Abstract
The Burkholderia cepacia complex (Bcc) comprises at least 17 closely-related species of the β-proteobacteria subdivision, widely distributed in natural and man-made inhabitats. Bcc bacteria are endowed with an extraordinary metabolic diversity and emerged in the 1980s as life-threatening and difficult-to-treat pathogens among patients suffering from cystic fibrosis. More recently, these bacteria became recognized as a threat to hospitalized patients suffering from other diseases, in particular oncological patients. In the present paper, we review these and other traits of Bcc bacteria, as well as some of the strategies used to identify and validate the virulence factors and determinants used by these bacteria. The identification and characterization of these virulence factors is expected to lead to the design of novel therapeutic strategies to fight the infections caused by these emergent multidrug resistant human pathogens.
1. The Burkholderia cepacia Complex—An Overview
Members of the Burkholderia cepacia complex (Bcc) are gram-negative bacteria of the β-proteobacteria subdivision and include plant, animal, and human pathogens, with a widespread distribution in natural and man-made inhabitats [1]. These bacteria exhibit an extraordinary metabolic versatility, allowing their adaptation to a wide range of environments. Among the Bcc bacteria, several strains of potential environmental application have been identified due to their ability to degrade pollutants in water and soils (e.g., crude oils, herbicides, recalcitrant aromatic compounds, and xenobiotics). A summary of Burkholderia strains capable of degrading recalcitrant xenobiotics is available at the Biodegradative Strain Database (http://bsd.cme.msu.edu/). Several Bcc strains are also able to produce antifungal compounds and to fix atmospheric nitrogen [2]. Recent evidence suggests that members of the Burkholderia genus are ancient nitrogen-fixing symbionts of Mimosa legumes particularly adapted to acidic infertile soils [3]. Due to the ability of some strains to promote plant growth, bacteria of the Bcc have attracted significant commercial interest as biocontrol, bioremediation, and plant-growth promoting agents, mainly due to their ability to colonize the rhizosphere of several crops of economical interest, like corn, maize, rice, pea, and sunflower [2]. However, these bacteria have also emerged as important human pathogens and the risks associated with the agricultural uses of Bcc strains remain unclear. There is a general consensus that the large-scale use of organisms of the Burkholderia genus is imprudent until more is known about the fate of biocontrol strains after their release in the environment. The pathogenic mechanisms and traits used by these bacteria, the clinical outcomes of infected patients, and the interaction of the introduced biocontrol strains with environmental and clinical strains need further studies [2]. Presently, there is some evidence that the environment can be a reservoir for the acquisition of novel Bcc infections. For example, the epidemic B. cenocepacia strain PHDC was recovered both from patients suffering from cystic fibrosis (CF) in the mid-Atlantic region of USA, as well as from agricultural soils [4].
2. Bcc as Opportunistic Pathogens in Humans
In the last 30 years, several epidemiological, taxonomic, and molecular biology studies of Bcc strains have been carried out by research groups worldwide, mainly due to the ability of these strains to cause chronic infections among CF patients. CF is the most frequent hereditary disorder among Caucasians. The disease results from mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), a cAMP-dependent chloride channel, mainly expressed in the apical membrane of epithelial cells [5]. The genetic defect results in multiple organ system impairment, being the respiratory tract the most affected. Chronic pulmonary infections, although caused by a limited number of bacterial species (e.g., Pseudomonas aeruginosa, Bcc, Staphylococcus aureus, Haemophilus influenza, and Stenotrophomonas maltophilia), remain the leading cause of death of these patients [5]. The large majority of respiratory infections among CF patients are due to P. aeruginosa strains [6]. Compared to this major pathogen, Bcc strains infect a relatively small fraction of CF patients. However, Bcc infections are particularly feared by CF patients and their caregivers since the clinical outcome is highly variable and unpredictable, ranging from asymptomatic carriage to the cepacia syndrome [1]. Additionally, in the vast majority of CF patients, pulmonary colonization with Bcc is associated with a worst prognosis, including an accelerated decline of the patients' clinical status and an increased risk of death [6].
Bcc bacteria are also important pathogens in other compromised patients, as is the case of patients suffering from chronic granulomatous disease (CGD) [7]. CGD is a rare hereditary disease that is caused by mutations in the subunits of the NADPH oxidase complex of the phagocytes, resulting in their inability to produce reactive oxygen species [7]. Invasive Bcc infections and pneumonia is the second leading cause of death of CGD patients [7]. There are also some reports of Bcc infections in immunocompromised patients such as cancer and HIV patients, and also among immunocompetent individuals [8, 9]. In immunocompetent individuals, Bcc strains have been isolated in cases of chronic suppurative otitis media, pharyngeal infections, and paediatric neck infections [9]. In recent years, an increasing number of bacteraemia cases caused by Bcc among non-CF hospitalized patients have been reported. Most of these patients have comorbidities such as chronic hemodialysis, diabetes mellitus, congestive heart failure, and malignancy. Among these hospitalized non-CF patients, hemodialysis, permanence in intensive care units, use of central venous catheters, indwelling urinary catheters, and endotracheal tubes are now recognized as risk factors contributing for Bcc acquisition. The accumulating reports of nosocomial outbreaks caused by Bcc led to the recognition of these bacteria as emergent nosocomial pathogens among non-CF patients, in particular among oncology patients [8].
The Bcc comprises at least seventeen distinct species, genetically distinct but phenotypically similar [10, 11] (Table 1). Strains from all the Bcc species have been isolated from CF patients and from the environment, however, their frequency of isolation is uneven [12]. While the majority of the isolates obtained from CF patients belong to the species B. cenocepacia and B. multivorans [12], the majority of the enviromental isolates belong to the species B. cepacia, B. ambifaria, B. cenocepacia, and B. pyrrocinia [2].
