Abstract
Using existing technology, it can be hard or impossible to determine whether two structural equation models that are being considered may be nested. There is also no routine technology for evaluating whether two very different structural models may be equivalent. A simple nesting and equivalence testing (NET) procedure is proposed that uses random sample and model-reproduced moment matrices to evaluate both model nesting and equivalence. The analysis is “local” rather than “global” in nature, but its use with simulation or bootstrapping can imply global conclusions. Two standard applications of NET are to verify whether or not two proposed models are equivalent, and whether a baseline model used in an incremental fit index is appropriately nested.
Keywords: Structural equation models, covariance structure models, nested models, equivalent models, NET
The basic ideas of model nesting and model equivalence in structural equation modeling (SEM) are widely known, already discussed in introductory texts (Byrne, 2006; Kline, 2005; Mulaik, 2009; Raykov & Marcoulides, 2006). Most SEM practitioners know that fixing one or more free parameters to yield a more restricted model will yield a nested model, and that changing the direction of one or more paths in a simple model may yield an equivalent structural equation model.
Model nesting is the easier concept. It is facilitated by SEM programs that allow a researcher to run any two models, whose test statistics output can then be used to compute a chi-square difference test. However, the difference test also can be computed when the models are not nested and such a test is meaningless. Although no researcher would do this on purpose, such a meaningless comparison sometimes is made in the context of widely used fit indices such as the comparative fit index (CFI, Bentler, 1990). To compute such an index, a SEM program may automatically generate the standard baseline or null model of uncorrelated variables and compute the fit index by comparing the fit of the current substantive model to that of this null model. However, as noted by Widaman and Thompson (2003), the model of uncorrelated variables often is not a nested subset of the model of interest, so the resulting fit index is inappropriate and meaningless. SEM programs, however, provide no information on the appropriateness of the model comparison used in computing incremental fit indices. A simple and automated method for evaluating model nesting would eliminate this problem.
Model equivalence is harder to evaluate, and tends to be overlooked in practice (e.g., Henley, Shook, & Peterson, 2006). Moment equivalent models are those “that, regardless of the data, yield identical (a) implied covariance, correlation, and other moment matrices when fit to the same data, which in turn imply identical (b) residuals and fitted moment matrices, (c) fit functions and chi-square values, and (d) goodness-of-fit indices based on fit functions and chi-square” (Hershberger, 2006, p. 15). What is especially hard for a practitioner to do is to evaluate whether models are nested or equivalent when, for example, the structures of the models being compared are very different or if the models are so large as to make overview difficult. SEM programs provide no help in evaluating equivalent models, and researchers are left to study the complicated rules of path replacement, recently summarized by Hershberger (2006), to see if models are equivalent.
This paper describes simple computations that can be added to any SEM program to obtain evidence on nesting or equivalence. Similarly, a researcher can perform their own nesting and equivalence tests using two SEM modeling runs. The first run fits the presumed more restricted model. The resulting implied moment matrix is fit in the second run by the presumed equal or more general model. Our proposed decision rules are then applied to standard output to yield the required conclusions.
We use the following notation. Let two models be designated as M1 and M2, where, if the models have different degrees of freedom (df), M1 is the model with the larger df. This implies that when models are nested, M1 is the more restricted model; or, if the df are equal, it may be equivalent to M2. When neither nesting nor equivalence holds, M1 and M2 are just two models of interest.
Methodology
Bentler and Bonett (1980) introduced the concepts of parameter nesting and covariance matrix nesting. The type of nesting typically considered is parameter nesting, where a free parameter in M2 is fixed in the more restricted model M1 or a free parameter is added into a constraint equation that reduces the effective number of free parameters. A result is that the df in M1 must be larger than those in M2. Parameter nesting is relatively easy to evaluate, especially in simple models, and hence a special technology to evaluate parameter nesting is typically not needed. However, verifying parameter nesting also can be difficult, or at least, subject to human error, and computerized assistance may be helpful.
Covariance matrix nesting, or, allowing mean structures as well, moment matrix nesting, is more difficult to verify. In such models, the mapping from parameters to mean and covariances matrices may take completely different forms in the two models being compared, and yet the set of possible moment matrices under M1 remains a subset of the set of possible mean and covariance matrices under M2. Consider for example the two models in Figure 1. These models look completely different -- they have a different number of equations; they have a different number of independent variables; and only one has latent variables. Yet, under standard identification conditions, they are nested. This can be seen easily since model M2 is a saturated model, and M1 is just some structural model. In more complicated models it can be even more difficult to establish moment matrix nesting. Computerized assistance would definitely be helpful.
Figure 1.
Two Nested Models
We will define model equivalence in terms of nesting. We will see that equivalent models are those with equal df in which M1 is nested in M2, and simultaneously, M2 is nested in M1. In terms of methodology, our approach is quite different from that of previous authors. The typical approach is based on abstract algebra (including computer algebra programs) associated with parameters, functions, and population model structures (e.g., Bekker, Merckens, & Wansbeek, 1994; Raykov & Penev, 1999), which is elegant but essentially impossible for an empirical scientist to implement. In contrast, the proposed approach uses sample data to investigate population nesting and equivalence. In order to assure that this methodology is statistically sound, we make several assumptions. First, nesting and equivalence are defined with regard to a given population with means μ and full rank covariance matrix Σ. Second, an empirical or simulated random sample from this population has been obtained, yielding sample means and full rank covariance matrix S. Third, it is assumed that the asymptotic variance matrix of the vector of sample moments is nonsingular. Fourth, the models Mi [Σi(θi) and μi(θi) if relevant] being considered are identified, and the vector-valued function that expresses the moments as a function of the free unconstrained parameters is continuously differentiable with Jacobian of full column rank in a neighborhood of population values. Fifth, no inequality constraints are involved in the definition of the models considered. These conditions1 are adopted to rule out pathological cases that can be devised to make standard methods break down, as further discussed below. At this point, we describe our method. A detailed rationale is provided subsequently.
Nesting and Equivalence Test (NET)
The sample means and covariance matrix S are used in the following.
Step 1. Do a normal SEM run on model M1. Save the df and the model reproduced covariances (and means, in a mean structure). Call these df1, (and if needed).
Step 2. Read in the (and if needed) as data to be analyzed in a SEM run with model M2 using the same estimation method. The output needed from this run is the df, say df2, and the minimum of the fit function (or the associated chi-square statistic).
Step 3. Compute d = df1−df2, and set ε, a small criterion value for (e.g., ε = .001).
- Step 4.
- If d > 0 and , the models are nested.
- If d = 0 and , the models are equivalent.
- If d < 0 or , M1 is not nested in, or equivalent to, M2.
