Abstract
Peroxisome proliferator-activated receptors (PPARs), which are known to regulate lipid homeostasis, are tightly controlled by nutrient availability, and they control nutrient handling. In this paper, we focus on how nutrients control the expression and action of PPARs and how cellular signaling events regulate the action of PPARs in metabolically active tissues (e.g., liver, skeletal muscle, heart, and white adipose tissue). We address the structure and function of the PPARs, and their interaction with other nuclear receptors, including PPAR cross-talk. We further discuss the roles played by different kinase pathways, including the extracellular signal-regulated kinases/mitogen-activated protein kinase (ERK MAPK), AMP-activated protein kinase (AMPK), Akt/protein kinase B (Akt/PKB), and the NAD+-regulated protein deacetylase SIRT1, serving to control the activity of the PPARs themselves as well as that of a key nutrient-related PPAR coactivator, PPARγ coactivator-1α (PGC-1α). We also highlight how currently applied nutrigenomic strategies will increase our understanding on how nutrients regulate metabolic homeostasis through PPAR signaling.
1. Introduction
1.1. PPARs: Nuclear Receptors Functioning as Metabolic Sensors
Energy homeostasis is mostly achieved by hormonal and nutrient-mediated control of the expression of genes encoding metabolic enzymes. Nuclear receptors are responsible for the transcriptional regulation of the vast majority of the aforementioned genes. These receptors are transcription factors that respond to small lipophilic hormones, vitamins, and metabolites. Among the nuclear receptors, the glucocorticoid, thyroid hormone, and estrogen receptors (GR, TR, and ER, resp.) are important regulators of genes involved in metabolic fuel homeostasis both during development and in response to metabolic stress, as well as in the regulation of cellular energy metabolism. Estrogen-related receptors (ERRs) also play critical roles in the regulation of cellular energy metabolism. Other nuclear receptors include the peroxisome proliferator-activated receptors (PPARs), liver X receptors (LXRs), the farnesoid X receptor (FXR), retinoid X receptor (RXR), and hepatocyte nuclear factor-4α (HNF-4α), all of which are activated by molecules of metabolic pathways, such as lipids and fatty acids (FAs), and thereby function as metabolic sensors. The PPARs take part in the genetic regulation of the complex pathways involved in mammalian metabolism, including fatty acid oxidation and lipogenesis that occur in response to nutritional and physiological stimuli. Taken together, PPARα or NR1C, PPARδ (also known as PPARβ) or NR1C2, and PPARγ or NR1C3 constitute group C in subfamily 1 of the superfamily of nuclear receptors [1].
1.2. PPARs: Structural Features and Interaction with Cofactors
Although the PPARs share high degrees of functional and structural similarities, they are encoded by distinct single-copy genes located on different chromosomes. Human PPARα is located on chromosome 22 [2, 3], PPARδ on chromosome 6 [4], and PPARγ on chromosome 3 [5]. The canonical PPAR response element (PPRE) is a DR1 motif (a direct repeat of the sequence separated by a single nucleotide, preferentially, and adenine [6]) formed by a 5′- and 3′-AGGTCA half-site [7]. This motif typically is present in the promoters of PPAR target genes, including those involved not only in nutrient handling but also in inflammation, cell growth, and differentiation [8, 9]. The formation of complexes between PPARs and other factors is required for the full transcriptional induction of PPAR targets in a variety of tissues. PPARs are predominantly located within the nucleus, where they generally heterodimerize with RXR. In order to achieve their nuclear activity, the PPARs, like all nuclear receptors, have distinct functional domains: an N-terminal domain involved in transcriptional activation, a DNA-binding domain consisting of two zinc-finger motifs, the second finger being involved in binding to the 5′-DR1 half-site as well as dimerization with RXR [6], a hinge region which allows for adequate rotation of the C-terminal domain for interaction with other proteins, and a C-terminal ligand-binding domain including the ligand-dependent activation function- (AF-2), important for RXR heterodimerization and interaction with cofactors [10, 11]. Again, like all nuclear receptors, PPARs act not alone but in association with cofactors that remodel the structure of chromatin in order to either permit or prevent transcription. In the absence of ligand (fatty acids or their derivatives), PPARs form complexes with corepressors such as NCoR, RIP140, or SMRT, which repress transcription through the recruitment of histone deacetylases [12–14]. In the presence of ligand, on the other hand, coactivators, such as p300, CBP, or SRC-1 (all being coactivators related to PPAR function but not primarily associated to nutrient regulation, indicated as coactivators in Figure 1), become bound to the amino terminal of PPARγ coactivator-1 (PGC-1, a key nutrient-modulated coactivator of the PPARs, discussed in Section 3), then acetylate and remodel chromatin, thus enhancing gene transcription via the relief of chromatin condensation (see Figure 1) [15–17]. Depending on the cell type, the ligand-induced conformational changes, and the sequence of the DNA-binding element, a specific complex is formed between the receptor and the coactivators or corepressors, thus allowing fine-tuning of the physiological response. This also explains the variety of changes in gene expression that occur when a nuclear receptor is activated by different ligands.
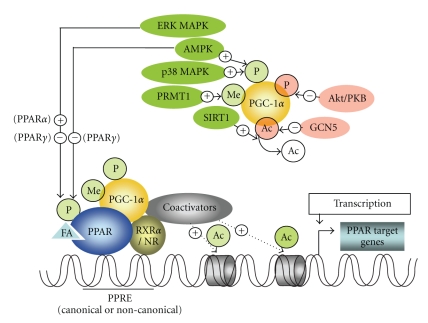
Figure 1.
