Abstract
The Fanconi Anemia pathway is activated in response to DNA damage, leading to monoubiquitination of the substrates FANCI and FANCD2 by the Fanconi Anemia core complex. Here we report the crystal structure of FANCL, the catalytic subunit of the Fanconi Anemia core complex at 3.2 Å. The structure reveals an architecture that is fundamentally different from previous sequence-based predictions. The molecule is composed of an N-terminal E2-like fold, which we term the ELF domain, a novel double-RWD (DRWD) domain, and a C-terminal RING domain predicted to facilitate E2 binding. Binding assays demonstrate that the DRWD domain, but not the ELF domain, is responsible for substrate binding.
Introduction
Fanconi Anemia (FA) is an autosomal recessive or X-linked inherited disorder associated with a range of skeletal abnormalities, short stature, microcephaly, bone marrow failure and a predisposition to a variety of cancers1. Patients with FA are susceptible to DNA damaging agents that induce interstrand crosslinks as they have mutations in a group of proteins involved in repairing DNA damage2-11. At the heart of this repair pathway is the FA core complex12, which monoubiquitinates the I/D2 complex13-15 to recruit downstream components required to repair the DNA damage by homologous recombination2,16,17.
At least 13 proteins are involved in Fanconi Anemia, 8 of which form the FA core complex required for the monoubiquitination of the I/D2 complex. However, it has recently been demonstrated that FANCL, FANCI and the E2 enzyme for the pathway, Ube2t, are sufficient for the site-restricted monoubiquitination of FANCD2 in vitro 18, which is the key event in initiating DNA repair16. A lack of structural information, however, has hindered a complete molecular understanding of this essential cascade. The catalytic core of FANCL, FANCI and the E2 enzyme is present in Drosophila melanogaster along with the upstream FANCM and the downstream FANCD2. Furthermore, loss of FANCL and FANCD2 makes Drosophila susceptible to DNA crosslinking agents in a manner analogous to that seen in higher organisms19. Drosophila therefore provides a model system to explore the conserved fundamental aspects of Fanconi Anemia. Based on this premise, we determined the crystal structure of FANCL from Drosophila at 3.2 Å to obtain insights into how FANCL is arranged to perform its functions of E2 binding, substrate binding and monoubiquitination of FANCD2.
Results
Overall structure of FANCL
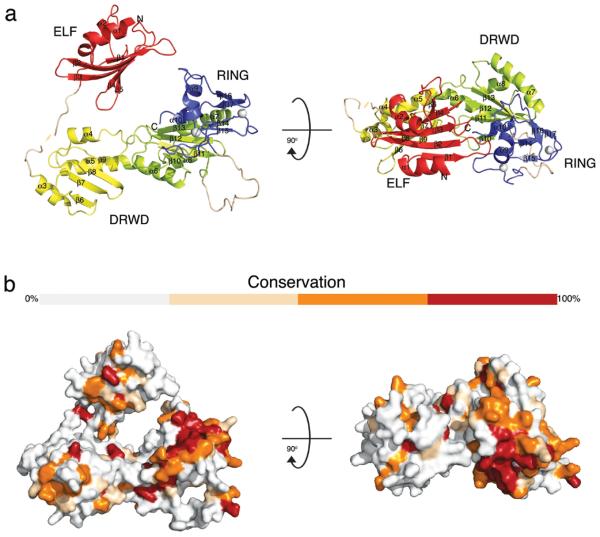
We purified and crystallised FANCL from Drosophila melanogaster. Using single wavelength anomalous dispersion methods from a gold derivative, we determined the 3-dimensional structure and refined it to 3.2 Å (Table 1). Previously, FANCL was classified as a member of the WD40 β-propeller superfamily, with a C-terminal PHD zinc-coordinating motif11. However, our structure reveals that FANCL encompasses 3 distinct domains (Fig. 1a). The N-terminus adopts an E2-like fold (ELF). The central domain comprises a tandem repeat of the RWD-like fold in a single domain with a clear hydrophobic core (Supplementary Fig. 1). This represents a novel domain, as no double RWD (DRWD) fold has previously been described or predicted. The C-terminus is a RING domain with an unusual arrangement of zinc-coordinating residues. This arrangement of domains is likely conserved in all FANCL homologues, given the conservation of the hydrophobic core residues across a wide range of divergent eukaryotes (Fig. 1b and Supplementary Fig. 2). The full structure is extended, with the ELF making no contacts with the rest of the protein.
