Abstract
We investigated the impact of parenteral antibiotic treatment in the early neonatal period on the evolution of bifidobacteria in the newborn. Nine babies treated with intravenous ampicillin/gentamicin in the first week of life and nine controls (no antibiotic treatment) were studied. Denaturing gradient gel electrophoresis was used to investigate the composition of Bifidobacterium in stool samples taken at four and eight weeks. Bifidobacteria were detected in all control infants at both four and eight weeks, while only six of nine antibiotic-treated infants had detectable bifidobacteria at four weeks and eight of nine at eight weeks. Moreover, stool samples of controls showed greater diversity of Bifidobacterium spp. compared with antibiotic-treated infants. In conclusion, short-term parenteral antibiotic treatment of neonates causes a disturbance in the expected colonization pattern of bifidobacteria in the first months of life. Further studies are required to probiotic determine if supplementation is necessary in this patient group.
1. Introduction
Up to 10% of newborn infants require admission to the neonatal intensive care unit (NICU) for ongoing medical care [1–3]. Many of these infants require treatment with parenteral antibiotics for variable periods of time. Antibiotic administration is known to perturb the composition of the intestinal microbiota, resulting in suppression of anaerobic bacteria (with the exception of clostridia, which remain at detectable levels) and increased numbers of potentially pathogenic bacteria such as Klebsiella, Enterobacter, Citrobacter, and Pseudomonas [4, 5]. Since the pioneering microbiota of infancy probably create perpetual habitats for themselves, defining the life-long composition of the gut microbiota and consequently contributing to host health and well-being [6, 7], the initial colonization of the infant gastrointestinal tract is of significant importance.
Bifidobacterium spp. have several reported health-promoting effects, including inhibition of growth of harmful bacteria, stimulation of the immune system, alleviation of constipation, and prevention of intestinal infections [8–13]. Species commonly associated with humans include Bifidobacterium infantis, B. longum, B. bifidum, B. breve, B. catenulatum, B. pseudocatenulatum, B. angulatum, B. gallicum, and B. adolescensis [8], of which the first four are prevalent in infants [14].
Suppression of the bifidobacteria population as occurred following antibiotic exposure in children [5, 15, 16] may have negative effects on host well-being and can be associated with higher susceptibility to enteropathogenic bacterial infection [17]. However, the impact, if any, of parenteral antibiotics prescribed shortly following birth on the developing bifidobacterial population has not been addressed. The aim of this study was therefore to determine the impact of intravenous antibiotic treatment in the neonatal period on the evolution of bifidobacterial colonization over time.
2. Material and Methods
2.1. Subjects
Faecal material was sampled, using swabs, from 18 infants, nine infants previously treated with antibiotics and nine healthy controls (Table 1). Approval for the study protocol was obtained from the Clinical Research Ethics Committee of the Cork Teaching Hospitals, Cork, Ireland. Inclusion criteria were defined as full or near term infants requiring parenteral antibiotic administration within the first 48 hours of life. Control subjects were eligible if they were full or near term infants who were otherwise well and did not require admission to the NICU. Infants placed on oral antibiotics, those kept nil by mouth or requiring surgery, infants with congenital anomalies, or those born before term were excluded from the study. The indications for antibiotic treatment were determined clinically, based on symptoms and/or signs of suspected sepsis, and independent of the study investigators. Written informed consent was obtained from parents of all infants. Stool samples were taken by the same clinician at four and eight weeks after birth and stored at −20°C pending analysis.
Table 1.
Description of infant samples.
| Sample | Sex* | Feeding** | Antibiotic treatment (days) | Type of Antibiotics† | Mode of delivery |
|---|---|---|---|---|---|
| A | M | B | + (9) | Amp. + Gent. | Caesarean section |
| B‡ | M | F + B | + (5) | Amp. + Gent. | Caesarean section |
| C‡ | M | B | + (2) | Amp. + Gent. | Caesarean section |
| D | M | F | + (2) | Amp. + Gent. | Vaginal delivery |
| E | F | F | + (5) | Amp. + Gent. | Caesarean section |
| F | F | B | + (2) | Amp. + Gent. | Vaginal delivery |
| G | F | B | + (2) | Amp. + Gent. | Vaginal delivery |
| H | M | F | + (2) | Amp. + Gent. | Caesarean section |
| I | M | F | + (2) | Amp. + Gent. | Vaginal delivery |
| J | M | F | — | — | Vaginal delivery |
| K | M | F | — | — | Vaginal delivery |
| L | F | F | — | — | Vaginal delivery |
| M | F | F | — | — | Vaginal delivery |
| N | M | B | — | — | Vaginal delivery |
| O | F | B | — | — | Vaginal delivery |
| P | M | B | — | — | Vaginal delivery |
| Q | M | F | — | — | Vaginal delivery |
| R | F | F | — | — | Vaginal delivery |
*M, male; F, female.**B, breast-feeding; F, formula feeding.†Amp. = ampicillin, Gent. = gentamicin. ‡ twins.
