Abstract
MicroRNAs (miRNAs) are a class of endogenous, noncoding, short RNAs directly involved in regulating gene expression at the posttranscriptional level. High conservation of miRNAs in plant provides the foundation for identification of new miRNAs in other plant species through homology alignment. Here, previous known plant miRNAs were BLASTed against the Expressed Sequence Tag (EST) and Genomic Survey Sequence (GSS) databases of Vigna unguiculata, and according to a series of filtering criteria, a total of 47 miRNAs belonging to 13 miRNA families were identified, and 30 potential target genes of them were subsequently predicted, most of which seemed to encode transcription factors or enzymes participating in regulation of development, growth, metabolism, and other physiological processes. Overall, our findings lay the foundation for further researches of miRNAs function in Vigna unguiculata.
1. Introduction
MicroRNAs (miRNAs) are a class of endogenous, small, noncoding, single-stranded RNAs that act as posttranscriptional regulators in eukaryotes [1]. They have been reported to be located mostly within noncoding regions of genomes, and usually transcribed from RNA polymerase II promoters [2, 3]. The generation of mature miRNA is a complicated enzyme-catalyzed process, from the initial transcript pri-miRNA to the precursor (pre-miRNA) with a characteristic hairpin structure, then a miRNA duplex (miRNA : miRNA*) [4]. In the end, it is assembled to the RNA-induced silencing complex (RISC) to direct its activity on a target mRNA, depending on the degree of base-pairing between the miRNA and the responsive element and results in either cleavage or translational repression of the target mRNA. Perfect complementarity generally results in cleavage, such as in plants, whereas imperfect base-pairing leads to translational repression [4, 5].
MiRNA genes represent about 1%-2% of the known eukaryotic genomes and constitute an important class of fine-tuning regulators that are involved in several physiological or disease-associated cellular processes [6]. For example, studies in plants have revealed the key roles of miRNAs in diverse regulatory pathways, including growth, development, and defense response against every sort of stress [7–17].
Considering the importance of miRNAs in gene regulation, two major categories of approaches have been applied for miRNA investigation [1]. Compared to the experimental approaches, computation (bioinformatics) methods have been proved to be faster, more affordable, and more effective, contributing mostly to today's plentiful storage in miRBase [1]. Different computational miRNA finding strategies have been developed based on a core principle of looking for conserved sequences between different species that can fold into extended hairpins [18]. The biogenesis of miRNAs suggests that it is possible to find miRNAs by searching Expressed Sequence Tags (ESTs) with known miRNAs. There have been more and more reports about the identification of miRNAs by mining the repository of available ESTs [19–26]. EST analysis makes it possible to rapidly study miRNAs and their functions in species whose genome sequences have not been well known [27].
Vigna unguiculata is a vital leguminous crop in tropical and subtropical areas of Asia, Africa, and Latin America, as well as parts of southern Europe and the USA [28]. High protein and carbohydrate contents make it not only important to the human diet, but also suitable as high protein feed and fodder to livestock. With its greater tolerance to heat, drought, and low soil fertility, Vigna unguiculata is a particularly valuable component of low-input farming systems of resource-poor farmers. Also, it is able to enhance soil fertility through nitrogen fixation [28, 29]. Until recently limited progress has been made in basic gene discovery and gene regulation in Vigna unguiculata, and only two new miRNAs of it have been reported [29, 30].
Despite the limited genome resources of Vigna unguiculata, publishing of EST and GSS databases in GenBank has provided the chance to get more genetic information. In this study, new miRNAs were mined for the purpose of understanding their roles in regulating growth, development, metabolism, and other physiological processes in Vigna unguiculata.
2. Materials and Methods
2.1. Sequences and Software
The known plant miRNA sequences from Arabidopsis, Brassica, Glycine, Saccharum, Sorghum, Vitis, Solanum, Oryza, Triticum, Chlamydomonas, and other plant species were downloaded from the miRNA database miRBase (http://www.mirbase.org/) (Release 14: Sept 2009). After removal of the repeated sequences, 2177 items were left as the reference set. The 187660 EST and 54194 GSS sequences of Vigna unguiculata were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/), Blast-2.2.21-ia3 was downloaded from NCBI and set up locally. RNA secondary structure and the free energy were calculated by web server mfold (http://mfold.bioinfo.rpi.edu/) [31]. The software MiRNAassist was applied to improve the analysis efficiency [20].
2.2. Prediction of Vigna unguiculata MiRNAs
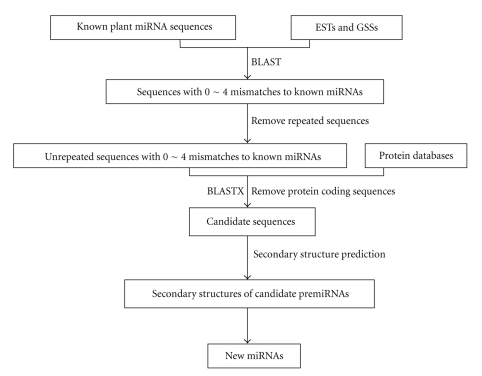
The prediction procedure was shown in Figure 1. The sequences of known plant miRNAs were used as query sequences for BLAST searches against the EST and GSS databases, with the BLAST parameters Evalue being 1000 and word-match size between the query and database sequences being 7. Mature miRNA sequences should be no less than 16 nt, and the mismatches should be less than 4. Wherever available, precursor sequences of 400 nt were extracted (200 nt upstream and 200 nt downstream to the BLAST hits) and used for the hairpin structure prediction. If the length of a sequence was less than 400 nt, the entire available sequence was used as a miRNA precursor sequence. These precursor sequences were then BLASTXed online to remove the protein coding sequences (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The retained precursor sequences underwent hairpin structure prediction through web server mfold. Only those meeting the following criteria were designated as miRNA homologs: (1) the RNA sequence folding into an appropriate stem-loop hairpin secondary structure, (2) a mature miRNA sequence located in one arm of the hairpin structure, (3) predicted mature miRNAs with no more than 3 nt substitutions as compared with the known miRNAs, (4) miRNAs having less than 6 mismatches with the opposite miRNA* sequence in the other arm, (5) no loop or break in miRNA* sequences, and (6) predicted secondary structures with higher minimal folding free energy (MFE) and minimal folding free energy index (MFEI), the MEFI usually being over 0.85 [32]. Also, the AU content of pre-miRNA within 30% to 70% was considered [20].
Figure 1.
