Abstract
Gene silencing by aberrant epigenetic chromatin alteration is a well-recognized event contributing to tumorigenesis. While genetically engineered tumor-prone mouse models have proven a powerful tool in understanding many aspects of carcinogenesis, to date few studies have focused on epigenetic alterations in mouse tumors. To uncover epigenetically silenced tumor suppressor genes (TSGs) in mouse mammary tumor cells, we conducted initial genome-wide screening by combining the treatment of cultured cells with the DNA demethylating drug 5-aza-2′-deoxycytidine (5-azadC) and the histone deacetylase inhibitor trichostatin A (TSA) with expression microarray. By conducting this initial screen on EMT6 cells and applying protein function and genomic structure criteria to genes identified as upregulated in response to 5-azadC/TSA, we were able to identify 2 characterized breast cancer TSGs (Timp3 and Rprm) and 4 putative TSGs (Atp1B2, Dusp2, FoxJ1 and Smpd3) silenced in this line. By testing a panel of ten mouse mammary tumor lines, we determined that each of these genes is commonly hypermethylated, albeit with varying frequency. Furthermore, by examining a panel of human breast tumor lines and primary tumors we observed that the human orthologs of ATP1B2, FOXJ1 and SMPD3 are aberrantly hypermethylated in the human disease while DUSP2 was not hypermethylated in primary breast tumors. Finally, we examined hypermethylation of several genes targeted for epigenetic silencing in human breast tumors in our panel of ten mouse mammary tumor lines. We observed that the orthologs of Cdh1, RarB, Gstp1, RassF1 genes were hypermethylated, while neither Dapk1 nor Wif1 were aberrantly methylated in this panel of mouse tumor lines. From this study, we conclude that there is significant, but not absolute, overlap in the epigenome of human and mouse mammary tumors.
INTRODUCTION
Tumorigenesis is a multi-step process stemming from increased oncogene activity in concert with constrained tumor suppressor gene (TSG) activity (Weinberg 1996). Such disease-producing changes in cellular phenotype result from dysregulation of gene expression and/or protein function attributable to genetic and/or epigenetic changes within the genome.
Several well-studied TSGs and other growth regulatory genes undergo de novo hypermethylation and consequential transcriptional silencing in human cancer cells. Aberrant cytosine methylation leading to gene silencing occurs at discreet, clustered 5′-CG-3′ dinucleotides (referred to as CpG islands) within the genome (Bird 1992). This cancer-associated CpG methylation is part of a complex set of epigenetic events that transform chromatin from a transcriptionally-active to an inactive state (Wolffe 2001) and epigenetic gene silencing is now widely recognized as either a causative or correlative event in tumor development (Jones and Baylin 2002; Robertson 2005) and is now widely regarded as one of the ‘hits’ in the Knudsen hypothesis leading to TSG inactivation (Jones and Laird 1999).
In recent years, numerous laboratories have developed tumor-prone mouse models owing to engineered over-expression of oncogenes or lost TSG expression. Clearly, these models have provided a powerful tool in studying tumorigenesis and unique insight into the molecular mechanisms that give rise to cancer in humans. For example, BRCA1, a TSG associated with breast and ovarian tumors in humans (Bishop 1999; Struewing et al., 1997), when conditionally deleted within the mammary epithelium of p53 heterozygous mice greatly increases mammary tumor incidence (Xu et al., 1999). This observation lends strong evidence as to the role BRCA1 plays in breast tumor suppression.
What remains less explored is the role that epigenetics plays during tumorigenesis in mice. While it seems straightforward to presume that gene silencing occurs in a parallel fashion in both human and mouse tumors, mice have not been widely utilized as a platform for either the discovery of epigenetically silenced genes or to study the role that gene silencing plays in the process of tumorigenesis. In one of the few studies to utilize a tumor-prone model system to dissect epigenetic events during disease progression, Yu et al (2005a) used Restriction Landmark Genomic Scanning (RLGS) to investigate aberrant DNA methylation events in a mouse model of T/natural killer acute lymphoblastic leukemia. These investigators observed nonrandom, aberrant DNA methylation occurring during tumorgenesis but was not present in benign preleukemic cells. Further, the inhibitor of DNA binding 4 (Idb4) gene was identified as a putative TSG silenced via promoter hypermethylation in mouse, and this gene was found epigenetically silenced in human leukemia as well. Similarly, Costello and colleagues found that SLC5A8, which encodes a sodium monocarboxylate cotransporter, was epigenetically silenced in both mouse and human brain tumors (Hong et al., 2005). These studies underscore the high potential of mice as a model system to understand the epigenetic basis of cancer and, potentially, other diseases as well.
A list of well-characterized TSG including BRCA1, CDKN2A, SFN, CDH1, and CST6 are targets for epigenetic silencing in breast cancer (Dobrovic and Simpfendorfer 1997; Esteller et al., 2000; Ferguson et al., 2000; Nass et al., 2000; Holst et al., 2003; Ai et al., 2006). However, at present, we are unaware of any genes that have been identified as targets for epigenetic silencing in mouse mammary tumors. To initially address this question, we conducted a screen for epigenetically silenced genes in a mouse mammary tumor line using an approach termed “pharmacological unmasking” (Esteller 2007). This often used approach examines changes in gene expression in response to the global DNA demethylating drug 5-aza-2′-deoxycytidine (5-azadC) and the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) via microarray. Our screen was conducted using a mouse mammary cell line and identified ~1,000 genes that were upregulated ≥ 5-fold in response to 5-azadC/TSA in this line. Based on gene function and genomic architecture in mouse and man, several candidates identified in the initial screen were selected for further study.
MATERIALS and METHODS
Cell Culture and Drug Treatment
Mouse mammary tumor lines 410.4, 410 and 66.1 were obtained from Dr. A. Fulton (University of Maryland, Baltimore, MD), D2F2 and 4T1 from Dr. F. Miller (Karamanos Cancer Foundation, Detroit, MI), LM2 and LM3 from Dr. E. Bal de Kier Joffé (Universidad de Buenos Aires, Argentina) and D1K2 and NeuT from Dr. B. Law (University of Florida, Gainesville, FL). The mouse mammary tumor line EMT6 was purchased from American Type Culture Collection (ATCC, Manassas, VA).
LM2 and LM3 were grown in Eagle’s MEM with 2 mM L-glutamine, 1.5g/L sodium bicarbonate, 0.1 mmol/L nonessential amino acids, and 1.0 mmol/L sodium pyruvate. 4T1 were cultured in RPMI 1640 with 2 mmol/L L-glutamine, 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, 10 mmol/L HEPES, and 1.0 mmol/L sodium pyruvate. 410, 410.4, 66.1, NeuT, D2F2, and D1K2 were maintained in DMEM with 2mM L-glutamine, 0.1 mmol/L nonessential amino acids. EMT6 was cultured in Waymouth’s MB 752/1 medium with 2mM L-glutamine. All cell lines were maintained in medium supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA) at 37°C in 5% CO2 humidified atmosphere. Human breast tumor cell lines were cultured as previously outlined (Ai et al., 2006).
Where indicated, cell lines were treated with 5 μM 5-azadC for 5 days (fresh drug was added every 24 hours) followed by a 24-hour treatment with 100 nM TSA. All drugs were purchased from Sigma.