Modulation of the actions of PPAR through phosphorylation by ERK MAPK or AMPK or through regulation of PGC-1α activity by various signaling events. Phosphorylation of the PPAR receptors can either increase or decrease their activity. SIRT1-mediated deacetylation activates PGC-1α, while acetylation by GCN5 inhibits PGC-1α-directed gene expression. Phosphorylation by AMPK or p38 MAPK increases the stabilization of PGC-1α, whereas Akt/PKB-mediated phosphorylation facilitates its degradation. PRMT1 activates PGC-1α through methylation at several arginine residues. Activation of PGC-1α that is recruited to ligand-bound PPAR, the latter being complexed with RXR and/or other nuclear receptors, allows the recruitment of coactivators that acetylate the chromatin, allowing the DNA encoding a particular PPAR target gene to be transcribed. Ac, acetyl group; ERK MAPK, extracellular signal-regulated kinases/mitogen-activated protein kinase; AMPK, AMP-dependent protein kinase; Akt/PKB, Akt/protein kinase B; p38 MAPK, p38 mitogen-activated protein kinase; FA, fatty acid or metabolite from nutrients binding to and activating PPAR; Me, methyl group; P, phosphate group; PGC-1α, peroxisome proliferator-activated receptor γ coactivator-1α; PPAR, peroxisome proliferator-activated receptor; PPRE, PPAR response element; PRMT1, protein arginine methyltransferase 1; RXR, retinoid X receptor; NR, nuclear receptor; SIRT1, sirtuin 2 ortholog 1; +, activation; −, inhibition.
1.3. Interaction between PPARs and Other Nuclear Receptors
PPARs can associate with other nuclear receptors, and these interactions can involve noncanonical PPREs. Although PPARs and TRs generally compete for interaction with RXR and thus inhibit their respective activity [18], synergism between PPAR and TR can also occur; an example of this is given in the context of nutrient handling, involving the regulation of the expression of the gene encoding uncoupling protein 3, playing a role in the handling of fatty acids within the mitochondria [19]. This interplay has been shown in rat skeletal muscle [20] as well as in cotransfection experiments in rat L6 myoblasts containing a reporter construct driven by the rat UCP3 promoter [20]. Activation of UCP3 gene transcription in vivo by thyroid hormone (T3) requires the presence of fatty acids (the natural ligands of PPARs; see Section 3), while in the absence of fatty acids, transcription can be restored by the PPARδ agonist L165041 [20]. The UCP3 gene promoter has been shown to contain a noncanonical thyroid hormone response element (TRE) termed TRE1 that is conserved from rodents to humans [20, 21], and this response element is also recognized by PPARs [21].
1.4. PPARs: Different Genes, Different Roles?
It was previously assumed that the three known isoforms, PPARα,δ, and γ—which display tissue-specific expression (see Table 1) and possess different gene-regulatory profiles—had clearly distinct roles. For instance, PPARγ, expressed predominantly in adipose tissue and the immune system, existing as two distinct proteins γ1 and γ2, which arise by differential transcription start sites and alternative splicing [5], was assigned the key role as regulator of adipose development, lipid mobilization, and adipose insulin sensitivity [22], whereas PPARα-regulated genes were considered to be associated with lipid oxidation in muscle and liver [23]. The PPARα target genes include carnitine palmitoyltransferase I (CPT I), which is involved in the transport of long-chain fatty acyl groups into the mitochondria, medium-chain acyl-CoA dehydrogenase (involved in β-oxidation), and (specifically in liver) mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase (the rate-limiting enzyme of ketogenesis), as well as peroxisomal acyl-CoA oxidase (peroxisomal β-oxidation) and microsomal cytochrome P450 (CYP) FA hydroxylases. Thus, PPARα would be expected to play a critical role in the maintenance of lipid homeostasis (oxidation and production). Although it seems clear that it is primarily involved in lipid metabolism, PPARα may also provide a link between dyslipidemia and diabetes. Exposure of insulin-sensitive tissues (in particular liver and skeletal muscle) to excess nonesterified FA and circulating triglycerides (triacylglycerol, TAG) induces insulin resistance [24], and this can be corrected by the administration of PPARα activators, the actions of which promote the removal of intracellular lipid through FA oxidation [25]. The PPARδ isoform is predominantly expressed in skeletal muscle (where it induces fatty acid oxidation and the expression of largely the same genes as does PPARα), but it is also expressed in brain, heart, liver, adipose tissue, and small intestine [26]. This receptor subtype, which is still under active study, is perhaps the most versatile of the three subtypes, to judge from its wide tissue distribution. PPARδ has been allocated a central role in the direction of fuel usage between different organs (for review, see [27]).
Table 1.
Tissue distributions of the various PPARs (RNA and protein) in adult rodents and humans. Abbreviations: GI, gastrointestinal; WAT, white adipose tissue; BAT, brown adipose tissue; CNS, central nervous system. Symbols: −, absent; ±, barely detectable; +, weak; ++, moderate; +++, high; ++++, very high (Taken from [26, 28, 45]).
| Tissue | Protein/mRNA | PPARα | PPARβ/δ | PPARγ |
|---|---|---|---|---|
| GI tract | ||||
| Mouse | Protein | ++++ | ||
| Human | mRNA | + or ++ | + to ++++ | ++ to ++++ |
| Mouse | mRNA | + or +++ | ||
| Rat | mRNA | ++ to ++++ | ++ to ++++ | + to ++ |
|
| ||||
| Liver | ||||
| Mouse | Protein | +++ | ||
| Rat | Protein | ++++ | ||
| Human | mRNA | + to ++++ | + or ++ | + to ++ |
| Mouse | mRNA | ++++ | ||
| Rat | mRNA | ++++ | ++ | − |
|
| ||||
| Kidney | ||||
| Mouse | Protein | +++ | ||
| Human | mRNA | ++ to ++++ | + to +++ | + to +++ |
| Mouse | mRNA | +++ | ||
| Rat | mRNA | +++ to ++++ | +++ | ± |
|
| ||||
| Heart | ||||
| Mouse | Protein | ++ | ||
| Human | mRNA | +++ to ++++ | + or +++ | ++ to +++ |
| Mouse | mRNA | ++ | ||
| Rat | mRNA | + or +++ | + | ± |
|
| ||||
| WAT | ||||
| Human | mRNA | + | ++ | ++++ |
| Rat | mRNA | + | ++ | +++ |
|
| ||||
| BAT | ||||
| Mouse | mRNA | +++ | ||
| Rat | mRNA | ++++ | ++ | ++++ |
|
| ||||
| CNS | ||||
| Rat | mRNA | + | + or +++ | ± |
|
| ||||
| Brain | ||||
| Mouse | Protein | ++ | ||
| Human | mRNA | ++ | + or +++ | |
| Rat | mRNA | + | +++ | |
|
| ||||
| Skeletal muscle | ||||
| Mouse | Protein | + | ||
| Human | mRNA | ++ to ++++ | ++ or ++++ | + or +++ |
| Mouse | mRNA | +++ | +++ | ± |
| Rat | mRNA | ++ | ++++ | ± |
|
| ||||
| Skin | ||||
| Mouse | Protein | +++ | ||
| Rat | mRNA | + | ± | |
|
| ||||
| Lung | ||||
| Mouse | Protein | ++ | ||
| Human | mRNA | + | + or +++ | ++++ |
| Rat | mRNA | + | ++ | + |
1.5. PPAR Cross-Talk and Fine-Tuning of Nutrient Handling
Because of the overlap in expression profiles between the PPARs [26, 28] (see Table 1), it is perhaps not surprising that there exists cross-talk among PPARs. Indeed, it has been shown that nonliganded PPARδ represses the transcriptional activity of PPARα and PPARγ [29, 30]. Contrasting evidence exists on a nongenomic action of PPARδ on regulation of PPARγ signaling. Using transient transfection studies, it has been shown that the ligand-binding domain of PPARδ, without binding to DNA, exerts ligand-dependent dominant-negative activity on PPARγ1 signaling [30], although in a previous study a non-DNA-binding PPARδ derivative failed to exert such an effect [29]. Since the PPARs act as RXR heterodimers, it is conceivable that, in analogy to ligand-dependent RXR competition between PPAR and liver X receptor (LXR) [31, 32], PPARδ and LXRα [33], PPARα and TR [34], as well as PPARγ and TR [35], competition for RXR could occur between the PPAR isoforms. Indeed, the nongenomic dominant-negative effect of PPARδ on PPARγ is likely to involve RXR sequestration, thus preventing PPARγ-RXR heterodimerization [30]. Interaction between PPARs and other nuclear receptors and PPAR cross-talk together allow for fine-tuning of interorgan nutrient handling, in concert with the effects of the various signaling molecules that are common to the nuclear receptors and that direct their actions.
