Abstract
Cross-reactive memory T cells induced by primary infection with one of the four serotypes of dengue virus (DENV) are hypothesized to play an immunopathological role in secondary heterologous DENV infection. To define the T cell response to heterologous serotypes, we isolated HLA-A*1101-restricted epitope-specific CD8+ T cell lines from primary DENV-immune donors. Cell lines exhibited marked cross-reactivity towards peptide variants representing the four DENV serotypes in tetramer binding and functional assays. Many clones responded similarly to homologous and heterologous serotypes with striking cross-reactivity between the DENV-1 and DENV-3 epitope variants. In vitro-stimulated T cell lines consistently revealed a hierarchical induction of MIP-1β > degranulation > TNFα > IFNγ, which depended on the concentration of agonistic peptide. Phosphoflow assays demonstrated peptide dose-dependent phosphorylation of ERK1/2, which correlated with cytolysis, degranulation, and induction of TNFα and IFNγ but not MIP-1β production. This is the first study to demonstrate significant DENV serotype-cross-reactivity of CD8+ T cells after naturally-acquired primary infection. We also demonstrate qualitatively different T cell receptor signaling after stimulation with homologous and heterologous peptides. Our data support a model whereby the order of sequential DENV infections influences the immune response to secondary heterologous DENV infection, contributing to varying disease outcomes.
Keywords: Antigens/Peptides/Epitopes, Human, Signal Transduction, T cells, Viral
INTRODUCTION
The World Health Organization estimates that 50 million dengue virus infections occur each year within the nearly two-fifths of the world population living in areas at risk for dengue transmission1. With increasing urbanization as well as international travel, the range of the principal mosquito vector of dengue virus (DENV), Aedes agypti, is expanding2. Co-circulation of the four serotypes of DENV, DENV 1-4, along with the increased risk for severe disease during secondary DENV infections3–5 represents a serious global health problem. With no reliable immunocompetent animal model available to mimic sequential human DENV infections, ex vivo studies on human samples are necessary to investigate the mechanisms for increased disease severity during heterologous secondary DENV infections.
Immunologic memory established by a primary DENV infection influences the response to a secondary heterologous DENV infection due to the significant (~70%) amino acid homology between the four DENV serotypes6. In particular, DENV-specific memory T and B cells can be reactivated during secondary heterologous DENV infection resulting in a more vigorous and cross-reactive secondary immune response. A number of studies have found increased markers of immune cell activation in patients with dengue hemorrhagic fever compared to patients with the less severe form of disease, dengue fever. These markers include interferon-gamma (IFNγ), tumor necrosis factor alpha (TNFα), soluble CD8, soluble IL-2 receptor, soluble TNF receptor, and CD697–10, which support a role for T cells in mediating immunopathology11.
Our laboratory and others have demonstrated the ability of DENV-specific T cells to recognize multiple DENV serotypes12–19. Most of these studies analyzed PBMC either from donors that received candidate live-attenuated monovalent vaccines or naturally-infected patients who experienced a secondary DENV infection. Few reports have described immune responses after naturally-occurring primary DENV infections20 and no published studies have reported on the memory CD8+ T cell repertoire after natural primary DENV infection, nor its subsequent response to homologous or heterologous variant epitopes. Studies of the ability of memory T cells generated by natural primary DENV infection to respond to heterologous serotypes are needed to understand how the order of sequential DENV infections can affect disease outcomes, as has been suggested epidemiologically3, 4.
Our study was designed to assess the cross-reactivity of the CD8+ T cell repertoire generated after primary DENV infection in both naturally-infected subjects as well as a vaccine recipient. HLA-A*1101 is a common haplotype found in DENV-endemic areas and has been shown to be associated with susceptibility to dengue disease21, so we focused on a previously described HLA-A*1101-restricted epitope. In order to model variability within the antigen-specific T cell response to secondary heterologous DENV exposure, we isolated antigen-specific CD8+ T cell lines from A*1101+ individuals exposed to a single DENV serotype and stimulated them in vitro with homologous and heterologous peptide variants representing the four DENV serotypes. We analyzed peptide-HLA binding, effector responses, and T cell receptor signaling in response to natural homologous and heterologous peptide variants.
RESULTS
Striking cross-reactivity of cell lines isolated from primary DENV-immune donors
We obtained convalescent PBMC from three HLA-A*1101+ individuals who had a single DENV infection (Table 1) and utilized three peptide variants of a previously identified HLA-A*1101-restricted epitope16 to expand epitope-specific cells in vitro. The DENV-2 peptide (pD2) and DENV-3 and -4 peptide (pD3/4) each differ from the DENV-1 peptide (pD1) by a single amino acid (Table 2). We also synthesized three peptide-MHC tetramers, each containing a different variant of the epitope, in order to measure the cross-reactivity of peptide-HLA binding by individual CD8+ T cells by flow cytometry. The specificity of our tetramers was verified by analysis of PBMC from HLA-A11+ DENV-naïve and HLA-A11− DENV-immune donors (Supplementary Figure 1).
Table 1.
Summary of donor information and clinical diagnosis
| Donor | Infection | Serotype (exposure) | Diagnosis1 | Age Range2 | Country of Origin | MHC Class I haplotype |
Time point of PBMC3 | ||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | |||||||
| 1 | Primary | DENV-1 (vaccine) | n/a4 | Adult | U.S.A. | 2,11 | 27,40 | 1,3 | 8 months |
| 2 | Primary | DENV-3 (natural) | DHF I | Child | Thailand | 2,11 | 13,38 | 7 | 6 months |
| 3 | Primary | DENV-3 (natural) | DHF II | Child | Thailand | 11,24 | 18,54 | 7,8 | 1 week |
According to WHO guidelines; DHF = dengue hemorrhagic fever
At time of infection
Post-defervescence
Subject experienced a mild dengue-like illness post-vaccination; see ref 38
Table 2.
Sequences of A11 epitope variants1
| Variant | Sequence2 | Designation |
|---|---|---|
| DENV-1 | G T S G S P I V N R | pD1 |
| DENV-2 | G T S G S P I V D R | pD2 |
| DENV-3, -4 | G T S G S P I I N R | pD3/4 |
This epitope and its variants were originally identified in ref 16
Amino acids that differ from pD1 are shown in bold
We generated bulk cultures from donor PBMC by in vitro stimulation with each of the epitope variants (Supplementary Figure 2A). After approximately two weeks in culture, tetramer staining revealed a modest enrichment of epitope-specific CD8+ T cells. Regardless of the donor or peptide variant used for stimulation, nearly all of the expanded tetramer+ cells bound the DENV-1 variant tetramer (Supplementary Figure 2B and data not shown). To increase our chances of isolating epitope-specific cell lines, we magnetically sorted the bulk cultures using the pD1 tetramer before performing limiting dilution cloning. Epitope-specific cell lines were selected on the basis of their ability to selectively lyse peptide-coated HLA-A*1101+ B-lymphoblastoid cell line (BLCL) target cells and were subsequently characterized with regard to peptide dose-dependent cytotoxicity as well as tetramer staining.
