Abstract
Discovery of diverse plant and animal viral proteins as suppressors of RNA silencing has provided strong support for an RNA-based viral immunity (RVI), which is now known to specifically destroy viral RNAs by RNA interference in fungi, plants and invertebrates. Here we review several recent studies that have revealed new mechanistic insights into plant and insect viral suppressors of RVI or suggested a role for RNA silencing suppression during mammalian viral infection.
Introduction
RNA silencing controls antiviral immunity in diverse eukaryotes including fungi, plants and invertebrates. In this RNA-based virus immunity (RVI), virus-specific dsRNA produced during infection is processed by a Dicer type III nuclease (RNase III) into siRNAs, which are then loaded in an Argonaute protein (AGO) to guide specific viral clearance by RNA silencing (Figure 1) (Ruiz-Ferrer and Voinnet, 2009). Consistent with an antiviral role for RNA silencing, many plant and animal viruses have been shown to encode a viral suppressor of RNA silencing (VRS). These VSRs often have important roles in viral pathogenesis before the identification of the VSR activity in assays of experimental RNA silencing induced by exogenous dsRNA or an over-expressing sense RNA transgene (Li and Ding, 2006). Recent studies have illustrated rescue of VSR-deficient mutant viruses in plant and insect host cells defective in RNA silencing, thereby demonstrating a specific role for active viral suppression of RVI during infection. In this short review, we highlight several VSRs whose host targets have been recently defined.
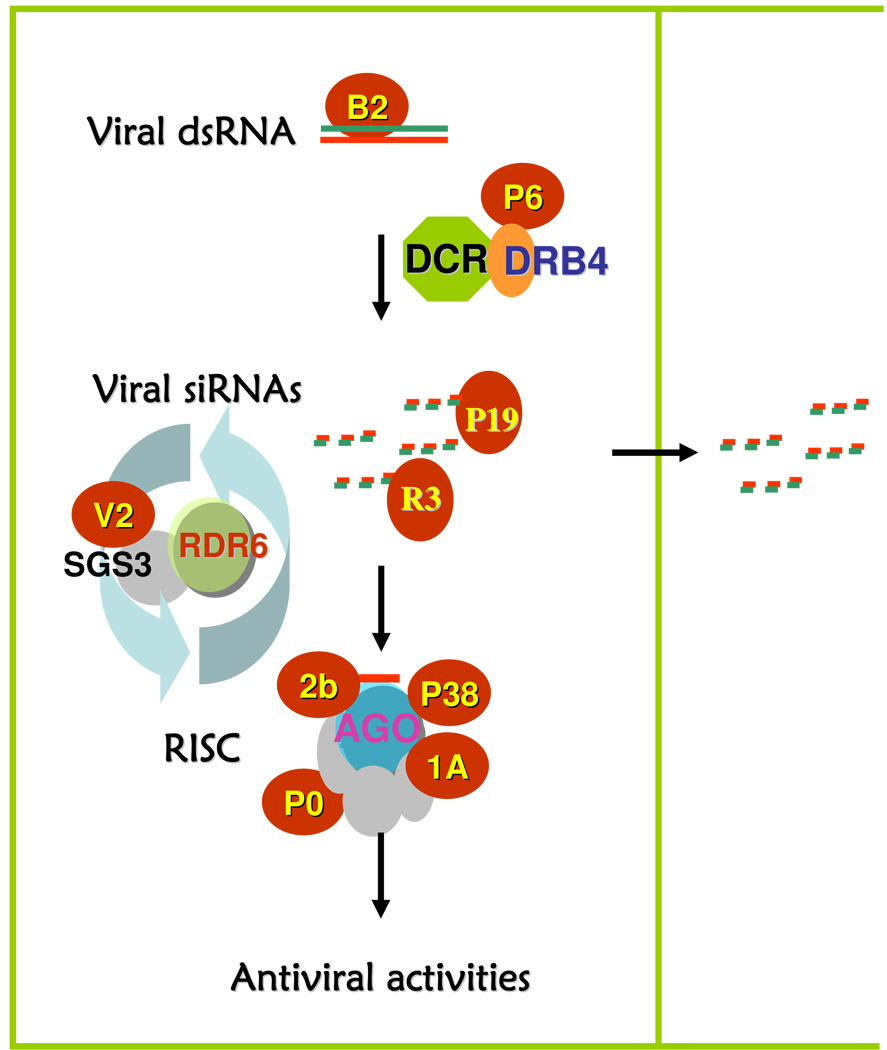
Fig. 1.