2. Nutritional Control of Expression and Activation of PPARs
2.1. Nutrients: Natural PPAR Ligands
PPARs have been shown to be under nutritional control. Dietary nutrients and their derivatives (or adipose-derived fatty acids during food deprivation) directly control PPAR activity since they are the natural ligands of PPARs. PPARs display the greatest preference for monounsaturated and polyunsaturated fatty acids (MUFAs and PUFAs, resp.), as demonstrated by means of various ligand-binding assays [36, 37]. The fact that each PPAR activates a different gene program, despite their overlapping expressions, would seem to suggest that PPARs display ligand specificity. Indeed, the structure of the ligand-binding pocket differs considerably among the various PPARs, as revealed by X-ray crystal-structure analysis [37, 38]. Despite this, natural fatty acids can be ligands of all three PPAR isoforms. Using comparative nutrigenomics analysis, it has been recently shown that, in response to high-fat diet, the diet-induced target genes of PPARα are conserved between yeast, mouse, rat, and man, underlining the importance of nutritional control of PPAR function [39]. A different nutrigenomic approach consisted of the use of synthetic triglycerides composed of one single fatty acid in combination with gene expression profiling to examine the effects of various individual dietary fatty acids on hepatic gene expression in mice. Results revealed that (i) increased fatty acid chain length and degree of unsaturation increased the number of genes being upregulated and that (ii) genes regulated by dietary unsaturated fatty acids remained unaltered in PPARα knockout mice, identifying PPARα as their target, and the same genes were upregulated in mice treated with the PPARα agonist WY14643 [40]. Since the binding of a ligand promotes a conformational change that is permissive for interactions with tissue-specific coactivator proteins (see Section 3), allowing nucleosome remodeling and activation of the transcription of cell type-specific target genes [37, 41], it is conceivable that upon binding to a ligand-binding pocket a given fatty acid induces conformational changes, which differ among the various PPAR subtypes. Given that the transcriptional activity induced by each PPAR subtype is cell type specific [42], the different conformations induced following ligand binding might confer cell specificity on the various PPARs (through heterodimerization with different receptors and binding to cell type-specific cofactors).
2.2. Role of PPARs in the Adaptation to Nutrient Deprivation
One widely employed way of studying how PPAR expression and function is controlled by nutrients in different tissues is by imposing nutrient deprivation. The fasting state influences the actions of all known PPARs. For instance, fasting is known to increase PPARα signaling in the liver, through increased mRNA levels of the coactivator PGC-1α [43], and thereby tightly to regulate hepatic gluconeogenesis and FA oxidation. Interestingly, and perhaps paradoxically, it has recently been shown that during fasting upregulation of gene expression by PPARδ, not PPARα, is sensitive to adipose-derived plasma FA, thus assigning a clear role for PPARδ as a plasma FA sensor in liver [44]. Given the central role of PPARδ in controlling skeletal muscle lipid utilization, fasting (which results in a greater reliance on fatty acids) would be expected to increase PPARδ expression and/or activity. In line with this, starvation has been shown to result in a dramatic but transient upregulation of PPARδ mRNA in rat gastrocnemius muscle [45]. This correlated with rapid nuclear accumulations of PPARδ and the coactivator PGC-1α after food deprivation [45]. It has also been shown that, in skeletal muscle, PPARδ and PGC-1α physically interact with each other within the nucleus [46]. This interaction would then lead both to increased fatty acid levels and to increased expression of genes such as those for myosin heavy chain I (MHC I), thioesterase I (MTE I) [47], and carnitine palmitoyl transferase 1 (CPT1), as well as uncoupling protein 3 (UCP3) [48, 49], thereby underlining the role of PPARδ as a key regulator of muscle-fiber switching and fatty acid metabolism. The transient nature of the increased PPAR expression that occurs during starvation (both the mRNA levels and the nuclear accumulation decreasing once serum FA levels increase [45]) would imply that excessive intracellular fat accumulation inhibits PPAR expression. Indeed, rat PPARα and PPARδ mRNA are each shown to be downregulated after a 48-hour fast [45] and conversely suppression of free fatty acids (using the antilipolytic drug nicotinic acid) was found to increase the mRNA levels of skeletal muscle PPARδ [50]. Both the early fasting state and nicotinic acid treatment cause increased phosphorylation of AMP-activated protein kinase [45], which is known to interact functionally with, and stimulate, PPAR (see Section 3). Once serum fatty acid levels become elevated during food deprivation, the raised muscle AMPK phosphorylation level falls. Taken together, these data indicate that the initial absence of burnable fatty acids during food deprivation triggers a process in which the myocyte is rapidly “converted” to a cell type dedicated to the uptake and burning of fat. Once the intracellular FA levels reach a certain value, the myocyte switches off AMPK and reduces PPAR expression throughout the remainder of the starvation period. Similarly, a downregulation of PPARδ mRNA levels in human skeletal muscle expression has been reported in healthy human subjects after a 48-hour fast [51]. Although data from shorter food-deprivation periods in humans are lacking, it seems likely that, after food deprivation, there is a rapid but transient increase in skeletal muscle PPAR expression in humans too. Activation of the white adipose tissue PPARγ promotes lipid synthesis and storage [52]. Therefore, during fasting, the action of PPARγ has to be inhibited. Indeed, in mature adipocytes, during fasting, its action is inhibited by a physical interaction with the protein sirtuin 1 (SIRT1) (the mammalian Sir2 ortholog), the result being lipolysis [53]. In contrast, the action of PPARγ is enhanced by direct binding to lipin-1, a protein which is known to promote triacylglycerol storage within adipocytes, and to be less expressed during fasting, resulting in lipolysis [54]. There exists cross-talk between PPARγ and PPARδ in this context: PPARδ inhibits the control of PPARγ expression [55], enhancing lipolysis.
