Abstract
Double-strand breaks (DSBs) are the most deleterious DNA lesions, which, if left unrepaired, may have severe consequences for cell survival, as they lead to chromosome aberrations, genomic instability, or cell death. Various physical, chemical, and biological factors are involved in DSB induction. Cells respond to DNA damage by activating the so-called DNA damage response (DDR), a complex molecular mechanism developed to detect and repair DNA damage. The formation of DSBs triggers activation of many factors, including phosphorylation of the histone variant H2AX, producing γH2AX. Phosphorylation of H2AX plays a key role in DDR and is required for the assembly of DNA repair proteins at the sites containing damaged chromatin as well as for activation of checkpoints proteins which arrest the cell cycle progression. In general, analysis of γH2AX expression can be used to detect the genotoxic effect of different toxic substances. When applied to clinical samples from cancer patients, evaluation of γH2AX levels may allow not only to monitor the efficiency of anticancer treatment but also to predict of tumor cell sensitivity to DNA damaging anticancer agents and toxicity of anticancer treatment toward normal cells.
1. DNA Double-Strand Breaks
DNA double-strand break (DSB) is a type of DNA damage in which two complementary strands of the double helix of DNA are damaged simultaneously, in locations close to each other. DSB is the most dangerous type of DNA damage, because it is believed that a single unrepaired DSB is sufficient for the induction of cell death process [1, 2]. Many different physical, chemical, and biological factors may lead to DSB formation. DSBs also occur in cells, for example, during differentiation of reproductive cells or lymphocytes [3, 4]. The factors leading to the formation of DSB include endogenous factors, that are associated with physiological processes occurring in the cell, and the exogenous ones [5, 6].
In living cells, DNA is subject to a constant process of oxidative damage by oxygen free radicals (reactive oxygen species—ROS) that are produced inside the cell as a result of metabolic processes [6, 7]. It is estimated that in a single cell cycle at least 5000 single-stranded DNA breaks can occur as a result of ROS production. Approximately 1% of these DNA breaks is converted into DSBs, mainly during DNA replication, while the remaining 99% is repaired. Thus, during the cell cycle in a single nucleus, about 50 so-called “endogenous” DSBs are formed. Accumulation of unrepaired DNA damage induced by ROS leads to cell aging and may be responsible for the induction of neoplastic transformation [6, 8].
One of the first exogenous factors involved in inducing DSBs was ionizing radiation. Among the various types of DNA damage caused by X-rays, formation of DSB seems to be the most important mechanism of reproductive cell damage or in changing genome integrity that leads to malignancy [9]. Similar changes may be induced by UV radiation [9, 10].
DNA damage is also induced by many anticancer drugs, among them the most effective appear to be inhibitors of topoisomerases. DNA topoisomerases are enzymes that regulate DNA over- and underwinding and remove knots and tangles from the genetic material by creating transient breaks in DNA double-helix [11, 12]. There are two classes of topoisomerases: type I enzymes introduce single-strand breaks in DNA and type II ones introduce double-strand breaks [11–13]. Type I topoisomerases modulate DNA under- and overwinding, but cannot remove knots and tangles from duplex DNA, while type II topoisomerases modulate DNA supercoiling and also remove DNA knots and tangles [14]. Since a single unrepaired DSB has potentially lethal consequences, type II topoisomerases, being necessary for cell survival, also have the capacity to fragment the genome [14]. When a nucleic acid tracking system, such as a replication or transcription complex, attempts to traverse the cleavable complex between topoisomerase and DNA, it may convert this transient enzyme-DNA intermediate to a permanent DSB in the genetic material, leading to DNA aberrations [15].
Inhibitors of DNA topoisomerase I (camptothecin and topotecan) and II (etoposide, doxorubicin, and mitoxantrone) belong to the most effective antitumor drugs. Their mode of action involves stabilization of cleavable complexes between topoisomerase and DNA. It leads to collisions of the progressing DNA replication forks or RNA polymerase with the drug-stabilized topoisomerase-DNA complexes and conversion them into DSBs, which trigger apoptosis [16, 17].
In eukaryotic cells, the efficient repair of DSB is essential for survival. Two major pathways have evolved to deal with these lesions, homologous recombination (HR), and non homologous end-joining (NHEJ). The mechanism of HR is based on using the genetic information from a corresponding undamaged region present on the second DNA molecule or homologous chromosomes and therefore is active mainly during S and G2 phases of cell cycle. NHEJ is based on a direct ligation of the two ends of damaged DNA molecules and repairs DSBs mainly in G1 phase [2].
2. H2AX Phosphorylation
It was reported several years ago, that in mammalian cells the phosphorylation of the subtype of histone H2A, called H2AX, in the position of Ser139 occurs in response to DSB formation. The phosphorylated form of H2AX is called γH2AX [18, 19]. Since then, many researchers focused on the explanation of mechanisms which induce phosphorylation of H2AX and its role in DNA damage signaling and repair.
