Abstract
Objective
The genetic basis for Autism Spectrum Disorder (ASD) symptoms in children with Attention-Deficit Hyperactivity Disorder (ADHD) was addressed using a genome-wide linkage approach.
Method
Participants of the International Multi-Center ADHD Genetics study comprising 1143 probands with ADHD and 1453 siblings were analyzed. The total and subscale scores of the Social Communication Questionnaire (SCQ) were used as quantitative traits for multipoint regression-based linkage analyses on 5407 autosomal single-nucleotide polymorphisms applying MERLIN-regress software, both without and with inclusion of ADHD symptom scores as covariates.
Results
The analyses without ADHD symptom scores as covariates resulted in 3 suggestive linkage signals, i.e., on chromosomes 15q24, 16p13, and 18p11. Inclusion of ADHD symptom scores as covariates resulted in additional suggestive loci on chromosomes 7q36 and 12q24, whereas the LOD score of the locus on chromosome 15q decreased below the threshold for suggestive linkage. The loci on 7q, 16p, and 18p were found for the SCQ restricted & repetitive subscale, that on 15q was found for the SCQ communication subscale, and that on 12q for the SCQ total score.
Conclusions
Our findings suggest that QTLs identified in this study are ASD specific, although the 15q QTL potentially has pleiotropic effects for ADHD and ASD. This study confirms that genetic factors influence ASD traits along a continuum of severity, as loci potentially underlying ASD symptoms in children with ADHD were identified even though subjects with autism had been excluded from the IMAGE sample, and supports the hypothesis that differential genetic factors underlie the three ASD dimensions.
Keywords: ADHD, autism spectrum disorder, linkage, comorbidity
Introduction
Children with Attention-Deficit/Hyperactivity Disorder (ADHD) often show symptoms of Autism Spectrum Disorders (ASD), with around one third of those children meeting criteria for a Pervasive Developmental Disorder according to the Diagnostic and Statistical Manual fourth edition (DSM-IV).1,2 Studies on the common etiology of ADHD and ASD in community twin samples have shown that a substantial part of genetic influences are shared between ADHD and ASD.3,4 Our own studies confirmed the familiality of ASD symptoms in children with ADHD and their siblings.5,6 Moreover, genetic overlap is suggested by genetic linkage studies for ADHD and ASD, in which overlapping sets of suggestive disease loci have been identified, 7–9 including 5p13 and 9q33.10–16 Both of these loci are supported by at least two independent studies of autism (although not necessarily genome wide significant), and at least one study of ADHD.
Although several linkage studies have been performed for ADHD and ASD separately, genome-wide linkage analysis has thus far never been applied to the investigation of the co-occurrence of ASD symptoms and ADHD. Addressing ASD symptoms in children with ADHD may aid in the search for susceptibility loci for the overlapping disorders, as this may reduce heterogeneity.5,6 A quantitative trait locus (QTL) approach is the most suitable method to find those loci, given that children diagnosed with ADHD according to DSM-IV criteria have ASD symptoms below diagnostic cut-offs. The QTL approach preserves all variation in ASD severity, which increases power of the analyses.17 The QTL approach follows the assumption that ASD traits lie along a continuum of severity, which is supported by findings of elevated levels of ASD symptoms in parents and siblings of cases compared to controls,18,19 and variation of ASD traits that has been found in the general population.20,21
In the present study, we aimed at identifying loci that underlie ASD symptoms in children with ADHD, all participating in the International Multicenter ADHD Genetics (IMAGE) project. We investigated both the total level of ASD symptoms as well as scores on the three ASD symptom domains, i.e., qualitative impairments in social interaction, in communication, and restricted repetitive and stereotyped patterns of behavior, thus taking into account the potential differential genetic origin of different ASD symptom domains (for a review, see Happé & Ronald 22). QTL linkage analyses for the different ASD domains were carried out using 5407 single nucleotide polymorphisms (SNPs) spanning the entire genome.
Method
Participants
The study was conducted in participants of the IMAGE project.23 This is an international collaborative study in seven European countries (Belgium, Germany, Ireland, Spain, Switzerland, the Netherlands, and the United Kingdom) and Israel that aims to identify genes that increase the risk of ADHD using QTL linkage and association strategies.24 Ethical approval was obtained from National Institutes of Health recognized local ethical review boards, and all families gave written informed consent prior to participation. To participate in IMAGE, probands had to have a clinical diagnosis of DSM-IV combined subtype of ADHD, as well as one or more full siblings and at least one biological parent available for ascertainment of clinical information and DNA collection. All children participating were aged 5–17 and of European Caucasian descent. Exclusion criteria were an IQ < 70, autism (see below for a detailed description of assessment), epilepsy, brain disorders, and any genetic or medical disorder associated with externalizing behaviors that might mimic ADHD.
The children were recruited from families referred to participating (academic) child psychiatric and pediatric outpatient clinics. All probands were included after having completed clinical evaluations by a pediatrician or child psychiatrist prior to the study. The clinical diagnosis of the ADHD probands and siblings (if possible) was verified with the Parental Account of Childhood Symptoms (PACS)25 by a trained interviewer. A standard algorithm for PACS was applied to all raw data to yield diagnoses based on operational DSM-IV criteria. For more detailed information see Brookes et al.24
Children were excluded from participation in IMAGE when classical or atypical autism was diagnosed. For this, both probands and siblings were screened using the Social Communication Questionnaire (SCQ)26 in conjunction with the prosocial scale of the Strengths and Difficulties Questionnaire (SDQ).27 Individuals falling above thresholds for the SCQ (i.e., ≥15) and SDQ (i.e., ≤4) were further evaluated using the autism spectrum disorder section of the PACS. Classical or atypical autism was regarded to be present in these children in case they showed ASD symptoms in at least two out of three DSM-IV autism domains, together with a developmental delay in at least one autism domain before the age of 3, according to DSM-IV definitions. Attention was given especially to symptoms that distinguish ASD from ADHD. Furthermore, a lack of relationships with peers alone was not sufficient for social impairment of the autistic type. The final sample investigated in this study included 1143 ADHD probands and 1453 siblings from 1143 families.