2.2. PCR -DGGE Analysis
Bacterial DNA was extracted from swabs using a QIAamp DNA stool minikit (Qiagen, Hilden, Germany) by following the manufacturer's instructions (lysis temperature, 95°C). To investigate the bifidobacterial population in the samples, PCR was performed as a nested approach. The first PCR applied primers Im26-f (5′- GATTCTGGCTCAGGATGAACG -3′) and Im3-r (5′- CGGGTGCTICCCCACTTTCATG -3′) described by Kaufmann et al. [18] amplified a 1,417 bp fragment of the bifidobacterial 16S rRNA gene. PCR volumes of 50 μL contained 5 μL of 10× PCR buffer, 5 μL of MgCl2 (2.5 mM), 8 μL of dNTPs (0,2 mM each), 1 μL of each primer (5 pmol), 0.5 μL of Taq polymerase (5 U/μL), 28.5 μL of sterile Milli-Q water, and 1 μL of DNA solution. PCR-reagents were obtained from Bioline (Taunton, MA, USA). The following PCR program was used: initial denaturation at 94°C for 5 min; 3 cycles of denaturation at 94°C for 45 s, annealing at 57°C for 2 minutes, and extension at 72°C for 1 min; 30 cycles of denaturation at 94°C for 20 s, annealing at 57°C for 1 min, and extension at 72°C for 1 min; and final extension at 72°C for 5 min followed by cooling to 4°C. The presence of PCR products was verified on a 1.5% (w/v) agarose gel. In order to eliminate the remaining oligonucleotides and original template DNA, purification of the amplicons was performed by use of the QIAquick PCR purification kit (Qiagen) according to the manufacturer's instructions. A second PCR was performed using the amplicons of the first PCR as template DNA. The primer set used (F357-GC and 518R) (5′GC-clamp- GCCTACGGAGGCAGCAG -3′ and 5′- ATTACCGCGGCTGCTGG -3′, resp.) amplifies the V3 region of the bacterial 16S rRNA gene [19]. The forward primer contained a GC clamp (5′- CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGG -3′) to facilitate separation of the amplicons in a DGGE gel. PCR volumes of 50 μL contained 5 μL of 10× PCR buffer, 2 μL of MgCl2 (1 mM), 8 μL of dNTPs (0.2 mM each), 2 μL of each primer (5 μM), 0.5 μL of Taq polymerase (5 U/μL), 29.5 μL of sterile Milli-Q water, and 1 μL of 10-fold diluted DNA solution (obtained from the first PCR). The following PCR program was used: initial denaturation at 94°C for 5 min, 30 cycles of denaturation at 94°C for 20 s, annealing at 58°C for 45 s, and extension at 72°C for 1 min, and final extension at 72°C for 7 min, following by cooling to 4°C. PCR products were analyzed on DGGE gels based on the protocol by Temmerman et al. [10]. A Dcode universal mutation detection system (Bio-Rad Laboratories, Hercules, CA, USA) was used, utilizing 16 cm by 16 cm by 1 mm gels. Eight percent polyacrylamide gels were prepared and run with 1× TAE buffer diluted from 50× TAE buffer (2 M Tris base, 1 M glacial acetic acid, and 50 mM EDTA). The denaturing gradient was formed with two 8% acrylamide (acrylamide-bis/acrylamide 40%) stock solutions (Severn Biotech Ltd, Worcestershire, UK). A 100% denaturing solution contained 40% (v/v) formamide and 7.0 M urea. The gels were poured from the top by using a gradient maker (CBS Scientific Linear Gradient Maker, Bio-Rad Laboratories) and a pump (Bio-Rad Laboratories) and gradients of 50% to 70% were used for the separation of the generated amplicons. Before polymerization of the denaturing gel (24 mL gradient volume), a 6 mL stacking gel without denaturing chemicals was added, and the appropriate comb was subsequently inserted. PCR amplicons were separated by electrophoresis at a constant voltage of 60 V in a 0.5 × TAE buffer at a constant temperature of 60°C for 16 h. Gels were stained in ethidium bromide for 30 min, allowing digital capturing of the DGGE band profiles under UV light.