Histone H2AX is a substrate of several phosphoinositide 3-kinase-related protein kinases (PIKKs), such as ATM (ataxia teleangiectasia mutated), ATR (ATM and Rad3-related), or DNA-dependent protein kinase (DNA-PK). ATM kinase is considered as a major physiological mediator of H2AX phosphorylation in response to DSB formation [20, 21]. ATM is activated by its autophosphorylation at Ser1981 position, which leads to dissociation of the inactive ATM dimers into single protein molecules with increased kinase activity [18–21]. A tri-protein complex called MRN complex (MRE11-RAD50-NBS1) recognizes DNA damage, recruits ATM to the site of damage and also functions in targeting ATM to initiate phosphorylation of the respective substrates [22–24]. It is also reported that ATM activation requires prior ATM acetylation, mediated by Tip60 histone acetyltransferase [24, 25]. Apart from H2AX, the target substrates phosphorylated by ATM are BRCA1, 53BP1, and MDC1 as well as checkpoint proteins, Chk1 and Chk2. These processes are aimed to stop the progression of the cell cycle and to activate proteins responsible for DNA repair, as is described in details below.
H2AX can also be phosphorylated by ATR and DNA-dependent protein kinases. ATR phosphorylates H2AX in response to single-stranded DNA breaks and during replication stress, such as replication fork arrest [26, 27]. DNA-PK mediates phosphorylation of H2AX in cells under hypertonic conditions and during apoptotic DNA fragmentation [28, 29]. However, DNA damage caused by ionizing radiation leads to phosphorylation of H2AX that is mediated by all PIKK kinases, ATM, ATR, and DNA-PK [30].
Recently, it was reported that some other events occur before H2AX phosphorylation in mammalian cells. Ayoub et al. [31] observed that DNA breaks mobilize heterochromatin protein 1β (HP1-β), a chromatin factor bound to histone H3 methylated on lysine 9 (H3K9me). In response to DNA damage, HP1-β was rapidly phosphorylated at threonine 51 (Thr51) by casein kinase 2 (CK2). This phosphorylation leads to releasing HP1-β from chromatin by disrupting hydrogen bonds that fold its chromodomain around H3K9me, resulting in its transient mobilization from chromatin. Although it is still not known what signals CK2 to phosphorylate HP1, the phosphorylation and mobilization of HP1 seems to be important for H2AX phosphorylation [31].
Based on the results of Ayoub et al. [31] and Goodarzi et al. [32, 33], it is expected that the loss of HP1 is beneficial to repair in heterochromatin. However, another recent study by Luijsterburg et al. [34], that also addressed the role of HP1 proteins in DNA damage response, suggested an apparently active and positive role for HP1 in DNA repair. These authors demonstrate that HP1 proteins accumulate de novo at sites of DNA damage, and this recruitment was independent of binding to H3K9me. It cannot be excluded that there is a partial HP1 dissociation from H3K9me at damaged sites, however the accumulation of HP1 reflects H3K9me-independent binding to damage sites rather than the rebinding of HP1 to H3K9me sites [34, 35]. Despite these controversies, the findings presented above illustrate the important role of HP1 proteins in DNA damage and open up a new direction of research to characterize the role of HP1 in H2AX phosphorylation and DNA damage response.
3. γH2AX and Its Role in DNA Damage Response
3.1. Accumulation of DNA Damage Signaling and Repair Proteins at DSBs
Unrepaired DSBs induce genome instability and promote tumorigenesis. Thus, cells have mechanisms responsible for recognition of DNA damage and activation of cell cycle checkpoints leading to DNA repair. The generation of DSBs triggers the relocalization of many DNA damage response (DDR) proteins such as MRE11/NBS1/RAD50, MDC1, 53BP1, and BRCA1 to nuclear foci where these proteins colocalize and interact with γH2AX [36–46]. Presumably, γH2AX foci specifically attract repair factors, leading to higher concentration of repair proteins surrounding a DSB site [47, 48]. Specific recognition of γH2AX by these repair factors requires the presence of protein domains, which bind to the phosphorylated carboxy terminus of γH2AX. So far, two domains, which are frequently found in proteins involved in DDR, have been described to specifically recognize phosphorylated amino acid residues. The forkhead-associated (FHA) domain recognizes phosphorylated threonine residues in a specific aminoacid sequence context [49]. In addition, two consecutive BRCT domains (BRCA1 C-terminal domain) can create a structural element with phospho–peptide binding capacity [50–63]. It was shown that the BRCT repeats of mediator of DNA damage checkpoint protein 1 (MDC1) build the predominant recognition module of γH2AX [39, 43, 54, 55]. Interaction between MDC1 and γH2AX can be recognized as the first step in which the site of the DSB is prepared for DNA damage signaling and repair. This is because there is experimental evidence that MDC1 also directly interacts in a highly dynamic manner with NBS1 [43, 56], which together with other proteins of the MRN complex is required for the activation of ATM [57, 58]. This interaction is mediated through phosphorylation of MDC1 by CK2 that, in turn, promotes phosphorylation-dependent interactions with NBS1, through its closely opposed FHA domain and two BRCA repeats [59]. In this way, a positive feed-back loop is generated that extends H2AX phosphorylation to large DNA regions (millions of base pairs).
