Abstract
Ultraspiracle (USP) is the invertebrate homologue of the mammalian retinoid X receptor (RXR). RXR plays a uniquely important role in differentiation, development, and homeostasis through its ability to serve as a heterodimeric partner to many other nuclear receptors. RXR is able to influence the activity of its partner receptors through the action of the ligand 9-cis retinoic acid. In contrast to RXR, USP has no known high-affinity ligand and is thought to be a silent component in the heterodimeric complex with partner receptors such as the ecdysone receptor. Here we report the 2.4-Å crystal structure of the USP ligand-binding domain. The structure shows that a conserved sequence motif found in dipteran and lepidopteran USPs, but not in mammalian RXRs, serves to lock USP in an inactive conformation. It also shows that USP has a large hydrophobic cavity, implying that there is almost certainly a natural ligand for USP. This cavity is larger than that seen previously for most other nuclear receptors. Intriguingly, this cavity has partial occupancy by a bound lipid, which is likely to resemble the natural ligand for USP.
Ultraspiracle (USP) was identified first as a transcriptional regulator in Drosophila (1). Since then, USP has been cloned from a variety of arthropods (2). Sequence and functional homology suggest that USP is the homologue of the mammalian retinoid X receptor (RXR), a member of the family of nuclear hormone receptors (3). Interestingly no USP–RXR homologue has been identified in the Caenorhabditis elegans genome (4). Like RXR, USP forms heterodimers with other nuclear receptors (5, 6). The best understood role of USP in Drosophila is to act as a heterodimeric partner to the ecdysone receptor that controls molting in response to the hormone 20-hydroxy ecdysone (7–9). In this context, it seems that USP enhances the affinity of the ecdysone receptor for its ligand (8). However, the mechanism by which this process is achieved is unclear. Unlike its mammalian counterpart, USP has no known high-affinity ligand, and it has been suggested that either this function has been lost in evolution (2) or RXRs have gained a ligand-binding activity lacked by the ancestral USP–RXR (10, 11). Other studies have suggested that USP might be a receptor for juvenile hormone (JH), a family of epoxymethylfarnesoate compounds (12, 13). This suggestion is based on the chemical similarity of JH to retinoic acid as well as direct binding and oligomerization assays. However, the reported binding affinity of USP for JH is rather weak. Furthermore, there are no reports that JH influences the transcriptional activity of USP. Thus the question of whether there is a natural ligand for USP remains open.
Structural studies of a number of nuclear receptor ligand-binding domains (LBDs) have yielded a relatively simple model for the mechanism by which ligands activate nuclear receptors. The LBDs of nuclear receptors share a common, predominantly helical, fold (14, 15). A cavity accommodates ligand in the core of the LBD. The short C-terminal helix of the LBD (termed helix 12 or AF2 helix) forms a cap covering this cavity and in most receptors contacts the ligand directly. The critical switch mechanism seems to reside in the position and dynamics of this C-terminal helix, which controls the ability of the receptor to bind coactivator proteins. Coactivator proteins are recruited to nuclear receptors through a conserved amphipathic interaction motif LxxLL (16). After binding to the receptor, this motif adopts a helical structure that interacts in a hydrophobic groove (17–19). A conserved glutamic acid residue in helix 12 and a conserved lysine form a “charge clamp” that interacts with the dipole at either end of the coactivator helix. Significantly, ligands that do not activate nuclear receptors (competitive antagonists) seem to exert their effect by displacing helix 12 from its “active” position so that it occupies the coactivator binding site, the so-called antagonist conformation, and thus prevents coactivator binding (19, 20).
To gain further insight into the structural, functional, and evolutionary relationship between USP and RXR and their ligand-binding ability, we have determined the 2.4-Å crystal structure of the USP LBD from Drosophila melanogaster (dmUSP). The structure reveals several surprising features. First, helix 12 occupies a position reminiscent of the antagonist-bound estrogen receptor (19, 20). This position is caused by an unusual conformation of the loop between helices 1 and 3. This conformation is a result of a conserved sequence motif characteristic of dipteran and lepidopteran USPs. The structure also shows that USP has an unusually large ligand-binding cavity, strongly suggesting that it is likely to have a natural ligand. The large size of the cavity is the result of the H1–H3 loop that influences the orientation of helix 3, so as to open a channel to the surface of the protein. Unexpectedly, we find that this large ligand-binding cavity has partial occupancy by a bound lipid molecule.
