Abstract
Triphenylphosphonium hydroethidine (TPP-HE) is a membrane-permeable probe that reacts with superoxide and forms hydroxytriphenylphosphonium ethidium (OH-TPP-E+), a fluorescent product that has been previously used in qualitative measurements of superoxide production. In order to develop quantitative methods to measure superoxide, it is necessary to take into consideration the principles that drive TPP-HE accumulation into various subcellular compartments. In the mitochondria matrix TPP-HE accumulation depends on the mitochondrial membrane potential, which varies from cell to cell. Here we address this issue by including rhodamine 123 (R123) as an internal mitochondrial membrane potential calibrant in chemical cytometry experiments. After loading with TPP-HE and R123, a single cell is lysed within a separation capillary and its contents are separated and detected by micellar electrokinetic capillary chromatography with laser-induced fluorescence detection (MEKC-LIF). Using theoretical arguments we show that the ratio [OH-TPP-E+]/[R123] is adequate to obtain a relative quantitation of mitochondrial matrix superoxide levels for each analyzed cell. We applied this method to single skeletal muscle myoblasts and determined that the steady state superoxide levels in the mitochondrial matrix is ~ (0.29 ± 0.10) × 10−12 M. The development of this quantitative method is a critical step toward establishing the importance of reactive oxygen species in biological systems, including those relevant to aging and disease.
Keywords: chemical cytometry, single cell, superoxide, membrane potential, rhodamine 123, tiron, carbonyl cyanide m-chlorophenylhydrazone, rotenone, antimycin A, micellar electrokinetic capillary chromatography
INTRODUCTION
Superoxide is a reactive oxygen species (ROS) that plays a decisive role in the generation of secondary reactive radicals,1–2 which could result in oxidative damage associated with multiple diseases and aging.2–4 As a probe to detect superoxide,5 triphenylphosphonium hydroethidine (TPP-HE, a.k.a. MitoSOX Red) has several advantages over commonly used probes such as dichlorodihydrofluorescein (H2DCF) and hydroethidine. TPP-HE is more selective than H2DCF,6 and forms the product hydroxytriphenylphosphonium ethidium (OH-TPP-E+) upon reaction with superoxide.5 Other reactive species also react with TPP-HE, but relative to the reaction with an equivalent level of superoxide the OH-TPP-E+ formed is a small fraction (i.e., ~0.4%, ~6% and ~4% for the reactions with hydrogen peroxide, hydroxyl radical and hypochlorite, respectively).7 In comparison with hydroethidine,8 TPP-HE accumulates in the mitochondrial matrix of cells according to the mitochondrial membrane potential,9 making it possible to detect superoxide in the matrix even in the presence of competing superoxide dismutase (SOD).5,7 Previous studies based on the use of TPP-HE are, however, only qualitative because the dependence of TPP-HE accumulation on the mitochondrial membrane potential was not taken into account.5,7,10–12 Variations in mitochondrial membrane potential between cells could cause different TPP-HE distributions and bias the measured ROS levels.7,13
Here we demonstrate that chemical cytometry can be used to quantitate superoxide levels in the mitochondrial matrix of single myoblasts. Single myoblasts were simultaneously treated with both probes, TPP-HE and R123, before the MEKC-LIF analysis. An intact cell was then introduced into a narrow-bore capillary and lysed therein. Subsequently, the R123 and OH-TPP-E+ contents released from each cell were analyzed by MEKC-LIF.7 Since both R123 and TPP-HE accumulate into mitochondria according to the mitochondrial membrane potential, the mole ratio of OH-TPP-E+/R123 is approximately proportional to the steady state superoxide concentration in the mitochondrial matrix of individual cells with polarized mitochondria. Thus, determination of this ratio make it possible to quantitate superoxide in single cells. In the future, this method could be used for investigating the role of superoxide in xenobiotic toxicity, aging and disease.
