Abstract
A number of brain imaging techniques have been developed in order to investigate brain function and to develop diagnostic tools for various brain disorders. Each modality has strengths as well as weaknesses compared to the others. Recent work has explored how multiple modalities can be integrated effectively so that they complement one another while maintaining their individual strengths. Bayesian inference employing Markov Chain Monte Carlo (MCMC) techniques provides a straightforward way to combine disparate forms of information while dealing with the uncertainty in each. In this paper we introduce methods of Bayesian inference as a way to integrate different forms of brain imaging data in a probabilistic framework. We formulate Bayesian integration of magnetoencephalography (MEG) data and functional magnetic resonance imaging (fMRI) data by incorporating fMRI data into a spatial prior. The usefulness and feasibility of the method are verified through testing with both simulated and empirical data.
Introduction
Since the demonstration of X-ray imaging over a century ago, medicine has exploited a series of physical methods to noninvasively characterize the structure and function of tissue that cannot be visualized directly. Anatomical and functional magnetic resonance imaging (fMRI), magnetoencephalography (MEG), electroencephalography (EEG), positron emission tomography (PET) and other methods are widely used for brain disease diagnosis and brain research. Although several techniques provide a measure of brain activation, these functional brain imaging techniques have different underlying physics and detect different physiological behaviors. These differences underlie the relative strengths and weakness of each technique. For example, MEG and EEG directly detect neural electrical currents and map brain temporal functional activity with millisecond resolution. fMRI measures the blood flow associated with brain electrical activity, limiting temporal resolution to one or a few seconds. However, spatial resolution of fMRI is better than that of MEG/EEG, and the method provides true tomographic images. Source localization by MEG/EEG requires the model-based solution of an inverse problem. PET detects positrons emitted by an injected radioactive tracer and can image the human body’s metabolism. It has intermediate resolution in both space and time compared to MEG/EEG and fMRI.
Many investigators have been seeking a unified framework within which to integrate different brain imaging modalities in a reasonable way (mathematically, computationally, and physically) so that each modality maintains its advantages while complementing each other. Several investigators described methods to combine anatomical MRI with MEG source localization (George et al., 1991; Wang et al., 1992). Dale and Sereno (1993) employed anatomical MRI as a constraint and suggested incorporating PETor fMRI information into their MEG/EEG linear estimation procedure. fMRI information was used to constrain dipole source locations (George et al., 1995; Fujimaki et al., 2002; Vanni et al., 2004; Korvenoja et al., 2001) or to seed multi-dipoles nonlinear optimization problems (Hillyard et al., 1997; Wagner et al., 2001). Liu et al. (1998) used fMRI weighting in a linear estimation procedure and found that an fMRI weighting of roughly 90% was a reasonable compromise between sensitivity to fMRI-visible and invisible sources. Babiloni et al. (2003) proposed a more generalized source correlation matrix defined on the basis of fMRI, which was applied to regions of interest.
Variational Bayesian methods have recently been employed for multi-modality integration. Daunizeau et al. (2005) investigated how the relevance of an fMRI prior in the EEG inverse problem can be assessed and in subsequent work (Daunizeau et al., 2007) proposed a symmetric information fusion technique for combining EEG and fMRI through a variational Bayesian learning scheme. Sato et al. (2004) introduced fMRI information into an MEG linear estimation procedure as a prior on the variance distribution rather than the variance itself. A variational Bayesian method approximating the original posterior distribution was used to reduce computing time. Recently, attempts to better understand the coupling between neuronal activity and BOLD responses have been reported (Sotero and Trujillo-Barreto, 2008; Vakorin et al., 2007), and in principle this information could be used to improve multi-modality integration.
Schmidt et al. (1999, 2001) proposed a novel Bayesian inference technique using Markov Chain Monte Carlo (MCMC) in which neural activity was modeled as an extended bounded region and uncertainty in the solution was explicitly addressed by sampling the posterior probability distribution. This idea was subsequently extended to a full spatiotemporal model. While the spatiotemporal analysis produced significant gains in accuracy, the MCMC sampling procedure was inefficient and slow to converge. In order to produce a more tractable and computationally practical method, Jun et al. (2005) and Jun et al. (2006a,b) developed spatiotemporal MEG Bayesian inference analysis for a multi-dipole source model. Conceptually, Bayesian inference provides a straightforward way to incorporate any prior information (such as a handcrafted map to test a hypothesis, or a region of interest derived from a probabilistic functional atlas) within a probabilistic framework. This led us to develop a combined MEG/EEG/fMRI Bayesian inference source analysis.
In this work, we develop an asymmetric Bayesian inference technique for combining MEG and fMRI by using fMRI as a spatial prior. Like our previous Bayesian inference techniques, this analysis yields many sample solutions from the posterior probability distribution, from which inferences are quantified. Most other methods yield one “best” solution, which may not be accurate, and for which equivalence classes of solutions cannot readily be determined. We formulate our procedure for a combined analysis within the Bayesian framework with appropriate assumptions that are explicitly stated. We test the feasibility and capability of the method with both simulated and empirical data.
Multi-modal Bayesian inference
Formulation
Bayesian inference analysis is a general procedure that exploits Bayes’ rule to construct a posterior probability distribution for parameters of interest from the measurements and the given prior probability distributions for all uncertain parameters. The most general version of the method employs a sampling technique to sample the posterior probability distribution. Even for complicated systems, this method is a conceptually simple and provides an easy tool to combine any prior information as well as any physical constraints for the given system. We begin with Bayes’ rule of probability:
Here θ represents parameters of interest, P(θ|Measurement) is a conditional probability distribution of θ given measurement information, P(Measurement|θ) is another conditional probability distribution of measurements given θ and commonly represents the physical relation between θ and measurement, and P(θ) is a prior probability distribution of θ.
We propose a combined spatiotemporal MEG/fMRI dipole analysis based on Bayesian inference. Assuming that sources of brain activity can be adequately modeled by equivalent point dipoles, we construct a model of neuronal currents assuming a variable number of current dipoles defined by their locations, dipole orientations, and current time courses representing dipole magnitudes over time. For most studies, we employed a fixed dipole model in which dipole locations and orientations are fixed, and dipole magnitudes vary over time. A rotating dipole model is discussed in Rotating source model section. A spherical head model based on Sarvas (1987) forward model is employed as the basis of the likelihood function,1 but this analysis could employ realistic forward models based on finite element, finite difference, or boundary element techniques that can be pre-computed and embedded in a matrix. Although the Sarvas model is specific for MEG, similar semi-analytic models exist for the EEG forward problem, and geometrically realistic forward models typically compute potentials due to currents as a first step toward computing magnetic field. Thus, the methods we describe are readily extensible for EEG.
For the given spatiotemporal measurements fMRI, MEG, and model m, our combined Bayesian formulation is as follows:
| (1) |
- MEG
T×L matrix representing observed spatiotemporal data. T is the time window size, and L is the number of channels
- fMRI
observed spatial fMRI data
- m
all model parameters including T, L, k, σi, C0, R0, α, β, η, and others. These are defined later
- N
a priori unknown number of dipole sources
- X=(X1, X2, …, XN)
vector of N dipole sources, with each Xi =(xi, yi, zi) representing the location of the ith dipole
- J=(J1, J2,…, JN)
vector of N current time courses, with each representing signed dipole moment magnitude over time of ith dipole. Negative sign means that dipole moment orientation is reversed
- Θ =(θ1, θ2, …, θN)
vector of N dipole moment orientations, with each θi representing a unit tangential direction of ith dipole
- C
TL×TL noise covariance matrix
fMRI data are transformed into statistical parametric maps and are used to inform the prior distribution of the source locations for the MEG analysis. This is not the only possible way to combine fMRI and MEG using Bayesian inference. The Discussion section addresses this point in more detail. Although fMRI and MEG are often assumed to arise from the same underlying sources,2 differences in the signal space representation can minimize the correlation observed at any particular putative source location. For our purposes, they are considered to have independent information. In spite of recent work to extract source dynamics from fMRI by employing a model of the hemodynamic response function, we assume that fMRI information conveys no temporal information on timescales relevant for MEG/EEG source dynamics and is correlated with source location only.