The key idea is that when , the M1 model-reproduced means and covariance matrix can be precisely reproduced by model M2. With nonnegative d, as in (a), this implies the models are nested. With d = 0, as in (b), this implies the models are equivalent. Negative degrees of freedom, as in (c), imply non-nesting of M1 in M2 and non-equivalence of the models. Note that if , i.e., if the restricted model reproduced means and covariances cannot be fit exactly by an equal or more general model, the models are neither nested or equivalent. The reason for picking some small ε as a criterion is that computations always involve some numerical approximations -- terminating computations by a convergence criterion depends on the specific choice of criterion, the computational accuracy of the computer, and so on. As computer precision improves, ε can be made very small indeed.2
In some situations, sample data may not be available. In such a case, we can create an artificial population and sample from it so that the moment matrix to be analyzed in NET corresponds to a true random matrix. We may do this in many ways; we mention three: (1) Choose some specific unstructured Σ (and μ when means are in included in the model) and draw a random sample from the corresponding multivariate normal distribution, obtaining the sample covariance matrix S (and sample means when means are included) and use NET as before. (2) As a variant of (1), instead of choosing a saturated model, use a restricted model with added random sampling. This can be done in two ways, (2a) choose values of the parameters in the equation/covariance specification of the model under consideration, generate the implied , and use this to define the artificial population, then proceed as in (1); or (2b) sample from a multinormal distribution to obtain sample values for the independent variables of the model, and use these with chosen values of the parameters in the model equations to obtain the values of the dependent variables. Obtain S (and if needed) from the full sample and proceed with Step 1. (3) Assign randomly chosen proper parameter values to the restricted model M1, generate the implied , , use these as , (with df1) in Step 1 of NET, then proceed directly to Step 2 and continue as above. The validity of each of these methods hinges on the randomness of its specified random aspect, either of the sample data from which S and are obtained, or the random choice of parameter values to obtain a moment matrix and vectors under the more restricted model. If the samples so generated create problems in model estimation, the simulation sample size can be increased to generate a tighter neighborhood in which these model comparisons are undertaken (see Appendix A). In a sense, these simulation methods are counterparts and extensions to standard SEM practice, in which the empirical rank of a Jacobian or information matrix is evaluated at some set of parameter estimates to determine local identification.
Next we illustrate how to do our nesting test with several small examples from the literature. Theoretical rationales are presented subsequently.
Illustrations
Nested Models
The first example involves the two models M1 and M2 of Figure 1. The sample covariance matrix S is the 3 by 3 covariance matrix given by Hershberger (2006, p. 15), namely,
Hershberger does not give the sample size, and we take it as N=100. Model M1 is his Model 1 (p. 17), which has 1 df (df1=1). In Step 1, we use maximum likelihood (ML) estimation, and obtain the model-reproduced covariance matrix based on the ML estimates as
This is identical to that reported by Hershberger (2nd matrix, top of p. 16). Next, we do Step 2, using as data to be analyzed using model M2 of Figure 1. The output from this run provides the df (df2=0) and the minimum of the ML fit function, . In step 3, we compute the difference in degrees of freedom as d = df1 − df2 = 1 − 0 = 1, and choose the default ε = .001. We see that the results favor part (a) of Step 4, that is, d > 0 and and hence we conclude that the models are nested, which is not surprising since M2 is a saturated model.
Model Equivalence
Figure 2 shows a model that Hershberger has shown to be equivalent to model M1 of Figure 1; it is his Model 3 (p. 17). We now use our NET procedures to evaluate the equivalence hypothesis. Step 1 gives the same results as previously. Using the output of Step 1 ( as shown above) as input to Step 2, we now run the model of Figure 2. The results show the df (df2=1) and the minimum of the ML fit function, . In step 3, difference in df is given as d = df1 − df2 = 1 − 1 = 0. With the same default ε as before, the results are consistent with part (b) of Step 4. Since d = 0 and , the models are equivalent.
Figure 2.

An Equivalent Model to Model M1 of Figure 1
Appropriateness of Fit Indices
Next we consider one of the examples provided by Widaman and Thompson (2003) on appropriate and inappropriate null models for fit indices. An inappropriate model is one in which the baseline model, typically taken by default as the model of uncorrelated variables, is not nested in the model of interest. They analyzed several psychometric test theory models on 4 variables taken from Votaw (1948). Based on 126 subjects, the means of these variables are 14.905, 15.484, 14.444, 15.123, and the covariance matrix is
Among the several models they considered is a restricted one-factor model with equal factor loadings, equal unique variances, and equal means (their model 1A). The ML test statistic TML of this substantive model, based on df=11, is TML = 115.266. Their standard baseline model (model 0C) had no factors, freely estimated unique variances, and freely estimated means. This model is clearly not nested in the substantive model since it frees up parameters rather than restricting them further. It yielded TML = 272.492 with 6 df. In this example, the smaller rather than greater number of df should clearly warn the user about the inappropriateness of this baseline model, and hence the inappropriateness of all incremental fit indices. Widaman and Thompson suggested that an acceptable baseline model for their substantive model would be a model that has no factors, equal unique variances, and equal means (model 0A). This yielded TML = 277.826 with 12 df. We now show how our NET methodology verifies that model 0C is not nested in the substantive model, while model 0A is appropriately nested. We use the χ2 statistics for this evaluation, rather than the minima of the fit function as before.
Our Step 1 model M1 for model 0C showed TML = 272.492 with 6 df (=df1). The model reproduced means are 14.905, 15.484, 14.444, and 15.123 and the unique variances are 25.070, 28.202, 22.739, and 21.871, i.e., these estimates correspond to the sample means and variances as would be expected. In Step 2, these are input into a new run with analysis based on M2, which is their model 1A (see above). The resulting TML = 5.334 with 11 df (=df2). In Step 3, we compute d = df1 − df2 = 6 − 11 = −5 and choose ε = .001 as before. In Step 4, since d < 0 and TML = 5.334 > ε, neither conditions (a) or (b) are met, while (c) is met, attesting to the conclusion that the models are not nested as hypothesized or equivalent. Repeating this procedure for model 0A, in Step 1, as noted TML = 277.826 with 12 df (=df1). The corresponding model reproduced means are 14.989, 14.989, 14.989, and 14.989, and the unique variances are 24.612, 24.612, 24.612, and 24.612. In Step 2, these are input into a new run with analysis based on model M2(model 1A), yielding TML = 0.000 with 11 df (=df1). In Step 3, d = df1 − df2 = 12 − 11 = 1, and we choose ε = .001. Using the results of Steps 2 and 3 in Step 4, we have d > 0 and TML < ε, and hence we conclude, correctly, that model 0A is nested in model 1A. Hence, incremental fit indices can be based on it.
Model Comparison without Data
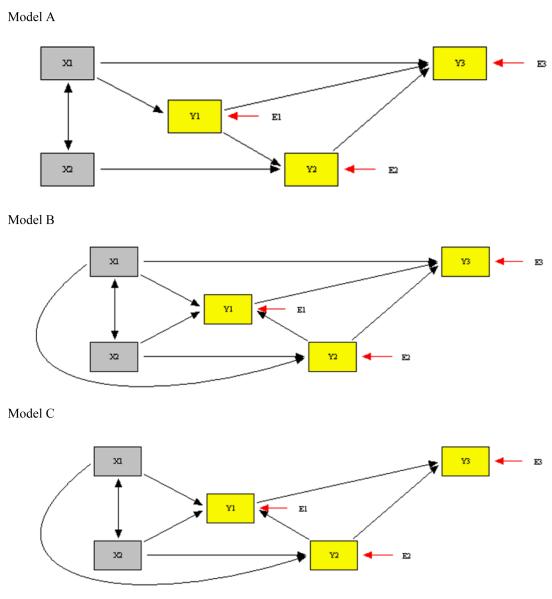
Next we show how to use the NET procedure when no sample data is available. We use the third method described above. The specific illustration is based on a model taken from educational psychology (Meece, Blumenfeld, & Hoyle, 1988) that was studied by MacCallum, Wegener, Uchino, and Fabrigar (1993) and shown by them to be equivalent to three further models as given in their figure 2. While we could demonstrate our method to evaluate model equivalence with the MacCallum et al. models, we consider nesting tests on models derived from two of the equivalent models: their “original model” and their model 2C. To achieve this, we suppress two paths of their original model, yielding our Model A as shown in the top part of Figure 3. We keep intact their model 2C and make it our Model B, shown in the middle of Figure 3. Finally, we suppress one path of model B (their 2C) to obtain our Model C, the bottom part of Figure 3. The three models are defined by equations as follows.3
- Model A
- Model B
- Model C
In all models, the variances and covariances of the exogenous X1 and X2, and the variances of the residuals E1, E2, E3 are unrestricted parameters of the model, with the Ei specified as uncorrelated. The df of models A, B and C are respectively 3, 1 and 2, implying that none of the models are equivalent. But are they nested?
