal (right). The single-stranded telomeric signal between ~9–100kb in each lane was normalized to the total telomeric DNA signal in the same region of that lane. The relative single-stranded signal was then determined with the lane containing vector only (no Cre) set to 100. (B) Quantification of relative single-stranded telomeric overhang signal with ApolloF/F MEFs. Values represent means for five independent experiments with SDs. For each rescuing allele (or cells infected with the empty vector) the normalized value was set at 100 for cells not treated with Cre and the post-Cre values are given as a percentage of this value. p-values were determined using paired student t-test. (C) Representative telomeric overhang analysis of TRF2F/− p53−/− MEFs expressing the indicated alleles without Cre and at 120h post Cre-infection, assayed as in (A). (D) Quantification of relative single-stranded telomeric signal with TRF2F/− p53−/− MEFs. Values represent means for three independent experiments with SDs. For each rescuing allele (or cells infected with the empty vector) the normalized value was set at 100 for cells not treated with Cre and the post-Cre values are given as a percentage of this value. p-values were determined using paired student t-test. (E) Representative telomeric overhang analysis of G1 and late S/G2 ApolloF/F MEFs without Cre and at 120 h after Cre. FUCCI-sorted cells were immediately embedded in agarose plugs for overhang analysis as in (A). The relative single-stranded signal was normalized to total TTAGGG signal and determined as a percentage of the signal in the lane containing G1 cells without Cre (set at 100). (F) Quantification of relative single-stranded telomeric signal in G1 and late S/G2 as assayed in (E). The single-stranded telomeric signal was normalized to total TTAGGG signal and determined as a percentage of the signal in G1 cells without Cre (set at 100). Values are the mean of three independent experiments and SDs. p-values were determined by paired student’s t-test. See also Figure S5.
In the presence of telomerase, the overhangs can be elongated transiently due to the uncoupling of telomerase action, which occurs throughout S phase, and C-strand fill-in, which is delayed until late S/G2 (Zhao et al., 2008). To exclude that the change in overhang signal was due to an effect of Apollo on telomerase, we generated TRF2F/F mTR−/− MEFs in order to assess the phenotype of the TRF2-F120A mutant in a telomerase-deficient setting. Cre treatment of SV40-LT immortalized TRF2F/F mTR−/− MEFs expressing TRF2-F120A again resulted in ~30% reduction in overhang signal (Fig. S5A-D). Furthermore, the cells showed the same induction of leading-end telomere fusions observed when TRF2-F120A replaced the endogenous TRF2 in telomerase-positive cells (Fig. S5E,F). This result indicates Apollo regulates the G-overhang in a manner that is independent of telomerase.
We next analyzed the overhang signal in Apollo null cells at different phases of the cell cycle using the FUCCI-sorting system described above. Both wild type and Apollo-deficient cells experienced a transient increase in overhang signal in late S phase. This Apollo-independent increase in overhang signal during progression from G1/early S to late S phase could arise from extension of the G-rich overhang by telomerase and/or resection of the C-rich strand by other nucleases. However, compared to wild type cells, Apollo-deficient cells showed a 40% reduction in telomeric overhang signal regardless of the cell cycle phase (Fig. 6E,F, Fig. S5G).
DISCUSSION
These data reveal that TRF2-bound Apollo functions at replicating telomeres, promoting the maintenance of the telomeric overhang, repressing S phase specific ATM signaling, and protecting leading-end telomeres from fusion. Based on these results, we propose a model in which TRF2 recruits the Apollo nuclease to process leading-end telomeres immediately after their replication. If this processing does not occur or is delayed, the leading-end telomeres would remain blunt, making them vulnerable to end-joining reactions and exposing the telomere end in a manner that activates the MRN/ATM pathway. These findings establish that, as was generally assumed, maintenance of the telomeric overhang is important for the protection of mammalian telomeres.