Priors
The parameters used in the prior distributions in this work are tabulated in Table 1
Table 1.
Priors for unknown parameters
| Parameter | Distribution type | Formula | |
|---|---|---|---|
| N | Uniform |
|
|
| Θ | Uniform |
|
|
| J | Gaussian |
|
|
| C | k-th order Inverse Wishart |
|
|
| X | Exponential |
|
We chose priors on the number of dipoles N and orientation Θ to be uniform distributions within regions of interest such as N ∈ [Nmin, Nmax] and Θ ∈ [0, 2π]. Nmin is commonly set to 0; Nmax is more problem-specific and we set it to up to 10 in this work. In particular, a Poisson distribution on N may be a useful alternative.
A Gaussian distribution with mean of 0 and temporal correlation of C̄ time courses J. In Table 1, σi represents the prior standard deviation of time-varying current magnitudes of ith dipole, and we set it to 20 nAm for all dipoles. A temporal correlation C̄ is parameterized using a control parameter β as . β is determined according to the size of temporal window. In the Experiment section, β was 2.0 for the 21-ms time window and 4.0 for the 60-ms time window.
A prior on noise covariance C is chosen as the kth order inverse Wishart distribution with asymptotic mean of C0. Wishart is a distribution typically used to describe the covariance matrix of multi-normal samples when the inverse of the covariance matrix C0 can be estimated from the obtained available noise data (Huizenga et al., 2002; Jun et al., 2006a,b; Plis et al., 2007a,b). Zc is a normalizing constant and k was set to 2LT + 1 which is slightly more than twice of the dimension of matrix C.
For the prior distribution of the source locations that depends on the fMRI data, we chose an exponential distribution as shown in Table 1. For the given fMRI t-statistics pX, the pX values are scaled to have a maximum of 1 and any value less than 0.1 (Background fMRI weighting) is set to 0.1. We used two control parameters (α and η) to give various weighting of fMRI. α is a scale factor controlling how strongly the fMRI information influences the analysis, and η is a control parameter describing sharpness of the fMRI distribution and controls the difference between fMRI weightings at highest fMRI spots (pX =1) and at background fMRI contrast (pX =0.1: background fMRI contrast thresholding parameter). Basically, higher α and η yield stronger weighting by fMRI. Zx is a normalizing constant.
We remark that location priors can be employed from other sources, including probabilistic atlases of neural function, or even assumptions or hypotheses regarding putative sources of observed activity. However, we note the caveat that unlike spatial filters, inappropriate priors might skew the analysis without producing obvious issues. We note that several recent papers attempt to extract source dynamics from fMRI data, typically by employing models of the hemodynamic response function (e.g. the balloon model (Buxton et al., 1998) or the Windkessel model (Friston et al., 2000)) However, these predict dynamics of the timescale of the hemodynamic response (hundreds of milliseconds to second) and produce little information of direct relevance for predicting MEG/EEG source dynamics. This problem is further exacerbated by differences between experimental and analytical protocols for MEG and fMRI used in these experiments (i.e. evoked response vs. steady state stimulation) and by much of the functional neuroimaging community.
Marginal posterior and sampling
We have observed that posterior distribution sampling techniques for this sort of complicated problem have more tendency to get trapped in local maxima (high complexity) and require substantial time to explore the whole range (slow convergence). To reduce these problems, noise covariance is treated as an uncertain parameter in Bayesian parlance and the formulation is marginalized along with the vector of current time courses and over the noise covariance matrix:
| (2) |
This marginalization allows us to greatly reduce the degrees of freedom and produces a smoother posterior, thereby producing more tractable sampling. Even though the current time course is marginalized in the above formulation, we separately sample the current time course from the approximate normal distribution for the sampled parameters {N, X, Θ} (Jun et al., 2005).
We used a Markov Chain Monte Carlo (MCMC) sampling technique to numerically estimate the posterior probability distribution (Gilks et al., 1996; Gelman et al., 2004). The transition probabilities in MCMC are designed so that the resulting probability distribution of samples converges to the targeted distribution as the number of samples approaches infinity (Green, 1995). In order to sample without prior determination of the number of dipoles, a trans-dimensional sampling strategy – jumps between subspaces of different dimensions – is needed. We adopted the reversible-jump MCMC combining classical Metropolis moves with reversible jump (RJ) moves and allowing movement between different parameter spaces that satisfies the detailed balance criterion (Green, 1995; Jun et al., 2005).
Although the MCMC sampling technique has important and unique strengths, it also has technical limitations that must be recognized for greatest utility. Theoretically, MCMC can be guaranteed to yield well-sampled inferences after long computation. But we experienced multi-modal behavior for complicated posterior distributions. This problem was often apparent as slow convergence, with long plateaus of suboptimal models, with abrupt transitions to a higher probability plateau. To address this problem, we explored the utility of systematically modulating the variance of update proposals in analogy to techniques such as simulated annealing. We are also considering the value of new types of trans-dimensional update proposals such as source splitting/combining. This strategy is motivated by our observation that nearby source locations with correlated time courses may be difficult to resolve on the basis of MEG-alone, especially by a random walk through the model parameter space.
Active time range
Each dipole source may have its own specific temporal behavior. Some have a short duration of activity relative to the total time window to be analyzed. Detectability, localization, and time course determination of such sources may be adversely affected when sources are allowed to be active during a much longer analysis. In an effort to improve detection and separation of such sources, we have introduced additional parameters into our Bayesian framework including the active time range—starting and ending time points ( ) for each dipole (Jun et al., 2006a,b). We incorporate these parameters tS,E={tS, tE}, into our combined analysis:
| (3) |
Prior on tS,E is chosen as a uniform distribution:
| (4) |
Throughout this work, we use the active time range information.
Rotating source model
In the above section, a fixed dipole source model was assumed to represent neural activity. However, a rotating source model –where locations are fixed over time, and orientation and dipole magnitudes vary over time – is applicable within the same computational framework. Assuming a rotating source model, dipole orientations Θ and current time courses J are merged into one parameter Jrot consisting of 2×T (or 3×T in realistic head model) matrices such as . So the final marginal posterior distribution is given as
| (5) |
In analogous fashion, the posterior distribution with Jrot can be sampled in our present formulation. We find that a rotating dipole model requires a 2 (or 3) times larger matrix to be inverted than calculating the posterior distribution for a fixed dipole model. This matrix inversion is associated with marginalizing over the current time courses in the MCMC procedure. Even though this causes an increased computational burden for rotating dipoles, this burden is still less than numerically sampling over the time courses. We have found that the increased degrees of freedom in a rotating model allow a better fit to empirical measurements with fewer dipoles than a fixed model. A comparative test between these two source models is discussed in the next section.
Noise covariance estimation
A diagonal noise covariance matrix, which assumes there are no temporal noise correlations and no cross correlations in space, is often employed in source analyses (Plis et al., 2007a,b). A diagonal noise covariance can be expressed as a Kronecker product of a diagonal matrix (spatial covariance) consisting of sensor variations and the identity matrix (temporal covariance). We have observed that using a diagonal noise covariance in empirical data analysis3 yielded many spurious dipole sources (Jun et al., 2005), which appear to model correlations typically present in empirical data. To overcome this difficulty, we developed a multi-pair Kronecker product covariance model for spatiotemporal noise analysis (Plis et al., 2007a,b). A multi-pair Kronecker product covariance model is composed of sum of pairs of Kronecker products of spatial covariance components and temporal covariance matrices, which allows easy inversion:
| (6) |
For more details, refer to Plis et al. (2007a,b). For the present work, we employed this multi-pair Kronecker product covariance model. We used a diagonal covariance model to test how our proposed combined analysis affects spurious dipole sources. For the cases of limited noise information case, a Toeplitz covariance model (Jun et al., 2006a,b) may be an acceptable alternative.
