Abstract
The AsiA protein is a T4 bacteriophage early gene product that regulates transcription of host and viral genes. Monomeric AsiA binds tightly to the σ70 subunit of Escherichia coli RNA polymerase, thereby inhibiting transcription from bacterial promoters and phage early promoters and co-activating transcription from phage middle promoters. Results of structural studies have identified amino acids at the protomer-protomer interface in dimeric AsiA and at the monomeric AsiA-σ70 interface and demonstrated substantial overlap in the sets of residues that comprise each. Here we evaluate the contributions of individual interfacial amino acid side chains to protomer-protomer affinity in AsiA homodimers, to monomeric AsiA affinity for σ70, and to AsiA function in transcription. Sedimentation equilibrium, dynamic light scattering, electrophoretic mobility shift and transcription activity measurements were used to assess affinity and function of site-specific AsiA mutants. Alanine substitutions for solvent-inaccessible residues positioned centrally in the protomer-protomer interface of the AsiA homodimer – V14, I17, and I40 – resulted in the largest changes in free energy of dimer association, whereas alanine substitutions at other interfacial positions had little effect. These residues also contribute significantly to AsiA-dependent regulation of RNA polymerase activity, as do additional residues positioned at the periphery of the interface (K20 and F21). Notably, the relative contributions of a given amino acid side chain to RNA polymerase inhibition and activation (MotA-independent) by AsiA are very similar in most cases. The mainstay for intermolecular affinity and AsiA function appears to be I17. Our results define the core interfacial residues of AsiA, establish roles for many of the interfacial amino acids, are in agreement with the tenets underlying protein-protein interactions and interfaces, and will be beneficial for a general, comprehensive understanding of the mechanistic underpinnings of bacterial RNA polymerase regulation.
Following infection of Escherichia coli by bacteriophage T4, phage encoded proteins bind tightly to the host RNA polymerase to direct and redirect transcription (1). One of these proteins, the AsiA protein, product of the asiA gene (2), associates tightly with the σ70 subunit of the polymerase holoenzyme (α2ββ′σ) (1, 3–5) and prevents recognition of the −10/−35 class promoters (2, 3, 6), which includes most host and early phage promoters (7, 8). The association of AsiA with σ70 also activates transcription of phage middle promoters, in concert with another phage early gene product, the MotA protein, which interacts with σ70 and recognizes the “Mot box” sequence centered near the −30 region of middle phage promoters (6, 9–12). Thus, AsiA functions as a molecular switch for promoter selection.
Previous results established a 1:1 stoichiometry for the AsiA-σ70 interaction (13) and the ability of AsiA to self-associate to form dimers (14, 15). Structural and functional studies indicate that the same surface of AsiA that interacts with σ70 is also responsible for AsiA dimerization (16–21). This interaction surface includes the exterior of a hydrophobic cleft on each protomer complementary to the hydrophobic C-terminal end of the N-terminal helix of the opposing protomer. Sidechains in the interior of the cleft form the stable hydrophobic core (22).
An interaction surface exposing substantial hydrophobic area to solvent is generally destabilizing yet introduces the potential for high affinity when buried by complex formation, as is often observed for obligate oligomers (23–27). Conversely, removing solvent exposed hydrophobic groups from the interaction surface tends to stabilize the monomer and decreases potential for high affinity dimer formation. Self-association of AsiA – via the same surface used for interaction with σ70 – protects the active hydrophobic surface from non-specific interactions with other proteins and perhaps precipitation, and potentially provides a large degree of stabilization. Self-association also buffers the concentration of the active monomer and provides a stable, protected reservoir of it. Because monomeric AsiA is a key regulator orchestrating the switch from early to middle viral transcription, its ability to self-associate (and thus become inactive) could be biologically significant when its local concentration is high.
Here we determine the contributions of amino acids of AsiA that, based on structural considerations, participate in intermolecular interactions in the AsiA homodimer and in the complex with σ70, to the AsiA self-association affinity, affinity for σ70, transcription inhibition and transcription activation. Ultimately, in order to understand how the bacteriophage use AsiA effectively to promote phage propagation, it will be necessary to understand the properties of AsiA (its structure, the amino acids important for activity and self-association) and how they dictate its ability to regulate the RNA polymerase.
EXPERIMENTAL PROCEDURES
General
Chemicals and media for protein production and purification were from reliable commercial sources and used without further purification. Molecular graphics were rendered with MOLMOL (28) and PyMOL (29) (http://www.pymol.org). PyMOL was used also used to generate computational mutants.
AsiA and Mutant AsiA Production
The cloning, overexpression, isotopic labeling and purification of AsiA have been described previously (14, 22). Mutant AsiA proteins were produced by mutating the codons for residues M1, E10, V14, I17, L18, K20, F21, V27, E39, I40 and V42 to alanine (a double mutant, I17A/I40A, was also produced) using standard PCR based methods (30–32). Oligonucleotide primer pairs for each mutation were designed and synthesized (MWG Biotech Inc., High Point, NC). The wild type asiA gene in pBluescript II SK (Stratagene, La Jolla, CA) was used as the PCR template. Mutated codons were incorporated into the asiA gene by PCR amplification using primers specific for that mutation. Purified PCR products were cloned into the expression vector pET-24b (Novagen, Madison, WI), and the sequences were confirmed by DNA sequencing (Iowa State University DNA Sequencing and Synthesis Facility, Ames, IA). For each mutant, one clone was selected and transformed into E. coli strain BL21(DE3) for expression of the full-length mutant asiA gene. A final purification step for AsiA and all mutant AsiA proteins was performed using reversed phase HPLC with an acetonitrile gradient containing 0.1% trifluoroacetic acid. Solvents were removed by lyophilization and dialysis. Protein concentrations were determined by quantitative amino acid analysis (Molecular Analysis Facility, University of Iowa).
RNA Polymerase Proteins
The RNA polymerase core, holoenzyme, σ70 subunit and the C-terminal truncated form (denoted σ565, lacking region 4.2 and remaining C-terminal residues) were produced as described (20). For His6-σ70 production, the E. coli σ70 gene was cloned into the pET-19 expression vector (Novagen, Madison, WI) and transformed into E. coli BL21(DE3) for expression. Cells from a 1 L growth in minimal medium (M9) were resuspended in lysis buffer (20 mM Tris-HCl, 500 mM NaCl, pH 7.9), lysed with lysozyme (150 µg/mL) and then sonicated. The sonication pellets were resuspended in 30 mL of denaturation buffer (20 mM Tris-HCl, 500 mM NaCl, and 6 M Urea, pH 7.9) and allowed to stir over night at 4 °C. An affinity column of Ni-NTA resin (GE Health Sciences, Piscataway, NJ) was equilibrated with denaturation buffer. The denatured His-σ70 was diluted to 150 mL with denaturation buffer and loaded onto the column. After washing with lysis buffer containing 20 mM imidazole, His-σ70 was refolded on the column by washing with a gradient of 6-0 M urea in 20 mM Tris-HCl, 500 mM NaCl, pH 7.9. The His-σ70 was eluted with a gradient of 0–500 mM imidazole in 20 mM Tris-HCl, 500 mM NaCl, pH 7.9. Fractions were collected and the purity of His-σ70 confirmed by SDS PAGE.
Circular Dichroism and Secondary Structure
Circular dichroism (CD) spectra of AsiA and the mutant AsiA proteins were acquired and analyzed to assess their respective secondary structures. The measurements were performed with a Jasco J-715 spectrapolarimeter using a 0.1 cm path length cell. Temperature was maintained at 25 °C with a Peltier device. The samples consisted of 10 µM AsiA or mutant AsiA in 10 mM potassium phosphate, 10 mM KCl, pH 7.0. Scans from 190 – 250 nm were recorded in 1 nm increments. Calculations of secondary structure contributions based on the CD spectra were performed using the CDSSTR algorithm (33–36) and the DICHROWEB server (37–39).
NMR Spectroscopy
NMR spectra were recorded with Varian INOVA spectrometers operating at 600 MHz (1H). Sample temperatures were maintained at 25 °C. Gradient sensitivity-enhanced 1H, 15N-HSQC spectra were collected as described previously (14, 40–43). Samples contained ~0.5 mM AsiA or mutant AsiA in 10 mM potassium phosphate, 10 mM potassium chloride, pH 6.5. Felix (Accelrys, San Diego, CA) was used for data processing and analysis. Na+DSS− in D2O (0.00 ppm) was used for referencing the 1H chemical shifts. The 15N and 13C chemical shifts were referenced indirectly (44).
Dynamic Light Scattering
Dynamic light scattering (DLS) measurements were performed with a Protein Solutions DynaPro99 instrument equipped with a Flex99 correlator. Scattered light was detected at a 90° angle from the incident light (λ = 825 nm). The initial delay time (τ) was 0.48 µs. The sample temperature was maintained at 20 °C. Each sample contained ~188 µM (2 mg/mL) AsiA or AsiA mutant protein in 10 mM potassium phosphate, pH 7.0, and from 50–200 mM potassium chloride (see legend to Table 1). Each sample was filtered through a 0.02 µm Whatman filter immediately before use. Data from 18–30 10 s acquisitions were averaged. Monomodal distributions were indicated for all samples (baseline ≤ 1.001, see Table 1). Translational diffusion coefficients (DT) were used to calculate the hydrodynamic radii (RH), assuming Brownian motion and spherical particles of standard density, using the Stokes-Einstein relationship:
| (1) |
where T is the temperature (°K), kb is the Boltzman constant, and η is the solution viscosity (set to 1.0 g/cm•sec for samples in aqueous solution). Molecular masses were estimated based on a standard curve of measured hydrodynamic radius versus mass for a set of globular proteins. All calculations were performed using the manufacturer’s software, with the exception of normal error propagation calculations.
Table 1.