Several lines of evidence suggest the critical role of H2AX phosphorylation at DSB sites for nuclear foci formation and induction of DSB repair.
H2AX-knockout cells manifested impaired recruitment of NBS1, 53BP1, and BRCA1 to irradiation-induced foci [45].
Both H2AX+/− and H2AX−/− mouse thymocytes show an increase in chromosomal aberrations [60–62].
Mouse embryonic stem (ES) cells deficient in H2AX phosphorylation have alterations in efficiency of DNA repair by NHEJ or HR [60–64]. As a result of these defects in DNA damage repair, such cells have increased sensitivity to DNA damage [60–65].
H2AX knock-out mice show male-specific infertility and reduced levels of secondary immunoglobulin isotypes, suggesting defects in class switch recombination (CSR) [45]. It was shown that efficient resolution of DSBs induced during CSR in lymphocytes requires H2AX [61, 63], and its absence is associated with chromosome abnormalities involving the immunoglobulin locus [61].
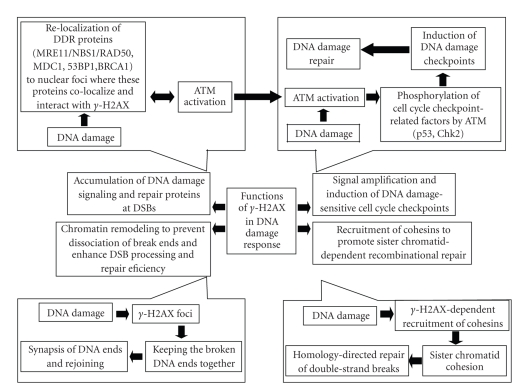
All these facts suggest that γH2AX might serve as a docking site for DNA damage/repair proteins and functions to promote DSB repair and genome stability ([60–68] and Figure 1).
Figure 1.
H2AX phosphorylation and its role in DNA damage response upon phosphorylation H2AX became critical player in DNA damage response. Activity of γH2AX could be summarized to following main points: accumulation of DNA damage signaling and repair proteins at DSBs The generation of DSBs triggers the relocalization of many DNA damage response (DDR) proteins such as MRE11/NBS1/RAD50, MDC1, 53BP1and BRCA1 to nuclear foci where these proteins colocalize and interact with γH2AX. Signal amplification and induction of DNA damage-sensitive cell cycle checkpoints γH2AX contribute to the recruitment of ATM to DSB sites and activation of ATM-dependent cell cycle checkpoints. Chromatin remodeling to prevent dissociation of break ends and enhance DSB processing and repair efficiency The γH2AX foci help keeping the broken DNA ends together and make successful and faithful repair more likely. Recruitment of cohesins to the site of DNA damage to promote sister chromatid-dependent recombinational repair DNA cohesion induced by double-strand DNA break and mediated by γH2AX has an important function during repair of double-strand breaks following DNA replication by holding the damaged chromatid close to its undamaged sister template.
3.2. Signal Amplification and Induction of DNA Damage-Sensitive Cell Cycle Checkpoints
One of most important kinases-activating cell cycle checkpoints following DNA damage is ATM. Mutations in the ATM gene results in the genomic instability and cancer predisposition syndrome Ataxia-Telangiectasia (AT) syndrome. In response to DSBs, ATM phosphorylates many cell cycle checkpoint-related factors such as p53, Chk2, SMC1 and NBS1 [69]. In the absence of DNA damage, ATM forms inactive homodimers, but chromatin remodeling following generation of DSBs leads to autophosphorylation of ATM at serine 1981 and dimer dissociation [18–21]. Next, phosphorylated ATM is recruited to DSB sites by its interaction with NBS1 and this recruitment is critical for phosphorylation of checkpoint proteins by ATM [57]. Mutations in the NBS1 gene result in Nijmegen breakage syndrome (NBS). Cells from both NBS and AT patients show similar phenotypes such as radio-resistant DNA synthesis, radiation hypersensitivity and genome instability [38]. As it was previously described, NBS1 forms a complex with hMRE11 and hRAD50. Since such a complex is involved in DNA double-strand break repair by homologous recombination (HR), one of the reasons of the genomic instability in NBS patients may be the defect in HR. Moreover, it was shown that this function of NBS1 requires DNA damage-induced focus formation of the NBS1/hMRE11/hRAD50 complex through direct interaction of NBS1 with γH2AX at DNA damage sites [37, 70].