Materials and Methods
Purification and Crystallization.
dmUSP LBD was expressed with an N-terminal 6-His tag in Escherichia coli [strain C41(DE3)] by using an isopropyl β-D-thiogalactoside-inducible pTac promoter. The protein was purified by using Ni-nitrilotriacetic acid affinity resin (Qiagen, Chatsworth, CA) and gel filtration using a Sephacryl S100 resin (Amersham Pharmacia). The monomeric homogeneous dmUSP LBD was concentrated to 10 mg/ml for crystallization. Crystals were grown at 18°C by using standard vapor-diffusion techniques with well buffer containing 130 mM ammonium sulfate, 160 mM sodium acetate (pH 4.6), and 25% polyethylene glycol (PEG) 2000. Larger crystals (maximum dimensions of 150 × 75 × 20 μm) were obtained by using streak-seeding techniques and by addition of ethylene glycol to the drop. After transfer to cryoprotectant (13 mM Tris, pH 7.0/32.5 mM sodium chloride/17.5 mM sodium acetate, pH 4.6/19.5 mM ammonium sulfate/2.5% PEG2000/30% glycerol), crystals were frozen in a stream of N2 at 100 K.
Data Collection and Structure Solution.
Data collection and refinement statistics are given in Table 1. Although the crystals diffract to beyond 2 Å, data were useful only to 2.4 Å because of poor data quality at higher resolution as a consequence of poor spot shape arising from crystal bending during freezing. The structure was solved by using the estrogen receptor LBD as a search model in the cns program suite (21). Iterative rounds of positional simulated annealing and B factor refinement were carried out by using CNS along with rounds of manual rebuilding in the molecular modelling program o (22).
Table 1.
Crystallographic data and refinement statistics
| X-ray Source/wavelength | SRS PX9.6/0.87 Å |
| Resolution (outer shell) | 34–2.4 (2.6–2.4) Å |
| Total/unique reflections | 175324 (60384) |
| Redundancy/completeness | 2.9/90% |
| Rmerge (outer shell) | 12.6% (30.5%) |
| I/σ (outer shell) | 4.4 (2.1) |
| Space group | P1 a = 37.98, b = 86.19, c = 137.00 |
| α = 85.57, β = 85.94, γ = 83.09 | |
| Rcryst/Rfree (5%) | 24.7%/28.4% |
| RMSD bonds/angles | 0.008 Å/1.2° |
| Average B factor | 35.1 Å2 |
| Ramachandran | |
| Favored/additional/generous | 92%/7.3%/0.7% |
Rmerge = Σ|I−〈I〉|/Σ〈I〉; Rcryst/Rfree = Σ|Fobs−Fcalc|/ΣFobs. RMSD, rms deviations from ideal values.
TLC.
Protein samples (typically 4 mg) and lipid standards were extracted with chloroform/methanol [2:1 (vol/vol)]. Samples were analyzed by using 0.25-mm TLC plates precoated with silica gel 60. Two TLC solvents were used [50:50 and 95:5 chloroform/methanol (vol/vol)] according to the run. Plates were dried and stained with iodine to visualize lipid.
Matrix-Assisted Laser Desorption Ionization (MALDI)/Time-of-Flight (TOF) Mass Spectrometry.
Lipid was extracted from the protein samples with chloroform/methanol [2:1 (vol/vol)]. The matrix used for all samples was 1 or 5 mg/ml α-cyano-4-hydroxy-trans-cinnamic acid in 60% aqueous acetonitrile with 0.1% trifluoroacetic acid. The matrix (0.5 μl) mixed with 0.6 μl of the lipid extract was applied directly to the sample plate and allowed to dry at ambient temperature. MALDI/TOF spectra were acquired on a Voyager-DE STR mass spectrometer (PerSeptive Biosystems, Framingham, MA) and calibrated with respect to standards deposited adjacent to each sample.
MALDI/Postsource Decay (PSD) was used to confirm the assignment of molecular-ion-peak 563.5 as diacylglycerol. The 563.5 PSD spectrum was compared with spectra of known lipid standards. The fragmentation pattern was similar to dimyristoyl (C14:0) glycerol + Na and was assigned tentatively as 1-palmitoyl-2-myristoyl-glycerol + Na.
Results
Structure Determination.
The dmUSP LBD (residues 230–508) was expressed and purified from E. coli. Diffraction-quality crystals were grown as described in Materials and Methods. The structure of the dmUSP LBD was solved by using molecular-replacement techniques. Initial attempts at determining the structure, by using either the apo or holo Homo sapiens RXRα (hRXRα) structure as a search model, were unsuccessful (23, 24). However, because efforts to prepare heavy atom derivatives also were unsuccessful, alternative molecular-replacement models were tested. Unexpectedly, a solution was obtained by using the liganded Homo sapiens estrogen receptor-α (holo hERα; ref. 20) as a search model.