Measures
To assess ASD symptoms, the life-time version of the SCQ was used. The SCQ is a parent-rated questionnaire that consists of 40 items based on the Autism Diagnostic Interview-Revised.28 Three subscales that relate directly to the three autism core domains as defined in the DSM-IV can be scored, i.e. the social (15 items), the communication (13 items), and the restricted & repetitive (8 items) scales. The SCQ appeared to have good discriminative validity for the discrimination of ASD from non-ASD in at-risk samples at the cut-off of >/= 15 established by Berument and colleagues.26 In the final sample, 233 children had SCQ scores of 15 or higher, 189 of whom were probands (for additional information on SCQ scores, see Table 1).
Table 1.
Phenotypic characteristics of the International Multi site ADHD Genetics (IMAGE) project sample
| Mean (sd) | ||
|---|---|---|
| probands (n=1143) | siblings (n=1453) | |
| Age in years | 10.9 (2.8) | 10.9 (3.4) |
| IQ | 100.6 (15.8) | 102.1 (14.1) |
| Gender (% female) | 13.2 | 48.4 |
| SCQ scores | ||
| SCQ total | 8.6 (6.2) | 4.5 (4.2) |
| social | 2.8 (2.8) | 1.3 (1.9) |
| communication | 3.4 (2.4) | 2.3 (2.0) |
| restricted & repetitive | 1.8 (1.9) | 0.7 (1.3) |
| Conners scores | ||
| inattentive | 19.4 (5.1) | 7.6 (7.4) |
| hyperactive impulsive | 17.7 (5.6) | 5.8 (6.4) |
Note. SCQ=Social Communication Questionnaire
The DSM-IV inattentive and hyperactive-impulsive scales of the Conners’ parent rating scale, long form, were used to assess ADHD symptom severity in both probands and siblings.29 The correlation between the Conners’ DSM-IV total score (calculated by adding up the Conners’ DSM-IV inattentive and hyperactive-impulsive scale scores) and the SCQ total score was .44 (p<0.01) in our sample.
Genotyping and Data Cleaning
An extensive description of DNA extraction and genotyping is provided elsewhere.24,30 Briefly, DNA was extracted from blood samples or immortalized cell lines at Rutgers University Cell and DNA Repository, New Jersey, USA. In a few cases mouth swabs were used to extract DNA at the Social Genetic and Developmental Psychiatry laboratories in London, UK. Illumina BeadArrayTM technology on a BeadLab system was used, which was provided by the Center for Inherited Disease Research (CIDR; http://www.cidr.jhmi.edu/). A total of 5,545 autosomal SNPs from the Illumina Linkage IVb SNP panel were successfully assayed with a call rate of 99.6% and a reproduction rate of 99.994%. The markers were ordered and placed on the physical map according to Genome Build 35. Interpolated genetic distances from the deCODE genetic map were used to estimate map distances. 31
Pedigree errors were identified and corrected by testing pairwise subject relationships with the program Relpair.32 Genotypes causing Mendelian inconsistencies were identified by PEDCHECK and removed by a custom script.33 Unlikely genotype combinations leading to double recombinations over short genetic distance in a few cases were removed by MERLIN.34,35 Following data cleaning, 5,407 autosomal SNPs with an average resolution of 1.66 SNP/centimorgan (cM) were entered into the linkage analyses.
Data analyses
Multivariate QTL linkage was examined for the SCQ total scale and each of the SCQ subscales. Age and gender were used as covariates. All analyses were repeated with scores on the ADHD/DSM-IV inattentive and hyperactive-impulsive scales of the Conners’ parent rating scale, long form, simultaneously added as additional covariates. The latter analyses served to investigate whether QTLs found in the first analyses were independent from ADHD (which would be the case if signals persisted after inclusion of ADHD scores), and also whether new QTLs would appear. Signals that would disappear could be suggestive of pleiotropic effects, while remaining and new QTLs could harbor genes uniquely contributing to ASD symptoms.
The linkage analyses were carried out using Merlin-regress software, which implements a regression based procedure using trait-squared sums and differences to predict Identity by Descent (IBD) sharing between relative pairs.34,36 With the population distribution parameters of mean, variance, and heritability specified, this method can be applied to selected samples with a statistical power similar to variance component linkage tests.30 Because treating tightly linked markers as independent markers can inflate LOD scores, we applied the criteria of r2 < 0.05 between SNPs to cluster SNPs into combined markers.30,37 For this study, heritability estimates were based on values previously reported for the broader ASD spectrum in twins from the general population by Constantino & Todd 38 and Ronald et al.39. Empirical p-values were derived using Merlin software by running 1,000 simulations under the null-hypothesis of no linkage, while preserving the original phenotypes, family structures, allele frequencies, LD structure, and missing data pattern.30,34 In each simulated data set, linkage was defined as peak LOD scores equal to or higher than the experimental LOD.
Results
In Table 1, sample characteristics are presented. Figures 1 to 5 and Table 2 and 3 show the results of the multivariate QTL linkage analyses.
Figure 1.