Table 1.
Data collection and refinement statistics
| SAD data collection | |
|---|---|
| Data collection | NaAu(CN)2 |
| Space group | H32 |
| Cell dimensions | |
| a, b, c (Å) | 188.68, 188.68, 259.36 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 3.2 |
| Rsym or Rmerge | 0.089 (0.479) |
| I / σI | 6.3 (1.6) |
| Completeness (%) | 100 (100) |
| Redundancy | 12.2 (12.5) |
| Refinement | |
| Resolution (Å) | 3.2 |
| No. reflections | 25772 |
| Rwork / Rfree | 20.5 %/ 24.7 % |
| No. atoms | 6088 |
| Protein | 5973 |
| Ligand/ion | 102 |
| Water | 13 |
| B-factors | |
| Protein | 99 |
| Ligand/ion | 107 |
| Water | 79 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.126 |
Values in parentheses are for highest-resolution shell.
Figure 1. Domain architecture of FANCL.
a. Ribbon diagram depicting the FANCL structure. The ELF, DRWD and RING domains are coloured red, yellow/lime and blue, respectively. Zinc atoms are represented by grey spheres and secondary structure elements are labelled. Two views are displayed related by a 90° rotation around the x-axis, as indicated. b. Surface representation of FANCL coloured by sequence conservation between divergent eukaryotic species. Grey indicates non-conserved residues, salmon semi-conservative substitutions, orange conservative substitutions and red conserved residues, shown in the alignment in Supplementary figure 2.
FANCL contains 3 E2-like folds
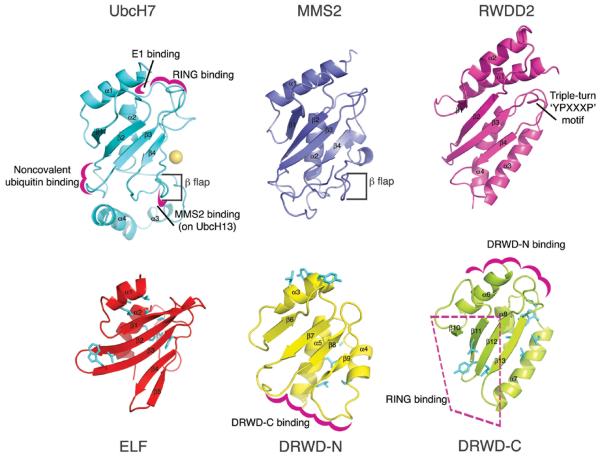
The ELF domain and the DRWD domain together form 3 folds of the ubiquitin-conjugating (UBC) superfamily. This arrangement encoded by FANCL is a novel structural motif, and its presence is intriguing. In order to determine how similar each fold is, we superposed ELF and the 2 lobes of the DRWD domain (Supplementary Fig. 3). Each fold forms a typical 4-stranded β-meander found in all UBC superfamily members (Fig. 2). The ELF domain has a 5th strand (β5) that packs against the meander. In catalytic E2s this 5th strand is replaced by the cysteine-containing β-flap that is important for ubiquitin loading (Fig. 2)20. The non-catalytic E2 variants (e.g. MMS2) exhibit the same structural feature, but lack cysteine. By contrast, the DRWD lobes both have a helix in this position, in common with the RWD subfamily members (Fig. 2 and Supplementary Fig. 3)21,22.
Figure 2. Comparison of UBC-superfamily folds with E2s and RWD proteins.
A catalytically active E2 (UbcH7) is shown in cyan, the inactive E2 MMS2 in blue, and RWD2-2 in magenta on the top row (pdb codes 1fbv23, 1j7d24 and 2daw, respectively). The catalytic cysteine of UbcH7 is represented as a sphere, structural features are indicated, and known protein–protein interaction surfaces are highlighted in pink. The bottom row shows the ELF domain in red, and the N-terminal and C-terminal lobes of the DRWD domain in yellow and lime, respectively. These folds are in the same orientation as the UBC-superfamily members, and protein–protein interaction surfaces are shaded in magenta. Residues highlighted in the structural analysis are coloured cyan.