Human Breast Tumor Specimens
Fifteen fresh-frozen archival breast tumors were obtained from the University of Florida Shands Cancer Center Molecular Tissue Bank. All specimens and pertinent patient information were treated in accordance with policies of the institutional review board (IRB) of the University of Florida Health Sciences Center. Tumors analyzed in this study were examined by a surgical pathologist and identified as invasive breast adenocarcinoma (stage II or III). Where indicated, matched normal breast tissue was obtained from disease-free surgical margin.
Microarray Analysis
Total RNA was extracted from EMT6 cells either untreated or treated with the global DNA demethylating agent 5-aza-2′-deoxycytidine (5-azadC) and the histone deacetylase inhibitor (HDAC) TSA. The quantity and purity of the extracted RNA was evaluated using a Nanodrop ND-100 spectrophometer (Nanodrop Technologies, Wilmington, DE) and its integrity measured using an Agilent Bioanalyzer. Five hundred nanograms (500 ng) of total RNA was amplified and labeled with a fluorescent dye (Cy3) using a Low RNA Input Linear Amplification Labeling kit (Agilent Technologies, Palo Alto, CA). Abundance and quality fluorescently labeled cRNA was also assessed using a Nanodrop ND-1000 spectrophotometer and an Agilent Bioanalyzer. The labeled cRNA (1.65 μg) was hybridized to an Agilent Mouse Whole Genome Oligo Microarray (Agilent Technologies, Inc., Palo Alto, CA) for 17 hrs prior to washing and scanning. Data was extracted from scanned images using Agilent’s Feature Extraction Software.
Gene Expression Analysis by RT-PCR
RT-PCR was carried out according to established protocols (Ai et al., 2006). Briefly, untreated and 5-azadC/TSA treated mouse mammary tumor cells were homogenized in Trizol (Invitrogen), and total RNA purified according to the manufacturer’s instructions. First-strand cDNA synthesis was carried out using SuperScript III reverse transcriptase (Invitrogen). Subsequently, this cDNA was used in PCR reactions using Timp3, Rprm, Smpd3, Atp1B2, Dusp2 and FoxJ1-specific oligonucleotide primers as listed in Supplemental Table I. Mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh) served as a control to assure equivalent RNA in each reaction. Following PCR, reactions were analyzed by electrophoresis on 10% polyacrylamide gels followed by staining with ethidium bromide.
Extraction and Sodium Bisulfite Modification of Genomic DNA
Genomic DNA (gDNA) was isolated from human breast tumor tissue samples and cultured human and mouse cell lines using a Qiagen Blood and Cell Culture DNA kit (Qiagen, Inc., Valencia, VA) and stored at −20°C before use. DNA was modified with EZ DNA Methylation kit (ZYMO Research Co., Orange, CA) according to the manufacturer’s instructions. Briefly, ~1.0 μg denatured gDNA was treated with sodium bisulfite at 50°C for ~16 hours in the dark. Following this, samples were applied to supplied columns, desulfonation conducted, and DNA was subsequently eluted in a volume of 20 μL with supplied elution buffer. One μL of bisulfite-converted gDNA solution was generally used in subsequent PCR reactions.
Methylation Specific-PCR
Primers used for Methylation Specific-PCR (MSP) analyses are listed in Supplemental Table II. MSP reactions were conducted using Qiagen HotStarTaq DNA polymerase and supplied 1X PCR buffer supplemented with 0.1 mmol/L dNTPs, 2.5 mmol/L MgCl2, and 0.5 μmol/L each of forward and reverse primers and bisulfite-converted gDNA. Following thermocycling, PCR products were loaded 10% polyacrylamide gels and visualized using ethidium bromide.
Human placental gDNA (Biochain Institute, Hayward, CA) and mouse gDNA isolated from tail snips of wildtype C57BL/6 mice were used in primer validation experiments of human and mouse MSP, respectively and where indicated DNA was methylated in vitro using SssI methylase (NEB, Ipswich, MA) as outlined previously (Vo et al., 2004).
Pyrosequencing
PCR reactions were conducted on bisulfite-converted gDNA as described above. Supplemental Table III summarizes pyrosequencing primers and sequence analyzed in each gene. Following PCR amplification using a biotinylated reverse primer, 5–20 μL of PCR product was mixed with 2 μL of streptavidin-coated sepharose beads (GE Heathcare Bio-Sciences AB, Uppsala, Sweden), 40ul of binding buffer (10 mM Tris-HCl, 2 M NaCl, 1mM EDTA, 0.1% Tween 20, pH 7.6), and brought up to 80 μL total volume with dH2O. This mixture was immobilized on streptavidin beads by shaking constantly at 1400 rpm for 10 minutes at RT. The beads were subsequently harvested using a vacuum preparation workstation and appropriate sequencing primer added to a final concentration of 330 nM. The sequencing primers were subsequently annealed to the template by incubating the samples at 80°C for 2 min and cooled at RT for 10 min prior to analysis. Pyrosequencing was conducted using a PyroMark MD system (Biotage AP, Uppsala, Sweden), and sequence analysis conducted using supplied software.
RESULTS
The focus of this investigation was to compare mouse and human mammary cancer for potential overlap in tumor epigenotype across these two species. Our screening strategy entailed utilizing the mouse mammary tumor line EMT6 in a pharmacological unmasking screen (Esteller 2007). Briefly, EMT6 cells were treated with the global DNA demethylating agent 5-aza-2′-deoxycytidine (5-azadC) and the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) to de-repress epigenetically silenced genes. After drug treatment, which was found to have minimal toxicity in EMT6 cells by MTT assay (data not shown), total RNA was harvested from two replicate cultures of drug treated and untreated EMT6 cells and used to probe Agilent Mouse Whole Genome microarrays.
Of the ~44,000 transcripts measured on these microarrays, an average of 54% were detected as significantly above background levels in each of the hybridizations (range: 51 to 57%). The intensity values from the replicate samples for both the treated and untreated cultures had Pearson correlation values greater than 0.97, suggesting that the reproducibility of the results was very high. Reproducibility of the within-group samples, along with the differences between the two groups, were also apparent when the z-score transformed intensity values for the 2,252 transcripts found to be differentially expressed (984 upregulated, 1,268 downregulated) using an absolute fold change criterion of 5-fold and a log ratio p-value of 0.001 were compared (Fig 1).
Figure 1. Hierarchical clustering of the differentially expressed transcripts in all of the biological replicate samples.
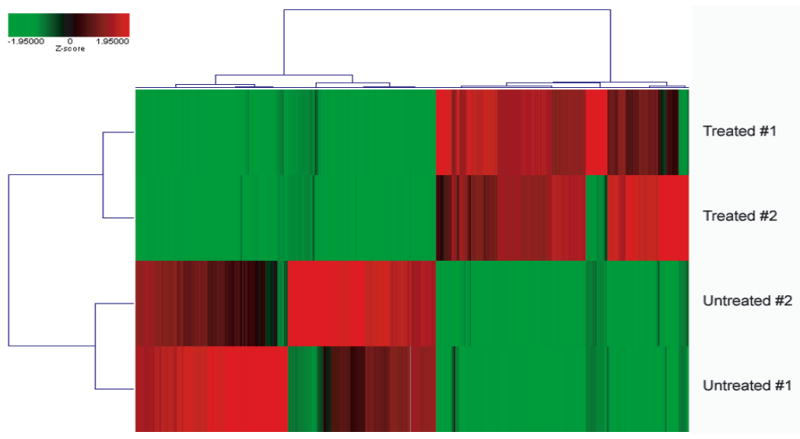
Two-way hierarchical clustering using Ward’s Minimum Variance as the heuristic criteria and Euclidean distance as the similarity metric was performed on all 4 samples using the z-score transformed expression values for the 2,339 transcripts that were identified as differentially expressed (fold-change > 5, log ratio p-value < 0.001) when the two groups were compared to each other (984 genes displayed increased expression, 1,355 displayed decreased expression in response to 5-azadC/TSA). The degree of relatedness between each sample is represented by the dendrogram (hierarchical tree) presented in this figure, wherein the height of each branch represents the distance between the 2 objects being connected. The relationship between the color intensity and the expression level is illustrated by the color bar.