As mentioned above, the recruitment of ATM to DSBs is dependent on NBS1 and NBS1 forms the complex with both ATM and γH2AX. Thus, both γH2AX and NBS1 contribute to the recruitment of ATM to DSB sites and its activation of cell cycle checkpoints. Molecular mechanism involved in ATM recruitment to DSBs was recently investigated in detail by Kobayashi and colleagues [38]. These authors provide a line of evidence that γH2AX plays an important role in the recruitment of ATM to DSB sites and the subsequent activation of ATM-related cellular response. Moreover, H2AX is a component of the complex containing ATM at DSBs and is important for activation of the ATM kinase. Importantly, H2AX-knockout cells displayed a defect in DSB-induced cell cycle checkpoint response. These results provide strong evidence that H2AX is one of essential components of the active ATM complex and participates in the activation of ATM-dependent cell cycle checkpoints. Moreover, Kobayashi and colleagues showed that ATM forms a complex with γH2AX via NBS1 or MDC1 and this might be crucial for the recruitment of ATM to DSB sites and induction of ATM-dependent checkpoints [38].
3.3. γ-H2AX and Chromatin Remodeling to Prevent Dissociation of Break Ends and Enhance DSB Processing and Repair Efficiency
During last few years several groups suggested that γH2AX has an important function as a docking site for protein complexes that bind to broken DNA ends and promote chromatin remodeling to keep broken chromosomal DNA ends together [60, 63, 71]. The critical role of γH2AX in chromatin reorganization following DNA damage and preventing the separation of DNA ends, thus facilitating DNA end rejoining, can be important not only in the repair of DSBs but also during V(D)J recombination. According to hypothesis put forward by Bassing and colleagues, γH2AX behaves like an anchor that facilitates the assembly of multiple DNA-protein interactions involving 53BP1, MDC1/NFBD1, and MRN complexes. Formation of such complexes prevents dissociation of broken DNA ends and subsequent error-prone repair of DSBs (reviewed in [71]). Moreover, this specific “anchoring” function of H2AX at sites containing DSBs, would inhibit the irreversible disassociation of broken DNA ends and promote chromatin compaction to facilitate error-free repair, thereby suppressing inappropriate translocations of chromatin fragments. In this way, γH2AX-mediated mechanisms prevent DNA ends from drifting apart, inappropriate rejoining of chromatin fragments, resulting in genetic translocations and other abnormalities (Figure 1 and reviewed in [71]).
3.4. Recruitment of Cohesins to the Site of DNA Damage to Promote Sister Chromatid-Dependent Recombinational Repair
γH2AX might be important for processes occurring further away from the break that are required for efficient repair. One of such processes can be sister chromatid cohesion, that in addition to its essential role in chromosome segregation, is important for efficient postreplicative double-strand DNA break repair [72]. Sister chromatid cohesion is established during S phase by cohesins. In the absence of cohesin complex, chromosomes cannot segregate properly. It was also shown that cohesins play an important role in DNA repair and recombination. Cells with mutated cohesins are oversensitive to irradiation and deficient in postreplicative DNA repair [72, 73]. Moreover, cohesins have been shown to accumulate at laser-induced DNA damage during S and G2 phases in a Mre11/Rad50- dependent manner [74]. In parallel, it was shown that DNA cohesion induced by double-strand DNA breaks and mediated by γH2AX is not only important to maintain sister chromatids. It is also likely to function during repair of double-strand breaks following DNA replication by holding the damaged chromatid close to its undamaged sister template [64]. The use of genetic information present in an undamaged sister chromatid to mediate error-free recombination repair of a DSB would prevent gross chromosomal alterations, which are frequently observed, for example, in cancer cells. Thus, this form of homology-directed DNA repair of double-strand breaks helps to maintain a high level of genome stability, even though immediate cell survival may be only minimally affected in its absence, because of other repair mechanisms (Figure 1 and reviewed in [75]).
4. Methods of γH2AX Detection
The quantitative assessment of DSBs was initially based on methods, such as pulse-field gel electrophoresis (PFGE), DNA elution tests, or the so-called “comet assay” (single cell gel electrophoresis—SCGE). Among these methods, the comet assay is a versatile, sensitive, and widely used one. In this method individual cells with damaged DNA embedded in agarose gels are subjected to an electric field and generate a characteristic pattern of DNA distribution forming a tail that, after staining with fluorescence dye, can be analyzed by fluorescence microscopy. The extent and length of the comet's tail correlates with the severity of DNA damage [76]. The sensitivity of the comet assay, however, depends on proper calibration and its specificity is not absolute [77, 78].
Development of an antibody, that is specific to γH2AX, made it possible to detect H2AX phosphorylation and thus detection of DNA damage and repair in situ in individual cells. The presence of γH2AX in chromatin can be detected shortly after induction of DSBs in the form of discrete nuclear foci. Since each focus represents a single DSB, their frequency reports the incidence of DSBs (reviewed in [79]). Compared with alternative methods of DNA damage assessment mentioned above, the immunocytochemical approach is less cumbersome and offers much greater sensitivity [79, 80]. The presence of γH2AX-containing nuclear foci can be measured by microscopy, flow cytometry, and possibly Western blotting of cell/tissue lysates, with normalization for the total H2AX levels. Measurement of γH2AX with the use of multiparameter flow or laser scanning cytometry seems to be particular advantageous as H2AX phosphorylation can be determined in individual cells with high sensitivity and accuracy and γH2AX expression in cell populations can be correlated with DNA content or induction of apoptosis [79, 80].
