Abstract
A transient inflammatory signal can initiate an epigenetic switch from non-transformed to cancer cells via a positive feedback loop involving NF-κB, Lin28, let-7, and IL6. We identify differentially regulated microRNAs important for this switch, and putative transcription factor binding sites in their promoters. STAT3, a transcription factor activated by IL6, directly activates miR-21 and miR-181b-1. Remarkably, transient expression of either microRNA induces the epigenetic switch. MiR-21 and miR-181b-1 respectively inhibit PTEN and CYLD tumor suppressors, leading to increased NF-κB activity required to maintain the transformed state. These STAT3-mediated regulatory circuits are required for the transformed state in diverse cell lines and tumor growth in xenografts, and their transcriptional signatures are observed in colon adenocarcinomas. Thus, STAT3 is not only a downstream target of IL6, but with miR-21, miR-181b-1, PTEN, and CYLD, is part of the positive feedback loop that underlies the epigenetic switch that links inflammation to cancer.
Keywords: microRNAs, transcription factors, inflammation, cancer, transformation, STAT3
INTRODUCTION
In human cells, there are ~1000 microRNAs that collectively regulate the expression of more than 30% of protein-coding genes at the post transcriptional and translational level (Bartel, 2009). Each microRNA represses multiple gene targets, and repression can occur by translational inhibition, mRNA cleavage, and mRNA decay initiated by miRNA-guided deadenylation. With respect to the regulation of gene expression patterns, microRNAs are analogous to DNA-binding transcription factors that directly regulate the expression of target genes. Thus, elucidation of transcriptional regulatory circuits requires the integration of transcription factors, microRNAs, and their direct targets into connected molecular pathways that are responsible for specific biological phenomena.
Bioinformatic programs can identify putative target genes for individual microRNAs, and many such microRNA-target interactions have been validated experimentally. However, there is limited information about how microRNAs are regulated at the transcriptional and post-transcriptional levels. The related RNA-binding proteins Lin28 and Lin28b that inhibit the Let-7 family of tumor suppressor microRNAs are the only known factors that mediate post-transcriptional regulation of microRNAs (Viswanathan et al., 2008; Hagan et al., 2009; Iliopoulos et al., 2009). Although examples of transcription factors directly regulating microRNA expression have been described (O’Donnell et al., 2005; Loffler et al., 2007; Chang et al., 2008; Lin et al., 2009), microRNA promoters and transcription start sites were largely unknown until recently, thereby making it difficult to study transcriptional regulation of microRNAs. However, promoter regions for 175 microRNAs were identified by combining nucleosome mapping and chromatin signatures, and the DNA sequence of the linker regions was used to predict transcription factors regulating microRNA (Ozsolak et al., 2008). In addition, candidate transcriptional start sites of microRNAs were identified by analyzing H3-K4 tri-methylation in multiple tissues at high resolution (Marson et al., 2008).
We recently described an inducible model of cellular transformation in order to identify transcriptional regulatory circuits important in oncogenesis (Iliopoulos et al., 2009). This model involves a non-transformed mammary epithelial cell line (MCF-10A) containing ER-Src, a derivative of the Src kinase oncoprotein (v-Src) that is fused to the ligand-binding domain of the estrogen receptor. Treatment of such cells with tamoxifen rapidly induces Src, and morphological transformation is observed within 36 hours. Unlike the parental cell line, the transformed cells form foci and colonies in soft agar, show increased motility and invasion, form mammospheres, and confer tumor formation in mouse xenografts. This model permits the opportunity to kinetically follow the pathway of cellular transformation in a manner similar to that used to study viral infection and other temporally ordered processes.
In this inducible transformation model, transient activation of Src triggers an inflammatory response that results in an epigenetic switch between non-transformed and transformed cells. The epigenetic switch is mediated by a positive feedback loop involving NF-κB, Lin28b, let-7 microRNA and IL6 (Iliopoulos et al., 2009). This regulatory circuit operates in other cancer cell lines, and its transcriptional signature is found in patient cancer tissues, indicating its importance in some human cancers. In addition to its role in the positive feedback loop, IL6 activates STAT3, a transcription factor that is critical for transformation. This observation is consistent with the conventional view that STAT3 is a downstream effector of IL6 (Frank, 2007; Yu et al., 2007; Yu et al., 2009), but not part of the central regulatory circuit that mediates the epigenetic switch.
Here, we use this inducible ER-Src model to study the regulation of microRNA expression throughout the process of cellular transformation. We identify differentially regulated microRNAs and show that many of them are important for transformation. Using computational algorithms, we identify individual and combinations of DNA sequence motifs that are overrepresented in the promoter regions of these differentially regulated microRNAs. We predict and experimentally validate transcription factor binding sites, and hence transcription factor-microRNA interactions that are likely to be involved in the oncogenic process.
In particular, we demonstrate that STAT3 directly activates transcription of miR-21 and miR-181b-1 microRNAs during the transformation process. Remarkably, transient expression of either miR-21 or miR-181b-1 is sufficient to induce a stable transformed state, and this occurs by direct targeting of the phosphatase and tensin homolog (PTEN) and cylindromatosis (CYLD) tumor suppressor genes respectively. The resulting inhibition of PTEN and CYLD expression leads to NF-κB activation, which is required to maintain the transformed state. Thus, STAT3 is not simply a downstream effector of IL6, but together with miR-21, miR-181b-1, PTEN, and CYLD, is part of the epigenetic switch that links inflammation to cancer.
RESULTS
Kinetic Analysis of Differentially Regulated MicroRNAs During the Process of Cellular Transformation
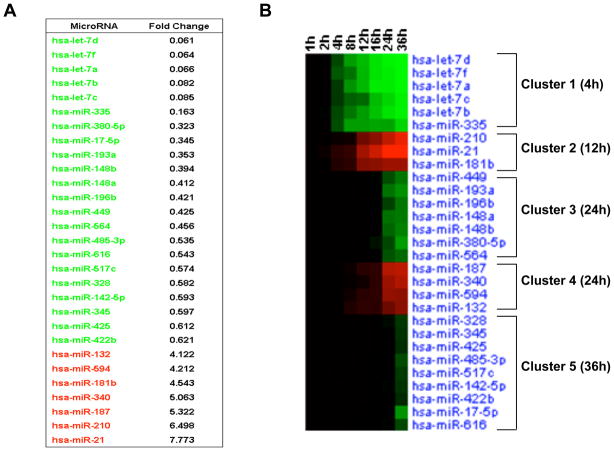
To identify differentially expressed microRNAs during transformation, we first measured the expression of 365 microRNAs in non-transformed and transformed (36h treatment with tamoxifen) MCF-10A-ER-Src cells by TaqMan microRNA analysis (Figure 1A). 29 differentially expressed microRNAs were identified, 22 of which are down-regulated and 7 of which are up-regulated during transformation. Highly down-regulated microRNAs include multiple members of the let-7 family (let-7d, let-7f, let-7a, let-7b, let-7c) and miR-335, while miR-21, miR-210 and miR-187 are highly up-regulated.
Figure 1. MicroRNA Dynamics during ER-Src cellular transformation.