We isolated three types of epitope-specific cell lines: pD1 serotype-specific, pD1-3/4 cross-reactive, and pD1-2-3/4 cross reactive. Data from representative cell lines are shown in Figure 1. Each cell line was stained with each individual tetramer in order to assess its ability to recognize the three peptide variants (Figure 1B). In general, tetramer binding reflected the ability to lyse target cells coated with the same peptide in 51Cr release assays (Figure 1C).
FIGURE 1. Three predominant patterns of serotype-cross-reactivity in HLA-A*1101-restricted T cell lines.
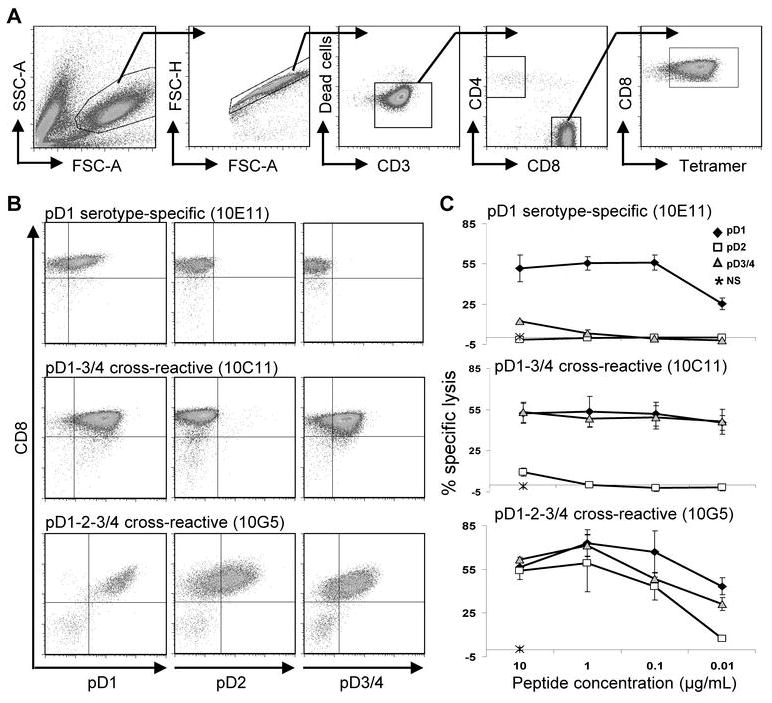
(A) Gating strategy used to identify tetramer+ cells. (B) Cell lines were stained with each of the three tetramer variants and reveal three types of serotype-cross-reactivity: pD1 serotype-specific, pD1-3/4 cross-reactive, pD1-2-3/4 cross-reactive. Cells were gated on live, CD3+ singlet lymphocytes. (C) 51Cr release assays demonstrate peptide dose-dependent cytolytic activity of representative cell lines at an E:T ratio of 10:1. NS = no stimulation.
Of the sixteen cell lines that were established, the majority demonstrated serotype-cross-reactivity by tetramer staining regardless of the donor or bulk culture from which they originated (Table 3). The extent of tetramer binding did not always predict the magnitude of its cytolytic response. For example, the cell line 10D6 demonstrated greatest binding to the pD1 tetramer while its cytolytic response was comparable toward targets coated with any of the three peptides. In addition, the lack of binding to a given tetramer did not necessarily indicate that the cell line could not mount a functional response against that particular peptide variant. For example, the cell line 10D7 did not bind the pD2 or pD3/4 tetramers yet demonstrated robust cytolysis of pD2 and pD3/4 peptide-coated target cells. Notably, there were no cases in which a cell line bound a given tetramer and did not respond in 51Cr release assays towards BLCL coated with that particular peptide variant. Thus, discrepancies between tetramer binding and cytolytic responses likely reflect lower avidity of a soluble peptide-MHC tetramer compared to the peptide-MHC complex presented on the surface of an antigen presenting cell.
Table 3.
Lytic activity and tetramer specificity of epitope-specific cell lines
| Donor | Peptide stimulation (bulk) | Cell line | Tetramer specificity1 |
% Specific Lysis2 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pD1 | pD2 | pD3/4 | [pD1]
|
[pD2]
|
[pD3/4]
|
||||||||||||
| 1 | pD13 | 10B8 | ++ | ++ | ++ | 59 | --- | --- | --- | 49 | 48 | 40 | 16 | 55 | 45 | 61 | 42 |
| 10D12 | +++ | −− | ++ | 53 | 54 | 57 | 38 | 24 | 10 | 2 | 1 | 53 | 51 | 47 | 45 | ||
| 10E11 | ++ | −− | −− | 51 | 55 | 55 | 25 | <0 | <0 | <0 | <0 | 12 | 3 | <0 | <0 | ||
| pD2 | 10A3 | +++ | +/− | + | 49 | 56 | 70 | 60 | 44 | 44 | 9 | 2 | 57 | 84 | 59 | 87 | |
| 10D7 | ++ | −− | −− | 50 | 55 | 62 | 37 | 37 | 27 | 8 | 1 | 49 | 78 | 45 | 65 | ||
| 10E12 | +++ | +/− | +++ | 76 | 66 | 57 | 43 | 61 | 57 | 24 | 3 | 97 | 86 | 86 | 81 | ||
| 10F9 | +++ | −− | +/− | 49 | 58 | 62 | 47 | 36 | 38 | 19 | 1 | 44 | 73 | 44 | 50 | ||
| 10H3 | +++ | +/− | +++ | 72 | 64 | 59 | 68 | 83 | 76 | 55 | 23 | 89 | 82 | 84 | 76 | ||
| 2 | pD3/4 | 10A6 | +++ | ++ | + | 54 | 61 | 45 | 71 | 66 | 38 | 42 | 5 | 37 | 62 | 39 | 23 |
| 10C6 | +++ | + | ++ | 65 | 58 | 65 | 72 | 70 | 42 | 31 | 4 | 56 | 72 | 48 | 23 | ||
| 10D6 | +++ | +/− | + | 33 | 41 | 43 | 70 | 50 | 46 | 40 | 4 | 48 | 48 | 61 | 36 | ||
| 10F5 | +++ | + | ++ | 68 | 71 | 60 | 92 | 48 | 31 | 28 | 5 | 56 | 52 | 50 | 29 | ||
| 10G5 | +++ | + | ++ | 57 | 73 | 67 | 43 | 54 | 59 | 43 | 8 | 62 | 71 | 48 | 31 | ||
| 10H5 | +++ | + | ++ | 62 | 64 | 47 | 66 | 48 | 23 | 31 | 4 | 54 | 45 | 50 | 28 | ||
| 3 | pD3/4 | 10C11 | +++ | −− | +++ | 53 | 54 | 53 | 46 | 10 | 1 | <0 | <0 | 54 | 49 | 50 | 47 |
| 10F12 | +++ | −− | +++ | 59 | 60 | 60 | 35 | <0 | 0 | 0 | 1 | 55 | 57 | 55 | 56 | ||
Based on single tetramer staining; −−, did not bind tetramer; +/−, <%30 bound tetramer; +, <%60 bound tetramer; ++, <%90 bound tetramer; +++, ≥ %90 bound tetramer
[peptide] = 10, 1, 0.1, 0.01μg/mL; numbers indicate average % specific lysis relative to negative (no peptide) control, n ≥ 3
This bulk culture was generated the 11mer variant of pD1, GTSGSPIVNRE, which was originally identified in ref 16
Peptide variants elicit a hierarchy of functional responses from epitope-specific T cells
To determine whether cytokine response profiles of individual CD8+ T cell lines correlated with lytic activity, we assessed degranulation and cytokine expression following stimulation with different concentrations of homologous and heterologous peptides in intracellular cytokine staining (ICS) assays (Figure 2). ICS of the pD1 serotype-specific cell line 10E11 revealed a robust response to high concentrations of homologous peptide; most cells up-regulated all four effector functions studied: degranulation (CD107a staining), and production of MIP-1β, TNFα, and IFNγ (Figure 2B). Conversely, stimulation with pD2 resulted in no detectable response. Stimulation with pD3/4 caused a low percentage of CD8+ T cells to respond. Of the responding T cells, most only produced MIP-1β, fewer cells were MIP-1β+CD107a+, and even fewer were MIP-1β+CD107a+TNFα+; no cells produced IFNγ.