Identification of candidate epigenetically silenced genes in the EMT6 mouse mammary tumor cell line
Following the initial screen by expression microarray, we established a series of bioinformatic criteria to narrow our search for epigenetically silenced tumor suppressor genes (TSGs) in the EMT6 cell line. From the entire list of genes measured by the microarray, we first excluded genes that showed a moderate change in expression in response to 5-azadC/TSA. Therefore, we required that a candidate gene demonstrate at least a 5-fold increase with a log ratio p-value of 0.001 to be considered further. This narrowed our focus to 984 genes of interest (Fig 2). (A listing of all 984 genes identified is given in Supplemental Table IV.)
Figure 2.
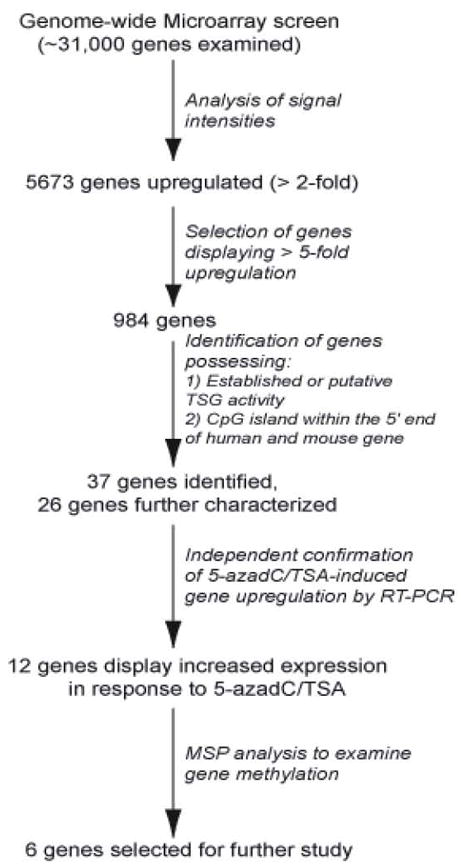
Flow chart summarizing various experimental steps used following initial microarray screen for epigenetically silenced genes in the EMT6 cell line.
Next, we used protein function and genomic structure criteria to further define potential TSGs that are targets for epigenetic silencing in EMT6 cells. We were principally interested in studying genes that encode proteins with an established tumor suppressor activity(ies), or a function consistent with tumor suppressive activity. Specifically, we included in our study gene products where previously published studies support a gene function that either antagonizes cell growth, activates apoptosis or suppresses anti-apoptotic processes, restrains cellular invasiveness, or inhibits angiogenic processes.
We next excluded from the list of putative/characterized TSGs those genes that do not possess a canonical CpG island within 5′ flanking region in both the human and mouse orthologs using the UC Santa Cruz genome browser (http://genome.ucsc.edu) to investigate genomic architecture of various candidate genes. We used the algorithm within the UCSC genome browser that scores the presence of CpG islands in genomic DNA using criteria originally established by Gardiner-Garden and Frommer (Gardiner-Garden and Frommer 1987). Our rationale for establishing these criteria is that genes which possess both features (i.e., putative TSG activity and the presence of a CpG island that overlaps the translational start site of the gene) are likely to identify high-value candidate targets for epigenetic silencing. Of the 984 genes initially identified as being upregulated ≥5-fold in our expression microarray screen, this bioinformatic investigation identified 37 genes meeting the outlined criteria (see Supplemental Table V). Of this, 26 genes were further investigated in this study.
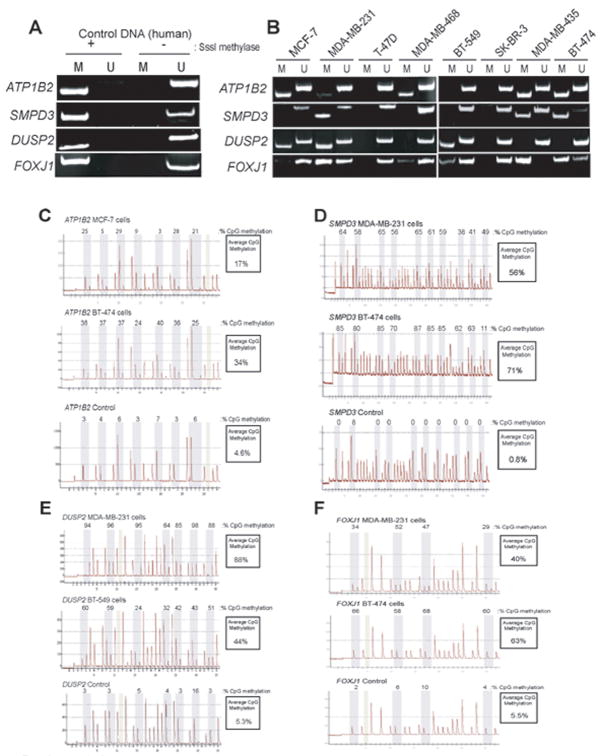
Our next test was to confirm 5-azadC/TSA-induced upregulation of gene expression by RT-PCR. This revealed 12 genes where the gene upregulation detected by microarray was confirmed by RT-PCR and 14 genes whose upregulation could not be independently confirmed using an independent set of RNA samples isolated from drug treated/untreated EMT6 cells. Methylation-Specific PCR (MSP), a common methodology to examine DNA methylation (Herman et al., 1996), was subsequently used to judge methylation within the CpG islands associated with remaining 12 candidate genes. This analysis revealed that six genes exhibited no methylation as judged by MSP. This may indicate that the 5-azadC/TSA-induced increase in expression of these genes is indirect in nature and/or that silencing of these genes is controlled by chromatin modifications other than CpG methylation (e.g., transcriptionally-repressive histone modifications). The later of these possibilities is quite possible since our drug treatment regimen included the histone deacetylase inhibitor TSA. As a result of these efforts our focus narrowed to six genes: Tissue Inhibitor of Metalloproteinase 3 (Timp3), TP53-dependent G2 Arrest Mediator Candidate also termed Reprimo (Rprm), Sphingomyelin Phosphodiesterase 3 Neutral (Smpd3), ATPase, Na+/K+ Transporting, Beta 2 Polypeptide (Atp1B2), Dual-Specificity Phosphatase 2 (Dusp2), and ForkheadboxJ1 (FoxJ1).
