Abstract
The exact mechanism by which cellular RNA polymerases translocate and maintain exceptionally high fidelity during transcription remains a major unresolved issue. Two recent structural studies of yeast RNA polymerase II in complex with its potent inhibitor, the fungal toxin α-amanitin, address this matter by describing critical and surprising details about the enzyme catalytic center dynamic organization.
Eukaryotic protein-coding genes are transcribed by a dedicated class of DNA-dependent RNA polymerases (RNAP), so called RNA polymerase II (pol II). This enzyme is exceptionally processive and faithful, synthesizing RNA molecules hundreds of thousand nucleotides long without a single mistake. Although many details of transcription mechanisms have already been revealed by structural, biochemical, and biophysical studies of pol II and related bacteria enzymes1, some fundamental issues, such as the RNAP driving forces and fidelity, remain to be elucidated. RNA synthesis occurs by simple two-metal catalysis facilitated by an absolutely conserved triad of acidic amino acids grouped together within a short peptide loop in the catalytic center of RNAP. Yet, remarkably, RNAP is comprised of several - in case of yeast pol II, twelve - subunits, thousands of amino acids and exhibits profound allosteric effects propagated over large distances. Two recent papers from the Kornberg and Cramer laboratories2,3 provide, in many aspects, complementary, insights into the structural and mechanochemical bases of pol II function and the mechanism of its inhibition by the powerful fungal toxin, α-amanitin.
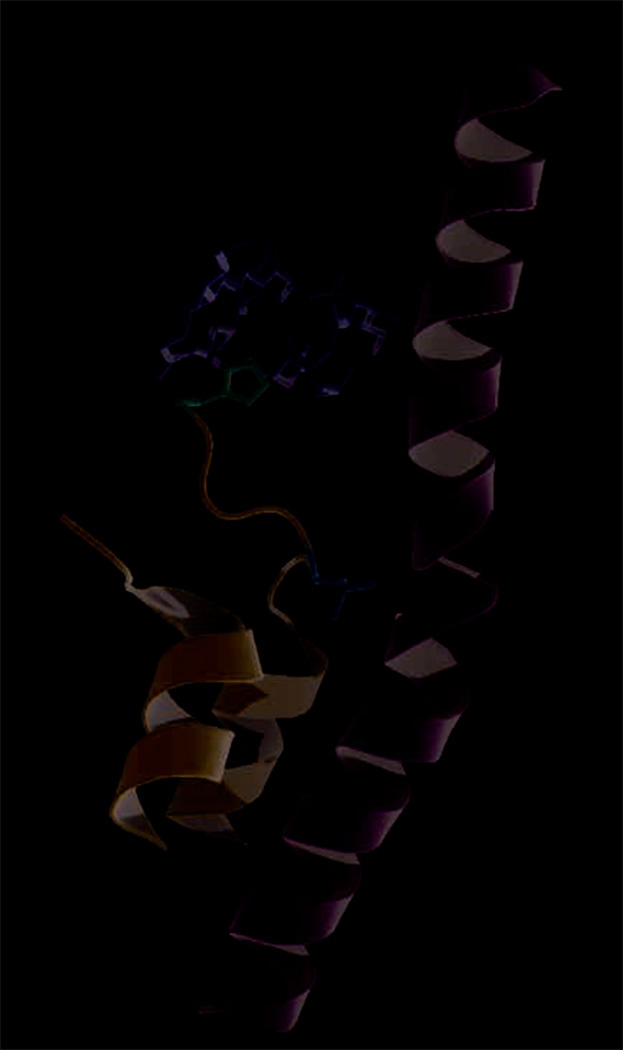
Kornberg and colleagues refined a previously published structure of the yeast pol II apo-enzyme co-crystallized with α-amanitin4 to reveal additional contacts between the enzyme and this inhibitor, most notably, specific complimentary hydrogen (H-) bonds with His1085 of Rpb1 (pol II largest subunit). His1085 is located within the so-called Trigger Loop (TL), a mobile structural element conserved in both eukaryotic and bacterial RNAPs (Fig.1). The paramount importance of TL in transcription elongation, control and fidelity was recently highlighted in a series of genetic, biochemical, and structural studies5–8. Substrate-dependent refolding of TL has been proposed to play a key role in substrate selection and catalysis9. TL was not resolved in the previously solved pol II co-crystal, but in the refined structure it appeared to adopt an ordered conformation, possibly stabilized by interaction with the α-amanitin. Structural analysis8 and molecular simulations10 show that His1085 (and its bacterial counterpart9) forms H-bonds with substrate (NTP) phosphate groups, facilitating formation of the phosphodiester bond and/or stabilizing a pyrophosphate group, a leaving group in the nucleotide addition cycle (NAC). Thus α-amanitin not only impedes the mechano-chemical changes accompanying NAC by binding in the space traversed by the mobile TL, but may also interfere with the basic chemistry of the nucleotide addition (Fig.2). Kornberg and colleagues further demonstrated that yeast pol II with Tyr in place of His1085 (H1085Y) was resistant to α-amanitin inhibition both in vivo (albeit it caused severe growth defects) and in vitro. It appeared that Tyr1085-dependent disruption of the network of imidazole-specific H-bonds formed by α-amanitin also caused other major impediments of pol II action, such as elongation, substrate selection (both base- and sugar-specific), and TFIIS-dependent RNA cleavage were substantially impaired in the H1085Y-substituted enzyme.