Mechanisms in the viral suppression of RNA-based viral immunity. VSRs target RNA and protein components of the RNA silencing pathway and inhibit siRNA production (B2, P6), RISC assembly (P0 and possibly P38), slicing activity of mature RISC (P1, 1A), amplification of viral secondary siRNAs (2b, RNaseIII (R3), V2), or intercellular movement of viral siRNAs (P19).
AGO-targeting VSRs
AGOs are a key player in RNA silencing because all three classes of small silencing RNAs, siRNAs, microRNAs (miRNAs) and Piwi-interacting RNAs, guide RNA silencing after specific binding to an AGO. Some AGOs cleave/slice the target RNA by the RNase H-like Piwi domain in RNA-induced silencing complex (RISC) whereas others recruit additional proteins into effector complexes to mediate either translational repression or transcriptional silencing. However, AGOs known to direct RVI in Arabidopsis thaliana (AGO1), Drosophila melanogaster (Ago-2), and Caenorhabditis elegans (RDE-1) all exhibit the slicer activity (Ruiz-Ferrer and Voinnet, 2009).
The first VSR shown to bind an AGO in vivo is the 2b protein of Cucumber mosaic virus (CMV), which contains a positive-strand (+) RNA genome (Zhang et al., 2006). Interaction of 2b with AGO1 of A. thaliana was demonstrated by coimmunoprecipitation in plants infected with CMV and was shown to inhibit in vitro slicing of the target RNA by siRNA-programmed AGO1 (Figure 1). Among the ten AGOs encoded by A. thaliana, AGO1 is the major AGO involved in cytoplasmic RNA silencing by both siRNAs and miRNAs and contributes to RVI targeting CMV because ago1 mutant plants are hypersusceptible to CMV and AGO1 immunoprecipitates contain CMV siRNAs (Ruiz-Ferrer and Voinnet, 2009; Zhang et al., 2006). However, 2b also binds long dsRNA and siRNA duplexes in vitro and in infected plants inhibits production of viral secondary siRNAs, which are processed from virus-specific dsRNA synthesized by host RNA-directed RNA polymerase 1 (RDR) or RDR6 and have been recently shown to play an essential role in RVI against CMV (Ruiz-Ferrer and Voinnet, 2009; Wang et al., 2010)(Wang et al., 2010). Thus, future studies will be necessary to determine how 2b binding to AGO1 and/or duplex RNA interferes with production of viral secondary siRNAs.
Suppression of RNA silencing by P0 encoded by (+)RNA genome of poleorviruses is associated with specific degradation of AGO1 in N. benthamiana plants (Baumberger et al., 2007; Bortolamiol et al., 2007). P0 is an F-box protein and interacts with the A. thaliana homologs of the S-phase kinase-related protein 1(SKP1) in a manner dependent on the integrity of the N-terminal F-box. However, P0-mediated AGO1 degradation is insensitive to inhibition of proteasome, which is inconsistent with the idea that AGO1 was targeted by P0 for ubiquitynation and proteasome-dependent degradation. P0 does not interact directly with AGO1 as shown by both yeast two-hybrid and coimmunoprecipitation (Bortolamiol et al., 2007; Csorba et al., 2010). However, both P0 and a P0 mutant that carries mutations in the F-box and is defective in silencing suppression, were found in the high molecular weight AGO1-containing RISC-like complex, indicating interaction of P0 with a component of RISC in an F-box independent manner (Figure 1) (Csorba et al., 2010). However, P0 exhibited VSR activity only when it is expressed before RISC assembly and did not inhibit the activity of mature RISC preloaded with either a host miRNA or viral siRNAs (Csorba et al., 2010). Csorba and colleagues therefore propose that P0 binding to a component of RISC prevents loading of siRNAs into AGO1 during RISC assembly and that AGO1 not assembled into RISC is readily degraded. However, this model does not include a possible role for the recognition of P0 by the SKP1 complex in the VSR activity and/or AGO1 degradation.