Silencing of Timp3 and Rprm in mouse mammary tumor cell lines
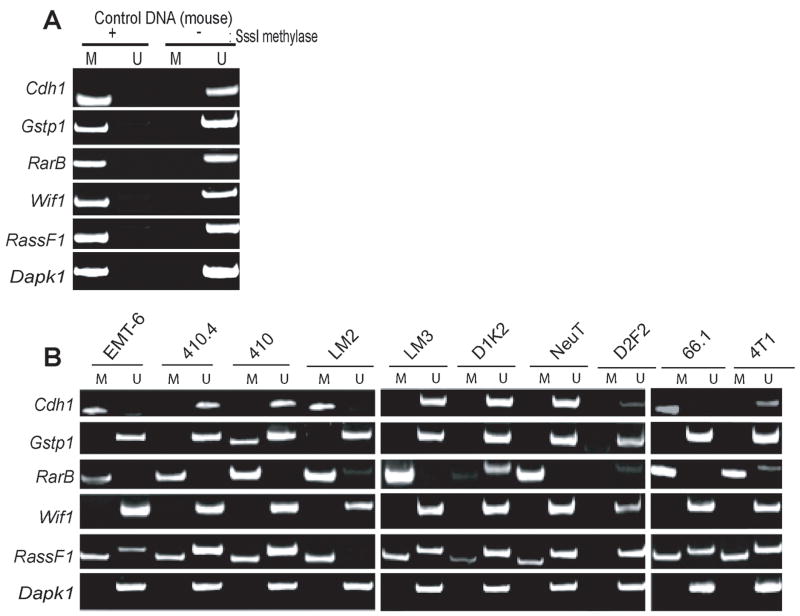
Timp3 and Rprm are well-characterized TSGs silenced in human breast cancer (Bachman et al., 1999; Takahashi et al., 2005). To test how commonly these genes are epigenetically silenced in mouse mammary tumor lines, we acquired 9 distinct cultured mouse mammary tumor lines (designated 410.4, 410, LM3, LM2, D1K2, NeuT, D2F2, 66.1 and 4T1), in addition to the EMT6 cell line. As outlined above, RT-PCR was used to confirm increased expression in response to 5-azadC/TSA; however, due to high-levels of toxicity to this drug cocktail, we found that only four of these cell lines (EMT6, 410.4, 410, and LM3) showed sufficiently low toxicity (>75% viability 48 hrs after 5 μM 5-azadC treatment) to be appropriate subjects for this experiment. These lines were cultured in the presence or absence of 5-azadC and TSA, total RNA harvested and subsequently cDNA synthesized. Results of this RT-PCR experiment clearly indicated that the abundance of Timp3 and Rprm transcripts increased substantially following treatment of each line with 5-azadC/TSA (Fig 3A). Parallel reactions using primers specific for the mouse form of Gapdh confirmed equivalent RNA abundance in each PCR reaction.
Figure 3. Analysis of Timp3 and Rprm silencing in cultured mouse mammary tumor cell lines.
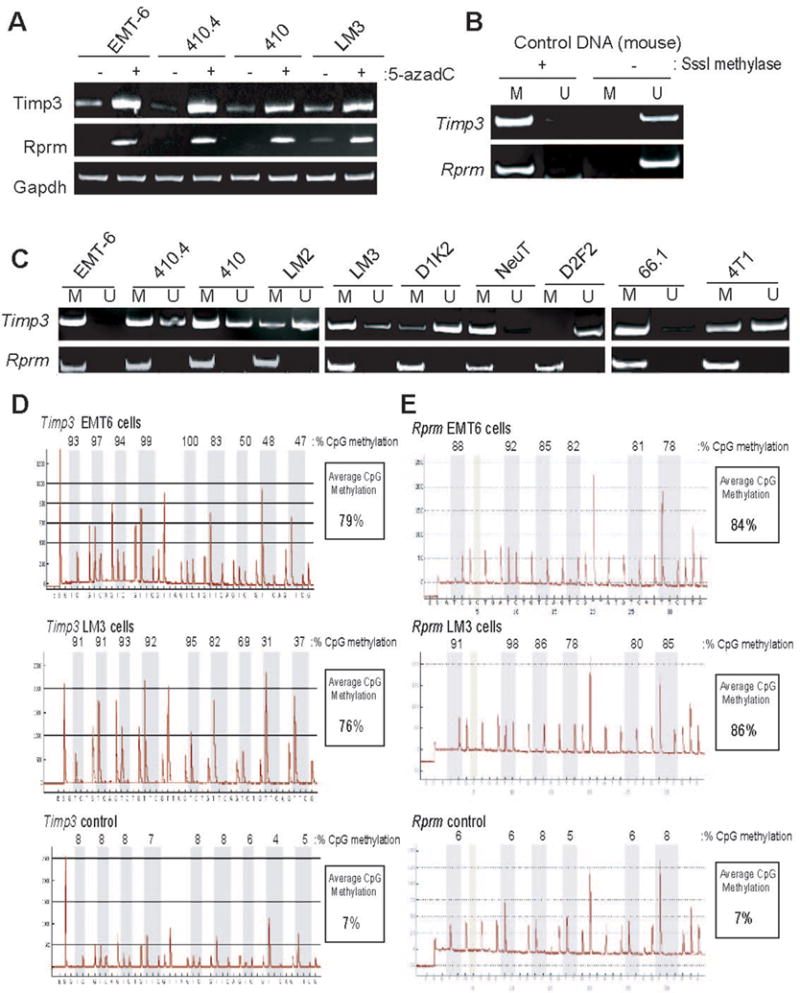
A Indicated mouse mammary tumor lines were either treated or not with a combination of 5-azadC and TSA. Subsequently, total RNA was harvested, cDNA synthesis performed, and PCR reactions conducted using primers specific for the Timp3 (top) or Rprm (middle) genes. Also, PCR reactions using primers specific for the Gapdh gene (bottom) were conducted to assure equivalent RNA abundance in each reaction. B. MSP primers were designed specific to the mouse Timp3 (top) and Rprm (bottom) genes. Primers designed to amplify methylated (M) or unmethylated (U) regions of the CpG island associated with each gene were tested in PCR reactions using mouse genomic DNA either incubated in the presence or absence of SssI methylase prior to bisulfite conversion. C. Genomic DNA harvested from mouse mammary tumor lines EMT6, 410.4, 410, LM3, LM2, NeuT, D2F2, 66.1, 4T1, D1K2 were bisulfite converted and used in MSP reactions with methylated-specific (M) or unmethylated-specific (U) primers. D. Pyrosequencing analysis was conducted on the Timp3 gene using bisulfite-modified genomic DNA from EMT6 and LM3 tumor lines, and mouse genomic DNA as a control. Shown are obtained pyrograms, measured methylation at CpG dinucleotides within the gene region analyzed, and the average CpG methylation within this region of the Timp3 gene. E. Pyrosequencing analysis of the Rprm gene was conducted on EMT6 and LM3 lines and mouse genomic DNA as outlined in D.
We next investigated the methylation status of Timp3 and Rprm in our full panel of mouse mammary tumor line by designing MSP primers using a publically available on-line primer design program (www.mspprimer.org/cgi-mspprimer/design.cgi). All primer sets designed for the analysis for DNA methylation status (both MSP and pyrosequencing analysis) were designed to assay DNA methylation at, or in close proximity to, the transcriptional start site (TSS) of candidate genes since this region is most likely to have undergone dense methylation during the process of epigenetic silencing (Bird 2002; Jones and Baylin 2002).