FIGURE 2. Hierarchical response of effector functions in epitope-specific T cells.
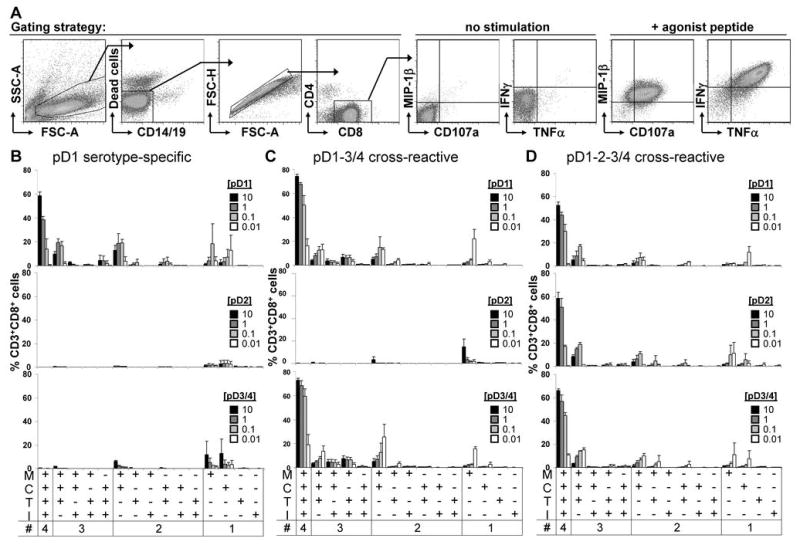
(A) Gating strategy and representative flow plots of a cell line in response to the absence or presence of agonist peptide. (B) pD1 serotype-specific, (C) pD1-3/4 cross-reactive, and (D) pD1-2-3/4 cross-reactive cell lines were stimulated with each of the peptide variants in intracellular cytokine staining assays. All possible combinations of the four effector functions (MIP-1β (M), TNFα (T), and IFNγ (I) production as well as degranulation (C), as determined by CD107a staining) are displayed across the x axis. Mean frequencies of responding CD8+ T cells of 10E11 (n = 2), 10C11 (n = 4), and 10B8 (n = 2) are shown. Peptide concentrations are shown in μg/mL.
Stimulation of pD1-3/4 cross-reactive cell lines (shown for the representative line 10C11 in Figure 2C) with either pD1 or pD3/4 induced nearly all CD3+CD8+ cells to respond, and a majority of cells in the culture expressed all four effector functions. On the other hand, pD2 stimulation induced a low percentage of cells to respond, most of which only produced MIP-1β. Akin to the cytolytic response, most cells of pD1-2-3/4 cross-reactive cell lines (represented by 10B8 in Figure 2D) responded with all four effector functions upon stimulation with any of the three peptide variants.
Regardless of a cell line’s recognition of different peptide variants its response to stimulation exhibited a hierarchical induction of MIP-1β > degranulation > TNFα > IFNγ, as evidenced by its peptide-dose response curve. With decreasing concentrations of agonist peptide (for example, pD1 and pD3/4 for the cell line 10C11), the effector response changed. As the overall frequency of responding cells decreased, their distribution shifted toward exhibiting fewer effector functions, in particular loss of IFNγ and TNFα expression to primarily producing MIP-1β. Below a critical concentration of agonistic peptide MIP-1β production also dropped off, as demonstrated by low concentrations of a partial agonist peptide (for example, pD2 for the cell line 10C11). These results suggest that a signal threshold is required to generate a polyfunctional effector response.
Phosphorylation of ERK1/2 correlates with cytolysis, degranulation, and induction of TNFα and IFNγ but not MIP-1β production
To assess differences in signaling after stimulation with each of the peptide variants, we utilized phosphoflow to measure the phosphorylation of ERK1/2, a member of the MAP kinase family located downstream of TCR signaling. Stimulation of T cell lines with full agonist peptides resulted in a dose-dependent increase of phosphorylated ERK1/2 (pERK1/2; Figure 3), which correlated with the induction of cytolysis, degranulation, and TNFα and IFNγ production. However, MIP-1β production was observed even in the absence of detectable ERK1/2 phosphorylation. For instance, pD1 or pD3/4 stimulation of the pD1-3/4 cross-reactive cell line 10C11 resulted in increased pERK1/2. However, pD2 stimulation of 10C11, which up-regulated MIP-1β in ICS assays, failed to phosphorylate ERK1/2.
FIGURE 3. Phosphorylation of ERK1/2 does not correlate with MIP-1β production.