Dynamic light scattering results for AsiA and mutant AsiA proteinsa
| DT (cm2/s)b | RH (nm)c | Pd (nm)d | % Pde | mass (kDa)f | baselineg | SSh | % monomeri | |
|---|---|---|---|---|---|---|---|---|
| WTj | 9.24 ± 0.07 × 10−7 | 2.27 ± 0.02 | 0.33 ± 0.17 | 14 ± 7 | 23.0 ± 0.4 | 1.001 ± 0.002 | 7 ± 2 | 3.0 |
| M1Aj | 9.41 ± 0.02 × 10−7 | 2.23 ± 0.04 | 0.37 ± 0.14 | 16 ± 6 | 22.1 ± 0.9 | 1.001 ± 0.002 | 8 ± 2 | 2.6 |
| V14Aj | 9.61 ± 0.03 × 10−7 | 2.19 ± 0.06 | 0.48 ± 0.21 | 22 ± 10 | 21.0 ± 1.3 | 1.000 ± 0.001 | 18 ± 5 | 5.5 |
| I17Aj | 9.30 ± 0.04 × 10−7 | 2.26 ± 0.08 | 0.43 ± 0.19 | 19 ± 8 | 22.7 ± 1.9 | 1.001 ± 0.001 | 10 ± 2 | 10.8 |
| K20Aj | 7.96 ± 0.11 × 10−7 | 2.64 ± 0.04 | 0.46 ± 0.20 | 17 ± 8 | 32.6 ± 1.1 | 1.001 ± 0.001 | 4 ± 1 | 3.5 |
| K20Ak | 8.68 ± 0.02 × 10−7 | 2.42 ± 0.05 | 0.45 ± 0.18 | 19 ± 7 | 26.7 ± 1.2 | 1.001 ± 0.001 | 9 ± 2 | 3.5 |
| K20Al | 9.14 ± 0.02 × 10−7 | 2.30 ± 0.06 | 0.64 ± 0.16 | 28 ± 7 | 23.6 ± 1.3 | 1.001 ± 0.001 | 23 ± 5 | 3.5 |
| F21Ak | 9.28 ± 0.02 × 10−7 | 2.27 ± 0.05 | 0.44 ± 0.21 | 19 ± 9 | 22.8 ± 1.2 | 1.001 ± 0.001 | 17 ± 4 | 2.3 |
| E39Ak | 9.03 ± 0.02 × 10−7 | 2.33 ± 0.05 | 0.40 ± 0.17 | 17 ± 7 | 24.3 ± 1.3 | 1.001 ± 0.001 | 9 ± 3 | 2.2 |
| I40Aj | 9.41 ± 0.03 × 10−7 | 2.23 ± 0.06 | 0.41 ± 0.20 | 18 ± 9 | 22.1 ± 1.4 | 1.001 ± 0.001 | 6 ± 3 | 8.1 |
| I17A/I40Aj | 10.98 ± 0.02 × 10−7 | 1.91 ± 0.03 | 0.48 ± 0.18 | 25 ± 9 | 15.4 ± 0.5 | 1.001 ± 0.001 | 15 ± 4 | 72.0 |
Errors are standard deviations of 18–30 measurements. DLS measurements were not performed on the E10A mutant.
Translational diffusion coefficient.
Mean hydrodynamic radius.
Polydispersity (standard deviation of the Gaussian particle size distribution about the mean).
Percent polydispersity (Pd/RH).
Mass calculated from RH as described in the text.
Deviation of correlation from ideality (unity).
Sum of squares of the differences between experimental and fitted correlation functions.
Percentage of total protein existing as a monomer under the conditions of the DLS measurements, calculated from Kd values determined by sedimentation equilibrium (Table 2).
Buffer was 10 mM potassium phosphate, 50 mM KCl, pH 7.0.
Buffer was 10 mM potassium phosphate, 100 mM KCl, pH 7.0.
Buffer was 10 mM potassium phosphate, 200 mM KCl, pH7.0.
Sedimentation Equilibrium Experiments
Sedimentation equilibrium experiments were performed with Beckman Optima XL-I and XL-A analytical ultracentrifuges using four-position AN-60-Ti rotors. The protein samples (in 10 mM potassium phosphate, 50 mM potassium chloride, pH 7.0) were centrifuged in six-sector epon/charcoal centerpieces. For analysis at 280 nm, three different concentrations (26 µM, 65 µM, and 104 µM, corresponding to 0.1, 0.25, and 0.4 absorbance units, respectively) of each AsiA sample were analyzed and the experiments were performed at 4°C and at a speed of 40,000 rpm as described previously (14). For analysis at 230 nm, three different concentrations (3 µM, 7 µM, and 12 µM, corresponding to 0.1, 0.25, and 0.4 absorbance units, respectively) of each AsiA sample were analyzed and the experiments were performed at 20 °C and at multiple speeds from 24,000 to 40,000 rpm. Scans were collected in a radial increment mode with 0.001 increments and 4 point averaging. Data points with absorbances higher than 0.9 were discarded. Data were analyzed with the UltraScan software versions 7.0 and 9.0 (45). The partial specific volume for each sample was calculated as implemented in the Ultrascan software (46, 47). For each species, global fitting methods were employed to estimate parameters using the sedimentation profiles for all protein concentrations at all rotor speeds (48). Standard free energies of association were calculated in the usual way:
| (2) |
where R is the gas constant (8.314 J/mol·K), T is the sample temperature (K), and Ka is the measured association constant. Monte Carlo simulations (10,000) were used to estimate confidence intervals (49).
Non-denaturing PAGE and Electrophoretic Mobility Shift Assays
Non-denaturing PAGE was used to assess the purity of AsiA and mutant AsiA preparations and qualitatively the affinity between σ70 and the AsiA proteins. Samples of AsiA (0.3 nmols, 0.6 nmols for the shift assays) in buffer (10 mM potassium phosphate, 10 mM potassium chloride, 5 mM DTT, pH 7.0) were subjected to electrophoresis for 3 hours at 150 volts in a 12% non-denaturing polyacrylamide gel. For the electrophoretic mobility shift assays, σ70 (20 µM) was incubated with increasing concentrations of AsiA or mutant AsiA (0–80 µM) for approximately 10 minutes in the same buffer at room temperature. The samples were subsequently subjected to the same electrophoretic conditions as above. The proteins were visualized by coomassie blue staining.
In vitro Transcription Assays
The ability of AsiA to inhibit transcription from −10/−35 early phage promoter T7A1 and, in the context of an RNA polymerase holoenzyme with a mutant σ70 truncated at residue 565 (σ565), to activate transcription from the T4 bacteriophage PrIIB2 middle promoter without MotA (henceforward denoted MotA-independent activation) was measured using in vitro abortive transcription assays (20). Briefly, reactions contained, in 10 µL of transcription buffer (50 mM Tris-HCI, pH 7.9, 10 mM MgCl2, 40 mM KCl, 1 mM β-mercaptoethanol), 20 nM of RNAP core enzyme combined with the same amount of sigma proteins and 80 nM-2.4 µM of wild type or mutant AsiA. Reactions were incubated for 10 minutes at 37 °C, followed by the addition of 10 nM of DNA fragments containing promoters (T7A1 or PrIIB2), 0.1 mM initiating dinucleotide CpA and 3 µCi (3000 Ci/mmol) of α–[32P]UTP or 0.1 mM GpA and α–[32P]GTP for T7A1 or PrIIB2 transcription reactions, respectively. Reactions proceeded for 10 minutes at 37 °C and were terminated by the addition of an equal volume of loading buffer containing 8 M urea. Reaction products were resolved by electrophoresis in denaturing (8 M urea) 20% (19:1) polyacrylamide gel, visualized by autoradiography and quantified by phosphorimagery (PhosphorImager, Molecular Dynamics).
Assays of MotA-dependent activation were also performed. Reactions contained, in 10 µL of transcription buffer (50 mM tris HCl (pH 7.9), 10 mM MgCl2, 40 mM KCl, 2 mM β-mercaptoethanol), 20 nM of RNAP holoenzyme, 100 nM of wild-type or mutant AsiA, 5 nM of MotA, and 10 nM of DNA fragment containing T4 promoter IIB2. Reactions were incubated for ten minutes at 37 °C, followed by the addition of 0.25 mM initiating dinucleotide GpA, 0.01 mM GTP, and 3 µCi (3000 Ci/mmol) of [α-32P]GTP. Reactions proceeded another ten minutes at 37 °C, and were terminated by the addition of an equal volume of loading buffer containing 8 M urea. Reaction products were resolved and quantified as described above.
Accessible Surface Area Calculations
Solvent accessible surface areas were calculated using the method of Lee and Richards (50) as implemented in the program NACCESS (51) using the default probe size (1.4 Angstroms). Areas for dimeric AsiA were calculated for each of the 25 deposited structures of the NMR ensemble in the PDB (1JR5) and averaged. Areas for each protomer in each of the structures were then calculated with the other protomer removed and averaged. The differences between the dimer and protomer areas were then calculated. Additionally, starting from the lowest energy structure of the NMR ensemble, we made two mutations computationally – F21A and M1A – and calculated the accessible surface areas for each of the corresponding mutant AsiA dimers.
RESULTS
AsiA Mutants
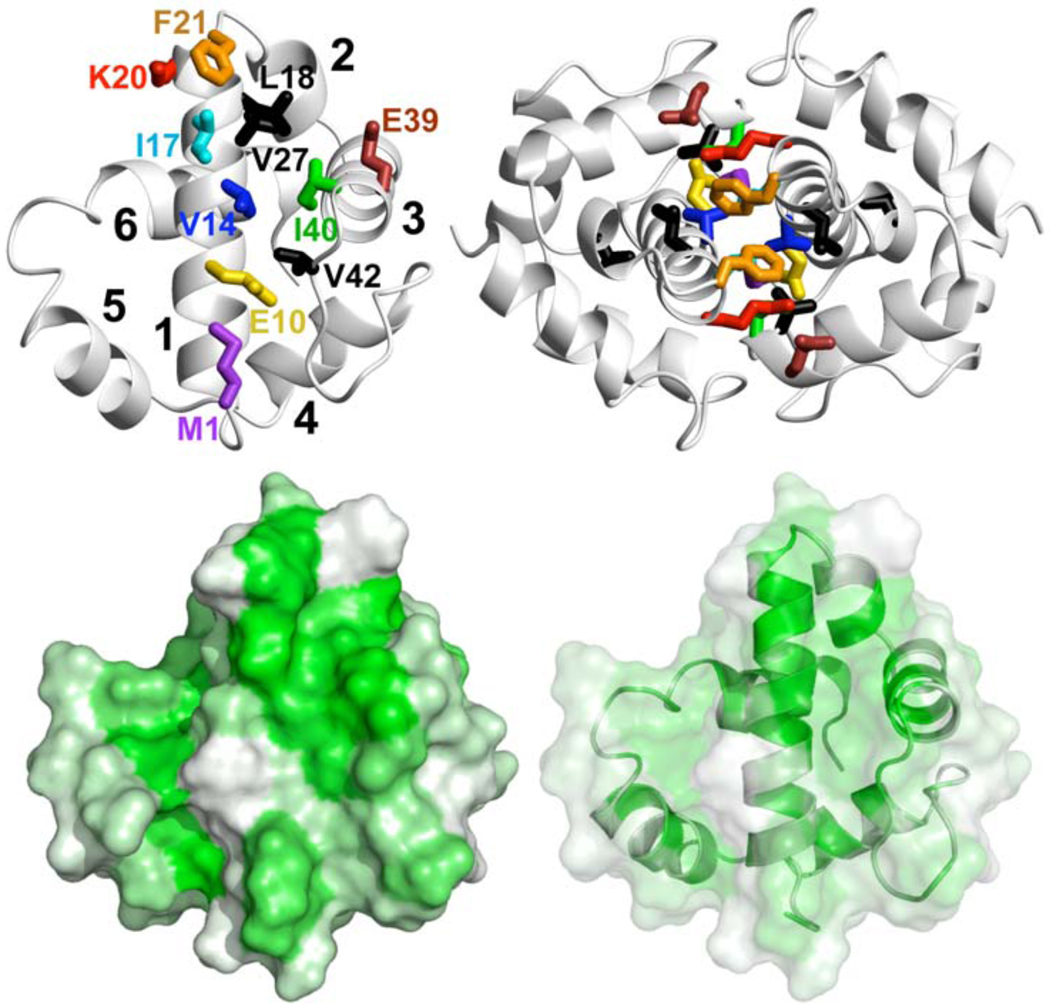
The high resolution solution structure of dimeric AsiA (PDB 1JR5)2 reveals amino acid side chains potentially important for protomer-protomer affinity (22) (Figure 1). These comprise the largely hydrophobic dimer interface and include the side chains of M1, V14, I17, L18, F21, I40, and V42, which appear to provide most of the hydrophobic character of the interaction surface (Figure 1) and most of the interprotomer contacts (22). The structure also suggests potential contributions to protomer-protomer affinity by charged side chains of certain residues such as E10, K20, and E39, the latter two forming a charge pair putatively capping the hydrophobic interface. Side chains of residues at or near the dimer interface that line the hydrophobic cleft formed by helices 1–3 could also ostensibly contribute to protomer-protomer affinity. Such residues, including L18, V27 and V42, are implicated by results of hydrogen-deuterium exchange studies as principal contributors to local and global stabilities (22). Finally, previous results demonstrate that many of the amino acids comprising the homodimer interface also participate in the heterodimer interface with σ70 (16, 18), and mutating some of these residues results in loss of function (19). In order to assess the contributions of side chains of AsiA residues at the homodimer interface to the affinity between the protomers in dimeric AsiA, to the interaction of AsiA with σ70, and to AsiA activity and stability, we mutated many of the putatively important AsiA residues (Figure 1) to Ala.