Flowchart of Vigna unguiculata miRNA prediction.
2.3. Computational Prediction of MiRNA Targets
MiRNA targets prediction was performed by aligning the predicted miRNA sequences with EST sequences of Vigna unguiculata through the BLAST program. The targets were screened according to these criteria: the number of mismatches should be less than 4, the minimal free energy of the pairing between miRNA and its target mRNA was lower than −28.2 kcal/mol [33]. After removal of the repeated sequences, the potential target genes were BLASTed against protein databases to predict their function (identity > 25%), since there were no functional annotations for mRNA sequences of Vigna unguiculata.
2.4. Phylogenetic Analysis of the New MiRNAs
Considering the conservation of miRNAs and their precursors, the precursor sequences of the novel and the known miRNAs in the same family were aligned and phylogenetically analyzed by ClustalW online to investigate their evolutionary relationships (http://www.clustal.org/).
3. Results and Discussion
3.1. Prediction of Vigna unguiculata MiRNAs
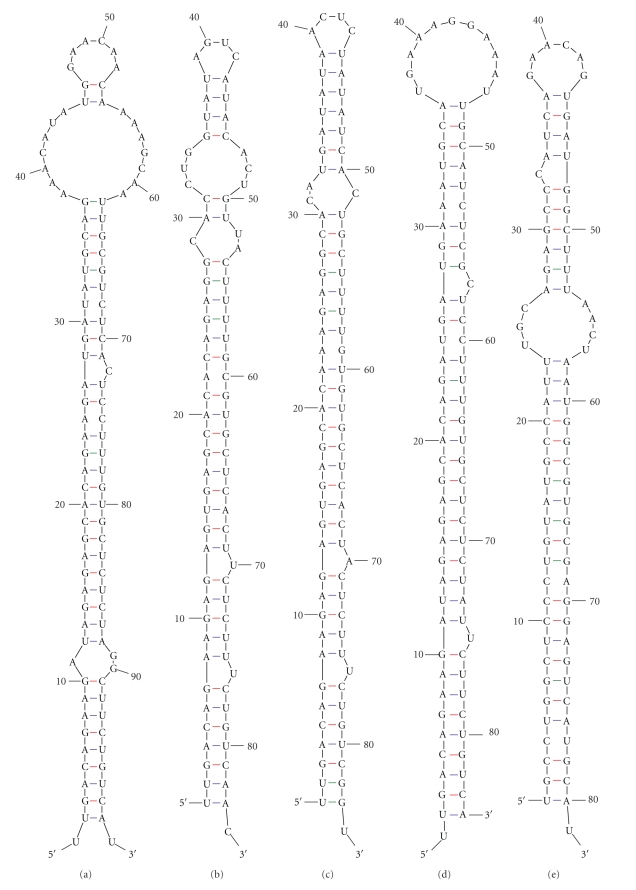
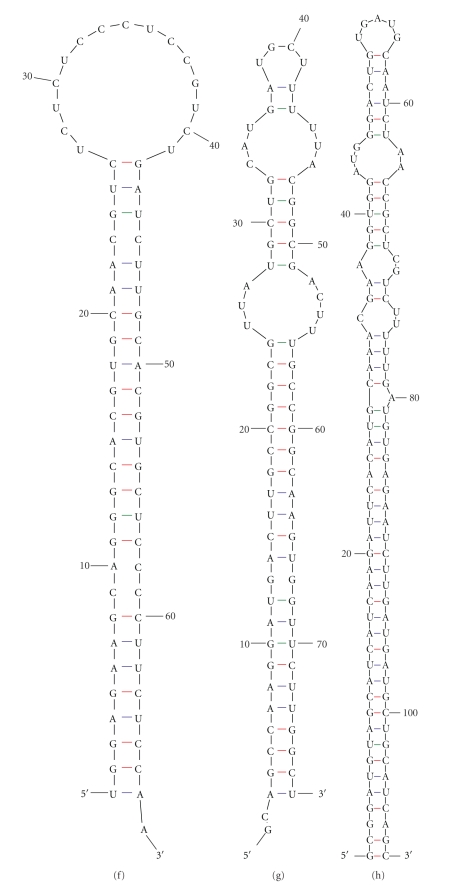
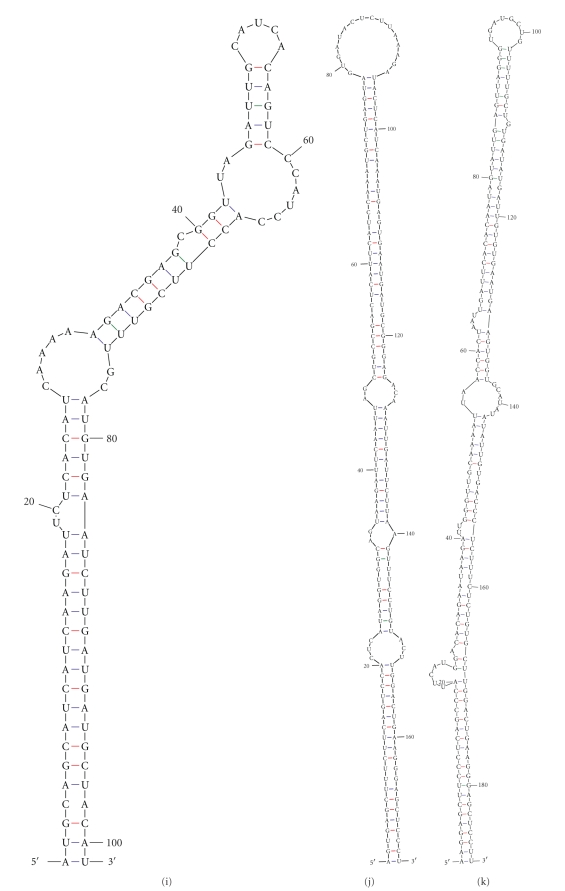
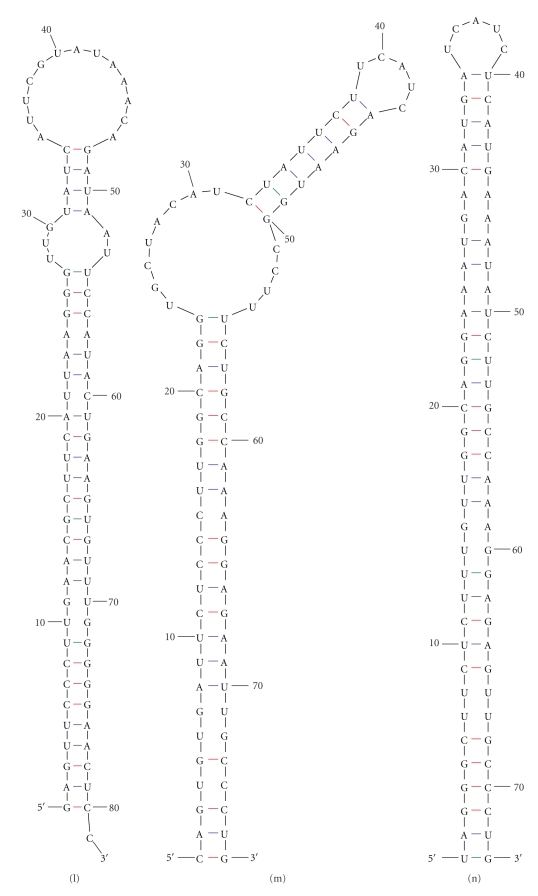
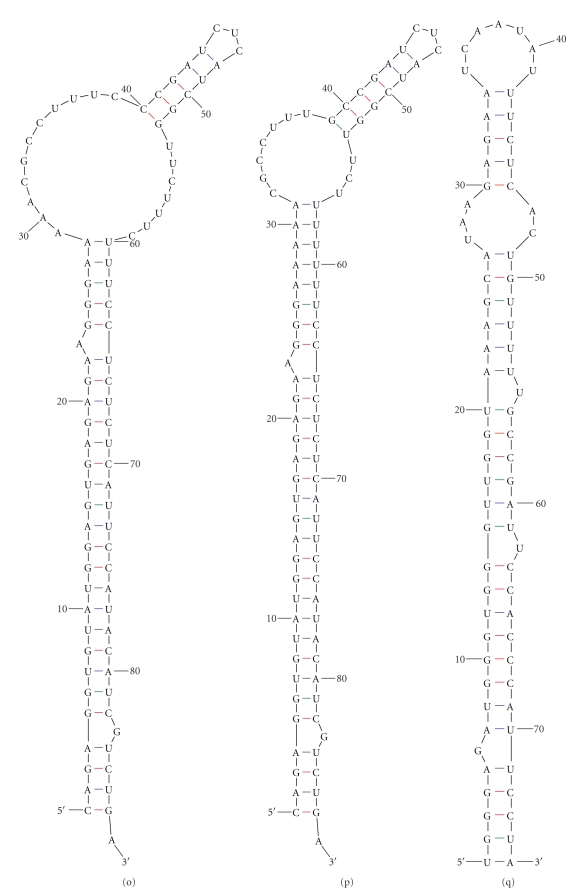
Sequence and structure homologies are the main theory behind the computer-based approach for miRNAs prediction. As described in Materials and Methods, after BLASTN searches, all blasted hits except coding sequences were maintained for secondary structure analysis, only those in line with the screening criteria were selected as candidates. In the end, 47 potential Vigna unguiculata miRNAs belonging to 13 miRNA families were identified, including two known miRNAs (vun-MIR1507a and vun-MIR1507b), and they were named according to Ambros [33]. Information on predicted Vigna unguiculata miRNAs, including names, lengths, sources, and other aspects, were listed in Table 1. The length of the 47 predicted miRNAs ranged from 19 nt to 24 nt, while the predicted precursor sequences ranged in length from 68 nt to 188 nt, all forming into typical stem-loop structures, with the mature miRNA either on the 5′ end or the 3′ end (Figure 2). All the MFEIs of these hairpin structures were over 0.85, which was thought to be the gold standard to differentiate miRNAs from other ones [32].