Previous studies have suggested that a YPXXXP motif is conserved in all RWD proteins, and indeed the FANCL homologues harbour one such motif and a related HPXXXP sequence (Supplementary Fig. 2). However, neither of these motifs is present in the Drosophila sequence, yet the structure clearly contains a DRWD domain. This suggests that a YPXXXP motif can predict an RWD domain, but is not essential. Rather, our analysis indicates that a helix in the position of the β-flap of catalytic UBC superfamily members marks out the subfamily of RWD-domain proteins (Fig. 2).
We also examined whether the ELF domain bears similarity to the Ube2t and Ube2w, E2 enzymes that interact with FANCL. There is 22 % sequence identity with each, and the ELF structure superposes on the core of Ube2t (pdb code 1yh2) with an rmsd of 2.2 Å2 over 52 atoms, comparable to ELF similarity with other E2s (Supplementary Tables 1 and 2). Given that the similarity is comparable for all the structurally characterised E2s, and given the lack of contacts between the ELF and the rest of the protein, we do not think that the ELF domain substitutes for Ube2t.
Structural analysis of FANCL domains
Surface analysis of the ELF domain reveals an exposed hydrophobic patch created by Val33 of β2 and Trp40 of β3 (Fig. 3a). There is also a hydrophobic groove formed by the loop connecting β3 and β4 and the loop connecting β5 to α2, with residues Leu53, Phe56 and Leu76 forming the base. The groove leads to a further pocket between α1 and the same two loops, formed by the hydrophobic residues Val7, Leu10 and Leu82 (Fig. 3a). Both the hydrophobic patch and the groove are buried in packing surfaces within the crystal, despite different orientations in the asymmetric unit, supporting the notion that these are important protein interaction surfaces. Furthermore, these regions are well conserved in FANCL homologues from divergent species (Supplementary Fig. 2). They are also implicated in protein interactions in other members of the UBC fold superfamily23,24, with the Val33-Trp40 patch located in the non-covalent ubiquitin-binding site of MMS224, and the Val7-Leu10-Leu82 and Leu53-Phe56-Leu76 patches in the E1- and RING-binding sites of E2s23,25 (Fig. 2).
Figure 3. Protein–protein interaction surfaces on FANCL.
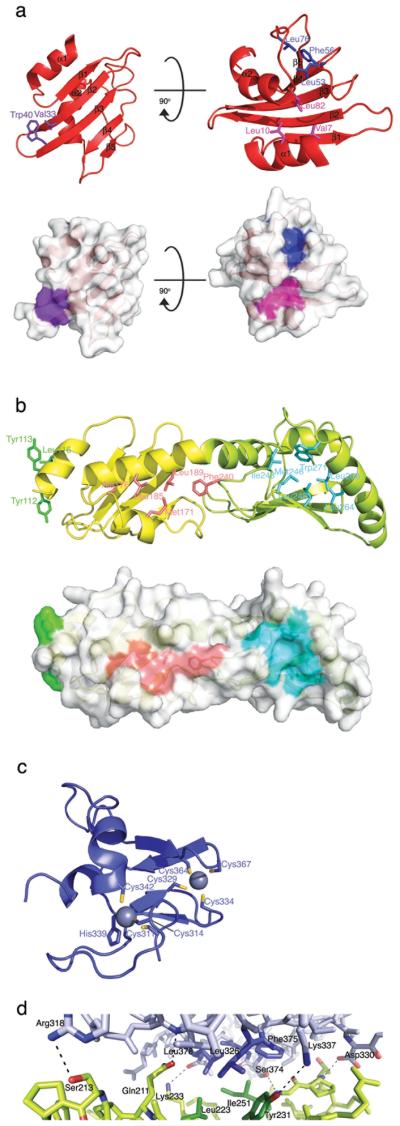
a. The ELF domain has 3 exposed hydrophobic patches coloured purple, pink and blue. Two views related by a 90° rotation around the x-axis are shown, represented as both surface patches and ball-and-stick residues. Individual residues are labelled. b. The DRWD domain has 3 hydrophobic patches shown in salmon, green and cyan. The view is 180° around the x-axis in relation to the left-hand panel in Figure 1a, represented as both surface patches and ball-and-stick residues. Individual residues are labelled. c. The RING domain coordinates 2 zinc atoms represented by grey spheres. Coordinating residues are labelled. d. The DRWD–RING interface. DRWD residues are indicated in lime, RING residues are in blue, and individual residues are labelled.