Figure 1.
Structural elements of yeast pol II critical for α-amanitin action. From the structure of the yeast pol II elongation complex bound to α-amanitin (2VUM)3: located within mobile Trigger Loop (cartoon, blue) are His1085 (sticks, pink) making H-bonds with α-amanitin (sticks, yellow)2,3, and Leu1081 (sticks, orange) “wedging” into Bridge Helix (cartoon, green)3.
Figure 2.
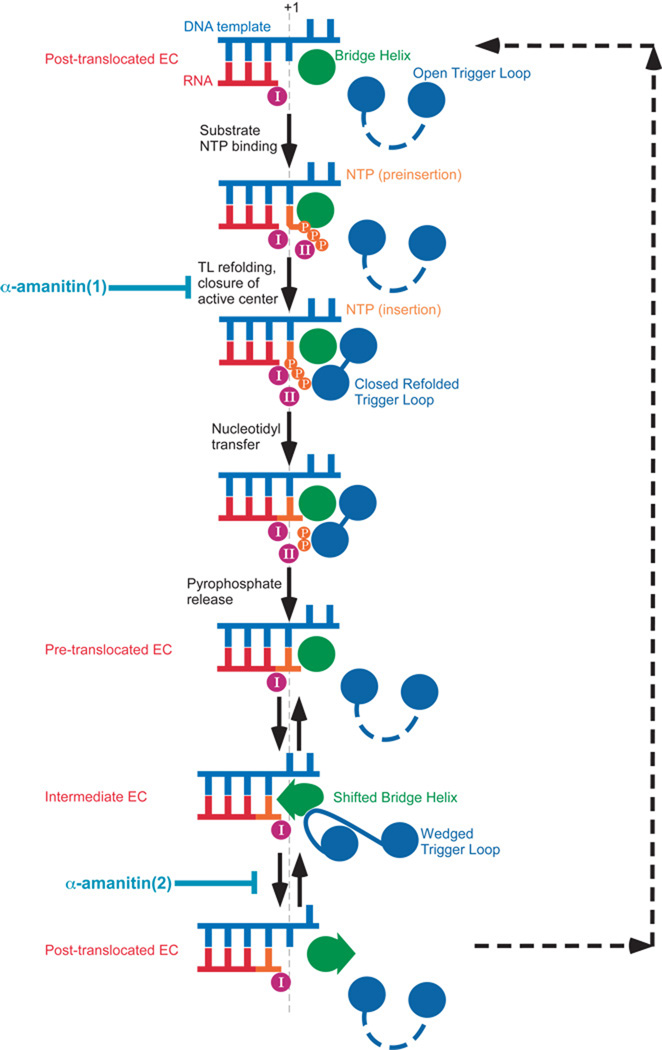
Model of RNA polymerase nucleotide addition cycle (NAC). Adopted from Brueckner and Cramer3 with modifications. α-amanitin(1) indicates the step where this inhibitor can interrupt Trigger Loop-dependent loading of a substrate2, α-amanitin(2) indicates the step where it can interfere with RNA polymerase translocation3. For further details, refer to the text.