Experiments: simulated data
In this section, we investigate how combined MEG/fMRI analysis performs in various cases, evaluating the effect of a spatial fMRI prior on MEG source localization accuracy. We acquired MEG and fMRI data for median nerve stimulation of the same subject (male, age 30–40). Both fMRI and MEG experiments were conducted using the same stimulation conditions, on the same day.
MEG data
Median nerve stimulation at the motor twitch threshold was applied using a block design of 30 s on, 30 s o ff for a total of 10 blocks for each of 8 runs. Data were acquired during both stimulation on and off epochs, the latter being used to construct the present noise data set. Stimuli alternated across runs, with four runs of left side stimulation and four runs of right side stimulation. The ISI (interstimulus interval) was randomized between 0.25 and 0.75 s. Data were collected with 1 kHz sampling on a 4 D Neuroimaging Neuromag-122 whole-head gradiometer system with 122 channels (Ahonen et al., 1993). One of 122 channels was discarded due to malfunction. Data were 1-median filtered. Sixty hertz of noise and harmonics was filtered out by removal of peaks in the spectrum and interpolation between adjacent spectrum points.
fMRI data
fMRI and structural MRI data were acquired on a Phillips 1.5T whole body scanner. Left and right median nerve stimulation at a strength sufficient to elicit a strong thumb twitch was applied using a block design of 30 s on, 30 s off with ISI varying randomly from between 0.25 and 0.75 s. fMRI volumes were acquired sagittally, at a 4×4 mm resolution with 6 m m slice thickness; 1 vol=21 slices, TR=3. Four runs were performed and stimulus was alternated between the left and right sides for each run.
Preprocessing and statistical analysis of the data were performed using SPM2b software (Wellcome Department of Cognitive Neurology, London). Preprocessing included realignment of each of the functional scans separately to remove movement artifact; coregistration of the functional scans to structural MRI and spatial smoothing with a 6-mm Gaussian kernel. As this is single subject data, there was no need to normalize the data which would warp it into the space of a template or composite brain. The data were modeled using a boxcar function, as an alternative to convolving with the canonical hemodynamic function. The first 3 acquisitions of each of the 30-s blocks were removed. In addition, the first five acquisitions prior to stimulus presentation were removed. In combination, this resulted in the use of 70 volumes per run. The between-condition comparison was implemented as a linear contrast. Student’s T-test maps were obtained for both stimulus conditions. The use of these T-test maps for MEG/fMRI integration is discussed below. To verify that fMRI results were reasonable, the statistical maps were thresholded at p>0.0001 (uncorrected for multiple comparisons) with an extent threshold of 4 voxels. The statistical maps show strong fMRI BOLD response activity in the contralateral S1 and S2 areas, as well as indications of activity in a few other regions.
In order to perform a Bayesian combined MEG/fMRI analysis, the fMRI T-test maps needed to be converted to a form usable in the combined analysis. The structural MRI (sMRI) scans obtained in conjunction with the fMRI scans were truncated such that the left and right MEG fiducial locations were not acquired. For this reason, the T-test maps were registered to a different sMRI of the same subject, which included the three required MEG fiducial locations. First, the truncated sMRI data were registered to the usable sMRI data using a surface matching procedure contained in MRIVIEW (Ranken et al., 2002). The transformation matrix obtained was applied to the T-test maps to obtain T-test volumes interpolated to 2 mm resolution and aligned to the usable sMRI. The MEG to sMRI homogeneous transformation matrix was also obtained using MRIVIEW, this time using the MEG fiducials. The MEG to sMRI transformation matrix could then be used to register MEG coordinates to the T-test volume coordinates.
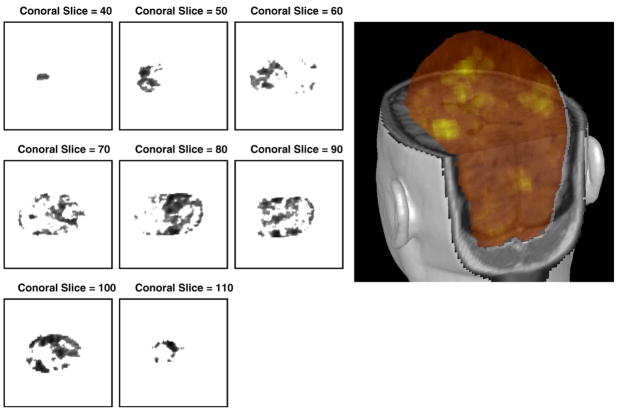
Each T-test volume was scaled so that positive T-test values, corresponding to an increased BOLD response, fell in the [0,1] interval. We call this a likelihood volume because it provides a measure of the likelihood that brain activity was present at a given voxel location, based on the fMRI analysis. Additional nonlinear scalings were applied to the voxel values of this likelihood volume in the combined analysis, as described in Priors section. We note that it would be possible to scale the entire range of T-values (i.e. to include negative values) in the T-test volume into [0,1]. This would create exclusion regions that would have a lower likelihood of brain activity in the combined MEG/fMRI analysis than they would in the corresponding MEG-only analysis. However, scaling the entire range of T-values was not investigated in this work. Fig. 1 shows the location prior distribution P(X|N, fMRI, m) (single dipole case) containing transformed fMRI volume probability for varying coronal view slices from bottom to top (40, 50,…, 110) and the corresponding 3D overlayed picture in MRIVIEW.
Fig. 1.
Location prior distribution P(X|N, fMRI, m) (single dipole case). Left: Coronal slice views from slice 40 to 110 (increment 10). Dark regions correspond to high probability. Right: 3D overlayed picture in MRIVIEW. Bright yellow regions correspond to high probability.
Simulation Study 1: localization performance by fMRI effect
Because the ground truth of source information on empirical data is not known, it is hard to estimate realistic comparative fMRI effects. For our purposes we generated simulated MEG data sets mimicking empirical MEG data in the following way:
Localize sources from empirical MEG data (time windows of 21 ms/60 ms right after 14 ms stimulation onset) through spatiotemporal MEG Bayesian inference dipole analysis (Jun et al., 2005).
Use the obtained localized source information (most probable) and generate simulated sensor activation through the spherical forward model (Sarvas, 1987).
Add empirical MEG noise (collected from prestimulus region) to the modeled sensor data.
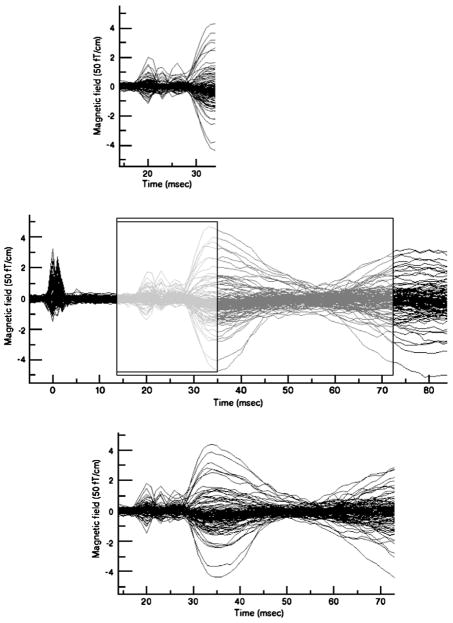
These simulated MEG data represent the best (maximum a posteriori) fits to empirical MEG data. We expect that like original empirical MEG data these simulated MEG data will be spatially correlated with empirical fMRI data. Fig. 2 depicts how the two simulated MEG data sets are very similar to the empirical MEG data within the corresponding time windows. Although we know the source locations of the simulated data, the MCMC algorithm does not. The algorithm is not initialized with a starting parameter set provided by the analyst as is common in nonlinear source localization. Initialization of the Markov Chain is stochastic and is performed by the algorithm itself.