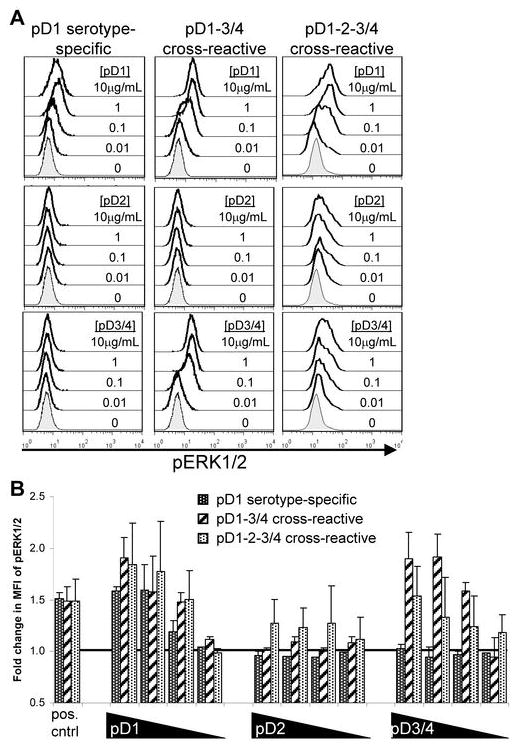
(A) Peptide dose-dependent phosphorylation of ERK1/2 after stimulation with epitope variants. (B) Average fold change in mean fluorescence intensity (MFI) of pERK1/2 in the cell lines 10E11 (n = 3), 10C11 (n = 4), and 10G5 (n = 2) after stimulation with decreasing concentrations (10, 1, 0.1, 0.01 μg/mL) of the three peptide variants. Fold change is relative to ‘no stimulation’ control.
MIP-1β is up-regulated in response to a short-lived, epitope-specific, TCR-mediated signal
We speculated that pD2 stimulation of the pD1-3/4 cross-reactive cell line 10C11 may induce a delayed pERK1/2 signal. Therefore we assessed pERK1/2 at 5, 10, 20, and 30 minutes after stimulation (Figure 4A). We were unable to detect up-regulation of pERK1/2, however, at any time point after pD2 stimulation. We next assessed phosphorylation of CD3ζ, a more proximal signal, to confirm that pD2 was specifically engaging the TCR. After three minutes of stimulation, we detected phosphorylated CD3ζ (pCD3ζ) in response to each of the three epitope variants (Figure 4B). The extent of pCD3ζ was comparable for all three dengue variants and was higher than that for a control HLA-A*1101-restricted peptide.
FIGURE 4. pD2 induces a short-lived activation signal in a pD1-3/4 cross-reactive cell line.
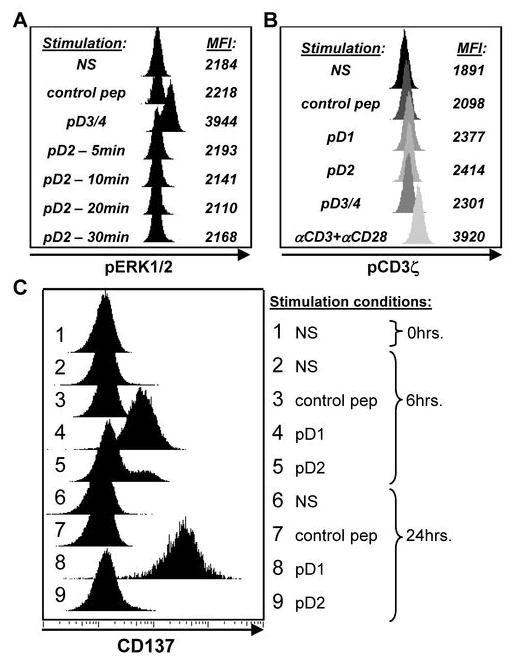
(A) Time course of pERK1/2 after pD2 stimulation of the cell line 10C11. One of 2 independent experiments is shown. (B) Phosphorylation of CD3ζ after stimulation of 10C11 with each of the three peptide variants. Histograms are shaded to emphasize the change in MFI. One of 3 independent experiments is shown. (C) Time course of CD137 expression after stimulation of 10C11 with pD1, pD2, and a control non-dengue HLA-A*1101-restricted viral peptide. The peptide concentration used for these experiments was 10μg/mL. NS = no stimulation.
We next analyzed CD137 expression as a further indicator of antigen-specific stimulation (Figure 4C). At 6 hours after stimulation, pD2 activated a low frequency of cells to up-regulate CD137, which correlated with its induction of MIP-1β. The pD2-induced CD137 expression was not sustained at 24 hours, however. In comparison, stimulation with a full agonist peptide (pD1) induced a robust up-regulation of CD137 at both 6 and 24 hours. These data indicate that pD2 induced a short-lived activating signal that was only strong enough to initiate MIP-1β production.
DISCUSSION
Plasma leakage, a hallmark of severe dengue disease, occurs late after infection and is coincident with viral clearance22, 23. This, together with evidence that increased disease severity is associated with secondary heterologous DENV infection3–5, suggests the involvement of cross-reactive DENV-specific memory T and B cells in contributing to an immunopathogenically-mediated clinical outcome.
We found that epitope-specific CD8+ T cell lines isolated from primary DENV immune donors were highly cross-reactive and demonstrated greater recognition of pD1 and pD3/4 variant peptides as compared to pD2. Dong, et al. also detected significant pD1-3/4 cross-reactivity within this A11 epitope-specific T cell population after secondary DENV infection13. Though a different DENV-2 epitope variant was used in that study, a majority of their ‘highly cross-reactive’ T cell clones recognized pD1 and pD3/4 with greater potency than the DENV-2 variant.
The predominance of pD1-3/4 cross-reactivity may be explained by the primary structure of these epitope variants. Classic A11-restricted epitopes have a small aliphatic residue at position 2 (P2) and a basic residue at the C terminus24–26. The epitope used in our study follows this motif since it contains a threonine at P2 as well as a C-terminal arginine. Recently, a secondary anchor residue for A11-restricted epitopes was identified at P6 or P7, which creates a double bulge of neighboring amino acids exposing likely sites of TCR contact27. Such bulging of the A11 dengue epitope would expose P9, the residue that differs between pD1 or pD3/4 and pD2. However, P9 of a 10-mer epitope from HIV was determined to be the C-terminal MHC anchor residue28. No matter which side of the MHC-TCR interface interacts with the P9 residue, the non-conservative amino acid change (and corresponding charge difference) between pD1 or pD3/4 (asparagine) and pD2 (aspartate) could drastically affect binding avidity. These epitope variants have been crystallized together with HLA-A*110129, which indicates tight binding of peptide and MHC. However, three different algorithms predicted that A11 binding for the three peptide variants reflects a hierarchy of pD3/4 > pD1 ≫ pD2 (Supplementary Table 1). We speculate that poor binding of pD2 (compared to pD1 and pD3/4) to either A11 or the TCR may explain its lower functional avidity apparent in our study.