Primers were designed specific for both the mouse Timp3 and Rprm genes and validated using mouse genomic DNA extracted from tail clippings of C57BL/6 mice. This mouse genomic DNA was either methylated in vitro with SssI methylase or not methylated prior to bisulfite conversion. MSP reactions conducted using this control DNA indicated that our designed primers discriminate between the Timp3 and Rprm genes in either a methylated or unmethylated state (Fig 3B). Subsequently, these MSP primer sets were used to probe the methylation status of the Timp3 and Rprm gene in our panel of 10 mouse mammary tumor cell lines (Fig 3C). With the exception of D2F2, a methylated-specific (M) amplicon was detected when the Timp3 gene was analyzed using bisulfite-converted genomic DNA isolated from each cell line. While amplification of both unmethylated and methylated-specific MSP products is a common finding in MSP analysis (Herman et al., 1996), potentially attributable to heterogeneous promoter methylation within the tumor cell population, this result indicates the presence of methylated CpG within the Timp3 gene in at least a subset of cells of each line except D2F2 where the Timp3 promoter appears to be unmethylated.
As an independent means of quantitatively analyzing DNA methylation, we conducted pyrosequencing on mouse Timp3 and Rprm genes in the EMT6 and LM3 cell lines. Pyrosequencing, like MSP, analyzes bisulfite-converted genomic DNA as a template for PCR; however, pyrosequencing is conducted on PCR products amplified using primers that do not contain internal CpG dinucleotides. Thus, amplification occurs in a manner that is unbiased by DNA methylation status. Subsequently, methylation at distinct CpGs can be determined quantitatively in a pool of genomic DNA (Colella et al., 2003; Tost et al., 2003).
The primer design developed for the mouse Timp3 gene allowed for analysis of 9 CpG dinucleotides within a ~45 bp portion of the CpG island associated with this gene. Pyrosequencing analysis, consistent with MSP results, indicated that Timp3 was methylated in both EMT6 and LM3 cell lines. Moreover, all the CpG dinucleotides investigated using this approach displayed methylation suggesting a high level of methylation density, although CpG methylation was not uniform over the region interrogated. The average CpG methylation measured in this region was 79% in EMT6 and 76% in LM3 (Fig 3D). The pyrosequencing protocol developed for Rprm analyzed 6 CpG dinucleotides over a ~35 bp region of the CpG island within this gene. Similar to Timp3 analysis, all CpG within this region of the Rprm gene exhibit significant methylation with average CpG methylation of 84% and 86% in EMT6 and LM3 cells, respectively (Fig 3E).
Pyrosequencing analysis was also conducted on control (mouse tail) DNA samples (Fig 3D,E). For both Timp3 and Rprm genes, average CpG methylation measured was 7%, indicating very low levels of DNA methylation in this control DNA sample. This result indicates that methylation of the CpG islands associated with the Timp3 and Rprm genes in the mouse genome is a tumor-associated event. Moreover, when our DNA methylation analysis is combined with the results of our RT-PCR experiments using the DNA demethylating agent 5-azadC, we conclude that the Timp3 and Rprm genes are targeted for epigenetic silencing in mouse mammary tumor lines through aberrant gene methylation. Considering most of the 10 distinct cell lines display this feature, silencing of these genes is apparently a common event in cultured mouse mammary tumor cells. Since both Timp3 and Rprm are also targets for epigenetic silencing in human breast tumor line and primary tumors, these results reinforce the validity of our expression microarray screen for epigenetically silenced genes in mouse mammary tumor lines and establish a point of epigenetic overlap between mouse and human mammary gland tumors.
The Atp1B2, Smpd3, Dusp2 and FoxJ1 genes are targets for epigenetic silencing in mouse mammary tumor cell lines
In addition to Timp3 and Rprm, our screen identified four genes (Atp1B2, Smpd3, Dusp2 and FoxJ1) that were strong candidates for epigenetic silencing in the EMT6 line. RT-PCR was conducted on EMT6, 410.4, 410, and LM3 cells cultured in the presence or absence of 5-azadC/TSA using primers designed specifically for the mouse orthologs of these genes. RT-PCR analysis of the Atp1B2 transcript in EMT6 cells showed a clear upregulation of this transcript in response to 5-azadC/TSA (Fig 4A). Conversely, 410.4 and LM3 cells displayed no detectable upregulation of Atp1B2 expression and analysis of 410 cells showed a slight (~2-fold) increase in Atp1B2 expression following 5-azadC/TSA treatment. RT-PCR results showed strong upregulation of Dusp2 expression in 5-azadC/TSA treated EMT6, 410 and 410.4 cells while LM3 cells showed no increased expression of this gene in drug treated LM3 cells. Smpd3 showed marked increases in gene expression in each cell line in response to 5-azadC/TSA. Similarly, FoxJ1 showed clear upregulation in drug treated EMT6 and LM3 cells while expression was more modestly increased in drug treated 410 and 410.4 cells.
Figure 4. Analysis of Atp1B2, Dusp2, Smpd3 and FoxJ1 silencing in mouse mammary tumor lines.
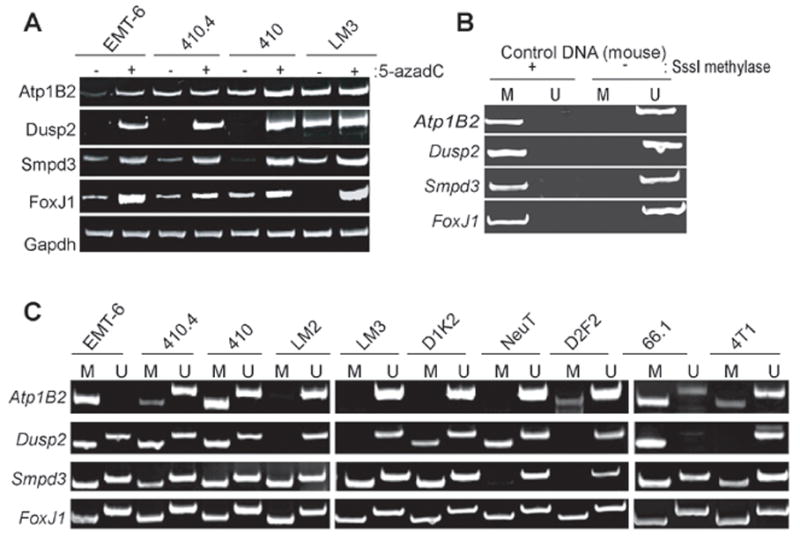
A RT-PCR was conducted on cDNA synthesized from indicated mouse mammary tumor lines treated and untreated with 5-azadC/TSA using primers specific for each indicated gene. B. MSP primers specific for each gene were designed and subsequently validated using bisulfite-modified mouse genomic DNA either methylated or not prior to bisulfite conversion. C. MSP analysis of these four genes using DNA harvested from the indicated mouse mammary cell lines was conducted.