THEORY
When cells are incubated with TPP-HE and R123, both probes accumulate in their mitochondria according to the mitochondrial membrane potential (ΔΨ).5,14 The relevant Nernst equations are:
| (1) |
| (2) |
Thus,
| (3) |
where [TPP-HE]inside and [R123]inside, and [TPP-HE]outside and [R123]outside are the concentrations of TPP-HE and R123 in the mitochondrial matrix and outside the mitochondria, respectively. In this study, [TPP-HE]outside is 10 μM and [R123]outside is 50 nM.
The reaction of TPP-HE with superoxide is
| (Reaction 1) |
| (4) |
| (5) |
where k is the kinetic rate of the reaction of TPP-HE with superoxide (~ 4 × 106 M−1s−1),5 T is the incubation time of the cells with TPP-HE (60 minutes), and is the integral of steady state superoxide concentration over the entire incubation time. If [ ] is constant during the incubation time,
| (6) |
where is the average steady state superoxide concentration over the length of the incubation time.
When a cell is lysed and analyzed by MEKC-LIF, the detected OH-TPP-E+ is the total from both the matrix and outside the mitochondria of the cell.7 Thus, the amount of OH-TPP-E+ in each individual cell is,
| (7) |
where mOH-TPP-E+, inside and mOH-TPP-E+, outside are the amounts of OH-TPP-E+ in the mitochondrial matrix and outside the mitochondria of a given cell, respectively. Based on the results from bulk analysis (Supporting Information, Part I; Figure S-1), the mOH-TPP-E+, outside is a small fraction of the mOH-TPP-E+, inside under basal conditions and upon treatments with rotenone and antimycin A. To simplify equation 7, the amount of OH-TPP-E+ in one cell can be expressed as
| (8) |
where x is the fraction of mOH-TPP-E+, outside relative to mOH-TPP-E+, inside. This fraction “x” represent the bias of the measurement due to the superoxide found outside the mitochondria. Based on bulk measurements, x is ~ 0.048 under basal conditions, ~ 0.063 upon treatment with rotenone, and ~ 0.071 upon treatment with antimycin A (Supporting Information, Part I). Note that equation 8 is not applicable when mitochondria are depolarized by carbonyl cyanide m-chlorophenylhydrazone (CCCP) treatment (Supporting Information, Part II).
By combining equations 5, 6, and 8,
| (9) |
where is the average of steady state superoxide concentration in the mitochondrial matrix of single cells over the incubation time.
Based on the MEKC-LIF calibration curves of OH-TPP-E+ and R123 yield the equations,
| (10) |
and
| (11) |
where the respective slopes are a and b, and the respective intercepts are m and n, and the respective peak areas in the electropherograms are AOH-TPP-E+and AR123.
Thus,
| (12) |
where .
EXPERIMENTAL SECTION
Chemicals
Rhodamine 123 (R123), TPP-HE (a.k.a. MitoSOX Red), and tetramethylrhodamine methyl ester (TMRM) were purchased from Invitrogen–Molecular Probes (Eugene, OR). OH-TPP-E+ was synthesized and purified following a previously described procedure.15 Its purity was verified by MEKC-LIF. Its concentration was determined spectrophotometrically using the reported molar extinction coefficient at 478 nm.5,15 OH-TPP-E+ is stable for at least one month when stored at −20 °C.15 All the other reagents were obtained from Sigma–Aldrich (St. Louis, MO). The MEKC running buffer contained 10 mM sodium borate and 1 mM cetyltrimethylammonium bromide (CTAB) (pH 9.3). All buffers were made with Milli-Q deionized water and filtered through 0.22-μm filters before use.
Cell Culture
Rat muscle L6 myoblast cell line was obtained from the American Tissue Culture Collection (Manassas, VA). The cells were cultured in Dulbecco’s modified eagle medium (DMEM) containing 10% (v/v) calf serum and 10 μg/ml gentamicin at 37 °C and 5% CO2. The cells were maintained by splitting them every 3–4 days.