Two recent study illustrate a new strategy for VSR binding to AGO via the characterized glycine/tryptophane (GW)/WG repetitive motif (Azevedo et al., 2010)(Giner et al., 2010). GW/WG motif is considered as the 'AGO hook' and encoded by cellular proteins such as GW182 and Tas3, recruited by Ago-1 of fruit fly and fission yeast in RISC and RNA-induced transcriptional silencing complex, respectively, to repress translation by miRNAs and to silence transcription near centromeres. Azevedo and colleagues show that the VSR P38 of turnip crinkle virus (TCV), a (+)RNA virus, contains two discrete GW repeats and interacts directly with AGO1 but not AGO4 or AGO7 in coimmunoprecipitation experiments (Azevedo et al., 2010). Gel filtration further provides evidence for the presence of P38 and AGO1 in the same high molecular weight complex. Direct AGO1 binding in vitro to the individual N-terminal and C-terminal GW motifs of P38 was also detected and the interaction was abolished by changing GW to GA. Several lines of evidence indicate that expression of P38 in TCV-infected plants inhibits the activities of AGO1 in A. thaliana plants (Figure 1). For example, GA mutation in both GW repeats of P38 abolished its VSR activity and the defect of the mutant TCV carrying the GA mutations (TCVGA2) in host infection was partially rescued in ago1-27 mutant. Moreover, the AGO1-dependent accumulation of host miRNAs was dramatically reduced in TCV-infected plants as occurs in hypomorphic ago1-27 mutant plants (Azevedo et al., 2010). However, no significant reduction in host miRNAs was previously observed in A. thaliana plants carrying an over-expressing P38 transgene (Ruiz-Ferrer and Voinnet, 2009). It is not clear if P38 binding of AGO1 acts to prevent AGO1 from being assembled into mature RISC or blocks the silencing activity of the AGO1-containing mature RISC. It is also not clear if the activity of P38 to bind siRNA and long dsRNA observed previously (Merai et al., 2006) plays a role in its bnding to AGO1. It is of interest to note that infection of TCVGA2 was rescued less effectively by ago1-27 mutation than by double loss-of-function muatations in both Dicer-like 2 (DCL2) and DCL4 (Azevedo et al., 2010), which produce 22- and 21-nucleotide viral siRNAs acting redundantly to guide RVI (Ruiz-Ferrer and Voinnet, 2009), suggesting participation of additional AGO (or AGOs) in RVI that is targeted by P38.
Giner and colleagues found the VSR of the (+)RNA virus sweet potato mild mottle virus (SPMMV), P1, contains three GW/WG repeats (Giner et al., 2010). Based on transient over-expression in Nicotiana benthamiana leaves, the authors show that P1 bound to A. thaliana AGO1 and co-fractionated with AGO1 and host miRNAs in a high molecular weight RISC-like complex (>670 kDa) in gel filtration experiments. W to A mutations in any two of the three GW/WG repeats in P1 abolished both the VSR activity and AGO1 binding although wildtype and mutant P1 were expressed at similar levels, indicating that AGO1 binding is essential for the VSR activity (Giner et al., 2010). Using two established assays based on co-expression of miRNA/siRNA-sensor constructs and VSR, it was further shown that P1 inhibited the silencing activity of mature RISC preloaded with either a host miRNA or viral siRNAs. As proposed by Giner and colleagues, high affinity binding of AGO1 by P1 may outcompete an essential endogenous GW/WG-containing component of RISC or prevent recognition of target RNA by complementary siRNA loaded in the RISC.