2.3. Influence of the Nutritional State on PPAR Action
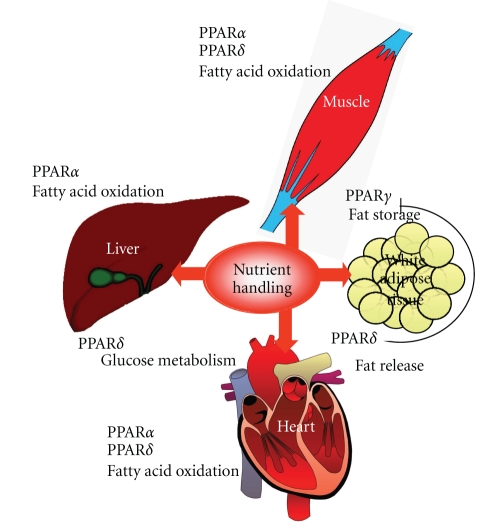
It is well known that in the liver PPARα controls fatty acid oxidation [23], but the role of PPARδ in the liver has up to now scarcely been assessed. A recent transcriptional profiling analysis using PPARα- versus PPARδ-depleted mice has revealed that PPARδ exerts a distinct role in the liver, namely in the control of glucose utilization and lipoprotein metabolism, as well as the suppression of inflammation [56], whereas PPARα mainly controls hepatic lipid homeostasis, which was especially revealed during fasting, causing drastic changes in the hepatic gene expression profile of the PPARα-depleted mice but not in that of the PPARδ-depleted mice [56]. It should, however, be noted that, as described above, the same group assigned a predominant role for PPARδ as FA sensor in liver during fasting, being activated in response to a rise in plasma FA levels, in contrast to PPARα [44]. It is conceivable that dietary FAs and adipose-derived FAs may activate different PPAR-related pathways, and more research is clearly necessary to gain more insight into the respective roles of PPARα and PPARδ in nutrient handling in liver and other organs. In Figure 2, an overview of PPAR action on nutrient handling in the different metabolically active organs, on the basis of the above data, is depicted.
Figure 2.
PPAR action in relation to nutrient handling in metabolically active tissues.
3. Nutrient Availability-Related Cytosolic Signaling Pathways Directly and Indirectly Modulating PPAR Activity
3.1. Control of PPAR Phosphorylation by Upstream Signaling Related to Fuel Use
Activation of cytosolic and nuclear signaling, modulating the activity of PPARs, is nutrient regulated. Recent data have revealed that kinases whose activity is modulated by the nutritional state can directly act on phosphorylation of PPARs. Insulin increases phosphorylation and transcriptional activity of PPARα, due to increased activity of the extracellular signal-regulated kinases/mitogen-activated protein kinase (ERK MAPK) pathway [57]. This same pathway inhibits adipogenesis through phosphorylation and inactivation of PPARγ [58]. Both actions are in favor of an increase in insulin sensitivity. Adenosine monophosphate-activated protein kinase (AMPK) is an energy sensor that is activated when the cell-energy level is low [59]. Once activated, AMPK stimulates both glucose uptake and lipid oxidation to produce energy, while turning off energy-consuming processes. Direct proof for phosphorylation of PPARα by AMPK has not been provided thus far, but it has been shown that phosphorylation of PPARγ by AMPK represses both the ligand-dependent and -independent transcriptional activation function of the receptor [60], thus counteracting adipogenesis and favoring lipid oxidation. PPARδ phosphorylation and activity are also likely to be directly controlled by kinase pathways, but to our knowledge evidence for this has not been obtained yet.
3.2. Nutrient-Related Control of PPAR Activity through Activation of PGC-1
The aforementioned kinases as well as other kinases and proteins, functionally associated with nutrient availability, do not only act directly on the PPARs but also modulate the activity of important regulators of the activity of PPARs, namely the PGC-1s, well known as key transcriptional coactivators involved in the control of nutrient and energy metabolism. Three isoforms of PGC-1 are known [namely, PGC-1α, PGC-1β, and PGC-1-related coactivator (PRC)]. They each control mitochondrial physiology and FA oxidation, while exerting an isoform-specific regulation of different metabolic pathways [61]. PGC-1α and PGC-1β are highly expressed in heart, skeletal muscle, and brown adipose tissue, where, via nuclear respiratory factors (NRFs), they induce expression of genes involved in the regulation of mitochondrial biogenesis [61]. PGC-1 family members can interact not only with PPARs but also with other members of the nuclear receptor superfamily, such as ERR, LXR, and HNF-4α [62–64], and with distinct transcription factors and regulatory elements, including cAMP response element-binding protein (CREB), the lipogenic transcription factor sterol regulatory element-binding protein-1c (SREBP-1c), and forkhead box O1 (FOXO1) [43, 65–68]. PPARs, ERR, HNF-4α, and GR commonly bind to LXXLL motifs present in the N-terminal domain of PGC-1, while other transcription factors bind to different regions of the protein, hence allowing a coordinated transcriptional response to nutrient and physiological signals. Although at least two clinical studies have identified a correlation between mutations of the gene encoding PGC-1α and either insulin resistance or diabetes [69, 70], basic research has produced contrasting results. For instance, overexpression of PGC-1 α, leading to increased PPARα expression in primary cultures of rat skeletal muscle cells, induces increased expression of the mammalian tribbles homolog TRB3, an inhibitor of Akt signaling [71], a result that implies that PGC-1α has the potential to cause insulin resistance through PPARα signaling. Moreover, PGC-1−/− mice are protected against the insulin resistance induced by a high-fat diet [72].