Fig. 2.
Two simulated MEG data sets mimicking empirical MEG data (Middle) with time windows of 21 ms (Top) and 60 ms (Bottom). The middle panel shows the empirical data.
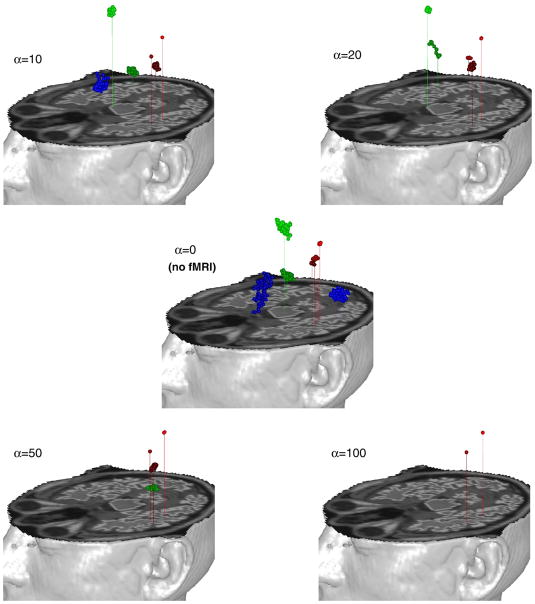
We used one of these simulated MEG data sets (time window of 21 ms) and the empirical fMRI data and applied the combined Bayesian inference analysis. In the analysis, we used various fMRI weightings, which are generated by varying α=0 (no fMRI information), 5, 10, 20, 50, 100 and η=0.5, 1.0, 1.5, 2.0. We note that the higher α and η yield the stronger fMRI effect in the analysis. For each case, we ran an MCMC run, generating 10000 samples4 for each. We discarded the first 2000 samples as a burn-in-period and collected the remaining 8000 samples for the analysis. For each case, we computed localization errors from averaged source locations and averaged current time courses for collected samples. The comparative localization performance was tabulated in Table 2. This shows that the fMRI prior clearly plays some role in this analysis. Interestingly, weak fMRI effects (α=5, 10, 20 and η=0.5 or 1.0) showed slightly better localization performance than no fMRI effect, while strong fMRI weights often degraded localization performance. This suggests an implicit correlation (but not identity) between sources of fMRI and MEG data. Our Bayesian inference technique exploits this imperfect correlation to produce synergy in the combined analysis. For another simulated MEG data set (time window of 60 ms), we did the same test and saw similar behavior to previous results. In Fig. 3, localized source blobs (100 samples per each) were shown for α=0 (red in color), 20 (blue), and 50 (green), keeping η=1.0. A set of dipole sources in the middle is expanded and used to investigate how log posterior probability in MEG-alone analysis and log location prior probability conditional on variation of fMRI volume probability behave on the line passing centers of all three colored blobs. We computed both probabilities as one dipole is moving over segment, keeping the other dipoles fixed. As expected, fMRI priors pushed dipole sources toward high fMRI contrast, and higher fMRI weighting attracted sources more strongly.
Table 2.
Localization results for the 21-ms time window data for varying α and η
| α | η | Location error (mm) | Current time course error (nAm) |
|---|---|---|---|
| 0 | – | 0.893 | 0.094 |
| 0.5 | 0.856 | 0.117 | |
| 1.0 | 0.558 | 0.078 | |
| 1.5 | 1.031 | 0.180 | |
| 5 | 2.0 | 2.732 | 0.401 |
| 0.5 | 0.710 | 0.102 | |
| 1.0 | 0.424 | 0.083 | |
| 1.5 | 1.526 | 0.214 | |
| 10 | 2.0 | 2.917 | 0.368 |
| 0.5 | 0.633 | 0.111 | |
| 1.0 | 1.978 | 0.306 | |
| 1.5 | 2.946 | 0.347 | |
| 20 | 2.0 | 3.281 | 0.360 |
| 0.5 | 1.056 | 0.203 | |
| 1.0 | 3.065 | 0.365 | |
| 1.5 | 3.903 | 0.566 | |
| 50 | 2.0 | 4.399 | 0.438 |
| 0.5 | 2.309 | 0.357 | |
| 1.0 | 3.475 | 0.301 | |
| 1.5 | 4.399 | 0.438 | |
| 100 | 2.0 | 5.613 | 0.652 |
Fig. 3.
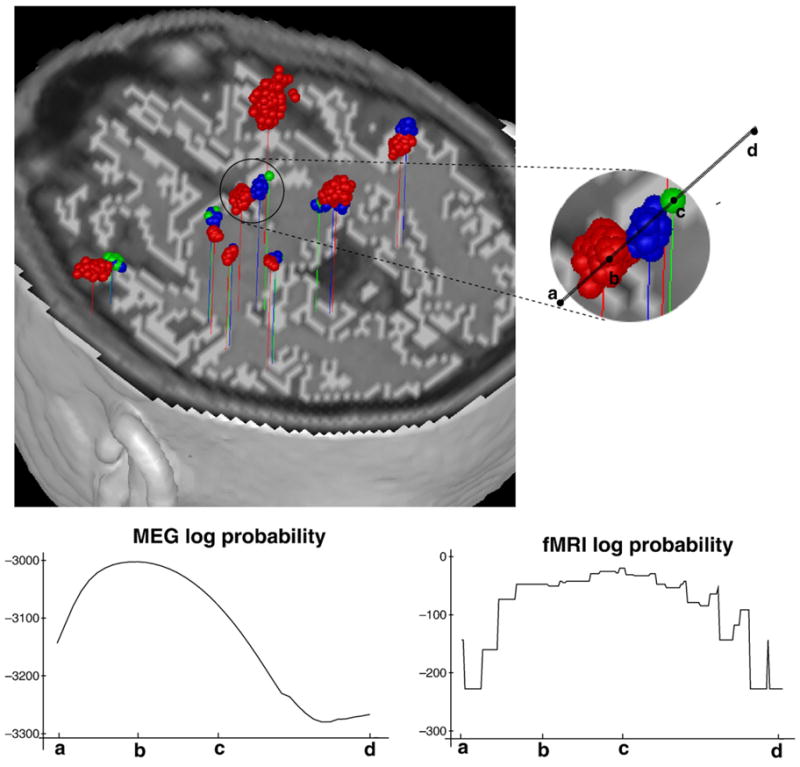
Top: Dipole localization result on 60 ms time window. MEG-alone analysis with no fMRI effect (α=0: red color) and combined analysis with fMRI effects (α=20, η=1.0: blue color, α=50, η=1.0: green color). Bottom: Posterior probability for MEG-alone analysis (left) and location prior probability conditional on variation of fMRI volume probability when α=50, η=1.0 (right) on segment from points a to d. Ending points “b” and “c” at colored blobs are the averaged locations of each blob.
The simulated MEG data were intended to provide a known “ground truth” for our analysis. Note that in this study, the algorithm is not initialized with a starting parameter set provided by the analyst as is common in nonlinear source localization. Initiation of the Markov chain is stochastic. In both simulated data and in analysis of the corresponding empirical data, across multiple runs (with different chains), we have never seen multi-modal behavior that lasted throughout the analysis. Eventually, the trans-dimensional jumps allow convergence on the known sources (in simulated data), and similar results were observed in the empirical data on which the simulations were based.