Cell Treatments
The cultured myoblasts were incubated with 10 μM TPP-HE and 50 nM R123 in DMEM at 37°C for 1 h. When needed, before incubation in the presence of TPP-HE and R123, cells were treated with either 1 mM tiron, 50 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP), 5 μM rotenone or 5 μM antimycin A. After treatments, the cells were washed two times with DMEM, trypsinized and resuspended in PBS for either whole cell lysate or single cell analysis.
MEKC-LIF
To do the whole cell lysate analysis, ~ 2 × 106 cells in PBS were centrifuged down at 600 g for 5 min. The cell pellet was then dissolved in running buffer, treated with 2mg/mL proteinase K and 400U/mL DNase 1, and analyzed by MEKC-LIF following previously described procedures.7 Briefly, separations were carried out at − 400 V/cm in MEKC running buffer.7 The 488-nm line (12 mW) of an Argon-ion laser (Melles Griot, Irvine, CA) was used for excitation, and fluorescence was selected with a bandpass filter transmitting in the 607–662 nm range (Omega Optical, Brattleboro, VT).
To do single-cell analysis, a cell was injected into a 150-μm-o.d., 30-μm-i.d. fused silica capillary (Polymicro Technologies, Phoenix, AZ) following previously reported procedures.16–17 After the cell lysis in the capillary, the MEKC-LIF procedure was done as described for bulk analysis. Upon completion of the separation, the capillary was washed for 2 min with 0.1 M NaOH and 5 min with MEKC running buffer between runs.
Separate calibration curves for OH-TPP-E+ and R123, (area in electropherogram versus amount injected) were built by injecting standards of these compounds. The resulting calibration curves for OH-TPP-E+ and R123 were:.
| (13) |
| (14) |
where m is given in attomoles, A is the peak area in the electropherogram, and the errors are standard deviations. These equations were used to carry out calculations described by the equations (10) and (11). The limits of detection (LOD at signal/noise = 3) were ~2 amol for OH-TPP-E+ and ~0.2 amol for R123. It would be possible to obtain a better LOD (e.g.zmol levels) for R123 by using a different bandpass filter (e.g. centered at 530nm). This alternative was not pursued here.
Data Analysis
The MEKC electropherograms were analyzed using Igor Pro 5.0 software (Wavemetrics, Lake Oswego, OR). Superoxide concentrations in single myoblasts under each condition were reported as mean ± standard deviation (SD). A student’s t-test was used to determine statistical significance of the data, with p values of < 0.05 considered significant. A F-test at a significance level of 0.05 was used to test significant differences between variances of two groups of data.
RESULTS AND DISCUSSION
Bulk Analysis of OH-TPP-E+ and R123 in Myoblasts
When the myoblasts are incubated with TPP-HE and R123, both probes cross the plasma membrane and accumulate in the cytosol and the matrix of mitochondria. In the latter, probe accumulation depends on the mitochondrial membrane potential.5,14 In both sites TPP-HE forms OH-TPP-E+ upon reaction with superoxide or TPP-E+ upon oxidation by other intracellular oxidative species such as cytochromes and oxidases.5 Since both OH-TPP-E+ and TPP-E+ exhibit similar fluorescence spectra,5 the superoxide specific OH-TPP-E+ cannot be distinguished from TPP-E+ by fluorescence imaging (Supporting Information, Figure S-2).
MEKC-LIF was adequate to separate OH-TPP-E+, TPP-E+ and R123, thereby preventing the problems arising from the fluorescence cross-talk observed in fluorescence microscopy. R123 was separated from OH-TPP-E+ and TPP-E+ with resolutions of 4.7 and 4.2, respectively (Figure 1). From the amount of OH-TPP-E+ in the whole cell lysate (c.f. equation 13), on average each cell contains ~ 7.7 amole. However, this bulk analysis is biased because it ignores that cells do not necessarily have the same membrane potential.18–20
Figure 1.