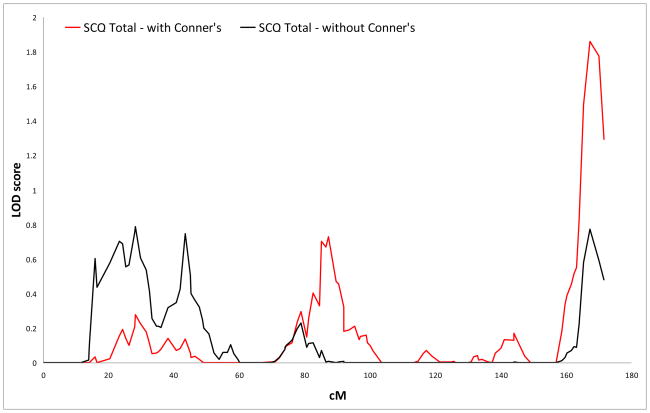
Logarithm of the odds (LOD) score graphs of the Social Communication Questionnaire (SCQ) total scale on chromosome 12.
Figure 5.
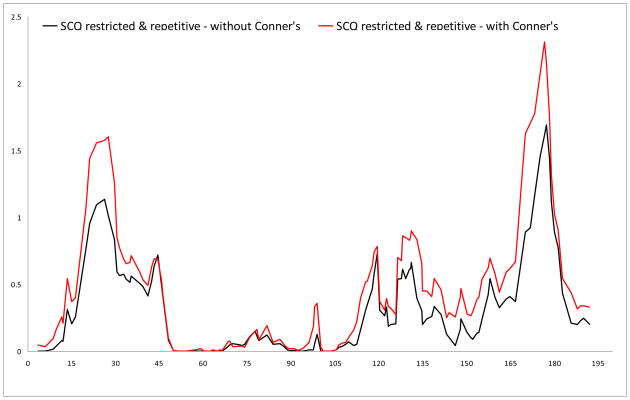
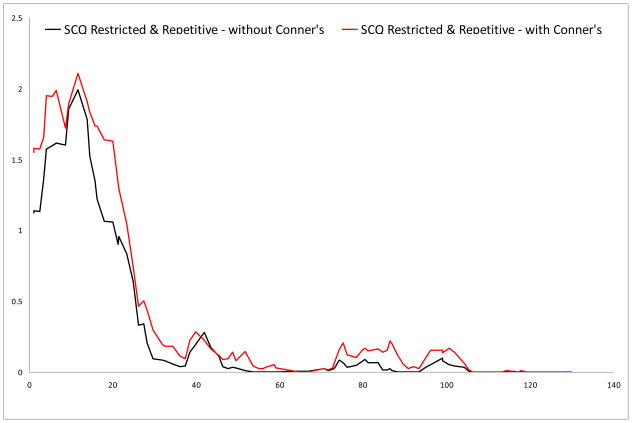
Logarithm of the odds (LOD) score graphs of the Social Communication Questionnaire (SCQ) restricted & repetitive scale on chromosome 18.
Table 2.
Thresholds for suggestive and significant linkage (Logarithm of the odds (LOD) scores)
| SCQ scale | Suggestive threshold | Significant threshold |
|---|---|---|
| Total | 1.85 | 3.76 |
| Social | 1.894 | 3.73 |
| Communication | 1.894 | 3.63 |
| Restricted & repetitive | 1.849 | 3.52 |
Note. LOD=Logarithm of the odds; SCQ=Social Communication Questionnaire
Table 3.
Linkage results for the total number of Autism Spectrum Disorder (ASD) Symptoms and the 3 ASD symptom domains
| SCQ scale | Chromosome | Position (cM)a | Locationb | Marker | LOD | No. of markersc |
|---|---|---|---|---|---|---|
| Total | 12 | 167.2 | 12q24.3 | rs1487602 | 0.772 | 1 |
| Social | . | . | . | . | . | |
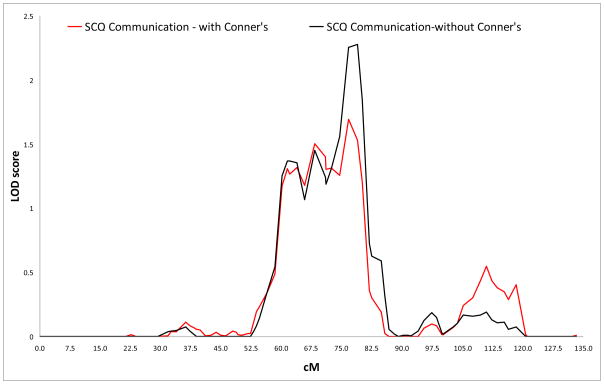
| Communication | 15 | 78.88 | 15q24.1 | rs896588 | 2.278 | 2 (2.1) |
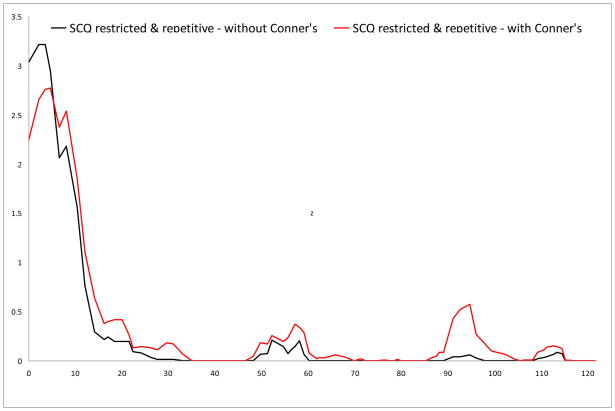
| Restricted & repetitive | 7 | 177.27 | 7q36.2 | rs1657290 | 1.689 | 2 (0.6) |
| 16 | 11.67 | 16p13.3 | rs859302 | 1.992 | 2 (2.3) | |
| 18 | 2.21 | 18p11.32 | rs1557299 | 3.216 | 6 (8.1) | |
|
Conners added to covariates | ||||||
| Total | 12 | 167.2 | 12q24.3 | rs1487602 | 1.86 | 1 |
| Social | . | . | . | . | . | |
| Communication | 15 | 76.79 | 15q24.1 | rs1348318 | 1.692 | 1 |
| Restricted & repetitive | 7 | 176.66 | 7q36.2 | rs306278 | 2.311 | 3 (2.0) |
| 16 | 11.67 | 16p13.3 | rs859302 | 2.105 | 3 (4.5) | |
| 18 | 4.64 | 18p11.32 | rs1371264 | 2.77 | 7 (10.5) | |
Note. LOD=Logarithm of the odds; SCQ=Social Communication Questionnaire.