Recently, FANCL was predicted to contain one RWD-like fold24. However, the structure reveals a DRWD domain, consisting of two β-meanders linked via a kinked helix (Fig. 3b). This is the first time that this tandem arrangement of RWD folds has been observed. The DRWD domain presents a surface-exposed strip of hydrophobic residues, Tyr112, Tyr113 and Leu116 (Fig. 3b). The location of these residues corresponds to the first helix in UBC family members that is frequently used in protein–protein interactions25(Fig. 2). These residues are also well conserved between different species (Supplementary Fig. 2). The hydrophobic groove between the proximal helices of lobe one forms a patch including residues Met171, Ala176, Leu185, Leu189 and Phe240 (Fig. 3b). A second patch, between helix α7 and the sheet, runs towards the first and involves Ile243, Met246, Leu248, Leu264, Leu268 and Trp271 (Fig. 3b). Conservation of hydrophobicity is, however, less marked in these regions (Supplementary Fig. 2).
The RING domain broadly exhibits the same fold characteristics of the cross brace seen in other RING structures (Fig. 3c)26. The conserved helix complete with tryptophan at the C-terminal end is present but the cysteines and histidines are arranged in the sequence as [Cys]4, His, [Cys]3, rather than the typical [Cys]3, His, [Cys]4, and the spacing between the third and fourth cysteines is an extended 4 residues rather than the usual 2 residues. This is also observed throughout the FANCL homologues (Supplementary Fig. 2). The RING domain interacts tightly with the C-terminal lobe of the DRWD domain (DRWD-C) with a hydrophobic core encompassing Leu326, Phe375 and Leu378 burying themselves against Leu223, Tyr231 and Ile251, and an additional hydrogen bonding network surrounding the core (Fig. 3d). Comparison of the RING with Rbx1 of the SCF (Skp1-Rbx1-Cullin-F-box) E3 ligase reveals that Rbx1 binds Cul1 through an N-terminal β-strand that forms part of a sheet with Cul127, while the RING domain of FANCL extensively interacts across the sheet of DRWD-C (Fig. 3d). The RING contains a hydrophobic patch comprising Ile313, Leu343, Pro365, Phe366 and the conserved Trp346. This hydrophobic patch is usually reserved for E2 binding in other RING proteins such as c-Cbl23 and cIAP228, and the RING domain is sufficient to pull down Ube2t29. As this patch is conserved in all the FANCL homologues, it is therefore likely to be the interaction surface for Ube2t.
The DRWD domain is required for substrate binding
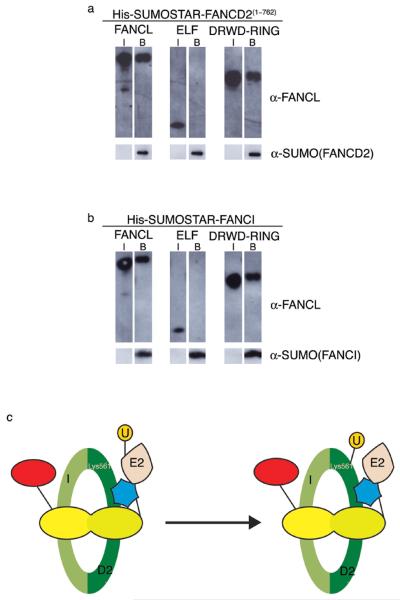
In order to determine which domains are involved in binding the substrates, FANCI and FANCD2, we generated deletion mutants of FANCL comprising either ELF or DRWD-RING. We performed pulldown binding assays with His-SUMOSTAR-FANCI or FANCD2(1–762). We find that the DRWD-RING construct binds as well as full-length FANCL, while the ELF domain cannot bind the substrates (Figs. 4a and 4b). These data suggest that the binding site(s) for FANCI and FANCD2 reside within the DRWD domain.
Figure 4. The DRWD domain recruits substrate.
a. Pulldown binding assays using His-SUMOSTAR-FANCD2(1–762) as bait. Input samples (I) of FANCL species and samples bound by His-SUMOSTAR-FANCD2(1–762) (B) are indicated. Samples are analysed by Western blotting using anti-FANCL or anti-SUMO antibodies. b. Pulldown binding assays using His-SUMOSTAR-FANCI as bait. Input samples (I) of FANCL species and samples bound by His-SUMOSTAR-FANCI (B) are indicated. Samples are analysed by Western blotting using anti-FANCL and anti-SUMO antibodies. c. A schematic representation of FANCD2 monoubiquitination. The I/D2 complex is represented in green, with Lys561 indicated. The ELF, DRWD and RING domains are red, yellow and blue, respectively. The E2 and ubiquitin (U) are indicated.