Electropherograms of TPP-HE oxidation products and R123 in the whole cell lysate (~ 2×106 myoblasts) and the blank buffer. Separations were performed in a 42-cm-long capillary at - 400 V/cm in MEKC running buffer. The 488-nm line of an Argon-ion laser was used for excitation, and a 607–663 nm bandpass filter was used for detection. Traces have been offset vertically for clarity. The respective electrophoretic mobilities of OH-TPP-E+ and R123 were - (6.89 ± 0.11) × 10−4 and - (6.41 ± 0.09) × 10 −4 cm2 V−1 s−1 (n=3). Separation efficiencies were calculated to be ~ 260,000 for OH-TPP-E+ and ~31,000 for R123.
It is important to bear in mind that superoxide dismutases (SODs) are present in the matrix (Mn-SOD) and outside the mitochondria (Cu, Zn-SOD).5 In general SODs transform superoxide into hydrogen peroxide (k ~ 2 × 109 M−1s−1),21–23 outcompeting the relatively slow reaction of superoxide with the TPP-HE probe (i.e. k ~ 4 × 106 M−1s−1).5 However, in the mitochondrial matrix, the mitochondrial membrane potential increases the TPP-HE concentration ~100-fold relative to the cytosol7 that partially compensates for its relative slow reaction rate constant with superoxide. The concentration of Mn-SOD in the matrix has been reported to be ~ 20 μM.24 Based on this value and the kinetic rate constants of TPP-HE and Mn-SOD with superoxide (and assuming that there are no other reactions consuming superoxide), the estimated rate of OH-TPP-E+ accumulation is ~ 10% of the formation rate of H2O2 by Mn-SOD. This simplified kinetic analysis argues that, at this relative rate of formation, the detected amounts of OH-TPP-E+ are adequate to calculate (equation 12), even in the presence of competing Mn-SOD.
On the other hand, outside the mitochondria there is no enhanced accumulation of TPP-HE. Given a concentration of Cu, Zn-SOD ~6 μM,25 the rate of OH-TPP-E+ accumulation outside the mitochondria is only ~ 0.3% of the rate of H2O2 formed by Cu, Zn-SOD. This calculation further confirms that OH-TPP-E+ produced outside the mitochondria does not contribute significantly to the total OH-TPP-E+ detected (i.e. “x” in equation 8 is small; c.f. Supporting Information, Part I).
Single Cell Superoxide Quantitative Analysis
We used chemical cytometry to analyze mitochondrial matrix superoxide levels in single cells (Figure 2). In order to correct for variations in mitochondrial membrane potentials observed among individual cells, cells were treated with TPP-HE and R123. The MEKC-LIF separation and detection of OH-TPP-E+ and R123 in single myoblasts provides the measurements needed to determine superoxide at the single-cell level (c.f. equation 12).
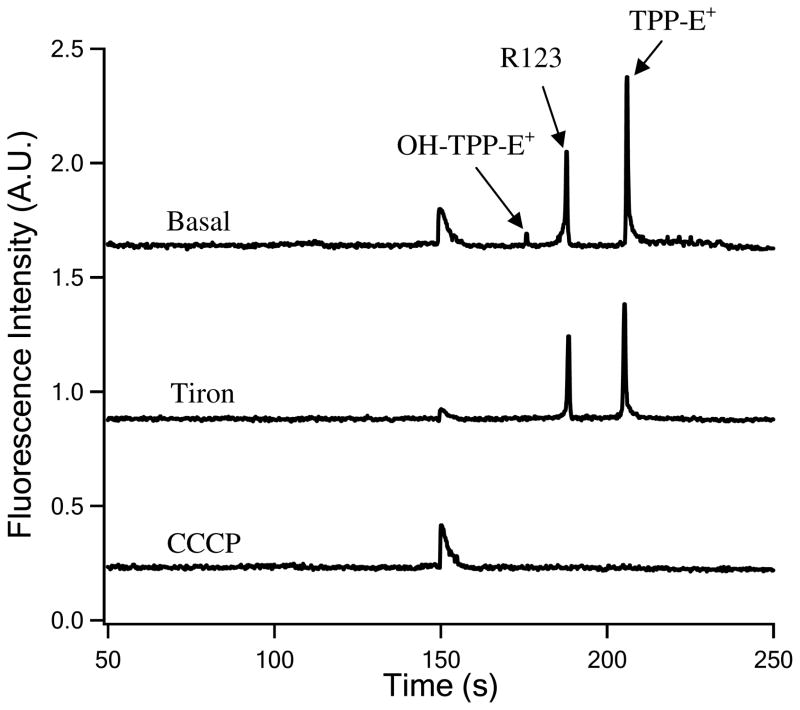
Figure 2.