Specific targeting of an antiviral AGO also was demonstrated for an insect VSR (Nayak et al., 2010), protein 1A of Cricket paralysis virus (CrPV) shown previously to suppress Ago2-mediated RVI in Drosophila S2 cells triggered by Flock house virus (FHV) replication (Wang et al., 2006). Direct interaction of the Drosophila Ago2 with 1A expressed from a transfected plasmid was detected in S2 cells by coimmunoprecipitation (Nayak et al., 2010). Binding of Ago-2 by 1A inhibited in vitro slicing of mRNA by a siRNA-programmed RISC without disrupting RISC assembly (Figure 1). Silencing activity of a preassembled RISC was also inhibited in S2 cells by 1A expressed from a cotransfected plasmid (Nayak et al., 2010). However, it should be pointed out that VSR activities of P1, P0 and 1A were all characterized following over-expression from a transgene and should ideally be verified in the context of virus infection in which expression of the VSR from the cognate viral genome is also targeted by RVI.
VSRs targeting other protein components of RNA silencing
Direct interaction was detected between the dsRNA-binding protein 4 (DRB4) of A. thaliana and the VSR P6 of cauliflower mosaic virus (CaMV), which contain circular dsDNA genome replicating through an RNA intermediate, 35S RNA (Haas et al., 2008). All of the four DCLs of A. thaliana participate in the biogenesis of CaMV-derived siRNAs although the predominant species of viral siRNAs are 24 and 21 nucleotides in length produced by DCL3 and DCL4, respectively (Ruiz-Ferrer and Voinnet, 2009). Interaction of DRB4 with DCL4 in A. thaliana is required for production of 21-nt siRNAs targeting the endogenous trans-acting siRNAs (ta-siRNAs) loci, silencing transgenes and CaMV, which is all effectively suppressed by P6 expressed either from a stably integrated transgene or an infecting CaMV (Haas et al., 2008). P6 encodes functional nuclear localization signals and mutations that prevent nuclear import abolish both the VSR activity and the translational activation of the viral polycistronic 35S RNA, an essential function previously assigned to P6. Notably, Haas and colleagues identified a three-residue substitution in P6’s nuclear localization signals that abolished CaMV infectivity without significantly altering P6’s activity in translational activation, indicating that CaMV infection requires a new function of P6. However, further studies will be necessary to determine if the VSR activity of P6 has a specific role in viral infection.
Both yeast two-hybrid and fluorescence resonance energy transfer (FRET) approaches detected in vivo interaction of the SGS3 protein from either A. thaliana or tomato with VSR V2 protein, encoded by tomato yellow leaf curl virus, a member of Geminiviridae containing a small circular ssDNA genome (Glick et al., 2008). SGS3 is essential for RDR6-dependent biogenesis of ta-siRNAs, transgene and viral siRNAs. A recent study found that both SGS3 and V2 selectively bind 5’-overhang-containing dsRNA and that V2 out-competes SGS3 for binding to the dsRNA substrate in vitro (Fukunaga and Doudna, 2009). Therefore, V2 may bind either SGS3 and/or a dsRNA intermediate produced during slicing and inhibit dsRNA synthesis by the RDR6 pathway (Figure 1).
VSRs targeting RNA components of RNA silencing
Binding to the long dsRNA and siRNA was once thought to be the main strategy of VSRs (Ruiz-Ferrer and Voinnet, 2009). VSR B2 protein of FHV contains a novel dsRNA domain and inhibits processing of long dsRNA into siRNAs in vitro, which explains why B2 suppresses RNAi only when it is expressed before long dsRNA is introduced (Figure 1). Both coimmunoprecipitation and pull-down experiments detected in vivo interaction of B2 with the viral dsRNA replicative intermediates (vRI-dsRNA) in the infected Drosophila cells (Aliyari et al., 2008). Production of viral siRNAs was dramatically suppressed by B2 during FHV infection, which is most likely because B2 is in close proximity to the nascent vRI-dsRNA following the recruitment of B2 to the viral replication complex by an interaction with the viral RdRP (Aliyari et al., 2008).