The dmUSP LBD is packed in the P1 crystal lattice as two interdigitated trimers. Consequently, the unit cell contains six copies of the LBD, each in a unique environment within the crystal lattice. An excellent initial electron-density map was obtained by averaging according to the six-fold noncrystallographic symmetry. Strict averaging was maintained in the early stages of refinement. Subsequently, the core LBD structure was restrained according to the noncrystallographic symmetry so as to optimize the refinement.
Overall Structure.
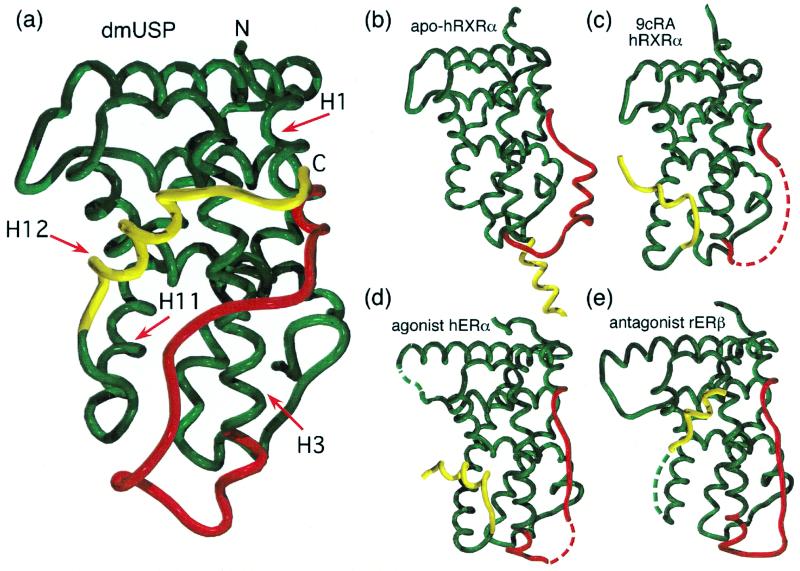
The LBD of dmUSP has a similar overall fold to other members of the nuclear hormone receptor family (Fig. 1a) and resembles a three-layered helical sandwich. To facilitate comparison with previous structures, the nomenclature used here is based on the apo-hRXRα structure (23). Helices 1 and 3 comprise one outer layer. Helices 4, 5, 8, and 9 form the central layer. Helices 7 and 10/11 form the other outer layer. The central layer is incomplete, leaving a cavity below helices 4 and 5 between helices 3, 7, and 10/11. Acting as β-sheet S1–S2 (between helices 5 and 6), helix 6 and helix 12 serve to seal off the edges of the cavity. dmUSP contains a glycine-rich insertion following helix 5. This region generally is disordered in the structure and 25 amino acids () could not be modeled. Similarly, residues 230–237 and 503–508 at the termini were not seen in the electron-density maps.
Figure 1.
The structure of USP compared with other nuclear receptors. The loop between helices 1 and 3 (red) and helix 12 (yellow) in dmUSP differ from other receptors.
In terms of sequence (and functional) similarity, dmUSP LBD is related most closely to the mammalian RXR. Alignment with hRXRα indicates 70% similarity (42% identity). Superposition of the dmUSP structure with those of the apo and holo forms of hRXRα (Fig. 1 b and c) give rms-deviation (RMSD) values of 1.33 and 1.22 Å (with 169 and 164 matched Cα atoms, respectively). Superposition of dmUSP on the hERα structure gives an RMSD of 1.12 Å with 172 matched Cα atoms. Thus, despite the lower sequence similarity of 58% (23% identity), the dmUSP structure is more similar to the holo hERα structure than the hRXRα structures, particularly in terms of the orientations/conformations of helices 1, 3, and 4.
USP Is Locked Into an Inactive Conformation.
Two regions of the dmUSP structure differ significantly from both the hRXRα and holo hERα structures: the loop joining helices 1 and 3; and helix 12 (shown in red and yellow in Fig. 1). This H1–H3 loop adopts an unusual position in dmUSP not seen in any previous nuclear receptor structure. It wraps over the top of helix 3 and lies between helices 3 and 11. In most of the nuclear receptor structures [e.g., apo-hRXRα, holo-H. sapiens peroxisome proliferator-activated receptor γ (18), and holo-H. sapiens thyroid receptor β (25)], this loop passes outside the β-strand (Fig. 1b, Right). An exception is the hERα structure in which the loop passes between the β-strand and helix 3 (Fig. 1 d and e).