5. Applications of γH2AX Detection
Assessment of H2AX phosphorylation as a reporter of DNA damage can be clinically useful. The most important clinical application of γH2AX measurements is to follow DSBs levels induced by radio- and chemotherapy as a marker of treatment efficacy [81, 82] and in dose/scheduling estimation as well as to determine the efficiency of DNA repair to predict possible tumor sensitivity or resistance to DNA damaging anticancer agents. By assessing DSB levels induced by anticancer treatment in normal cells, one may also predict toxicity of anticancer treatment.
Exposure to endogenous or environmental mutagens results in DNA damage which in turn activates the DDR cascade [83, 84]. Once activated, the DDR machinery functions as an “anticancer barrier” and delays or suppresses tumor development by inducing cell death or replicative senescence [83, 85]. However, prolonged activation of DDR may sometimes lead to survival of malignant cells if only these cells are able to bypass cell growth barrier imposed by the DDR pathway [79]. Thus, activated components of the DDR can be used as cancer biomarkers, with γH2AX being the most sensitive one. In this way, γH2AX level measurements may help to detect precancerous lesions or cancer at its early stages [86–88]. In this case, increased background levels of γH2AX or the presence of DSBs on telomere ends may be indicative for replicative senescence, including premature senescence induced by anticancer drugs [89] or cancer progression. Finally, other possible practical applications of γH2AX include screening for effects of environmental pollutants (air- or food-borne etc.) or follow-up of DNA lesions induced by mutagenic substances at work places, including nurses distributing anticancer drugs [90]. Similarly, γH2AX may help to evaluate effects of chemopreventive agents.
Endogenous γH2AX levels may also reflect genomic instability phenotype since optimal protection against cancer requires full functionality of both alleles of H2AX gene (H2AFX). Several different tumor types have been reported to possess mutations, including single gene polymorphism, amplification, or deletion in the region 11q23 that maps for H2AFX [62, 91]. Other groups reported alterations in H2AX gene copy number or changes in its promoter region to be associated with the risk of sporadic breast cancer and lymphomas [81, 92]. In this way, one may also relate H2AFX mutations or deletions with the susceptibility of individuals to tumors, such as lymphoma, acute myeloid leukemia, and head and neck cell carcinomas for tumor risk assessment studies [92–95].
6. Conclusions
As discussed above, determination of γH2AX levels is not only important from the fundamental perspective as a sensitive marker of the DNA damage response, including DNA repair processes. It has also many potential practical applications in monitoring effects induced by anticancer therapy and assessing tumor risk as well as in determination of DNA lesions resulting from exposure of animals and humans to environmental pollutants. However, it is important to discuss not only advantages but also potential limitations and problems of the application of γH2AX in clinical practice. An obvious advantage of γH2AX compared to other DNA damage markers is its extremely high sensitivity that enables one to detect very small changes in genome integrity on a single cell level. However, the lower limit of γH2AX detection depends on many factors, including background levels of nuclear foci observed in unstressed cells, which are associated with DNA replication and progression through mitosis [96, 97]. In addition, background γH2AX levels may differ between various tumor cells and, more importantly, between tumor and normal cells due to changed functionality of H2AXF discussed above. This can make it more complicated to predict the toxicity of anticancer treatment in patients based on the direct comparison of DSBs induced in tumor versus normal cells.
Timing of γH2AX measurements after drug treatment may also be critical for the appropriate data analysis and interpretation. Induction of H2AX phosphorylation is, at least for some agents, very rapid and this requires quick methods of patient sample acquisition and analysis. In some situations, the choice between of different sample material may be crucial for the assay (PBMLs versus tumor cells—leukemia, normal tissue, and tumor samples for solid tumors etc.). Moreover, anticancer treatment may lead to increased phosphorylation of H2AX due to death-associated DNA fragmentation, especially after long time periods after drug treatment or in case of agents which induce rapid apoptosis. However, cell death-related H2AX phosphorylation can be discriminated from that induced by anticancer treatment based on focal and diffuse or soluble γH2AX signals [98]. This requires selection of appropriate diagnostic methods and application of biparametric flow cytometry, scanning cytometry, or fluorescence microscopy to estimate γH2AX levels.
Acknowledgments
This work was supported by a research Grant from Polish Ministry of Science and Higher Education (Grant no. N402 208535) and statutory funds from the Faculty of Chemistry, Gdansk University of Technology.