The VSR P19 of plant tombusviruses such as Cymbidium ringspot virus (CymRSV) selectively binds short dsRNA and can sequester 21-nt siRNA duplexes from being assembled into holo-RISC in Drosophila cells (Lakatos et al., 2006). However, P19-deficient mutant of CymRSV accumulates to levels similar to wildtype virus in both protoplasts and the inoculated leaves and only exhibits defects in spreading out of the vasculature bundles to invade the surrounding tissues in the systemically infected leaves (Havelda et al., 2003), indicating that P19 does not prevent RISC-mediated degradation of viral RNAs. Dunoyer and colleagues found recently that P19 could specifically sequester DCL4-dependent 21-nt transgene siRNAs in silencing-incipient cells and prevent them from spreading cell-to-cell to induce RNA silencing in the neighboring recipient cells (Figure 1) (Dunoyer et al., 2010). These findings support an earlier hypothesis that P19 promotes systemic viral infection by sequestering viral siRNA duplexes from spreading out of the vasculature to immune neighboring cells against CymRSV (Li and Ding, 2006).
A VSR encoded by sweet potato chlorotic stunt virus (SPCSV) is a class 1 RNase III consisting of an RNase domain and a canonical dsRNA-binding doamin (Cuellar et al., 2009). In addition to long dsRNA substrates, the viral RNase III also cleaved siRNA duplexes of 21 to 24 nucleotides long into 14 bp fragments in vitro. In transgenic plants, the viral RNaseIII suppresses RNA silencing induced by the RDR6-dependent pathway, but is inactive when induction of RNA silencing does not depend on de novo dsRNA synthesis by RDR6. Cleavage of viral siRNAs has been proposed as the mechanism for the VSR of SPCSV. However, available data do not exclude long dsRNA as possible targets.
Does VSR activity play a role in mammalian viral infection?
Many mammalian viruses encode a VSR (Li and Ding, 2006). However, it has not been clear if the VSR activity has a specific role in mammalian viral infection because rescue of VSR-deficient mutant viruses in mammalian host cells defective in RNA silencing is not yet demonstrated. Using an indirect approach, two recent studies (Qian et al., 2009; Schnettler et al., 2009) investigated the role of the VSR activity during infection of human immunodeficiency virus (HIV). HIV was reported to encode a VSR, Tat (Bennasser et al., 2005), but this conclusion has been debated by a subsequent study carried out by another lab (Lin and Cullen, 2007). A known function of Tat is to enhance transcription of HIV RNA from the integrated proviral DNA by binding to an internal stem-loop structural element of HIV RNA. However, HIV gene expression in the infected cells required a transcriptional enhancer-independent activity of Tat and this activity could be substituted by two distinct plant VSRs: the tombusviral P19 and the NS3 protein of rice hoja blanca virus (Qian et al., 2009; Schnettler et al., 2009). NS3 exhibited the same affinity as P19 for siRNA duplexes and neither sequesters long dsRNA, which is recognized as a pathogen-associated molecular pattern in vertebrates to activate multiple innate immunity pathways. Notably, NS3/P19 mutants defective in siRNA binding also were unable to rescue HIV gene expression in infected mammalian cells (Qian et al., 2009; Schnettler et al., 2009).
These findings indicate that HIV infection requires suppression of small RNA-directed gene silencing, which is consistent with previous observations that knockdown of Dicer enhances virus accumulation in mammalian host cells (Matskevich and Moelling, 2007; Otsuka et al., 2007; Triboulet et al., 2007). Future studies will be necessary to determine if Tat and other mammalian VSRs target RNA silencing induced by small RNAs of either viral or host origin. It is known that mammalian viral infection can induce production of virus-derived miRNAs or alter the expression profile of cellular miRNAs (Skalsky and Cullen, 2010). Moreover, although early studies based on standard RNA sequencing protocols were not successful, a recent survey in a wide range of mammalian host systems by deep sequencing has identified low abundant virus-derived small RNAs from several distinct RNA viruses (Parameswaran et al., 2010). Notably, the newly cloned viral small RNAs contain a subpopulation with features of siRNAs similar to those detected in plant and invertebrate hosts, including approximately equal positive and negative strands ratios, pairs of siRNA duplexes with one or two unpaired nucleotides at the 3’-ends and association with AGO proteins in vivo (Parameswaran et al., 2010). These studies provide experimental systems for future rigorous exploration on the role of mammalian VSRs and virus-derived small RNAs in the RNA-based virus immunity.