Regardless of their serotype-cross-reactivity, the functional response of all the CD8+ T cell lines we tested revealed a hierarchical induction of MIP-1β > degranulation > TNFα > IFNγ. This is in line with a recent report from our laboratory characterizing PBMC from DENV vaccinees in response to in vitro stimulation of homologous and heterologous HLA-A*0201-restricted epitope variants12. Imrie et al., however, found that a larger proportion of CD8+ T cell clones specific for a B*5502-restricted epitope on the DENV NS5 protein produced IFNγ compared to TNFα14. Studies of HIV-specific T cells demonstrated a hierarchical induction of MIP-1β > degranulation > IFNγ > TNFα > IL-230, 31. Patients who were better able to control their HIV viral loads had higher numbers of T cells exhibiting multiple effector functions in both PBMC30 as well as rectal mucosa31, suggesting a protective role for polyfunctional CD8+ T cells. While the hierarchical induction of MIP-1β > degranulation > cytokines in virus-specific CD8+ T cells was observed in all of these studies, it is unclear what factors affect the particular order of cytokine production.
Previous studies have shown that varying degrees of TCR-ligand interaction elicit a hierarchical order of response thresholds for different effector functions32–37. It was determined that, regardless of ligand potency, the extent of TCR engagement as measured by TCR down-regulation dictated the functional response elicited from the T cell. Specifically, cytotoxicity was elicited after very low levels of TCR occupancy by peptide-MHC whereas cytokine responses required higher occupancy for induction. Our data support these findings in that cytotoxicity (measured by target cell lysis and degranulation) was induced at the lowest concentrations of full agonist peptides used as well as high concentrations of partial agonist peptides. TNFα and IFNγ production was only seen after stimulation with full agonist peptides, and the proportion of cells producing these cytokines increased with increasing concentrations of those peptides, supporting the idea that greater TCR occupancy triggers cytokine production. Additionally, phosphoflow assays performed in our study confirm that low TCR occupancy qualitatively alters peptide-induced signaling pathways, which helps explain differential effector responses. This is the first time phosphoflow has been utilized to demonstrate variations in peptide-induced signaling in T cells and has proven to be an effective tool for measuring such differences. Altogether our data argues that production of the chemokine MIP-1β has an even lower signal threshold than cytotoxicity, supporting other studies which suggest MIP-1β is a more sensitive measure of antigen-specific cells than IFNγ30,38.
In addition to lifelong immunity to the currently-infecting serotype, primary DENV infection induces transient but protective immunity to heterologous DENV serotypes in patients for at least two months after illness39. Our patient samples were collected eight months or less after primary infection. It is possible that the cross-reactive T cell responses we observed might not represent stable, long term T cell memory. Analysis of memory T cell responses after longer intervals would be complicated by the substantial potential for re-exposure to DENV in individuals living in DENV-endemic areas. However, we have detected cross-reactive T cell responses in vaccine recipients several years after immunization12, 40, 41 suggesting that primary exposure to the virus generates a long-lived cross-reactive memory T cell response.
Our findings support epidemiological evidence which indicates that the order of DENV serotypes with which a particular individual is infected plays a critical role in the outcome of the secondary immune response. The data suggest that, for the response to this A11-restricted epitope, DENV-1 infection followed by DENV-3 infection, or vice versa, would trigger the activation of cross-reactive T cells that undergo a vigorous polyfunctional response involving IFNγ production. On the other hand, a secondary DENV-2 infection may elicit cross-reactive, poorly responding T cells that are skewed toward MIP-1β production. Increased MIP-1β production at the site of infection may augment IFNγ and TNFα production by T cells42 activated by other viral epitopes and increase TNFα production by monocytes43 elevating the circulating levels of these cytokines and ultimately contributing to immunopathology. Secondary DENV-2 infection has been observed as a risk factor for increased disease severity in epidemiological studies in Asia3, 4.
Our study characterized effector functions in CD8+ T cells specific for a single epitope, whereas the overall T cell response would reflect responses to multiple epitopes with varying patterns of serotype-cross-reactivity. The clinical outcome in a given DENV-infected individual would also be expected to depend on other host characteristics including age, prior infection history, HLA alleles, and other genetic variables11. Although our study was limited to three patients, the subjects varied with regard to the infecting serotype, age, illness grade as well as genetic background. We isolated many cell lines from each patient that showed similar patterns of serotype-cross-reactivity to this epitope. Further studies on individuals infected with other DENV serotypes, experiencing different illnesses and with different genetic backgrounds than our donors will add to our understanding of the contribution of this cross-reactive epitope-specific T cell population to disease.
This study is the first to demonstrate substantial cross-reactivity of antigen-specific T cells generated in response to naturally-acquired primary DENV infection. We provide evidence that the DENV-immune T cell repertoire exhibits altered functional capabilities that are dependent on the particular serotype of infection. Investigations into the role of heterologous epitope variants in triggering memory T cell responses in donors with primary DENV infection help to define parts of the preexisting immune response that may predispose a person towards developing severe disease. An effective vaccine or treatment regimen will have to consider the effects of prior flavivirus immunity on disease outcome and will hopefully lead to the eradication of this widespread viral threat.
METHODS
Subjects and specimens
Convalescent peripheral blood samples were obtained from three HLA-A*1101+ DENV-immune donors: two were naturally infected with DENV and the third was immunized with an experimental live-attenuated DENV vaccine44 (Table I). All three donors had serologically confirmed primary DENV infection. PBMC were cryopreserved for approximately 19 years (donor 1) or 13 years (donors 2 and 3) until use; cell viability was >95%, 60% and 75%, respectively. HLA typing was performed at the University of Massachusetts Medical School or the Department of Transfusion Medicine, Siriraj Hospital, as previously described15, 21. Written informed consent was obtained from each subject and/or his/her parent or guardian and the study design was approved by the Institutional Review Boards of the Thai Ministry of Public Health, the Office of the US Army General and the University of Massachusetts Medical School.
Peptides
Peptides were synthesized at >90% purity from AnaSpec, Inc. (San Jose, CA). Sequences are as shown in Table II. The epitope was originally identified by Mongkolsapaya et al.16.
Generation of T cell lines
Cryopreserved PBMC from each donor were thawed and stimulated in vitro with 10μg/mL peptide in complete RPMI 1640 medium supplemented with 10% heat-inactivated FBS and 5ng/mL IL-7. Cells were fed with RPMI/10% FBS medium containing 50U/mL IL-2 on day 3 and every 2–3 days thereafter. After 2 to 3 weeks, pD1 tetramer-positive cells were positively selected using MACS α-allophycocyanin (APC) microbeads (Miltenyi Biotec). T cell lines were then generated by limiting dilution (3 or 10 cells/well) and maintained by biweekly re-stimulation with α-CD3, allogeneic irradiated PBMC, and 50U/mL IL-2.