(A) Differentially expressed microRNAs between transformed (TAM 36h) and untransformed (0h) MCF10A ER-Src cells. (B) Heatmap representation of differentially expressed microRNAs in different time points (1, 2, 4, 8, 12, 16, 24, 36h) during ER-Src transformation. Up-regulated microRNAs are shown in red, while down-regulated microRNAs in green.
To follow kinetically microRNA expression levels during cellular transformation, microRNA profiling was performed in cells isolated at various times (1, 2, 4, 8, 12, 16, 24 and 36h) after tamoxifen treatment. This analysis reveals 5 distinct microRNA clusters (Figure 1B; Figure S1). The first cluster includes 5 microRNAs (let-7d, let-7f, let-7a, let-7c, let-7b, miR-335) down-regulated very early (4h) during transformation. The second cluster consists of 3 early (12h) up-regulated microRNAs (miR-210, miR-21, miR-181b-1; interestingly, miR-181a and miR-181b-2 are not differentially regulated). The third and fourth clusters consist of microRNAs down-regulated and up-regulated, respectively, 24h post tamoxifen treatment. The last cluster consists of 9 down-regulated microRNAs 36h post treatment. Overall, this kinetic analysis reveals 2 clusters of early microRNA responders and 3 clusters of late microRNA responders during transformation. Interestingly, microRNAs that show the earliest responses generally show the highest degree of differential regulation at later times.
Differentially Regulated MicroRNAs are Important for Transformation, Tumorigenicity, and Chemotactic Cell Invasion
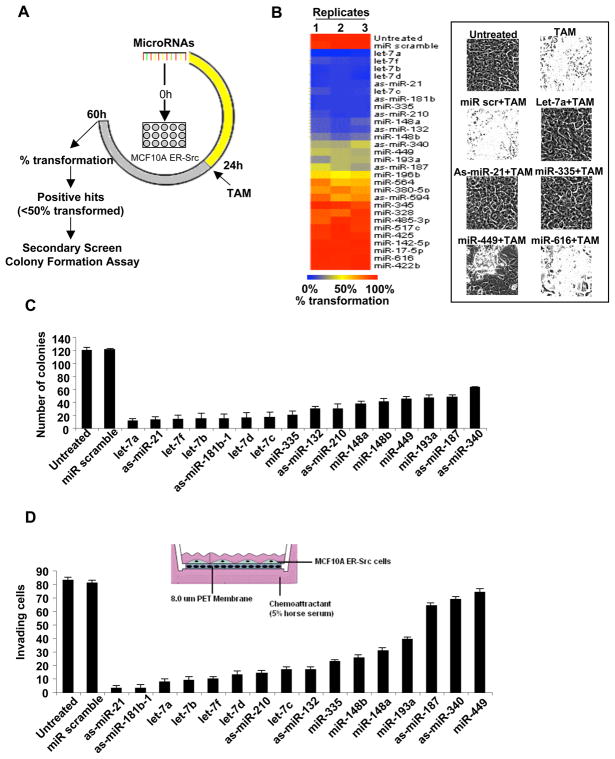
To assess the functional role of these differentially regulated microRNAs in transformation, we tested whether inhibition of up-regulated microRNAs or down-regulation of up-regulated microRNAs blocks the transformation process. Non-transformed cells were transfected with antisense microRNAs for the 7 up-regulated microRNAs and with microRNA precursors for the 22 down-regulated microRNAs. Tamoxifen was added 24h post transfection, and transformation efficiency was assayed by morphological changes 36h later (60h from the beginning of the experiment) (Figure 2A). 6 out of 7 antisense microRNAs (as-miR-21, as-miR-181b-1, as-miR-210, as-miR-132, as-miR-340, as-miR-187) and 10 out of 22 microRNA precursors (let-7a, let-7f, let-7b, let-7d, let-7c, miR-335, miR-148a, miR-148b, miR-449, miR-193a) show >50% inhibition of transformation efficiency (Figure 2B). Interestingly, microRNAs differentially expressed very early during transformation have the most significant effects on transformation, whereas late responding microRNAs have little effect.
Figure 2. MicroRNAs Important for ER-Src transformation.
(A) Strategy for identifying microRNAs regulating ER-Src transformation. (B) Heatmap representation of transformation efficiency (% transformed cells as assayed by cell morphology) after transfection of microRNAs or antisense microRNAs in ER-Src cells. (C) Soft agar colony assay and (D) invasion assay in ER-Src cells transfected with sense or antisense microRNAs. Experiments performed in triplicate and data are shown as mean ± SD.
To extend these results, we used the same experimental procedure to test the 16 microRNAs that affected morphological transformation for effects on tumorigenicity and metastatic potential. All 16 microRNAs affected tumorigenicity, as assayed by the ability to form colonies in soft agar (Figure 2C). To analyze metastatic potential, we measured the infiltration of these cells through Matrigel in a modified Boyden chamber assay using 5% horse serum as a chemo attractant (Figure 2D). Under these conditions, 13 out of 16 tested microRNAs inhibit >50% of the invasive ability of transformed cells. Taken together, our results identify at least 13 microRNAs that are differentially regulated during the process of cellular transformation, and are important for morphological alterations, tumorigenicity, and metastatic potential.
Enrichment of Transcription Factor Binding Sites in MicroRNA Promoters
To identify putative transcription factor binding sites associated with the 13 microRNAs described above, we used the Lever algorithm (Warner et al., 2008) to scan the flanking sequences (2kb up- and 7kb down-stream) from the microRNA transcription start sites for over-represented DNA sequence motifs. As shown in Table S1, this analysis was performed on 466 human transcription factor binding site motifs from the TRANSFAC database and on motifs derived by universal protein binding microarrays for 272 mouse transcription factors (Berger et al., 2008; Badis et al., 2009b). DNA sequence motifs for STAT3 and Myc are among the top ranked motifs enriched in these microRNA promoter regions (Figure 3A), and both STAT3 and Myc are up-regulated during cellular transformation (Figure 3B).
Figure 3. STAT3 and Myc are important regulators of transformation.
(A) Lever algorithm analysis to identify transcription factor binding site motifs over-represented in promoter areas of differentially expressed microRNAs. Motifs with AUC score higher than 0.7 and z-score higher than 2 are considered statistically significant (red color) with STAT3 an Myc indicated. (B) Pairs of motifs (connected by lines) that are differentially regulated (up-regulated in red and down-regulated in blue) during transformation and over-represented in microRNA promoters. (C) STAT3 and Myc mRNA expression levels in ER-Src cells during transformation. (D) Soft agar colony assay (mean ± SD) in TAM-treated (36h) cells transfected with siRNAs against STAT3, Myc, or control. (E) Soft agar colony assay (mean ± SD) in transformed ER-Src cells (for 10 days) transfected with siRNAs against STAT3, Myc, or control.
Putative STAT3 and Myc binding sites were further ranked according to their PhylCRM score (Warner et al., 2008) that combines binding affinity and evolutionary conservation across all sequenced mammalian species. We identified putative STAT3 binding sites in the promoters of miR-148a, miR-21, miR-132, miR-181b-1, miR-148b, miR-193a, miR-340, miR-335, miR-210, miR-187 (ordered based on the maximum window PhylCRM score, Tables S2–3). We also identified putative Myc binding sites in miR-181b-1, miR-148a, miR-335, and let-7a3 promoter regions (Tables S4–5), although PhylCRM scores for the Myc sites are generally lower than those for the STAT3 sites. As assayed by chromatin immunoprecipitation, 9 out of 18 putative STAT3 sites and 3 out of 5 putative Myc sites with PhylCRM scores > 1 are bound by their cognate proteins in vivo (Tables S3, S5).