Simulation Study 2: apparent in fMRI but not in MEG
In addition, we investigated the detectability of source dipoles located around fMRI hot spots (high likelihood of activation) that are not detected by the MEG analysis alone. For this purpose, we generated different MEG simulated data as follows:
Select region identified during fMRI analysis as being the most significantly activated and generate information assuming a single dipole in that region which is very weak.
Combine this simulated, under-threshold, dipole information with the localization source information obtained in MEG data section for the empirical MEG data within a time window of 21 ms in order to calculate a simulated sensor activation.
Add empirical MEG noise (collected from prestimulus region) to the calculated sensor activation.
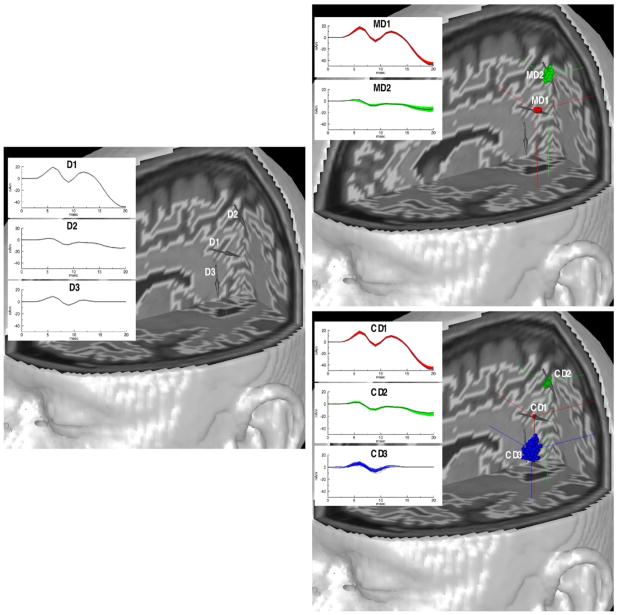
We employed this simulated MEG data for the MEG analysis and the proposed combined MEG/fMRI analysis. Fig. 4 shows the original three dipole sources problem (left) and two reconstructed results (right) for both analyses. D1 and D2 are detectable dipole sources apparent in MEG, and D3 is a dipole source too weak to be apparent in MEG, but apparent in fMRI. Although the MEG-only analysis did not localize the D3 dipole (i.e. produced no peak in the source probability volume), the combined analysis localized all three dipole sources (CD1, CD2, and CD3) successfully. This shows that the combined MEG/fMRI analysis has the ability to improve the detectability of source dipoles that are apparent in fMRI, but invisible in MEG.
Fig. 4.
Left: Original three dipole problem. Right: The MEG analysis result with no fMRI data (upper) in which the third source is not found and the combined analysis result with fMRI weighting, α=10 and η=1.0 (lower) in which the third source is found. Gray arrows and gray current time courses in figures represent original source information. Reconstructed red blobs (MD1 and CD1), green blobs (MD2 and CD2), and blue blob (CD3) correspond to original sources D1, D2, and D3, respectively.
Regarding case of being apparent in MEG but not in fMRI, it is clear that the asymmetry in our analysis procedure tends to minimize this problem. The analysis is fundamentally driven by the electromagnetic data, and if the MEG evidence is strong it generally produces a source. This is the reason that we employ a finite background probability in the fMRI prior. It is possible that the absence of fMRI evidence for a source apparent in MEG, coupled with a nearby fMRI hot spot with no corresponding MEG evidence will produce special problems, but we consider this to be a case of an effectively ”misplaced fMRI source”, which clearly can introduce errors in the analysis.
Simulation Study 3: two nearby sources apparent in fMRI
We investigated the separability of two nearby source dipoles in the combined analysis. According to previous work on MEG dipole analysis (Jun et al., 2006a,b), two nearby dipoles with a separation distance of 12 mm or less are typically difficult for MEG analysis to discriminate, and these sorts of two dipole problems are more likely to be localized into one equivalent current dipole located between the two dipole sources.
In this simulation, we chose two dipole sources that are closely located (interdistance of 10 mm) and temporally correlated. fMRI information was synthetically generated to yield two discrete (disconnected) high contrast nearby regions. Each fMRI contrast region contains a dipole source, which means two sources are apparent in fMRI. Fig. 5 depicts such a two dipole source problem (left) and comparative analysis results (right) showing source blobs (100 sample per each) and the corresponding averaged current time courses in the right lower inset. It shows that the combined analysis could separate the two dipoles well, while the MEG analysis alone could not separate them. The MEG analysis alone typically yielded one equivalent dipole located between the two sources and the corresponding current time course very similar to the vector sum of two original current time courses. From this result, it is clear that the combined analysis can improve separability of two nearby sources when they are discretely apparent in fMRI.
Fig. 5.
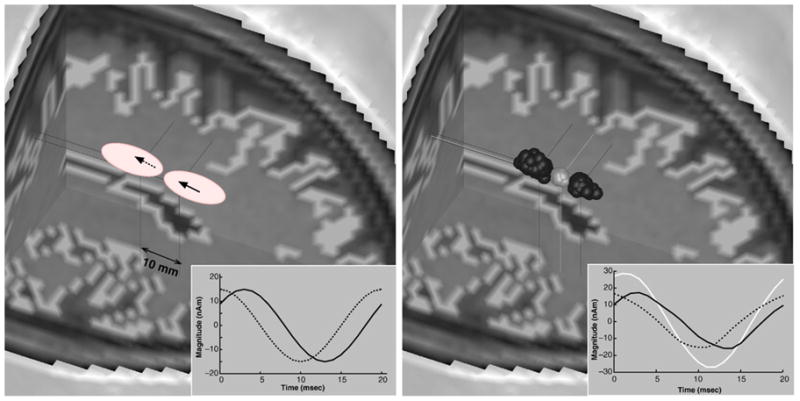
Left: Original two dipole problem with closely located sources. Right: Comparison of MEG analysis (bright gray) and combined MEG/fMRI analysis (dark gray) results. Insets describe current time courses for original problem (left) and estimated time courses (right, bright gray: MEG-alone analysis, dark gray: combined analysis).
Experiments: empirical data
Next we applied our combined analysis of MEG and fMRI data for right hand median nerve stimulation, which was acquired as described in the previous section. As depicted in the middle of Fig. 2, a 21-ms time window was selected for analysis. We compared MEG analysis (with no fMRI weighting) and the combined analysis with various fMRI weightings.
In Fig. 6, results for MEG analysis (red) and for the combined analysis (blue and green) are compared. Two sources are consistently localized and are closely located for all cases. Just like the results for simulated data, we see that the higher fMRI weighting yields sources moving slightly farther toward locations where high fMRI contrast is located.
Fig. 6.
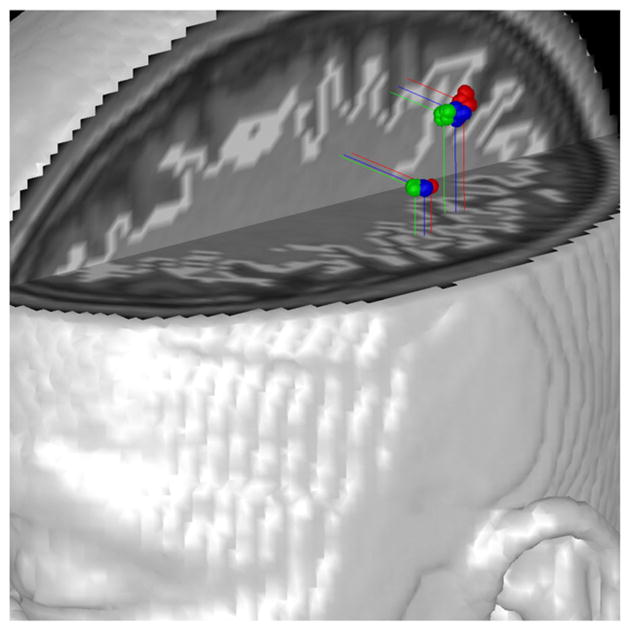
Empirical data (21 ms window). Analysis results for MEG analysis alone (red) and the combined analysis (blue: α=20, η=1.0; green: α=50, η =1.0).