Discussion
Our structure of FANCL reveals a very different domain architecture than predicted from primary sequence analyses or secondary structure predictions. Previous mutational analyses based on the predicted WD40-propeller structure of FANCL identified non-functioning mutants30. Our structure now provides a molecular explanation for these data. We mapped the ‘WD40’ deletions onto the structure, which predicts each deletion to result in unfolded protein (Supplementary Fig. 4). Point mutations Y111E, W201A and W275A (human numbering) each lead to a marked decrease in Fanconi Anemia core complex (FACC) assembly, and a diminished but appreciable level of FANCD2 monoubiquitination30. Tyr111 corresponds to Tyr113 of DmFANCL (Supplementary Fig. 4), and is part of an exposed hydrophobic patch (Fig. 3b). It is possible that this patch is involved in complex assembly even though it is conserved in DmFANCL, but it is unlikely to be required for substrate and/or E2 binding given the continued monoubiquitination of FANCD2. W201A corresponds to Tyr197, and this mutation would disrupt the DRWD domain core to some extent, but maintain the hydrophobic nature (Supplementary Fig. 4). However, the ELF and RING domains should remain intact. Gurtan et al. also mutated a conserved arginine residue (Arg226, human numbering) to a glutamate, and found that this mutation was completely deficient in both FACC assembly and FANCD2 monoubiquitination. Arg222 (Drosophila numbering) forms a buried salt bridge in the core of the DRWD domain with the conserved Asp204 (Supplementary Figs. 1 and 4). The residues surrounding this salt bridge are well conserved (Supplementary Fig. 2), and form the core of the DRWD domain. R222E would render the protein unable to fold since a buried electrostatic repulsion would be intolerable. We predict that this would disrupt the tertiary structure of FANCL and hence abolish both FACC assembly and activity, as observed30. Trp275 corresponds to Trp271 and in our structure contributes to an exposed hydrophobic patch (Fig. 3b and Supplementary Fig. 4). Mutation to alanine would be unlikely to disrupt the overall fold of the domain, and therefore could play in role in FACC assembly, although it is conserved in the Drosophila protein. Finally, W341G mutation has no impact on FACC assembly, but does completely abolish FANCD2 monoubiquitination. W341G corresponds to Trp346 and is required for E2 binding30. The W341G mutation suggests that the RING is required for FANCD2 monoubiquitination, but not FACC assembly.
Comparisons of Drosophila and human FANCL sequences based on a structural alignment reveal 3 sites of significant difference. First, the loop connecting the ELF to the DRWD is conserved in length, but not sequence. Second, the region comprising β9-α4 is not well conserved (Fig. 1b and Supplementary Fig. 2). Finally, the loop linking the DRWD and RING domains has a 7-residue insertion in Drosophila (Supplementary Fig. 2). The human FACC involves more proteins than the Drosophila system seems to require. These differences may be sufficient for human FANCL to require additional proteins for function. Drosophila FANCL is the most divergent with sequence identity ranging from 19 % with zebrafish FANCL to 22 % with bovine FANCL. Human, mouse, cow, chicken, Xenopus and zebrafish have sequence identities ranging from 58% to 88% (Supplementary Tables 3-6). However, sequence similarity between Drosophila and human FANCL is ~60 %. The ELF, DRWD-C and RING domains are 67, 68 and 72 % similar respectively, while the least similarity exists within the N-terminal half of the DRWD domain (52 %). Clearly, the differences between human and Drosophila FANCL impact the solubility of the protein as we can express and purify milligramme quantities of Drosophila but not human FANCL. However, given the similarity between the FANCLs from divergent species, it is also possible that Drosophila have as-yet-unidentified FACC components. Indeed, a recent report suggests that the slime-mould Dictyostelium discoideum has at least a FANCE homologue31. Finally, it is also conceivable that the complex nature of mammalian cell-cycle control and DNA repair requires more subtle regulation than necessary in Drosophila.