DeCODE Genetic map position
Most likely cytogenic location
Number of consecutive SNP markers with LODs above thresholds for suggestive linkage and (in brackets) the genetic distance they span.
No results surpassed the criteria for genome-wide significance, but we did find several suggestive linkage signals. In the primary analyses (i.e., Conners’ scores not included as covariates), the highest LOD score (LOD 3.216) was found for rs1557299 on 18p11.32 for the SCQ restricted & repetitive subscale. Other regions that showed suggestive linkage were found on 15q24 and 16p13, for the communication and the restricted & repetitive subscale, respectively.
For the analyses corrected for Conners inattentive and hyperactive-impulsive scores, suggestive linkage was again found for 18p11.32 and 16p13 for the restricted & repetitive scale. The LOD for the region on 15q24 (LOD 1.692) just dropped below the empirically derived value for suggestive linkage. Additional regions that showed increased significance and surpassed thresholds for suggestive linkage in this analysis were found on 7q36.2 for the restricted & repetitive scale and on 12q24 for the total scale.
Discussion
In the current study, multivariate QTL linkage analysis was performed on ASD symptom domains in 1143 children with ADHD and 1453 of their siblings from 1143 families, using an autism screening questionnaire, i.e., the SCQ. We identified 5 suggestive quantitative trait loci, with the highest overall LOD scores identified for the SCQ restricted & repetitive subscale on 18p11 (LOD 3.216). Additional suggestive QTLs were 7q36, 16p13 (both for the restricted & repetitive scale), 15q24 (for the communication scale), and 12q24 (for the total scale). Our findings suggest that linkage studies regarding the occurrence of ASD symptoms in children with ADHD may assist in teasing apart the genetics underlying ADHD and ASD. Furthermore, they support the assumption that ASD traits are influenced by genetic risk factors along a continuum of severity, as loci potentially underlying ASD symptoms in children with ADHD were identified that correspond with identified loci or genes for clinical PDD, even though autistic cases had been excluded from the IMAGE sample. Additionally, our findings appear to confirm the hypothesis that the different ASD symptom domains have partially different genetic origins.22
Among the suggestive loci identified in the present study, the QTL on chromosome 15q could potentially have pleiotropic effects on ADHD and ASD, given that its LOD score decreased below the threshold for suggestive linkage after including ADHD symptom scores in the analyses. The other suggestive QTLs (i.e., on chromosome 7q, 12q, 16p, and 18p) appear to be primarily associated with ASD symptoms, independent of ADHD symptom scores, as LOD scores did not change substantially, and in the case of chromosome 12q24 even appeared, after inclusion of Conners’ scores as covariates. Supporting the ASD specificity of our results, there is no overlap between the suggestive QTLs we found and those observed in the IMAGE sample for ADHD symptoms as quantitative traits, i.e., genome-wide significant linkage to 1p36, and suggestive linkage findings for 9p23 and 11q21.30
The most prominent result in terms of significance was the signal we found on chromosome 18p11, which only just failed to reach genome-wide significance in the analyses uncorrected for ADHD symptoms. This region has never been reported in previous linkage studies for ADHD, but has been found to be a possible locus for ASD, in particular Asperger’s disorder, in one study.40 Here, suggestive linkage for marker D18S59, at 0 cM (184 kb from our peak SNP) was reported. The SNPs that showed the highest LOD scores in our study lie within an intergenic region, but are flanked by an interesting gene approximately 19 kb upstream, called adenylate cyclase activating polypeptide 1 (ADCYAP1). This gene encodes a neuropeptide, and in animal studies has been shown to be associated with hyperactivity, increased exploratory behavior, and abnormal social behavior.41,42 Investigating the role of ADCYAP1 in ADHD, autism, and the overlap between the two may be an interesting target for future studies. The nearest gene downstream of the linked SNPs on 18p11 lies at a large distance from our peak (1.6 Mb), and is a gene of unknown function called Methyltransferase-like 4 (METTL4), which may also prove to be associated with ADHD or ASD.
Another interesting result is the signal we found for chromosome 7q36. This lies within the most replicated linkage region for autism, namely 7q21.2–36.2 (for reviews see 9,43), and our results once more confirm the importance of this region for ASD symptoms. The peak SNP lies within the paired box interacting protein 1 (PAXIP1) gene, which encodes a nuclear protein, important for genome stability, chromatin condensation, and progression through mitosis. Its role in psychiatric disease has never been investigated thus far, but it has been reported as a candidate gene for Alzheimer’s disease.44 An interesting and replicated autism gene in the vicinity of our 7q peak is Engrailed 2 (EN2), located at ~180 cM.45,46 Our study is the first to suggest the relevance of the 7q36 region for ASD symptoms in a non-ASD sample, making this region and the genes herein, including PAXIP1 and EN2, even more interesting targets for further ASD research.