FIGURE 1.
The dimer interface of AsiA. (Top) Site directed amino acid substitutions in AsiA. Eleven Ala mutants of AsiA were produced to evaluate the functions of the replaced side chains. These side chains are shown on ribbon models of monomeric AsiA (left) and dimeric AsiA (right). Most of these are at the homodimer interface and include M1 (purple), E10 (gold), V14 (blue), I17 (cyan), K20 (red), F21 (orange), E39 (brown), and I40 (green). The double mutant I17A/I40A (magenta) was also produced. Unstable proteins resulted from mutation of the amino acids with side chains colored black (L18, V27, V42) to Ala. The six helices of AsiA are numbered from the N-terminus. (Bottom) The hydrophobic character of the interface region of AsiA. The hydropathies of the individual residues are plotted on the Connolly surface of one protomer of dimeric AsiA, with green representing the most hydrophobic and white the least. The large hydrophobic cleft in AsiA is apparent, formed by the side chains of residues from helices 1–3 including V14, I17, L18, V27, F21, I40, and V42. The dimer interface is constructed by aligning the C-terminal end of helix 1 of each protomer with the hydrophobic cleft of the other.
Production of the AsiA Mutants
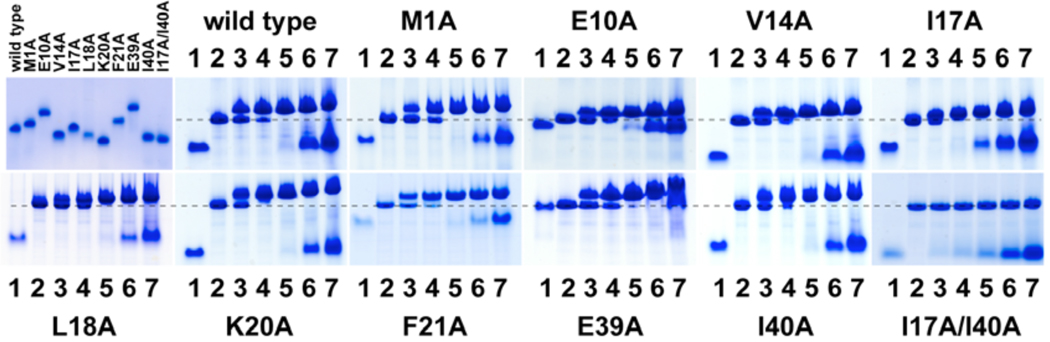
Most of the mutant AsiA proteins, including the I17A/I40A double mutant, were readily produced and purified to homogeneity (Figure 2). The exceptions were the L18A, V27A, and V42A mutants, which formed inclusion bodies when overexpressed. A fraction of the L18A and V42A proteins could be solubilized from the inclusion bodies, refolded and purified with low yield. The L18A mutant protein precipitated steadily with time, and the stabilities of the L18A, V27A and V42A mutants were deemed too equivocal for detailed studies, the exception being a single study of σ70 binding to the L18A mutant. Thus, a total of nine AsiA mutants were used for the detailed structural and functional studies. The basis for apparent instability of mutants that were excluded from further analysis is elaborated below.
FIGURE 2.
AsiA and mutant AsiA binding to σ70. Electrophoretic mobility shift assays using non-denaturing PAGE were used to evaluate the apparent affinities of AsiA and the mutant AsiA proteins for σ70 by monitoring formation of the AsiA-σ70 complexes. Non-denaturing PAGE was also used to confirm the purities of the AsiA proteins and to demonstrate their relative electrophoretic mobilities, as shown in the first (upper left) panel. The remaining panels show the results when increasing concentrations of AsiA or mutant AsiA were incubated with a fixed concentration of σ70. Electrophoresis of the samples was performed as described in the text. Lane 1, AsiA or mutant AsiA only (20 µM). Lane 2, σ70 only (20 µM). Lanes 3–7, 20 µM σ70 and 5, 10, 20, 40, or 80 µM AsiA or mutant AsiA, respectively.
Structural Integrity of the AsiA Mutants
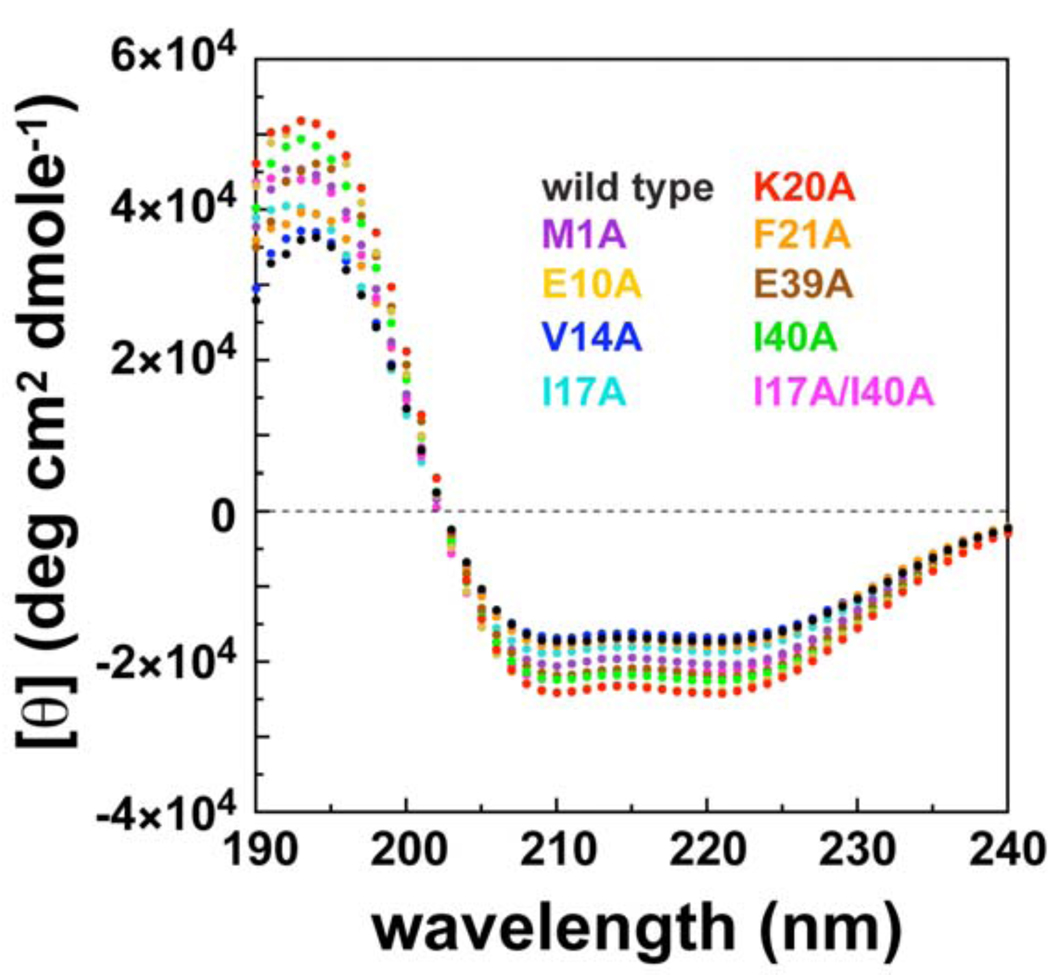
The secondary and tertiary structures of the nine mutant AsiA proteins were assessed using circular dichroism (CD) spectrapolarimetry and NMR spectroscopy. The far-UV CD spectra of AsiA and the mutant AsiA proteins (Figure 3) all exhibit the distinct double minimum at 208 and 222 nm indicative of α-helical structure. The spectra of the mutant proteins are all similar to the wild type spectrum, and the results indicate that the mutations did not result in a decrease in secondary structure (helical) content. In fact, results of secondary structure calculations based on the CD spectra (not shown) suggest that the helical content of most of the mutant proteins is slightly higher than wild type, as can be deduced from the more negative signals for (most of) the mutants at 208 and 222 nm and the more positive signals at 190 nm (Figure 3).
FIGURE 3.
Circular dichroism spectra for AsiA and the AsiA mutants. The far-UV circular dichroism spectra of the mutant proteins are similar to one another and to wild type and clearly indicate no loss of helical secondary structure in the mutants. Shown are spectra for wild type AsiA (black) and mutants M1A (purple), E10A (gold), V14A (blue), I17A (cyan), K20A (red), F21A (orange), E39A (brown), I40A (green), and the double mutant I17A/I40A (magenta).
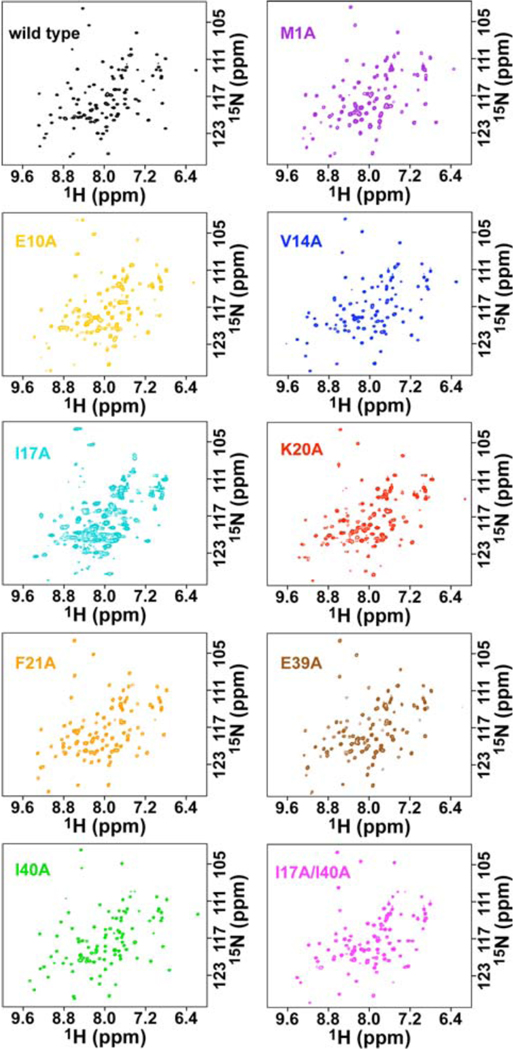
Two-dimensional 1H, 15N-HSQC NMR spectra were recorded for each of the AsiA mutants to evaluate tertiary structure. The spectra of most of the proteins indicate folded proteins with defined tertiary structure. The chemical shift dispersions are consistent with those observed typically in spectra of other folded helical proteins. These indicate that the mutations did not result in protein unfolding. Except for the K20A and I17A mutants, the line widths and numbers of observed signals suggest a lack of slow conformational averaging. The line widths in the NMR spectrum of the K20A mutant suggest a limited slow conformational averaging. The NMR spectrum of I17A displays similar but somewhat more marked symptoms, although the chemical shift dispersion is not affected.