References
- 1.Jackson SP. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23(5):687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- 2.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair. 2006;5(9-10):1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 3.Smider V, Chu G. The end-joining reaction in V(D)J recombination. Seminars in Immunology. 1997;9(3):189–197. doi: 10.1006/smim.1997.0070. [DOI] [PubMed] [Google Scholar]
- 4.Jackson SP. Detecting, signalling and repairing DNA double-strand breaks. Biochemical Society Transactions. 2001;29(6):655–661. doi: 10.1042/0300-5127:0290655. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka T, Halicka HD, Huang X, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation, the reporters of DNA damage by endogenous oxidants. Cell Cycle. 2006;5(17):1940–1945. doi: 10.4161/cc.5.17.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka T, Huang X, Halicka HD, et al. Cytometry of ATM activation and histone H2AX phosphorylation to estimate extent of DNA damage induced by exogenous agents. Cytometry Part A. 2007;71(9):648–661. doi: 10.1002/cyto.a.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vilenchik MM, Knudson AG. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(22):12871–12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi A, Ohnishi T. Does γH2AX foci formation depend on the presence of DNA double strand breaks? Cancer Letters. 2005;229(2):171–179. doi: 10.1016/j.canlet.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T, Kajstura M, Halicka HD, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation are strongly amplified during mitogenic stimulation of lymphocytes. Cell Proliferation. 2007;40(1):1–13. doi: 10.1111/j.1365-2184.2007.00417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuadrado M, Martinez-Pastor B, Fernandez-Capetillo O. ATR activation in response to ionizing radiation: still ATM territory. Cell Division. 2006;1, article 7:1–4. doi: 10.1186/1747-1028-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JC. DNA topoisomerases. Annual Review of Biochemistry. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 12.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Reviews Molecular Cell Biology. 2002;3(6):430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 13.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nature Reviews Cancer. 2009;9(5):327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Research. 2009;37(3):738–748. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClendon AK, Osheroff N. DNA topoisomerase II, genotoxicity, and cancer. Mutation Research. 2007;623(1-2):83–97. doi: 10.1016/j.mrfmmm.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pommier Y. DNA topoisomerase I Inhibitors: chemistry, biology, and interfacial inhibition. Chemical Reviews. 2009;109(7):2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nature Reviews Cancer. 2006;6(10):789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 18.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. Journal of Biological Chemistry. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 19.Helt CE, Cliby WA, Keng PC, Bambara RA, O’Reilly MA. Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. Journal of Biological Chemistry. 2005;280(2):1186–1192. doi: 10.1074/jbc.M410873200. [DOI] [PubMed] [Google Scholar]
- 20.Kastan MB, Lim D-S. The many substrates and functions of ATM. Nature Reviews Molecular Cell Biology. 2000;1(3):179–186. doi: 10.1038/35043058. [DOI] [PubMed] [Google Scholar]
- 21.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 22.Dupré A, Boyer-Chatenet L, Gautier J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nature Structural and Molecular Biology. 2006;13(5):451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava N, Gochhait S, de Boer P, Bamezai RNK. Role of H2AX in DNA damage response and human cancers. Mutation Research. 2009;681(2-3):180–188. doi: 10.1016/j.mrrev.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 24.Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(37):13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Xu Y, Roy K, Price BD. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Molecular and Cellular Biology. 2007;27(24):8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ward IM, Chen J. Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. Journal of Biological Chemistry. 2001;276(51):47759–47762. doi: 10.1074/jbc.C100569200. [DOI] [PubMed] [Google Scholar]
- 27.Ward IM, Minn K, Chen J. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. Journal of Biological Chemistry. 2004;279(11):9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee B, Kessinger C, Kobayashi J, et al. DNA-PK phosphorylates histone H2AX during apoptotic DNA fragmentation in mammalian cells. DNA Repair. 2006;5(5):575–590. doi: 10.1016/j.dnarep.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Reitsema T, Klokov D, Banáth JP, Olive PL. DNA-PK is responsible for enhanced phosphorylation of histone H2AX under hypertonic conditions. DNA Repair. 2005;4(10):1172–1181. doi: 10.1016/j.dnarep.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 30.Wang H, Wang M, Wang H, Böcker W, Iliakis G. Complex H2AX phosphorylation patterns by multiple kinases including ATM and DNA-PK in human cells exposed to ionizing radiation and treated with kinase inhibitors. Journal of Cellular Physiology. 2005;202(2):492–502. doi: 10.1002/jcp.20141. [DOI] [PubMed] [Google Scholar]
- 31.Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-β mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453(7195):682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 32.Goodarzi AA, Noon AT, Deckbar D, et al. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Molecular Cell. 2008;31(2):167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- 33.Goodarzi AA, Noon AT, Jeggo PA. The impact of heterochromatin on DSB repair. Biochemical Society Transactions. 2009;37(3):569–576. doi: 10.1042/BST0370569. [DOI] [PubMed] [Google Scholar]