Thus far, we have used a fixed dipole source model for all experiments. A number of investigators prefer to use a rotating dipole source model rather than a fixed dipole source model. A rotating dipole allows the same source to account for more general activity in a given region even if the detailed distribution of activity varies across time. In this paper, we do not investigate which model is more reasonable or useful for our analysis, but we check to see if both source models work adequately with the combined analysis. With our analysis, we tested two dipole source models with the same empirical MEG data used in the previous work. Fig. 7 shows the comparative results for the two source models. Two sources are consistently localized and only minor differences in locations and orientations were seen. This suggests that for the empirical MEG data (over a relatively short time range), our analysis does not show any significant differences between source models. However, analysis over a longer time interval data produced more complicated dipole source distributions. Some of these dipoles largely canceled each other and may represent noise. We found that using a rotating dipole source model could reduce such phenomena, thereby reducing the number of localized dipole sources. However, a rotating dipole source model requires a bigger matrix inversion and thus requires more computational time than a fixed dipole source model. Nevertheless, we expect our combined analysis to be applicable with either a fixed dipole model or a rotating dipole mode.
Fig. 7.
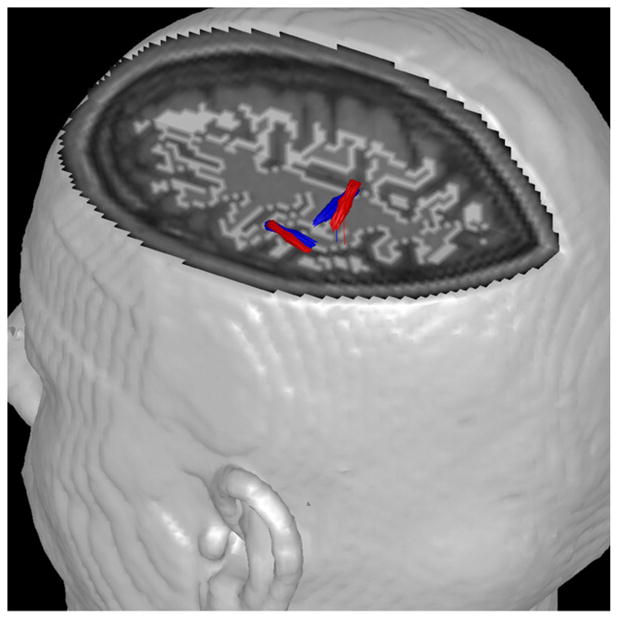
Empirical data (21 ms window). Analysis results for the combined analysis with fMRI weighting using α=20, η=1.0 (red: fixed dipole source model; blue: rotating dipole source model). The orientations for the rotating model are from the peak amplitude time slice. Many arrows are seen closely overlapped.
Effects of noise model
Thus far, we have used a multi-pair Kronecker product spatiotemporal noise covariance model for MEG analysis and the combined analysis. As described in the Noise covariance estimation section, when we use a conventional diagonal covariance in MEG analysis alone rather than a multi-pair Kronecker product covariance model, we observed many spurious dipole sources (Jun et al., 2005). We investigated whether this phenomenon is observed in the combined analysis. We employ the same empirical MEG data (21 ms time window) and use a diagonal noise covariance estimation in both the MEG analysis alone and the combined analysis.
Fig. 8 shows the results for MEG analysis alone (α=0) and the combined analysis with various fMRI weights (α=10, 20, 50, 100, η=1.0). The MEG analysis localized up to 7 different dipole sources, while the combined analysis localized 2 to 6 dipole sources. Among localized dipole sources, 2 or 3 dipole sources in red (dark and light) in the figures are seemingly consistent with results of Fig. 6, which are believed to be associated with right hand median nerve stimulations. The remaining broader clustered dipole sources (blue and green) are spread over the brain and their corresponding current time courses (not shown here) jitter around zero over time, suggesting these are spurious dipole sources arising from the mismatch between empirical noise structure and the noise covariance model. Interestingly, higher fMRI weighting showed less spurious dipoles. This experiment suggests that fMRI data can play a significant role in filtering out spurious sources in the combined analysis.
Fig. 8.
Empirical data (21 ms window). Analysis results using diagonal noise covariance estimation for MEG analysis (α=0, no fMRI effect) and the combined analysis with various fMRI weightings (α=10, 20, 50, 100, keeping η=1.0). Red (dark and light) blobs in each figure seem to be associated with right hand median nerve stimulations. Those locations are consistent with the results of Fig. 6. Green (dark and light) and blue (dark and light) blobs are likely to be spurious sources arising from mismatch between empirical noise and noise model.
Discussion
In this paper, we describe a technique for combined MEG/fMRI Bayesian inference multiple dipole analysis. Although both MEG and fMRI are spatiotemporal data, MEG has greater temporal resolution. Also, the steady state stimulation protocol and block design analysis we employ (and used by many investigators) sacrifices the dynamic resolution available in event-related fMRI analysis in favor of more reliable source localization. This time scale discrepancy leads us to treat fMRI as spatial data in the analysis. The degree of spatial correlation between MEG and fMRI is a matter of active investigation (Marrelec et al., 2005; Singh et al., 2003; Kober et al., 2001); investigators debate how and to what degree they are correlated. Due to the incomplete understanding of the correlation between the methods, in our analysis we treat fMRI and MEG data as independent information.
We appreciate the appeal of symmetrical treatment of multiple data sources (Daunizeau et al., 2007) and believe that this work takes a more balanced approach than previous work by ourselves and others (e.g. using fMRI location constraints or dipole seeding). However, we acknowledge that the methods have inherently asymmetrical strengths and analytical requirements. For example, we do not believe that fMRI response dynamics contribute meaningfully to the estimation of MEG/EEG source dynamics. Source localization (and subsequent time course estimation) by MEG and EEG requires a model-based solution to an inverse problem, that is the core of our Bayesian method. fMRI produces statistical parametric maps that are a starting input for our procedure. Attempts to account for uncertainties in fMRI source localization would deal with empirical issues of vascular anatomy and function rather than physical issues that are readily modeled. Given these fundamental differences between the methods, a more symmetrical treatment is not easily developed or justified. On the other hand, a symmetrical approach to combined analysis of MEG and EEG might better exploit the complementary strengths of these methods.
There are other ways to generate fMRI weightings than our proposed exponential distribution prior. However, our strategy enabled us to easily incorporate fMRI prior into the Bayesian formulation and yielded a concise marginal posterior distribution. In the Simulation study 1 section, we investigated comparative localization performance over various fMRI weightings controlled by two parameters α and η. Even though optimal parameters may be dependent on the MEG data to be analyzed, we have continued to use α=10 and η=1.0 for other experiments since they yielded the best localization performance in the simulation study. Alternatively, these fMRI weighting parameters can be estimated by incorporating them as hyperparameters into the Bayesian framework to produce a weighting that is optimal by some criterion. However, our previous experience with a similar approach (Plis et al., 2007a,b) cautions us that simultaneous optimization of linked parameters can increase uncertainty without providing significant improvement in model quality.
Liu et al. (1998) claimed that for their combined analysis a fMRI weighting of roughly 90% was a good compromise between sensitivities of fMRI-visible and invisible sources. This weighting is more closely related to thresholding probability on the background of fMRI, which was set to 0.1 in our work. The work of Liu et al. (1998) did not consider the variation of fMRI contrast over neocortex. They considered fMRI as a constant weighting over superthreshold regions of cortical activation in their analysis. In our combined analysis, higher fMRI contrasts had higher probability values. Thus, equivalent current dipoles are more attracted to higher fMRI contrasts, as depicted in Fig. 3.