Cytotoxicity assay
Cytotoxicity was assessed as previously described15. Briefly, HLA-A*1101+ B-lymphoblastoid cell line (BLCL) targets were labeled with 51Cr for 1 hour, washed, and pulsed with 0.01–10μg/mL of the indicated peptide. Target cells were cultured with T cells at an effector-to-target ratio of 10:1. After 4 hours, supernatants were harvested and 51Cr content was measured in a gamma counter. Percent specific lysis was calculated by the following formula: % lysis = (experimental 51Cr release − minimum 51Cr release)/(maximum 51Cr release − minimum 51Cr release) × 100.
Tetramer staining
HLA-A*1101 tetramers for all three epitope variants were generated at the University of Massachusetts Medical School Tetramer Core. Each variant was conjugated to a different fluorochrome to allow for tetramer staining with all three variants simultaneously (APC-pD1, PE-Cy7-pD2, PE-pD3/4). Approximately 0.1–1×106 cells were stained with a dead cell marker (LIVE/DEAD® Aqua; Molecular Probes, Invitrogen Corp.) according to the manufacturer’s instructions. After washing, cells were stained with 0.5–1μL of each tetramer for 20 minutes at 4°C before addition of mAb directed at the surface markers CD3 (clone UCHT1, Alexa700), CD8 (clone SK1, PerCP-Cy5.5; BD Biosciences), and CD4 (clone OKT4, Pacific Blue; eBioscience) for 30 minutes at 4°C. Cells were washed and placed in BD™ Cytofix or BD™ Stabilizing Fixative until analysis. Data were collected on a BD FACSAria™ and analyzed using FlowJo version 7.2.2 or 7.2.4.
Intracellular cytokine staining
T cells (2×105) were cultured with 2×104 HLA-A*1101+ BLCL and 0.01–10μg/mL peptide, FITC-αCD107a (clone H4A3), brefeldin A and monensin (BD GolgiPlug™ and BD GolgiStop™; BD Biosciences) for 6 hours. Cells were washed and stained with the viability marker LIVE/DEAD® Aqua before subsequent staining with APC-H7-αCD3 (clone SK7), PE-Cy7-αCD19/14 (clones SJ25C1 and M5E2), PerCP-Cy5.5-αCD8 (clone SK1; BD Biosciences), and PacBlue-αCD4 (clone OKT4; eBioscience) at 4°C for 30 minutes. After washing, the cells were fixed and permeabilized (BD Cytofix/Cytoperm™; BD Biosciences) before staining with Alexa700-αIFNγ (clone B27), PE-αMIP-1β (clone D21-1351), and APC-αTNFα (clone Mab11; BD Biosciences). Cells were washed again and placed in fixative until data collection.
Phosphoflow signaling assay
T cell receptor-induced signaling events were analyzed by flow cytometry as described45 and following the methods detailed by BD Biosciences. Briefly, 4×105 HLA-A*1101+ BLCL and 2×105 T cells were pulsed with 0.01–10μg/mL peptide for 30 minutes on ice. Cells were then washed and placed in 1% FBS in PBS for 3 or 10 minutes in a 37°C water bath. Cells were immediately fixed (Phosflow Fix Buffer I; BD Biosciences), washed, and then permeabilized (Phosflow Perm Buffer III; BD Biosciences) on ice for 30 minutes. After washing, cells were blocked with mouse IgG for 10 minutes prior to addition of PerCP-Cy5.5-αCD3 (clone SK7), APC-αCD4 (clone RPA-T4), PE-αCD8 (clone RPA-T8), and Alexa488-αpCD3ζ (clone K25-407.69) or -αpERK1/2 (clone 20A; BD Biosciences) for an additional 30 minutes at room temperature. Cells were washed and placed in fixative until data collection.
CD137 expression analysis
Approximately 2×105 T cells were cultured together with 2×104 HLA-A*1101+ BLCL and 10μg/mL peptide in 10% FBS in RPMI at 37°C for 0–24 hours. At T-30min, cells were washed in PBS and stained with LIVE/DEAD® Aqua as above, then fixed (Phosflow Fix Buffer I; BD Biosciences) and placed at 4°C until all time points were collected. At the end of the time course all of the cells were washed and stained with antibodies for CD3 (clone UCHT1, V450), CD8 (clone RPA-T8, Alexa700), CD4 (clone RPA-T4, APC), and CD137 (clone 4B4-1, PE; BD Biosciences) for 30 minutes at room temperature. Finally, cells were washed and placed in fixative until data collection.
Peptide-MHC binding prediction analysis
MHC class I binding prediction scores were calculated for each of the four peptides used in this study according to three prediction models provided by the Immune Epitope Database and Analysis Resource website (http://tools.immuneepitope.org/main/html/tcell_tools.html).
Supplementary Material
SUPPLEMENTARY FIGURE 1. Specificity of tetrameric variants. Peptide-MHC tetramers specific for each of the epitope variants were generated. Tetramer specificity was verified by staining PBMC from HLA-A*1101+ DENV-naïve, HLA-A*1101− DENV-immune, and HLA-A*1101+ DENV-immune subjects as well as an epitope-specific cell line. Cells were gated on live, CD3+ singlet lymphocytes.
SUPPLEMENTARY FIGURE 2. Generation of epitope-specific cell lines. (A) Protocol used to generate and isolate epitope-specific CD8+ T cells lines from the PBMC of three DENV-immune donors. (B) Tetramer staining of bulk cultures two weeks after initial peptide stimulation of PBMC from donor 3. Cells were stained simultaneously with all three tetramer variants, and each row shows two flow plots from the same bulk culture staining sample. Cells were gated on live, CD3+CD4−CD8+ singlet lymphocytes.
Acknowledgments
This work was funded by the National Institutes of Health, grants P01 AI34533 and U19 AI57319. Core resources supported by the Diabetes Endocrinology Research Center grant P30 DK032520 were used. We would like to thank the donors who generously provided PBMC for use in our studies. We thank Drs. Suchitra Nimmannitya and David Vaughn, as well as the staffs of the Queen Sirikit National Institute for Child Health, the Department of Virology, Armed Forces Research Institute of Medical Sciences, and the Department of Transfusion Medicine, Siriraj Hospital, for patient recruitment, blood collection, and clinical, virology, and HLA information. We would also like to thank Joyce Pepe, Jim Coderre, Thomas Greenough, and the UMass Tetramer Core for donating their time and effort to generate the tetramers used here, Pam Pazoles and the UMass Flow Cytometry Core for help with flow cytometry data collection, and Kim West for help with cell culture.