2.3. Reference Ladder
A mixture of five different species of bifidobacteria which are the most commonly isolated species of bifidobacteria from the infant gastrointestinal tract (GIT) [14, 20, 21] was used to create a reference ladder in order to enable a visual comparison of the bands. DNA extraction from the reference strains, Bifidobacterium breve, B. bifidum, B. infantis, B. adolescentis, and B. longum, was performed according to the method described by Hoffman and Winston [22].
3. Results
Nine subjects were recruited to both control and antibiotic-treated groups in this study. Their pertinent demographic data are summarized in Table 1. While the gender distribution and feeding history was similar in both groups, the antibiotic treated infants were more likely to be born by caesarean section (5/9 infants) and all control infants were born by spontaneous vaginal delivery. Ampicillin and gentamicin were the only antibiotics used and the median duration of therapy was 2 days (range 2–9 days).
3.1. Development of the Bifidobacterium Population at Four Weeks of Age
In order to investigate the population of bifidobacteria in the samples, the Bifidobacterium-specific primers Im-3 and Im-26 were used. Amplicons obtained using these primers served as template DNA for the V3 primer combination V3R and V3F, during a second PCR step. The mobility of the PCR products obtained by these primers in DGGE was compared with the PCR pattern of bifidobacteria reference strains. Because of the high G+C content of bifidobacteria, the conventional 35% to 70% denaturing gradient was replaced with a 50% to 70% denaturing gradient.
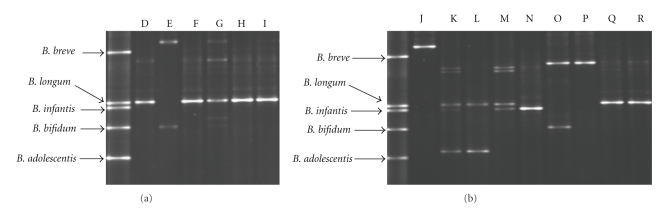
At four weeks of age, species of bifidobacteria were detected in six out of nine infants treated with antibiotics (subjects A–I) and in all infants in the control group (subjects J–R). The samples from the antibiotic treated infants exhibited a lower number of bands in the DGGE gel (Figure 1(a)) compared with controls (Figure 1(b)) at four weeks of age, representing a less diverse population of bifidobacteria at this age. Five of six infants in the antibiotic-treated group showed a single dominant band, which corresponded to B. longum (Figure 1(a)). All five infants had been treated with intravenous ampicillin/gentamicin for two days and there were both breast fed (subject F, G) and formula fed (subject D, H, I) infants within this group. The sample lacking a band in the position of B. longum was obtained from an infant that had received antibiotics for five days (subject E). According to the band pattern of this subject, some Bifidobacterium species were able to colonize this infant despite antibiotic treatment for five days; that is, the lower band appears to be B. bifidum, while the upper band is not comparable to any of the species within the reference ladder. All samples from the formula fed controls (K, L, M, Q, R) exhibited a band in the same position as B. longum at four weeks of age, except for one infant (subject J) (Figure 1(b)). In contrast, samples from the breast fed infants in the control group (N, O, P) showed bands in the positions of B. infantis and B. bifidum. These two species were only found in one of the formula fed infants. While all vaginally born infants in the antibiotic-treated group harboured bifidobacteria by four weeks of age, two out of five infants born by caesarean section harboured bifidobacteria (B. bifidum and B. longum) at this time point.
Figure 1.
DGGE of bifidobacterial PCR-products (V3-region) from stool samples taken at four weeks of age from infants treated with antibiotics D–I (a) and controls J–R (b). The mobility of the PCR products obtained in DGGE was compared to the PCR pattern of reference strains obtained with the same primer set.