Figure 3.
Models to Illustrate Nesting or Nonnesting with Artificial Data
If Models A and B are nested, Model A must be the more restricted model since Model B has fewer df, i.e., more free parameters. In fact, we know from MacCallum et al. that Model A must be nested in Model B, since model A is just one of their equivalent models with some paths suppressed. Yet nesting is not obvious from the figures. Comparing the figures, or equivalently, the equations, it is easy to see that the paths X1 to Y2(γ21) and X2 to Y1(γ12) in Model B are set to zero to yield Model A, implying nesting. However, at the same time, the path from Y2 to Y1 also is reversed, implying non-nesting. To avoid the extensive required algebra to see if the models are nested, let us apply NET.
Using the above third-mentioned method for dealing with absence of empirical data, we used an integer-based uniform random number generator to obtain random parameter values for Model A, the more restricted model in both comparisons. For greater plausibility when compared with real data, the generated random parameters were limited to appropriate ranges for coefficients, variances, and covariances. Distributions also could be specified, but we did not do so. For coefficients, we limited the random numbers to the range 1-200 and computed γij or βij as . To keep the correlation among independent variables from being too large, we limited random to the range 1-150 and computed it as . To keep variances in a small range around 1, we use in the range 80-120 and computed them as . Standard deviations were taken as square roots of variances; together with a correlation, they yielded a covariance. The implied model covariance matrix from this procedure was
Applying NET, the minimum when Model B is fit to this matrix was .00000, indicating that Model A is nested in Model B. With Model C, minimum , indicating that A is not nested in C. This methodology was repeated several times to generate different random , and the nesting conclusions were identical each time. A formal simulation was not undertaken since it would just verify our derivations.
In this example, we could anticipate the conclusions. Without MacCallum et al., we would have had to do tedious algebra to obtain the results; with NET, the conclusions were reached with a few computer runs. The R code in Appendix C can be used to do these runs.
Rationale for NET Methodology
Consider a situation in which the set of possible mean and covariance matrices under M1 is equal to, or is a subset of, the set of possible mean and covariance matrices under M2. This moment matrix nesting requires that all possible sets of parameters θ1 that could generate means μ1(θ1) and covariance matrices Σ1(θ1) must generate sets of means and covariances that also are found among the means μ2(θ2) and covariance matrices Σ2(θ2). As a consequence, fitting the more general model {μ2(θ2), Σ2(θ2)} to the restricted means and covariances {μ1(θ1), Σ1(θ1)} must yield a perfect fit. In practice, the restricted model is estimated to obtain , and the more general model {μ2(θ2), Σ2(θ2)} is then fit to with some estimation methodology. If the fit is perfect and the fitted moments under the more restricted model are based on a random sample, or on a random choice of parameter values under the null hypothesis, the models are nested. Furthermore, if both models have the same degrees of freedom, they are equivalent. The mathematical and statistical basis for this methodology is given in Appendix B.
The Role of Random Sampling
The importance random sampling in our NET methodology can be visualized in a simple example. Consider the following three models for two variables X and Y
where a, b, and c are labels for free parameters. Note that M is a saturated model. We associate the names of the model with the set of covariance matrices reproduced when varying the parameters of the model, e.g., M1 is the set of covariance matrices σ1 reproduced as σxx = 1 and only b and c are varied.
In Figure 4 we represent M by unrestricted points in three-dimensional Euclidean space (a cone, if we restrict the covariance matrix to be non-negative definite); M1 is the plane defined by a = 1; M2 is the plane defined by c = 1. Note that both planes intersect on a line, say M12 = M1 ⋂ M2, which is a proper subset of both M1 and M2 where this intersection M12 is a non-empty set. Here it is clearly visible that M1 is not nested within M2, and that both M1 and M2 are nested within M. We denote nesting of A within B by A<B.
Figure 4.
M1 and M2 as Planes with Intersection M12
It is clear that NET will yield incorrect conclusions on global equivalence of M1 and M2 if a sample covariance matrix S happens to contain variances sxx = syy = 1. Although this is true, it does not impair NET because, as we have noted, NET must be evaluated on a sample moment matrix, not on a given matrix. That is, we are not allowed to choose S, which must be obtained randomly, and when it is, NET will yield the correct conclusion even if σxx =σyy = 1 are common points in both M1 and M2.
To verify that NET does not break down, suppose that we have a true random sample S from M. In Step 1 of NET, we fit M1 :σ1(1,b,c) = {σxx =1,σxy = b,σyy = c} to S and obtain with df1 = 1. Because the sampling variability of each element of S is absolutely continuous, and any single point has probability mass zero, the probability that syy = 1 is equal to zero. In Step 2, we fit M2 :σ2(a,b,1) = {σxx = a, σxy = b, σyy =1} to with df2 =1. Since s is not equal to 1, we will obtain . In Step 3 of NET we compute d = df1−df2=0. In Step 4, we see that since , we conclude that M1 is not nested in, or equivalent to, M2. Next assume that we have no sample S. To proceed, we first have to define the population M :σ(a,b,c) = {σxx = a,σxy = b,σyy = c}. To make this more challenging, we choose σxx = σyy = 1 and σxy = .4, conceivably hastening the breakdown of NET. We choose a sample of size 100, and use method (1) to draw a random sample and obtain
In Step 1 of NET, we fit model M1 to S and obtain with
In Step 2 of NET, we fit M2 to and obtain . In step 3 of NET, we compute d = df1−df2=0. In Step 4, we see again that since , M1 is not nested in, or equivalent to, M2. This procedure was repeated many times, and the NET conclusion was identical each time. Notice that the correct conclusion is reached by NET whether or not we have a sample S at hand and even when sampling from a population which is in the intersection of the two models.
With regard to M1 < M2, clearly, only if is in M12 = M1 ⋂ M2 will χ2 ≤ ε. This would require that contains the free parameter c whose sample estimate happens to be precisely 1.0. The probability of this occurring, when S is an empirical random sample, is zero! As Appendix B shows, the set M12 = M1 ⋂ M2 has Lebesgue measure zero in M1 and thus, when using a random sample, there is zero probability that falls into M12 = M1 ⋂ M2.