We extended our computational analysis by searching for over-represented pair-wise combinations of motifs within the microRNA promoter regions. We identified 267 PBM motif pairs and 620 TRANSFAC motif pairs with z score higher than 2.3 and AUC >0.7 (Table S1). A subset of these pairs contains a motif for one transcription factor differentially regulated during transformation (Figure S2), and there are 30 pairs (including STAT3 and Myc) in which both factors are differentially expressed (Figure 3B). Interestingly, one of the top scoring motif pairs is for Max and RXRα and this motif pair is predicted in the promoter regions of miR-148a, miR-132, miR-181b-1, miR-210, miR-193a, miR-148b, miR-335 and miR-340 (Tables S6–7). Chromatin immunoprecipitation experiments validate the binding of both Max and RXRα to the miR-148a, miR-181b-1, and miR-148b promoter regions (Figure S2).
STAT3 and Myc are Important for Maintenance of the Transformed State
During the transformation process, STAT3 expression is strongly induced during transformation while Myc expression is induced to a lesser extent (Figure 3C). In previous work, we showed that STAT3 is important for cellular transformation in the ER-Src model (Iliopoulos et al., 2009). To address the role of STAT3 and Myc, we inhibited STAT3 and Myc expression by 24h pre-treatment with siRNA (Figure S2), added tamoxifen for 36h to induce transformation, and then plated the resulting cells in soft agar to evaluate colony formation 15 days later. Inhibition of Myc results in a modest decrease in colony formation, whereas inhibition of STAT3 in parallel experiments confers a 10-fold decrease (Figure 3D). Similar results were obtained when siRNAs against STAT3 and Myc were added 10 days after cells were stably transformed, and the resulting cells tested for colony formation (Figure 3E). Thus, STAT3 and (to a significantly lesser extent) Myc are important for cellular transformation and maintenance of the transformed phenotype.
STAT3- and Myc-Regulated MicroRNAs During Transformation
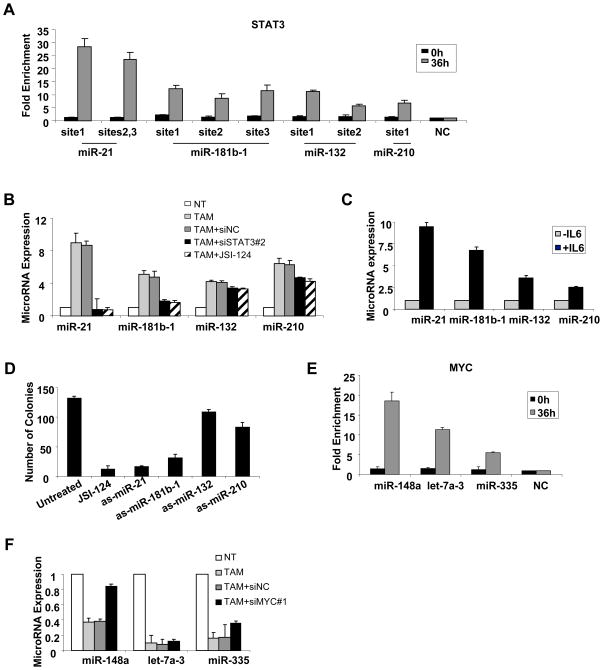
In transformed cells, STAT3 binds to three sites in the miR-21 promoter, three sites in the miR-181b-1 promoter, two STAT3 sites in the miR-132 promoter, and one STAT3 site in the miR-210 promoter during transformation (Figure 4A, Table S2). STAT3 binding to the targets sites in the miR-21 promoter upon IL6 induction has been reported previously (Loffler et al., 2007). STAT3 binding to these sites is induced upon transformation, because it is not detected in non-transformed cells. Inhibition of STAT3 by siRNA or by a pharmacological inhibitor (JSI-124) strongly reduces miR-21 and miR-181b-1 expression levels, but has only a very modest effect on miR-132 and miR-210 expression levels (Figure 4B). Conversely, STAT3 activation by IL6 treatment in MCF-10A cells (i.e. lacking the ER-Src construct) results in up-regulation of these microRNAs (Figure 4C, Figure S3). Interestingly, inhibition of miR-21 and miR-181b-1 had a major effect while miR-132 and miR-210 had a minor effect on colony formation ability of the ER-Src cells (Figure 4D). These results strongly suggest that STAT3 function in transformation involves transcriptional activation miR-21 and miR-181b-1.
Figure 4. STAT3 and MYC-regulated microRNAs during ER-Src transformation.
(A) STAT3 occupancy (fold-enrichment) at the indicated miR loci in cells that were or were not treated with TAM as determined by chromatin immunoprecipitation. (B) MicroRNA expression levels in TAM-treated cells ER-Src cells transfected in the presence of STAT3 inhibitors (siSTAT3#2 or 8 μM JSI-124). (C) MicroRNA expression levels in MCF-10A cells treated with 50 ng/ml IL6. (D) Soft agar colony assay in MCF10A-IL6 transformed cells transfected with the indicated antisense-microRNAs. (E) Myc occupancy (fold enrichment) at the indicated miR loci that were or were not treated with TAM. (F) MicroRNA expression levels in TAM-treated ER-Src cells transfected with siRNA negative control or siMyc. In all experiments, data are presented as mean ± SD of 3 independent experiments.
Chromatin immunoprecipitation revealed one Myc binding site in the let-7a3 promoter, one site in the miR-335 promoter and one site in the miR-148a promoter (Figure 4E). Inhibition of Myc expression by siRNA did not affect significantly let-7a-3 and miR-335 expression, but it did result in increased levels of miR-148a (Figure 4F). These data suggest that Myc directly inhibits miR-148a expression levels during transformation.
Transient expression of either miR-21 or miR-181b-1 is sufficient to induce the epigenetic switch to a stable transformed state
STAT3 is phosphorylated in response to IL6 during cellular transformation (Frank, 2007), and in our experimental system this occurs only at later time points after IL6 super-activation (Iliopoulos et al., 2009). Based on these observations, we proposed that STAT3 is a downstream effector of IL6 required for transformation, but not part of the epigenetic switch that is needed to convert a non-transformed cell into a stable transformed line (Iliopoulos et al., 2009). If STAT3 and its target microRNAs are simply downstream effectors of IL6, they should be insufficient for cellular transformation, because they would not affect the function of NF-κB, Lin28, or Let-7, all of which are required for the process.