Acknowledgments
Work in my laboratory is supported by grants from the National Institutes of Health and USDA. Because of space limitations we have cited reviews rather than primary research papers. I apologize to those investigators whose original papers have not been cited.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aliyari R, Wu Q, Li HW, Wang XH, Li F, Green LD, Han CS, Li WX, Ding SW. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo J, Garcia D, Pontier D, Ohnesorge S, Yu A, Garcia S, Braun L, Bergdoll M, Hakimi MA, Lagrange T, et al. Genes Dev. 2010;24:904–915. doi: 10.1101/gad.1908710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Tsai C-H, Lie M, Havecker E, Baulcombe DC. Curr. Biol. 2007;17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V. Curr. Biol. 2007;17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- Csorba T, Lozsa R, Hutvagner G, Burgyan J. Plant J. 2010;62:463–472. doi: 10.1111/j.1365-313X.2010.04163.x. [DOI] [PubMed] [Google Scholar]
- Cuellar W, Kreuze J, Rajam ki M, Cruzado K, Untiveros M, Valkonen J. Proc Natl Acad Sci USA. 2009;106:10354. doi: 10.1073/pnas.0806042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P, Schott G, Himber C, Meyer D, Takeda A, Carrington JC, Voinnet O. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Doudna J. EMBO J. 2009;28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giner A, Lakatos L, Garcia-Chapa M, López-Moya J, Burgyan J. PLoS Pathog. 2010 doi: 10.1371/journal.ppat.1000996. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y. Proc Natl Acad Sci USA. 2008;105:157–161. doi: 10.1073/pnas.0709036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O. EMBO J. 2008;27:2102–2112. doi: 10.1038/emboj.2008.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelda Z, Hornyik C, Crescenzi A, Burgyan J. J Virol. 2003;77:6082–6086. doi: 10.1128/JVI.77.10.6082-6086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, Lopez-Moya JJ, Burgyan J. EMBO J. 2006;25:2768–2780. doi: 10.1038/sj.emboj.7601164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ding SW. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Cullen BR. J Virol. 2007;81:12218–12226. doi: 10.1128/JVI.01390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matskevich AA, Moelling K. J Gen Virol. 2007;88:2627–2635. doi: 10.1099/vir.0.83103-0. [DOI] [PubMed] [Google Scholar]
- Merai Z, Kerenyi Z, Kertesz S, Magna M, Lakatos L, Silhavy D. J Virol. 2006;80:5747–5756. doi: 10.1128/JVI.01963-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak A, Berry B, Tassetto M, Kunitomi M, Acevedo A, Deng C, Krutchinsky A, Gross J, Antoniewski C, Andino R. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1810. doi:10.1038/nsmb.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R, et al. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W, et al. PLoS Pathog. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Zhong X, Yu L, Ding B, de Haan P, Boris-Lawrie K. Proc Natl Acad Sci U S A. 2009;106:605–610. doi: 10.1073/pnas.0806822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ferrer V, Voinnet O. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- Schnettler E, de Vries W, Hemmes H, Haasnoot J, Kormelink R, Goldbach R, Berkhout B. EMBO Rep. 2009;10:258–263. doi: 10.1038/embor.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalsky RL, Cullen BR. Viruses, microRNAs, and Host Interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Marurin T, Barbry P, Baillat V, et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, Atkinson P, Ding SW. Science. 2006;312:452–454. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH. Genes Dev. 2006;20:3255–3268. doi: 10.1101/gad.1495506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XB, Wu Q, Ito T, Cillo F, Li WX, Chen X, Yu JL, Ding SW. Proc Natl Acad Sci U S A. 2010;107:484–489. doi: 10.1073/pnas.0904086107. [DOI] [PMC free article] [PubMed] [Google Scholar]