Chemical shift changes are delicate indicators of structural changes. Unfortunately, chemical shift assignments for the mutant AsiA proteins are not available. However, conservative estimates for chemical shift changes resulting from mutation can be calculated using the minimal chemical shift perturbation method, given the wild type chemical shift assignments (53). These minimal chemical shift changes can assist in localizing the structural changes resulting from mutation. In the Supporting Information (Figure S1), sequence-specific minimal chemical shift perturbations for main chain 1HN, 15N pairs are shown for each mutant AsiA protein, as are averages at each position. In general, the largest minimal chemical shifts shown are at the ends of helices and between helices. In conjunction with the CD results noted above, this would suggest that the most significant structural changes accompanying the mutations are reorientation or repositioning of helices, including protomer-protomer reorientation in the dimers, rather than changes in secondary structure or helical character. The minimal chemical shift perturbations are largest overall for the double mutant, which might suggest that the combined effect of the two substitutions results in larger structural changes. The minimal chemical shift perturbations are smallest overall for the I17A mutant. For this mutant, because of the conformational averaging, there are many signals in the spectra, generally increasing the probability for a small minimal chemical shift. On the other hand, a simple explanation would be that one of the conformers present is structurally very similar to wild type. Finally, it is important to note that there are relatively large minimal perturbations observed throughout each of the mutant AsiA proteins. This indicates that structural changes are not localized to the sites of the amino acid substitutions, and that in this relatively small protein compensatory structural changes occur throughout each protomer in dimeric AsiA. Such behavior has been noted previously in N-terminal His-tagged AsiA. This AsiA species is largely monomeric in solution, and its high resolution solution structure reveals that, although the helices remain intact, there are significant changes in helix positioning (16).
Dimerization of the AsiA Mutants
Dynamic light scattering (DLS) and sedimentation equilibrium studies were performed to provide an initial assessment of the propensity for self-association of the AsiA mutants to form dimers at protein concentrations known to favor dimer formation by wild type AsiA (results are not available for the E10A mutant, see below). For each protein, the DLS data fit well to a monomodal distribution (Table 1). For wild type AsiA, the mass derived from the hydrodynamic radius (23 kDa) is consistent with an AsiA dimer (21.2 kD). The translational diffusion coefficient (9.72 × 10−7 cm2/s) and Stokes radius (2.21 nm) for the AsiA dimer calculated from the known structure of dimeric AsiA (22) using HYDROPRO (54–56) are likewise similar to the values determined by DLS (9.24 × 10−7 cm2/s and 2.27 nm, respectively). The observed polydispersity is not unexpected and must be due to the presence of some monomeric protein. The results with the M1A, V14A, I17A and I40A mutants are essentially identical to wild type and indicate that these proteins are mostly dimeric under the conditions (~188 µM AsiA or mutant species) of the measurements.
Initial DLS results with the K20A, F21A, and E39A mutant AsiA proteins suggested that species larger than dimers were present. Increasing the ionic strength with additional KCl (Table 1) promoted a reduction of the respective hydrodynamic radii and masses to values consistent with dimers. The effect was most pronounced with the K20A mutant (Table 1). We thus attribute the results at lower ionic strength to non-specific electrostatic aggregation.
DLS analysis of the I17A/I40A double mutant indicates that this protein is mostly monomeric under the conditions of the experiment. The observed mass is somewhat higher than the actual monomeric mass due to the presence of some dimer and perhaps to deviation of the I17A/I40A monomer structure from an ideal sphere, both of which would decrease the observed translational diffusion coefficient and increase the observed mass.
An initial set of sedimentation equilibrium experiments was also performed to address dimer formation by the AsiA mutants. These experiments were conducted at a single speed and at protein concentrations higher than those used to determine association constants. The absorbance at 280 nm was monitored. For wild type AsiA, and for each mutant, the data fit well to a single species model with random residuals (not shown). In all cases, the single species molecular mass was equivalent to that of an AsiA dimer, with the exception of the I17A/I40A double mutant, for which the single species mass was equivalent to that of an AsiA monomer. Thus, these results, and those from the DLS measurements, indicate that the mutant proteins retain the ability to self-associate to form dimers. The results also suggest that the association constant lower limits are not dramatically decreased by the mutations. The exception is the I17A/I40A double mutant, for which a significantly lower association constant is indicated, as is a substantial combined contribution of the I17 and I40 side chains to the free energy of AsiA dimer formation.
Free Energies of Dimer Formation for AsiA Mutants
Using sedimentation equilibrium methods we demonstrated previously the ability of wild type AsiA monomers to self-associate in solution to form homodimers (14); subsequently an association constant was determined using similar methods (21). In order to establish the contributions of individual amino acid side chains at the dimer interface to the free energy of AsiA dimer formation, we conducted additional sedimentation equilibrium experiments with each of the mutant AsiA proteins to determine the free energy changes resulting from the mutations.
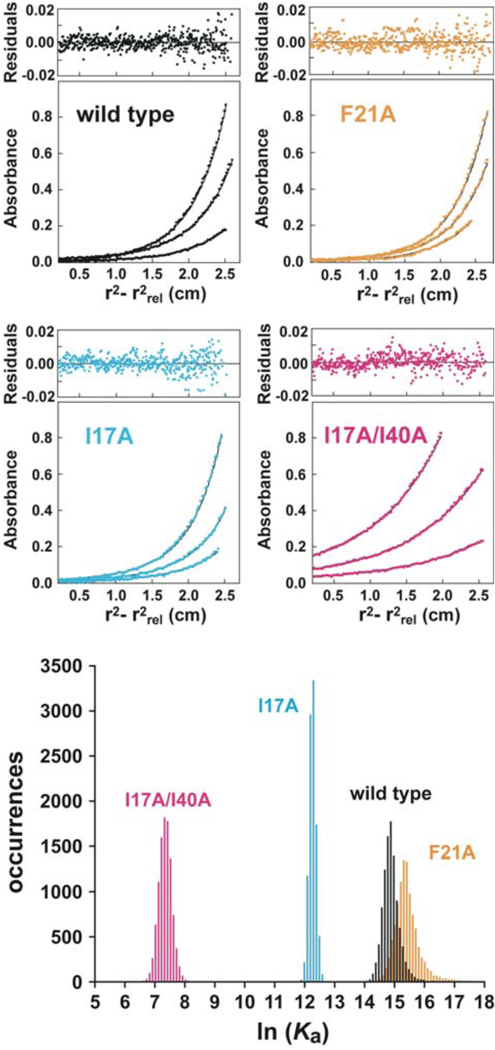
In order to measure the association constants for homodimer formation by the mutant AsiA proteins, the sedimentation equilibrium experiments were performed at lower concentrations (3 µM to 12 µM), the absorbance was monitored at 230 nm, and multiple speeds (24,000, 30,000, and 40,000 rpm) were used. For wild type AsiA and for all AsiA mutants, the results were best fit by a monomer-dimer equilibrium (self-association) model. The resulting monomer molecular masses were consistent with the monomeric AsiA mass, and the residuals from the fits were random (representative results are shown in Figure 5). The values for the association constants and free energies of association were determined (Table 2), with confidence intervals estimated from results of Monte Carlo simulations (Figure 5).
FIGURE 5.
Sedimentation equilibrium results for AsiA and AsiA mutant proteins. (Top) For each protein, sedimentation equilibrium data were acquired at three protein concentrations and three centrifugation speeds. Shown are representative results at 40,000 rpm for 3.0, 7.0, and 12.0 µM protein (wild type, I17A, F21A, and the I17A/I40A double mutant) monitored at 230 nm. In each case, the best fit from the global analysis (shown as a black line through the data points) was to a monomer-dimer equilibrium model. Residuals are shown at the top of each panel. (Bottom) Distributions of values for ln Ka from Monte Carlo simulations for wild type AsiA and the I17A, F21A, and I17A/I40A mutants.
Table 2.
Sedimentation equilibrium measurements of protomer-protomer affinities for AsiA and mutant AsiA proteinsa
| Ka (M−1) | Kd (µM) | ΔG° (kJ/mol)b | mass (kDa)c | |
|---|---|---|---|---|
| WT | 2.8 ± 0.8 × 106 | 0.35 ± 0.10 | −36.8 ± 0.7 | 10.90 ± 0.08 |
| M1A | 3.8 ± 1.0 × 106 | 0.26 ± 0.07 | −37.6 ± 0.6 | 12.33 ± 0.08 |
| V14A | 8.2 ± 1.2 × 105 | 1.2 ± 0.2 | −33.7 ± 0.4 | 10.23 ± 0.07 |
| I17A | 2.0 ± 0.2 × 105 | 4.9 ± 0.6 | −30.3 ± 0.3 | 13.49 ± 0.13 |
| K20A | 2.1 ± 0.4 × 106 | 0.48 ± 0.10 | −36.1 ± 0.5 | 12.62 ± 0.09 |
| F21A | 4.7 ± 1.9 × 106 | 0.21 ± 0.09 | −38.0 ± 1.0 | 11.87 ± 0.11 |
| E39A | 5.7 ± 2.0 × 106 | 0.18 ± 0.06 | −38.5 ± 1.0 | 12.58 ± 0.09 |
| I40A | 3.6 ± 0.5 × 105 | 2.7 ± 0.4 | −31.7 ± 0.3 | 10.72 ± 0.09 |
| I17A/I40Ad | 1.4 ± 0.3 × 103 | 694 ± 0.149 | −18.0 ± 0.5 | 11.47 ± 0.27 |
Fits are to a monomer-dimer self-association model and are from data acquired at three speeds for three protein concentrations monitored at 230 nm unless otherwise noted. Error limits are standard deviations from Monte Carlo simulations.
Free energy of association.
Fitted monomer molecular mass. The actual AsiA molecular mass is 10.59 kDa.
Based on data collected at a single speed at higher concentrations monitored at 280 nm (see text).
The association constant for AsiA self-association to form dimers is consistent with that determined previously (21) and corresponds to a free energy of association of −36.8 kJ/mol (−8.8 kcal/mol). The M1A, K20A, and F21A substitutions did not significantly change this free energy, whereas the E39A substitution only nominally (if significantly) decreased it (Table 2). Thus, the sidechains of M1, K20, F21 and E39, and the putative electrostatic interaction between K20 and E39, do not apparently promote affinity between the protomers in dimeric AsiA.
The results with the E10A mutant were ambiguous. Several data sets were collected for this mutant, and in each case, fits to monomer-dimer self-association models indicated association constants substantially larger than wild type. The data also fit well to a single species model, with the fitted mass consistent with a dimeric species. However, the indicators of the quality of the data and fits suggested that the results were unreliable. Therefore, we can only speculate that this mutation promotes an increase in protomer-protomer affinity in dimeric AsiA relative to wild type. Additional studies with E10A were not pursued.