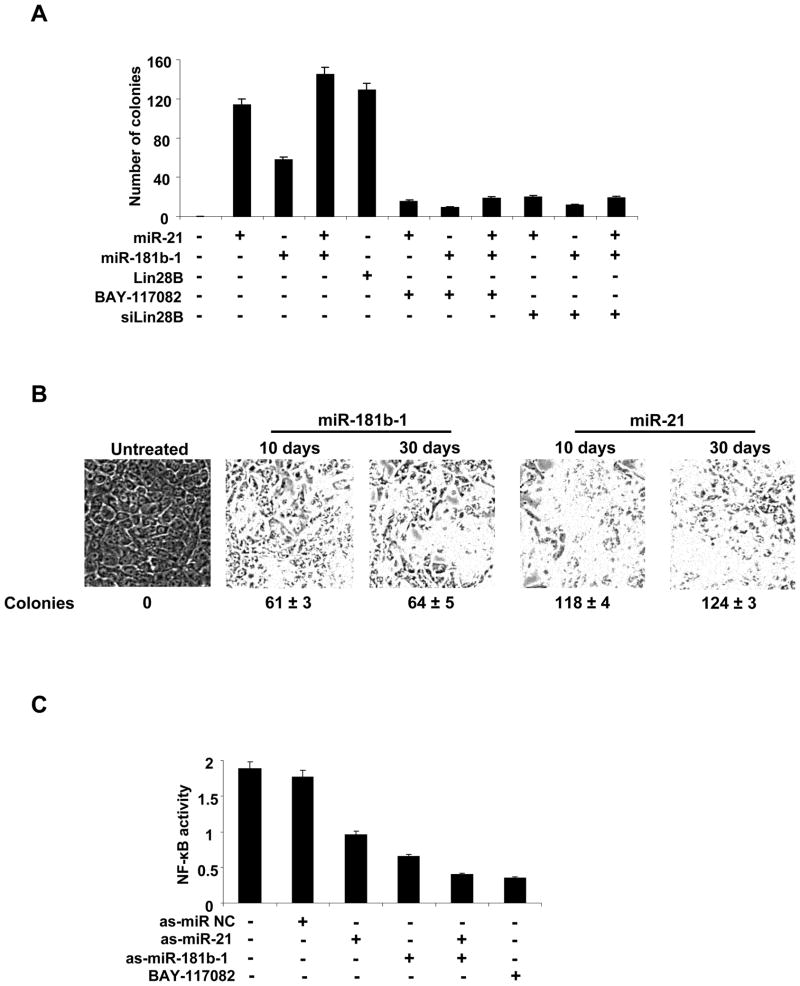
In striking contrast to our previous model, individual expression miR-21 of miR-181b-1 is sufficient for cellular transformation (Figure 5A). As assayed by colony formation, miR-21 is ~2-fold more effective than miR-181b-1, and of comparably efficacy to that occurring upon overexpression of Lin28b. Remarkably, even though transiently transfected microRNAs are likely to be present only for a few days, the transformed cells can be passaged for at least 30 days without losing their ability to form colonies in soft agar, indicating that cells are stably transformed (Figure 5B).
Figure 5. MiR-21 and miR-181b-1 induce transformation of MCF-10A cells.
(A) Colony formation assay of MCF-10A cells treated with miR-21, miR-181b-1, Lin28b, BAY-117082 (5μM) or siRNA against Lin28B. (B) Cells treated with miR-21 or miR-181b-1 were propagated for 10 or 30 days and tested for cell morphology and colony formation. (C) NF-κB activity in MCF-10A cells treated with as-miR-NC, as-miR-21, as-miR-181b-1, or BAY-117082. The data are presented as mean ± SD of three independent experiments.
These observations indicate that transient expression of either miR-21 or miR-181b-1 is sufficient to induce the epigenetic switch to the stable transformed state. In accord with this suggestion, treatment with BAY-117082 (NF-κB inhibitor) or siRNA against Lin28b reduces the tumorigenicity of cells transformed by miR-21 and/or miR-181b-1 transformed cells. Furthermore, ER-Src transformed cells treated with antisense RNAs against miR-21 and/or miR-181b-1 show reduced levels of NF-κB activation (Figure 5C) that are roughly comparable to those obtained upon inhibition by BAY-117082. The ability of miR-21 and miR-181b-1 to induce a stable transformed state in a manner dependent on NF-κB and Lin28b indicates that these microRNAs and STAT3 are part of the positive feedback loop mediating the epigenetic switch, and are not simply downstream effectors of IL6.
miR-21 Targets the PTEN Tumor Suppressor Gene that Functions Through the Akt Pathway
The ability of miR-21 or miR-181b-1 to induce a stable transformed state indicates that these microRNAs must, in some manner, activate NF-κB to sustain the inflammatory positive feedback loop. In accord with a previous study identifying the PTEN tumor suppressor gene as a direct target of miR-21 (Meng et al., 2007), expression levels of miR-21 and PTEN mRNA expression levels are inversely correlated during ER-Src transformation (Figure S4A). In addition, inhibition of miR-21 expression results in up-regulation of PTEN mRNA expression (Figure S4B), suggesting that PTEN expression is regulated by miR-21 in ER-Src cells. PTEN is an upstream regulator of Akt kinase pathway that is involved in oncogenesis (Yuan and Cantley, 2008). In this regard, Akt activity (assayed by phosphorylation) increases during ER-Src transformation in a manner that depends upon miR-21 expression (Figure S4C), and inhibition of Akt expression reduces tumorigenicity of ER-Src transformed cells (Figure S4D). Lastly, the PTEN/Akt pathway regulates NF-κB activity in cancer cells, PTEN deletion results in increased NF-κB activity (Garcia et al., 2009; Guigon et al., 2009), and inhibition of Akt expression reduces NF-κB activity and IL6 production (Figure S4E, F). Taken together, these results are consistent with a pathway in which STAT3-activated miR-21 directly inhibits PTEN, thereby resulting in increased Akt activity and subsequent activation of NF-κB that is required for the positive feedback loop that maintains the epigenetically stable transformed state.
MiR-181b-1 Transforms Cells via Direct Targeting of the CYLD Tumor Suppressor Gene
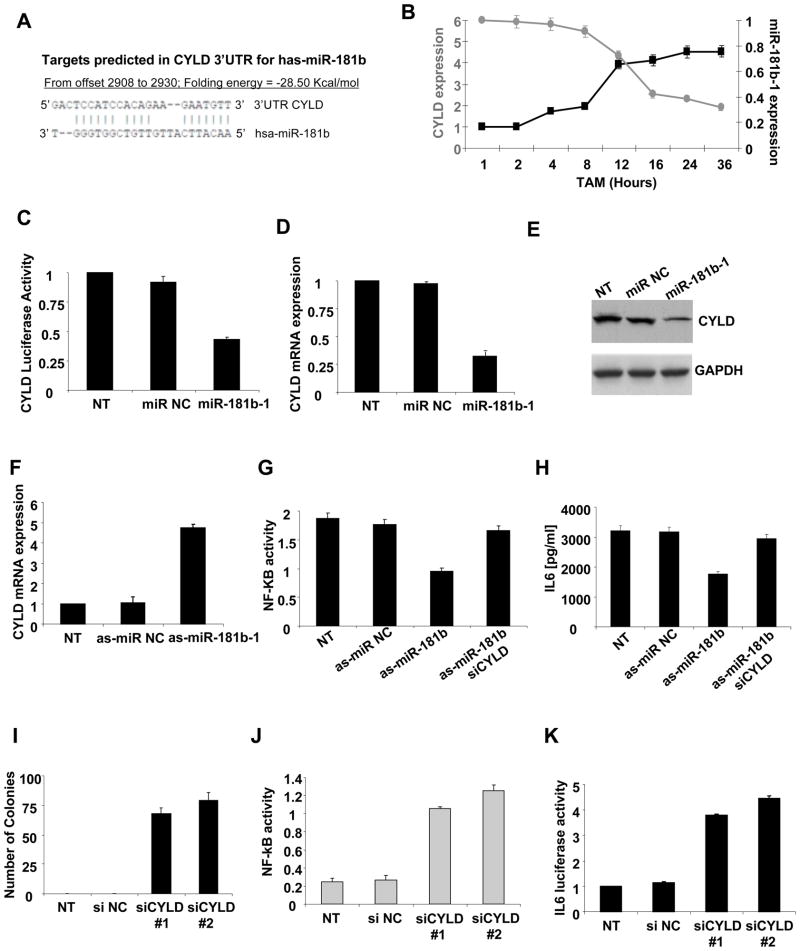
Unlike the case for miR-21, direct targets and potential pathways for miR-181b-1 are unknown. Interestingly, bioinformatic analysis reveals sequence complementarity between miR-181b-1 and the cylindromatosis tumor suppressor gene (CYLD) 3’UTR (Figure 6A). During ER-Src transformation, miR-181b-1 and CYLD expression levels are inversely correlated (Figure 6B), and miR-181b-1 overexpression inhibits activity of a luciferase reporter construct containing the CYLD 3’UTR (Figure 6C), as well as CYLD mRNA (Figure 6D) and protein expression (Figure 6E) levels. Conversely, suppression of miR-181b-1 expression results in up-regulation of CYLD expression (Figure 6F), suggesting that miR-181b-1 regulates directly CYLD expression during transformation.