The action of Apollo at telomeres requires its association with TRF2, as an allele of Apollo lacking the TRF2 binding site is non-functional, and cells expressing a TRF2 allele that does not bind Apollo have the same telomere phenotypes as Apollo-deficient cells. We also conclude that the nuclease activity of Apollo is required for its function, since the Apollo null phenotypes are not rescued by Apollo-ND, in which essential residues of the nuclease domain have been mutated. Although Apollo-ND appears to be a null allele with regard to overhang maintenance and repression of ATM signaling, its ability to repress the leading-end telomere fusions, while weaker than for the wild type Apollo, is not nil. In contrast, a second nuclease-deficient allele of Apollo, lacking the HxHxDH motif in the metallo β-lactamase domain, completely fails to protect leading-end telomeres from fusions. One explanation for this discrepancy is that the Apollo-ND allele has residual nuclease activity that is sufficient to protect leading-end telomeres during the short time period in S/G2 when they are vulnerable to fusion. This residual nuclease activity would have to be very minor because there is no overt difference in the overhang signal compared to Apollo deletion. Another possibility is that the Apollo protein itself protects the leading-end telomeres from fusion. Such protection could conceivably involve the nuclease domain in a manner that is destroyed by deletion of amino acids 31–37 but preserved in Apollo-ND despite the point mutations. It will therefore be of interest to study the nuclease and end-binding activities of TRF2-Apollo complexes in vitro.
As Apollo has been implicated in ICL repair (Demuth et al., 2004; Bae et al., 2008), the appearance of S-phase TIFs in the absence of Apollo could alternatively be explained by an inability to repair lesions encountered during telomere replication. We do not favor this explanation because the major phenotypes observed in the absence of Apollo are not associated with aphidicolin-induced replication stress or deletion of the shelterin component TRF1. Replication fork stalling prior to collapse is often associated with accumulation of single-stranded DNA and activation of the ATR kinase, which we do not observe in Apollo null cells. Furthermore, deletion of Apollo in mouse cells does not induce the fragile telomere phenotype associated with telomere replication problems (Sfeir et al., 2009). Although we have not formally excluded contributions of telomeric Apollo in repairing lesions encountered by the replication machinery, the major phenotypes observed at telomeres lacking Apollo are inconsistent with the prevention of replication stress being a primary function of Apollo at telomeres.
The role of Apollo at replicating telomeres is distinct from previously characterized functions of the core components of shelterin. The predominance of leading-end telomere fusions in the Apollo knockout cells contrasts with both the TRF2 and POT1a/b knockout phenotypes. Although TRF2 deletion induces occasional chromatid-type fusions, most of the fusions in TRF2 null cells occur in G1 and manifest as chromosome-type fusions in the subsequent metaphase (Celli and de Lange, 2005; Konishi and de Lange, 2008). POT1a/b deletion results in sporadic chromosome-type fusions and post-replicative fusions involving sister telomeres (Hockemeyer et al., 2006). Neither chromosome-type fusions nor sister fusions are observed in Apollo null cells. Thus, whereas TRF2 and POT1a/b function at both leading- and lagging-end telomeres, Apollo acts specifically in the protection of leading-end telomeres.
Our results suggest that the protective role of Apollo is limited to S phase. In the absence of Apollo, telomeres experience a transient DNA damage response mainly in S phase despite a persistent overhang defect throughout the cell cycle. Furthermore, we do not observe the chromosome-type fusions that would result if leading-end telomeres continued to be vulnerable to fusion in daughter cells. How the protected state is restored as cells progress through mitosis remains unknown. One possibility is that intrinsic properties of telomeres allow protection of unprocessed ends in G1 but not in S phase. For instance, leading-end telomeres with short overhangs might still form t-loops at a slower rate. Another possibility is that additional Apollo-independent processing in late S/G2 could be sufficient to restore end protection. Indeed, the overhang signal transiently increases in late S/G2 in both wild type and Apollo-deficient cells. This is presumably due to 5′ resection by other nucleases or elongation of the overhang by telomerase, which is uncoupled from fill-in synthesis of the C-rich strand (Zhao et al., 2009). Finally, it is possible that the milieu of G1 and S phase cells exposes telomeres to different threats, requiring distinct protective measures to ensure telomere protection in these different stages of the cell cycle.