3.2. Development of the Bifidobacterium Population at Eight Weeks of Age
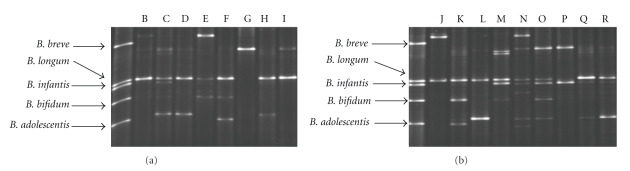
At eight weeks of age, eight out of nine antibiotic-treated infants and all controls had detectable levels of bifidobacteria in their stools. Interestingly, the only sample in which bifidobacteria was not detected was from an infant that had received antibiotics for nine days (subject A) (the other infants were treated with antibiotics for 2–5 days). Between four and eight weeks, an increase in bifidobacteria diversity in both groups was observed as evidenced by the increased number of bands (Figure 2). Nonetheless, while both groups were similar at eight weeks, samples from antibiotic-treated infants continued to display a less diverse population of bifidobacteria when compared with controls (Figure 2). The samples obtained from the breast-fed infants in the control group had in general a more diverse population of bifidobacteria compared with formula-fed infants (7-8 bands compared with 2–5 bands, resp.). However, no differences between the samples from the breast-fed and formula-fed infants in the antibiotic-treated groups were observed. Subject G exhibited a decrease in diversity from four to eight weeks of age. Moreover, the band with highest intensity at four weeks, identified as B. longum, was replaced by another dominant strain, identified as B. breve, after eight weeks. Four of the five infants in the antibiotic-treated group born by caesarean section harboured bifidobacteria by week eight. As found at week four, all vaginally born infants harboured bifidobacteria at eight weeks of age, with a higher diversity observed compared with four weeks of age. Overall, B. longum appeared to be the most commonly detected Bifidobacterium species in the infants at eight weeks of life (present in 15 out of 17 infants harbouring bifidobacteria) (Figure 2).
Figure 2.
DGGE of bifidobacterial PCR-products (V3-region) from stool samples taken at eight weeks of age from infants treated with antibiotics B–I (a) and controls J–R (b). The mobility of the PCR products obtained in DGGE was compared to the PCR pattern of reference strains obtained with the same primer set.
4. Discussion
Bifidobacterium and Lactobacillus species are considered among the most important beneficial bacteria in the human GIT and their presence in large numbers in the microbiota is regarded as beneficial [23, 24]. Suppression of these beneficial components could facilitate growth of potentially pathogenic bacteria such as Klebsiella and Clostridium and have consequences for the well-being of the host. This study demonstrates that even brief parenteral antibiotic treatment alters the pattern of bifidobacterial evolution over time. Infants treated with antibiotics showed a reduction in both colonisation and diversity of bifidobacteria compared with controls. While control and antibiotic-treated groups were similar by eight weeks of age, a number of control subjects continued to display a more diverse population of bifidobacteria. Mackie et al. [25] reported that the colonisation of bifidobacteria in infants is normally well established by one week of age. It is tempting to speculate that the observed difference in detectable levels of bifidobacteria between the antibiotic-treated infants and controls in our study reflects the early antibiotic exposure. Favier et al. [26] previously described a significant difference in the microbiota of a single infant receiving antibiotics compared to four nonantibiotic treated infants, using similar methodology that is, PCR-DGGE and Bifidobacterium-specific primers as used in our study. Bifidobacterium spp. were absent in the antibiotic-treated infant at all time points assessed, from five days to five months of age. However, in that study, the antibiotic-treated subject received prolonged antibiotic therapy with both parenteral and oral agents (thirteen days of parenteral coamoxiclav followed by oral cotrimoxazole) [26]. In contrast, the majority of participants in our study had relatively brief periods of intravenous antibiotic administration and yet differences were still observed compared with controls.
B. longum was the dominant species in both the antibiotic-treated infants and in the control group. This finding is consistent with the study by Grönlund et al. [27] which found B. longum to be the most common Bifidobacterium species present in infants. B. longum was also the most prevalent species in the antibiotic-treated infants. This result correlates with a recent finding by Mättö et al. [14], demonstrating that B. longum has a higher minimum inhibitory concentration (MIC) for ampicillin compared with other Bifidobacterium species. Thus, it appears that the antibiotic treatment itself may have selected for this species. One infant showed a decrease in numbers of species from three species at four weeks of age to one species at eight weeks of age. The species that showed the highest intensity in the DGGE gel after four weeks was replaced by another dominant species after eight weeks. These results are in agreement with a study by Favier et al. [26] in which the initially dominant bifidobacteria in two infants were replaced following the emergence of new species within the study time period.
Although less than half of the infants in this study were breast-fed, our study confirms previous reports that breast-feeding is positively associated with a more diverse bifidobacterial population compared with formula-fed infants [16, 28, 29]. Bifidobacterium species are considered among the most important beneficial bacteria in the human GIT [18, 19, 24]. A higher density of beneficial bacteria in infants may confer a degree of protection against childhood diseases. For example, bifidobacterial colonisation has been associated with a lower risk of atopy [21, 30, 31]. In a prospective study, children with lower numbers of fecal bifidobacteria and increased numbers of clostridia from the first weeks of life were more likely to become sensitized to common allergens [23]. In addition, it has been reported that differences in the gut microbiota during infancy may precede the onset of obesity. In this study by Kalliomäki et al. [32], the numbers of Bifidobacterium spp. were higher during infancy in children who remained at normal weight than those in children who became overweight. Factors such as breast-feeding, vaginal delivery, and the absence of antibiotic exposure would promote microbial diversity with higher numbers of bifidobacteria.