Figure 6. MiR-181b-1 regulates CYLD expression during ER-Src transformation.
(A) Sequence complementarity between miR-181b-1 and the 3’UTR of CYLD gene. (B) MiR-181b-1 and CYLD mRNA expression levels at the indicated times during ER-Src transformation. (C) Luciferase activity of a reporter containing the 3’UTR of CYLD 24h after transfection with miR-181b-1 or miR negative control. (D) CYLD mRNA levels after treatment with miR-181b-1 or miR negative control. (E) CYLD protein levels after treatment with miR-181b-1 or miR negative control. (F) CYLD mRNA levels after treatment with as-miR-181b-1 or as-miR-NC. (G) NF-κB activity (ELISA assay) in ER-Src cells untreated (NT) or treated with as-miR-181b-1, si-CYLD, or as-miR-NC. (H) IL6 production (ELISA assay) in ER-Src cells treated with as-miR-181b-1, si-CYLD or as-miR-NC. (I) Number of colonies, (J) NF-κB activity assessed by ELISA assay and (K) IL6 luciferase activity in ER-Src cells treated with two different siRNAs against CYLD. The data are presented as mean ± SD of three independent experiments.
CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activity (Brummelkamp et al., 2003; Trompouki et al., 2003). In ER-Src cells, inhibition of miR-181b-1 expression results in reduced NF-κB activity (Figure 6G) and reduced production of IL6, a direct NF-κB target gene (Figure 6H). Importantly, this effect is blocked by concurrent inhibition of CYLD, suggesting that miR-181b-1 regulates NF-κB through CYLD. In accord with the previously characterized tumor suppressor function of CYLD, MCF10A cells acquire the ability to form colonies in soft agar after siRNA inhibition of CYLD expression (Figure 6I). Furthermore, CYLD inhibition results in increased NF-κB (Figure 6J) and IL6 luciferase activity (Figure 6K) and mRNA expression (Figure S5A). Lastly, CYLD suppression increases the motility of ER-Src cells in a manner that depends on NF-κB (Figure S5B). Taken together, these observations are consistent with a pathway in which miR-181b-1 directly targets CYLD, leading to increased NF-κB activity and maintenance of the inflammatory feedback loop necessary for the transformed states.
STAT3-regulated MicroRNAs are Important for Tumorigenicity of Cancer Cell Lines of Diverse Developmental Origin
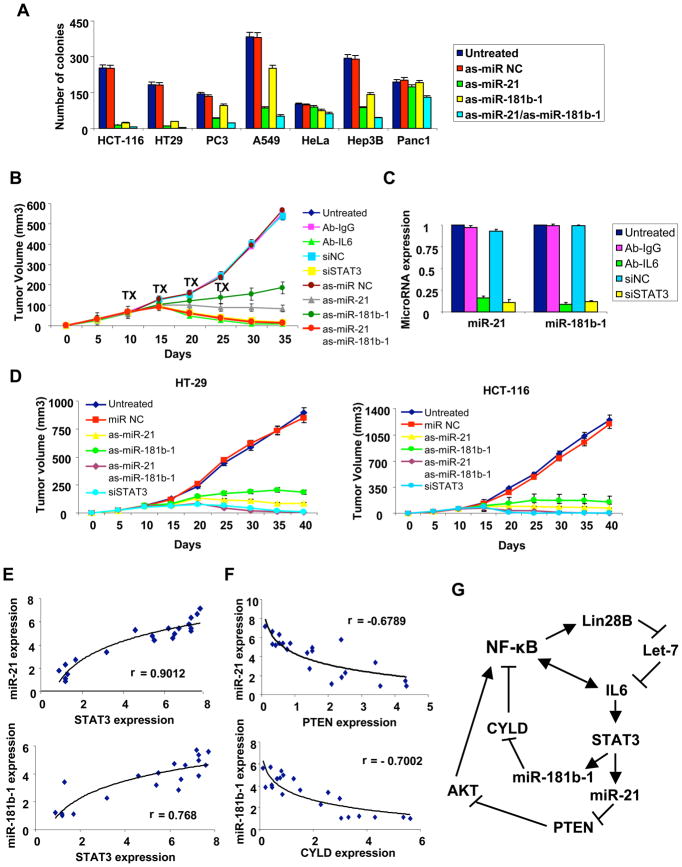
The results above indicate that STAT3-induced microRNAs (miR-21 and miR-181b-1) are important for transformation and tumorigenicity of breast cells. To generalize these observations beyond breast cells, we asked whether miR-21 and miR-181b-1 are important for tumorigenicity of cancer cell lines of diverse developmental origin (Figure 7A). Inhibition of miR-21 dramatically reduces colony formation in soft agar of colon (HCT-116, HT-29) cancer cells, and to a lesser extent, prostate (PC3), lung (A549), and hepatocellular (Hep3B) cancer cells. Inhibition of miR-181b-1 yields similar results, although the effects are often less pronounced. Neither microRNA affects colony formation in cervical (HeLa) or pancreatic (Panc1) cells. Thus, miR-21 and miR-181b-1 are important for tumorigenicity of many, but not all, developmentally unrelated cancer cell lines.
Figure 7. STAT3-regulated microRNAs in cancer cells, xenografts and cancer patients.