We previously proposed that the F120 site in TRF2 not only provides a docking site for Apollo but also recruits additional shelterin accessory factors, such as Nbs1, XPF, PARP1, ATM, and ATR (Chen et al., 2008; Palm and de Lange, 2008). There would be no competition for this docking site because TRF2 is very abundant at telomeres, whereas most shelterin accessory factors are not (Takai et al., 2010). The versatility of the F120 docking site was supported by the identification of two proteins, PNUTS and MCPH1, which can bind to TRF2 using the YxLxP motif (Kim et al., 2009). However, our current data now cast doubt on the importance of the F120 docking site beyond the interaction with Apollo. The phenotype of the TRF2-F120A mutant is mild, showing the limited DNA damage response phenotype and telomere fusions associated with Apollo loss but no additional telomere dysfunction. Although it is possible that the other F120 interacting factors are dispensable for telomere protection (for instance, due to redundancy), our data warrant further testing of the concept of the common F120 docking site.
The main conclusion from the observations on Apollo-deficient telomeres concerns the role of the telomeric overhang in telomere protection. The data are consistent with the long-held (but previously unsubstantiated) view that the overhang has a protective role. In particular, leading-end telomeres appear to become resistant to telomere fusions and can avoid activating the ATM kinase pathway if an overhang is formed rapidly. We imagine that the overhang would help in forming the t-loop structure which is expected to block binding of the MRN DSB sensor in the ATM pathway and prevent loading of the NHEJ factor Ku70/80. In addition, the loading of POT1 proteins on the overhang may be crucial to prevent telomere fusion immediately after telomere replication. Prior data indicated that POT1a/b protects leading-end telomeres (as well as lagging-end telomeres) from fusion. The current data argue that the generation of the appropriate POT1a/b binding sites at leading-end telomeres is mediated by Apollo.
The findings on Apollo are relevant to the Hoyeraal-Hreidarsson syndrome, a severe variant of the telomere dysfunction disease dyskeratosis congenita (DC). DC is a bone marrow failure syndrome that can be caused by mutations in telomerase components and the shelterin component TIN2, but shelterin accessory factors had not been implicated in the disease (Mason et al., 2005; Savage et al., 2008; Walne et al., 2008). A recent study identified a single Hoyeraal-Hreidarsson syndrome patient expressing a dominant negative allele of Apollo that lacks the TRF2 binding domain and induces an ATM-dependent telomere damage signal (Touzot et al., 2010). However, the Apollo locus appeared to lack a disease-causing mutation. Our findings that Apollo deficiency is specifically associated with defective overhang maintenance and leading-end telomere fusions may facilitate the identification of additional patients with Hoyeraal-Hreidarsson syndrome or other forms of DC that are caused by defects in Apollo.
EXPERIMENTAL PROCEDURES
Apollo gene targeting
The targeting vector for modification of the mouse Apollo locus was generated in pSL301 by standard cloning techniques using Bac-derived DNA fragments extending from the NheI site upstream of the Apollo gene to the SacI site in the 3′ UTR. A TK-neomycin cassette flanked by FRT sites and carrying one loxP site was inserted into the PacI site in the third intron of the Apollo gene. A second loxP site was introduced into the NseI site in intron 1 by insertion of a loxP oligonucleotide that also contained an NsiI site used in genomic analysis of targeted ES cells. ES clones with the correct integration were identified by genomic blotting of NsiI-digested DNA using a probe upstream of the NheI site and the presence of a single neo integration was confirmed. Targeted ES cells were used to generate chimeras and offspring with the targeted genotype. The neo cassette was removed using the FLPe deleter mouse strain (Jackson labs). The resulting ApolloF/+ genotype was maintained on a mixed background (129/C57Bl/6J).
Cell lines, plasmids, and shRNAs
ApolloF/F MEFs were isolated from E13.5 embryos from ApolloF/+ intercrosses and immortalized at passage 2 with pBabeSV40-LT (a gift from Greg Hannon). Cells were maintained thereafter in DMEM/10–15%FBS supplemented with non-essential amino acids, glutamine, and penicillin/streptomycin. Cre recombinase was introduced into immortalized MEFs using 2–4 rounds of infection (12 hour intervals) with Hit&Run-Cre or pWZL-Cre as described previously (Celli and de Lange, 2005). For analysis of different time points after infection, t=0 refers to 12 hours after the first infection with Cre retrovirus.