The position of this loop in dmUSP is significant because it prevents helix 12 from adopting the position seen in either the unliganded or 9-cis retinoic acid (9cRA)-bound structures of hRXRα (Fig. 1 b and c). As a consequence, helix 12 adopts a position that closely resembles that seen in the structure of estrogen receptor bound to an antagonist (Fig. 1e; refs. 19 and 20). However, in contrast to estrogen receptor, helix 12 is locked firmly in this inactive position by making extensive contacts to the H1–H3 loop (Fig. 2 a and b).
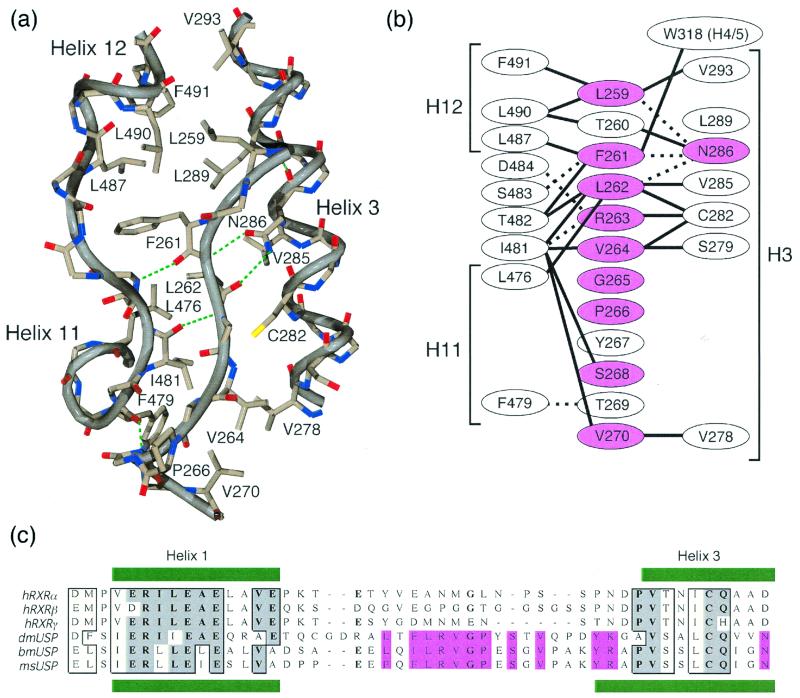
Figure 2.
The conserved H1–H3 loop locks USP in an inactive conformation. (a) Interactions between the H1–H3 loop and adjacent parts of the structure. Green dashes indicate hydrogen bonds. (b) Schematic view of the interactions illustrated in a. Solid and dashed lines indicate nonpolar van der Waals interactions and hydrogen bonds, respectively. Residues conserved in all dipteran and lepidopteran USPs are colored pink. (c) Sequence alignment of dipteran/lepidopteran USPs and mammalian RXRs. Residues conserved in the holometabolous insect orders (pink) correspond to those residues highlighted in b. dm, Drosophila melanogaster; bm, Bombyx mori; ms, Manduca sexta.
Sequence comparison of the various USP proteins reveals a sequence motif within the H1–H3 loop that is conserved in both diptera (flies) and lepidoptera (moths) USP proteins but not in hymenoptera (honey bee, GenBank accession no. AF263459), orthoptera (migratory locust; ref. 2), or other arthropods such as crab and tick (highlighted in Fig. 2c; refs. 26 and 27). The structure shows that every residue within this conserved motif plays an architectural role in mediating interactions with helices 3, 4/5, 11, and 12 as well as the loop between helices 11 and 12.
This sequence conservation and extensive pattern of interactions clearly indicate that the position of the H1–H3 loop and of helix 12 are bona fide features of the structure that are not the result of crystal-packing interactions. This finding is supported further by the evidence that all six copies of the dmUSP in the crystal have identical conformations in this respect.
An Unexpected Ligand for Ultraspiracle.
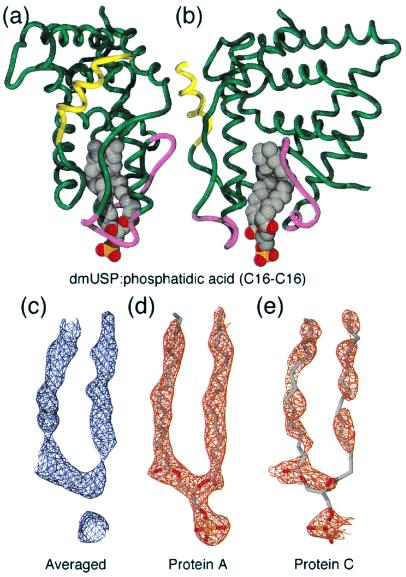
One of the goals of determining the structure of USP was to ascertain whether it has a ligand-binding cavity and hence whether its activity is likely to be regulated by ligand. The structure reveals that dmUSP has an unusually large hydrophobic cavity bounded by helices 3, 5, 6, 7, and 11 as well as the β-sheet S1–S2 (Fig. 3a). Furthermore, in the earliest electron-density maps (averaged according to the noncrystallographic symmetry) it was apparent that this cavity contained additional electron density that could not be accounted for by the protein (Fig. 3c). This density resembled a lipid molecule with two fatty acyl chains and a relatively electron-dense phosphate head group.