References
- 1.Pinheiro FP, Corber SJ. Global situation of dengue and dengue haemorrhagic fever, and its emergence in the Americas. World Health Stat Q. 1997;50:161–169. [PubMed] [Google Scholar]
- 2.Wilder-Smith A, Gubler DJ. Geographic expansion of dengue: the impact of international travel. Med Clin North Am. 2008;92:1377–1390. x. doi: 10.1016/j.mcna.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Thein S, Aung MM, Shwe TN, Aye M, Zaw A, Aye K, et al. Risk factors in dengue shock syndrome. Am J Trop Med Hyg. 1997;56:566–572. doi: 10.4269/ajtmh.1997.56.566. [DOI] [PubMed] [Google Scholar]
- 4.Guzman MG, Kouri GP, Bravo J, Soler M, Vazquez S, Morier L. Dengue hemorrhagic fever in Cuba, 1981: a retrospective seroepidemiologic study. Am J Trop Med Hyg. 1990;42:179–184. doi: 10.4269/ajtmh.1990.42.179. [DOI] [PubMed] [Google Scholar]
- 5.Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 6.Irie K, Mohan PM, Sasaguri Y, Putnak R, Padmanabhan R. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain) Gene. 1989;75:197–211. doi: 10.1016/0378-1119(89)90266-7. [DOI] [PubMed] [Google Scholar]
- 7.Kurane I, Innis BL, Nimmannitya S, Nisalak A, Meager A, Janus J, et al. Activation of T lymphocytes in dengue virus infections. High levels of soluble interleukin 2 receptor, soluble CD4, soluble CD8, interleukin 2, and interferon-gamma in sera of children with dengue. J Clin Invest. 1991;88:1473–1480. doi: 10.1172/JCI115457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Libraty DH, Endy TP, Houng HS, Green S, Kalayanarooj S, Suntayakorn S, et al. Differing influences of virus burden and immune activation on disease severity in secondary dengue-3 virus infections. J Infect Dis. 2002;185:1213–1221. doi: 10.1086/340365. [DOI] [PubMed] [Google Scholar]
- 9.Green S, Pichyangkul S, Vaughn DW, Kalayanarooj S, Nimmannitya S, Nisalak A, et al. Early CD69 expression on peripheral blood lymphocytes from children with dengue hemorrhagic fever. J Infect Dis. 1999;180:1429–1435. doi: 10.1086/315072. [DOI] [PubMed] [Google Scholar]
- 10.Green S, Vaughn DW, Kalayanarooj S, Nimmannitya S, Suntayakorn S, Nisalak A, et al. Early immune activation in acute dengue illness is related to development of plasma leakage and disease severity. J Infect Dis. 1999;179:755–762. doi: 10.1086/314680. [DOI] [PubMed] [Google Scholar]
- 11.Mathew A, Rothman AL. Understanding the contribution of cellular immunity to dengue disease pathogenesis. Immunol Rev. 2008;225:300–313. doi: 10.1111/j.1600-065X.2008.00678.x. [DOI] [PubMed] [Google Scholar]
- 12.Bashyam HS, Green S, Rothman AL. Dengue virus-reactive CD8+ T cells display quantitative and qualitative differences in their response to variant epitopes of heterologous viral serotypes. J Immunol. 2006;176:2817–2824. doi: 10.4049/jimmunol.176.5.2817. [DOI] [PubMed] [Google Scholar]
- 13.Dong T, Moran E, Vinh Chau N, Simmons C, Luhn K, Peng Y, et al. High pro-inflammatory cytokine secretion and loss of high avidity cross-reactive cytotoxic T-cells during the course of secondary dengue virus infection. PLoS ONE. 2007;2:e1192. doi: 10.1371/journal.pone.0001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imrie A, Meeks J, Gurary A, Sukhbataar M, Kitsutani P, Effler P, et al. Differential functional avidity of dengue virus-specific T-cell clones for variant peptides representing heterologous and previously encountered serotypes. J Virol. 2007;81:10081–10091. doi: 10.1128/JVI.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew A, Kurane I, Green S, Stephens HA, Vaughn DW, Kalayanarooj S, et al. Predominance of HLA-restricted cytotoxic T-lymphocyte responses to serotype-cross-reactive epitopes on nonstructural proteins following natural secondary dengue virus infection. J Virol. 1998;72:3999–4004. doi: 10.1128/jvi.72.5.3999-4004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mongkolsapaya J, Dejnirattisai W, Xu XN, Vasanawathana S, Tangthawornchaikul N, Chairunsri A, et al. Original antigenic sin and apoptosis in the pathogenesis of dengue hemorrhagic fever. Nat Med. 2003;9:921–927. doi: 10.1038/nm887. [DOI] [PubMed] [Google Scholar]
- 17.Mongkolsapaya J, Duangchinda T, Dejnirattisai W, Vasanawathana S, Avirutnan P, Jairungsri A, et al. T cell responses in dengue hemorrhagic fever: are cross-reactive T cells suboptimal? J Immunol. 2006;176:3821–3829. doi: 10.4049/jimmunol.176.6.3821. [DOI] [PubMed] [Google Scholar]
- 18.Moran E, Simmons C, Vinh Chau N, Luhn K, Wills B, Dung NP, et al. Preservation of a critical epitope core region is associated with the high degree of flaviviral cross-reactivity exhibited by a dengue-specific CD4+ T cell clone. Eur J Immunol. 2008;38:1050–1057. doi: 10.1002/eji.200737699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zivna I, Green S, Vaughn DW, Kalayanarooj S, Stephens HA, Chandanayingyong D, et al. T cell responses to an HLA-B*07-restricted epitope on the dengue NS3 protein correlate with disease severity. J Immunol. 2002;168:5959–5965. doi: 10.4049/jimmunol.168.11.5959. [DOI] [PubMed] [Google Scholar]
- 20.Sierra B, Garcia G, Perez AB, Morier L, Rodriguez R, Alvarez M, et al. Long-term memory cellular immune response to dengue virus after a natural primary infection. Int J Infect Dis. 2002;6:125–128. doi: 10.1016/s1201-9712(02)90073-1. [DOI] [PubMed] [Google Scholar]
- 21.Stephens HA, Klaythong R, Sirikong M, Vaughn DW, Green S, Kalayanarooj S, et al. HLA-A and -B allele associations with secondary dengue virus infections correlate with disease severity and the infecting viral serotype in ethnic Thais. Tissue Antigens. 2002;60:309–318. doi: 10.1034/j.1399-0039.2002.600405.x. [DOI] [PubMed] [Google Scholar]
- 22.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis. 2000;181:2–9. doi: 10.1086/315215. [DOI] [PubMed] [Google Scholar]
- 23.Vaughn DW, Green S, Kalayanarooj S, Innis BL, Nimmannitya S, Suntayakorn S, et al. Dengue in the early febrile phase: viremia and antibody responses. J Infect Dis. 1997;176:322–330. doi: 10.1086/514048. [DOI] [PubMed] [Google Scholar]