Table 1.
New miRNAs identified in Vigna unguiculata.
| New miRNA | Gene ID | Source | miRNA sequence (5′3′) | NM/nt | LM/nt | LP/nt | Location | A + U (%) | MFEI |
|---|---|---|---|---|---|---|---|---|---|
| vun-MIR156a | 146512093 | GSS | UGACAGAAGAUAGAGAGCAC | 0 | 20 | 100 | 5 | 0.58 | 1.06 |
| vun-MIR156b | 146512093 | GSS | CAGAAGAUAGAGAGCACAGA | 2 | 20 | 92 | 5 | 0.58 | 1.06 |
| vun-MIR156c | 146512093 | GSS | UUGACAGAAGAUAGAGAGCAC | 0 | 21 | 100 | 5 | 0.59 | 1.06 |
| vun-MIR156d | 146512093 | GSS | UGACAGAAGAUAGAGAGCACA | 1 | 21 | 100 | 5 | 0.58 | 1.06 |
| vun-MIR156e | 190469749 | EST | CAGAAGAUAGAGAGCACAGA | 2 | 20 | 77 | 5 | 0.58 | 1.18 |
| vun-MIR156f | 190510876 | EST | UGACAGAAGAGAGUGAGCACA | 0 | 21 | 82 | 5 | .055 | 1.09 |
| vun-MIR156g | 190510876 | EST | UUGACAGAAGAGAGUGAGCAC | 3 | 21 | 84 | 5 | 0.55 | 1.07 |
| vun-MIR156h | 190510876 | EST | UGACAGAAGAGAGUGAGCAC | 0 | 20 | 82 | 5 | 0.55 | 1.09 |
| vun-MIR156i | 190416464 | EST | UGACAGAAGAGAGUGAGCACA | 0 | 21 | 82 | 5 | 0.57 | 1.29 |
| vun-MIR156j | 190416464 | EST | UUGACAGAAGAGAGUGAGCAC | 3 | 21 | 84 | 5 | 0.58 | 1.30 |
| vun-MIR156k | 190416464 | EST | UGACAGAAGAGAGUGAGCAC | 0 | 20 | 82 | 5 | 0.57 | 1.29 |
| vun-MIR157a | 190469749 | EST | UUGACAGAAGAUAGAGAGCAC | 1 | 21 | 84 | 5 | 0.60 | 1.32 |
| vun-MIR157b | 190469749 | EST | UGACAGAAGAUAGAGAGCAC | 0 | 20 | 84 | 5 | 0.60 | 1.31 |
| vun-MIR157c | 190469749 | EST | UGACAGAAGAUAGAGAGCACA | 1 | 21 | 84 | 5 | 0.60 | 1.31 |
| vun-MIR159 | 182406416 | EST | CUUGGACUGAAGGGAGCUCCU | 1 | 21 | 186 | 3 | 0.60 | 1.00 |
| vun-MIR160a | 190509770 | EST | UGCCUGGCUCCCUGUAUGCCA | 0 | 21 | 81 | 5 | 0.48 | 1.05 |
| vun-MIR160b | 190509770 | EST | UGCCUGGCUCCCUGUAUGCC | 0 | 20 | 81 | 5 | 0.48 | 1.05 |
| vun-MIR164 | 146508076 | GSS | UGGAGAAGCAGGGCACGUGCA | 0 | 21 | 68 | 5 | 0.41 | 0.90 |
| vun-MIR169a | 190499818 | EST | CAGCCAAGGAUGACUUGCCGGC | 2 | 22 | 76 | 5 | 0.46 | 1.04 |
| vun-MIR169b | 190499818 | EST | CAGCCAAGGAUGACUUGCCGG | 0 | 21 | 76 | 5 | 0.46 | 1.04 |
| vun-MIR169c | 190499818 | EST | GCAGCCAAGGAUGACUUGCCG | 1 | 21 | 77 | 5 | 0.46 | 1.02 |
| vun-MIR172a | 190415307 | EST | AGAAUCUUGAUGAUGCUGCAUCAG | 2 | 24 | 109 | 3 | 0.55 | 1.02 |
| vun-MIR172b | 190415307 | EST | UGAAUCUUGAUGAUGCUACAU | 0 | 21 | 101 | 3 | 0. 57 | 0.89 |