Figure 3.
An unexpected ligand for USP. (a and b) Orthogonal views of phosphatidic acid bound to dmUSP. Helix 12 is colored yellow. Magenta loops indicate mobile regions in some LBDs (see text) (c–e) 2Fo − Fc electron-density maps of the ligand before and after refinement (contoured at 1σ).
TLC of the purified USP confirmed the presence of several classes of lipid exhibiting different mobilities in the various solvents assayed. The more polar species migrates in a similar position to that of phosphatidyl glycerol, and the less polar species shows mobility consistent with diacylglycerol.
MALDI/time-of-flight mass spectra also suggested that there was a mixture of lipids bound to the protein. The observed masses are consistent with diacylglycerol, phosphatidic acid, and phosphatidyl ethanolamine with various acyl chain lengths and oxidation states. One intense peak that was seen in all samples tested (but not in the negative controls) correlates with a mass of 563.5 Da, which is consistent with diacylglycerol (C14:0/C16:0). MALDI/postsource-decay fragmentation of this species supported this assignment.
In conclusion, it seems that a mixture of lipid ligands is bound to the dmUSP LBD including diacylglycerol and various phospholipids. The most common lipids in E. coli are phosphatidyl ethanolamine, phosphatidyl glycerol, and cardiolipin (28) with acyl chains of C16:0 (43%), C16:1 (33%), and C18:1 (24%) (29). To model this mixture of lipids best, phosphatidic acid with C16 acyl chains was built into the averaged electron density (Fig. 3 a and b) and included in subsequent refinement.
Lipid Binding Affects the Dynamics of USP.
During the later stages of the refinement, it became apparent that the occupancy of the lipid is markedly lower in three of the six LBDs. This difference is clear in the 2Fo − Fc electron-density maps (Fig. 3 d and e). The differences in ligand occupancy can be explained by differences in the environment of the six LBDs in the crystal. Significantly, the lower ligand occupancy is not correlated with any major structural differences in the LBDs. The only notable difference is that two regions of the structure (magenta in Fig. 3 a and b) exhibit elevated temperature factors in those LBDs with reduced ligand occupancy (62 vs. 39 Å2 and 50 vs. 34 Å2). The first region comprises the turn before helix 3 (residues 266–275) and the second region comprises part of the β-sheet and helix 6 (residues 367–379).
These regions form a gateway to the ligand-binding pocket. One might expect therefore that they are mobile in an unliganded LBD so as to facilitate the entry of a rather large ligand. Once the ligand is bound, this gateway is stabilized by hydrophobic side chains packing against the lipid. The change in mobility of these two regions would seem to be the only difference between the apo and holo forms of the LBD.
An Unusually Large Ligand-Binding Cavity.
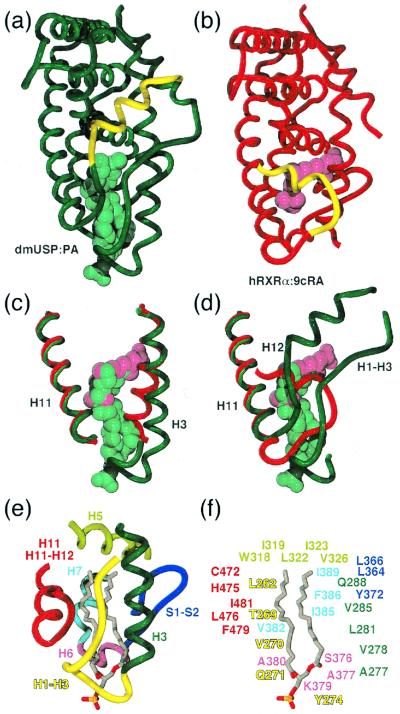
Fig. 4 a and b shows a comparison of dmUSP bound to phosphatidic acid and hRXRα bound to 9cRA. The position of the ligand in dmUSP is notably different from that in hRXRα, and that in the ligand-binding cavity of dmUSP is far larger. These differences are the result of several concerted changes in the protein structure. In hRXRα, the amino terminus of helix 3 is curved around the ligand and in toward the main body of the receptor (Fig. 4c). In dmUSP, helix 3 is straighter and has an additional turn at the amino terminus that serves to open a channel from the core of the dmUSP ligand-binding pocket to the outside of the protein.