Unfortunately, an unambiguous interpretation of the effects of the H1085Y and other substitutions (including multiple amino acid changes and deletions) within TL is difficult due to the fact that each can affect one or more of the TL properties, such as the specific contacts with substrate and/or other pol II structural elements, especially the so called Bridge Helix (BH) (Fig.1), TL movement and/or substrate-dependent refolding, emphasizing the need for more structural data and extensive molecular dynamics simulations. Nevertheless, biochemical analysis of a large set of TL substitutions and the effect of α-amanitin of NTP usage reported by Kornberg and colleagues provide ample evidence for the direct and critical involvement of pol II TL in substrate selection (fidelity of templated NTP addition), and potentially also in catalysis and translocation.
Cramer’s group took a different approach to examine the effects of α-amanitin on pol II translocation. They first created crystals of yeast pol II elongation complexes (EC) on a nucleic acid scaffold where the post- and pre-translocated forms of EC were at equilibrium as demonstrated by the position of a reporter group, bromo-uracil (BrU), in the template DNA strand3. Without α-amanitin BrU was found in registers −4 and −3 relative to the nucleotide incorporation site, indicating that, as predicted by the Brownian ratchet model5, pol II EC oscillates between the two states without input of energy other than thermal energy. By various estimates, the free energy difference between these states ranges from −0.2 to 3.4 kBT for perfectly base-paired nucleic acids11. Mismatch at the +2 position appeared to fortuitously aid the establishment of the equilibration between the states in the crystal. After such crystals were soaked in a solution of α-amanitin, BrU was found only in the −3, post-translocated register of the RNA-DNA hybrid. Solved at 3.4Å (the highest resolution of pol II EC to date), the crystals displayed unambiguous electronic densities for the α-amanitin, nucleic acid scaffold, and previously disordered regions of TL. However, the lack of a substrate prevented the structure from providing information concerning the NTP-dependent refolding of TL, described for the eukaryotic and bacterial ECs8,9. This caveat notwithstanding, the new structure offers insights into the translocation mechanism of yeast pol II and the mechanism of α-amanitin inhibition of transcription. Similar to the refined structure of the apo-enzyme co-crystallized with α-amanitin, the new EC-amanitin co-crystals reveal additional interactions between the inhibitor and the enzyme, in particular, those involving the same H-bonds to Rpb1 His1085 Fig.1). In addition, another TL residue, Leu1081, was noted to “wedge” itself between residues of two other structural modules, Val829 of the Bridge Helix (BH) and Pro1099 of helix α37 (Fig.1). A mobile RNAP element adjacent to the TL, the BH appeared to adopt a new conformation, wherein its central portion (from Asp826 to Glu833) was shifted by 2.5Å in the direction of the RNA-DNA hybrid, occluding the position of the +1 templating nucleotide, effectively shifting DNA to a previously unobserved position above the BH, designated as “pretemplating”. Brueckner and Cramer observed that RNA-DNA hybrid in this new structural arrangement corresponded to the post-translocated state while the mobile protein modules surrounding the pol II active site (particularly BH and TL) were in somewhat intermediary positions. They reasoned that this represented an intermediate translocation state trapped by the bound α-amanitin (Fig.2). Indeed, in several EC structures of Thermus RNAPs Met1238 (whose position in the TL corresponds to that of Leu1081) appeared to intrude in a similar fashion between the BH and helix G’, while the former adopts a flipped-out conformation in the area that is shifted in the yeast enzyme12,13, emphasizing the overall congruity of the BH-TL rearrangements. They also highlighted the profound (11-fold) effect of substitution of the corresponding residue in E. coli RNAP TL (Met932) on transcriptional pausing7, as the evidence of the importance of this residue and, by extension, the effect of the “wedged” TL conformation on transcription. It must be noted, however, that it is possible that the particular configuration of the EC trapped by α-amanitin observed by Brueckner and Cramer may represent an off-pathway translocation state (yeast pol II can add nucleotides and translocate even in the presence of α-amanitin, albeit slowly and with reduced accuracy2), or even an artifact caused by the particular scaffold design, absence of NTP, or other factors. The importance of Met932 for bacterial transcription can not be unequivocally associated with its role in “wedging” of TL into BH, because its counterpart Met1238 was also reported to stack on the substrate base in the T. thermophilus EC structure9, and its substitution may have unknown effects on TL mobility and/or refolding. Similar substitution of a nearby Thr934 had even more drastic effect (22-fold) on pausing without any direct involvement in the formation of the “wedge”7.