| vun-MIR172c | 190415307 | EST | AGAAUCUUGAUGAUGCUGCAU | 0 | 21 | 101 | 3 | 0.57 | 1.03 |
| vun-MIR172d | 190415307 | EST | UGAAUCUUGAUGAUGCUACA | 2 | 20 | 100 | 3 | 0.57 | 0.87 |
| vun-MIR172e | 190415307 | EST | AGAAUCUUGAUGAUGCUGCA | 1 | 20 | 101 | 3 | 0.56 | 0.99 |
| vun-MIR319a | 182650666 | EST | UUGGACUGAAGGGAGCUCCCU | 0 | 21 | 172 | 3 | 0.58 | 0.99 |
| vun-MIR319b | 182650666 | EST | CUUGGACUGAAGGGAGCUCCC | 0 | 21 | 174 | 3 | 0.59 | 1.03 |
| vun-MIR319c | 182650666 | EST | UUGGACUGAAGGGAGCUCCC | 0 | 20 | 174 | 3 | 0.59 | 1.03 |
| vun-MIR319d | 182650666 | EST | CUUGGACUGAAGGGAGCUCC | 0 | 20 | 167 | 3 | 0.58 | 1.01 |
| vun-MIR319e | 182650666 | EST | UUGGACUGAAGGGAGCUCC | 0 | 19 | 167 | 3 | 0.58 | 1.01 |
| vun-MIR319f | 182406416 | EST | UUGGACUGAAGGGAGCUCCUU | 0 | 21 | 188 | 3 | 0.59 | 1.00 |
| vun-MIR319g | 182406416 | EST | CUUGGACUGAAGGGAGCUCC | 0 | 20 | 184 | 3 | 0.58 | 1.00 |
| vun-MIR319h | 182406416 | EST | UUGGACUGAAGGGAGCUCCU | 0 | 20 | 186 | 3 | 0.59 | 1.00 |
| vun-MIR319i | 182406416 | EST | UUGGACUGAAGGGAGCUCC | 0 | 19 | 184 | 3 | 0.58 | 0.98 |
| vun-MIR395a | 146525281 | GSS | CUGAAGUGUUUGGGGGAACUCC | 0 | 22 | 81 | 3 | 0.59 | 1.04 |
| vun-MIR395b | 146525281 | GSS | CUGAAGUGUUUGGGGGAACUC | 0 | 21 | 80 | 3 | 0.60 | 1.04 |
| vun-MIR399a | 182643512 | EST | UGCCAAAGGAGAAUUGCCCUG | 0 | 21 | 77 | 3 | 0.52 | 0.91 |
| vun-MIR399b | 182643512 | EST | UGCCAAAGGAGAAUUGCCC | 0 | 19 | 77 | 3 | 0.52 | 0.91 |
| vun-MIR399c | 182648023 | EST | UGCCAAAGGAGAGUUGCCCUG | 0 | 21 | 73 | 3 | 0.56 | 1.16 |
| vun-MIR399d | 182648023 | EST | UGCCAAAGGAGAGUUGCCC | 1 | 19 | 69 | 3 | 0.55 | 1.11 |
| vun-MIR482 | 146504713 | GSS | UCUUCCCAAUUCCGCCCAUUCCUA | 0 | 24 | 80 | 3 | 0.56 | 0.96 |
| vun-MIR1507a | 182400468 | EST | UCUCAUUCCAUACAUCGUCUGA | 0 | 22 | 88 | 3 | 0.53 | 1.03 |
| vun-MIR1507b | 182400468 | EST | UCUCAUUCCAUACAUCGUCUG | 0 | 21 | 87 | 3 | 0.53 | 1.03 |
| vun-MIR1507c | 190551234 | EST | UCUCAUUCCAUACAUCGUCUGA | 0 | 22 | 88 | 3 | 0.55 | 1.18 |
| vun-MIR1507d | 190551234 | EST | UCUCAUUCCAUACAUCGUCUG | 0 | 21 | 87 | 3 | 0.54 | 1.16 |
| vun-MIR2118 | 190540631 | EST | UUGCCGAUUCCACCCAUUCCUA | 1 | 22 | 75 | 3 | 0.57 | 1.11 |
NM: Number of mismatch; LM: Length of mature miRNA; LP: Length of precursor; MFEI: Minimal folding free energy index.
Figure 2.