Figure 4.
An unusual ligand-binding cavity. (a and b) dmUSP (green) compared with hRXRα (red). 9cRA (magenta) is smaller than the phosphatidic acid (cyan). Helix 12 is colored yellow. Compared with hRXRα:9cRA, helix 3 is splayed out of the ligand-binding cavity (c), and helix 12 is displaced by the H1–H3 loop (d). (e) Regions of the LBD contributing to the ligand-binding cavity: H1–H3 (yellow), H3 (dark green), H5 (light green), S1–S2 (blue), H6 (magenta), H7 (cyan), and H11 (red). (f) Side chains in contact with ligand; coloring is as in e.
Whereas helix 3 is one-turn longer in dmUSP, helix 11 is shorter by one turn (Fig. 4d). The loop after helix 11 adopts an extended conformation making several backbone hydrogen bonds and many nonpolar contacts with the H1–H3 loop. Together, the H11–H12 and H1–H3 loops form a wall on one side of the channel. On the other side, the cavity is substantially open to solvent between helix 3 and the β-sheet (see Fig. 3b).
The ligand-binding cavity in dmUSP is formed from four regions of the protein (Fig. 4e): the H1–H3 loop and helix 3; helix 5; the β-sheet S1–S2 along with helices 6 and 7; and also helix 11 and the H11–H12 loop. The interior surface of the ligand-binding cavity is almost entirely nonpolar. The only polar contact with the ligand is a hydrogen bond between the amide of Q271 and a carbonyl of the lipid. After binding ligand, 31 residues become shielded from solvent (Fig. 4f). Despite this, the cavity is not filled completely by the ligand, and there remains a small unoccupied volume adjacent to helix 3.
The residues that surround the 9cRA in the holo hRXRα ligand-binding cavity are essentially all conserved in dmUSP (in size and character if not identity) with the exception of the arginine that forms a salt bridge with the carboxyl of the retinoic acid. In dmUSP, this arginine is a cysteine, and because of its shorter side chain it does not participate in the ligand-binding pocket. It is potentially possible that USP could bind 9cRA if this cysteine were mutated to an arginine, although the enlarged ligand-binding cavity would be far from filled.
Discussion
A large number of nuclear hormone receptors are called “orphan receptors,” because a cognate ligand has yet to be identified (30, 31). It seems likely that many of these orphan receptors do not have cognate ligands and that they are constitutive activators or repressors of transcription. Indeed it has been proposed that the ligand-binding ability of nuclear receptors has been acquired during evolution and that the ancestral nuclear receptor was an orphan receptor (10, 11). Furthermore, it has been suggested that the RXR/USP family of receptors represents a good example of this development, because the RXRs seem to have acquired an ability to bind retinoids, which is an ability that is lacked by USPs (10). This idea however is not accepted universally, and other proposals suggest that USPs have either lost ligand-binding activity or have changed ligand specificity during evolution (2).
To understand more about the function of USP and how it differs from mammalian RXRs, we have determined the structure of the dmUSP LBD expressed and purified from E. coli. Remarkably, it seems that just a few critical residues in the H1–H3 loop modulate the structure, and hence activity, of the USP in a very significant fashion. It is interesting also that these residues are conserved in the higher holometabolous insect orders of diptera and lepidoptera but not in the hemimetabolous insect orders or other arthropods. Previous analyses have suggested also that the dipteran and lepidopteran USPs form a group distinct from other arthropod USPs that are more similar to vertebrate RXRs (2). This finding suggests that the H1–H3 loop is an evolutionary acquisition specific to the dipteran and lepidopteran insect orders and gives a remarkable insight into the mechanisms of evolution whereby residues in the H1–H3 loop have gained a new structural and functional role.
The finding that USP is bound to a lipid ligand raises several questions. What is the natural ligand for USP? Is the “antagonist” conformation a consequence of the ligand? What might be the role of the natural ligand?