Roles of Parameter and Moment Structure Nesting
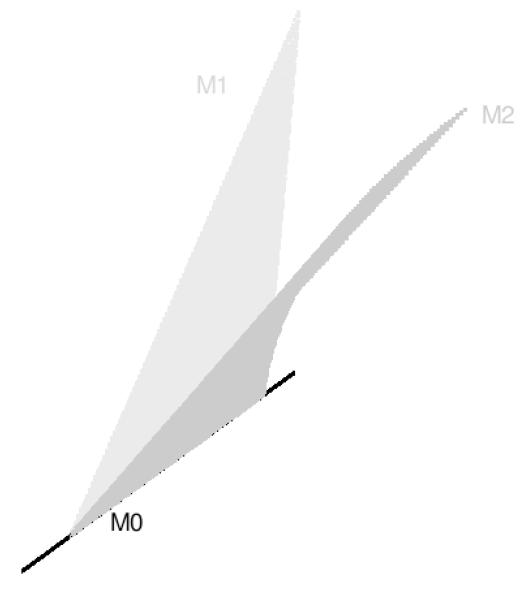
Parameter nesting is usually an obvious feature when comparing models, while moment nesting, the method used in NET, can be difficult to understand since, as a reviewer noted, it is not widely discussed in the SEM literature. We use a simple example that allows a graphical view to illustrate these types of nesting. Consider three restricted regression models M1, M2 and M0 involving just two observable variables X and Y which, for simplicity of exposition, are considered to be centered. (1) M1 is Y = γX + ey, where ey is the residual in the regression with E(Xey) = 0, γ, ϕxx = E(X2) and are scalar parameters, and E is the expectation operator. We restrict the regression so that ϕxx = ψyy. (2) M2 is X = βY + ex, where ex is the residual in the regression with E(Yex) = 0, and β, ϕyy = E(Y2) and are scalar parameters. We restrict the regression so that ϕyy=ψxx. (3) The final model M0 is X = ex, with E(Yex) = 0.
Model M0 clearly is parameter nested in M2, since it can be obtained by setting β = 0. More difficult tasks are to see whether M1 and M2 are nested or equivalent and whether M0 is also nested in M1. Using the NET procedure outlined above without data, we generate random data of sample size n = 500 from a 2 -variate standard normal distribution with arbitrary correlation .36. Its sample covariance matrix S had variances 1.017 and 1.029 and covariance .374 (to 3 decimals). When M1 was fitted to S, we obtained and the fitted covariance matrix with variances 1.081 and .955 and covariance .347. We then fitted M2 to and obtained . So, according to NET, M1 is not nested in M2.
To assess whether M0 is nested in M1, we repeated the process of generating a sample moment matrix S. M0 was then fitted to this S, yielding a fitted and . Fitting M2 to yielded a perfect fit with . Hence, according to NET, M0 is nested in M1. Finally, we repeated the same process but now with respect to M2, and, as we would require from parameter nesting, NET also concluded that M0 is nested in M2.
Covariance algebra can be given to verify these NET results. A picture may be even more informative. Figure 5 shows the two models M1 and M2 as two manifolds, and well as the model M0. Clearly, M0 is in the intersection of the two manifolds, meaning that it is moment nested in both M1 and M2 as NET had shown. The figure also shows that M1 is not nested in M2, and similarly, M2 is not nested in M1.
Figure 5.
Nesting Relations Among Three Models
In this case, the nesting results are global, as we verified empirically by obtaining identical results from NET analysis regardless of a wide variation of the population covariance matrix used to generate the sample matrix S. We discuss this general issue more fully next.
Local vs Global NET
Although the requirements for NET are described above and in Appendix B, their implications may not be obvious and so we discuss further one key issue: Are NET results guaranteed to hold globally for all conceivable sets of parameters and moment matrices, or do they hold only locally in some small neighborhood of the current randomly drawn data from μ, Σ ? In general, the topological structure (on a p-dimensional Euclidean space) of matrices reproduced by variation on the parameters of the model can be too complex for addressing results of global equivalence and nesting. Thus NET gives results that apply to model equivalence and nesting on a neighborhood of the sample covariance matrix (and sample means where relevant). This requirement is not unusual, since it is, in fact, within such a neighborhood that standard statistical inference and χ2 difference testing operates (see e.g., Satorra, 1989).
The issue of nesting and equivalence in the pathological case of set of Lebesgue measure zero has a parallel in the area of model identification. For example, the four variable 2-factor confirmatory factor (CFA) model with factor loadings (λ1,λ2,0,0) for factor 1, loadings of (0,0,λ3,λ4) for factor 2, factor variances of 1, and factor correlation ϕ12 is identified almost everywhere. But there are special situations where it is not identified, e.g., when ϕ12 = 0. This limitation is far less serious than it might seem. In contrast to traditional theoretical discussions of equivalence and nesting that utilize only sets of population moment matrices and their relations, we make use of the properties of simple random sampling from a continuous population to almost always rule out pathological cases.4 We assume that researchers know the neighborhood where testing is to be done, after all, they should be able to at least conceptually specify the population μ, Σ from which the random sample , S is drawn. While a pathological sample, e.g., a sample where identification breaks down, is possible, this will occur with probability zero. For example, in the 4-variable CFA model just discussed, if the population ϕ12 ≠ 0, the probability is zero that a random sample S has exactly 0.0 values for all four cross-covariances of variables {1,2} with {3,4}, or values where . This is not to say that it is impossible to observe a sample from such a pathological set of Lebesgue measure zero, but it will occur with probability zero since we require S to be randomly drawn from the relevant population.
There are indeed situations where global and local results will differ depending on regions of the parameter or data space considered. An example is with three variables X, Y, and Z, and two models M1 and M2. The equations for M1 are Z = X + e, Y = γX + ε, with uncorrelated independent variables with variances , , . M2 contains a phantom variable F (Rindskopf, 1984) and has equations Z = αX + d, F = βX, Y =βF + δ with uncorrelated independent variables with variances , , . Is M1 is nested in M2 ? Suppose we consider the parameter space under the data restriction σxy ≥ 0. This covariance is reproduced by γ in M1 and by β2 in M2, so the data restriction implies the parameter inequality γ > 0. This inequality can be met. We can set γ = β2, 1 = α, , and model M1 is nested in M2 because the free parameter α in M2 is restricted to 1.0 in M1. However, if we consider the parameter space where σxy < 0, in M1 we have γ = σxy while in M2 we have the imaginary number (now the square root of a negative number!). Model M2 with nonimaginary parameters will not reproduce the population covariances. So we can conclude that (a) with real parameters, the models are not nested; (b) with σxy ≥ 0 and real parameters, the models are nested; (c) with σxy < 0 and real parameters, the models are not nested; and (d) allowing imaginary parameters, the models are nested. For the typical case of real parameters, the global conclusion is that the models are not nested. The local conclusion depends on the sign of σxy and, in a sample, on the sign of its sample equivalent sxy. Note that even though this example with its inequality constraints contradicts one of the requirements of our method, NET asymptotically will yield the correct local answer (b) or(c), depending on the sign of σxy.
Bootstrap Net
Conclusions from NET will represent nesting and equivalence in the neighborhood of the population μ, Σ if the sample size is large enough, e.g., in the example above, if the sign of sxy is the same as the sign of σxy. If there is a question on whether NET might yield different results for different samples, NET can be applied repeatedly to bootstrap resamples from the data (see e.g., Yung & Bentler, 1996). Varying results would imply that conclusions are not invariant across regions of the sample space; we would expect this to occur rarely with standard models.