Secondary structures of Vigna unguiculata new miRNA precursors. (a) vun-MIR156 precursor 1 (vun-MIR156a (2 to 21); vun-MIR156b (5 to 24); vun-MIR156c (1 to 21); vun-MIR156d (2 to 22)). (b) vun-MIR156 precursor 2 (vun-MIR156f (2 to 22); vun-MIR156g (1 to 21); vun-MIR156h (2 to 21)). (c) vun-MIR156 precursor 3 (vun-MIR156i (2 to 22); vun-MIR156j (1 to 21); vun-MIR156k (2 to 21)). (d) vun-MIR157 precursor (vun-MIR157a (1 to21); vun-MIR157b (2 to 21); vun-MIR157c (2 to 22)). (e) vun-MIR160 precursor (vun-MIR160a (1 to 21); vun-MIR160b (1 to 20)). (f) vun-MIR164 precursor (vun-MIR164 (1 to 21)). (g) vun-MIR169 precursor (vun-MIR169a (2 to 23); vun-MIR169b (2 to 22); vun-MIR169c (1 to 21)). (h) vun-MIR172 precursor 1 (vun-MIR172a (85 to 108); vun-MIR172c (85 to 105); vun-MIR172e (85 to 104)). (i) vun-MIR172 precursor 2 (vun-MIR172b (81 to 101); vun-MIR172d (81 to 100)). (j) vun-MIR319 precursor 1 (vun-MIR319a (152 to 172); vun-MIR319b (151 to 171); vun-MIR319c (152 to 171); vun-MIR319d (151 to 170); vun-MIR319e (152 to 170)). (k) vun-MIR319 precursor 2 (vun-MIR159 (167 to 187); vun-MIR319f (168 to 188); vun-MIR319g (167 to 186); vun-MIR319h (168 to 187); vun-MIR319i (168 to 186)). (l) vun-MIR395 precursor (vun-MIR395a (60 to 81); vun-MIR395b (60 to 80)). (m) vun-MIR399 precursor 1 (vun-MIR399a (57 to 77); vun-MIR399b (57 to 75)). (n) vun-MIR399 precursor 2 (vun-MIR399c (53 to 73); vun-MIR399d (53 to 71)). (o) vun-MIR1507 precursor 1 (vun-MIR1507a (67 to 88); vun-MIR1507b (67 to 87)). (p) vun-MIR1507 precursor 2 (vun-MIR1507c (67 to 88); vun-MIR1507d (67 to 87)). (q) vun-MIR2118 precursor (vun-MIR2118 (54 to 75)). (r) vun-MIR482 precursor (vun-MIR482 (57 to 80)). The position of each miRNA was shown in parentheses.
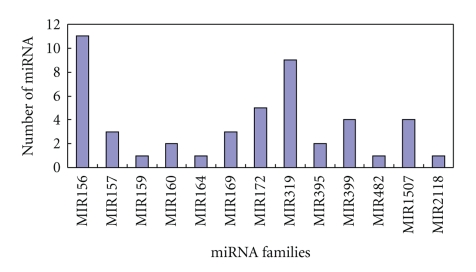
Vigna unguiculata miRNA precursors were diverse in structure and size, even if they were from the same family, such as those from MIR 156, MIR 172, MIR 319, and MIR 399 families (Figure 2). The distribution of miRNAs in each famiy was different too, some miRNA families have more than 10 members, such as miRNA156 family whereas only one was predicted in other families, such as in miRNA164, miRNA482 and miRNA2118 families (Figure 3). That was consistent with the diversity of miRNAs in other plants [27].
Figure 3.
The distribution of new miRNAs in different miRNA families.
With the availability of sequence resources, computer-based miRNA identification is becoming more and more important than experimental approaches due to its advantages of low cost and high efficiency [1]. At present, three kinds of databases, genome, GSS, and EST, are mainly used for plant miRNA prediction. Considering the unavailability of genome sequences of Vigna unguiculata, both EST and GSS databases were searched for miRNA identification. The number and sorts of miRNAs predicted in this work showed that this software-based approach was as feasible and effective as in other work [19–26].
According to Zhang [27], about 10000 ESTs contained one miRNA, so about 20 miRNAs should be predicted theoretically from the total of 241854 ESTs and GSSs in this work. Why as many as 47 were predicted? Generally, plant miRNA clusters have not been frequently observed, so only one plant miRNA can come from the same transcript. In this work, different length of mature miRNAs from the same precursor were regarded as different ones, considering they corresponded to different target genes.
3.2. Prediction of Vigna unguiculata MiRNA Targets
Based on the complementarity between miRNAs and their target genes in plants, the Vigna unguiculata EST database was searched for homology to the new miRNA sequences with a BLASTN algorithm for the discovery of miRNA targets. A total of 30 potential targets for 47 Vigna unguiculata miRNAs were identified, and the miRNA target site could be found in 5′UTR, 3′UTR or open reading frame. The miRNAs and their putative targets with known functions were listed in Table 2. These potential miRNA targets belonged to a number of gene families that had different biological functions.
Table 2.
The potential targets of newly identified miRNAs in Vigna unguiculata.
| miRNA | Targeted gene | Targeted protein | Possible function | |
|---|---|---|---|---|
| MIR156c/MIR157a | 190524055(1) | squamosa promoter-binding protein | transcription factor | 3′UTR |
| 190515614(0) | squamosa promoter-binding protein | transcription factor | ORF | |
| 190473822(0) | squamosa promoter-binding protein | transcription factor | 5′UTR | |
| 190427748(1) | LIGULELESS1 protein | transcription factor | ORF | |
| MIR156j//MIR156g | 190416464(0) | rho-related protein racG | signal transduction | 5′UTR |
| MIR160b | 190447817(0) | auxin response factor | stress response | ORF |
| MIR164 | 182656787(2) | NAC domain protein | transcription factor | ORF |
| 182647558(3) | NAC domain protein | transcription factor | ORF | |
| MIR172b | 182399040(3) | protein kinase family protein | metabolism | ORF |
| 190449132(3) | trehalose-6-phosphate synthase | metabolism | ORF | |
| MIR172c | 182652950(2) | transcription factor AHAP2 | transcription factor | ORF |
| 182404391(2) | AP2 domain-containing transcription factor | transcription factor | ORF | |
| MIR172e | 190524349(3) | alpha/beta hydrolase | metabolism | ORF |
| 190501430(2) | transcription factor APETALA2 | transcription factor | ORF | |
| 182389603(2) | protein AINTEGUMENTA, putative | transcription factor | ORF | |
| MIR319e/MIR319i | 190555544(3) | taxadien-5-alpha-ol O-acetyltransferase | metabolism | ORF |
| 182406172(n) | 182402586(3) | ATP synthase alpha chain, mitochondrial | metabolism | 3′UTR |
| MIR319h | 190541283(3) | TCP family transcription factor | transcription factor | ORF |
| MIR399b | 190527737(3) | delta-aminolevulinic acid dehydratase | metabolism | ORF |
| 190504725(3) | 50S ribosomal protein L22, | metabolism | ORF | |
| 190478794(2) | arginine biosynthesis protein argJ 1ornithine acetyltransferase (OAT) family | metabolism | ORF | |
| 190452752(1) | cytochrome | metabolism | 5′UTR | |
| 182643288(3) | aldehyde dehydrogenase | metabolism | ORF | |
| MIR2118 | 190540632(0) | cytoplasmic tRNA adenylyltransferase 1chromatin assembly factor 1 subunit p90 ATP-dependent RNA helicase DBP3 | translation | ORF |
Parentheses indicate the number of mismatches between the miRNA and mRNA; ORF: open reading frame; UTR: untranslated region.