(A) Colony formation assay in colon (HCT-116, HT-29), prostate (PC3), lung (A549), cervical (HeLa), hepatocellular (Hep3B) and pancreatic (Panc1) cancer cell lines treated with antisense microRNA negative control, antisense-miR-21, antisense-miR-181b-1 or their combination. The data are presented as mean ± SD of three independent experiments. (B) Tumor growth (mean ± SD) of ER-Src cells after i.p treatment (days 10, 15, 20, 25) with Ab-IgG, Ab-IL6, siRNA negative control, siRNA against STAT3, antisense miR-NC, antisense-miR-21 and/or antisense-miR-181b-1. (C) MiR-21 and miR-181b-1 expression levels from tumors derived from the experiment described above. (D) Tumor growth (mean ± SD) of HCT-116 and HT-29 colon cancer cells after intraperitoneal treatment (days 10, 15, 20, 25) with microRNA negative control or antisense-miR-21 or antisense-miR-181b-1 or siRNA against STAT3. (E) MiR-21, miR-181b-1 and STAT3 mRNA expression levels in colon adenocarcinomas, with each data point represents an individual sample and a correlation coefficients (r) indicated. (F) MiR-21, miR-181b-1, PTEN and CYLD mRNA expression levels in colon adenocarcinomas. (G) Model of the inflammatory positive feedback loop that mediates the epigenetic switch between non-transformed and transformed cells.
STAT3 and STAT3-regulated MicroRNAs are Important for Tumor Growth in Mouse Xenografts
To examine the ability of miR-21 and miR-181b-1 to regulate tumor growth in mouse xenografts, ER-Src transformed cells were injected subcutaneously in nu/nu mice to generate tumors with a size of 60mm3. The mice were randomly separated into 9 groups and treated intraperitoneally with IL6 antibodies, siRNA against STAT3, antisense RNAs against miR-21 and miR-181b-1, or appropriate negative controls for 4 cycles every 5 days (Figure 7B). Inhibition of miR-21 or (to a lesser extent) miR-181b-1 reduces tumor growth with respect to control experiments, although tumors are not eliminated after 40 days. In contrast, the combined inhibition of miR21 and miR-181b-1 completely suppresses tumor growth, as is also observed upon inhibition of STAT3 or IL6. As expected, tumors treated with IL6 antibody siRNA against STAT3 had much reduced levels of miR-21 and miR-181b-1 microRNAs (Figure 7C). These results indicate that STAT3 activation of both miR-21 and miR-181b-1 is important for tumor growth in mouse xenografts, with miR-21 having a more pronounced effect.
Given the importance of miR-21 and miR-181b-1 microRNAs for tumorigenicity in colon cancer cells in vitro (Figure 7A), we performed similar experiments in which HCT-116 and HT-29 colon cancer cells were injected subcutaneously into nu/nu mice. After the completion of the second cycle of treatments in HCT-116 cells and the third cycle of treatments in HT-29 cells there was significant suppression of tumor growth in all treated mice (Figure 7D). As observed with transformed ER-Src cells, reduced growth of tumors generated by both colon cancer cell lines was more pronounced upon miR-21 inhibition than miR-181b-1 inhibition, and complete suppression of tumor growth occurred with simultaneous inhibition of both microRNAs or inhibition of STAT3.
STAT3, miR-21-PTEN, miR-181b-1-CYLD RNA Levels in Human Cancer Tissues
To address whether the above observations in cancer cell lines are relevant to human cancer, we examined the relationship between STAT3 and miR-21 or miR-181b-1 expression levels in colon adenocarcinomas. There is a striking positive correlation between STAT3 and miR-21 expression levels (r=0.9012) and STAT3 and miR-181b-1 expression levels (r=0.768) in the same cancers (Figure 7E). These data suggest a mechanistic relationship between STAT3, miR-21 and miR-181b-1 expression levels in at least one type of human cancer. In addition, we found an inverse correlation between miR-21 and PTEN expression levels (r=-0.678) and miR-181b-1 and CYLD expression levels (r=-0.7) in colon adenocarcinomas (Figure 7F), strongly suggesting that both the miR-21/PTEN and miR-181b-1/CYLD pathways are relevant for human cancer.
DISCUSSION
Kinetic Profiling Identifies MicroRNAs that are Important at Distinct Stages of Cellular Transformation and are Linked to Human Cancer
Using an inducible model of cellular transformation involving breast epithelial cells, we perform a kinetic profile of microRNA expression and identify 29 microRNAs that are differentially regulated during this process (22 up-regulated and 7 down-regulated). In general, microRNAs whose expression is affected early in the transformation process are also more highly regulated (fold-induction or fold-repression) at later times. This progressive enhancement of differential microRNA expression suggests that the transition between non-transformed and transformed cells occurs in a continuous fashion rather than in a set of distinct stages. In a functional sense, these early- and high-regulated microRNAs are more important for cellular transformation than late- and weakly-regulated microRNAs. Of particular interest are microRNAs that are rapidly and dramatically lost (Let-7 family and miR-335) or induced (miR-21, miR-181b-1, and miR-210) during cellular transformation.
MicroRNAs that are rapidly and dramatically regulated during the process of cellular transformation in our ER-Src model have been previously linked to cancer. Let-7 family members are down-regulated in multiple types of cancer, and they directly target oncogenes such as HMGA2, KRAS and IL6 (Johnson et al., 2005; Mayr et al., 2007; Iliopoulos et al., 2009). In our model, the Let-7 family is a key component of the epigenetic switch that mediates transformation, and Let-7 down-regulation is due to the rapid induction of the Lin28 processing factor, which is a direct target of NF-κB (Iliopoulos et al., 2009). Similar to the let-7 family, miR-335 is rapidly and dramatically down-regulated during transformation, although it does not appear that Lin28 is involved. This tumor-suppressor role of miR-335 during the early stages of tumorigenesis is unexpected, as this microRNA was previously characterized as being down-regulated in metastatic tumors (Tavazoie et al., 2008).
Important microRNAs induced early during the transformation process in ER-Src cells (miR-21, miR-181b-1, and mir-210) are also associated with human cancer. MiR-21 has been characterized as an anti-apoptotic microRNA that directly regulates PDCD4 and PTEN expression and is up-regulated in many cancer types (Meng et al., 2007; Frankel et al., 2008; Yan et al., 2008). Overexpression of miR-21 and miR-181b-1 is associated with the progression of leukoplakia to oral carcinoma (Cervigne et al., 2009) and prognosis and therapeutic outcome in colon cancer (Schetter et al., 2008), although down-regulation of miR-181b-1 occurs in other types of cancer (Shi et al., 2008; Conti et al., 2009). miR-210, a key player in the response to hypoxia, directly inhibits the Myc antagonist MNT, and it is up-regulated in a variety of solid tumors including breast cancer (Zhang et al., 2009). The observation that microRNAs critical for transformation in ER-Src cells are relevant for cancer provides additional validation for our experimental model, and it permits mechanistic analysis of these microRNAs in a well-defined system.
Identification of Transcription Factor Binding Sites Associated with Differentially Regulated microRNAs During Transformation
MicroRNAs and transcription factors play critical regulatory roles in oncogenesis, but there are few examples of transcription factors directly regulating the expression of microRNAs that are relevant for this process. As a first step to identify such transcription factor-microRNA interactions on a more global basis, we performed a computational analysis to identify transcription factor binding site motifs that are both over-represented and evolutionarily conserved in the putative promoter regions of differentially regulated microRNAs. Many individual motifs and pairwise combinations of motifs meet these criteria at varying levels of stringency, and binding of the cognate transcription factor to some of these sites in vivo was validated by chromatin immunoprecipitation. Furthermore, in some cases, the transcription factors associated with the microRNA promoters are important for regulating the expression of the microRNAs.