Table 1.
Burkholderia cepacia complex species and strains with their genome sequences finished or in progress. The Bcc genomes here described are summarized in the Burkholderia genome database (http://www.burkholderia.com/viewAllGenomes.do).
| Bcc Species | Sources and Relevant Characteristics | Strains sequenced | Unfinished genomes | References |
|---|---|---|---|---|
| B. cepacia | Infections in Humans (CF and non-CF); bioremediation and biocontrol agent | [13] | ||
|
| ||||
| B. multivorans | Infections in Humans (CF and non-CF) | ATCC17616 | CGD1, CGD2, CGD2M | [13] |
|
| ||||
| B. cenocepacia | Infections in Humans (CF and non-CF); biocontrol agent | J2315, AU1054, HI2424, MCO-3, PC184 | Bu72 | [14] |
|
| ||||
| B. stabilis | Infections in Humans (CF and non-CF) | [15] | ||
|
| ||||
| B. vietnamiensis | Infections in Humans (CF and non-CF); bioremediation and biocontrol agent | G4 | [13] | |
|
| ||||
| B. dolosa | Infections in CF patients | AU0158 | [16] | |
|
| ||||
| B. ambifaria | Infections in Humans (CF and non-CF); biocontrol agent | AMMD, MC40-6, MEX-5, IOP40-10 | [17] | |
|
| ||||
| B. pyrrocinia | Infections in CF patients; biocontrol agent | [18] | ||
|
| ||||
| B. anthina | Infections in Humans (CF and non-CF) | [18] | ||
|
| ||||
| B. ubonensis | Nosocomial infection | Bu | [10] | |
|
| ||||
| B. latens | Infections in CF patients | [10] | ||
|
| ||||
| B. diffusa | Infections in Humans (CF and non-CF), isolated from water and soil | [10] | ||
|
| ||||
| B. arboris | Infections in Humans (CF and non-CF), environmental sources | [10] | ||
|
| ||||
| B. seminalis | Infections in Humans (CF and non-CF), environmental sources | [10] | ||
|
| ||||
| B. metallica | Infections in CF patients | [10] | ||
|
| ||||
| B. contaminans | Infections in CF patients and in animals | [11, 19] | ||
|
| ||||
| B. lata | Isolates from forest soil | 383 | [11] | |
A considerable phenotypic variability has been found for all the Bcc species [13], even within sequential clinical isolates of the same strain [20]. This phenotypic variability difficults the correct identification of Bcc strains by diagnostic microbiology laboratories [17]. Several phenotypic and genetic methods have been used for the identification of Bcc species, including whole-cell protein profile, fatty acid analysis, and 16S rRNA and recA gene restriction and sequencing analysis [2]. However, the genetic methods have proven to be the most effective for the correct identification of Bcc strains. Nowadays, the multilocus sequence typing scheme (MLST) is considered the golden standard method for the identification of Bcc species [21]. MLST analysis compares the nucleotide sequence of seven house-keeping genes of Bcc and the information obtained for each strain sequence type (ST) is stored in a public database (http://pubmlst.org/bcc/), thus allowing its use worlwide [21]. Recently, this method was redesigned in a nested-PCR MLST format that can be used for the accurate identification of Bcc strains directly from sputum samples [19]. This approach allowed the identification of Bcc strains in 23 sputum samples obtained from 17 CF patients, of which 8 samples where culture was negative [19]. In addition, the performance of MLST directly with sputum samples also allowed the identification of Bcc strains from CF patients with mixed Bcc infections or co-infected with P. aeruginosa strains, without the need for strain isolation [19].
3. Burkholderia cepacia Complex Infections in Cystic Fibrosis Patients
Bcc bacteria emerged as important CF pathogens during the 1980s, when some infected patients exhibited a rapid clinical deterioration due to necrotizing pneumonia and sepsis, resulting in early death [22]. This fatal decline in the patient's clinical condition became known as the cepacia syndrome and was not observed for patients infected with any of the other CF pathogens. The key determinants associated with the cepacia syndrome are not completely understood, and both bacterial and host factors are thought to play important roles in determining this dramatic clinical outcome [23, 24]. Several strains of the species B. multivorans, B. cenocepacia, B. cepacia, and B. dolosa have been shown to be highly transmissible among CF patients through social contact [25, 26]. In particular, highly epidemic lineages of the B. cenocepacia species have been described, including the Electrophoretic Type 12 (ET12), the Philadelphia-District of Columbia (PHDC), and the MidWest epidemic lineages [27, 28]. These epidemic strains can have an international impact, as is the case of the highly transmissible ET12 lineage. This epidemic lineage spread among individuals with CF from Canada, UK, and other European countries, being able to replace B. multivorans and causing a high mortality due to its ability to cause the cepacia syndrome [14, 23, 29]. Due to the easy transmission of highly virulent strains among CF patients, segregation measures of Bcc-infected patients have been successfully implemented and led to the reduction of the transmission of Bcc strains [30].
The prevalence of Bcc species varies geographically, being B. cenocepacia the most predominant species in CF centers in North America, while B. multivorans is the most common species in European CF centers [31]. However, outbreaks caused by other Bcc species have occurred worldwide. For instance, in the major Portuguese CF centre, B. cepacia is the most prevalent Bcc species. In addition, an outbreak of B. cepacia was reported and associated with the use of nonsterile saline solutions for nasal application [32]. Bcc outbreaks among non-CF populations, mainly due to strains of the species B. cenocepacia, B. cepacia, and B. multivorans are also well documented [33]. Accumulating evidence points out contaminated pharmaceuticals, cosmetics, disinfectants, and preservative products as major sources of Bcc bacteria [9, 34]. This is due to their ability to survive in these products. In hospital settings, these pathogens have been recovered from tap and distilled water, dialysis machines, nebulisers, catheters, blood gas analysers, thermometers, ventilator temperature sensors, solutions, and intravenous fluids [9].