An Equivalence Example with Non-regular Points
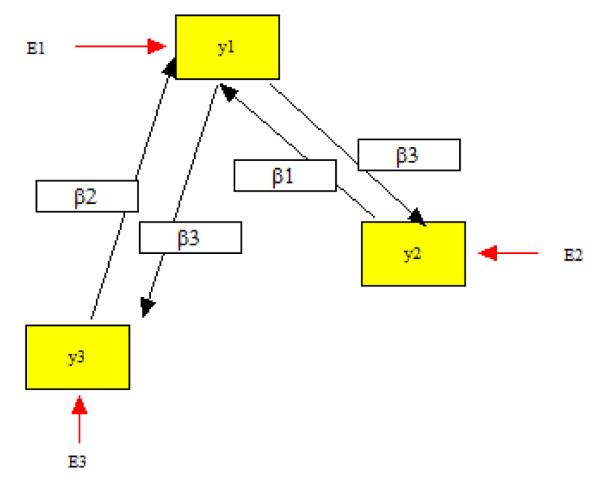
An important question is whether an empirical methodology such as NET can handle models that are evaluated at points close to empirical nonidentification.5 This example shows that NET can draw correct conclusions in such a case. We use a nonrecursive model that was analyzed in mathematical detail by Bekker, Merckens and Wansbeek (1994, p. 163, eq. 7.3.1), using the rank of elaborated Jacobian matrices in symbolic form. The model is
where the disturbances E1, E2, E3 are uncorrelated with zero expectations and variances , , . A path diagram is given in Figure 6. The model has six parameters and zero df. Bekker et al. show that although this model is identified in general, at the point {β1 = β2 = 0} the Jacobian is singular and the parameters are not identified. We use this knowledge to generate a multivariate normal population with {β1 = β2 = 0, β3 = 0.5, }, and draw a sample of N=300 from it using our method (2a) to deal with absence of a sample covariance matrix, namely, we choose parameter values and then sample from the corresponding model (see also Appendix A).6 This sample is used to evaluate the equivalence of two submodels A and B, where Model A is defined by β2 = 0 and β1, β3, , , as free parameters, and Model B is defined by β1 = 0 and β2, β3, , , free. Note that it is not at all obvious from Figure 6 that these are equivalent models, although Bekker et al. prove that they are locally equivalent. The question is whether the results from NET agree with the technical mathematical proof, even when sampling from a point that is not regular for this model with zero df (i.e., a point where the model is not identified).
Figure 6.
A Nonrecursive Model
The sample covariance matrix we obtained is given by
In Step 1 of NET, Model A was fit to this matrix with ML, yielding the goodness-of-fit . The parameter estimates {, , , , } were used to obtain , which was then used in Step 2 of NET as the input covariance matrix to be analyzed by Model B. Model B yielded . Applying Steps 3 and 4 of NET, the difference in d = 1−1 = 0 and , so Models A and B are locally equivalent. This example is a small model but it illustrates the type of complexities that may arise in practice. Even when starting with a non-regular population, NET yields the same conclusions as the elaborate mathematics of Bekker et al.
Discussion
In principle, the approach outlined here for evaluating model nesting and equivalence applies to a wide variety of related modeling situations such as multiple group models or higher moment structures in SEM, log-linear models in categorical data situations, and so on. Since incremental fit indices are almost universally reported in SEM, one of the simplest yet most important areas of application is that of verifying the nestedness of a baseline or null model for the computation of incremental fit indices. Although Bentler and Bonett (1980) had emphasized that an appropriate null model for such indices must be a nested model relative to the substantive model of interest, they also noted that “there are cases when it is not a simple matter to verify that M0 [the null model] is a special case of Mk, Ml [substantive models], or Ms [the saturated model]. Nonetheless, this is a fundamental requirement of M0” (p. 596). Such a difficulty may be inevitable when the researcher uses prior results (Sobel & Bohrnstedt, 1985), an equal-correlation baseline model (Rigdon, 1998), or some untested ideas to specify the null model. The only standard default used in SEM programs that we know about is the uncorrelated variables model as the baseline model, yet, as Widaman and Thompson (2003) show, this can be an inappropriate model choice when it is not nested in the model of interest. As illustrated in our example, the methodology provided here can be easily implemented to routinely evaluate nesting for incremental fit indices, especially when the null model is provided by a program default.
It may seem that missing data could provide difficulties for NET, but any methodology that permits a stochastically derived empirical covariance matrix S (and a vector of means when needed) can be used in the step 1 of NET. For example, the two-stage maximum likelihood method described by Yuan and Bentler (2000), Savalei and Bentler (2009), Cai (2008), and Yuan and Lu (2008) can be used with the proposed NET framework. In the two-stage approach, a saturated model (sometimes called the EM means and covariance matrix) is first estimated. These saturated means and covariances are then taken as the sample means and covariances to be modeled in the subsequent step. For the NET procedure, the saturated moments are taken as the data to be analyzed in Step 1 above, and model M1 is fit to it. Then Steps 2-4 are completed as usual.
A useful application is to evaluate nestedness to determine whether a χ2 difference test is appropriate when nesting is not obvious. For example, consider the 4-variable CFA model M2 with two factors F1 and F2. Suppose the F1 loadings are (1,λ2,0,0) and the F2 loadings are (0,0,1,λ4), with λi, , , and covariance ϕ12 as free parameters. Compare this to the 1-factor model M1 with free loadings (λ1,λ2,λ3,λ4) and . The 1-factor model can not be obtained by simply fixing some free parameters in the 2-factor model, i.e., the models are not parameter-nested. They are covariance-matrix nested; after all, M1 can be obtained from a new model by parameter nesting, where is the equivalent model to M2 obtained by freeing its loadings of 1 and fixing the factor variances at 1. Setting ϕ12 =1 in leads to M1. NET would prove this directly by fitting M1 to a random S, and generating . A fit of M2 to would yield .7
NET can also provide a theoretical justification of the typical method for determining whether a given model Mk is a saturated model. It is well-known that a saturated model is one with 0 df that perfectly reproduces any sample covariance matrix S. Hence, running Mk and getting a perfect fit with 0 df implies saturation. NET justifies this conclusion, since all saturated models are equivalent, and a well-known saturated model model Ms is the one whose free parameters are the components of the moment vector. In the language of NET, Mk will be saturated if Ms is nested or equivalent to Mk. Step 1 of NET is not needed, as the fitted moment matrix is the same as S ; Step 2 fits Mk to S, concluding nesting (i.e., saturation) or not as in standard NET practice.
Traditional approaches to nesting and equivalence do not use data at all; they are concerned with algebraic and set-theoretic relations holding among classes of population means and covariance matrices. Our approach is unique in that nesting and equivalence are evaluated on a random sample of data from the population. We substitute probability theory for mathematical certainty. As a result, the NET methodology theoretically can fail when a truly bizarre sample is chosen. However, the theorem given in Appendix B verifies that this will occur with a probability approaching zero. If there is suspicion about the results, simulated random samples from the population or bootstrap resamples from a given sample can be drawn and the NET methodology applied as a verification. Although it may be tempting to create an artificial example where the NET methodology breaks down, it should be remembered that NET cannot be used on an artificial covariance matrix. It should only be applied to a sample covariance matrix.
An immediate question, then, is how to apply the proposed NET procedure when there are no data at hand, i.e., when no sample covariance matrix S (and means, when relevant) is available for Step 1 of our method. The solution is to generate a randomly simulated sample covariance matrix S or a model-implied covariance matrix . This can be done in various ways using simulation and random data generation as was discussed and illustrated earlier and in Appendix A. While our NET procedure suggests sampling from the true population Σ, we have found that drawing a random sample near Σ works as well. Surprisingly, and though we do not recommend it, we have found that it often works when data is randomly drawn from a possibly widely different population, such as one with covariance matrix Σ = I. Clearly, further research is needed to understand the boundaries of NET.