Our structure provides the first molecular insight into FANCD2 monoubiquitination, the key event in the Fanconi Anemia pathway. It is also the first full-length FA protein to be structurally characterised. Taken together with our biochemical pulldown data, we propose a model in which the DRWD domain with multiple protein interaction surfaces binds substrate(s) with the RING domain recruiting the E2 enzyme, enabling FANCL to bind its protein substrate and facilitate the monoubiquitination of FANCD2 (Fig. 4c).
Methods
Protein purification
We cloned Drosophila melanogaster FANCL into pET28b (Invitrogen), with an N-terminal His-SUMO tag, and expressed in E. coli in LB media with antibiotics and 0.5 mM ZnCl2. We cultured cells at 37°C to an OD 600nm of 0.8, induced with 0.25 mM IPTG, and cultured overnight at 16°C. We lysed cells by sonication in 100 mM Tris pH 8.0, 500 mM NaCl, 250 μM Tris-carboxy-ethyl-phosphine (TCEP) and purified via Ni2+-affinity chromatography. We cleaved His-SUMO-FANCL overnight at 4°C with SUMO protease, Ulp132 at a ratio of 1:30 Ulp1:His-SUMO-FANCL and purified by size exclusion chromatography (SuperDex200), eluting at ~ 40 kDa. We analysed the purified protein by Dynamic Light Scattering analysis and assessed it as monodisperse. We concentrated FANCL to ~ 30 mg ml−1 in 50 mM ammonium citrate, 50 mM bis-tris-propane pH 8.0, 500 μM TCEP and 100 μM ZnCl2, flash-froze in liquid nitrogen and stored at −80°C.
We generated ELF (residues 1–104), ELF-DRWD (1–294), DRWD (105–294), DRWD-RING (105–381), RING (310–381) using restriction-free cloning with the His-SUMO-FANCL plasmid, and purified proteins using Ni2+-affinity chromatography, followed by cleavage with Ulp1 and subsequent size-exclusion chromatography. The ELF-DRWD, DRWD and RING constructs were unstable, expressed with significant quantities of chaperones, and aggregated during purification. This is due to the exposure of the interface between DRWD and RING.
We cloned D. melanogaster FANCI and FANCD2(1–762) into the SUMOstar vector (LifeSensors Inc), with an N-terminal His-SUMOstar fusion tag, and transposed DH10bac cells (Invitrogen) to create bacmids. We generated baculoviruses in Sf9 cells, expressed protein in Sf9 cells, and harvested after 3 days. We sonicated cells in 100 mM Tris pH 8.0, 500 mM NaCl, 250 μM TCEP and purified proteins via Ni2+-affinity chromatography, eluted with 300 mM imidazole, and dialysed into 100 mM Tris pH 8.0, 500 mM NaCl, 250 μM TCEP.
Structure determination
We grew crystals in 3 days by hanging drop vapour diffusion at 4°C in spacegroup H32, with cell dimensions a=b=188.7 Å, c=259.4 Å, α=β=90°, γ=120°. Solvent content estimates suggested 2 monomers per asymmetric unit with a solvent content of 72 %. Crystallisation conditions were 1 M ammonium citrate, 100 mM bis-tris-propane, pH 8.0, with 1 mM TCEP, and 50 μM ZnCl2. Crystals were harvested and cryo-cooled after protection with 35 % maltose. We obtained derivative crystals for the single anomalous dispersion experiment using 1mM gold cyanide soaks for 5 hours prior to cryo-cooling. We collected data at the DIAMOND synchrotron light source, beamline IO4, at the gold absorption edge (1.04 Å). Data were processed and scaled using Mosflm and Scala 33. Phasing was performed using SHELXD34 and SHARP35 with 13 gold sites in the asymmetric unit. The phasing power of the anomalous signal was 1.18, the R-cullis 0.87, and the overall figure of merit was 0.39. Electron density modification and solvent flattening with PIRATE31 improved the maps, generating interpretable density. The model was built manually using the graphics program COOT32, and refined using PHENIX36, with 98.5% of residues in the allowed regions of the Ramachandran plot. Details of geometry and refinement statistics are in Table 1. Figures were made using PyMol37.