One of the major problems associated with Bcc infection is their intrinsic resistance to most of the clinically available antimicrobials, including aminoglycosides, quinolones, polymyxins, and β-lactams [35]. The multiresistance of Bcc bacteria appears to result from various efflux pumps that efficiently remove antibiotics from the cell, decreased contact of antibiotics with the bacterial cell surface due to their ability to form biofilms, and changes in the cell envelope that reduce the permeability of the membrane to the antibiotic [36]. Bcc bacteria are also resistant to neutrophil-mediated non-oxidative killing and to the antimicrobial peptides produced by airway epithelial cells, including lysozyme, lactoferrin, and phospholipase A2 [37]. Therefore, CF patients chronically infected with Bcc are difficult to treat and, although current treatment strategies use double or triple antibiotic combinations to achieve bactericidal activity, they rarely result in the eradication of the pathogen, particularly in the case of chronic infection [36].
4. Organization of Bcc Genomes
In 2003, the Wellcome Trust Sanger Institute sequenced the first genome of a Bcc strain. The strain chosen was the type strain of the ET12 epidemic lineage, the B. cenocepacia strain J2315 (http://www.sanger.ac.uk/Projects/B_cenocepacia/). Presently, the genomic sequences of 18 strains from 7 Bcc species are publicly available (http://pathema.jcvi.org/cgi-bin/Burkholderia/PathemaHomePage.cgi). The genomes of Bcc bacteria are organized in three circular chromosomal replicons and one to five megaplasmids, ranging from 6.2 to 8.7 Mbp in size, with a GC content of about 67%. The large size and repartition of the genomes of Bcc is thought to increase their flexibility to acquire and lose genes. In a recent bioinformatics study, Cooper et al. (2010) suggested that the genes located in secondary chromosomes exhibit a weaker codon usage bias than those located in primary chromosomes, being subject to a faster evolutionary rate [38]. Several evidences point out that more than 10% of the Bcc genomes have been acquired by horizontal gene transfer, contributing to the genomic plasticity and metabolic diversity of these bacteria. For example, in the case of the B. cenocepacia strain J2315, 14 genomic islands, most probably arisen from horizontal gene transfer, have been identified based on their distinct GC content percentage [39]. The acquisition of genomic islands appears to play a crucial role in the evolution of this particular epidemic lineage, introducing new functions that promoted survival and pathogenesis in the CF lung. This is the case of the 31.7 kb cci pathogenicity island, which appears to be unique to B. cenocepacia strains [40]. This pathogenicity island encodes both virulence and metabolism-associated genes, including the CciIR quorum sensing system, a fatty acid biosynthesis operon, transcriptional regulators, and genes involved in the metabolism of amino acids [40]. In addition, the genome of B. cenocepacia J2315 contains 79 insertion sequence (IS) elements that are most probably involved in genomic rearrangements, replicon fusion, activation/silencing of gene expression, mobilization of DNA, and recruitment of foreign genes [39]. Another feature of the genomes of Bcc bacteria is the presence of multiple pathways with related functions, and gene redundancy due to the occurrence of paralogous genes.
Sequencing of several Bcc genomes, followed by comparative genomics, is a powerful tool for the identification of virulence-associated genes of Bcc bacteria, including new genes encoding proteins with no predicted function. In the sequenced Bcc genomes, the percentage of protein encoding genes with unknown function varies between 13 and 35% (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). It is quite possible that a significant percentage of these genes of unknown function might be involved, either directly or indirectly, in the pathogenesis of Bcc bacteria.
5. Strategies to Discover New Bcc Virulence Factors and Determinants
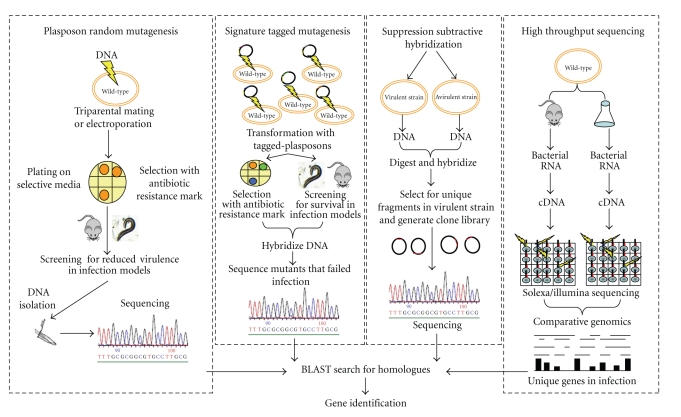
Different strategies have been designed to identify pathogenicity-related genes from Bcc bacteria, including the generation of mutant libraries with transposons and plasposons, systematic gene-by-gene inactivation and high-throughput sequencing, as illustrated in Figure 1. Our research group has been using a strategy based on the generation of mutant libraries from B. cenocepacia and B. cepacia strains by random mutagenesis with plasposons [41], followed by rescue of the interrupted genes, sequencing and comparison of the nucleotide sequence of the interrupted genes with the available genome sequences of Bcc strains, combined with the virulence assessment in the Bcc infection models X-CGD mice and/or Caenorhabditis elegans. A mutant library derived from B. cepacia IST408 allowed the identification of the bce-I gene cluster that encodes proteins and enzymes involved in the biosynthesis of the exopolysaccharide (EPS) Cepacian [42]. Cepacian is composed of a branched acetylated heptasaccharide repeat unit with D-glucose, D-rhamnose, D-mannose, D-galactose, and D-glucuronic acid, in the ratio 1 : 1 : 1 : 3 : 1, respectively [43]. Several studies have shown that Cepacian interfered with the phagocytosis