- 34.Luijsterburg MS, Dinant C, Lans H, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. Journal of Cell Biology. 2009;185(4):577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinant C, Luijsterburg MS. The emerging role of HP1 in the DNA damage response. Molecular and Cellular Biology. 2009;29(24):6335–6340. doi: 10.1128/MCB.01048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Current Biology. 2000;10(15):886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi J, Tauchi H, Sakamoto S, et al. NBS1 localizes to γ-H2AX foci through interaction with the FHA/BRCT domain. Current Biology. 2002;12(21):1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi J, Antoccia A, Tauchi H, Matsuura S, Komatsu K. NBS1 and its functional role in the DNA damage response. DNA Repair. 2004;3(8-9):855–861. doi: 10.1016/j.dnarep.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 39.Stewart GS, Wang B, Bigneli CR, Taylor AMR, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421(6926):961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 40.Stucki M, Jackson SP. γH2AX and MDC1: anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair. 2006;5(5):534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. Journal of Biological Chemistry. 2003;278(22):19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 42.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. Journal of Cell Biology. 2000;151(7):1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123(7):1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 44.Lou Z, Minter-Dykhouse K, Franco S, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Molecular Cell. 2006;21(2):187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 45.Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296(5569):922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minter-Dykhouse K, Ward I, Huen MSY, Chen J, Lou Z. Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. Journal of Cell Biology. 2008;181(5):727–735. doi: 10.1083/jcb.200801083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nature Structural and Molecular Biology. 2007;14(11):1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Molecular Cell. 1999;4(3):387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- 50.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302(5645):636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 51.Yu X, Chini CCS, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302(5645):639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 52.Clapperton JA, Manke IA, Lowery DM, et al. Structure and mechanism of BRCA1 BRCT domain recognition of phosphorylated BACH1 with implications for cancer. Nature Structural and Molecular Biology. 2004;11(6):512–518. doi: 10.1038/nsmb775. [DOI] [PubMed] [Google Scholar]
- 53.Williams RS, Lee MS, Hau DD, Glover JNM. Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nature Structural and Molecular Biology. 2004;11(6):519–525. doi: 10.1038/nsmb776. [DOI] [PubMed] [Google Scholar]
- 54.Lukas C, Melander F, Stucki M, et al. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO Journal. 2004;23(13):2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez M, Yu X, Chen J, Songyang Z. Phosphopeptide Binding Specificities of BRCA1 COOH-terminal (BRCT) Domains. Journal of Biological Chemistry. 2003;278(52):52914–52918. doi: 10.1074/jbc.C300407200. [DOI] [PubMed] [Google Scholar]
- 56.Goldberg M, Stucki M, Falck J, et al. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421(6926):952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 57.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434(7033):605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- 58.You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Molecular and Cellular Biology. 2005;25(13):5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Reports. 2008;9(8):795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Celeste A, Difilippantonio S, Difilippantonio MJ, et al. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell. 2003;114(3):371–383. doi: 10.1016/s0092-8674(03)00567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petersen S, Casellas R, Reina-San-Martin B, et al. AID is required to initiate Nbs1/γ-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414(6864):660–665. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bassing CH, Chua KF, Sekiguchi J, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(12):8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reina-San-Martin B, Difilippantonio S, Hanitsch L, Masilamani RF, Nussenzweig A, Nussenzweig MC. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. Journal of Experimental Medicine. 2003;197(12):1767–1778. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xie A, Puget N, Shim I, et al. Control of sister chromatid recombination by histone H2AX. Molecular Cell. 2004;16(6):1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kao J, Milano MT, Javaheri A, et al. γ-H2AX as a therapeutic target for improving the efficacy of radiation therapy. Current Cancer Drug Targets. 2006;6(3):197–205. doi: 10.2174/156800906776842957. [DOI] [PubMed] [Google Scholar]
- 66.Franco S, Gostissa M, Zha S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Molecular Cell. 2006;21(2):201–214. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 67.Ramiro AR, Jankovic M, Callen E, et al. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440(7080):105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Research. 2008;36(17):5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsuoka S, Ballif BA, Smogorzewska A, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316(5828):1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 70.Sakamoto S, Iijima K, Mochizuki D, et al. Homologous recombination repair is regulated by domains at the N- and C-terminus of NBS1 and is dissociated with ATM functions. Oncogene. 2007;26(41):6002–6009. doi: 10.1038/sj.onc.1210428. [DOI] [PubMed] [Google Scholar]