4. Nutrient-Related Factors Controlling PGC-1 Activity
In response to nutrient signaling, (e.g., in the fasting/fed state, discussed in Section 2), PGC-1 isoforms regulate their own transcription, but a posttranslational regulation also occurs. In particular, (i) phosphorylation, (ii) reversible acetylation, and (iii) methylation are key mechanisms by which the function of PGC-1α function is maintained. (i) Three protein kinases directly phosphorylate PGC-1α. P38 mitogen-activated kinase (P38 MAPK) phosphorylates PGC-1α [73], leading to a more active and stable protein that is unable to bind the p160 corepressor. Also, AMPK phosphorylates and activates PGC-1 [59] (Figure 1). Activation of PGC-1α by phosphorylation through p38 MAPK and AMPK occurs in muscle [74, 75] whereas, in contrast, phosphorylation of PGC-1α in the liver by Akt/protein kinase B (Akt/PKB, which is a central kinase in the insulin-signaling cascade) leads to decreased stability and activity (see [76]; Figure 1). It is noteworthy that AMPK is constitutively activated in the muscles of transgenic mice harboring an activated form of PPARδ [77], while skeletal muscle cells exposed to a pharmacological PPARδ activator show increased AMPK activity [78, 79]. (ii) By analogy with the antagonistic phosphorylation induced by AMPK and Akt/PKB, the protein deacetylase SIRT1 opposes the action of GCN5, an acetyl transferase. GCN5 inhibits PGC-1 transcriptional activity by acetylating PGC-1 at several lysine residues [80, 81] while SIRT1 activates PGC-1 by deacetylating it and induces expression of PGC-1 gene targets (see [82, 83]; Figure 1). SIRT1, located within the cell nucleus, modulates gene expression in ways that depend on the cellular energy state, which it senses through the cell's NAD+ levels. SIRT1 is an important regulator of those metabolic processes that are initiated by a rise in the intracellular NAD+/NADH ratio when the energy supply is low. Recently, SIRT1-mediated deacetylation of PGC-1α was reported to play a critical role in the regulation of hepatic FA oxidation: nutrient signaling involving SIRT1 and PGC-1α activated gluconeogenic and fatty acid oxidation genes in the fasting liver [82]. Such nutrient signaling gives rise to increases in pyruvate and NAD+ levels, resulting in increases in the amount and enzymatic activity of SIRT1. Loss of SIRT1 from hepatocytes impairs PPARα signaling, resulting in decreased fatty acid oxidation and leading to the development of hepatic steatosis on a high-fat diet, whereas overexpression of SIRT1 induces expression of PPARα gene targets [84]. SIRT1 induces PPARα signaling through deacetylation of PGC-1α without affecting the formation of the PPARα-PGC-1α complex: in SIRT-knockdown hepatocytes, PGC-1α is still recruited to the PPAR response element (PPRE) of FA oxidation genes [84], but it remains acetylated and thus unable to induce transcription of PPARα gene targets. SIRT1 has been identified as a functional regulator of PGC-1α that induces a metabolic gene transcription program of muscle mitochondrial function and fatty acid oxidation during fasting [82]. Thus, consistent with a switch from glucose to fatty acid oxidation that occurs in nutrient-deprivation states, SIRT1 is required for induction and maintenance of fatty acid oxidation in response to low glucose concentrations. Importantly, the action of SIRT1 on PGC-1 differs among stimuli, deacetylating PGC-1α only in response to nutrient signaling, not to glucagon [82, 85]. In addition, whereas SIRT1 overexpression protects against both the hepatic steatosis and glucose intolerance induced by high-fat feeding [86, 87], oligonucleotide knockdown of hepatic SIRT1 in a rat model of T2DM has been reported to reduce hyperglycemia by normalizing basal hepatic glucose production and increasing hepatic insulin sensitivity, leading to the suggestion that novel SIRT1 inhibitors targeted at the liver might prove beneficial in the treatment of T2DM [88]. (iii) Protein arginine methyltransferase I (PRMT1; Figure 1) coactivates nuclear receptors [89] and has been reported to induce the PGC-1 α function through methylation at several arginine residues in the C-terminal region [89]. Further, it has recently been demonstrated that impaired PRMT1 activity may be implicated in glucose intolerance in nonobese diabetic Goto-Kakizaki rats through disturbed hepatic glucose metabolism and insulin secretion [90]. An overview of the above discussed nutrition-related kinases and enzymes modulating PPAR activity directly or via PGC-1 is given in Figure 1.
5. Conclusions
The beginning of this century saw a rapid expansion of research on the obesity-counteracting potential of PPARs. It is important to consider that dietary changes can drastically interfere with interorgan signaling, and that PPARs can play an important role in this context. Excess fat supply and nutrient deprivation, followed by analysis utilizing comparative nutrigenomics and transcriptional profiling, have rapidly increased our knowledge on nutrient-mediated regulation of PPAR action, and has revealed that nutrient-based regulation of PPAR action is conserved from yeast to man. PPAR action is under tight control of upstream signaling through factors such as SIRT1 and AMPK, having PGC-1 as a target, and this signaling is also influenced by the nutritional state. The further application of potent transcriptomic profiling techniques under different dietary conditions will help to establish which natural ligands might activate each PPAR in a given cellular context. This research will be important since it will allow us to assess how even subtle changes in our daily diet can cause drastic variations in PPAR-mediated metabolic homeostasis. Normalization of lipid and glucose metabolism may be achieved via pharmacological modulation of PPAR activity but the obtained results are not always beneficial (not discussed in this review), which indicates that our knowledge on the intricate network controlling PPAR action is still far from complete. The challenge for future research is to unravel the complex nutrient-influenced metabolic signaling involving the aforementioned factors, in order to be able to “safely” interfere with these processes, as an important step to relieve the burden of obesity and its related disorders.