The V14A, I17A, I40A, and I17A/I40A mutants all exhibited protomer-protomer affinities significantly lower than wild type AsiA. For the single mutants, the affinities are lowest for the I17A and I40A, demonstrating that the sidechains of both I17 and I40 are important for protomer-protomer affinity. Furthermore, substituting both I17 and I40 with alanine produced a dramatic decrease in affinity (18.8 kJ/mol), signifying the importance of the combined contributions of the sidechains of these two amino acids to the affinity between the AsiA protomers.
Affinity of AsiA Mutants for σ70
A qualitative assessment of the relative affinities of AsiA and the mutant AsiA proteins for σ70 was derived from electrophoretic mobility shift assays using non-denaturing PAGE (Figure 2). With the exception of the I17A/I40A double mutant, under the conditions of the experiment, most of the mutant proteins were able to bind to σ70 with apparent affinities similar to the affinity of wild type AsiA for σ70. This conclusion is also true for the L18A mutant that was not used in the biophysical experiments described above. The apparent affinity of the I17A mutant for σ70, however, appears somewhat lower, as indicated by a large amount of unbound I17A observed at equimolar concentrations of σ70 and I17A (lane 5). The affinity of the I17A/I40A double mutant for σ70 is nominal at best, indicating that the affinity of AsiA for σ70 results largely from the combined interactions of the sidechains of I17 and I40 with σ70.
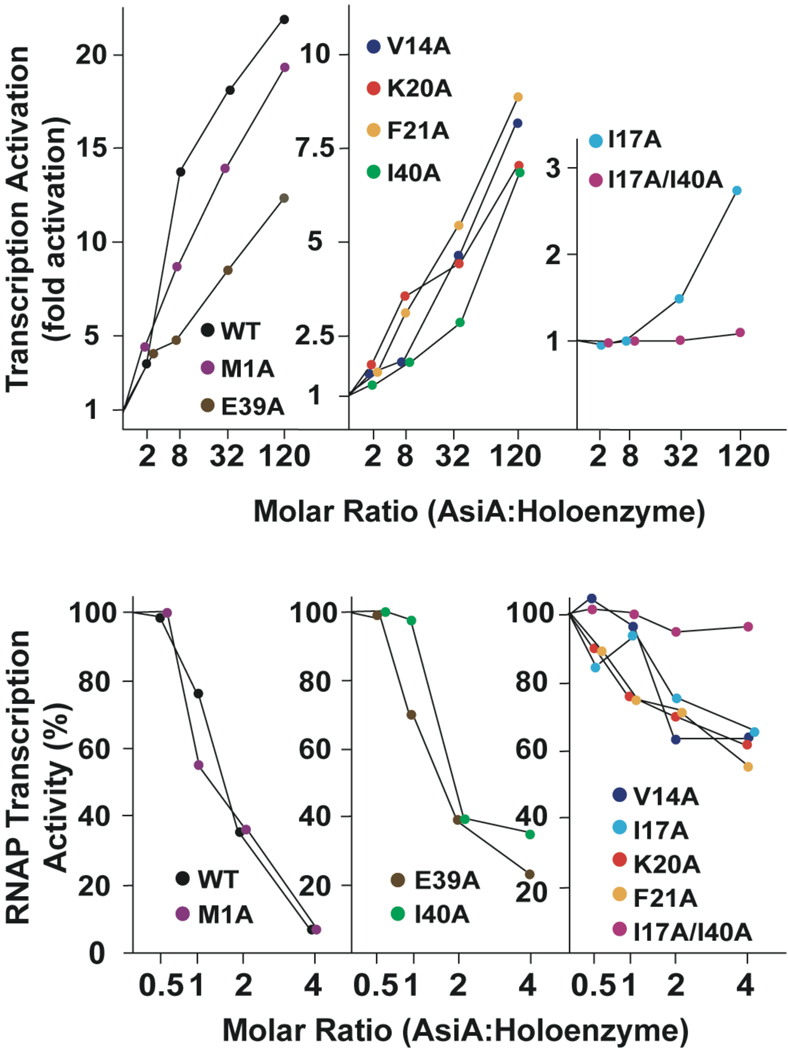
Transcription Inhibition and Activation by AsiA Mutants
The effects of AsiA and AsiA mutants on both inhibition and MotA-independent activation of transcription by the E. coli RNA polymerase were measured using in vitro transcription assays (Figure 6). Wild type AsiA inhibited transcription more than 90% at a ratio of 4:1 (AsiA:holoenzyme). Inhibition of transcription by the M1A mutant was similar to wild type. Both the E39A and I40A mutants also proved to be relatively effective inhibitors. A group of mutants including V14A, I17A, K20A, and F21A proved to be less effective inhibitors, with only ~40% transcription inhibition attained at a 4:1 ratio of AsiA to holoenzyme. The I17A/I40A double mutant demonstrated little ability to inhibit transcription.
FIGURE 6.
Transcription inhibition and activation by AsiA and mutant AsiA proteins. The ability of AsiA and mutant AsiA proteins to inhibit or activate transcription by E. coli RNA polymerase are shown. (Top) Activation of transcription from the T7 bacteriophage early promoter A1. (Bottom) Inhibition of transcription from the T4 bacteriophage PrIIB2 middle promoter.
As shown previously (20), when σ70 in the RNA holoenzyme is replaced with a mutant lacking region 4.2 (truncated at residue 565, denoted σ565 or σ70 Δ565–613), AsiA activates transcription from the T4 phage middle promoter PrIIb2 in a MotA-independent manner. This is a convenient assay, despite the decreased affinity of AsiA for this mutant σ70 and the correspondingly higher molar ratios of AsiA necessary for activation to be observed. As shown (Figure 6), wild type AsiA is able to activate MotA-independent transcription by more than 20-fold at a high molar ratio (120:1) of AsiA to holoenzyme. Similar results are observed for the M1A mutant, whereas the E39A mutant is able to activate MotA-independent transcription by ~12-fold under these conditions. A group of mutants including V14A, K20A, F21A and I40A are less effective, and activate MotA-independent transcription 7–9 fold at the 120:1 molar ratio. The I17A mutant is a poor transcription activator (~3 fold), and the I17A/I40A mutant shows little MotA-independent activation ability.
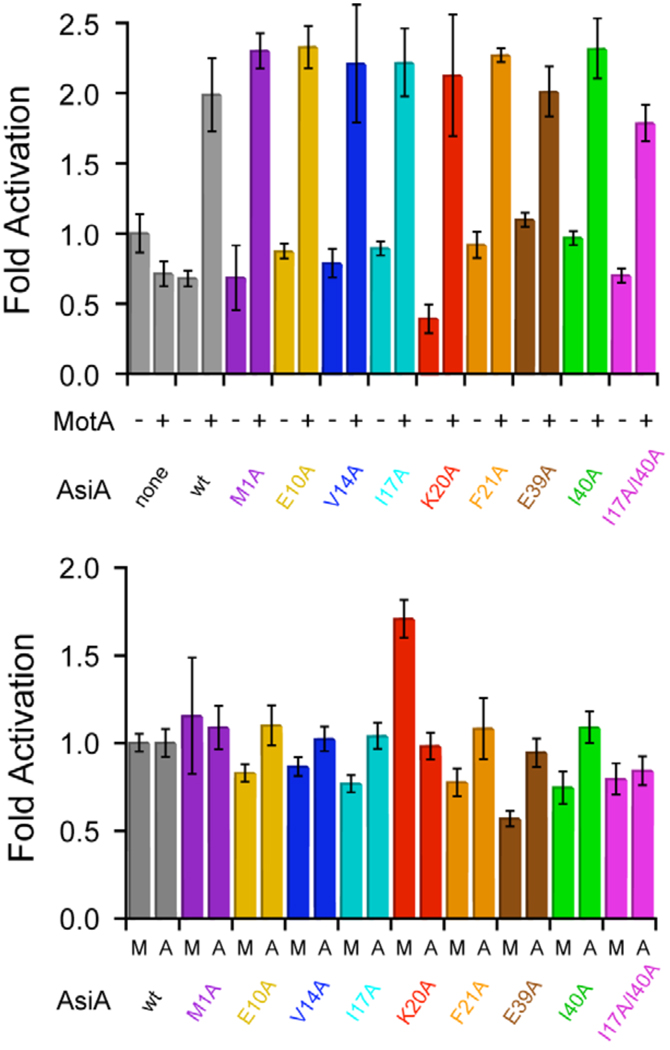
The affinity and activity measurements suggest that impaired AsiA function upon mutation is due to decreased affinity for the RNA polymerase (σ70) and not due to nonproductive binding or essential roles for any of the amino acids in question in transcription activation or inhibition. We therefore performed an experiment at saturating AsiA (and mutant AsiA) concentrations, measuring MotA-dependent RNA polymerase activation by AsiA, to determine of any of the mutant AsiA proteins were essential for activation or full activation. This is a more convenient assay for this purpose, as saturating AsiA concentrations for the MotA-independent assay would be prohibitively high. In the presence of MotA, saturating concentrations of AsiA or mutant AsiA proteins produced similar degrees of transcription activation (Figure 7, top) compared to activity in the absence of both MotA and AsiA, with an average fold activation of ~2.15. At the 95% confidence level (error limits shown are 68% confidence intervals), degrees of activation by all AsiA species were statistically identical. When co-activation by MotA and co-activation by AsiA and AsiA mutants are compared, with normalization to activity with wild type AsiA (Figure 7, bottom), again no remarkable differences are noted, with the possible exception of MotA activation with the K20A AsiA mutant protein. These results indicate that none of the mutant AsiA proteins are defective for MotA-dependent co-activation and none of the side chains under consideration are essential for this activation process. So, the individual side chains in question mediate affinity, but none appear essential for activity.
FIGURE 7.
MotA-dependent transcription co-activation by AsiA and mutant AsiA proteins. (Top) Fold activation with or without MotA in the presence of saturating wild type AsiA or mutant AsiA, normalized to the activity in the absence of both MotA and AsiA. (Bottom) Fold activation by MotA or wild type AsiA and AsiA mutants normalized to wild-type activation. “M” on the abscissa represents activation by MotA in the presence of saturating wild type AsiA or mutant AsiA, with activation normalized to wild type AsiA. “A” on the abscissa represents activation by wild type AsiA (or mutant AsiA) in the presence of MotA, normalized to wild type AsiA. Error limits are ± 1 standard deviation.