- 24.Kubo RT, Sette A, Grey HM, Appella E, Sakaguchi K, Zhu NZ, et al. Definition of specific peptide motifs for four major HLA-A alleles. J Immunol. 1994;152:3913–3924. [PubMed] [Google Scholar]
- 25.Sidney J, Grey HM, Kubo RT, Sette A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol Today. 1996;17:261–266. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 26.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Bouvier M. Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue. J Immunol. 2004;172:6175–6184. doi: 10.4049/jimmunol.172.10.6175. [DOI] [PubMed] [Google Scholar]
- 28.Couillin I, Culmann-Penciolelli B, Gomard E, Choppin J, Levy JP, Guillet JG, et al. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J Exp Med. 1994;180:1129–1134. doi: 10.1084/jem.180.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chotiyarnwong P, Stewart-Jones GB, Tarry MJ, Dejnirattisai W, Siebold C, Koch M, et al. Humidity control as a strategy for lattice optimization applied to crystals of HLA-A*1101 complexed with variant peptides from dengue virus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2007;63:386–392. doi: 10.1107/S1744309107013693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Betts MR, Nason MC, West SM, De Rosa SC, Migueles SA, Abraham J, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–4789. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferre AL, Hunt PW, Critchfield JW, Young DH, Morris MM, Garcia JC, et al. Mucosal immune responses to HIV-1 in elite controllers: a potential correlate of immune control. Blood. 2009;113:3978–3989. doi: 10.1182/blood-2008-10-182709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh Y, Germain RN. Single cell analysis reveals regulated hierarchical T cell antigen receptor signaling thresholds and intraclonal heterogeneity for individual cytokine responses of CD4+ T cells. J Exp Med. 1997;186:757–766. doi: 10.1084/jem.186.5.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valitutti S, Muller S, Dessing M, Lanzavecchia A. Different responses are elicited in cytotoxic T lymphocytes by different levels of T cell receptor occupancy. J Exp Med. 1996;183:1917–1921. doi: 10.1084/jem.183.4.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hemmer B, Stefanova I, Vergelli M, Germain RN, Martin R. Relationships among TCR ligand potency, thresholds for effector function elicitation, and the quality of early signaling events in human T cells. J Immunol. 1998;160:5807–5814. [PubMed] [Google Scholar]
- 35.La Gruta NL, Turner SJ, Doherty PC. Hierarchies in cytokine expression profiles for acute and resolving influenza virus-specific CD8+ T cell responses: correlation of cytokine profile and TCR avidity. J Immunol. 2004;172:5553–5560. doi: 10.4049/jimmunol.172.9.5553. [DOI] [PubMed] [Google Scholar]
- 36.Betts MR, Price DA, Brenchley JM, Lore K, Guenaga FJ, Smed-Sorensen A, et al. The functional profile of primary human antiviral CD8+ T cell effector activity is dictated by cognate peptide concentration. J Immunol. 2004;172:6407–6417. doi: 10.4049/jimmunol.172.10.6407. [DOI] [PubMed] [Google Scholar]
- 37.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 38.De Rosa SC, Lu FX, Yu J, Perfetto SP, Falloon J, Moser S, et al. Vaccination in humans generates broad T cell cytokine responses. J Immunol. 2004;173:5372–5380. doi: 10.4049/jimmunol.173.9.5372. [DOI] [PubMed] [Google Scholar]
- 39.Sabin AB. Research on dengue during World War II. Am J Trop Med Hyg. 1952;1:30–50. doi: 10.4269/ajtmh.1952.1.30. [DOI] [PubMed] [Google Scholar]
- 40.Mangada MM, Rothman AL. Altered cytokine responses of dengue-specific CD4+ T cells to heterologous serotypes. J Immunol. 2005;175:2676–2683. doi: 10.4049/jimmunol.175.4.2676. [DOI] [PubMed] [Google Scholar]
- 41.Zivny J, DeFronzo M, Jarry W, Jameson J, Cruz J, Ennis FA, et al. Partial agonist effect influences the CTL response to a heterologous dengue virus serotype. J Immunol. 1999;163:2754–2760. [PubMed] [Google Scholar]
- 42.Lillard JW, Jr, Singh UP, Boyaka PN, Singh S, Taub DD, McGhee JR. MIP-1alpha and MIP-1beta differentially mediate mucosal and systemic adaptive immunity. Blood. 2003;101:807–814. doi: 10.1182/blood-2002-07-2305. [DOI] [PubMed] [Google Scholar]
- 43.Nath A, Chattopadhya S, Chattopadhyay U, Sharma NK. Macrophage inflammatory protein (MIP)1alpha and MIP1beta differentially regulate release of inflammatory cytokines and generation of tumoricidal monocytes in malignancy. Cancer Immunol Immunother. 2006;55:1534–1541. doi: 10.1007/s00262-006-0149-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green S, Kurane I, Edelman R, Tacket CO, Eckels KH, Vaughn DW, et al. Dengue virus-specific human CD4+ T-lymphocyte responses in a recipient of an experimental live-attenuated dengue virus type 1 vaccine: bulk culture proliferation, clonal analysis, and precursor frequency determination. J Virol. 1993;67:5962–5967. doi: 10.1128/jvi.67.10.5962-5967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perez OD, Mitchell D, Campos R, Gao GJ, Li L, Nolan GP. Multiparameter analysis of intracellular phosphoepitopes in immunophenotyped cell populations by flow cytometry. Curr Protoc Cytom. 2005;Chapter 6(Unit 6):20. doi: 10.1002/0471142956.cy0620s32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY FIGURE 1. Specificity of tetrameric variants. Peptide-MHC tetramers specific for each of the epitope variants were generated. Tetramer specificity was verified by staining PBMC from HLA-A*1101+ DENV-naïve, HLA-A*1101− DENV-immune, and HLA-A*1101+ DENV-immune subjects as well as an epitope-specific cell line. Cells were gated on live, CD3+ singlet lymphocytes.
SUPPLEMENTARY FIGURE 2. Generation of epitope-specific cell lines. (A) Protocol used to generate and isolate epitope-specific CD8+ T cells lines from the PBMC of three DENV-immune donors. (B) Tetramer staining of bulk cultures two weeks after initial peptide stimulation of PBMC from donor 3. Cells were stained simultaneously with all three tetramer variants, and each row shows two flow plots from the same bulk culture staining sample. Cells were gated on live, CD3+CD4−CD8+ singlet lymphocytes.