of bacteria by human neutrophils and, inhibits neutrophil chemotaxis, and the production of reactive oxygen species [44, 45]. The ability to produce this EPS was also associated with persistence of infection in the BALB/c and X-CGD mice models of infection [44, 46]. Studies performed with cepacian-defective mutants have also shown that cepacian is required for the formation of thick and mature biofilms [47]. Biofilm formation in vitro is a common trait of Bcc strains and has been associated with the persistence of Bcc infections [48]. In addition, bacteria of the Bcc growing in biofilms have been found to be more resistant to antimicrobials than those growing plancktonically. It is also worth to mention that in a recent study by Dales et al. (2009), biofilms formed by Bcc were found to be more resistant to antibiotics compared to P. aeruginosa biofilms [49]. Remarkably, a mutant producing about one half of the amount of the EPS was recently found to carry a plasposon insertion interrupting a gene encoding the RNA chaperone Hfq [50]. Hfq proteins are global regulators of metabolism, acting as RNA chaperones involved in the riboregulation of target mRNAs by small regulatory noncoding RNAs (sRNAs), facilitating the interaction with their target mRNAs [51]. The B. cepacia hfq mutant was shown to be more susceptible to stress conditions, particularly to those that mimicked the lung environment of the CF host, indicating that Hfq plays a major role in the survival of Bcc bacteria under those stress conditions [50]. In addition, the hfq mutants from B. cepacia, B. dolosa, and B. ambifaria exhibited a reduced ability to colonize and kill the nematode C. elegans, indicating that Hfq is an important virulence determinant of Bcc bacteria. In agreement with the roles played by Hfq in other bacteria, Sousa et al. (2010) have also shown that the B. cepacia IST408 Hfq is able to bind to sRNAs. Recently, 213 putative sRNAs were identified within the genome of B. cenocepacia J2315, based on the combination of comparative genomics and prediction of their secondary structures [52]. Work in progress envisages the identification and characterization of sRNAs from Bcc to gain clues on their possible contributions to virulence.
Figure 1.
Strategies used to identify Bcc virulence factors and determinants.
Another mutant library, derived from the highly epidemic strain B. cenocepacia J2315, allowed the identification of a gene encoding an acyl carrier protein (ACP) [53]. Bacterial ACPs play a central role in metabolism, being the donors of the acyl moiety that is required for the biosynthesis of fatty acids, phospholipids, endotoxins, glycolipids, and signalling molecules that are necessary for growth and pathogenesis [54]. The acp mutant exhibited an increased ability to form biofilms in vitro, a more hydrophobic cell surface, and reduced ability to colonize and kill the nematode C. elegans, indicating that ACP protein is a virulence determinant for Bcc bacteria [53]. In addition, the amino acid sequence and structural differences between the ACP proteins from bacteria and humans make this protein an attractive target for the development of novel antimicrobial compounds [55].
A mutant library from B. cenocepacia K56-2 was also constructed. The screen of this library for mutants impaired in their ability to kill the nematode C. elegans allowed the identification of the regulatory protein Pbr. The pbr mutant exhibited a pleiotropic phenotype, being unable to produce phenazines, exhibited a reduced resistance to stresses such as oxidative and osmotic stress, and a reduced ability to survive prolonged nutrient starvation periods [56].
A signature-tagged mutagenesis (STM) strategy was used by Hunt et al. to identify putative virulence factors of Bcc [57] (Figure 1). STM is a comparative hybridization technique that uses a collection of transposons, each modified by the incorporation of a DNA sequence tag [57]. The pool of mutants is inoculated into a chronic pulmonary infection animal model and the bacteria recovered after infection are identified due to the tags. Mutants containing a transposon insertion in genes required for survival will fail to pass through the in vivo selection, thus allowing the identification of these genes. This strategy led to the identification of several B. cenocepacia K56-2 genes that were required for bacterial survival in a rat model of chronic lung infection, including genes involved in cellular metabolism, global regulation, DNA replication and repair, cell surface proteins, and polysaccharide production [57].
A suppression-subtractive hybridization (SSH) strategy was used to identify genes that are unique to the B. cenocepacia and/or to ET12 epidemic lineage strains [58] (Figure 1). Recently, a high-throughput sequencing strategy was used to compare the transcriptional response of clinical and environmental strains of B. cenocepacia [59] (Figure 1). This strategy revealed a large number of regulatory differences between environmental and clinical strains, which might result from specific adaptations to each of the different niches, despite their high degree of DNA sequence similarity. Genes that encode for molecular chaperones and iron acquisition proteins were found to be particularly induced in the clinical strain [59].