Accessible Surface Areas
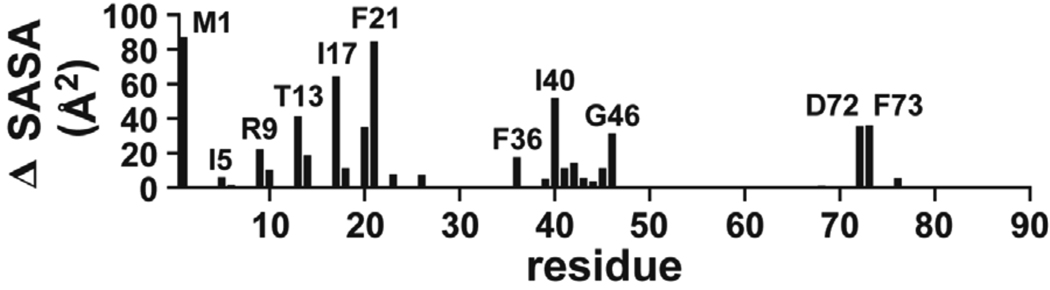
To approximate the changes in solvent accessible surface areas for residues at the protomer-protomer interface of AsiA upon dimer dissociation, these areas were calculated for dimeric AsiA and for the individual protomers with the opposing protomer removed from the structure (Figure 8). Overall, the protomer-protomer interface in dimeric AsiA is approximately 2012 ± 102 Å2 (1253 ± 102 Å2 of this is non-polar), with 1741 ± 74 Å2 contributed by amino acid side chains (error limits are standard deviations computed from the ensemble of 25 deposited NMR structures). The interface size is not unusual (23), but it is somewhat larger than a typical homodimeric interface relative to the monomeric mass (24). At the residue level, the increase in solvent accessible hydrophobic surface area upon dimer dissociation is largest for M1 and F21 followed by I17 and I40 (Figure 7). The side chains of I14, I17 and I40 are essentially completely buried in the AsiA dimer (100%, 100% and 97% buried, respectively), and somewhat exposed in the monomer (83%, 53% and 60% buried, respectively). Therefore, the changes in hydrophobic surface areas for these amino acids upon AsiA dimer dissociation are moderate (I14) to quite large (I17, I40). Although the side chains of M1 and F21 are more exposed in the dimer (91% and 88% buried), they are also highly exposed in the monomer (both 36% buried), resulting in large absolute changes of exposed hydrophobic surface areas upon dimer association/dissociation. With the exception of M1, the changes in exposed hydrophobic surface areas upon dimer dissociation are smaller for residues near the N-terminal end of helix 1 of AsiA and larger towards the C-terminal end (note, in Figure 8, the periodicity of helix 1 reflected in these changes), indicating an overall tighter packing of the interfacial residues on one protomer that interact with the hydrophobic cleft formed by helix 1 and 3 of the opposing protomer.
FIGURE 8.
Solvent accessible surface area differences between AsiA dimers and protomers. The differences shown are residue localized increases in non-polar solvent accessible surface areas for AsiA protomers (from the dimer structures, but in the absence of the opposing protomer) relative to dimers. The protomer-protomer interface in dimeric AsiA is approximately 2012 ± 102 Å2 (1253 ± 102 Å2 of this is non-polar), with 1741 ± 74 Å2 contributed by amino acid side chains (error limits are standard deviations computed from the ensemble of 25 deposited NMR structures).
In monomeric AsiA, the side chains of L18, V27, F33, F36, L37 and V42 are nearly completely buried (92%, 99%, 98%, 89%, 100%, and 88%, respectively), and all are 98–100% buried in the dimer. This suggests important roles for these residues in stabilizing monomeric AsiA, as is indicated by the instability of the L18A, V27A and V42A mutants.
DISCUSSION
Monomeric AsiA binds tightly to σ70 and mediates the promoter specificity of the RNA polymerase holoenzyme. Structural studies indicate that many of the same residues comprising the surface that interacts with σ70 also contribute to the protomer-protomer interface in dimeric AsiA. Presumably, this surface/interface has been optimized for phage survival and propagation by a sapient balance between protomer-protomer affinity in the homodimer, affinity for σ70 in the AsiA-σ70 heterodimer, and stability. Here we explored the contributions of interfacial residues to protomer-protomer affinity in homodimeric AsiA, to AsiA function, and, to an extent, AsiA stability.
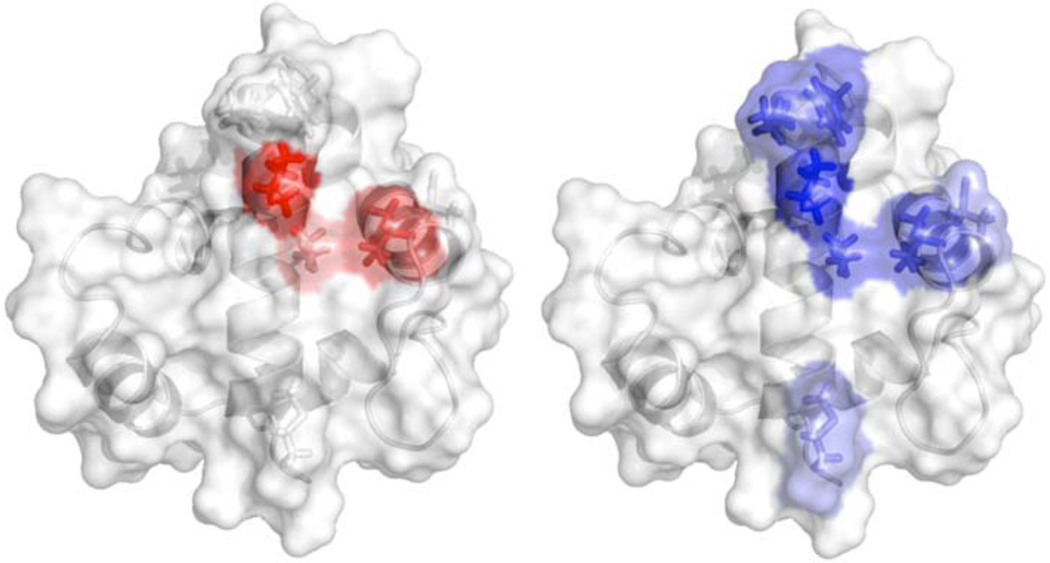
Of the residues suggested by structural studies to be important for AsiA homodimer affinity and the AsiA-σ70 heterodimer interaction, V14, I17, and I40 clearly contribute substantially to protomer-protomer affinity in dimeric AsiA (Table 2). Structurally, the side chains of these residues interact with one another within a given protomer and between protomers (Figure 9). The combined effect of the I17A and I40A substitutions, in the I17A/I40A double mutant, increases the observed homodimer dissociation constant by approximately 2000 fold and decreases the total free energy of homodimer formation by just over one-half compared to the wild type protein. Also, as discussed above, these residues are essentially completely buried in the dimer. Our results are consistent with recent suggestions that solvent inaccessibility in the complex is a necessary, but not sufficient, prerequisite for residues that contribute significantly to affinity (23, 57). Thus, the side chains of these residues comprise a core hydrophobic assemblage or “hot spot” (58) maintaining the affinity and structural integrity of the protomer-protomer interface in dimeric AsiA (Figure 9).
FIGURE 9.
Functional epitopes of AsiA maintaining (left) protomer-protomer affinity in dimeric AsiA and (right) AsiA-σ70 affinity. Color intensity is a qualitative indicator of relative contribution.
It is worthwhile noting that the effects of the I17A and I40A substitutions, for instance, are not independent, and their effects on the free energy of AsiA dimer formation/dissociation are not additive. For residues such as I14, I17, and I40, such apparent coupling is not unexpected (59–63), given the intra- and intermolecular interactions between the side chains of these amino acids evident from structural studies of the homodimer (Figure 9). Also relevant is the fact that substitution of a single AsiA residue constitutes a double mutation of the symmetrical AsiA homodimer. With these points in mind, it appears that compensatory structural changes underlie a resilient AsiA homodimer interface that accommodates single alanine substitutions of hot spot residues with relatively modest protomer-protomer affinity changes. Alanine substitution of more than one of these residues is less readily accommodated, and relatively much more deleterious to interface structure and affinity. A more exhaustive analysis would be necessary to test these assertions thoroughly.
Residues M1, K20, F21 and E39 are positioned near the edges of the AsiA homodimer interface. Mutating any one of these residues to alanine does not alter significantly the free energy of dimer dissociation relative to wild type. Although the changes in the solvent accessible hydrophobic surface areas upon homodimer association/dissociation for the side chains of M1 and F21 are large, these side chains are significantly solvent exposed in the homodimer. When these residues are substituted computationally to alanine, there is little if any significant change in solvent accessibility of residues constituting the interior of the interface. These results are consistent with “O-ring” (57) and similar (23) models of interfaces and results of experimental (64, 65) and computational (57) “alanine shaving” experiments. Thus, the side chains of these residues provide little contribution directly to the affinity between the protomers of dimeric AsiA, but evidently the main chain and beta positions assist in occluding solvent from the hot spot. Although K20 and E39 appear to form a charge pair in the homodimer structure, the results indicate that these two amino acids do not contribute directly to protomer-protomer affinity in the homodimer. We also note that previous, preliminary results suggested a very high dissociation constant for the K20A mutant. We postulated that the tighter association of this mutant with σ70 in bacterial two-hybrid analyses stemmed from decreased competition from homodimerization (17). This is not the case, as indicated herein. Nevertheless, this mutant is very effective at increasing the sensitivity of bacterial two-hybrid analyses of AsiA binding, has been utilized effectively in this capacity and is clearly a useful tool for these types of studies (17, 66).
Representatives of a group of residues at or near the AsiA homodimer interface suspected to contribute to the intrinsic stability of monomeric AsiA were also mutated to alanine. For AsiA, the global and site-resolved stabilities are known from hydrogen exchange studies (22), which indicate that the most stable part of the AsiA protein is the hydrophobic cleft formed by residues of helices 1–3, and the global stability is about 7 kcal/mol (determined in 90% D2O, which may differ from the value in H2O, 67, 68). Residues with hydrophobic side chains in this cleft include A15, L18, V27, F33, F36, L37 and V42. Of these, the amide hydrogens of A15, L18, and F36 exchange only from the globally unfolded state, and therefore they define the global stability (22, 67, 68). Although some of the side chains of residues in this cleft contribute interprotomer contacts in homodimeric AsiA and in the AsiA-σ70 heterodimer, the number of these contacts is very small compared to the number of intraprotomer contacts that they contribute. Furthermore, most of these residues are completely or nearly completely solvent-inaccessible in monomeric AsiA. Given these facts, it would be expected that alanine substitution, eliminating the intraprotomer contacts conferred by atoms beyond the beta carbon position, would result in a significant destabilization of the AsiA monomer, but would have a smaller impact on the affinity for σ70. This appears to be the case for the L18A, V27A and V42A mutants. All of these formed inclusion bodies when overproduced. Both the V42A and L18A inclusion bodies were solubilized and refolding was attempted, but yields of purified proteins were low and the proteins precipitated easily and were not amenable to detailed study. However, we were able to demonstrate that the L18A mutant still binds tightly with σ70, indicating that the intrinsic ability to do so has not been lost due to the alanine substitution and suggesting that any significant loss of function that might be observed for this mutant would be due to loss of the active conformation resulting from destabilization of the AsiA monomer.