Two other peaks identified in the present study, i.e., those on chromosome 15q and 16p, are also in the vicinity of regions identified for ASD in other studies. Our peak SNP on chromosome 15q24 lies at a 2.6 Mb distance of a peak SNP (rs1372828) identified for autism by Szatmari et al., 12 and is located within a locus known to harbor microdeletions in mental retardation.47 Also, Marshall and colleagues identified structural variations in this region in individuals with ASD.48 The SNP with the highest LOD score we identified on chromosome 16p13 lies in a brain-expressed gene of provisional status called ankyrin repeat and sterile alpha motif domain containing 3 (ANKS3). Furthermore, at respectively 5Mb and 7 Mb from our peak SNP,49–51 suggestive linkage peaks have been found for ASD. Additionally, trisomy of 16p has been reported in individuals with autistic traits.52
Correspondence to previous linkage studies for ADHD was found for the chromosome 12q and the 16p QTL. In two independent samples, suggestive linkage at, respectively, 129 kb and 4Mb from our peak SNP at 12q24 was found.10,53–55 Smalley and colleagues found genome-wide significant linkage for a broad region on 16p13, with the most significant SNP (D16S3114) at 7 Mb from the SNP with the highest LOD score in the present study (rs859302).56 No previous studies report linkage to 15q24 for ADHD, which would have supported the pleiotropic effects of this region that our findings suggest.
In summary, our results for 7q, 18p, 15q and 16p show overlap with previous studies for ASD, which suggests that these findings may not be confined to ASD symptoms within the context of ADHD. One hypothesis worth testing is whether our 18p result may indicate the existence of a unique ADHD subtype associated with hyperactivity, increased exploratory behavior, and abnormal social behavior. Furthermore, our results for 12q and 16p correspond to previous ADHD findings. From the QTLs identified in this study, those on 12q, 16p, and 18p may be the most likely to explain the ASD comorbidity in ADHD, given their identification in previous ADHD (linkage) studies, and given their possible role in ASD symptoms as found in the current study and in previous autism linkage studies (16p and 18p). However, it cannot be precluded that these previous ADHD linkage results were driven by ASD symptoms, as subtle ASD symptoms may still have been present in the ADHD samples analyzed even when ASD cases were excluded. In line with recent discussions in neurocognitive ADHD and ASD research,57 our results suggest that subsequent ADHD genetic studies may need to consider correcting for ASD symptoms.
Notably, three regions we found to coincide with previously reported linkage peaks for ASD were all identified for the SCQ restricted & repetitive scale, and one was found for the SCQ communication scale. This may indicate domain-specific effects for these loci, which appears to be in line with the apparent genetic independence of the three ASD symptom domains,22 and with findings suggesting that restricted and repetitive behavior may have a relatively high heritability compared to other ASD traits.5,58 As far as we know, only one previous autism linkage study has specifically addressed restricted behaviors,59 but the regions identified in that study do not overlap with our suggestive linkage peaks. Similarly, several ASD linkage studies have addressed the communication domain (e.g., Alarcon et al.60), but none has reported linkage to 15q. Notably, in two independent samples of individuals with autism, genome wide significance was found for a quantitative trait for language acquisition at 7q36, whereas we found suggestive linkage to this region for restricted and repetitive behavior.15,60 Clearly, our results need replication in independent samples.
A limitation of our study may have been that the most severe ASD cases were excluded from participation in the IMAGE project. However, the fact that we did find overlap with previous linkage studies for ASD affection status, even though autistic cases were excluded from our sample, appears to confirm that similar genetic factors influence ASD symptoms along a continuum of severity, and supports the usefulness of the SCQ to measure this continuum. It should be remembered, however, that the study mostly excluded SCQ scores in the higher range. Nevertheless, further studies are needed to investigate whether indeed our findings pertain to narrowly defined autism as well. As another potential limitation, currently the SCQ has not yet been demonstrated to validly measure ASD symptoms as a continuous trait. Furthermore, the heritability figures used for the SCQ subscales were estimations based on previous ASD research, whereas ideally, these should be derived from twin pair SCQ scores. However, it is unlikely that the heritability estimates have influenced the results importantly, as LOD scores were stable across a range of heritabilities (0.20–0.95; data not shown).
Lastly, we did not adjust for testing multiple ASD dimensions, since a Bonferroni correction would have been overly conservative due to the high correlation between the ASD symptom scales. However, our simulation approach allowed us to take into account multiple testing resulting from analyzing several thousands of markers, and produced conservative LOD scores for suggestive linkage. Replicating the QTLs identified in the present study with genome-wide significance will likely require larger samples, since our data suggest effects are not large. Ideally, in these samples extremes of both the ADHD and ASD spectra would be represented, in which parent reports as well as a detailed developmental history and observational data would be collected to assess autistic and ADHD symptomatology.
To our knowledge, our findings represent the first attempt to identify QTLs underlying the occurrence of ASD symptoms in children with ADHD. The strengths of this study are the use of a well-defined combined type ADHD sample, and the relatively large sample size. This resulted in the identification of suggestive QTLs on chromosome 7q, 12q, 15q, 16p, and 18p.
Figure 2.
Logarithm of the odds (LOD) score graphs of the Social Communication Questionnaire (SCQ) communication scale on chromosome 15.
Figure 3.
Logarithm of the odds (LOD) score graphs of the Social Communication Questionnaire (SCQ) restricted & repetitive scale on chromosome 7.
Figure 4.
Logarithm of the odds (LOD) score graphs of the Social Communication Questionnaire (SCQ) restricted & repetitive scale on chromosome 16.
Acknowledgments
We thank all the families who kindly participated in this research.
The IMAGE project is a multi-site, international effort supported by NIH grants R01MH081803 and R01MH62873 to S.V. Faraone.
ZONMW = "Nederlandse organisatie voor gezondheidsonderzoek en zorginnovatie". (English: Dutch organisation for health research and healthcare innovation)
Footnotes
Site Principal Investigators are Philip Asherson, Tobias Banaschewski, Jan Buitelaar, Richard P. Ebstein, Stephen V. Faraone, Michael Gill, Ana Miranda, Fernando Mulas, Robert D. Oades, Herbert Roeyers, Aribert Rothenberger, Joseph Sergeant, Edmund Sonuga-Barke, and Hans-Christoph Steinhausen. Senior coinvestigators are Margaret Thompson, Pak Sham, Peter McGuffin, Robert Plomin, Ian Craig and Eric Taylor. Chief Investigators at each site are Rafaela Marco, Nanda Rommelse, Wai Chen, Henrik Uebel, Hanna Christiansen, Ueli Mueller, Marieke Altink, Barbara Franke, Lamprini Psychogiou.