- 71.Bassing CH, Alt FW. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004;3(2):149–153. doi: 10.4161/cc.3.2.689. [DOI] [PubMed] [Google Scholar]
- 72.Sjögren C, Nasmyth K. Sister chromatid cohesion is required for postreplicative double-strand break repair in Saccharomyces cerevisiae. Current Biology. 2001;11(12):991–995. doi: 10.1016/s0960-9822(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 73.Sonoda E, Matsusaka T, Morrison C, et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Developmental Cell. 2001;1(6):759–770. doi: 10.1016/s1534-5807(01)00088-0. [DOI] [PubMed] [Google Scholar]
- 74.Kim J-S, Krasieva TB, LaMorte V, Taylor AM, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. Journal of Biological Chemistry. 2002;277(47):45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- 75.Lowndes NF, Toh GW-L. DNA repair: the importance of phosphorylating histone H2AX. Current Biology. 2005;15(3):R99–R102. doi: 10.1016/j.cub.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 76.Ostling O, Johanson KJ. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochemical and Biophysical Research Communications. 1984;123(1):291–298. doi: 10.1016/0006-291x(84)90411-x. [DOI] [PubMed] [Google Scholar]
- 77.Collins AR, Oscoz AA, Brunborg G, et al. The comet assay: topical issues. Mutagenesis. 2008;23(3):143–151. doi: 10.1093/mutage/gem051. [DOI] [PubMed] [Google Scholar]
- 78.McArt DG, McKerr G, Howard CV, Saetzier K, Wasson GR. Modelling the comet assay. Biochemical Society Transactions. 2009;37(4):914–917. doi: 10.1042/BST0370914. [DOI] [PubMed] [Google Scholar]
- 79.Huang X, Halicka HD, Traganos F, Tanaka T, Kurose A, Darzynkiewicz Z. Cytometric assessment of DNA damage in relation to cell cycle phase and apoptosis. Cell Proliferation. 2005;38(4):223–243. doi: 10.1111/j.1365-2184.2005.00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Darzynkiewicz Z, Huang X, Okafuji M. Detection of DNA strand breaks by flow and laser scanning cytometry in studies of apoptosis and cell proliferation (DNA replication) Methods in Molecular Biology. 2006;314:81–93. doi: 10.1385/1-59259-973-7:081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Downs JA. Chromatin structure and DNA double-strand break responses in cancer progression and therapy. Oncogene. 2007;26(56):7765–7772. doi: 10.1038/sj.onc.1210874. [DOI] [PubMed] [Google Scholar]
- 82.Taneja N, Davis M, Choy JS, et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. Journal of Biological Chemistry. 2004;279(3):2273–2280. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- 83.Bartkova J, Hořejší Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 84.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 85.Campisi J. Suppressing cancer: the importance of being senescent. Science. 2005;309(5736):886–887. doi: 10.1126/science.1116801. [DOI] [PubMed] [Google Scholar]
- 86.Kuo LJ, Yang L-X. γ-H2AX—a novel biomaker for DNA double-strand breaks. In Vivo. 2008;22(3):305–310. [PubMed] [Google Scholar]
- 87.Sedelnikova OA, Bonner WM. γH2AX in cancer cells: a potential biomarker for cancer diagnostics, prediction and recurrence. Cell Cycle. 2006;5(24):2909–2913. doi: 10.4161/cc.5.24.3569. [DOI] [PubMed] [Google Scholar]
- 88.Gorgoulis VG, Vassiliou L-VF, Karakaidos P, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 89.Roninson IB. Tumor cell senescence in cancer treatment. Cancer Research. 2003;63(11):2705–2715. [PubMed] [Google Scholar]
- 90.Rekhadevi PV, Sailaja N, Chandrasekhar M, Mahboob M, Rahman MF, Grover P. Genotoxicity assessment in oncology nurses handling anti-neoplastic drugs. Mutagenesis. 2007;22(6):395–401. doi: 10.1093/mutage/gem032. [DOI] [PubMed] [Google Scholar]
- 91.Bassing CH, Suh H, Ferguson DO, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114(3):359–370. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- 92.Novik KL, Spinelli JJ, Macarthur AC, et al. Genetic variation in H2AFX contributes to risk of non-Hodgkin lymphoma. Cancer Epidemiology Biomarkers and Prevention. 2007;16(6):1098–1106. doi: 10.1158/1055-9965.EPI-06-0639. [DOI] [PubMed] [Google Scholar]
- 93.Lu J, Wei Q, Bondy ML, et al. Genetic variants in the H2AFX promoter region are associated with risk of sporadic breast cancer in non-Hispanic white women aged ≤55 years. Breast Cancer Research and Treatment. 2008;110(2):357–366. doi: 10.1007/s10549-007-9717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thirman MJ, Gill HJ, Burnett RC, et al. Rearrangement of the MLL gene in acute lymphoblastic and acute myeloid leukemias with 11q23 chromosomal translocations. New England Journal of Medicine. 1993;329(13):909–914. doi: 10.1056/NEJM199309233291302. [DOI] [PubMed] [Google Scholar]
- 95.Parikh RA, White JS, Huang X, et al. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes and Cancer. 2007;46(8):761–775. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- 96.Ichijima Y, Sakasai R, Okita N, Asahina K, Mizutani S, Teraoka H. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochemical and Biophysical Research Communications. 2005;336(3):807–812. doi: 10.1016/j.bbrc.2005.08.164. [DOI] [PubMed] [Google Scholar]
- 97.McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Molecular Biology of the Cell. 2005;16(10):5013–5025. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu Y, Tseng M, Perdreau SA, et al. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Research. 2007;67(6):2685–2692. doi: 10.1158/0008-5472.CAN-06-3497. [DOI] [PubMed] [Google Scholar]