The interactions between σ70 and the mutant AsiA proteins were evaluated by measuring the ability of the AsiA mutants to regulate transcription and by non-denaturing electrophoretic mobility shift assays. Although the mobility shift experiments cannot reveal subtle differences in affinities, they did reveal that the I17A/I40A double mutant displays little measurable affinity for σ70, and that the affinity between the I17A mutant and σ70 is somewhat attenuated relative to wild type and the other mutants. These residues appear to be integral to the affinity between AsiA and σ70, in analogy with the AsiA homodimer. This is consistent with the structural studies (16, 18) and is supported by the results of the activity assays. At concentrations less than saturating, the I17A/I40A double mutant is a poor transcription regulator, apparently reflecting the very poor affinity for σ70. The likewise poor affinity of the I17A mutant renders it a poor transcriptional regulator at sub-saturating concentrations. The V14A mutant also shows low activity. The I40A mutant is also a very poor activator (of MotA-independent activity), but relatively less ineffectual as an inhibitor. Nevertheless, under conditions where the mutant AsiA proteins are saturating, none of the mutant proteins are defective for MotA-dependent activation, indicating that none of these sidechains are essential for activity, and indicating that affinity governs activity. Because of the structural differences implicit in the transcriptional complexes for early versus middle phage promoters (16, 18), there is no reason that the contribution of a particular amino acid in AsiA to affinity for σ70 need be the same for activation and inhibition. Analogously, affinity and activity need not be fully coupled, so that a particular mutation may affect one more than the other. Nonetheless, for each of the mutant AsiA proteins, the correspondence between relative activation and inhibition abilities are remarkably similar. Finally, the activity assays indicate that residues such as K20 and F21, which do not contribute directly to homodimer affinity, are clearly important for AsiA activity, as is suggested by the solution structure of the AsiA-σ70 region 4 complex (16), which shows many interactions between the side chains of these residues of AsiA and region 4. Interactions between E39 and both positively charged and hydrophobic groups in region 4.2 of σ70 are also observed in the structure, but, based on the results of the transcription inhibition assays with E39A, these apparently do not contribute substantially to polymerase inhibition by AsiA.
The functional roles of some of the AsiA residues that comprise the homo- and heterodimer interface have been addressed previously. For instance, V14, L18, and I40 were suggested to be important for forming or maintaining the AsiA-σ70 complex (19). The side chain of L18A makes contacts with at least one residue in region 4 of σ70 in the structural model of this complex (16). L18S and L18F substitutions render AsiA defective for binding to σ70, and this observation was interpreted to indicate that L18 was important for tight binding to σ70 (19). However, substituting the L18 side chain with alanine does not decrease the apparent affinity of AsiA for σ70 noticeably, indicating that contacts between L18 and σ70 are not critical in this respect. Because the alanine substitution apparently decreases the stability of monomeric AsiA, which is consistent with the nearly complete burial of L18 in the AsiA monomer, it seems most likely that the steric perturbations and polarity changes introduced by the L18F and L18S mutations destabilize monomeric AsiA and disrupt the interface, leading to the observed effects. In contrast, V14 and I40 certainly contribute to the affinity of AsiA for σ70, and results of studies using mutations likely to perturb the interface (V14D and I40N) are consistent with this conclusion (19). Previous studies have also addressed the role of E10 (69) demonstrating that the E10K substitution eliminated both AsiA toxicity to E. coli and the interaction with σ70, although E10 substitutions to leucine, serine, glutamine, tyrosine and alanine did not affect toxicity. It was concluded that the negative charge of the E10 side chain is not important for the interaction with σ70.
The linchpin for AsiA activity and intermolecular affinity appears to be I17. In the AsiA homodimer, the side chain of I17 from one protomer is positioned centrally at the interface, between I40 and I17 of the other protomer (Figure 9) and is a key contributor to the affinity between the protomers. When one of the protomers is displaced by σ70 to form the AsiA-σ70 heterodimer, the I17 side chain is positioned in a hydrophobic pocket formed by side chains of residues from regions 4.1 and 4.2 (16). Interestingly, in the heterodimer, the F563 side chain of σ70 occupies the central position between I17 and I40 of AsiA occupied previously by the I17 side chain of the displaced protomer. It has likewise been demonstrated that F563 is pivotal to the interaction of σ70 with both AsiA and the beta flap (16, 17, 66, 70). These facts serve to underscore the fundamental importance of I17 in AsiA structure and function.
The relative abilities of the AsiA mutants to inhibit transcription (from a phage early promoter) are remarkably similar to their relative abilities to activate MotA-independent transcription (from a middle promoter). We assayed AsiA-dependent, MotA-independent activation of transcription from the T4 bacteriophage PrIIB2 middle promoter with RNA polymerase holoenzyme incorporating a truncated σ70 (σ565) that lacks the C-terminus, which includes conserved region 4.2 (20). This proves a useful means to monitor activation by AsiA as it does not require MotA. Activation without region 4.2 is possible, in part, because the interaction of AsiA with σ70 occurs via both conserved regions 4.1 and 4.2 of σ70 (14). As demonstrated previously, AsiA binds tightly in vitro to short peptides corresponding to regions 4.1 and 4.2 of σ70 both individually, to form binary complexes, or simultaneously to form a ternary complex (14). The exchange behavior of the complexes and observed NOE-based intermolecular contacts indicate that the binary complexes are similar structurally. Either peptide, in the absence of the other, can occupy one or more sites in the hydrophobic cleft between helices 1 and 3 of AsiA. However, in the ternary complex, regions 4.1 and 4.2 are each restricted to a single defined site on AsiA (18). The different patterns of intermolecular contacts from NOE spectra reveal directly the structural differences between binary and ternary complexes and the different contacts between σ70 and regions 4.1 and 4.2 in these contexts. This latter fact has been corroborated recently by remarkable results demonstrating that both AsiA and MotA interactions with region 4.1 are perturbed by specific mutations in region 4.1; however, these mutations do not affect the ability of AsiA and MotA to coactivate transcription from the T4 phage PuvsX middle promoter (66). Furthermore, the AsiA-σ70 interface must reorganize markedly following formation of the initial encounter complex through the course of subsequent events, including binding to the core polymerase, MotA, and (middle) promoters (9, 16, 18, 20, 66, 70, 71). However, even in the context of a draconian truncation of σ70, the relative contributions of the AsiA amino acids at the AsiA-σ70 interface to transcription activation are similar to the contributions to inhibition when wild type σ70 is present. These facts together serve to illustrate the robust structural and functional plasticity in the AsiA-σ70 interaction (18, 20) and partly how it is accomplished.
Supplementary Material
FIGURE 4.
NMR spectra of the AsiA mutants. The 1H, 15N-HSQC spectra of wild type AsiA and the stable mutant AsiA proteins are shown. Although some slight (K20A) to moderate (I17A) conformational averaging is observed, the chemical shifts and chemical shift dispersion observed in the spectra of the mutants are similar to wild type and indicate that the mutations do not eliminate tertiary structure.
ACKNOWLEDGEMENT
We thank Dr. John Brewer for his assistance in many aspects of the work and for many beneficial conversations. We thank Dr. Borries Demeler for his expert advice on sedimentation equilibrium data collection and analysis and for the software (Ultrascan) to analyze the sedimentation equilibrium data. We thank Dr. Aaron Cowley for performing a preliminary sedimentation equilibrium experiment on one of the mutant proteins.
Abbreviations
- CD
circular dichroism
- DLS
dynamic light scattering
- DSS
2,2-dimethyl-2-silapentane-5-sulfonate
- DTT
dithiothreitol
- HPLC
high performance liquid chromatography
- HSQC
heteronuclear single quantum coherence
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- SDS
sodium dodecyl sulfate
Footnotes
This work was supported by the NIH (GM54998 to J.L.U. and GM59295 to K.S.) and the office of the Vice President for Research at the University of Georgia.
An incorrect structure, determined independently by other investigators (21), has since been retracted (52).
SUPPORTING INFORMATION AVAILABLE
One figure detailing NMR minimal chemical shift perturbation values for all mutant AsiA proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Stevens A. New small polypeptides associated with DNA-dependent RNA polymerase of Escherichia coli after infection with bacteriophage T4. Proc Natl Acad Sci U S A. 1972;69:603–607. doi: 10.1073/pnas.69.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orsini G, Ouhammouch M, Le Caer JP, Brody EN. The asiA gene of bacteriophage T4 codes for the anti-sigma 70 protein. J Bacteriol. 1993;175:85–93. doi: 10.1128/jb.175.1.85-93.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stevens A. Inhibition of DNA-enzyme binding by an RNA polymerase inhibitor from T4 phage-infected Escherichia coli. Biochim Biophys Acta. 1977;475:193–196. doi: 10.1016/0005-2787(77)90355-0. [DOI] [PubMed] [Google Scholar]
- 4.Stevens A, Rhoton JC. Characterization of an inhibitor causing potassium chloride sensitivity of an RNA polymerase from T4 phage-infected Escherichia coli. Biochemistry. 1975;14:5074–5079. doi: 10.1021/bi00694a007. [DOI] [PubMed] [Google Scholar]
- 5.Stevens A. Deoxyribonucleic acid dependent ribonucleic acid polymerases from two T4 phage-infected systems. Biochemistry. 1974;13:493–503. doi: 10.1021/bi00700a015. [DOI] [PubMed] [Google Scholar]
- 6.Ouhammouch M, Adelman K, Harvey SR, Orsini G, Brody EN. Bacteriophage T4 MotA and AsiA proteins suffice to direct Escherichia coli RNA polymerase to initiate transcription at T4 middle promoters. Proc Natl Acad Sci U S A. 1995;92:1451–1455. doi: 10.1073/pnas.92.5.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosig G, Hall DH. In: Molecular Biology of Bacteriophage T4. Karam JD, editor. Washington, D. C.: ASM Press; 1994. pp. 127–131. [Google Scholar]
- 8.Harley CB, Reynolds RP. Analysis of E. coli promoter sequences. Nucleic acids research. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pande S, Makela A, Dove SL, Nickels BE, Hochschild A, Hinton DM. The bacteriophage T4 transcription activator MotA interacts with the far-C-terminal region of the sigma70 subunit of Escherichia coli RNA polymerase. J Bacteriol. 2002;184:3957–3964. doi: 10.1128/JB.184.14.3957-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adelman K, Brody EN, Buckle M. Stimulation of bacteriophage T4 middle transcription by the T4 proteins MotA and AsiA occurs at two distinct steps in the transcription cycle. Proc Natl Acad Sci U S A. 1998;95:15247–15252. doi: 10.1073/pnas.95.26.15247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinton DM, March-Amegadzie R, Gerber JS, Sharma M. Bacteriophage T4 middle transcription system: T4-modified RNA polymerase; AsiA, a sigma 70 binding protein; and transcriptional activator MotA. Methods Enzymol. 1996;274:43–57. doi: 10.1016/s0076-6879(96)74007-7. [DOI] [PubMed] [Google Scholar]