Regardless of the extent to which this “wedged” conformation represents an in-pathway translocation intermediate, Brueckner and Cramer made a number of important contributions to our understanding of how RNAP functions. The new structure of yeast pol II EC allows for more rigorous molecular dynamics simulations of transcription elongation, which, together with structural studies, bulk and single-molecule biochemistry, and kinetic analysis are needed to generate an explicit physical model of transcription. Direct observation of the equilibrium between pre- and post-translocated ECs lends additional support to the concept of RNAP as a Brownian ratchet. This work also attempts a synthesis of BH-centric and TL-centric models of elongation by emphasizing the concerted movements of BH and TL during translocation. Broad acceptance of the Brownian ratchet mechanism of multi-subunit RNAPs (as opposed to power-stroke mechanism) began with the BH-centric model of Bar-Nahum et al.5 They demonstrated that mutations in TL affected BH conformations as well as enzyme translocation, fidelity and response to regulatory signals and factors5. BH conformational changes in that work were observed directly using crosslinking approaches. However, the particular structural manifestations of the BH-centric thermal ratchet (bending and straightening of the BH) appeared to be a result of an earlier crystallographic aberration, which in turn, called into question the significance of the BH element in RNAP function (the extreme TL-centric model discounted the importance of BH altogether7). The work of Brueckner and Cramer restores BH to a crucial role in RNAP translocation and provides a more accurate view of the structural changes (shifting of the central portion of BH) that accompany the repositioning of the DNA template to a new, “pre-templating” position in an intermediate translocation state. The movement of the BH in and out of the pre-templating position may depend on the “wedged” TL, which can push BH into position, stabilize the shifted BH or act passively as a boundary condition directing the distortion of BH by a force arising elsewhere. Thus, BH oscillation could still be a structural manifestation of the Brownian ratchet in translocation. Swinging of the refolded substrate-bound TL, which takes the shape of an α-helical hairpin packed against BH, toward the insertion site could provide the initial velocity and directionality of the nucleophilic attack of the RNA 3’-OH on the α-phosphate of the substrate NTP. Indeed, molecular dynamics simulations of the yeast pol II NAC suggest that the direction of the attack is reversed in such a way that it is the substrate bound to the mobile TL that attacks the static nucleophile (RNA)14. Thus, BH and TL can switch between “leading” and “assisting” roles during translocation and catalysis. An integrated NAC model (Fig.2) incorporates these views and indicates the steps where α-amanitin could interfere with RNA synthesis. The work of Cramer and Kornberg laboratories reveal previously unobserved conformations and states of the RNAP trapped by the transcriptional inhibitor α-amanitin. They provide a new understanding for the role of the catalytic center mobile elements in the nucleotide addition cycle and pave the way for future investigations.
Acknowledgments
This work was supported by grants from the NIH.
Reference list
- 1.Borukhov S, Nudler E. Trends Microbiol. 2008;16:126–134. doi: 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Kaplan CD, Larsson KM, Kornberg RD. Mol. Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brueckner F, Cramer P. Nat. Struct. Mol. Biol. 2008 doi: 10.1038/nsmb.1458. Advanced online publication. [DOI] [PubMed] [Google Scholar]
- 4.Bushnell DA, Cramer P, Kornberg RD. Proc. Natl. Acad. Sci. USA. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar-Nahum G, et al. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 6.Kireeva ML, et al. Mol. Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toulokhonov I, Zhang J, Palangat M, Landick R. Mol. Cell. 2007;27:406–419. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg R. Cell. 2007;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vassylyev DG, et al. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 10.Zhu R, et al. Theor. Chem. Account. 2008;120:479–489. [Google Scholar]
- 11.Bai L, Fulbright RM, Wang MD. Phys. Rev. Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- 12.Vassylyev DG, et al. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 13.Campbell EA, et al. EMBO J. 2005;24:674–682. doi: 10.1038/sj.emboj.7600499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu R, Salahub DR. AIP Conf. Proc. 2007;963:104–110. [Google Scholar]