An important theoretical point, and one that implies further research, is the local nature of our NET procedure. As is made precise in Appendix B, by “local” we mean that we restrict attention to moment matrices in a neighborhood of a given moment matrix σ0 that has non-null intersection with model M1. NET provides results that are valid in a local sense, based on assumptions parallel to those that underlie the usual χ2 difference test. Of course, we concentrate on a region of the parameter space where it is likely that the true population value lies, which justifies taking a sample covariance matrix in this region. Even though the results hold locally, depending on the class of models, the topological local structure is often the same as the global one, in which case local nesting or equivalence implies the global one. Yet this is not always true, as our example with an inequality constraint demonstrates. It is a matter of further research to investigate the topological local versus global nature of moment spaces for different families of SEM models, which will have implications for model identification as well as nesting and equivalence. Even abstract studies on identification and equivalent models have focused primarily on local identification and only occasionally on global identification (Bekker et al., 1994, p. 19).
Throughout this paper, we emphasized the importance of using random matrices to ensure that NET results apply almost everywhere on a neighborhood of a chosen point. Points of the parameter space that are NET non-regular (in the sense that nesting or equivalence relations differ from what applies almost everywhere in its neighborhood) will thus not be captured, since such points arise with probability zero when the input to Step 1 is indeed a true random matrix. This is similar as in the 2-factor, 2-indicator model that is identified almost everywhere except on the set of points where the correlation between factors is exactly zero. We now note that NET can indeed also be used to address results of nesting or equivalence at a particular point, such as the non-regular points that may exist in the parameter space. However, when we choose a specific moment matrix in Step 1, we have to be clear that NET conclusions then no longer apply almost everywhere in the neighborhood of the chosen matrix. In some instances, this non-standard use of NET may provide interesting information.
With regard to model equivalence, we should note that our goals have been modest. We proposed a way to evaluate whether any two models that a researcher nominates might be equivalent moment structure models. Making such a comparison is critical to ruling out potentially competing explanations of a phenomenon (MacCallum, Wegener, Uchino, & Fabrigar, 1993; Stelzl, 1986). However, our methods do not address the more complicated problem of starting with only one model and generating an entire class of models that might be equivalent to a given model. Rules for specifying some equivalent models have been developing for over two decades since Stelzl (1986) first pointed out that alternative causal hypotheses could yield identical indices of model fit (e.g., Lee & Hershberger, 1990; Luijben, 1991; Hershberger, 1994, 2006). These rules can be complex and difficult to implement with complete accuracy, so our NET procedure can make a contribution by providing a method for evaluating models that are considered to be candidates for equivalence through use of these rules. Since there may be a lot of equivalent models (Raykov & Marcoulides, 2001, 2007; Markus, 2002), a computer algebra surely will need to be incorporated into SEM programs to help the researcher generate and evaluate such candidates. Our methods also do not address the complicated issue of whether models might be equivalent in the broader sense of observational equivalence, implying that their generating probability distributions are equivalent. This topic may require studying individual case scores and residuals (e.g., Raykov & Penev, 2001).
Acknowledgments
Research supported by grants 5K05DA000017-32 and 5P01DA001070-35 from the National Institute on Drug Abuse and grants SEJ2006-13537 and PR2007-0221 from the Spanish Ministry of Science and Technology. The helpful and challenging comments of the Associate Editor and reviewers on earlier drafts are gratefully acknowledged.
Appendix A
NET requires convergence in model fit of the algorithms used. This may be a limitation in some cases, where the model is very complex and/or large deviations of sample moments from the hypothesized model arise. Such a problem frequently occurs, for example, with non-recursive structural equation models. A practical solution is to tighten up the neighborhood where NET applies. One approach to doing this is to assure that the specified population moment vector σ0 is in M1 and the analyzed moment vector s is close enough to σ0. This can be done by using methods (2a) or (2b) described in the text. In both of these methods, enlarging the sample size will tighten the neighborhood around σ0 as much as is needed for reaching convergence in computations required for the NET procedure. At some large enough sample size, the models will be locally identified, the algorithm will converge, and NET will give a Yes/No answer regarding nesting and equivalence. If doubts remain, alternative sample input covariance matrices based on various sample sizes can be studied to determine if they yield identical conclusions.
Appendix B
Let σ ≡ [μ’,vech(Σ)’]’ and consider two moment structural models M1 :σ = σ1(θ1), θ1 ∈ Θ1 and M2 :σ = σ2(θ2), θ2 ∈ Θ2 defined on the space of unconstrained moments C = {σ ∈ Rp |σ ∈ Θ}, where Θ1 ⊂ Rq1, Θ2 ⊂ Rp are open sets, and σ1(.) and σ2(.) are continuously differentiable functions.8 Consider the subsets C1 and C2 of C associated to the two models, and their intersection C12 = C1 ⋂ C2. It holds that (see, e.g., Satorra, 1989)
Result 1. For any σ0 ∈Cj locally9 around σ0 there is a differentiable vector-valued function hj of full row rank where Cj = {σ ∈ Rp | hj(σ) = 0}.
Assume that s is a sample realization from an absolutely continuous distribution in the set C .10 Consider the fit of M1 to s and let be the fitted moment vector. Consider the fit of M2 to and let F12 be the minimum of the fitting function. Assume that the Jacobians of h1 and of h = (h1’, h2’)’ are regular at . Under this set-up, we have the following theorem.
Theorem. Let s be a sample moment vector and F12 the associated fit value of Step 3 of NET. Then locally at
F12 = 0 implies with probability one that M1 is nested in M2
F12 > 0 implies with probability one that M1 is not nested in M2.
Proof: Current assumptions and Result 1 imply that C1 , C2 and C12 are manifolds, with the dimension of C12 smaller than or equal to the dimensions of C1 . If the dimension of C12 is strictly smaller than the dimension of C1 then Sard’s Theorem11 implies that C12 is of Lebesgue measure zero in C1 and thus there is probability zero of , hence of F12 = 0 .12 Note that the distribution of is absolutely continuous in C1 with nonsingular covariance matrix, since the Jacobian of h1 is of full row rank. So, if we observe F12 = 0 , with probability one the dimension ofC12 equals the dimension of C1 , and thus the two models are nested. Clearly, if M1 is locally nested in M2 , necessarily F12 = 0 ; so, F12 > 0 implies that M1 is not locally nested in M2 . Q.E.D.
This theorem can be illustrated with a graph such as the one in Figure B1. The graph shows model M2 of dimension 2, and two models M1 (models M1a and M1b) both of dimension 1. With regard to the intersection M12 of the two models, we have the following situation: when M1 = M1a , M12 is of dimension one less than M1 ( M12 is a point); when M1 = M1b , M12 is of dimension equal to the dimension of M1 ( M12 equals M1). Thus, M1 = M1a is a case of M1 non-nested in M2 , and M1 = M1b is a case of M1 nested in M2 . Note that a sample moment s would be standing in the tridimensional space with probability one deviating from the manifolds. Step 1 of NET projects s into M1 obtaining ; clearly, in the case of M1 = M1a , the projected point has probability zero of standing also in M2 (since the intersection of M12 is a point), and so F12 > ò); while in the case of M1 = M1b , also would be standing in M2 , and so F12 < ò .
Figure B1.