All these strategies have allowed the identification of several genes putatively involved in the virulence of Bcc strains. However, the characterization of knockout mutants in these genes is hampered by limited available genetic tools and the inherent resistance of Bcc strains to the most common antibiotics used for genetic selection. In this context, some research groups have been developing molecular tools to genetically manipulate Bcc strains. For instance, Lefebre and Valvano (2002) constructed several expression vectors that contain the dhfr gene, encoding the dihydrofolate reductase enzyme required for trimethoprim resistance, together with either the constitutive promoter of the S7 ribossomal protein gene from Burkholderia sp LB400, or the arabinose-inducible BAD promoter from Escherichia coli [60]. However, the concentration of arabinose required for maximal gene expression [2% (w/v) or higher] causes a change in cell volume typical of osmotic stress [61]. As a consequence, the full complementation of a given mutation using these vectors is seldom achieved, limiting its use [50]. Therefore, those authors have constructed another expression vector, containing the rhamnose-regulated PrhaB promoter of E. coli that allows maximal gene expression at low concentrations of rhamnose [61]. Another limitation derives from the fact that the B. cenocepacia J2315 strain is a poor recipient of DNA in transformation and conjugation. As an example, transformation of B. cepacia IST408 with plasposon pTnMod was 104-fold more efficient than B. cenocepacia J2315 (S. A. Sousa and J. H. Leitão, unpublished results). Recently, the electroporation procedure for this strain was modified to increase its transformation efficiency [62]. Factors that contribute to this improvement include the addition of glycine to the growth medium to weaken the thick cell wall, demethylation of transforming DNA by extraction from a E. coli dam dcm host strain to escape to the J2315 restriction system specific for methylated GATC sites, the inclusion of the Ocr protein in the transformation mixture to act as a decoy to inhibit Type I restriction endonuclease attack of entering DNA, and the use of spermine to reduce the resistance of the B. cenocepacia J2315 strain to several antibiotics [62]. Other strategies used to effectively generate Bcc mutants rely on the lambda red recombinase system, as proposed by Datsenko and Wanner (2000) [63]. This strategy uses linear DNA transformations and has allowed the successful homologous expression of a lipase gene in B. cepacia [64], as well the construction of an insertion mutant in B. cenocepacia J2315 BCAL1538 (C. G. Ramos, S. A. Sousa, J. H. Leitão, unpublished results).
A combination of one or more of the previous approaches has allowed the identification of several potential virulence factors, including the cable pili and various adhesins [65], flagella [66], a type III and a type VI secretion systems [67, 68], lipopolysaccharide [69], four types of iron-chelating siderophores (salicylic acid, ornibactin, pyochelin, cepaciachelin, and cepabactin) [70], production of extracellular proteins, like proteases, lipases, and haemolysins [71, 72], quorum-sensing systems [73, 74], and others (recently reviewed in [75]). However, not all strains produce each of these virulence factors, and none of these factors has been clearly demonstrated to be a major contributor to human disease. In fact, contrasting with other pathogens, the pathogenicity of Bcc bacteria does not rely on a single gene. Accumulating evidences point out that Bcc virulence is polygenic, involving genes related to survival under stress conditions [50, 76–78]. Nevertheless, the Bcc genome is equipped with the known crucial genes for colonization and initiation of chronic infection in the respiratory tract, which are involved in motility, adhesion, and host tissue damage. Another important feature of some Bcc strains is their ability to invade and survive inside eukaryotic cells, including soil-dwelling amoebae, human macrophages, and airway epithelial cells [79].
6. Concluding Remarks
Members of the Bcc have emerged in the last decades as important pathogens to human, animals, and plants. The pathogenicity of these bacteria is polygenic, and thus involves a multitude of known and unknown virulence factors and determinants. Several strategies have been successfully used by several research groups to reveal novel and unknown virulence factors and determinants. The knowledge of the molecular mechanisms employed by Bcc bacteria for virulence and pathogenesis is of crucial importance to identify new targets for the rational design of novel strategies and/or molecules to combat Bcc infections, since their resistance to most of the clinically-relevant antimicrobials renders the infections untreatable. In order to be regarded as a potential drug target, a given gene or gene product must be essential for survival of the pathogen in the host and should be conserved in the various strains of the pathogen, while presenting little or no conservation in humans. Genome-based strategies, including genome sequencing, microarray-based expression technology, and large-scale mutagenesis studies, are expected to contribute, in the near future, for the development of new strategies and/or antimicrobials molecules to fight the devastating and presently difficult-to-treat infections caused by Bcc strains.
Acknowledgments
This work was funded by FEDER and Fundação para a Ciência e Tecnologia (FCT), Portugal, through Contract PTDC/EBB-BIO/098352/2008, and a postdoctoral grant to S. A. Sousa. A doctoral grant from Fundação Calouste Gulbenkian (FCG) to C. G. Ramos is also acknowledged.
References
- 1.Mahenthiralingam E, Urban TA, Goldberg JB. The multifarious, multireplicon Burkholderia cepacia complex. Nature Reviews Microbiology. 2005;3(2):144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 2.Chiarini L, Bevivino A, Dalmastri C, Tabacchioni S, Visca P. Burkholderia cepacia complex species: health hazards and biotechnological potential. Trends in Microbiology. 2006;14(6):277–286. doi: 10.1016/j.tim.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Bontemps C, Elliott GN, Simon MF, et al. Burkholderia species are ancient symbionts of legumes. Molecular Ecology. 2010;19(1):44–52. doi: 10.1111/j.1365-294X.2009.04458.x. [DOI] [PubMed] [Google Scholar]