The combined Bayesian inference analysis proposed here has two unique points to be highlighted. First, our combined Bayesian inference analysis has a different strategy to deal with fMRI from others. Most MEG/fMRI or EEG/fMRI combined analysis in the literature are MEG/EEG analyses using fMRI to seed dipole locations, or to constrain the allowed region of source space, or employ fMRI as a source correlation or source amplitude prior. Our combined Bayesian inference analysis treats fMRI as a prior probability distribution on source location and incorporates it into the Bayesian inference analysis. Our analysis also exploits fMRI contrast variation.
Second, unlike other methods (Hämäläinen et al., 1993) giving the best one solution based on various norms, or optimization criteria, the combined Bayesian inference analysis yields the probability distribution of likely solutions. This is a good strategy to overcome the inherent ill-posed nature of the electromagnetic inverse problem. By considering the probability distribution of likely solutions, better understanding and statistical inference is possible.
The MCMC sampling technique implemented in this work uses two kinds of jump movements. One is a trans-dimensional jump consisting of dipole birth and dipole death. The other is a fixed dimensional jump consisting of dipole location change, dipole orientation change, and active time range change. Through numerous supervised analysis tests (essentially trial and error), we tried to optimize MCMC sampling parameters such as jump probabilities for each jump and proposal distributions for each jump. Sampling performance got worse as the time window to be analyzed got larger. Presumably the pattern of brain functionality is more complex in the longer time window, and the final posterior distribution becomes too rugged to be adequately sampled. We continue to seek the best conditions and jumping strategies for optimal performance of MCMC sampling in the combined analysis.
As in Jun et al. (2005), in order to speed up the evaluation of the final marginal posterior distribution, the formulation in spatiotemporal source space is transformed from original time variables to eigentime variables,5 keeping the significant ones and neglecting the insignificant ones. MCMC is run in this truncated spatio-eigentime space and then the formulation is transformed back into original time variables.
In Simulation study 3 section, we investigated the utility of the combined analysis for separating nearby sources, which are closely located (interdistance of 10 mm) and strongly temporally correlated. This simulation was done using the background fMRI thresholding probability of 0.1 and sources were successfully separated by the combined analysis. We believe this separability is more related to background fMRI thresholding probability rather than to other parameters. The smaller background fMRI thresholding probability improves separability of nearby sources. However, it has little effect on the detection of invisible fMRI sources apparent in MEG.
Our proposed combined analysis was able to eliminate spurious sources in some cases, a natural advantage when background noise and fMRI information are not correlated. However, weak fMRI weighting often produced better localization performance. In any case, sophisticated noise covariance estimation produced clear benefits for the combined analysis.
In order to focus on our primary objective of demonstrating and evaluating our Bayesian approach to multi-modality integration, we adopted the strategy of limiting our analysis to a relatively small, specified temporal window. Although analogous techniques are routinely used for MEG/EEG source localization through nonlinear spatiotemporal optimization, the approach violates our preferred strategy of allowing the data and the algorithm to determine the details of the model. The practical need for a restricted temporal analysis window is not due to a simple implementation or numerical issue; it probably reflects the fact that a small transient source in a long window carries relatively little statistical power relative to other physiological and noise sources. We are considering alternative analysis strategies such as rolling windows that achieve the same effect without the arbitrary specification of time ranges. Even more attractive is the idea of defining a source as an entity limited in both spatial and temporal extent, so that temporal limits are a natural part of the source definition.
Acknowledgments
This work was supported by NIH grant 2 R01 EB000310-05, the Mental Illness and Neuroscience Discovery (MIND) Institute, the Gwangju Institute of Science and Technology (GIST) Faculty Start-up Fund, the BioImaging Research Center at GIST, and the Korea Meteorological Administration Research and Development Program under the Grant CATER 2007-5108.
Footnotes
Our analysis includes a likelihood function coming from the MEG Sarvas model. We do not include a term in the likelihood function for fMRI based on a hemodynamic response model. Instead, fMRI information is incorporated into a location prior. As the coupling between the BOLD response and neural electric current is more clearly understood, it will be interesting to include fMRI hemodynamic response as a likelihood function.
The correspondence between MEG and fMRI sources is a common starting assumption that appears true to first order(Sanders et al., 1996). It is almost certainly not true in detail since the vagaries of cerebral vasculature anatomy, patterns of parenchymal flow, draining veins, regional differences in oxygen extraction, etc., can all effect the correspondence. Also, experimental work (Logothetis et al., 2001) suggests that the BOLD response is more closely correlated with gamma band oscillatory behavior than with evoked potentials or changes in spike rate. While this may not affect source localization, it could in principle influence reconstructed MEG/EEG dynamics based on fMRI.
Since a diagonal noise covariance assumes no temporal correlation, we used no temporal prior by setting β (explained in the Priors section) to zero even though empirical data has temporal correlation in both signal and noise. We found more spurious dipole sources which try to model correlations in noise when we used a temporal prior with nonzero β.
Each sample was randomly chosen among 10 iterations, so we required 100000 iterations for each run.
Temporal correlation matrix C̄ in the current time courses prior is used to decompose a total spatiotemporal source space into significant spatio-eigentime space and less significant (negligible) spatio-eigentime space.
References
- Ahonen AI, Hämäläinen MS, Knuutila JET, Kajola MJ, Laine PP, Lounasmaa OV, Parkkonen LT, Simola JT, Tesche CD. 122-channel SQUID instrument for investigating the magnetic signals from the human brain. Phys Scr. 1993;T49:198–205. [Google Scholar]
- Babiloni F, Babiloni C, Carducci F, Romani GL, Rossini PM, Angelone LM, Cincotti F. Multimodal integration of high-resolution EEG and functional magnetic resonance imaging data: a simulation study. NeuroImage. 2003;19:1–15. doi: 10.1016/s1053-8119(03)00052-1. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Wong EC, Frank LR. Dynamics of blood flow and oxygenation changes during brain activation: the balloon model. Magn Reson Med. 1998;39:855–864. doi: 10.1002/mrm.1910390602. [DOI] [PubMed] [Google Scholar]
- Dale A, Sereno M. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction. J Cogn Neurosci. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Daunizeau J, Grova C, Marrelec G, Mattout J, Jbabdi S, Pelegrini-Issac M, Lina J, Benali H. Symmetrical event-related EEG/fMRI information fusion in a variational bayesian framework. Neuro-Image. 2007;36:69–87. doi: 10.1016/j.neuroimage.2007.01.044. [DOI] [PubMed] [Google Scholar]