Model M2 and Two Different Nested Models M1a and M1b
This result suggests the procedure of the Nesting and Equivalence Test given in the text, where F12 is designated as , concluding that M1 < M2 when F12 < ò , for ò a small constant; model equivalence when in addition to F12 < ò , the two models have the same number of degrees of freedom ( d = 0 ) ; or non-equivalence and non- nesting of M1 in M2 when F12 > ò.
Appendix C
A NET analysis can be undertaken using the R function net1.R given below. This makes use of the R interface REQS (Mair, Wu, & Bentler, in press) between R (Development Core Team, 2009) and EQS (Bentler, 2006)13 that contains functions to read EQS script files and import the results into R, call EQS script files from R, run EQS script files from R, and import the results after computation.
An illustration is the NET evaluation of models A and B of Figure 3. Models have to be specified in *.eqs files along with the number of variables p . The function call net(“MA.eqs”,“MB.eqs”, p= 5) implies that “MA.eqs” (any name with extension .eqs) is the EQS file for M1 and “MB.eqs“ is the EQS file for M2 , and p = 5 . Based on the appropriate *.eqs input files, R produces the result
[1] “M1 is nested to M2” $FitToModel1 [1] 80.442 3.000 $FitToModel2 [1] 0 1
This reports the conclusion first, here, that model A is nested in model B, or more generally, whether or not the model in the first *.eqs file (MA.eqs) is nested in the model of the second *.eqs file (MB.eqs). The numerical output then gives the χ2 and df for the fit of M1 to a randomly generated s , and finally provides the results of the fit of M2 to the fitted matrix . In this case, with 1 df.
The analysis implemented in net1.r does not require a data matrix. It generates simulated data from a spherical normal distribution. An R user can easily modify this function to accommodate other simulation options as described in the text.
Function net1.R for NET Computations in R and EQS
library(“gtools”)
library(“REQS”)
library(“mvtnorm”)
net= function(model1,model2, p){
setwd(“/A/UCLA2008research/NestingSEM/Revisions/Revision_II/REQSrunsNET/RNET”)
#----------------------- begin NET -------------------------------
### machine accuracy parameter
epsilon= .00001
n <- 500 #total sample size
Sigma1 <- matrix(0, p, p) #VC matrix component 1 (3 variables)
diag(Sigma1) <- 1
X1 <- rmvnorm(n, mean = rep(0, p), sigma = Sigma1)
S1 = cov(X1)
write.table(S1, file = “factorcov.dat”, col.names = FALSE, row.names = FALSE)
###EQS under M1
res.F1 <- run.eqs(EQSpgm = “/A/MACEQS/maceqs”, EQSmodel = model1, serial = “1234”)
Fit1 <- res.F1$fit.indices[c(“CHI”),]; DF1=res.F1$model.info[5,]
‘Fit and DF of S under M1’
FITM1= c(round(Fit1,3),round(DF1))
FITM1
FS1 = res.F1$sigma.hat
###EQS under M2
write.table(FS1, file = “factorcov.dat“, col.names = FALSE, row.names = FALSE)
### un EQS under M1
res.F1 <- run.eqs(EQSpgm = “/A/MACEQS/maceqs”, EQSmodel = model2, serial = “1234”)
### end EQS under M2
Fit1 <- res.F1$fit.indices[c(“CHI”),]; DF1=res.F1$model.info[5,]
‘Fit and DF of S1 under M2’
FITM2= c(round(Fit1,3),round(DF1))
FITM2
if (Fit1 < epsilon){ print(“M1 is nested into M2”)}
if (Fit1 > epsilon){ print(“M1 is NOT nested into M2”)}
list(FitToModel1 = FITM1 ,FitToModel2= FITM2)
#-----------------------end NET -------------------------------
}Footnotes
Further technical details on required regularity conditions are given e.g., Steiger, Shapiro, and Browne (1985) and Satorra (1989).
Our proposed methods are rooted in mathematical statements that presuppose we have a machine with infinite precision; that is, that we are able to assess whether the fit value is exactly zero or not (in examples, we report to only 5 decimal places). The precision of computers in current use require, however, that we set a bound ò > 0 asserting that F = 0 whenever F < ò. As commented by a reviewer, the introduction of such a potential “rounding” error opens the door for nonzero probability (in a single try) of concluding nesting when in fact the models are not nested. This probability of an incorrect conclusion has limit zero as ò → 0. For a fixed ò, i.e. with today’s computer precision, we can make the probability of an incorrect conclusion also to go to zero by the simple procedure of replicating the analysis under the given model (with or without a data set). Notice that replication here is very easily performed using an automatic version of NET as the one proposed in Appendix C. No cost of collecting new data is required, since replication is just a matter of simulation -- it is similar to bootstrap evaluation of standard errors, where precision can be gained just at the expense of more computer CPU time.
Substantively, the variables Y1,Y2,Y3,X1,X2 correspond to “Task Mastery Goals”, “Ego-Social Goals”, “Active Cognitive Engagement”, “Intrinsic Motivation” and “Science Attitudes.” To save space, we do not emphasize the substantive meaning of our example.
“In probability theory, a probability distribution is called continuous if its cumulative distribution function is continuous. That is equivalent to saying that for random variables X with the distribution in question, Pr[X = a] = 0 for all real numbers a, i.e.: the probability that X attains the value a is zero, for any number a.” (http://en.wikipedia.org/wiki/Continuous_probability_distribution)
This would occur when we are close to non-regular points of the Jacobian of the model. A regular point θ0 is one where the rank of a matrix function is constant in an open neighborhood of θ0 (e.g., Bekker et al., 1994, p. 19). At a non-regular point, the Jacobian and information matrix may become singular, so the parameters become underidentified.
We avoided parameter values that would yield a nonconvergent or unstable nonrecursive model (see Bentler & Freeman, 1983). Bekker et al. did not discuss whether their analysis and conclusions would change in such a case.
A reviewer raised the question of whether we could reverse the procedure to disconfirm a special case of a general model. Here we might first fit M2 to get , and then fit M1 to , to see if we could disconfirm that the two factors correlate 1.0. After all, would be obtained if the two factors underlying were correlated 1.0. However, the NET requirement that M2 is fit to a random S will guarantee that the probability is zero that these two factors will correlate 1.0. We would certainly obtain .
Vech vectorizes a matrix, suppressing the redundant elements due to symmetry.
Locally means that the statement is restricted to an open ball C(σ0,δ) = {σ ∈ Rp | ∥σ−σ0∥<δ} where ∥.∥ denotes Euclidian norm and σ0 and δ(> 0) are respectively the center and radius of the ball.
That the distribution is absolutely continuous with nonsingular covariance matrix is essential, but is not restrictive, since it encompasses all structural equation models except for the case of a degenerate distribution. It excludes the rare case of a distribution of the sample moments that is discrete, not continuous.
A simplified version of Sard’s Theorem is “If f : A → Rp is continuously differentiable and A → Rp is open, then the set has measure zero in Rp” (Spivak, 1965, Thm. 3-14). Here, refers to the determinant of the Jacobian.
For purposes of the present paper, we only require Result 5-8 of Spivak (1965 p. 115) which follows from Sard’s theorem : if M is a k-dimensional manifold in Rn and k < n then M has measure zero. In our application: when the dimension of C12 is strictly smaller than the dimension of C1 then C12 is of Lebesgue measure zero in C1.
Bentler acknowledges a financial interest in EQS and its distributor, Multivariate Software.