In conclusion, the present study demonstrates that short-term parenteral antibiotic treatment of neonates causes a disturbance in the expected colonization pattern of bifidobacteria in the first months of life. Whether a delay in bifidobacterial colonisation could adversely influence health status, either in infancy or later life remains unknown. However, adequately powered prospective studies that document changes in the microbiota and incorporate long-term health status outcomes are needed to further our understanding of the sequelae, if any, of early neonatal antibiotic administration. Until such data are available, supplementation with bifidobacteria in this patient group cannot be justified.
Acknowledgments
This work was funded in part by Science Foundation Ireland (SFI) funds, Enterprise Ireland, and the Irish Government under the National Development Plan 2000-2006.
Abbreviations
- DGGE:
Denaturing gradient gel electrophoresis
- GIT:
Gastrointestinal tract
- MIC:
Minimum inhibitory concentration
- NICU:
Neonatal intensive care unit.
References
- 1.McDonald M, Moloney A, Clarke TA, Matthews TG. Blood cultures and antibiotic use in a neonatal intensive care unit. Irish Journal of Medical Science. 1992;161(1):3–4. doi: 10.1007/BF02984666. [DOI] [PubMed] [Google Scholar]
- 2.Spitzer AR, Kirkby S, Kornhauser M. Practice variation in suspected neonatal sepsis: a costly problem in neonatal intensive care. Journal of Perinatology. 2005;25(4):265–269. doi: 10.1038/sj.jp.7211252. [DOI] [PubMed] [Google Scholar]
- 3.Carbajal R, Rousset A, Danan C, et al. Epidemiology and treatment of painful procedures in neonates in intensive care units. Journal of the American Medical Association. 2008;300(1):60–70. doi: 10.1001/jama.300.1.60. [DOI] [PubMed] [Google Scholar]
- 4.Adlerberth I. Establishment of a normal intestinal microflora in the newborn infant. In: Hanson LA, Yolken RH, editors. Probiotics, Other Nutritional Factors and Intestinal Microflora. Philadelphia, Pa, USA: Lippincott-Raven; 1999. pp. 63–78. (Nestlè Nutrition Workshop Series). [Google Scholar]
- 5.Bennet R, Eriksson M, Nord CE. The fecal microflora of 1-3-month-old infants during treatment with eight oral antibiotics. Infection. 2002;30(3):158–160. doi: 10.1007/s15010-002-2140-z. [DOI] [PubMed] [Google Scholar]
- 6.Ballongue J. Bifidobacteria and probiotic action. In: Salminen S, von Wright A, editors. Lactic Acid Bacteria, Microbiology and Functional Aspects. 2nd edition. New York, NY, USA: Marcel Dekkers; 1998. pp. 519–587. [Google Scholar]
- 7.Thompson-Chagoyán OC, Maldonado J, Gil A. Colonization and impact of disease and other factors on intestinal microbiota. Digestive Diseases and Sciences. 2007;52(9):2069–2077. doi: 10.1007/s10620-006-9285-z. [DOI] [PubMed] [Google Scholar]
- 8.Satokari RM, Vaughan EE, Akkermans ADL, Saarela M, De Vos WM. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Applied and Environmental Microbiology. 2001;67(2):504–513. doi: 10.1128/AEM.67.2.504-513.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germond J-E, Mamin O, Mollet B. Species specific identification of nine human Bifidobacterium spp. in feces. Systematic and Applied Microbiology. 2002;25(4):536–543. doi: 10.1078/07232020260517670. [DOI] [PubMed] [Google Scholar]
- 10.Temmerman R, Masco L, Vanhoutte T, Huys G, Swings J. Development and validation of a nested-PCR-denaturing gradient gel electrophoresis method for taxonomic characterization of bifidobacterial communities. Applied and Environmental Microbiology. 2003;69(11):380–385. doi: 10.1128/AEM.69.11.6380-6385.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marteau P, Shanahan F. Basic aspects and pharmacology of probiotics: an overview of pharmacokinetics, mechanisms of action and side-effects. Bailliere’s Best Practice and Research in Clinical Gastroenterology. 2003;17(5):725–740. doi: 10.1016/s1521-6918(03)00055-6. [DOI] [PubMed] [Google Scholar]
- 12.Brown AC, Valiere A. Probiotics and medical nutrition therapy. Nutrition in Clinical Care. 2004;7(2):56–68. [PMC free article] [PubMed] [Google Scholar]