- 4.LiPuma JJ, Spilker T, Coenye T, Gonzalez CF. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet. 2002;359(9322):2002–2003. doi: 10.1016/S0140-6736(02)08836-0. [DOI] [PubMed] [Google Scholar]
- 5.Ratjen F, Döring G. Cystic fibrosis. Lancet. 2003;361(9358):681–689. doi: 10.1016/S0140-6736(03)12567-6. [DOI] [PubMed] [Google Scholar]
- 6.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clinical Microbiology Reviews. 2002;15(2):194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston RB., Jr. Clinical aspects of chronic granulomatous disease. Current Opinion in Hematology. 2001;8(1):17–22. doi: 10.1097/00062752-200101000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Mann T, Ben-David D, Zlotkin A, et al. An outbreak of Burkholderia cenocepacia bacteremia in immunocompromised oncology patients. Infection. 2010;38(3):187–194. doi: 10.1007/s15010-010-0017-0. [DOI] [PubMed] [Google Scholar]
- 9.Marioni G, Rinaldi R, Ottaviano G, Marchese-Ragona R, Savastano M, Staffieri A. Cervical necrotizing fasciitis: a novel clinical presentation of Burkholderia cepacia infection. Journal of Infection. 2006;53(5):e219–e222. doi: 10.1016/j.jinf.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 10.Vanlaere E, LiPuma JJ, Baldwin A, et al. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov., and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. International Journal of Systematic and Evolutionary Microbiology. 2008;58(7):1580–1590. doi: 10.1099/ijs.0.65634-0. [DOI] [PubMed] [Google Scholar]
- 11.Vanlaere E, Baldwin A, Gevers D, et al. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2009;59(1):102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- 12.Mahenthiralingam E, Baldwin A, Dowson CG. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. Journal of Applied Microbiology. 2008;104(6):1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 13.Vandamme P, Holmes B, Vancanneyt M, et al. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. International Journal of Systematic Bacteriology. 1997;47(4):1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 14.Vandamme P, Holmes B, Coenye T, et al. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Research in Microbiology. 2003;154(2):91–96. doi: 10.1016/S0923-2508(03)00026-3. [DOI] [PubMed] [Google Scholar]
- 15.Vandamme P, Mahenthiralingam E, Holmes B, et al. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) Journal of Clinical Microbiology. 2000;38(3):1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermis K, Coenye T, LiPuma JJ, Mahenthiralingam E, Nelis HJ, Vandamme P. Proposal to accommodate Burkholderia cepacia genomovar VI as Burkholderia dolosa sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2004;54(3):689–691. doi: 10.1099/ijs.0.02888-0. [DOI] [PubMed] [Google Scholar]
- 17.Coenye T, Vandamme P, Govan JRW, Lipuma JJ. Taxonomy and identification of the Burkholderia cepacia complex. Journal of Clinical Microbiology. 2001;39(10):3427–3436. doi: 10.1128/JCM.39.10.3427-3436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandamme P, Henry D, Coenye T, et al. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunology and Medical Microbiology. 2002;33(2):143–149. doi: 10.1111/j.1574-695X.2002.tb00584.x. [DOI] [PubMed] [Google Scholar]
- 19.Drevinek P, Vosahlikova S, Dedeckova K, Cinek O, Mahenthiralingam E. Direct culture-independent strain typing of Burkholderia cepacia complex in sputum samples from patients with cystic fibrosis. Journal of Clinical Microbiology. 2010;48(5):1888–1891. doi: 10.1128/JCM.02359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen GY, Stull TL, Burns JL. Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. Journal of Clinical Microbiology. 1993;31(4):788–792. doi: 10.1128/jcm.31.4.788-792.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baldwin A, Mahenthiralingam E, Thickett KM, et al. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. Journal of Clinical Microbiology. 2005;43(9):4665–4673. doi: 10.1128/JCM.43.9.4665-4673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isles A, Maclusky I, Corey M. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. Journal of Pediatrics. 1984;104(2):206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 23.Goven JRW, Brown PH, Maddison J, et al. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet. 1993;342(8862):15–19. doi: 10.1016/0140-6736(93)91881-l. [DOI] [PubMed] [Google Scholar]
- 24.Jones AM, Dodd ME, Govan JRW, et al. Burkholderia cenocepacia and Burkholderia multivorans: influence on survival in cystic fibrosis. Thorax. 2004;59(11):948–951. doi: 10.1136/thx.2003.017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipuma JJ, Dasen SE, Nielson DW, Stern RC, Stull TL. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet. 1990;336(8723):1094–1096. doi: 10.1016/0140-6736(90)92571-x. [DOI] [PubMed] [Google Scholar]
- 26.Biddick R, Spilker T, Martin A, LiPuma JJ. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiology Letters. 2003;228(1):57–62. doi: 10.1016/S0378-1097(03)00724-9. [DOI] [PubMed] [Google Scholar]
- 27.Johnson WM, Tyler SD, Rozee KR. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. Journal of Clinical Microbiology. 1994;32(4):924–930. doi: 10.1128/jcm.32.4.924-930.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JS, Witzmann KA, Spilker T, Fink RJ, LiPuma JJ. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. Journal of Pediatrics. 2001;139(5):643–649. doi: 10.1067/mpd.2001.118430. [DOI] [PubMed] [Google Scholar]
- 29.Speert DP, Henry D, Vandamme P, Corey M, Mahenthiralingam E. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerging Infectious Diseases. 2002;8(2):181–187. doi: 10.3201/eid0802.010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Festini F, Buzzetti R, Bassi C, et al. Isolation measures for prevention of infection with respiratory pathogens in cystic fibrosis: a systematic review. Journal of Hospital Infection. 2006;64(1):1–6. doi: 10.1016/j.jhin.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 31.Govan JRW, Brown AR, Jones AM. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiology. 2007;2(2):153–164. doi: 10.2217/17460913.2.2.153. [DOI] [PubMed] [Google Scholar]