Publisher's Disclaimer: The following manuscript is the final accepted manuscript. It has not been subjected to the final copyediting, fact-checking, and proofreading required for formal publication. It is not the definitive, publisher-authenticated version. The American Psychological Association and its Council of Editors disclaim any responsibility or liabilities for errors or omissions of this manuscript version, any version derived from this manuscript by NIH, or other third parties. The published version is available at www.apa.org/pubs/journals/met
Contributor Information
Peter M. Bentler, University of California, Los Angeles
Albert Satorra, Universitat Pompeu Fabra.
References
- Bekker PA, Merckens A, Wansbeek TJ. Identification, equivalent models, and computer algebra. Academic; London: 1994. [Google Scholar]
- Bentler PM. Comparative fit indexes in structural models. Psychological Bulletin. 1990;107:238–246. doi: 10.1037/0033-2909.107.2.238. [DOI] [PubMed] [Google Scholar]
- Bentler PM. EQS 6 structural equations program manual. Multivariate Software; Encino, CA: 2006. [Google Scholar]
- Bentler PM, Bonett DG. Significance tests and goodness of fit in the analysis of covariance structures. Psychological Bulletin. 1980;88:588–606. [Google Scholar]
- Bentler PM, Freeman EH. Tests for stability in linear structural equation systems. Psychometrika. 1983;48:143–145. [Google Scholar]
- Byrne BM. Structural equation modeling with EQS: Basic concepts, applications, and programming. 2nd ed Erlbaum; Mahwah, NJ: 2006. [Google Scholar]
- Cai L. SEM of another flavour: Two new applications of the supplemented EM algorithm. British Journal of Mathematical and Statistical Psychology. 2008;61:309–329. doi: 10.1348/000711007X249603. [DOI] [PubMed] [Google Scholar]
- Henley AB, Shook CL, Peterson M. The presence of equivalent models in strategic management research using structural equation modeling: Assessing and addressing the problem. Organizational Research Methods. 2006;9:516–535. [Google Scholar]
- Hershberger SL. The specification of equivalent models before the collection of data. In: von Eye A, Clogg CC, editors. Latent variables analysis. Sage; Thousand Oaks, CA: 1994. pp. 68–108. [Google Scholar]
- Hershberger SL. The problem of equivalent structural models. In: Hancock GR, Mueller RO, editors. Structural equation modeling: A second course. Information Age; Greenwich, CN: 2006. pp. 13–41. [Google Scholar]
- Kline RB. Principles and practice of structural equation modeling. Guilford; New York: 2005. [Google Scholar]
- Lee S, Hershberger S. A simple rule for generating equivalent models in covariance structure modeling. Multivariate Behavioral Research. 1990;25:313–334. doi: 10.1207/s15327906mbr2503_4. [DOI] [PubMed] [Google Scholar]
- Luijben TC. Equivalent models in covariance structure analysis. Psychometrika. 1991;56:653–665. [Google Scholar]
- MacCallum RC, Wegener DT, Uchino BN, Fabrigar LR. The problem of equivalent models in applications of covariance structure analysis. Psychological Bulletin. 1993;114:185–199. doi: 10.1037/0033-2909.114.1.185. [DOI] [PubMed] [Google Scholar]
- Mair P, Wu E, Bentler PM. EQS Goes R: Simulations for SEM using the package REQS. Structural Equation Modeling. (in press) [Google Scholar]
- Markus K. Statistical equivalence, semantic equivalence, eliminative induction, and the Raykov-Marcoulides proof of infinite equivalence. Structural Equation Modeling. 2002;9:503–522. [Google Scholar]
- Meece JL, Blumenfeld PC, Hoyle RH. Students’ goal orientations and cognitive engagement in classroom activities. Journal of Educational Psychology. 1988;80:514–523. [Google Scholar]
- Mulaik SA. Linear causal modeling with structural equations. Chapman & Hall/CRC; Boca Raton: 2009. [Google Scholar]
- Raykov T, Marcoulides GA. Can there be infinitely many models equivalent to a given covariance structure model? Structural Equation Modeling. 2001;8:142–149. [Google Scholar]
- Raykov T, Marcoulides GA. A first course in structural equation modeling. Erlbaum; Mahwah, NJ: 2006. [Google Scholar]
- Raykov T, Marcoulides GA. Equivalent structural equation models: A challenge and responsibility. Structural Equation Modeling. 2007;14:695–700. [Google Scholar]
- Raykov T, Penev S. On structural equation model equivalence. Multivariate Behavioral Research. 1999;34:199–244. doi: 10.1207/S15327906Mb340204. [DOI] [PubMed] [Google Scholar]
- Raykov T, Penev S. The problem of equivalent structural equation models: An individual residual perspective. In: Marcoulides GA, Schumacker RE, editors. New developments and techniques in structural equation modeling. Erlbaum; Mahwah, NJ: 2001. pp. 297–322. [Google Scholar]
- R Development Core Team R: A language and environment for statistical computing. 2009 Available from http://www.R-project.org (ISBN 3-900051-07-0)
- Rigdon EE. The equal correlation baseline model for comparative fit assessment in structural equation modeling. Structural Equation Modeling. 1998;5:63–77. [Google Scholar]
- Rindskopf D. Using phantom and imaginary latent variables to parameterize constraints in linear structural models. Psychometrika. 1984;49:37–47. [Google Scholar]
- Satorra A. Alternative test criteria in covariance structure analysis: A unified approach. Psychometrika. 1989;54:131–151. [Google Scholar]
- Savalei V, Bentler PM. A two-stage approach to missing data: Theory and application to auxiliary variables. Structural Equation Modeling. 2009;16:477–497. [Google Scholar]
- Sobel ME, Bohrnstedt GW. Use of null models in evaluating the fit of covariance structure models. In: Tuma NB, editor. Sociological methodology. Jossey Bass; San Francisco: 1985. pp. 152–178. [Google Scholar]
- Spivak M. Calculus on manifolds. Perseus Books; Reading, MA: 1965. [Google Scholar]
- Steiger JH, Shapiro A, Browne MW. On the multivariate asymptotic distribution of sequential chi-square statistics. Psychometrika. 1985;50:253–264. [Google Scholar]
- Stelzl I. Changing a causal hypothesis without changing the fit: Some rules for generating equivalent path models. Multivariate Behavioral Research. 1986;21:309–331. doi: 10.1207/s15327906mbr2103_3. [DOI] [PubMed] [Google Scholar]
- Votaw DF., Jr. Testing compound symmetry in a normal multivariate distribution. Annals of Mathematical Statistics. 1948;19:447–473. [Google Scholar]
- Widaman KF, Thompson JS. On specifying the null model for incremental fit indices in structural equation modeling. Psychological Methods. 2003;8:16–37. doi: 10.1037/1082-989x.8.1.16. [DOI] [PubMed] [Google Scholar]
- Yuan K-H, Bentler PM. Three likelihood-based methods for mean and covariance structure analysis with nonnormal missing data. Sociological Methodology. 2000:165–200. [Google Scholar]
- Yuan K–H, Lu L. SEM with missing data and unknown population distributions using two-stage ML: Theory and its application. Structural Equation Modeling. 2008;43:621–652. doi: 10.1080/00273170802490699. [DOI] [PubMed] [Google Scholar]
- Yung Y–F, Bentler PM. Bootstrapping techniques in analysis of mean and covariance structures. In: Marcoulides GA, Schumacker RE, editors. Advanced structural equation modeling techniques. LEA; Hillsdale, NJ: 1996. pp. 195–226. [Google Scholar]