Electropherograms of TPP-HE oxidation products and R123 in individual myoblasts under basal conditions and upon treatments with tiron and CCCP. Separations and detection conditions were same to those described for Figure 1.
The detection of mitochondrial matrix superoxide at the single cell level was further confirmed by treatment with tiron and CCCP. Tiron is an SOD mimetic that scavenges intracellular superoxide in cultured cells.6,26 As expected, OH-TPP-E+ was not detected in single myoblasts treated with tiron (Figure 2). The R123 peak was still detected showing that tiron treatment does not compromise the mitochondrial membrane potential. CCCP is a proton ionophore that dissipates the mitochondrial membrane potential.27–28 Upon CCCP treatment both R123 and TPP-HE concentrations in the matrix decrease ~100-fold relative to untreated cells (Supporting Information, Parts II and III). The expected R123 amount in a CCCP-treated cell is ~ 0.2 amol R123 (Supporting Information, Part III), which is undetectable in the MEKC-LIF system used in this study (i.e. limit of detection ~ 0.2 amol R123). Furthermore, the low TPP-HE concentration in CCCP-treated cells significantly decreases the rate of formation of OH-TPP-E+ inside the mitochondria (c.f. Equation 5; Supporting Information, Part II). The expected amount of OH-TPP-E+ formed in a CCCP-treated myoblast is ~0.4 amol, which is undetectable in the MEKC-LIF that was used in this study (i.e. limit of detection ~ 2 amol OH-TPP-E+). Thus, the concomitant disappearance of the R123 and the OH-TPP-E+ peaks in CCCP-treated myoblasts (Figure 2) demonstrates that the mitochondrial membrane potential is vital to the accumulation of TPP-HE and formation of OH-TPP-E+ in the mitochondrial matrix.5
Based on fluorescence microscopy, individual myoblasts display ~39% variation in mitochondrial membrane potential (Supporting Information, Part V; Figure S-3). In MEKC-LIF the ratio of the areas of OH-TPP-E+ and R123 corrects for variations in mitochondrial membrane potential between individual cells (c.f. equation 12). This calculation is based on the assumptions that both R123 and TPP-HE accumulate into the mitochondria according to a Nernstian behavior (c.f. equations 1 and 2) and that the amount of superoxide product outside the mitochondria is only a small fraction of that in the mitochondrial matrix (c.f. equations 7 and 8; Supporting Information, Part I). Based on MEKC-LIF, the steady state superoxide concentration in the mitochondrial matrix of single myoblasts under basal conditions was ~ (0.29 ± 0.10) × 10−12 M with a RSD of 35% (n=12) (Figure 4). This variance is statistically different from that obtained if the signal of OH-TPP-E+ is not corrected by membrane potential (RSD ~69%) (F-test: p < 0.05). In summary, the simultaneous monitoring of OH-TPP-E+ and R123 provides a correction for variations in membrane potential and provides the means to quantitate mitochondrial matrix superoxide levels in single cells.
Figure 4.
Effects of rotenone and antimycin A on the superoxide levels detected in the mitochondrial matrix of single myoblasts. Data of individual cells as well as means ± SD are presented for each condition (n = 12 cells for basal, 10 for rotenone and 12 for antimycin A). * p < 0.05 vs basal.