The above analysis identifies many potential transcription factors, individually or in combination, that may be important for cellular transformation as well as potential regulatory sites through which they act. While our identification of Myc and STAT3 was not surprising given their well-known roles in transformation, the combination of Max and RXRα was unexpected. However, as the computational analysis only identifies candidate protein-DNA interactions, it is critical to establish their physiological relevance through chromatin immunoprecipitation and transcription measurements. In addition, as many motifs are recognized by a family of transcription factors, experimental validation is necessary to determine which family member(s) is physiologically relevant. Lastly, it is likely that transcription factors identified here via regulation of microRNAs will also be important for regulation of mRNAs during transformation.
STAT3 Activation of miR-21 and miR-181b-1 is Important for Cellular Transformation and Associated with Some Human Cancers
While transcription factors and microRNAs are associated with human cancer, regulatory pathways directly connecting them are largely unknown. Previously, we described a regulatory circuit in which the transcription factor NF-κB is required for inhibition of the Let-7 microRNA family, but this regulation is indirect and involves Lin28 as an intermediary protein (Iliopoulos et al., 2009). In addition, we and others have identified STAT3 as an oncogenic transcription factor activated by inflammatory responses (Iliopoulos et al., 2009) (Bromberg et al., 1999; Grivennikov et al., 2009), but microRNA targets are unknown.
Here, we provide strong evidence that STAT3 activation of miR-21 and miR-181b-1 is important for cellular transformation. First, in a transformation-specific manner, STAT3 directly binds multiple sites in the miR-21 and miR-181b-1 promoter regions and is required for transcriptional induction of these microRNAs. Second, STAT3, miR-21, and miR-181b-1 are required for cellular transformation, tumorigenicity, and maintenance of the transformed state in vitro as well as tumor growth in vivo. Third, overexpression of either miR-21 or miR-181b-1 is sufficient to transform cells, indicating that these microRNAs can act as oncogenes. While overexpression of some microRNAs is correlated with transformed or tumor cells, to our knowledge, these are the first examples in which transient expression of oncogenic microRNAs can convert non-transformed cells to transformed cells. Taken together, our observations strongly suggest that STAT3 oncogenic function depends on miR-21 and miR-181b-1.
The importance of STAT3 activation of miR-21 and miR-181b-1 for cellular transformation is not limited to our experimental model or to breast cells. Indeed, miR21 and (to a lesser extent) miR-181b-1 are important for tumorigenicity (colony formation in soft agar) of a variety of cell lines of diverse developmental origin. In addition, these microRNAs are important for growth of tumors in xenografts generated by colon cancer cells. However, this pathway does not appear to be involved in all types of cancer cells, as cervical and pancreatic cancer cell lines are unaffected by depletion of either microRNAs.
In accord with previous work, RNA levels of STAT3 (Iliopoulos et al., 2009) (Bromberg et al., 1999; Grivennikov et al., 2009), miR-21, and miR-181b-1 (Schetter et al., 2008; Yan et al., 2008; Cervigne et al., 2009) are often increased in cancer tissues relative to normal tissues. More importantly, we observe a striking direct relationship between STAT3, miR-21, and miR-181b-1 levels in a set of colon adenocarcinomas, strongly arguing for a mechanistic relationship. Furthermore, in this same set of tumor samples, there is a striking inverse relationship between miR-21 and PTEN as well as between miR-181b-1 and CYLD. These observations indicate that the pathway in which STAT3 directly activates miR-21 and miR-181b-1, which in turn inhibit PTEN and CYLD, is relevant for at least some forms of human cancer.
STAT3 Activation of miR-21 and miR-181b-1, via PTEN and CYLD, is Part of the Epigenetic Switch from Non-transformed to Transformed Cells
In the ER-Src transformation model, a transient inflammatory signal (mediated by Src) activates a positive feedback loop involving NF-κB, Lin28, Let-7 microRNAs, and IL6 that is responsible for an epigenetic switch between stable non-transformed and transformed states (Iliopoulos et al., 2009). The epigenetic switch, and hence transformation, can be started by the appropriate manipulation of any component within the loop. In addition, IL6 is required for activating the STAT3 pathway at later times during transformation, and we imagined that this function and the STAT3 pathway itself was required for transformation, but not part of the positive feedback loop necessary for the epigenetic switch (Iliopoulos et al., 2009).
In contrast to this view, our observation that overexpression of miR-21 or miR-181b-1 is sufficient for transformation strongly argues that STAT3 and these microRNAs not simply downstream effectors, but are actually part of the regulatory circuit that constitutes the epigenetic switch (Figure 7G). To put it differently, miR-21 and miR-181b-1 are not merely up-regulated in association with oncogenesis, but rather can act as oncogenic microRNAs when transiently overexpressed. Importantly, transformation by these transiently transfected microRNAs requires NF-κB and Lin28, and the transformed state is sufficiently stable for formation of colonies in soft agar and mammospheres, even though the introduced microRNAs are unlikely to be present. To achieve such a stable transformed state, STAT3 and its microRNA targets must feedback to induce NF-κB and start the inflammatory feedback loop. At a minimum, miR-21 directly targets PTEN, which acts through the Akt pathway, and miR-181b-1 targets CYLD, and each of these microRNA pathways is sufficient to induce NF-κB. Thus, STAT3 activation of miR-21 and miR-181b-1 induces two positive feedback loops that are part of the larger inflammatory feedback loop that constitutes the epigenetic change from non-transformed to transformed cells.
More generally, our results suggest that the link of inflammation to cancer is not a simple linear pathway, but rather a complex, self-reinforcing regulatory circuit. In this sense, the distinction between non-transformed and transformed cells is analogous to a switch in normal development in which a given stable state is converted to a different stable state. As discussed previously (Iliopoulos et al., 2009), activation of this inflammatory regulatory circuit is insufficient to trigger transformation of normal cells. Instead, this inflammatory circuit is likely to be relevant in “pre-disposed” cells that are genetically altered to be at an intermediate stage in the transition between a primary cell and a cancer cell. In such predisposed cells, anything that activates the positive factors (NF-κB, Lin28, IL6, STAT3, miR-21, mir-181b-1) or inhibits the negative factors (Let-7, PTEN, CYLD) will trigger the inflammatory feedback loop that induces and maintains the transformed state. As such, the triggering event does not have to be an inflammatory signal (e.g. Src) per se, but can be any genetic or environmental change that affects the activity of the factors in the inflammatory feedback loop.
MATERIALS AND METHODS
Cell Culture and Transformation Assays
MCF10A cells containing the ER-Src fusion protein were grown in DMEM/F12 medium supplemented with 5% donor horse serum (HS), 20ng/ml epidermal growth factor (EGF), 10 μg/ml insulin, 100 μg/ml hydrocortisone, 1 ng/ml cholera toxin, 50 units/ml pen/step, with the addition of puromycin (Iliopoulos et al., 2009; Hirsch et al., 2010). To induce transformation, the Src oncogene was activated by the addition of 1 μM Tamoxifen (Sigma) to confluent cell cultures. Cancer cell lines (HCT116, HT29, PC3, A549, HeLa, Hep3B, Panc1) were grown in DMEM, 10% FBS and pen/step.
MicroRNA Expression Analysis
Expression levels of 365 microRNAs at various times after TAM addition were evaluated with microRNA profiling assays (TLDA human miRNA v1.0) in the Dana Farber Molecular Diagnostics Facility. Validation of these results was performed using the mirVana qRT–PCR miRNA Detection Kit and qRT–PCR Primer Sets, according to the manufacturer’s instructions (Ambion Inc, TX, USA). RNU48 expression was used as an internal control.