At first sight, it would seem unlikely that the lipid bound to the bacterially expressed USP is the authentic ligand for USP, especially because it seems that the USP is bound to a mixture of different lipids. However, the hydrophobic cavity revealed in the structure implies that the USP is very likely to have a natural ligand in Drosophila. This implication raises the question: What might this ligand be? Importantly, there is only one polar side chain, Q288, that is in a position to interact with the ligand and suggests that the ligand would need to be largely nonpolar in character. It has been suggested that JH might serve as a ligand for USP (13). Although this may make biological sense, such a small ligand would leave most of the cavity empty and therefore does not seem to be the most likely possibility. Recently it has been reported that RXR is activated in vivo by docosahexaenoic acid (a long chain fatty acid; ref. 32). Furthermore, a mutant mouse RXR also has been observed to bind oleic acid and is presumed to be activated by this ligand (33). Could the natural USP ligand be a fatty acid? USP does not have the basic arginine needed to interact with the carboxyl group, and therefore a fatty acid ligand for USP is perhaps not so likely. On balance, it seems that a diacylglycerol-based ligand is well suited to binding in the cavity seen in the USP structure. It is striking that the protein has retained the ligand through multiple chromatographic purifications, suggesting that the dissociation constant, or at least the off rate, must be very low. Further insight into the natural ligand or ligands will require investigation beyond the scope of this present study.
The observation that the crystal lattice contains three LBDs with significantly lower ligand occupancy, which does not correlate with any changes in the conformation of helix 12 or the H1–H3 loop, strongly suggests that lipid binding has little effect on the overall structure of the USP LBD. Of course it is not possible to exclude the possibility that a cognate ligand might cause significant structural perturbation. However, the large number of backbone–backbone and side-chain–side-chain contacts between H12, H1–H3, and H3 would not be consistent with a substantial rearrangement after binding any ligand.
Because it seems that the USP is locked into an antagonized conformation irrespective of bound ligand, a role for a USP ligand is not immediately obvious. As discussed above, the consensus view is that nuclear receptor ligands cause dissociation of any bound corepressor and facilitate binding of LxxLL-containing coactivators (34). This effect is achieved by modulating the position and dynamics of helix 12. Clearly this mechanism cannot operate in USP, because not only would the position of helix 12 prevent binding of LxxLL-containing coactivators but also because ligand binding does not influence the position of helix 12. These conclusions are consistent with the fact that no direct-activation activity of USP has been reported so far and that indeed coactivators of the p160 family have not been identified in Drosophila.
There are however several possible mechanisms through which a dmUSP ligand might regulate transcription independently of helix 12 and p160 coactivators. Such mechanisms require that a cofactor protein recognize a region on the surface of USP that is different from the canonical LxxLL-binding pocket. If this surface was to involve the exposed portions of the ligand or a region of which stability is influenced by ligand (e.g., the regions shown in magenta in Fig. 3 a and b), then the surface could serve readily to distinguish liganded from unliganded receptor. In principle, such a cofactor could be one of a range of proteins including different coactivators or corepressors, components of the general transcriptional machinery, or even a partner receptor. It is even possible that ligand binding influences the half-life of the receptor. Indeed, there is precedent for receptors interacting with the basal transcriptional machinery (35); for ligand binding influencing receptor half-life (36, 37) and for communication between heterodimeric partners (38). In particular it has been shown that the ecdysone receptor absolutely requires its USP partner for activity (8). Understanding whether and how USP ligands regulate transcription remains to be established.
Evidently this structure does not provide all of the answers concerning the mechanism of USP action. However, it seems clear that USP almost certainly must have a ligand, but the mode of action of that ligand may well differ from the canonical model. The structure also gives a remarkable view of the mechanisms through which protein structure and function can be changed through evolution.
As this manuscript was being sent for review, the crystal structure of the USP LBD from the lepidoptera Heliothis virecens was reported (39). This structure shows many of the same features as the structure described here, including the antagonist conformation of helix 12 and the bound lipid. However, in the H. virecens crystals, there is only one LBD in the asymmetric unit and it is fully occupied by lipid. Accordingly, it is suggested that the antagonist conformation may be a consequence of the bound lipid, and the authors are unable to discuss the nature of unliganded USP.
Acknowledgments
We thank the following for help, advice, and encouragement: Ester Banayo, Tony Crowther, Louise Fairall, Matthew Freeman, John Gooch, Chuan Li, James Love, Laszlo Nagy, Thomas Perlmann, Daniela Rhodes, and Brian Wimberly.