Effects of Mitochondrial Respiratory Inhibitors
Mitochondria is considered a major source of superoxide in cells, where superoxide is mainly released by complex I and III in the mitochondrial electron transport chain (ETC).1–2 Rotenone and antimycin A are two respiratory inhibitors that block electron transfer through complex I and III, respectively, thereby stimulating superoxide production.29–30 Although recent studies have utilized the TPP-HE probe to investigate the effects of rotenone and antimycin A on intracellular superoxide release in single cells by fluorescent microscopy and/or flow cytomery,5,10–12 these studies are qualitative because they have not considered the effect of mitochondrial membrane potential.
Here we demonstrated that chemical cytometry is suitable to quantify mitochondrial matrix superoxide levels in individual myoblasts upon their treatment with either rotenone or antimycin A (Figure 3). Some studies have reported such inhibitors at elevated concentrations may alter the membrane potential of isolated mitochondria.31–34 In this report bulk analyses showed that the treatments with rotenone and antimycin A did appear to alter the mitochondria membrane potentials because there were no significant changes in the ratio of mOH-TPP-E+, outside to mOH-TPP-E+, inside compared to basal conditions (Supporting Information, Part I). Thus, equation 12 is adequate to determine mitochondrial matrix superoxide levels following inhibitor treatments.
Figure 3.
Electropherograms of TPP-HE oxidation products and R123 in individual myoblasts under basal conditions and upon treatments with rotenone and antimycin. Separations and detection conditions were same to those described for Figure 1.
In single myoblasts treated with rotenone or antimycin A, the steady state superoxide concentrations in the mitochondrial matrix were (0.97 ± 0.61) × 10−12 M (n=10) and (2.15 ± 1.20) × 10−12 M (n=12), respectively (Figure 4). These values are ~3.5- and 8.2-fold higher compared to basal levels. The magnitude of such changes are comparable with the ~ 2 to 3-fold change observed in aged cells or cells associated with diabetic hyperglycemia relative to their respective controls.35–37 Therefore, the use of inhibitors demonstrates that chemical cytometry is a suitable approach to quantitate mitochondrial superoxide levels expected in aging and disease related studies.
CONCLUDING REMARKS
Here we report the quantitation of mitochondrial matrix superoxide levels in single cells using a MEKC-based chemical cytometry method that is not biased by the mitochondrial membrane potential. Based on equation 12, the ratio of OH-TPP-E+ and R123 signals corrects for variation in membrane potential of mitochondria among individual cells. According to this method the mitochondrial matrix superoxide levels of single myoblasts are in the picomolar range. The method could also be extended to investigate other cellular systems such as cultured single skeletal muscle fibers,38–39 whose superoxide levels were analyzed qualitatively in our earlier study.40 The method may also be modified to monitor superoxide released outside the mitochondria, which would be particularly useful to investigate cell lines derived from Cu, Zn-SOD-deficient animal models.41–42 Long-term application of chemical cytometry to quantify superoxide levels may find wide applications to the fields of toxicology, aging and oxidative-stress related disease.4,43–44
Supplementary Material
Acknowledgments
The National Institutes of Health supported this work through grant R01-AG-20866. We thank Dr. Margaret Donoghue for providing comments on the manuscript.