Acknowledgment
This work was supported in part by the following Grant: MIUR-COFIN 2008, protocol no. 20089SRS2X.
References
- 1.Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97(2):161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- 2.Sher T, Yi H-F, McBride OW, Gonzalez FJ. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry. 1993;32(21):5598–5604. doi: 10.1021/bi00072a015. [DOI] [PubMed] [Google Scholar]
- 3.Auboeuf D, Rieusset J, Fajas L, et al. Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator-activated receptors and liver X receptor-α in humans: no alteration in adipose tissue of obese and NIDDM patients. Diabetes. 1997;46(8):1319–1327. doi: 10.2337/diab.46.8.1319. [DOI] [PubMed] [Google Scholar]
- 4.Skogsberg J, Kannisto K, Roshani L, et al. Characterization of the human peroxisome proliferator activated receptor delta gene and its expression. International Journal of Molecular Medicine. 2000;6(1):73–81. doi: 10.3892/ijmm.6.1.73. [DOI] [PubMed] [Google Scholar]
- 5.Fajas L, Auboeuf D, Raspé E, et al. The organization, promoter analysis, and expression of the human PPARγ gene. Journal of Biological Chemistry. 1997;272(30):18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 6.IJpenberg A, Jeannin E, Wahli W, Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. Journal of Biological Chemistry. 1997;272(32):20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 7.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358(6389):771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalik L, Wahli W. Peroxisome proliferator-activated receptors: three isotypes for a multitude of functions. Current Opinion in Biotechnology. 1999;10(6):564–570. doi: 10.1016/s0958-1669(99)00030-0. [DOI] [PubMed] [Google Scholar]
- 9.Tontonoz P, Hu E, Devine J, Beale EG, Spiegelman BM. PPARγ2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Molecular and Cellular Biology. 1995;15(1):351–357. doi: 10.1128/mcb.15.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolte RT, Wisely GB, Westin S, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ . Nature. 1998;395(6698):137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 11.Schulman IG, Shao G, Heyman RA. Transactivation by retinoid X receptor-peroxisome proliferator-activated receptor γ (PPARγ) heterodimers: intermolecular synergy requires only the PPRAγ hormone-dependent activation function. Molecular and Cellular Biology. 1998;18(6):3483–3494. doi: 10.1128/mcb.18.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinzel T, Lavinsky RM, Mullen T-M, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387(6628):43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 13.Cavailles V, Dauvois S, L’Horset F, et al. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO Journal. 1995;14(15):3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JD, Umesono K, Evans RM. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(15):7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Onate SA, Tsai SY, Tsai M-J, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270(5240):1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 16.Gelman L, Zhou G, Fajas L, Raspé E, Fruchart J-C, Auwerx J. p300 Interacts with the N- and C-terminal part of PPARγ2 in a ligand-independent and -dependent manner, respectively. Journal of Biological Chemistry. 1999;274(12):7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- 17.Heinlein CA, Ting H-J, Yeh S, Chang C. Identification of ARA70 as a ligand-enhanced coactivator for the peroxisome proliferator-activated receptor γ . Journal of Biological Chemistry. 1999;274(23):16147–16152. doi: 10.1074/jbc.274.23.16147. [DOI] [PubMed] [Google Scholar]
- 18.Lu C, Cheng S-Y. Thyroid hormone receptors regulate adipogenesis and carcinogenesis via crosstalk signaling with peroxisome proliferator-activated receptors. Journal of Molecular Endocrinology. 2010;44(3):143–154. doi: 10.1677/JME-09-0107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cioffi F, Senese R, de Lange P, Goglia F, Lanni A, Lombardi A. Uncoupling protection: a complex journey to function discovery. BioFactors. 2009;35(5):417–428. doi: 10.1002/biof.54. [DOI] [PubMed] [Google Scholar]
- 20.de Lange P, Feola A, Ragni M, et al. Differential 3,5,3′-triiodothyronine-mediated regulation of uncoupling protein 3 transcription: role of fatty acids. Endocrinology. 2007;148(8):4064–4072. doi: 10.1210/en.2007-0206. [DOI] [PubMed] [Google Scholar]
- 21.Solanes G, Pedraza N, Iglesias R, Giralt M, Villarroya F. Functional relationship between MyoD and peroxisome proliferator-activated receptor-dependent regulatory pathways in the control of the human uncoupling protein-3 gene transcription. Molecular Endocrinology. 2003;17(10):1944–1958. doi: 10.1210/me.2002-0395. [DOI] [PubMed] [Google Scholar]
- 22.He W, Barak Y, Hevener A, et al. Adipose-specific peroxisome proliferator-activated receptor γ knockout causes insulin resistance in fat and liver but not in muscle. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger J, Moller DE. The mechanisms of action of PPARs. Annual Review of Medicine. 2002;53(1):409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- 24.Friedman J. Fat in all the wrong places. Nature. 2002;415(6869):268–269. doi: 10.1038/415268a. [DOI] [PubMed] [Google Scholar]
- 25.Ye J-M, Doyle PJ, Iglesias MA, Watson DG, Cooney GJ, Kraegen EW. Peroxisome proliferator-activated receptor (PPAR)-α activation lowers muscle lipids and improves insulin sensitivity in high fat-fed rats. Comparison with PPAR-γ activation. Diabetes. 2001;50(2):411–417. doi: 10.2337/diabetes.50.2.411. [DOI] [PubMed] [Google Scholar]
- 26.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142(10):4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 27.de Lange P, Lombardi A, Silvestri E, Goglia F, Lanni A, Moreno M. Peroxisome proliferator-activated receptor delta: a conserved director of lipid homeostasis through regulation of the oxidative capacity of muscle. PPAR Research. 2008;2008:7 pages. doi: 10.1155/2008/172676. Article ID 172676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abbot BD. Review of the expression of peroxisome proliferators-activated receptors alpha (PPARalpha), beta (PPARbeta), and gamma (PPARgamma) in rodent and human development. Reproductive Toxicology. 2009;75(1):72–77. doi: 10.1016/j.reprotox.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gustafsson MCU, Knight D, Palmer CNA. Ligand modulated antagonism of PPARγ by genomic and non-genomic actions of PPARδ . PLoS ONE. 2009;4(9) doi: 10.1371/journal.pone.0007046. Article ID e7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ide T, Shimano H, Yoshikawa T, et al. Cross-talk between peroxisome proliferator-activated receptor (PPAR) α and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. II. LXRs suppress lipid degradation gene promoters through inhibition of PPAR signaling. Molecular Endocrinology. 2003;17(7):1255–1267. doi: 10.1210/me.2002-0191. [DOI] [PubMed] [Google Scholar]