Disclosure: Dr. Asherson has served as a consultant and on advisory boards for Eli Lilly, Shire, Janssen Cilag and Flynn Pharma. He received a research grant from Shire and an educational grant from Janssen-Cilag.
Dr. Banaschewski served as an advisor or consultant for Desitin, Eli Lilly, Medice, Novartis, Pfizer, Shire, UCB, Viforpharma. He received conference attendance support and conference support from or served on the speakers’ bureau for Eli Lilly, Janssen McNeil, Medice, Novartis, Shire, UCB. He is involved in clinical trials conducted by Eli Lilly, Shire, and a study on ADHD care management conducted by Novartis. The present study is unrelated to the above grants and relationships, and there are no conflicts of interest of any type concerning this article.
Dr. Buitelaar has served as a consultant and on the advisory board for Shire, Janssen Cilag, Eli Lilly, Pfizer, Organon, UCB, Servier, and Otsuka. He has served on the speakers’ bureau for Janssen Cilag and Eli Lilly. He has received research funding from Shire.
Dr. Faraone has received consulting fees and has served on the advisory boards for Eli Lilly and Shire and has received research support from Eli Lilly, Pfizer, Shire, and the National Institutes of Health in the past year. In previous years, Dr. Faraone has received consulting fees or has been on advisory boards or has been a speaker for Shire, McNeil, Janssen, Novartis, Pfizer, and Eli Lilly. In previous years he has received research support from Eli Lilly, Shire, Pfizer and the National Institutes of Health.
Dr. Hoekstra has received funding through ZONMW as well as honoraria for presentations or advice over the past two years from the university of Frankfurt, Germany, and the pharmaceutical companies Desitin, Shire, and Eli Lilly.
Dr. Oades has received research support from UCB GmbH, Janssen Cilag and Shire.
Dr. Sergeant has served on the advisory board of Eli Lilly and Shire. He served on the speakers’ bureau for Eli Lilly, Janssen-Cilag, and Shire. He has received research funding from Eli Lilly.
Dr. Sonuga-Barke has served on the speakers’ bureau and as a consultant for Shire and UCB. He has received research support from Janssen Cilag, Shire, Flynn, and Qbtech. He has served on the advisory board for Shire, Flynn, UCB, and Astra Zeneca. He has received conference support from Shire.
Dr. Steinhausen has served on the advisory board and the speakers’ bureau for Janssen Cilag, Eli Lilly, Medice, Novartis, Shire, and UCB. He has received research funding from Eli Lilly and Novartis.
Dr. Roeyers has served as an advisor to Shire and received research support from Shire and Eli Lilly, and conference attendance support from Eli Lilly. The present study is unrelated to these relationships.
Dr. Rothenberger has served on the advisory board and on the speakers’ bureau for Eli Lilly, Shire, Medice, and Novartis. He has received research funding from Shire, German Research Society, and Schwaabe, and travel support from Shire.
Drs. Nijmeijer, Arias-Vásquez, Rommelse, Altink, Anney, Buschgens, Fliers, Gill, Minderaa, Poustka, Franke, Ebstein, Miranda, Mulas, and Hartman report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Judith S. Nijmeijer, University Medical Center Groningen, Groningen, The Netherlands.
Alejandro Arias-Vásquez, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands.
Nanda N.J. Rommelse, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands, Karakter Child and Adolescent Psychiatry University Medical Center, Nijmegen, The Netherlands.
Marieke E. Altink, Karakter Child and Adolescent Psychiatry University Medical Center, Nijmegen, The Netherlands.
Richard J.L. Anney, Trinity Centre for Health Sciences, St. James’s Hospital, Dublin, Ireland.
Philip Asherson, Institute of Psychiatry, King’s College London, London, United Kingdom.
Tobias Banaschewski, Central Institute of Mental Health, University of Heidelberg, Mannheim, Germany.
Cathelijne J.M. Buschgens, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands.
Ellen A. Fliers, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands, Parnassia-Bavo-Group, Rotterdam, The Netherlands.
Michael Gill, Trinity Centre for Health Sciences, St. James’s Hospital, Dublin, Ireland.
Ruud B. Minderaa, University Medical Center Groningen, Groningen, The Netherlands.
Luise Poustka, Central Institute of Mental Health, University of Heidelberg, Mannheim, Germany.
Joseph A. Sergeant, Vrije Universiteit University Amsterdam, Amsterdam, The Netherlands.
Jan K. Buitelaar, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands, Karakter Child and Adolescent Psychiatry University Medical Center, Nijmegen, The Netherlands.
Barbara Franke, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands.
Richard P. Ebstein, Sarah Herzog Memorial Hospital, Jerusalem, Israel.
Ana Miranda, University of Valencia, Valencia, Spain.
Fernando Mulas, La Fe University Hospital, Valencia, Spain.
Robert D. Oades, University of Duisburg-Essen, Essen, Germany.
Herbert Roeyers, Ghent University, Ghent, Belgium.
Aribert Rothenberger, University of Goettingen, Goettingen, Germany.
Edmund J.S. Sonuga-Barke, School of Psychology, Southampton, United Kingdom.
Hans-Christoph Steinhausen, University of Aarhus, Aalborg, Denmark.
Stephen. V. Faraone, SUNY Upstate Medical University, Syracuse, New York.
Catharina A. Hartman, University Medical Center Groningen, Groningen, The Netherlands.