References
- 1.St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. J Biol Chem. 2002;277:44784. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 2.Orrenius S, Gogvadze V, Zhivotovsky B. Annu Rev Pharmacol Toxicol. 2007;47:143. doi: 10.1146/annurev.pharmtox.47.120505.105122. [DOI] [PubMed] [Google Scholar]
- 3.Ashok BT, Ali R. Exp Gerontol. 1999;34:293. doi: 10.1016/s0531-5565(99)00005-4. [DOI] [PubMed] [Google Scholar]
- 4.Beckman KB, Ames BN. Physiol Rev. 1998;78:547. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 5.Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Proc Natl Acad Sci U S A. 2006;103:15038. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McArdle F, Pattwell DM, Vasilaki A, McArdle A, Jackson MJ. Free Radic Biol Med. 2005;39:651. doi: 10.1016/j.freeradbiomed.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Xu X, Arriaga EA. Free Radical Biology and Medicine. 2009;46:905. doi: 10.1016/j.freeradbiomed.2008.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao H, Joseph J, Fales HM, Sokoloski EA, Levine RL, Vasquez-Vivar J, Kalyanaraman B. Proc Natl Acad Sci U S A. 2005;102:5727. doi: 10.1073/pnas.0501719102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross MF, Kelso GF, Blaikie FH, James AM, Cocheme HM, Filipovska A, Da Ros T, Hurd TR, Smith RAJ, Murphy MP. Biochemistry-Moscow. 2005;70:222. doi: 10.1007/s10541-005-0104-5. [DOI] [PubMed] [Google Scholar]
- 10.Mukhopadhyay P, Rajesh M, Yoshihiro K, Hasko G, Pacher P. Biochem Biophys Res Commun. 2007;358:203. doi: 10.1016/j.bbrc.2007.04.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Z, Varadharaj S, Giedt RJ, Zweier JL, Szeto HH, Alevriadou BR. J Pharmacol Exp Ther. 2009;329:94. doi: 10.1124/jpet.108.145557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe N, Zmijewski JW, Takabe W, Umezu-Goto M, Le Goffe C, Sekine A, Landar A, Watanabe A, Aoki J, Arai H, Kodama T, Murphy MP, Kalyanaraman R, Darley-Usmar VM, Noguchi N. Am J Pathol. 2006;168:1737. doi: 10.2353/ajpath.2006.050648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zielonka J, Kalyanaraman B. Free Radical Biology and Medicine. 2010;48:983. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scaduto RC, Grotyohann LW. Biophysical Journal. 1999;76:469. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zielonka J, Vasquez-Vivar J, Kalyanaraman B. Nature Protocols. 2008;3:8. doi: 10.1038/nprot.2007.473. [DOI] [PubMed] [Google Scholar]
- 16.Krylov SN, Starke DA, Arriaga EA, Zhang Z, Chan NW, Palcic MM, Dovichi NJ. Analytical Chemistry. 2000;72:872. doi: 10.1021/ac991096m. [DOI] [PubMed] [Google Scholar]
- 17.Anderson AB, Gergen J, Arriaga EA. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;769:97. doi: 10.1016/s1570-0232(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 18.Johnson LV, Walsh ML, Bockus BJ, Chen LB. J Cell Biol. 1981;88:526. doi: 10.1083/jcb.88.3.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ludovico P, Sansonetty F, Corte-Real M. Microbiology. 2001;147:3335. doi: 10.1099/00221287-147-12-3335. [DOI] [PubMed] [Google Scholar]
- 20.Heerdt BG, Houston MA, Augenlicht LH. Cancer Res. 2005;65:9861. doi: 10.1158/0008-5472.CAN-05-2444. [DOI] [PubMed] [Google Scholar]
- 21.Klug D, Rabani J, Fridovich I. J Biol Chem. 1972;247:4839. [PubMed] [Google Scholar]
- 22.Pick M, Rabani J, Yost F, Fridovich I. J Am Chem Soc. 1974;96:7329. doi: 10.1021/ja00830a026. [DOI] [PubMed] [Google Scholar]