Based on the positive RT-PCR results outlined above, we designed MSP primers for the mouse forms of the Atp1B2, Smpd3, Dusp2 and FoxJ1 genes. As shown (Fig 4B), validation experiments using in vitro methylated/unmethylated mouse genomic DNA confirmed that MSP primer sets were capable of determining the methylation state of each gene. In EMT6 and 410 cells, MSP analysis of the Atp1B2 gene indicated abundant M-specific amplicon indicating that this gene is hypermethylated in these cell lines. This result is consistent with the increased expression of this gene observed in RT-PCR experiments. In 410.4 cells, slight amplification of the M-specific band was noted and no M-specific amplicon was detected when genomic DNA isolated from LM3 cells was assayed, indicating that methylation of this gene is not a prominent feature in this cell line. MSP analysis of the other lines in our panel of mouse mammary tumor cells indicated that only 66.1 cells display prominent amplification of the M-specific product from the Atp1B2 gene. Reduced amplification of the M-specific product was observed in 4T1 and D2F2 cells while no M-specific product was obtained from the other 3 lines (LM2, D1K2 and NeuT) in our panel.
MSP analysis of the Dusp2 gene in EMT6, 410.4 and 410 cells indicated strong amplification of the M-specific product (Fig 4C). When combined with the clear upregulation of the Dusp2 transcript when each of these lines were treated with 5-azadC/TSA, this result indicates that this gene is epigenetically silenced in these lines. Conversely, no aberrant methylation of the Dusp2 gene in LM3 cells was observed. In the remaining six lines in our panel, methylation of the Dusp2 gene was observed in D1K2, NeuT and 66.1 cells and no M-specific amplicon was observed when LM2, D2F2 and 4T1 cells were assayed by MSP.
When the Smpd3 gene was analyzed by MSP, methylation of this gene was observed in all of the cell lines in our panel with the exception of NeuT and D2F2 (Fig 4C). Since we observed upregulation of the Smpd3 in each of the four cell lines treated with 5-azadC/TSA, MSP results are consistent with epigenetic silencing of the Smpd3 gene in cultured mammary tumor lines. Similarly, all ten cultured mammary tumor lines displayed methylation of the FoxJ1 gene as judged by MSP. Again, this is consistent with our observation of increased FoxJ1 expression in 5-azadC/TSA treated cells and indicates that the FoxJ1 is commonly targeted for epigenetic silencing in cultured mouse mammary tumor lines. In sum, these lines of experimentation indicate that the Atp1B2, Dusp2, Smpd3 and FoxJ1 genes are epigenetically silenced in mouse mammary tumor lines; however, the occurrence of these aberrant events is variable within this panel of cultured lines.
Analysis of ATP1B2, DUSP2, SMPD3 and FOXJ1 hypermethylation in human breast cancer lines and primary breast tumors
The ATP1B2, DUSP2, SMPD3 and FOXJ1 genes have not previously been characterized as targets for epigenetic silencing in human breast tumors. Thus, we examined the methylation status of each of these genes in a panel of eight cultured human breast cancer cell lines by designing MSP primer sets for the human ortholog of each gene. Validation experiments using in vitro methylated/unmethylated human placental genomic DNA indicated that each of these primer sets can discriminate between methylated and unmethylated forms of these four genes (Fig 5A). MSP analysis of the ATP1B2 gene in our panel of human breast tumor lines indicated robust amplification of the methylation-specific product in MCF-7, MDA-MB-435 and BT-474 cells while MDA-MB-468 cells show moderate accumulation of the M amplicon and low level amplification of the M product was observed in MDA-MB-231. Other lines in our panel (T-47D, BT-549 and SKBR3) displayed no evidence of ATP1B2 methylation. MSP analysis of the SMPD3 gene determined that MDA-MB-231, MDA-MB-435 and BT-474 cells display methylation of this gene, while the other cell lines show no evidence of SMPD3 methylation. Similarly, DUSP2 was found to be methylated in MCF7, MDA-MB-231 and BT-549 cells but not in T-47D, MDA-MB-468, SKBR3, MDA-MB-435 and BT-474 cells. Finally, MSP analysis of the FOXJ1 gene indicates strong amplification of the M-specific amplicon in MDA-MB-231, MDA-MB-435 and BT-474 cells. Weak amplification of the M-specific FOXJ1 amplicon was observed in MCF-7, MDA-MB-468 and BT-549. No amplification of the FOXJ1 M-specific product was observed in either T-47D or SKBR3 cells.
Figure 5. Analysis of ATP1B2, DUSP2, SMPD3 and FOXJ1 methylation in human breast tumor cell lines.
A MSP primers were designed to the human ortholog of each indicated gene and validated using human placental DNA that was incubated in the presence or absence of SssI methylase prior to bisulfite conversion. B. MSP analysis of indicated genes was conducted on indicated human breast tumor cell lines. C. Pyrosequencing analysis of the ATP1B2 gene was conducted using genomic DNA harvested from MCF-7, BT-474, and human placenta (control). D. Pyrosequencing analysis of the SMPD3 gene in MDA-MB-231, BT-474 and control DNA. E. Pyrosequencing analysis of the DUSP2 gene in MDA-MB-231, BT-549 and control DNA. F. Pyrosequencing analysis of the FOXJ1 gene in MDA-MB-231, BT-474 and control DNA samples.
To confirm the MSP results obtained when the methylation status of the ATP1B2, DUSP2, SMPD3 and FOXJ1 genes was scored by MSP, we conducted pyrosequencing. The pyrosequencing assay developed for the human ATP1B2 gene scores methylation of seven CpG dinucleotides within a ~30 nt region of the CpG island within the 5′ end of the human ATP1B2 gene. When genomic DNA harvested from MCF-7 cells was analyzed by pyrosequencing, we observed low (3%) to moderate (29%) levels of CpG methylation within this cell line in the ATP1B2 gene (Fig 5C). Overall, average CpG methylation measured within the ATP1B2 gene was 17% in this cell line. In BT-474 cells, we observed markedly higher methylation within the ATP1B2 gene with average CpG methylation of 34%. Pyrosequencing of control (human placental) DNA showed a low level of CpG methylation (4.6%) in this gene.
Pyrosequencing analysis of the SMPD3 gene showed high average levels of CpG methylation (56% and 71%) within the SMPD3 gene in MDA-MB-231 and BT-474 cells, respectively (Fig 5D). Conversely, low levels of CpG methylation (0.8%) of the SMPD3 gene was measured in control DNA. Consistent with MSP results, we observed methylation of the CpG island associated with the human DUSP2 gene in MDA-MB-231 (88%) and BT-549 (44%) cells with low (5.5%) CpG methylation measured in control DNA (Fig 5E). We also observed CpG methylation within the human FOXJ1 gene in MDA-MB-231 (40%) and BT-474 (63%) cells and control DNA displayed low (5.5%) CpG methylation within FOXJ1 (Fig 5F). Taken together, both MSP and pyrosequencing analyses indicate that the ATP1B2, DUSP2, SMPD3 and FOXJ1 are aberrantly methylated, albeit at differing frequencies, in cultured human breast tumor lines.
Next, we conducted MSP analysis to examine ATP1B2, DUSP2, SMPD3 and FOXJ1 gene methylation in primary breast tumor/normal mammary tissue samples. Fifteen frozen human breast tumors were obtained and patient-matched disease-free breast tissue were obtained with two of these tumor samples (Tumor72 and Tumor75). Genomic DNA was isolated, bisulfite modified, and used in MSP reactions with primers specific for the ATP1B2, DUSP2, SMPD3 and FOXJ1 genes. In all normal breast tissue samples, we observed exclusive amplification of the unmethylated-specific (U) amplicon, indicating that these genes are not methylated in normal mammary tissue (Fig 6A). When patient-matched breast tumor samples were analyzed by MSP, we observed methylation of the SMPD3 gene in both Tumor72 and Tumor75. Further, methylation of the FOXJ1 gene was observed in Tumor72. No methylation of either DUSP2 or ATP1B2 was observed in either Tumor72 or Tumor75.