Apollo alleles with an N-terminal FLAG-[HA]2 tag were generated by PCR-mediated mutagenesis and expressed using the pLPC puromycin-selectable retroviral vector. Phoenix packaging cells were transfected with the plasmids, and the retroviral supernatant was used for two infections of SV40-LT ApolloF/F cells (12 hour intervals). Cells were selected for 3 days prior to Cre infection.
TRF2F/− p53−/− MEFs were previously described (Celli and de Lange, 2005). TRF2F/F mTR−/− MEFs were generated from TRF2F/+ mTR+/− intercrosses and immortalized with pBabeSV40-LT at passage 2. N-terminal myc-tagged TRF2-F120A was generated by site directed mutagenesis and cloned in a pLPC puromycin-selectable retroviral expression vector. TRF2 alleles were introduced into TRF2F/− p53−/− or SV40-LT TRF2F/F mTR−/− MEFs using two infections at 12 hr intervals, followed by 3 days of selection prior to Cre infection.
shRNAs for ATR kinase (shATR3-1; (Lazzerini Denchi and de Lange, 2007)) and ATM kinase (ggaagtcaaggaacaacaacta) were introduced in four infections at 12 hr intervals using the pSuperior puromycin retroviral vector.
PCR genotyping and RT-PCR transcript analysis
PCR with the following primers was used to monitor Apollo deletion on isolated genomic DNA before and after introduction of Cre: F: ACATCTCCTCATCTT GTCTG, R1: CCTATCATGATAATCCCAGC, R2: CTTGAGGGTTTCTTTTGGAG. RT-PCR was performed with the oligo-dT ThermoScript RT-PCR system (Invitrogen). RNA was isolated from approximately 106 cells with the Qiagen RNAeasy kit. Two to three micrograms RNA was reverse transcribed with the ThermoScript RT-PCR system (Invitrogen) by using oligo dT priming and the protocol provided by the manufacturer. The primers used for PCR after cDNA synthesis are as follows: Apollo RT1 (forward CACGGTGGGTTTGTCTAGC, reverse GTTGCTCCAGCAGTGATTC), Apollo RT2 (forward CTCCCATCA CTGCTTGCCTC, reverse GCAACTGTACCAACTCCAGG), GAPDH (forward TGAAGGTCGGTGTGAACGGATTTGGC, reverse CATGTAGGCCATGAGGT CCACCAC), Ap4b1 RT1 (forward GACGATGCCATACCTTGGCTC, reverse GTTCAGTACTTCAGCCTG), and Ap4b1 RT2 (forward GACGATGCCATACC TTGGCTC, reverse CTGCTCTTGAGATAGCTGTC).
Immunofluorescence, immunoblotting, and immunoprecipitation
For IF/IF-FISH, immunoblotting, and co-IP, previously described methods were used ((Celli and de Lange, 2005; Dimitrova et al., 2008; Chen et al., 2008)) with the following antibodies: HA.11, Covance; myc, 9B11 (IB, IF) and 9E10, Sigma (IP); FLAG, M2; α-tubulin, Sigma; γ-tubulin, GTU88; Chk2, BD Transduction; TRF1 (mouse), #644; TRF2, #647; 53BP1, 100–304, Novus; ATM, Mat3, Sigma; ATR, FRP goat, Santa Cruz N-19. For IF detection of Apollo, cells were extracted for 90 s with Triton X-100 buffer (0.5% Triton X-100, 20 mM Hepes-KOH pH 7.9, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose) prior to fixation with 3% paraformaldehyde/2% sucrose.
Analysis of telomeric DNA by in-gel hybridization, FISH, CO-FISH
Telomeric overhang signals and telomeric restriction fragment patterns were determined by in-gel hybridization with an end-labeled 32P-(AACCCT)4 telomeric oligonucleotide as previously described (Celli and de Lange, 2005). CO-FISH and FISH for telomeric DNA was performed with C- and G-strand PNA probes on methanol/acetic acid-fixed metaphase spreads as previously described (Celli et al., 2006).