- 13.Vlková E, Nevoral J, Jencikova B, et al. Detection of infant faecal bifidobacteria by enzymatic methods. Journal of Microbiological Methods. 2005;60(3):365–373. doi: 10.1016/j.mimet.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 14.Mättö J, van Hoek AHAM, Domig KJ, et al. Susceptibility of human and probiotic Bifidobacterium spp. to selected antibiotics as determined by the Etest method. International Dairy Journal. 2007;17(9):1123–1131. [Google Scholar]
- 15.Gewolb IH, Schwalbe RS, Taciak VL, Harrison TS, Panigrahi P. Stool microflora in extremely low birthweight infants. Archives of Disease in Childhood. 1999;80(3):F167–F173. doi: 10.1136/fn.80.3.f167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penders J, Thijs C, Vink C, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 17.Collier-Hyams LS, Neish AS. Innate immune relationship between commensal flora and the mammalian intestinal epithelium. Cellular and Molecular Life Sciences. 2005;62(12):1339–1348. doi: 10.1007/s00018-005-5038-y. [DOI] [PubMed] [Google Scholar]
- 18.Kaufmann P, Pfefferkorn A, Teuber M, Meile L. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Applied and Environmental Microbiology. 1997;63(4):1268–1273. doi: 10.1128/aem.63.4.1268-1273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muyzer G. DGGE/TGGE a method for identifying genes from natural ecosystems. Current Opinion in Microbiology. 1999;2(3):317–322. doi: 10.1016/S1369-5274(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 20.Matsuki T, Watanabe K, Tanaka R, Fukuda M, Oyaizu H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Applied and Environmental Microbiology. 1999;65(10):4506–4512. doi: 10.1128/aem.65.10.4506-4512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malinen E, Mättö J, Salmitie M, Alander M, Saarela M, Palva A. PCR-ELISA: II: analysis of Bifidobacterium populations in human faecal samples from a consumption trial with Bifidobacterium lactis Bb-12 and a galacto-oligosaccharide preparation. Systematic and Applied Microbiology. 2002;25(2):249–258. doi: 10.1078/0723-2020-00117. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases automomous plasmids for transformation of Escherichia coli . Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 23.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. Journal of Allergy and Clinical Immunology. 2001;107(1):129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 24.Chen C-C, Walker WA. Probiotics and prebiotics: role in clinical disease states. Advances in Pediatrics. 2005;52:77–113. doi: 10.1016/j.yapd.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. American Journal of Clinical Nutrition. 1999;69(5):1035–1045. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 26.Favier CF, De Vos WM, Akkermans ADL. Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe. 2003;9(5):219–229. doi: 10.1016/j.anaerobe.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 27.Grönlund M-M, Gueimonde M, Laitinen K, et al. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clinical and Experimental Allergy. 2007;37(12):1764–1772. doi: 10.1111/j.1365-2222.2007.02849.x. [DOI] [PubMed] [Google Scholar]
- 28.Harmsen HJM, Wildeboer-Veloo ACM, Raangs GC, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. Journal of Pediatric Gastroenterology and Nutrition. 2000;30(1):61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Mountzouris KC, McCartney AL, Gibson GR. Intestinal microflora of human infants and current trends for its nutritional modulation. British Journal of Nutrition. 2002;87(5):405–420. doi: 10.1079/BJNBJN2002563. [DOI] [PubMed] [Google Scholar]
- 30.Björkstén B, Sepp E, Julge K, Voor T, Mikelsaar M. Allergy development and the intestinal microflora during the first year of life. Journal of Allergy and Clinical Immunology. 2001;108(4):516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe S, Narisawa Y, Arase S, et al. Differences in fecal microflora between patients with atopic dermatitis and healthy control subjects. Journal of Allergy and Clinical Immunology. 2003;111(3):587–591. doi: 10.1067/mai.2003.105. [DOI] [PubMed] [Google Scholar]
- 32.Kalliomäki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. American Journal of Clinical Nutrition. 2008;87(3):534–538. doi: 10.1093/ajcn/87.3.534. [DOI] [PubMed] [Google Scholar]