Many of these targets were transcription factors that controlled plant development and phase change from vegetative growth to reproductive growth. Several studies indicated that MIR156/157 targeted squamosa promoter binding protein (SBP)-like (SPL) genes, a plant-specific family of transcription factors involved in early flower development and vegetative phase changes [34, 35]. Auxin response factors (ARF), a plant-specific family of DNA binding proteins, were involved in hormone signal transduction [36]. AP(2)APETALA2 transcription factors were involved in several developmental processes in Arabidopsis thaliana embryo, endosperm, and seed coat development [37]. TCP family transcription factors have been reported to play roles in various aspects of plant development [38, 39]. Heat shock transcription factor was reported to participate in heat shock response to environmental stress [40].
In addition to the transcription factors, another important part of the predicted targets were various kinds of enzymes such as trehalose-6-phosphate synthase, taxadien-5-alpha-ol O-acetyltransferase, delta-aminolevulinic acid dehydratase, porphobilinogen synthase, and alpha/beta hydrolase, which might play roles in various metabolic pathways [41–44].
Identification of genes targeted by miRNAs is widely believed to be an important step toward understanding the role of miRNAs in gene regulatory networks. EST database searches play a vital role for the discovery of miRNA targets in plants based on the homology between miRNA and its target sequences [45]. The protein databases applied to predict the functions of the target genes in this work included nonredundant protein sequences, reference protein sequences and so on. Though this method may be error-prone, it provides a fast way to know something about the gene function.
Our prediction of target genes for the 47 miRNAs also revealed that not just one gene might be regulated by individual miRNA (Table 2). This result was consistent with recent findings in other plant species. Wu et al. also reported that multiple miRNAs could target the same gene. All these suggested that miRNA research should focus on networks more than on individual connections between miRNA and strongly predicted targets [46].
3.3. Phylogenetic Analysis of the New MiRNAs
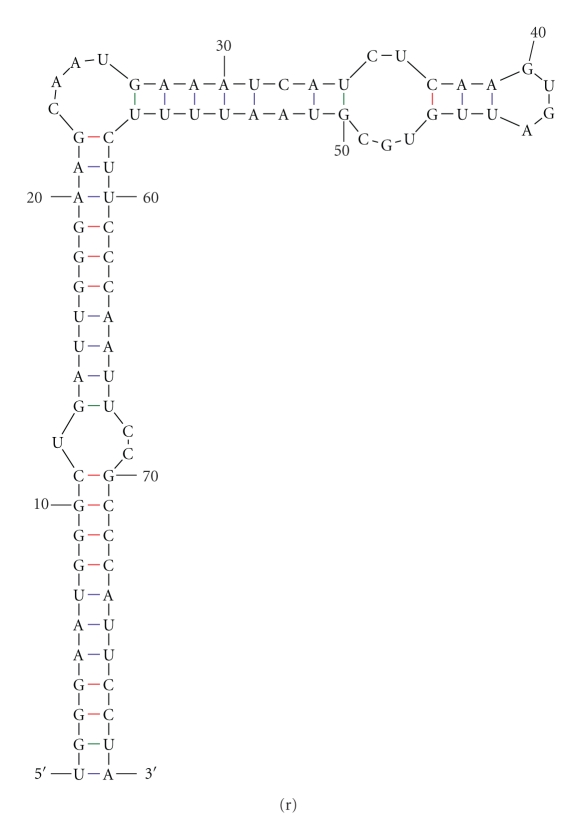