Figure 6. Analysis of ATP1B2, DUSP2, SMPD3 and FOXJ1 methylation in primary human breast tumors.
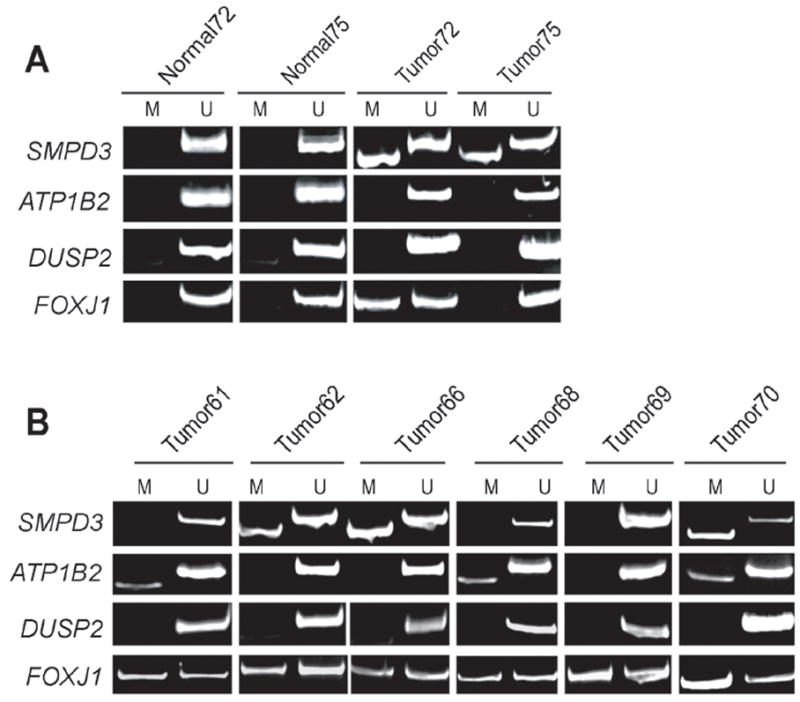
A MSP analysis of the indicated genes was conducted on human primary breast tumors and patient-matched normal breast tissue. B. Representative MSP analysis of six additional human primary breast tumors.
Figure 6B shows a representative subset of additional breast tumors analyzed by MSP. We observed methylation of the SMPD3 gene in Tumor 62, 66 and 70. Overall, we observed SMPD3 methylation in 60% of breast tumors investigated (9 out of 15). We observed methylation of the ATP1B2 gene in Tumor 61, 68 and 70 and, overall, detected a 20% rate (3/15) of ATP1B2 methylation in breast tumors. In contrast, FOXJ1 was found to be commonly methylated in human breast tumors; specifically, we observed that 11 out of 15 tumors surveyed (73%) displayed aberrant FOXJ1 methylation as scored by MSP. Taken together, MSP analysis suggests that promoter hypermethylation the ATP1B2, SMPD3 and FOXJ1 occur in primary human breast tumors with varying occurrence.
Despite the fact that we documented hypermethylation of DUSP2 in cultured human breast cancer lines, this gene was found to be unmethylated in all human breast tumors examined (Fig 6B). DUSP2 encodes a phosphatase (also known as PAC1) that dephosphorylates and inactivates mitogen-activated protein (MAP) kinases (Chu et al., 1996) and is a transcriptional target of p53 in signalling apoptosis (Yin et al., 2003). These properties make this gene an excellent candidate TSG and DUSP2 hypermethylation has been reported in prostate cancer (Yu et al., 2005b). However, we did not observe methylation of this gene in primary tumors leading us to conclude that epigenetic silencing of DUSP2 in human breast cancer is likely a rare event.
Analysis of methylation status of characterized targets for epigenetic silencing in human breast tumors in cultured mouse mammary tumor lines
Finally, we determined the methylation status of well-defined TSGs known to be targets for epigenetic silencing in human breast cancer in our panel of cultured mouse mammary tumor lines. We designed MSP primer sets to assay methylation within the CpG islands associated with the mouse orthologs of the Cdh1, RarB, Gstp1, Dapk1, RassF1 and Wif1 genes. Validation experiments using in vitro methylated/unmethylated mouse genomic DNA indicate that each of these primer sets can delineate gene methylation status (Fig 7A).
Figure 7. Analysis of genes characterized as targets for epigenetic silencing in human breast tumors in a panel of mouse mammary tumor lines.
A MSP primers were designed to the mouse orthologs of the Cdh1, Gstp1, RarB, Wif1, RassF1 and Dapk1 genes. Primer sets were validated using control mouse genomic DNA that was either methylated/unmethylated in vitro prior to bisulfite conversion. B. MSP analysis of indicated genes was conducted on bisulfite-converted DNA isolated from indicated mouse mammary tumor lines. gDNA was harvested from the cell lines and bisulfite-modified. These DNA samples were used in MSP analyses.
Our MSP analyses indicate that RassF1 and RarB were commonly methylated in our panel of mouse mammary tumor lines with 90% and 80% showing significant methylation, respectively (Fig 7B). In contrast, methylation of Cdh1 and Gstp1 were far less commonly encountered in this panel of cell lines with 30% and 10%, respectively, displaying methylation of these genes. In all mouse mammary cell lines, we observed exclusive amplification of the unmethylated-specific products for Dapk1 and Wif1 genes indicating that neither gene is methylated in this panel of mouse cell lines.
DISCUSSION
Comparative epigenomics of mouse and human mammary tumors
By coupling genome-wide screening with bioinformatics analyses and inspection of publically available mouse and human genome sequence, we were able to characterize 6 candidate genes as targets for epigenetic silencing in the EMT6 line. These genes are: Timp3, Rprm, Smpd3, Dusp2, Atp1B2, and FoxJ1. Of these, Timp3 and Rprm have already been characterized as TSGs and targets for epigenetic silencing in human breast and other tumor types (Esteller et al., 2001; Takahashi et al., 2005; Kim et al., 2006;). The fact that our initial screening identified these TSGs as silencing candidates in EMT6 underscores the successful nature of our initial screen. Moreover, since analysis of 10 distinct mouse mammary tumor lines shows that both Timp3 and Rprm are commonly silenced in cell lines of this type, we conclude that both human and mouse mammary tumors share silencing of these tumor suppressor genes in common.
A combination of RT-PCR and MSP confirmed that Smpd3, Dusp2, Atp1B2, and FoxJ1 are silenced in mouse mammary tumor lines. Our findings indicate that Atp1B2 is a target, although likely an uncommon one, for silencing in mouse mammary cell lines. The other 3 genes identified in our screen appear to be more commonly hypermethylated in mouse mammary tumor lines. We observed significant amplification of the M-specific amplicon specific for Dusp2 in 60% of the cell lines tested, Smpd3 in 80%, and FoxJ1 in 100%. When a panel of eight human breast tumor cell lines were assayed for methylation by pyrosequencing and/or MSP, we observed substantial methylation of the ATP1B2 gene in 50% (4/8) cell lines assayed. Also, SMPD3, DUSP2, and FOXJ1 methylation was detected in 38% (3/8) of the cell lines surveyed. Of note, methylation of each of these genes, with the exception of DUSP2, was observed in human primary breast tumors and MSP analysis of normal breast tissue showed methylation of these genes to be a disease-associated event. The fact that DUSP2 methylation was observed in cultured human cell lines but not primary tumors strongly suggests that silencing of this gene is likely a rare event in human breast tumors.