FACS
Cell cycle analyses were performed using standard techniques to evaluate BrdU incorporation and propidium iodide (PI) staining of DNA content. Cells were pulsed with 10 μM BrdU for 30 minutes, then fixed and stained with FITC-conjugated anti-BrdU antibody (BD Biosciences) and PI. Flow cytometry was performed on FACSCalibur-1 (Becton Dickinson), and data were analyzed using FlowJo 8.7.1 software.
For the FUCCI sort experiments, SV40LT-immortalized ApolloF/F MEFs were transduced with three infections of mKO2-Cdt1 30/120 (lentiviral) followed by three infections of mAG-Geminin 1/110 (lentiviral) at 6 hour intervals (gift from A. Miyawaki, (Sakaue-Sawano et al., 2008)). Cdt1+Gem+ cells were collected by FACS, replated, and infected with two rounds of Hit&Run Cre. Sorting of G1 and S phase cells according to levels of Cdt1 and Geminin was performed on BD FACSAria-1 and Aria-2 cell sorters (BD Biosciences) with excitation by the 488 nm and 561 nm lasers. Cells were collected in PBS and immediately plated on coverslips or embedded in agarose for DNA analysis.
Supplementary Material
Acknowledgments
The authors thank Devon White for expert mouse husbandry and Shaheen Kabir for the TRF2-F120A allele. The Rockefeller University Gene Targeting Facility and Svetlana Mazel, Cris Bare, Aliza Lloyd, and Lily Li of the RU Flow Cytometry Resource Center are thanked for technical assistance. We also thank members of the de Lange lab for helpful discussions and comments on the manuscript. P.W. was supported by the NIA/NIH Ruth L. Kirschstein NRSA Individual Fellowship F30AG034744 and NIH MSTP grant GM07739 to the Weill Cornell/RU/MSKCC Tri-Institutional MD-PhD Program. This work was supported by NIH grants CA76027 and GM49046.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attwooll CL, Akpinar M, Petrini JH. The mre11 complex and the response to dysfunctional telomeres. Mol Cell Biol. 2009;29:5540–5551. doi: 10.1128/MCB.00479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JB, Mukhopadhyay SS, Liu L, Zhang N, Tan J, Akhter S, Liu X, Shen X, Li L, Legerski RJ. Snm1B/Apollo mediates replication fork collapse and S Phase checkpoint activation in response to DNA interstrand cross-links. Oncogene. 2008;27:5045–5056. doi: 10.1038/onc.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SM, Cornforth MN, Kurimasa A, Chen DJ, Goodwin EH. Strand-specific Postreplicative Processing of Mammalian Telomeres. Science. 2001;293:2462–2465. doi: 10.1126/science.1062560. [DOI] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009 doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Moshous D, Mornon JP, de Villartay JP. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002;30:3592–3601. doi: 10.1093/nar/gkf470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli G, de Lange T. DNA processing not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Celli GB, Lazzerini Denchi E, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang Y, van Overbeek M, Donigian JR, Baciu P, de Lange T, Lei M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science. 2008;319:1092–1096. doi: 10.1126/science.1151804. [DOI] [PubMed] [Google Scholar]
- de Lange T. How telomeres solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villartay JP. V(D)J recombination deficiencies. Adv Exp Med Biol. 2009;650:46–58. doi: 10.1007/978-1-4419-0296-2_4. [DOI] [PubMed] [Google Scholar]
- de Villartay JP, Shimazaki N, Charbonnier JB, Fischer A, Mornon JP, Lieber MR, Callebaut I. A histidine in the beta-CASP domain of Artemis is critical for its full in vitro and in vivo functions. DNA Repair (Amst) 2009;8:202–208. doi: 10.1016/j.dnarep.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Demuth I, Digweed M, Concannon P. Human SNM1B is required for normal cellular response to both DNA interstrand crosslink-inducing agents and ionizing radiation. Oncogene. 2004;23:8611–8618. doi: 10.1038/sj.onc.1207895. [DOI] [PubMed] [Google Scholar]
- Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T. Cell cycle dependent role of MRN at dysfunctional telomeres: ATM signaling-dependent induction of NHEJ in G1 and resection-mediated inhibition of NHEJ in G2. Mol Cell Biol. 2009;29:5552–5563. doi: 10.1128/MCB.00476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- Dronkert ML, de Wit J, Boeve M, Vasconcelos ML, van Steeg H, Tan TL, Hoeijmakers JH, Kanaar R. Disruption of mouse SNM1 causes increased sensitivity to the DNA interstrand cross-linking agent mitomycin C. Mol Cell Biol. 2000;20:4553–4561. doi: 10.1128/mcb.20.13.4553-4561.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Counter CM. The protein hSnm1B is stabilized when bound to the telomere-binding protein TRF2. J Biol Chem. 2008;283:23671–23676. doi: 10.1074/jbc.M800388200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell. 1987;51:887–898. doi: 10.1016/0092-8674(87)90576-9. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Hemann MT, Greider CW. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 1999;27:3964–3969. doi: 10.1093/nar/27.20.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- Huffman KE, Levene SD, Tesmer VM, Shay JW, Wright WE. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J Biol Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- Kim H, Lee OH, Xin H, Chen LY, Qin J, Chae HK, Lin SY, Safari A, Liu D, Songyang Z. TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat Struct Mol Biol. 2009;16:372–379. doi: 10.1038/nsmb.1575. [DOI] [PubMed] [Google Scholar]
- Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosawa A, Koyama H, Takayama S, Miki K, Ayusawa D, Fujii M, Iiizumi S, Adachi N. The requirement of Artemis in double-strand break repair depends on the type of DNA damage. DNA Cell Biol. 2008;27:55–61. doi: 10.1089/dna.2007.0649. [DOI] [PubMed] [Google Scholar]
- Lazzerini Denchi E, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- Mason PJ, Wilson DB, Bessler M. Dyskeratosis congenita -- a disease of dysfunctional telomere maintenance. Curr Mol Med. 2005;5:159–170. doi: 10.2174/1566524053586581. [DOI] [PubMed] [Google Scholar]
- McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, Hockemeyer D, Kibe T, de Lange T. Functional dissection of human and mouse POT1 proteins. Mol Cell Biol. 2009;29:471–482. doi: 10.1128/MCB.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Ann Rev Genetics. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pannicke U, Ma Y, Hopfner KP, Niewolik D, Lieber MR, Schwarz K. Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J. 2004;23:1987–1997. doi: 10.1038/sj.emboj.7600206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Savage SA, Giri N, Baerlocher GM, Orr N, Lansdorp PM, Alter BP. TINF2, a component of the shelterin telomere protection complex, is mutated in dyskeratosis congenita. Am J Hum Genet. 2008;82:501–509. doi: 10.1016/j.ajhg.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010;285:1457–1467. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzot F, Callebaut I, Soulier J, Gaillard L, Azerrad C, Durandy A, Fischer A, de Villartay JP, Revy P. Function of Apollo (SNM1B) at telomere highlighted by a splice variant identified in a patient with Hoyeraal-Hreidarsson syndrome. Proc Natl Acad Sci U S A. 2010;107:10097–10102. doi: 10.1073/pnas.0914918107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M, de Lange T. Apollo, an Artemis-related nuclease, interacts with TRF2 and protects human telomeres in S phase. Curr Biol. 2006;16:1295–1302. doi: 10.1016/j.cub.2006.05.022. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Walne AJ, Vulliamy T, Beswick R, Kirwan M, Dokal I. TINF2 mutations result in very short telomeres: analysis of a large cohort of patients with dyskeratosis congenita and related bone marrow failure syndromes. Blood. 2008;112:3594–3600. doi: 10.1182/blood-2008-05-153445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Deng Y, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hoshiyama H, Shay JW, Wright WE. Quantitative telomeric overhang determination using a double-strand specific nuclease. Nucleic Acids Res. 2008;36:e14. doi: 10.1093/nar/gkm1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.