- 32.Cunha MV, Pinto-de-Oliveira A, Meirinhos-Soares L, et al. Exceptionally high representation of Burkholderia cepacia among B. cepacia complex isolates recovered from the major Portuguese cystic fibrosis center. Journal of Clinical Microbiology. 2007;45(5):1628–1633. doi: 10.1128/JCM.00234-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reik R, Spilker T, LiPuma JJ. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. Journal of Clinical Microbiology. 2005;43(6):2926–2928. doi: 10.1128/JCM.43.6.2926-2928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Geftic SG, Heymann H, Adair FW. Fourteen year survival of Pseudomonas cepacia in a salts solution preserved with benzalkonium chloride. Applied and Environmental Microbiology. 1979;37(3):505–510. doi: 10.1128/aem.37.3.505-510.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leitão JH, Sousa SA, Cunha MV, et al. Variation of the antimicrobial susceptibility profiles of Burkholderia cepacia complex clonal isolates obtained from chronically infected cystic fibrosis patients: a five-year survey in the major Portuguese treatment center. European Journal of Clinical Microbiology and Infectious Diseases. 2008;27(11):1101–1111. doi: 10.1007/s10096-008-0552-0. [DOI] [PubMed] [Google Scholar]
- 36.George AM, Jones PM, Middleton PG. Cystic fibrosis infections: treatment strategies and prospects. FEMS Microbiology Letters. 2009;300(2):153–164. doi: 10.1111/j.1574-6968.2009.01704.x. [DOI] [PubMed] [Google Scholar]
- 37.Baird RM, Brown H, Smith AW, Watson ML. Burkholderia cepacia is resistant to the antimicrobial activity of airway epithelial cells. Immunopharmacology. 1999;44(3):267–272. doi: 10.1016/s0162-3109(99)00122-8. [DOI] [PubMed] [Google Scholar]
- 38.Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Computational Biology. 2010;6(4) doi: 10.1371/journal.pcbi.1000732. Article ID e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden MTG, Seth-Smith HMB, Crossman LC, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. Journal of Bacteriology. 2009;91(1):261–277. doi: 10.1128/JB.01230-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldwin A, Sokol PA, Parkhill J, Mahenthiralingam E. The Burkholderia cepacia epidemic strain marker is part of a novel genomic island encoding both virulence and metabolism-associated genes in Burkholderia cenocepacia . Infection and Immunity. 2004;72(3):1537–1547. doi: 10.1128/IAI.72.3.1537-1547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennis JJ, Zylstra GJ. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Applied and Environmental Microbiology. 1998;64(7):2710–2715. doi: 10.1128/aem.64.7.2710-2715.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreira LM, Videira PA, Sousa SA, Leitão JH, Cunha MV, Sá-Correia I. Identification and physical organization of the gene cluster involved in the biosynthesis of Burkholderia cepacia complex exopolysaccharide. Biochemical and Biophysical Research Communications. 2003;312(2):323–333. doi: 10.1016/j.bbrc.2003.10.118. [DOI] [PubMed] [Google Scholar]
- 43.Cescutti P, Bosco M, Picotti F, et al. Structural study of the exopolysaccharide produced by a clinical isolate of Burkholderia cepacia . Biochemical and Biophysical Research Communications. 2000;273(3):1088–1094. doi: 10.1006/bbrc.2000.3059. [DOI] [PubMed] [Google Scholar]
- 44.Conway B-AD, Chu KK, Bylund J, Altman E, Speert DP. Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. Journal of Infectious Diseases. 2004;190(5):957–966. doi: 10.1086/423141. [DOI] [PubMed] [Google Scholar]
- 45.Bylund J, Burgess L-A, Cescutti P, Ernst RK, Speert DP. Exopolysaccharides from Burkholderia cenocepacia inhibit neutrophil chemotaxis and scavenge reactive oxygen species. Journal of Biological Chemistry. 2006;281(5):2526–2532. doi: 10.1074/jbc.M510692200. [DOI] [PubMed] [Google Scholar]
- 46.Sousa SA, Ulrich M, Bragonzi A, et al. Virulence of Burkholderia cepacia complex strains in gp91phox−/− mice. Cellular Microbiology. 2007;9(12):2817–2825. doi: 10.1111/j.1462-5822.2007.00998.x. [DOI] [PubMed] [Google Scholar]
- 47.Cunha MV, Sousa SA, Leitão JH, Moreira LM, Videira PA, Sá-Correia I. Studies on the involvement of the exopolysaccharide produced by cystic fibrosis-associated isolates of the Burkholderia cepacia complex in biofilm formation and in persistence of respiratory infections. Journal of Clinical Microbiology. 2004;42(7):3052–3058. doi: 10.1128/JCM.42.7.3052-3058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conway B-AD, Venu V, Speert DP. Biofilm formation and acyl homoserine lactone production in the Burkholderia cepacia complex. Journal of Bacteriology. 2002;184(20):5678–5685. doi: 10.1128/JB.184.20.5678-5685.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dales L, Ferris W, Vandemheen K, Aaron SD. Combination antibiotic susceptibility of biofilm-grown Burkholderia cepacia and Pseudomonas aeruginosa isolated from patients with pulmonary exacerbations of cystic fibrosis. European Journal of Clinical Microbiology and Infectious Diseases. 2009;28(10):1275–1279. doi: 10.1007/s10096-009-0774-9. [DOI] [PubMed] [Google Scholar]
- 50.Sousa SA, Ramos CG, Moreira LM, Leitão JH. The hfq gene is required for stress resistance and full virulence of Burkholderia cepacia to the nematode Caenorhabditis elegans . Microbiology. 2010;156(3):896–908. doi: 10.1099/mic.0.035139-0. [DOI] [PubMed] [Google Scholar]
- 51.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Molecular Microbiology. 2004;51(6):1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 52.Coenye T, Drevinek P, Mahenthiralingam E, et al. Identification of putative noncoding RNA genes in the Burkholderia cenocepacia J2315 genome. FEMS Microbiology Letters. 2007;276(1):83–92. doi: 10.1111/j.1574-6968.2007.00916.x. [DOI] [PubMed] [Google Scholar]