Plant miRNAs are highly conserved among distantly related plant species, both in terms of primary and mature miRNAs [27]. Comparison of the precursor sequences of the predicted miRNAs with other members in the same family showed that most members could be found to have a high degree of sequence similarity with others, for example, the precursor sequence similarity between vun-MIR156 and other MIR156 members was over 60%, that between vun-MIR157 and ath-MIR157a, gra-MIR157a, bol-MIR157a, or sly-MIR157b was over 80% (data not shown).
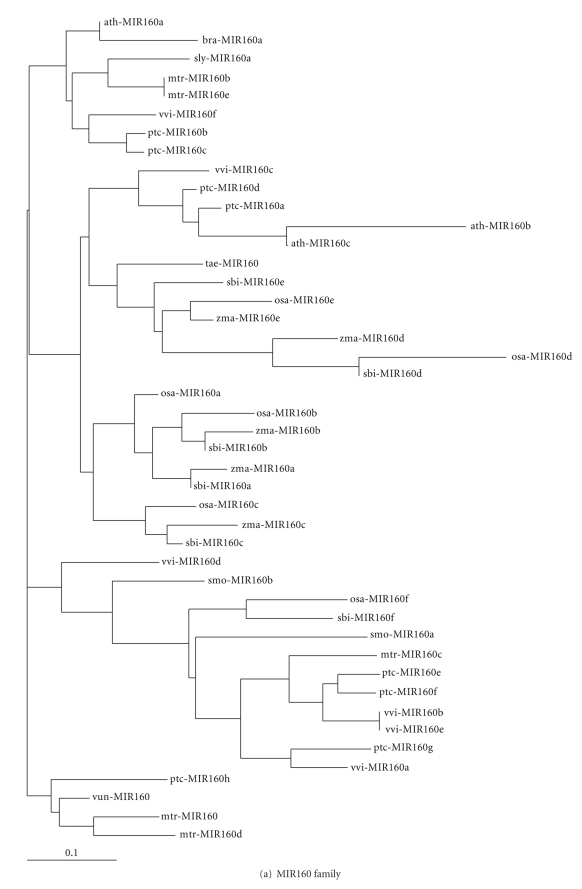
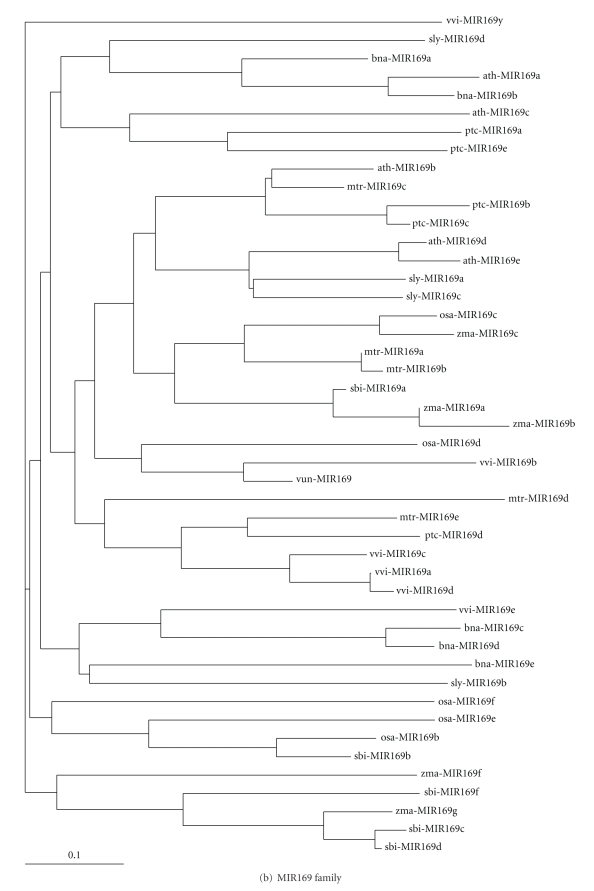
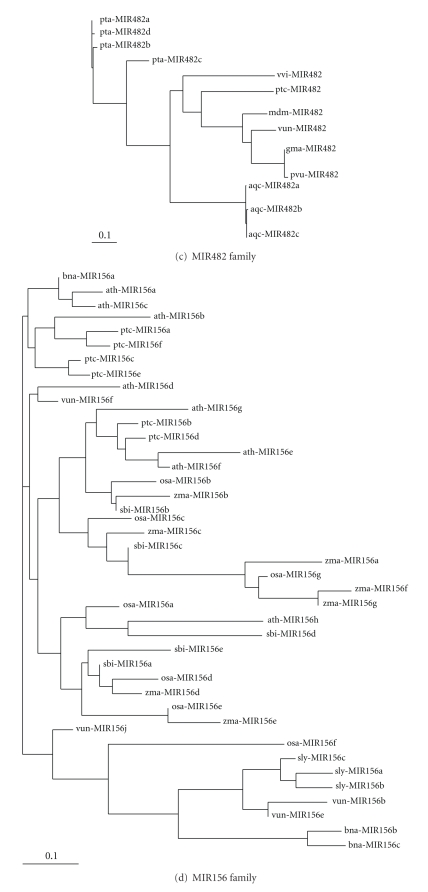
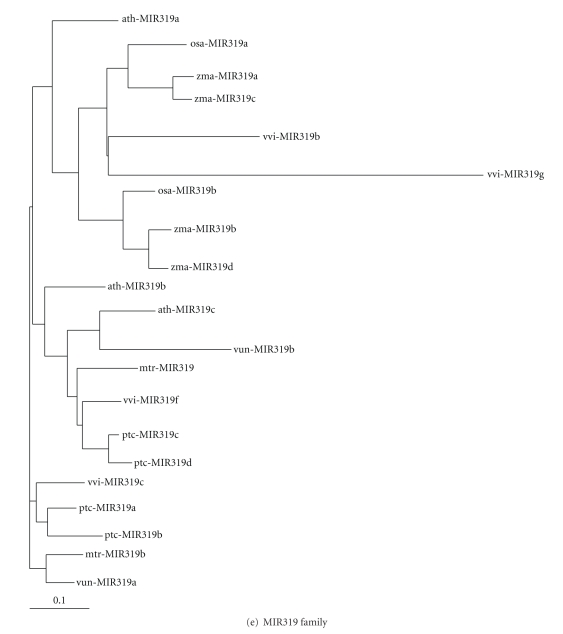
The conservation of mature miRNAs and premiRNAs provides the chance to investigate their evolutionary relationships. Based on the premiRNA sequence comparisons, the evolutionary relationships of Vigna unguiculata miRNAs with other members from the same families were analyzed using the ClustalW online. It could be seen from the phylogenetic trees that in different families, the evolutionary relationships of Vigna unguiculata miRNAs with other species were different; for example, in MIR160 family, vun-MIR160 and mtr-MIR160 were on the same branch, while in MIR169 family, vun-MIR169 had close relationship with wi-MIR169, and in MIR482 family, vun-MIR482 was closely related to pvu-MIR482 and gma-MIR482 (Figures 4(a)–4(c)). Also, it could be seen that different Vigna unguiculata miRNA members in the same family were often distantly related, such as in MIR156 and MIR319 families (Figures 4(d) and 4(e)). These results suggested that different miRNAs might evolve at different rates not only within the same plant species, but also in different ones.
Figure 4.
Phylogenetic analysis of miRNAs in different families.
4. Conclusions
In this paper, with a bioinformatic method, 47 miRNAs were identified from the EST and GSS databases of Vigna unguiculata; also about 30 potential targets of them were predicted, which appeared to be related to the development, growth, metabolism, and other physiological processes such as stress response of Vigna unguiculata. The findings from this study will contribute to further researches of miRNAs function and regulatory mechanisms in Vigna unguiculata.
Acknowledgments
This work was supported by the Grants from Key Projects in the National Science & Technology Pillar Program in the Eleventh Five-year Plan Period of China (no. 2008BADB1B01-3). The authors thank Professor Z. M. Yang, Nanjing Agricultural University, for the generous gift of MiRNAassist software.