- 12.Ouhammouch M, Orsini G, Brody EN. The asiA gene product of bacteriophage T4 is required for middle mode RNA synthesis. J Bacteriol. 1994;176:3956–3965. doi: 10.1128/jb.176.13.3956-3965.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adelman K, Orsini G, Kolb A, Graziani L, Brody EN. The interaction between the AsiA protein of bacteriophage T4 and the sigma70 subunit of Escherichia coli RNA polymerase. J Biol Chem. 1997;272:27435–27443. doi: 10.1074/jbc.272.43.27435. [DOI] [PubMed] [Google Scholar]
- 14.Urbauer JL, Adelman K, Urbauer RJ, Simeonov MF, Gilmore JM, Zolkiewski M, Brody EN. Conserved regions 4.1 and 4.2 of sigma(70) constitute the recognition sites for the anti-sigma factor AsiA, and AsiA is a dimer free in solution. J Biol Chem. 2001;276:41128–41132. doi: 10.1074/jbc.M106400200. [DOI] [PubMed] [Google Scholar]
- 15.Minakhin L, Camarero JA, Holford M, Parker C, Muir TW, Severinov K. Mapping the molecular interface between the sigma(70) subunit of E. coli RNA polymerase and T4 AsiA. J Mol Biol. 2001;306:631–642. doi: 10.1006/jmbi.2001.4445. [DOI] [PubMed] [Google Scholar]
- 16.Lambert LJ, Wei Y, Schirf V, Demeler B, Werner MH. T4 AsiA blocks DNA recognition by remodeling sigma70 region 4. Embo J. 2004;23:2952–2962. doi: 10.1038/sj.emboj.7600312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gregory BD, Nickels BE, Garrity SJ, Severinova E, Minakhin L, Urbauer RJ, Urbauer JL, Heyduk T, Severinov K, Hochschild A. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc Natl Acad Sci U S A. 2004;101:4554–4559. doi: 10.1073/pnas.0400923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simeonov MF, Bieber Urbauer RJ, Gilmore JM, Adelman K, Brody EN, Niedziela-Majka A, Minakhin L, Heyduk T, Urbauer JL. Characterization of the interactions between the bacteriophage T4 AsiA protein and RNA polymerase. Biochemistry. 2003;42:7717–7726. doi: 10.1021/bi0340797. [DOI] [PubMed] [Google Scholar]
- 19.Pal D, Vuthoori M, Pande S, Wheeler D, Hinton DM. Analysis of regions within the bacteriophage T4 AsiA protein involved in its binding to the sigma70 subunit of E. coli RNA polymerase and its role as a transcriptional inhibitor and co-activator. J Mol Biol. 2003;325:827–841. doi: 10.1016/s0022-2836(02)01307-4. [DOI] [PubMed] [Google Scholar]
- 20.Minakhin L, Niedziela-Majka A, Kuznedelov K, Adelman K, Urbauer JL, Heyduk T, Severinov K. Interaction of T4 AsiA with its target sites in the RNA polymerase sigma70 subunit leads to distinct and opposite effects on transcription. J Mol Biol. 2003;326:679–690. doi: 10.1016/s0022-2836(02)01442-0. [DOI] [PubMed] [Google Scholar]
- 21.Lambert LJ, Schirf V, Demeler B, Cadene M, Werner MH. Flipping a genetic switch by subunit exchange. Embo J. 2001;20:7149–7159. doi: 10.1093/emboj/20.24.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urbauer JL, Simeonov MF, Urbauer RJ, Adelman K, Gilmore JM, Brody EN. Solution structure and stability of the anti-sigma factor AsiA: implications for novel functions. Proc Natl Acad Sci U S A. 2002;99:1831–1835. doi: 10.1073/pnas.032464699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 24.Jones S, Thornton JM. Principles of protein-protein interactions. Proc Natl Acad Sci U S A. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janin J, Miller S, Chothia C. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 26.Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- 27.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J Mol Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 28.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. Journal of molecular graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 29.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA: USA; 2002. [Google Scholar]
- 30.Weiner MP, Costa GL, Schoettlin W, Cline J, Mathur E, Bauer JC. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene. 1994;151:119–123. doi: 10.1016/0378-1119(94)90641-6. [DOI] [PubMed] [Google Scholar]
- 31.Weiner MP, Costa GL. Rapid PCR site-directed mutagenesis. PCR methods and applications. 1994;4:S131–S136. doi: 10.1101/gr.4.3.s131. [DOI] [PubMed] [Google Scholar]
- 32.Hemsley A, Arnheim N, Toney MD, Cortopassi G, Galas DJ. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic acids research. 1989;17:6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Analytical biochemistry. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 34.Sreerama N, Venyaminov SY, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Analytical biochemistry. 2000;287:243–251. doi: 10.1006/abio.2000.4879. [DOI] [PubMed] [Google Scholar]
- 35.Manavalan P, Johnson WC., Jr Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Analytical biochemistry. 1987;167:76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- 36.Compton LA, Johnson WC., Jr Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Analytical biochemistry. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 37.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic acids research. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics (Oxford, England) 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 39.Lobley A, Wallace BA. Dichroweb: A website for the analysis of protein secondary structure from circular dichroism spectra. Biophysical Journal. 2001;80:373A–373A. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 40.Cowley AB, Urbauer RJ, Urbauer JL. 1H, 13C and 15N NMR assignments for AlgH, a putative transcriptional regulator from Pseudomonas aeruginosa. J Biomol NMR. 2005;33:74. doi: 10.1007/s10858-005-1271-9. [DOI] [PubMed] [Google Scholar]
- 41.Anbanandam A, Bieber Urbauer RJ, Bartlett RK, Smallwood HS, Squier TC, Urbauer JL. Mediating molecular recognition by methionine oxidation: conformational switching by oxidation of methionine in the carboxyl-terminal domain of calmodulin. Biochemistry. 2005;44:9486–9496. doi: 10.1021/bi0504963. [DOI] [PubMed] [Google Scholar]
- 42.Bartlett RK, Bieber Urbauer RJ, Anbanandam A, Smallwood HS, Urbauer JL, Squier TC. Oxidation of Met144 and Met145 in calmodulin blocks calmodulin dependent activation of the plasma membrane Ca-ATPase. Biochemistry. 2003;42:3231–3238. doi: 10.1021/bi026956z. [DOI] [PubMed] [Google Scholar]
- 43.Urbauer JL, Adelman K, Brody EN. Main-chain NMR assignments for AsiA. J Biomol NMR. 1997;10:205–206. doi: 10.1023/a:1018343223888. [DOI] [PubMed] [Google Scholar]
- 44.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- 45.Demeler B. Ultrascan 7.0: An Integrated Data Analysis Software Package for Sedimentation Experiments. Department of Biochemistry, University of Texas Health Science Center at San Antonio, TX. 2005 [Google Scholar]
- 46.Durchschlag H. Specific volumes of biological macromolecules and some other molecules of biological interest. In: Hinz H-J, editor. Thermodynamic Data for Biochemistry and Biotechnology. New York: Springer-Verlag; 1986. pp. 45–128. [Google Scholar]
- 47.Cohn EJ, Edsall JT. Proteins, Amino Acids and Peptides as Ions and Dipolar Ions. Reinhold, New York, NY. 1943 [Google Scholar]
- 48.Johnson ML, Correia JJ, Yphantis DA, Halvorson HR. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demeler B, Brookes E. Monte Carlo analysis of sedimentation experiments. Colloid and Polymer Science. 2008;286:129–137. [Google Scholar]
- 50.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 51.Hubbard SJ, Thornton JM. ‘NACCESS’, Computer Program. Department of Biochemistry and Molecular Biology, University College London; 1993. [Google Scholar]
- 52.Lambert LJ, Schirf V, Demeler B, Cadene M, Werner MH. Corrigendum: Flipping a genetic switch by subunit exchange. Embo J. 2004;23:3186. doi: 10.1093/emboj/20.24.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farmer BT, 2nd, Constantine KL, Goldfarb V, Friedrichs MS, Wittekind M, Yanchunas J, Jr, Robertson JG, Mueller L. Localizing the NADP+ binding site on the MurB enzyme by NMR. Nature structural biology. 1996;3:995–997. doi: 10.1038/nsb1296-995. [DOI] [PubMed] [Google Scholar]
- 54.de la Torre JG, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophysical Journal. 2000;78:719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carrasco B, de la Torre JG. Hydrodynamic properties of rigid particles: Comparison of different modeling and computational procedures. Biophysical Journal. 1999;76:3044–3057. doi: 10.1016/S0006-3495(99)77457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de la Torre JG, Bloomfield VA. Hydrodynamic Properties of Complex, Rigid, Biological Macromolecules - Theory and Applications. Quarterly Reviews of Biophysics. 1981;14:81–139. doi: 10.1017/s0033583500002080. [DOI] [PubMed] [Google Scholar]
- 57.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 58.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 59.Naider F, Becker JM, Lee YH, Horovitz A. Double-mutant cycle scanning of the interaction of a peptide ligand and its G protein-coupled receptor. Biochemistry. 2007;46:3476–3481. doi: 10.1021/bi602415u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schreiber G, Fersht AR. Energetics of protein-protein interactions: analysis of the barnase-barstar interface by single mutations and double mutant cycles. J Mol Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- 61.Horovitz A, Fersht AR. Strategy for analysing the co-operativity of intramolecular interactions in peptides and proteins. J Mol Biol. 1990;214:613–617. doi: 10.1016/0022-2836(90)90275-Q. [DOI] [PubMed] [Google Scholar]
- 62.Horovitz A. Non-additivity in protein-protein interactions. J Mol Biol. 1987;196:733–735. doi: 10.1016/0022-2836(87)90045-3. [DOI] [PubMed] [Google Scholar]
- 63.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 64.Clackson T, Ultsch MH, Wells JA, de Vos AM. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J Mol Biol. 1998;277:1111–1128. doi: 10.1006/jmbi.1998.1669. [DOI] [PubMed] [Google Scholar]
- 65.Jin L, Wells JA. Dissecting the energetics of an antibody-antigen interface by alanine shaving and molecular grafting. Protein Sci. 1994;3:2351–2357. doi: 10.1002/pro.5560031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baxter K, Lee J, Minakhin L, Severinov K, Hinton DM. Mutational analysis of sigma70 region 4 needed for appropriation by the bacteriophage T4 transcription factors AsiA and MotA. J Mol Biol. 2006;363:931–944. doi: 10.1016/j.jmb.2006.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huyghues-Despointes BM, Pace CN, Englander SW, Scholtz JM. Measuring the conformational stability of a protein by hydrogen exchange. Methods in molecular biology (Clifton, N.J. 2001;168:69–92. doi: 10.1385/1-59259-193-0:069. [DOI] [PubMed] [Google Scholar]
- 68.Huyghues-Despointes BM, Scholtz JM, Pace CN. Protein conformational stabilities can be determined from hydrogen exchange rates. Nature structural biology. 1999;6:910–912. doi: 10.1038/13273. [DOI] [PubMed] [Google Scholar]
- 69.Sharma UK, Praveen PV, Balganesh TS. Mutational analysis of bacteriophage T4 AsiA: involvement of N- and C-terminal regions in binding to sigma(70) of Escherichia coli in vivo. Gene. 2002;295:125–134. doi: 10.1016/s0378-1119(02)00831-4. [DOI] [PubMed] [Google Scholar]
- 70.Pineda M, Gregory BD, Szczypinski B, Baxter KR, Hochschild A, Miller ES, Hinton DM. A family of anti-sigma70 proteins in T4-type phages and bacteria that are similar to AsiA, a Transcription inhibitor and co-activator of bacteriophage T4. J Mol Biol. 2004;344:1183–1197. doi: 10.1016/j.jmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 71.Gerber JS, Hinton DM. An N-terminal mutation in the bacteriophage T4 motA gene yields a protein that binds DNA but is defective for activation of transcription. J Bacteriol. 1996;178:6133–6139. doi: 10.1128/jb.178.21.6133-6139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.