- 32.Yoshikawa T, Ide T, Shimano H, et al. Cross-talk between peroxisome proliferator-activated receptor (PPAR) α and liver X receptor (LXR) in nutritional regulation of fatty acid metabolism. I. PPARS suppress sterol regulatory element binding protein-1c promoter through inhibition of LXR signaling. Molecular Endocrinology. 2003;17(7):1240–1254. doi: 10.1210/me.2002-0190. [DOI] [PubMed] [Google Scholar]
- 33.Matsusue K, Miyoshi A, Yamano S, Gonzalez FJ. Ligand-activated PPARβ efficiently represses the induction of LXR-dependent promoter activity through competition with RXR. Molecular and Cellular Endocrinology. 2006;256(1-2):23–33. doi: 10.1016/j.mce.2006.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu R, Madison LD, Lin Y, et al. Thyroid hormone (T3) inhibits ciprofibrate-induced transcription of genes encoding β-oxidation enzymes: cross talk between peroxisome proliferator and T3 signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(25):11593–11597. doi: 10.1073/pnas.92.25.11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ying H, Araki O, Furuya F, Kato Y, Cheng S-Y. Impaired adipogenesis caused by a mutated thyroid hormone α1 receptor. Molecular and Cellular Biology. 2007;27(6):2359–2371. doi: 10.1128/MCB.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krey G, Braissant O, L’Horset F, et al. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Molecular Endocrinology. 1997;11(6):779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 37.Itoh T, Fairall L, Amin K, et al. Structural basis for the activation of PPARγ by oxidized fatty acids. Nature Structural and Molecular Biology. 2008;15(9):924–931. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu HE, Lambert MH, Montana VG, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Molecular Cell. 1999;3(3):397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 39.Cavalieri D, Calura E, Romualdi C, et al. Filling gaps in PPAR-alpha signaling through comparative nutrigenomics analysis. BMC Genomics. 2009;10(1):596–612. doi: 10.1186/1471-2164-10-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanderson LM, de Groot PJ, Hooiveld GJEJ, et al. Effect of synthetic dietary triglycerides: a novel research paradigm for nutrigenomics. PLoS ONE. 2008;27(2) doi: 10.1371/journal.pone.0001681. Article ID e1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surapureddi S, Yu S, Bu H, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor α-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11836–11841. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen R, Grøntved L, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Molecular and Cellular Biology. 2006;26(15):5698–5714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoon JC, Puigserver P, Chen G, et al. Control of hepatic gluconeogenesis through the transcriptional coaotivator PGC-1. Nature. 2001;413(6852):131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 44.Sanderson LM, Degenhardt T, Koppen A, et al. Peroxisome proliferator-activated receptor β/δ (PPARβ/δ) but not PPARα serves as a plasma free fatty acid sensor in liver. Molecular and Cellular Biology. 2009;29(23):6257–6267. doi: 10.1128/MCB.00370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Lange P, Farina P, Moreno M, et al. Sequential changes in the signal transduction responses of skeletal muscle following food deprivation. The FASEB Journal. 2006;20(14):2579–2581. doi: 10.1096/fj.06-6025fje. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y-X, Lee C-H, Tiep S, et al. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113(2):159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 47.Stavinoha MA, Ray Spellicy JW, Essop MF, et al. Evidence for mitochondrial thioesterase 1 as a peroxisome proliferator-activated receptor-α-regulated gene in cardiac and skeletal muscle. American Journal of Physiology. 2004;287(5):E888–E895. doi: 10.1152/ajpendo.00190.2004. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y-X, Zhang C-L, Yu RT, et al. Regulation of muscle fiber type and running endurance by PPARδ . PLoS Biology. 2004;2(10, article e294) doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka T, Yamamoto J, Iwasaki S, et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(26):15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watt MJ, Southgate RJ, Holmes AG, Febbraio MA. Suppression of plasma free fatty acids upregulates peroxisome proliferator-activated receptor (PPAR) α and δ and PPAR coactivator 1α in human skeletal muscle, but not lipid regulatory genes. Journal of Molecular Endocrinology. 2004;33(2):533–544. doi: 10.1677/jme.1.01499. [DOI] [PubMed] [Google Scholar]
- 51.Tsintzas K, Jewell K, Kamran M, et al. Differential regulation of metabolic genes in skeletal muscle during starvation and refeeding in humans. Journal of Physiology. 2006;575(1):291–303. doi: 10.1113/jphysiol.2006.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kubota N, Terauchi Y, Miki H, et al. PPARγ mediates high-fat diet-induced adipocyte hypertrophy and insulin resistance. Molecular Cell. 1999;4(4):597–609. doi: 10.1016/s1097-2765(00)80210-5. [DOI] [PubMed] [Google Scholar]
- 53.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ . Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koh Y-K, Lee M-Y, Kim J-W, et al. Lipin1 is a key factor for the maturation and maintenance of adipocytes in the regulatory network with CCAAT/enhancer-binding protein α and peroxisome proliferator-activated receptor γ2. Journal of Biological Chemistry. 2008;283(50):34896–34906. doi: 10.1074/jbc.M804007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grimaldi PA. The roles of PPARs in adipocyte differentiation. Progress in Lipid Research. 2001;40(4):269–281. doi: 10.1016/s0163-7827(01)00005-4. [DOI] [PubMed] [Google Scholar]
- 56.Sanderson LM, Boekschoten MV, Desvergne B, Müller M, Kersten S. Transcriptional profiling reveals divergent roles of PPARα and PPARβ/δ in regulation of gene expression in mouse liver. Physiological Genomics. 2010;41(1):42–52. doi: 10.1152/physiolgenomics.00127.2009. [DOI] [PubMed] [Google Scholar]
- 57.Shalev A, Siegrist-Kaiser CA, Yen PM, et al. The peroxisome proliferator-activated receptor α is a phosphoprotein: regulation by insulin. Endocrinology. 1996;137(10):4499–4502. doi: 10.1210/endo.137.10.8828512. [DOI] [PubMed] [Google Scholar]