Structural analysis
There are 2 conformations of FANCL in the asymmetric unit, with the ELF domain mobile, rotating ~60° to maintain packing restraints within the crystal. The monomers superpose with an rmsd of 0.4 Å2 over 274 atoms, starting from residue 107. The difference is due to the rotation of the ELF domain. Monomer A is almost complete, lacking density for residues 1–4 and 103. Monomer B lacks residues 1–5 and 95–106. Consequently, the structural depictions and analyses refer to monomer A. The arrangement of the 2 monomers is intriguing: ELF-A interfaces with RING-B and vice versa. The 2 interfaces are not identical due to the breaking of symmetry induced by the mobile ELF domain. We performed a protein interfaces, surfaces and assemblies (PISA) analysis38 to ascertain whether an oligomer is likely. The most stable oligomer is a hexameric arrangement of 3 dimers achieved through the ELF-A-RING-B and ELF-B-RING-A asymmetric pairing we see in the asymmetric unit, and the hexamer formed through consecutive DRWD domains. 12% of each monomer is buried in the AB interface, and the interfaces across the DRWD domain involve ~3% of residues. During purification we observe a broad shoulder preceding the main peak. This material is neither monodisperse nor crystallisable, and we see no evidence of a distinct, higher order species. The monomer peak runs true when re-passed over the gel-filtration column. Taken together with the purification of a soluble, stable, monodisperse monomer in solution, it is unlikely that full-length Drosophila FANCL oligomerises.
We performed sequence analyses using ClustalW alignment and manual adjustment. We assessed sequence similarity manually as the number of residues that maintained a similar size and/or chemical property (Supplementary Tables 3-6).
We superposed structures using the lsq commands in O39, and made structural comparisons with E2s using the DALI server to identify top hits (Supplementary Tables 1 and 2), followed by manual analysis in O.
Binding assays
We diluted 10 μg of each FANCL construct in a total volume of 150 μL buffer (100 mM Tris pH 8, 400 mM NaCl, 250 μM TCEP, 10 μM ZnCl2). We added 10 μL of this to 1 mL of the same buffer, and added 100 μL of 10 ng/μL His-SUMOSTAR-FANCI or His-SUMOSTAR-FANCD2(1–762) (Figs. 4a and 4b). Controls were FANCL constructs alone (input), and FANCL constructs with His-SUMO added at the same level as the His-SUMOstar-FANCD2(1–762) or FANCI proteins (data not shown). Assays were incubated for 1 hour at 4°C on a roller. 50 μL of Ni2+-resin was added (100 μL of a 50 % slurry), and incubated for a further hour. We applied each assay to a disposable column, and washed with 10 mL assay buffer, resuspended in 100 μL of buffer and added to 50 μL 2x SDS loading buffer. 5 μL were run on a gel and analysed by Western blotting using Drosophila FANCL and SUMO (AbCam) antibodies (Figs. 4a and 4b).
Supplementary Material
Acknowledgements
We thank M. Way and F. Pinto for help with improving the manuscript, and V. Chaugule for critical comments, discussion and technical assistance. We thank L. Wood for assistance with sequence analysis and S. Kjaer and S. Kisakye-Nambozo of the Protein Production Facility for generation of the baculoviruses and subsequent Sf9 infection. All authors are funded by Cancer Research UK.
Footnotes
Accession code
Coordinates and structure factors for FANCL have been deposited in the Protein Data Bank with accession code 3K1L.
Supplementary information accompanies this paper.