Pieter J. Hoekstra, University Medical Center Groningen, Groningen, The Netherlands.
Reference List
- 1.American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4. Washington DC: 1994. [Google Scholar]
- 2.Reiersen AM, Constantino JN, Volk HE, Todd RD. Autistic traits in a population-based ADHD twin sample. J Child Psychol Psychiatry. 2007;48(5):464–472. doi: 10.1111/j.1469-7610.2006.01720.x. [DOI] [PubMed] [Google Scholar]
- 3.Reiersen AM, Constantino JN, Grimmer M, Martin NG, Todd RD. Evidence for shared genetic influences on self-reported ADHD and autistic symptoms in young adult Australian twins. Twin Res Hum Genet. 2008;11(6):579–585. doi: 10.1375/twin.11.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ronald A, Simonoff E, Kuntsi J, Asherson P, Plomin R. Evidence for overlapping genetic influences on autistic and ADHD behaviours in a community twin sample. J Child Psychol Psychiatry. 2008;49(5):535–542. doi: 10.1111/j.1469-7610.2007.01857.x. [DOI] [PubMed] [Google Scholar]
- 5.Nijmeijer JS, Hoekstra PJ, Minderaa RB, et al. PDD Symptoms in ADHD, an Independent Familial Trait? J Abn Child Psychol. 2009;32(3):443–453. doi: 10.1007/s10802-008-9282-0. [DOI] [PubMed] [Google Scholar]
- 6.Mulligan A, Anney RJ, O’Regan M, et al. Autism symptoms in Attention-Deficit/Hyperactivity Disorder: A Familial trait which Correlates with Conduct, Oppositional Defiant, Language and Motor Disorders. J Autism Dev Disord. 2009;39(2):197–209. doi: 10.1007/s10803-008-0621-3. [DOI] [PubMed] [Google Scholar]
- 7.Faraone SV, Perlis RH, Doyle AE, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 8.Waldman ID, Gizer IR. The genetics of attention deficit hyperactivity disorder. Clin Psychol Rev. 2006;26(4):396–432. doi: 10.1016/j.cpr.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Yang MS, Gill M. A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. Int J Dev Neurosci. 2007;25(2):69–85. doi: 10.1016/j.ijdevneu.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Jain M, Palacio LG, Castellanos FX, et al. Evidence of Pleiotropy and New Susceptibility Loci. Biol Psychiatry. 2007;61(12):1329–1339. doi: 10.1016/j.biopsych.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Ogdie MN, MacPhie IL, Minassian SL, et al. A genomewide scan for attention-deficit/hyperactivity disorder in an extended sample: suggestive linkage on 17p11. Am J Hum Genet. 2003;72(5):1268–1279. doi: 10.1086/375139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39(3):319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yonan AL, Alarcon M, Cheng R, et al. A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet. 2003;73(4):886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Nyholt DR, Magnussen P, et al. A genomewide screen for autism susceptibility loci. Am J Hum Genet. 2001;69(2):327–340. doi: 10.1086/321980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schellenberg GD, Dawson G, Sung YJ, et al. Evidence for multiple loci from a genome scan of autism kindreds. Mol Psychiatry. 2006;11(11):1049–60. 979. doi: 10.1038/sj.mp.4001874. [DOI] [PubMed] [Google Scholar]
- 16.Romanos M, Freitag C, Jacob C, et al. Genome-wide linkage analysis of ADHD using high-density SNP arrays: novel loci at 5q13.1 and 14q12. Mol Psychiatry. 2008;13(5):522–530. doi: 10.1038/mp.2008.12. [DOI] [PubMed] [Google Scholar]
- 17.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bishop DV, Maybery M, Maley A, Wong D, Hill W, Hallmayer J. Using self-report to identify the broad phenotype in parents of children with autistic spectrum disorders: a study using the Autism-Spectrum Quotient. J Child Psychol Psychiatry. 2004;45(8):1431–1436. doi: 10.1111/j.1469-7610.2004.00849.x. [DOI] [PubMed] [Google Scholar]
- 19.Bolton PF, Pickles A, Murphy M, Rutter M. Autism, affective and other psychiatric disorders: patterns of familial aggregation. Psychol Med. 1998;28(2):385–395. doi: 10.1017/s0033291797006004. [DOI] [PubMed] [Google Scholar]
- 20.Hoekstra RA, Bartels M, Verweij CJ, Boomsma DI. Heritability of autistic traits in the general population. Arch Pediatr Adolesc Med. 2007;161(4):372–377. doi: 10.1001/archpedi.161.4.372. [DOI] [PubMed] [Google Scholar]
- 21.Constantino JN, Todd RD. Autistic traits in the general population: a twin study. Arch Gen Psychiatry. 2003;60(5):524–530. doi: 10.1001/archpsyc.60.5.524. [DOI] [PubMed] [Google Scholar]
- 22.Happé F, Ronald A. The ‘fractionable autism triad’: a review of evidence from behavioural, genetic, cognitive and neural research. Neuropsychol Rev. 2008;18(4):287–304. doi: 10.1007/s11065-008-9076-8. [DOI] [PubMed] [Google Scholar]
- 23.Kuntsi J, Neale BM, Chen W, Faraone SV, Asherson P. The IMAGE project: methodological issues for the molecular genetic analysis of ADHD. Behav Brain Funct. 2006;2:27. doi: 10.1186/1744-9081-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brookes K, Xu X, Chen W, et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry. 2006;11(10):934–953. doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- 25.Taylor EA. Childhood hyperactivity. Br J Psychiatry. 1986;149:562–573. doi: 10.1192/bjp.149.5.562. [DOI] [PubMed] [Google Scholar]
- 26.Berument SK, Rutter M, Lord C, Pickles A, Bailey A. Autism screening questionnaire: diagnostic validity. Br J Psychiatr. 1999;175:444–451. doi: 10.1192/bjp.175.5.444. [DOI] [PubMed] [Google Scholar]
- 27.Goodman R. The Strengths and Difficulties Questionnaire: a research note. J Child Psychol Psychiatry. 1997;38(5):581–586. doi: 10.1111/j.1469-7610.1997.tb01545.x. [DOI] [PubMed] [Google Scholar]
- 28.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24(5):659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 29.Conners K. Rating scales in ADHD. Durham, NC: Duke University Medical Center; 1996. [Google Scholar]