Morphological, Soft Agar Colony, and Invasion Assays for Cellular Transformation
ER-Src cells were treated with 100 nM of microRNAs or antisense microRNAs for 24h, whereupon TAM was added and cellular transformation was assessed 36h later (total time 60h). Cell morphology was assessed by phase contrast microscopy (10X objective), and percentage of transformed ER-Src cells was calculated by evaluation of cell morphology by Metamorph v5.0 software. The soft agar colony and MATRIGEL invasion assays were performed as described previously (Iliopoulos et al., 2009; Hirsch et al., 2010). In all cases, experiments were repeated thrice, and the statistical significance was calculated using Student’s t test.
Lever and PhylCRM Analysis
The Lever and PhylCRM algorithms were described previously (Warner et al., 2008). MicroRNA promoters to be evaluated by Lever for each pair of transcription factor binding site motifs were rank-ordered according to their maximum window PhylCRM score. In PhylCRM analysis, we evaluated evaluated evolutionary conservation across 12 mammals: mouse, rat, human, rabbit, chimp, macaque, cow, dog, armadillo, tenrec, opossum and elephant using the MultiZ genome alignment as described in Supplementary Methods. PBM data were for 104 TFs (transcription factors) from 22 DNA-binding domain structural classes (Badis et al., 2009a) and for 168 additional homeodomains (Berger et al., 2008). Position weight matrix (PWM) representations of the PBM data derived using the “Seed-and-Wobble” algorithm (Berger et al., 2006) were used in the Lever analysis. The TRANSFAC PWMs were downloaded from the TRANSFAC human database. To annotate the TSS for the miRNAs, we incorporated the findings from (Ozsolak et al., 2008) and (Marson et al., 2008), and mapped the genome coordinates to the hg18 version of the human genome by using liftOver program from UCSC. We note that some of the identified promoters were more than 50 kb from the miRNA gene. For microRNAs whose predicted TSS locations differed substantially in the two studies, we used the ones identified by (Ozsolak et al., 2008). For each miRNA gene, the Lever and PhylCRM analyses were applied to the sequence from 2kb upstream to 7kb downstream of the annotated TSS. See Supplementary Methods for details.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was carried out as described previously (Yang et al., 2006). Briefly, the chromatin fragments, derived from untreated and TAM-treated (36h) MCF10A ER-Src cells, were immunoprecipitated with 6 μg of antibody against STAT3 (9139, Cell signaling), Myc (ab56, Abcam Inc), Max (ab53570, Abcam Inc) and RXRα (sc553, Santa Cruz Biotechnology Inc). DNA extraction was performed using Qiagen Purification Kit. The samples were analyzed by quantitative PCR in real time using primers listed in supplemental methods.
RNA Analysis
Equal amounts of purified RNA samples from untransformed and transformed MCF10A ER-Src cells were reverse-transcribed to form cDNA, which was subjected to SYBR Green based real-time PCR analysis as described previously (Iliopoulos et al., 2009; Hirsch et al., 2010). To analyze patient samples, RNAs from 21 colon adenocarcinomas (Origene and Biochain Inc) were analyzed for levels of STAT3, miR-21 and miR-181b-1. Each sample was run in triplicate and the data represent the mean ± SD. Correlation coefficient represents correlation between STAT3 and miR-21 or miR-181b-1 expression levels in these colon adenocarcinomas.
siRNA Experiments
MCF10A ER-Src cells were seeded in 6-well plates and were transfected with different siRNAs (100nM) from Ambion Inc. against STAT3 (s23967 and s23969), CYLD (s590), Myc (s9129 and s9130), Max (s8538 and s224031) and RXRα (s12385 and s12386) using siPORT NeoFX transfection agent. Transfection with 100 nM siRNA control (AM4611, Ambion Inc) was used as a control. No cell toxicity was detected due to the transfection agent. The resulting cells were analyzed for transformation or RNA levels as described above.
Xenograft Experiments
5×106 MCF10A-IL6 transformed cells or HCT-116 and HT-29 colon cancer cells were injected subcutaneously in the right flank of athymic nude mice (Charles River Laboratories). Tumor growth was monitored every five days and tumor volumes were calculated by the equation V(mm3)=axb2/2, where a is the largest diameter and b is the perpendicular diameter. When the tumors reached a size of ~65mm3 mice were randomly distributed in 6 groups (4 mice/group) and treated intraperitoneally with Ab-IgG (2ug/ml), Ab-IL6 (2ug/ml), siNC (100nM), siSTAT3 (100nM), as-miR NC (120nM), as-miR-21 (120nM), as-miR-181b-1 (120nM), or combinations. These treatments were repeated in a weekly basis for 4 cycles (days 10, 15, 20, 25). In addition, RNA was extracted from tumors treated with Ab-IgG, Ab-IL6, siNC and siSTAT3. Real-time PCR analysis was performed for miR-21 and miR-181b-1. Nude mice were maintained in accordance with Tufts Institutional Animal Care and Use Committee procedures and guidelines.
Supplementary Material
Acknowledgments
We thank Philip N. Tsichlis for providing facilities for performing the xenograft experiments and George Q. Daley for Lin28B expression vector. This work was supported by grants from the National Institutes of Health to M.L.B. (HG 3985) and K.S. (CA 107486).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009a;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009b doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Cervigne NK, Reis PP, Machado J, Sadikovic B, Bradley G, Galloni NN, Pintilie M, Jurisica I, Gilbert R, Gullane P, et al. Identification of a microRNA signature associated with progression of leukoplakia to oral carcinoma. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp446. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti A, Aguennouz M, La Torre D, Tomasello C, Cardali S, Angileri FF, Maio F, Cama A, Germano A, Vita G, et al. miR-21 and 221 upregulation and miR-181b downregulation in human grade II–IV astrocytic tumors. J Neurooncol. 2009;93:325–332. doi: 10.1007/s11060-009-9797-4. [DOI] [PubMed] [Google Scholar]
- Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Garcia MG, Alaniz LD, Cordo Russo RI, Alvarez E, Hajos SE. PI3K/Akt inhibition modulates multidrug resistance and activates NF-kappaB in murine lymphoma cell lines. Leuk Res. 2009;33:288–296. doi: 10.1016/j.leukres.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigon CJ, Zhao L, Willingham MC, Cheng SY. PTEN deficiency accelerates tumour progression in a mouse model of thyroid cancer. Oncogene. 2009;28:509–517. doi: 10.1038/onc.2008.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Joshi A, Zhang Y, Jaeger SA, Bulyk M, Tsichlis PN, Liu XS, Struhl K. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17:348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-κB, lin 28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28:3157–3170. doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-myc-regulated micrornas modulate e2f1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Cheng Z, Zhang J, Li R, Zhao P, Fu Z, You Y. hsa-mir-181a and hsa-mir-181b function as tumor suppressors in human glioma cells. Brain Res. 2008;1236:185–193. doi: 10.1016/j.brainres.2008.07.085. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs’ target genes and functions. Nat Methods. 2008;5:347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, Burchard J, Dai X, Chang AN, Diaz RL, et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.