- 53.Sousa SA, Ramos CG, Almeida F, et al. Burkholderia cenocepacia J2315 acyl carrier protein: a potential target for antimicrobials’ development? Microbial Pathogenesis. 2008;45(5-6):331–336. doi: 10.1016/j.micpath.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Byers DM, Gong H. Acyl carrier protein: structure-function relationships in a conserved multifunctional protein family. Biochemistry and Cell Biology. 2007;85(6):649–662. doi: 10.1139/o07-109. [DOI] [PubMed] [Google Scholar]
- 55.White SW, Zheng J, Zhang Y-M, Rock CO. The structural biology of type II fatty acid biosynthesis. Annual Review of Biochemistry. 2005;74:791–831. doi: 10.1146/annurev.biochem.74.082803.133524. [DOI] [PubMed] [Google Scholar]
- 56.Ramos CG, Sousa SA, Grilo AM, Eberl L, Leitão JH. The Burkholderia cenocepacia K56-2 pleiotropic regulator Pbr, is required for stress resistance and virulence. Microbial Pathogenesis. 2010;48(5):168–177. doi: 10.1016/j.micpath.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 57.Hunt TA, Kooi C, Sokol PA, Valvano MA. Identification of Burkholderia cenocepacia genes required for bacterial survival in vivo. Infection and Immunity. 2004;72(7):4010–4022. doi: 10.1128/IAI.72.7.4010-4022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bernier SP, Sokol PA. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia . Journal of Bacteriology. 2005;187(15):5278–5291. doi: 10.1128/JB.187.15.5278-5291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoder-Himes DR, Chain PSG, Zhu Y, et al. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3976–3981. doi: 10.1073/pnas.0813403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lefebre MD, Valvano MA. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Applied and Environmental Microbiology. 2002;68(12):5956–5964. doi: 10.1128/AEM.68.12.5956-5964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cardona ST, Valvano MA. An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia . Plasmid. 2005;54(3):219–228. doi: 10.1016/j.plasmid.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 62.Dubarry N, Du W, Lane D, Pasta F. Improved electrotransformation and decreased antibiotic resistance of the cystic fibrosis pathogen Burkholderia cenocepacia strain J2315. Applied and Environmental Microbiology. 2010;76(4):1095–1102. doi: 10.1128/AEM.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jia B, Yang J-K, Liu W-S, Li X, Yan Y-J. Homologous overexpression of a lipase from Burkholderia cepacia using the lambda Red recombinase system. Biotechnology Letters. 2010;32(4):521–526. doi: 10.1007/s10529-009-0189-9. [DOI] [PubMed] [Google Scholar]
- 65.Urban TA, Goldberg JB, Forstner JF, Sajjan US. Cable pili and the 22-kilodalton adhesin are required for Burkholderia cenocepacia binding to and transmigration across the squamous epithelium. Infection and Immunity. 2005;73(9):5426–5437. doi: 10.1128/IAI.73.9.5426-5437.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Urban TA, Griffith A, Torok AM, Smolkin ME, Burns JL, Goldberg JB. Contribution of Burkholderia cenocepacia flagella to infectivity and inflammation. Infection and Immunity. 2004;72(9):5126–5134. doi: 10.1128/IAI.72.9.5126-5134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tomich M, Griffith A, Herfst CA, Burns JL, Mohr CD. Attenuated virulence of a Burkholderia cepacia type III secretion mutant in a murine model of infection. Infection and Immunity. 2003;71(3):1405–1415. doi: 10.1128/IAI.71.3.1405-1415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aubert DF, Flannagan RS, Valvano MA. A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia . Infection and Immunity. 2008;76(5):1979–1991. doi: 10.1128/IAI.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vinion-Dubiel AD, Goldberg JB. Lipopolysaccharide of Burkholderia cepacia complex. Journal of Endotoxin Research. 2003;9(4):201–213. doi: 10.1179/096805103225001404. [DOI] [PubMed] [Google Scholar]
- 70.Thomas MS. Iron acquisition mechanisms of the Burkholderia cepacia complex. BioMetals. 2007;20(3-4):431–452. doi: 10.1007/s10534-006-9065-4. [DOI] [PubMed] [Google Scholar]
- 71.Kooi C, Subsin B, Chen R, Pohorelic B, Sokol PA. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infection and Immunity. 2006;74(7):4083–4093. doi: 10.1128/IAI.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hutchison ML, Poxton IR, Govan JRW. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infection and Immunity. 1998;66(5):2033–2039. doi: 10.1128/iai.66.5.2033-2039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sokol PA, Malott R, Riedel K, Eberl L. Communication systems in the genus Burkholderia: global regulators and targets for novel antipathogenic drugs. Future Microbiology. 2007;2(5):555–563. doi: 10.2217/17460913.2.5.555. [DOI] [PubMed] [Google Scholar]
- 74.Deng Y, Boon C, Eberl L, Zhang L-H. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. Journal of Bacteriology. 2009;191(23):7270–7278. doi: 10.1128/JB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leitão JH, Sousa SA, Ferreira AS, Ramos CG, Silva IN, Moreira LM. Pathogenicity, virulence factors, and strategies to fight against Burkholderia cepacia complex pathogens and related species. Applied Microbiology and Biotechnology. 2010;87(1):31–40. doi: 10.1007/s00253-010-2528-0. [DOI] [PubMed] [Google Scholar]
- 76.Flannagan RS, Aubert D, Kooi C, Sokol PA, Valvano MA. Burkholderia cenocepacia requires a periplasmic HtrA protease for growth under thermal and osmotic stress and for survival in vivo. Infection and Immunity. 2007;75(4):1679–1689. doi: 10.1128/IAI.01581-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maloney KE, Valvano MA. The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages. Infection and Immunity. 2006;74(10):5477–5486. doi: 10.1128/IAI.00798-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saldías MS, Lamothe J, Wu R, Valvano MA. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infection and Immunity. 2008;76(3):1059–1067. doi: 10.1128/IAI.01167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saldías MS, Valvano MA. Interactions of Burkholderia cenocepacia and other Burkholderia cepacia complex bacteria with epithelial and phagocytic cells. Microbiology. 2009;155(9):2809–2817. doi: 10.1099/mic.0.031344-0. [DOI] [PubMed] [Google Scholar]