- Daunizeau J, Grova C, Mattout J, Marrelec G, Clonda D, Goulard B, Pelegrini-Issac M, Lina JM, Benali H. Assessing the relevance of fMRI-based prior in the EEG inverse problem: A Bayesian model comparison approach. IEEE Trans Signal Process. 2005;53:3461–3472. [Google Scholar]
- Friston KJ, Mechelli A, Turner R, Price CJ. Nonlinear responses in fMRI: The balloon model, volterra kernels, and other hemodynamics. NeuroImage. 2000;12:466–477. doi: 10.1006/nimg.2000.0630. [DOI] [PubMed] [Google Scholar]
- Fujimaki N, Hayakawa T, Nielsen M, Knosche TR, Miyauchi S. An fMRI-constrained MEG source analysis with procedures for dividing and grouping activation. NeuroImage. 2002;17:324–343. doi: 10.1006/nimg.2002.1160. [DOI] [PubMed] [Google Scholar]
- Gelman A, Carlin JB, Stern HS, Rubin DB. Bayesian Data Analysis. Chapman and Hall/CRC; 2004. [Google Scholar]
- George J, Aine C, Mosher J, Schmidt D, Ranken D, Wood C, Lewine J, Sanders J, Belliveau J. Mapping function in the human brain with magnetoencephalography, anatomical magnetic resonance imaging, and functional magnetic resonance imaging. J Clin Neurophys. 1995;12:406–431. doi: 10.1097/00004691-199509010-00002. [DOI] [PubMed] [Google Scholar]
- George JS, Lewis PS, Ranken D, Kaplan L, Wood C. Anatomical constraints for neuromagnetic source models. SPIE: Med Imaging V—Image Phys. 1991;1443:37–51. [Google Scholar]
- Gilks WR, Richardson S, Spiegelhalter DJ. Markov Chain Monte Carlo in Practice. Chapman and Hall/CRC; 1996. [Google Scholar]
- Green PJ. Reversible jump Markov Chain Monte Carlo computation and Bayesian model determination. Biometrika. 1995;82:711–732. [Google Scholar]
- Hämäläinen M, Hari R, Ilmoniemi RJ, Knuutila J, Lounasmaa OV. Magnetoencephalography—theory, instrumentation, and applications to noninvasive studies of the working human brain. Rev Mod Phys. 1993;65:413–497. [Google Scholar]
- Hillyard SA, Hinrichs H, Tempelmann C, Morgan ST, Hansen JC, Scheich H, Heinze HJ. Combining steady-state visual evoked potentials and fMRI to localize brain activity during selective attention. Hum Brain Mapp. 1997;5 (4):287–292. doi: 10.1002/(SICI)1097-0193(1997)5:4<287::AID-HBM14>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Huizenga HM, de Munck JC, Waldorp LJ, Grasman RPPP. Spatiotemporal EEG/MEG source analysis based on a parametric noise covariance model. IEEE Trans Biomed Eng. 2002;49:533–539. doi: 10.1109/TBME.2002.1001967. [DOI] [PubMed] [Google Scholar]
- Jun SC, George JS, Plis SM, Ranken DM, Schmidt DM, Wood CC. Improving source detection and separation in spatiotemporal Bayesian inference dipole analysis. Phys Med Biol. 2006a;51:2395–2414. doi: 10.1088/0031-9155/51/10/004. [DOI] [PubMed] [Google Scholar]
- Jun SC, Plis SM, Ranken DM, Schmidt DM. Spatiotemporal noise covariance estimation from limited empirical MEG data. Phys Med Biol. 2006b;51:5549–5564. doi: 10.1088/0031-9155/51/21/011. [DOI] [PubMed] [Google Scholar]
- Jun SC, George JS, Paré-Blagoev J, Plis SM, Ranken DM, Schmidt DM, Wood CC. Spatiotemporal Bayesian inference dipole analysis for MEG neuroimaging data. NeuroImage. 2005;28:84–98. doi: 10.1016/j.neuroimage.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Kober H, Nimsky C, Mller M, Hastreiter P, Fahlbusch R, Ganslandt O. Correlation of sensorimotor activation with functional magnetic resonance imaging and magnetoencephalography in presurgical functional imaging: a spatial analysis. NeuroImage. 2001;14:1214–1228. doi: 10.1006/nimg.2001.0909. [DOI] [PubMed] [Google Scholar]
- Korvenoja A, Aronen HJ, Ilmoniemi RJ. Functional MRI as a constraint in multi-dipole models of MEG data. Int J Bioelectromagn. 2001:3. [Google Scholar]
- Liu AK, Belliveau JW, Dale AM. Spatiotemporal imaging of human brain activity using functional MRI constrained magnetoencephalography data: Monte Carlo simulations. Proc Natl Acad Sci U S A. 1998;95:8945–8950. doi: 10.1073/pnas.95.15.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412 (6843):150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Marrelec G, Daunizeau J, Pelegrini-Issac M, Doyon J, Benali H. Conditional correlation as a measure of mediated interactivity in fMRI and MEG/EEG. IEEE Trans Signal Process. 2005;53:3503–3516. [Google Scholar]
- Plis SM, George JS, Jun SC, Pare-Blagoev J, Ranken DM, Schmidt DM, Wood CC. Modeling spatiotemporal covariance for MEG/EEG source analysis. Phys Rev, E. 2007a;75:011928. doi: 10.1103/PhysRevE.75.011928. [DOI] [PubMed] [Google Scholar]
- Plis SM, George JS, Jun SC, Ranken DM, Volegov PL, Schmidt DM. Probabilistic forward model for electroencephalography source analysis. Phys Med Biol. 2007b;52 (17):5309–5327. doi: 10.1088/0031-9155/52/17/014. [DOI] [PubMed] [Google Scholar]
- Ranken DM, Best E, Stephen JM, Schmidt DM, George JS, Wood CC, Huang M. MEG/EEG forward and inverse modeling using MRIVIEW. Biomag 2002: 13th International Conference on Biomagnetism Jena; Germany. 2002. pp. 785–787. http://www.lanl.gov/p/p21/mriview.shtml. [Google Scholar]
- Sanders JA, Lewine JD, Orrison WW. Comparison of primary motor cortex localization using functional magnetic resonance imaging and magnetoencephalography. Hum Brain Mapp. 1996;4:47–57. doi: 10.1002/(SICI)1097-0193(1996)4:1<47::AID-HBM3>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Sarvas J. Basic mathematical and electromagnetic concepts of the biomagnetic inverse problem. Phys Rev D Part Fields. 1987;32:11–22. doi: 10.1088/0031-9155/32/1/004. [DOI] [PubMed] [Google Scholar]
- Sato M, Yoshioka T, Kajihara S, Toyama K, Goda N, Doya K, Kawato M. Hierarchical Bayesian estimation for meg inverse problem. NeuroImage. 2004;23:806–826. doi: 10.1016/j.neuroimage.2004.06.037. [DOI] [PubMed] [Google Scholar]
- Schmidt DM, George JS, Ranken DM, Wood CC. Spatial-temporal Bayesian inference for MEG/EEG. Biomag 2000: 12th International Conference on Biomagnetism Espoo; Finland. 2001. pp. 671–673. [Google Scholar]
- Schmidt DM, George JS, Wood CC. Bayesian inference applied to the electromagnetic inverse problem. Hum Brain Mapp. 1999;7:195–212. doi: 10.1002/(SICI)1097-0193(1999)7:3<195::AID-HBM4>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Kim S, Kim T. Correlation between bold-fMRI and EEG signal changes in response to visual stimulus frequency in humans. Magn Reson Med. 2003;49:108–114. doi: 10.1002/mrm.10335. [DOI] [PubMed] [Google Scholar]
- Sotero RC, Trujillo-Barreto NJ. Biophysical model for integrating neuronal activity, EEG, fMRI and metabolism. NeuroImage. 2008;39:290–309. doi: 10.1016/j.neuroimage.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Vakorin VA, Krakovska OO, Borowsky R, Sarty GE. Inferring neural activity from bold signals through nonlinear optimization. Neuro-Image. 2007;38:248–260. doi: 10.1016/j.neuroimage.2007.06.033. [DOI] [PubMed] [Google Scholar]
- Vanni S, Warnking J, Dojat M, Delon-Martin C, Bullier J, Segebarth C. Sequence of pattern onset responses in the human visual areas: an fMRI constrained VEP source analysis. NeuroImage. 2004;21:801–817. doi: 10.1016/j.neuroimage.2003.10.047. [DOI] [PubMed] [Google Scholar]
- Wagner M, Fuchs M, Kastner J. fMRI-constrained dipole fits and current density reconstructions. Biomag 2001: 12th International Conference on Biomagnetism Espoo; Finland. 2001. pp. 785–788. [Google Scholar]
- Wang JZ, Williamson S, Kaufman L. Magnetic source images determined by a leadfield analysis: the unique minimum norm least squares estimation. IEEE Trans Biomed Eng. 1992;39:665–675. doi: 10.1109/10.142641. [DOI] [PubMed] [Google Scholar]