- 23.Behar D, Czapski G, Rabani J, Dorfman LM, Schwarz HA. J Phys Chem. 1970;74:3209. [Google Scholar]
- 24.Quijano C, Hernandez-Saavedra D, Castro L, McCord JM, Freeman BA, Radi R. J Biol Chem. 2001;276:11631. doi: 10.1074/jbc.M009429200. [DOI] [PubMed] [Google Scholar]
- 25.Chang LY, Slot JW, Geuze HJ, Crapo JD. J Cell Biol. 1988;107:2169. doi: 10.1083/jcb.107.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada J, Yoshimura S, Yamakawa H, Sawada M, Nakagawa M, Hara S, Kaku Y, Iwama T, Naganawa T, Banno Y, Nakashima S, Sakai N. Neurosci Res. 2003;45:1. doi: 10.1016/s0168-0102(02)00196-7. [DOI] [PubMed] [Google Scholar]
- 27.Heytler PG, Prichard WW. Biochem Biophys Res Commun. 1962;7:272. doi: 10.1016/0006-291x(62)90189-4. [DOI] [PubMed] [Google Scholar]
- 28.Lim ML, Minamikawa T, Nagley P. FEBS Lett. 2001;503:69. doi: 10.1016/s0014-5793(01)02693-x. [DOI] [PubMed] [Google Scholar]
- 29.Grivennikova VG, Vinogradov AD. Biochim Biophys Acta. 2006;1757:553. doi: 10.1016/j.bbabio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Muller FL, Liu YH, Van Remmen H. Journal of Biological Chemistry. 2004;279:49064. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 31.Petit PX, O’Connor JE, Grunwald D, Brown SC. Eur J Biochem. 1990;194:389. doi: 10.1111/j.1432-1033.1990.tb15632.x. [DOI] [PubMed] [Google Scholar]
- 32.Kataoka M, Fukura Y, Shinohara Y, Baba Y. Electrophoresis. 2005;26:3025. doi: 10.1002/elps.200410402. [DOI] [PubMed] [Google Scholar]
- 33.Tzung SP, Kim KM, Basanez G, Giedt CD, Simon J, Zimmerberg J, Zhang KY, Hockenbery DM. Nat Cell Biol. 2001;3:183. doi: 10.1038/35055095. [DOI] [PubMed] [Google Scholar]
- 34.de Bari L, Atlante A, Guaragnella N, Principato G, Passarella S. Biochem J. 2002;365:391. doi: 10.1042/BJ20020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Cangello D, Benson S, Folmer J, Zhu H, Trush MA, Zirkin BR. Experimental Gerontology. 2001;36:1361. doi: 10.1016/s0531-5565(01)00118-8. [DOI] [PubMed] [Google Scholar]
- 36.Klamt F, Gottfried C, Tramontina F, Dal-Pizzol F, Da Frota ML, Jr, Moreira JC, Dias RD, Moriguchi E, Wofchuk S, Souza DO. Neuroreport. 2002;13:1515. doi: 10.1097/00001756-200208270-00005. [DOI] [PubMed] [Google Scholar]
- 37.Nishikawa T, Edelstein D, Du XL, Yamagishi S-i, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes H-P, Giardino I, Brownlee M. Nature. 2000;404:787. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- 38.Pye D, Palomero J, Kabayo T, Jackson MJ. J Physiol. 2007;581:309. doi: 10.1113/jphysiol.2006.125930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palomero J, Pye D, Kabayo T, Spiller DG, Jackson MJ. Antioxidants & Redox Signaling. 2008;10:1463. doi: 10.1089/ars.2007.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu X, Thompson LV, Navratil M, Arriaga EA. Analytical Chemistry. 2010 doi: 10.1021/ac100577q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, Wilcox HM, Flood DG, Beal MF, Brown RH, Jr, Scott RW, Snider WD. Nat Genet. 1996;13:43. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 42.Veerareddy S, Cooke CL, Baker PN, Davidge ST. Am J Physiol Heart Circ Physiol. 2004;287:H40. doi: 10.1152/ajpheart.01179.2003. [DOI] [PubMed] [Google Scholar]
- 43.Thompson LV. Experimental Gerontology. 2009;44:106. doi: 10.1016/j.exger.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amacher DE. Curr Med Chem. 2005;12:1829. doi: 10.2174/0929867054546663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.