- 30.Zhou K, Asherson P, Sham P, et al. Linkage to chromosome 1p36 for attention-deficit/hyperactivity disorder traits in school and home settings. Biol Psychiatry. 2008;64(7):571–576. doi: 10.1016/j.biopsych.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31(3):241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 32.Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67(5):1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63(1):259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 35.Wigginton JE, Abecasis GR. PEDSTATS: descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21(16):3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- 36.Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71(2):238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet. 2005;77(5):754–767. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Constantino JN, Todd RD. Genetic structure of reciprocal social behavior. Am J Psychiatry. 2000;157(12):2043–2045. doi: 10.1176/appi.ajp.157.12.2043. [DOI] [PubMed] [Google Scholar]
- 39.Ronald A, Happé F, Bolton P, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45(6):691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 40.Ylisaukko-Oja T, Nieminen-von Wendt T, Kempas E, et al. Genome-wide scan for loci of Asperger syndrome. Mol Psychiatry. 2004;9(2):161–168. doi: 10.1038/sj.mp.4001385. [DOI] [PubMed] [Google Scholar]
- 41.Nicot A, Otto T, Brabet P, Dicicco-Bloom EM. Altered social behavior in pituitary adenylate cyclase-activating polypeptide type I receptor-deficient mice. J Neurosci. 2004;24(40):8786–8795. doi: 10.1523/JNEUROSCI.1910-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka K, Shintani N, Hashimoto H, et al. Psychostimulant-induced attenuation of hyperactivity and prepulse inhibition deficits in Adcyap1-deficient mice. J Neurosci. 2006;26(19):5091–5097. doi: 10.1523/JNEUROSCI.4376-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9(5):341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rademakers R, Cruts M, Sleegers K, et al. Linkage and association studies identify a novel locus for Alzheimer disease at 7q36 in a Dutch population-based sample. Am J Hum Genet. 2005;77(4):643–652. doi: 10.1086/491749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benayed R, Gharani N, Rossman I, et al. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet. 2005;77(5):851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gharani N, Benayed R, Mancuso V, Brzustowicz LM, Millonig JH. Association of the homeobox transcription factor, ENGRAILED 2, 3, with autism spectrum disorder. Mol Psychiatry. 2004;9(5):474–484. doi: 10.1038/sj.mp.4001498. [DOI] [PubMed] [Google Scholar]
- 47.Sharp AJ, Selzer RR, Veltman JA, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum Mol Genet. 2007;16(5):567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 48.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.International Molecular Genetic Study of Autism Consortium. A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet. 2001;69(3):570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lauritsen MB, Als TD, Dahl HA, et al. A genome-wide search for alleles and haplotypes associated with autism and related pervasive developmental disorders on the Faroe Islands. Mol Psychiatry. 2006;11(1):37–46. doi: 10.1038/sj.mp.4001754. [DOI] [PubMed] [Google Scholar]
- 51.Philippe A, Martinez M, Guilloud-Bataille M, et al. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet. 1999;8(5):805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- 52.Finelli P, Natacci F, Bonati MT, et al. FISH characterisation of an identical (16)(p11.2p12.2) tandem duplication in two unrelated patients with autistic behaviour. J Med Genet. 2004;41(7):e90. doi: 10.1136/jmg.2003.016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hebebrand J, Dempfle A, Saar K, et al. A genome-wide scan for attention-deficit/hyperactivity disorder in 155 German sib-pairs. Mol Psychiatry. 2006;11(2):196–205. doi: 10.1038/sj.mp.4001761. [DOI] [PubMed] [Google Scholar]
- 54.Fisher SE, Francks C, McCracken JT, et al. A genomewide scan for loci involved in attention-deficit/hyperactivity disorder. Am J Hum Genet. 2002;70(5):1183–1196. doi: 10.1086/340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ogdie MN, Fisher SE, Yang M, et al. Attention deficit hyperactivity disorder: fine mapping supports linkage to 5p13, 6q12, 16p13, and 17p11. Am J Hum Genet. 2004;75(4):661–668. doi: 10.1086/424387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smalley SL, Kustanovich V, Minassian SL, et al. Genetic linkage of attention-deficit/hyperactivity disorder on chromosome 16p13, in a region implicated in autism. Am J Hum Genet. 2002;71(4):959–963. doi: 10.1086/342732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geurts HM, Corbett B, Solomon M. The paradox of cognitive flexibility in autism. Trends Cogn Sci. 2009;13(2):74–82. doi: 10.1016/j.tics.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sung YJ, Dawson G, Munson J, Estes A, Schellenberg GD, Wijsman EM. Genetic investigation of quantitative traits related to autism: use of multivariate polygenic models with ascertainment adjustment. Am J Hum Genet. 2005;76(1):68–81. doi: 10.1086/426951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buxbaum JD, Silverman J, Keddache M, et al. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol Psychiatry. 2004;9(2):144–150. doi: 10.1038/sj.mp.4001465. [DOI] [PubMed] [Google Scholar]
- 60.Alarcon M, Cantor RM, Liu J, Gilliam TC, Geschwind DH. Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet. 2002;70(1):60–71. doi: 10.1086/338241. [DOI] [PMC free article] [PubMed] [Google Scholar]