References
- 1.Alter BP. Fanconi's anemia and malignancies. Am J Hematol. 1996;53:99–110. doi: 10.1002/(SICI)1096-8652(199610)53:2<99::AID-AJH7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 2.Howlett NG, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- 3.Strathdee CA, Duncan AM, Buchwald M. Evidence for at least four Fanconi anaemia genes including FACC on chromosome 9. Nat Genet. 1992;1:196–198. doi: 10.1038/ng0692-196. [DOI] [PubMed] [Google Scholar]
- 4.Lo Ten Foe JR, et al. Expression cloning of a cDNA for the major Fanconi anaemia gene, FAA. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 5.Meetei AR, et al. X-linked inheritance of Fanconi anemia complementation group B. Nat Genet. 2004;36:1219–1224. doi: 10.1038/ng1458. [DOI] [PubMed] [Google Scholar]
- 6.Timmers C, et al. Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell. 2001;7:241–248. doi: 10.1016/s1097-2765(01)00172-1. [DOI] [PubMed] [Google Scholar]
- 7.de Winter JP, et al. The Fanconi anaemia gene FANCF encodes a novel protein with homology to ROM. Nat Genet. 2000;24:15–16. doi: 10.1038/71626. [DOI] [PubMed] [Google Scholar]
- 8.de Winter JP, et al. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 9.Levitus M, et al. Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood. 2004;103:2498–2503. doi: 10.1182/blood-2003-08-2915. [DOI] [PubMed] [Google Scholar]
- 10.Levitus M, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37:934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 11.Meetei AR, et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat Genet. 2003;35:165–170. doi: 10.1038/ng1241. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Higuera I, Kuang Y, Naf D, Wasik J, D'Andrea AD. Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol Cell Biol. 1999;19:4866–4873. doi: 10.1128/mcb.19.7.4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Higuera I, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 14.Smogorzewska A, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims AE, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–567. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Andreassen PR, D'Andrea AD. Functional interaction of monoubiquitinated FANCD2 and BRCA2/FANCD1 in chromatin. Mol Cell Biol. 2004;24:5850–5862. doi: 10.1128/MCB.24.13.5850-5862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakanishi K, et al. Interaction of FANCD2 and NBS1 in the DNA damage response. Nat Cell Biol. 2002;4:913–920. doi: 10.1038/ncb879. [DOI] [PubMed] [Google Scholar]
- 18.Alpi AF, Pace PE, Babu MM, Patel KJ. Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol Cell. 2008;32:767–777. doi: 10.1016/j.molcel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Marek LR, Bale AE. Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst) 2006;5:1317–1326. doi: 10.1016/j.dnarep.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton KS, et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 21.Nameki N, et al. Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 2004;13:2089–2100. doi: 10.1110/ps.04751804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burroughs AM, Jaffee M, Iyer LM, Aravind L. Anatomy of the E2 ligase fold: implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J Struct Biol. 2008;162:205–218. doi: 10.1016/j.jsb.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 24.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 25.Huang DT, et al. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8's E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 26.Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 27.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 28.Mace PD, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–31640. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 29.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23:589–596. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 30.Gurtan AM, Stuckert P, D'Andrea AD. The WD40 repeats of FANCL are required for Fanconi anemia core complex assembly. J Biol Chem. 2006;281:10896–10905. doi: 10.1074/jbc.M511411200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang XY, et al. Xpf and not the Fanconi anaemia proteins or Rev3 accounts for the extreme resistance to cisplatin in Dictyostelium discoideum. PLoS Genet. 2009;5:e1000645. doi: 10.1371/journal.pgen.1000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project, Number 4 The CCP4 Suite: Programs for Protein Crystallography. Acta. Cryst. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta. Cryst. 2002;D58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 35.de La Fortelle E, Bricogne G. Maximum-Likelihood Heavy-Atom Parameter Refinement for the Multiple Isomorphous Replacement and Multiwavelength Anomalous Diffraction Methods. Meth. Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta. Cryst. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 37.DeLano WL. The PyMol molecular graphics system. DeLano Scientific LLC, Palo Alto, CA, USA. 2008 [Google Scholar]
- 38.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 39.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Cryst. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 40.Sundquist WI, et al. Ubiquitin recognition by the human TSG101 protein. Mol. Cell. 2004;13:783–789. doi: 10.1016/s1097-2765(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita A, Maeda K, Maeda Y. Crystal structure of CapZ: structural basis for actin filament barbed end capping. EMBO J. 2003;22:1529–1538. doi: 10.1093/emboj/cdg167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vedadi M, et al. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol. Biochem. Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 43.Zhang M, et al. Chaperoned ubiquitylation—crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 44.Moraes TF, et al. Crystal structure of the human ubiquitin conjugating enzyme complex hMms2-hUbc13. Nat. Struct. Biol. 2001;8:669–673. doi: 10.1038/90373. [DOI] [PubMed] [Google Scholar]
- 45.Wei RR, et al. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–1009. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat. Struct. Mol. Biol. 2009;16:658–666. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teo H, Veprintser D, Williams RL. Structural insights into endosomal sorting complex required for transport (ESCRT-I) recognition of ubiquitinated proteins. J. Biol. Chem. 2004;279:28689–28696. doi: 10.1074/jbc.M400023200. [DOI] [PubMed] [Google Scholar]
- 49.Lewis MJ, Saltibus LF, Hau DD, Xiao W, Spyracopolous L. Structural basis for non-covalent interaction between ubiquitin and the ubiquitin conjugating enzyme human MMS2. J. Biomol. NMR. 2006;34:89–100. doi: 10.1007/s10858-005-5583-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.