Abbreviations
- USP
ultraspiracle
- RXR
retinoid X receptor
- JH
juvenile hormone
- LBD
ligand-binding domain
- dmUSP
Drosophila melanogaster USP
- MALDI
matrix-assisted laser desorption ionization
- hRXRα
Homo sapiens RXRα
- hERα
Homo sapiens estrogen receptor-α
- 9cRA
9-cis retinoic acid
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1hg4).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.041611298.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.041611298
References
- 1.Oro A E, McKeown M, Evans R M. Nature (London) 1990;347:298–301. doi: 10.1038/347298a0. [DOI] [PubMed] [Google Scholar]
- 2.Hayward D C, Bastiani M J, Trueman J W, Truman J W, Riddiford L M, Ball E E. Dev Genes Evol. 1999;209:564–571. doi: 10.1007/s004270050290. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sluder A E, Mathews S W, Hough D, Yin V P, Maina C V. Genome Res. 1999;9:103–120. [PubMed] [Google Scholar]
- 5.Yao T P, Segraves W A, Oro A E, McKeown M, Evans R M. Cell. 1992;71:63–72. doi: 10.1016/0092-8674(92)90266-f. [DOI] [PubMed] [Google Scholar]
- 6.Sutherland J D, Kozlova T, Tzertzinis G, Kafatos F C. Proc Natl Acad Sci USA. 1995;92:7966–7970. doi: 10.1073/pnas.92.17.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas H E, Stunnenberg H G, Stewart A F. Nature (London) 1993;362:471–475. doi: 10.1038/362471a0. [DOI] [PubMed] [Google Scholar]
- 8.Yao T P, Forman B M, Jiang Z, Cherbas L, Chen J D, McKeown M, Cherbas P, Evans R M. Nature (London) 1993;366:476–479. doi: 10.1038/366476a0. [DOI] [PubMed] [Google Scholar]
- 9.Hall B L, Thummel C S. Development (Cambridge, UK) 1998;125:4709–4717. doi: 10.1242/dev.125.23.4709. [DOI] [PubMed] [Google Scholar]
- 10.Escriva H, Safi R, Hanni C, Langlois M C, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laudet V. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- 12.Jones G, Sharp P A. Proc Natl Acad Sci USA. 1997;94:13499–13503. doi: 10.1073/pnas.94.25.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones G, Jones D. Insect Biochem Mol Biol. 2000;30:671–679. doi: 10.1016/s0965-1748(00)00038-2. [DOI] [PubMed] [Google Scholar]
- 14.Wurtz J M, Bourguet W, Renaud J P, Vivat V, Chambon P, Moras D, Gronemeyer H. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 15.Weatherman R V, Fletterick R J, Scanlan T S. Annu Rev Biochem. 1999;68:559–581. doi: 10.1146/annurev.biochem.68.1.559. [DOI] [PubMed] [Google Scholar]
- 16.Heery D M, Kalkhoven E, Hoare S, Parker M G. Nature (London) 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 17.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 19.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 20.Brzozowski A M, Pike A C, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J A, Carlquist M. Nature (London) 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 21.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 24.Egea P F, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. EMBO J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. Nature (London) 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 26.Chung A C, Durica D S, Clifton S W, Roe B A, Hopkins P M. Mol Cell Endocrinol. 1998;139:209–227. doi: 10.1016/s0303-7207(98)00056-2. [DOI] [PubMed] [Google Scholar]
- 27.Guo X, Xu Q, Harmon M A, Jin X, Laudet V, Mangelsdorf D J, Palmer M J. Mol Cell Endocrinol. 1998;139:45–60. doi: 10.1016/s0303-7207(98)00073-2. [DOI] [PubMed] [Google Scholar]
- 28.Neidhardt F C, Umbarger H E. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. [Google Scholar]
- 29.Cronan J E, Rock C O. In: Escherichia coli and Salmonella. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. [Google Scholar]
- 30.Blumberg B, Evans R M. Genes Dev. 1998;12:3149–3155. doi: 10.1101/gad.12.20.3149. [DOI] [PubMed] [Google Scholar]
- 31.Giguere V. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- 32.de Urquiza A M, Liu S, Sjoberg M, Zetterstrom R H, Griffiths W, Sjovall J, Perlmann T. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 33.Bourguet W, Vivat V, Wurtz J M, Chambon P, Gronemeyer H, Moras D. Mol Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 34.Glass C K, Rosenfeld M G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 35.Schulman I G, Chakravarti D, Juguilon H, Romo A, Evans R M. Proc Natl Acad Sci USA. 1995;92:8288–8292. doi: 10.1073/pnas.92.18.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boudjelal M, Wang Z, Voorhees J J, Fisher G J. Cancer Res. 2000;60:2247–2252. [PubMed] [Google Scholar]
- 37.Dace A, Zhao L, Park K S, Furuno T, Takamura N, Nakanishi M, West B L, Hanover J A, Cheng S. Proc Natl Acad Sci USA. 2000;97:8985–8990. doi: 10.1073/pnas.160257997. . (First Published July 25, 2000; 10.1073/pnas.160257997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulman I, Li C, Schwabe J, Evans R. Genes Dev. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- 39.Billas I M, Moulinier L, Rochel N, Moras D. J. Biol. Chem. 2000. [DOI] [PubMed] [Google Scholar]