- 58.Fuentes P, Acuña MJ, Cifuentes M, Rojas CV. The anti-adipogenic effect of angiotensin II on human preadipose cells involves ERK1,2 activation and PPARG phosphorylation. Journal of Endocrinology. 2010;206(1):75–83. doi: 10.1677/JOE-10-0049. [DOI] [PubMed] [Google Scholar]
- 59.Yun CL, Zierath JR. AMP-activated protein kinase signaling in metabolic regulation. Journal of Clinical Investigation. 2006;116(7):1776–1783. doi: 10.1172/JCI29044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leff T. AMP-activated protein kinase regulates gene expression by direct phosphorylation of nuclear proteins. Biochemical Society Transactions. 2003;31(1):224–226. doi: 10.1042/bst0310224. [DOI] [PubMed] [Google Scholar]
- 61.Sugden MC, Caton PW, Holness MJ. PPAR control: it's SIRTainly as easy as PGC. Journal of Endocrinology. 2010;204(2):93–104. doi: 10.1677/JOE-09-0359. [DOI] [PubMed] [Google Scholar]
- 62.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): transcriptional coactivator and metabolic regulator. Endocrine Reviews. 2003;24(1):78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 63.Nagai Y, Yonemitsu S, Erion DM, et al. The role of peroxisome proliferator-activated receptor γ coactivator-1 β in the pathogenesis of fructose-induced insulin resistance. Cell Metabolism. 2009;9(3):252–264. doi: 10.1016/j.cmet.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J, Sanders Williams R, Kelly DP. Bcl3 interacts cooperatively with peroxisome proliferator-activated receptor gamma (PPARγ) coactivator 1α to coactivate nuclear receptors estrogen-related receptor α and PPAR. Molecular and Cellular Biology. 2009;29(15):4091–4102. doi: 10.1128/MCB.01669-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamagata K, Furuta H, Oda N, et al. Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384(6608):458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 66.Nakae J, Biggs WH, III, Kitamura T, et al. Regulation of insulin action and pancreatic β-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nature Genetics. 2002;32(2):245–253. doi: 10.1038/ng890. [DOI] [PubMed] [Google Scholar]
- 67.Puigserver P, Rhee J, Donovan J, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423(6939):550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 68.Gupta RK, Vatamaniuk MZ, Lee CS, et al. The MODY1 gene HNF-4α regulates selected genes involved in insulin secretion. Journal of Clinical Investigation. 2005;115(4):1006–1015. doi: 10.1172/JCI200522365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ek J, Andersen G, Urhammer SA, et al. Mutation analysis of peroxisome proliferator-activated receptor-γ coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to Type II diabetes mellitus. Diabetologia. 2001;44(12):2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 70.Hara K, Tobe K, Okada T, et al. A genetic variation in the PGC-1 gene could confer insulin resistance and susceptibility to type II diabetes. Diabetologia. 2002;45(5):740–743. doi: 10.1007/s00125-002-0803-z. [DOI] [PubMed] [Google Scholar]
- 71.Mortensen OH, Frandsen L, Schjerling P, Nishimura E, Grunnet N. PGC-1α and PGC-1β have both similar and distinct effects on myofiber switching toward an oxidative phenotype. American Journal of Physiology. 2006;291(4):E807–E816. doi: 10.1152/ajpendo.00591.2005. [DOI] [PubMed] [Google Scholar]
- 72.Leone TC, Lehman JJ, Finck BN, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biology. 2005;3(4, article e101) doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roux PP, Blenis J. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiology and Molecular Biology Reviews. 2004;68(2):320–344. doi: 10.1128/MMBR.68.2.320-344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Puigserver P, Rhee J, Lin J, et al. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Molecular Cell. 2001;8(5):971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 75.Irrcher I, Ljubicic V, Hood DA. Interactions between ROS and AMP kinase activity in the regulation of PGC-1α transcription in skeletal muscle cells. American Journal of Physiology. 2009;296(1):C116–C123. doi: 10.1152/ajpcell.00267.2007. [DOI] [PubMed] [Google Scholar]
- 76.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature. 2007;447(7147):1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 77.Narkar VA, Downes M, Yu RT, et al. AMPK and PPARδ agonists are exercise mimetics. Cell. 2008;134(3):405–415. doi: 10.1016/j.cell.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kitz Krämer D, Al-Khalili L, Perrini S, et al. Direct activation of glucose transport in primary human myotubes after activation of peroxisome proliferator—activated receptor δ . Diabetes. 2005;54(4):1157–1163. doi: 10.2337/diabetes.54.4.1157. [DOI] [PubMed] [Google Scholar]
- 79.Krämer DK, Al-Khalili L, Guigas B, Leng Y, Garcia-Roves PM, Krook A. Role of AMP kinase and PPARδ in the regulation of lipid and glucose metabolism in human skeletal muscle. Journal of Biological Chemistry. 2007;282(27):19313–19320. doi: 10.1074/jbc.M702329200. [DOI] [PubMed] [Google Scholar]
- 80.Lerin C, Rodgers JT, Kalume DE, Kim S-H, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α . Cell Metabolism. 2006;3(6):429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 81.Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α . EMBO Journal. 2007;26(7):1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 83.Kelly TJ, Lerin C, Haas W, Gygi SP, Puigserver P. GCN5-mediated transcriptional control of the metabolic coactivator PGC-1β through lysine acetylation. Journal of Biological Chemistry. 2009;284(30):19945–19952. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metabolism. 2009;9(4):327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Letters. 2008;582(1):46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Banks AS, Kon N, Knight C, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metabolism. 2008;8(4):333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschöp MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(28):9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Erion DM, Yonemitsu S, Nie Y, et al. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(27):11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes and Development. 2005;19(12):1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iwasaki H. Impaired PRMT1 activity in the liver and pancreas of type 2 diabetic Goto-Kakizaki rats. Life Sciences. 2009;85(3-4):161–166. doi: 10.1016/j.lfs.2009.05.007. [DOI] [PubMed] [Google Scholar]