References
- 1.Unver T, Namuth-Covert DM, Budak H. Review of current methodological approaches for characterizing MicroRNAs in plants. International Journal of Plant Genomics. 2009;2009:11 pages. doi: 10.1155/2009/262463. Article ID 262463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Megraw M, Hatzigeorgiou AG. MicroRNA promoter analysis. Methods in Molecular Biology. 2010;592:149–161. doi: 10.1007/978-1-60327-005-2_11. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO Journal. 2004;23(20):4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301(5631):336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 6.Barbato C, Arisi I, Frizzo ME, Brandi R, Da Sacco L, Masotti A. Computational challenges in miRNA target predictions: to be or not to be a true target? Journal of Biomedicine and Biotechnology. 2009;2009:9 pages. doi: 10.1155/2009/803069. Article ID 803069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF. Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development. 2010;137(1):103–112. doi: 10.1242/dev.043067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J-W, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana . Cell. 2009;138(4):738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 9.Mathieu J, Yant LJ, Mürdter F, Küttner F, Schmid M. Repression of flowering by the miR172 target SMZ. PLoS Biology. 2009;7(7) doi: 10.1371/journal.pbio.1000148. Article ID e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornara F, Coupland G. Plant phase transitions make a SPLash. Cell. 2009;138(4):625–627. doi: 10.1016/j.cell.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 11.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303(5666):2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134(9):1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 13.Ding Y-F, Zhu C. The role of microRNAs in copper and cadmium homeostasis. Biochemical and Biophysical Research Communications. 2009;386(1):6–10. doi: 10.1016/j.bbrc.2009.05.137. [DOI] [PubMed] [Google Scholar]
- 14.Lu S, Sun Y-H, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive MicroRNAs in Populus trichocarpa that are absent from Arabidopsis . Plant Cell. 2005;17(8):2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pothof J, Verkaik NS, Van IJcken W, et al. MicroRNA-mediated gene silencing modulates the UV-induced DNA-damage response. EMBO Journal. 2009;28(14):2090–2099. doi: 10.1038/emboj.2009.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annual Review of Plant Biology. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 17.Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends in Plant Science. 2007;12(7):301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Bonnet E, Wuyts J, Rouzé P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliania and Oryza sativa identifies important target genes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(31):11511–11516. doi: 10.1073/pnas.0404025101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Research. 2005;15(5):336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- 20.Xie FL, Huang SQ, Guo K, et al. Computational identification of novel microRNAs and targets in Brassica napus . FEBS Letters. 2007;581(7):1464–1474. doi: 10.1016/j.febslet.2007.02.074. [DOI] [PubMed] [Google Scholar]
- 21.Yin Z, Li C, Han X, Shen F. Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum) Gene. 2008;414(1-2):60–66. doi: 10.1016/j.gene.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 22.He P-A, Nie Z, Chen J, et al. Identification and characteristics of microRNAs from Bombyx mori . BMC Genomics. 2008;9, article 248 doi: 10.1186/1471-2164-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunkar R, Jagadeeswaran G. In silico identification of conserved microRNAs in large number of diverse plant species. BMC Plant Biology. 2008;8, article 37 doi: 10.1186/1471-2229-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song C, Fang J, Li X, Liu H, Thomas Chao C. Identification and characterization of 27 conserved microRNAs in citrus. Planta. 2009;230(4):671–685. doi: 10.1007/s00425-009-0971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Luo Y, Gong X, Zeng W, Li S. Computational identification of 48 potato microRNAs and their targets. Computational Biology and Chemistry. 2009;33(1):84–93. doi: 10.1016/j.compbiolchem.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Han YS, Luan FL, Zhu HL, et al. Computational identification of microRNAs and their targets in wheat (Triticum aestivum L.) Science in China, Series C. 2009;52(11):1091–1100. doi: 10.1007/s11427-009-0144-y. [DOI] [PubMed] [Google Scholar]
- 27.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant Journal. 2006;46(2):243–259. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 28.Muchero W, Diop NN, Bhat PR, et al. A consensus genetic map of cowpea [Vigna unguiculata (L) Walp.] and synteny based on EST-derived SNPs. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(43):18159–18164. doi: 10.1073/pnas.0905886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pule-Meulenberg F, Belane AK, Krasova-Wade T, Dakora FD. Symbiotic functioning and bradyrhizobial biodiversity of cowpea (Vigna unguiculata L. Walp.) in Africa. BMC Microbiology. 2010;10, article 89 doi: 10.1186/1471-2180-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Li P, Cao X, Wang X, Zhang A, Li X. Identification and expression analysis of miRNAs from nitrogen-fixing soybean nodules. Biochemical and Biophysical Research Communications. 2009;378(4):799–803. doi: 10.1016/j.bbrc.2008.11.140. [DOI] [PubMed] [Google Scholar]
- 31.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang BH, Pan XP, Cox SB, Cobb GP, Anderson TA. Evidence that miRNAs are different from other RNAs. Cellular and Molecular Life Sciences. 2006;63(2):246–254. doi: 10.1007/s00018-005-5467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277–279. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miRr156 and its target SPL3. Development. 2006;133(18):3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J-W, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana . Cell. 2009;138(4):738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Guilfoyle TJ, Hagen G. Auxin response factors. Current Opinion in Plant Biology. 2007;10(5):453–460. doi: 10.1016/j.pbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 37.Ohto M-A, Floyd SK, Fischer RL, Goldberg RB, Harada JJ. Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis . Sexual Plant Reproduction. 2009;22(4):277–289. doi: 10.1007/s00497-009-0116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nag A, King S, Jack T. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America. 2009;106(52):22534–22539. doi: 10.1073/pnas.0908718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martín-Trillo M, Cubas P. TCP genes: a family snapshot ten years later. Trends in Plant Science. 2010;15(1):31–39. doi: 10.1016/j.tplants.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Pirkkala L, Nykänen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB Journal. 2001;15(7):1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 41.Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis . Plant Physiology. 2008;146(1):97–107. doi: 10.1104/pp.107.107441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker K, Ketchum REB, Hezari M, et al. Partial purification and characterization of acetyl coenzyme A: taxa- 4(20),11 (12)-dien-5α-ol O-acetyl transferase that catalyzes the first acylation step of taxol biosynthesis. Archives of Biochemistry and Biophysics. 1999;364(2):273–279. doi: 10.1006/abbi.1999.1125. [DOI] [PubMed] [Google Scholar]
- 43.Jaffe EK. The porphobilinogen synthase catalyzed reaction mechanism. Bioorganic Chemistry. 2004;32(5):316–325. doi: 10.1016/j.bioorg.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Shoolingin-Jordan PM, Spencer P, Sarwar M, et al. 5-aminolaevulinic acid dehydratase: metals, mutants and mechanism. Biochemical Society Transactions. 2002;30(4):584–590. doi: 10.1042/bst0300584. [DOI] [PubMed] [Google Scholar]
- 45.Fahlgren N, Carrington JC. miRNA target prediction in plants. Methods in Molecular Biology. 2010;592:51–57. doi: 10.1007/978-1-60327-005-2_4. [DOI] [PubMed] [Google Scholar]
- 46.Peter ME. Targeting of mRNAs by multiple miRNAs: the next step. Oncogene. 2010;29(15):2161–2164. doi: 10.1038/onc.2010.59. [DOI] [PubMed] [Google Scholar]