As part of our study we also examined the methylation status of 6 genes known to be targets of epigenetic silencing in human breast tumors in our panel of cultured mouse mammary tumor lines. In this investigation the mouse ortholog of the Cdh1, Gstp1, RarB, Wif1, RassF1, and Dapk1 genes were assayed by MSP. We observed, to various extents, that Cdh1, Gstp1, RarB, and RassF1 were hypermethylated in our panel of mouse tumor lines. However, we obtained no evidence that either the Wif1 or Dapk1 genes are aberrantly methylated in any of these lines.
When the results of our studies on various epiegentically silenced genes in human and mouse cultured mammary tumor cell lines and human primary breast tumors are considered collectively, we conclude that there is significant overlap in the mammary tumor cell epigenome between these two species. Of the twelve genes studied, only Wif1 and Dapk1 were found not be silenced in both species. While Dusp2 was found to be commonly hypermethylated in mouse cell lines, methylation of this gene was less commonly observed in cultured human lines and no evidence for hypermethylation was observed in primary human breast tumors. These results indicate that some genes targeted for epigenetic silencing in human breast tumors are also aberrantly methylated in mouse mammary tumor lines demonstrating considerable but not absolute overlap in the mammary tumor epigenome between these two species.
SMPD3, ATP1B2 and FOXJ1 as novel targets for epigenetic silencing in human breast cancer
Sphingomyelinases (SMases) belong to a multigene family of phosphodiesterases. Each of the three well-characterized SMases (SMPD1, SMPD2, and SMPD3) hydrolyze sphingomyelin to ceramide and phosphocholine. The acid sphingomyelinase SMPD1 is located in the endolysosomal compartment, while the neutral SMases (nSMases) SMPD2 and SMPD3 are located within the endoplasmic reticulum membranes and in the Golgi apparatus, respectively (Stoffel et al., 2007). nSMases are a major intracellular regulator of ceramide, a bioactive lipid.
Several lines of evidence support the view that the ability of nSMases contribute to proliferation of cancer cells and/or to their invasiveness. Of note, the SMPD3 locus is present, as well as six members of the cadherin family, with chromosome 16q22.1, and genetic alterations in this region have been implicated in the progression of various cancers, notably breast cancer (Chalmers et al., 2001; Driouch et al., 1997). Moreover, it has recently been shown that reduction of ceramide levels due to inhibition of nSMase in Hs578T breast adenocarcinoma cells overexpressing caveolin-1 results in a significant enhancement in Akt activation a well-characterized pro-growth, pro-survival mechanism (Wu et al., 2007). Ample evidence also supports a role for ceramide in the activation of apoptosis. For example, Luberto et al (2002) showed inhibition of TNF-α-induced cell death in MCF7 cells using a novel inhibitor of nSMase activity. Data obtained in this study indicates that ceramide induces conformational changes in Bax and activates mitochondrial phosphatase 2A, which, in turn, dephosphorylates Bcl-2. These two events clearly link ceramide and nSMases to the activation of cell death. Similarly, studies link ceramide to the regulation of cell cycle progression by inducing growth arrest by promoting dephosphorylation of the Rb tumor suppressor (Alesse et al., 1998).
In this report, we show that the SMPD3 gene in both mouse and man is a target for epigenetic silencing in mammary tumors. To our knowledge, this is the first report of the SMPD3 gene being targeted for silencing during tumorigenesis. Functionally, this aberrant event will result in decreased ceramide production in response to normal physiological cues promoting dysregulation of both cellular death and growth programs. Thus, this event has clear implications in the process of breast tumorigenesis.
The protein encoded by ATP1B2 is a member of the Na+/K+ ATPase beta chain protein family. Na+/K+ ATPases, which are composed of two subunits (a large catalytic subunit (alpha) and a smaller glycoprotein subunit (beta)), are integral membrane proteins responsible for establishing and maintaining the electrochemical gradients of Na+ and K+ ions across the plasma membrane. This process is essential for osmoregulation, for sodium-coupled transport of a variety of organic and inorganic molecules, and for electrical excitability of nerve and muscle (Dobretsov and Stimers 2005).
The adhesion molecule on glia (AMOG) protein was originally identified as a neural cell adhesion molecule and controls neuron–astrocyte adhesion as well as neural cell migration in vitro (Antonicek et al., 1987), was later found to be encoded by the ATP1B2 gene (Gloor et al., 1990). Others (van den Boom et al., 2006) recently showed that the ATP1B2 gene was targeted for epigenetic silencing via aberrant gene methylation in malignant glioma cells. This finding, supported by the observation that reduced ATP1B2 expression is associated with increased tumor grade (Senner et al., 2003), led these investigators to propose that ATP1B2 silencing could be a significant event in glioma development. Our results indicate that Atp1B2 is epigenetically silenced in a subset of cell lines in our panel of mouse mammary tumor lines and its human ortholog is subject to gene hypermethylation in both human breast tumor cell lines and primary tumors. Given the role that this gene product plays in controlling glial migration, these findings suggest that silencing of ATP1B2 may influence breast tumor cell migration as well. This compelling notion requires further investigation.
FOXJ1 is a member of the fork head gene family of transcription factors originally identified in Drosophila, and a large family of FOX genes are currently characterized in humans (Katoh 2004). In mammalian cells, FOXJ1 has been shown to suppress NFκB transcriptional activity in vitro and suppress inflammation in vivo (Lin et al., 2004). While NFκB is a key regulator of immune response, dysregulation of NFκB often drives deregulated growth, resistance to apoptosis, and increased metastatic potential (Lu and Stark 2004). Since FOXJ1 functions as an antagonist to NFκB activity it is reasonable to suggest that this molecule functions as a TSG, and others have commented that FOXJ1 is an excellent candidate for anti-cancer therapy (Chen 2004). While the role of FOXJ1 in cancer suppression remains to be explored, our finding that this gene is a common target for aberrant hypermethylation in both cultured cells and primary tumors strongly suggests that FOXJ1 silencing is potentially an important event in breast tumorigenesis.
In conclusion, our study has documented considerable, but not absolute, overlap exists between the epigenome of mouse and human mammary tumors. This finding supports the possible utility of using mouse as a model system to understand early epigenetic events during tumorigenesis as well as the general impact of aberrant chromatin remodeling events in cancer. Moreover, our study revealed several putative TSG that are novel targets for epigenetic silencing in human breast cancer.
Table I.
RT-PCR Primers
| Primer Designation | Human/Mouse | Primer Sequence (5′ ➝ 3′) | Orientation (Forw/Rev) | Annealing Temp (°C) | Thermocycles | Product Size (bp) |
|---|---|---|---|---|---|---|
| Gapdh-RT | M | AACGACCCCTTCATTGAC | F | 57 | 30 | 191 |
| TCCACGACATACTCAGCAC | R | |||||
| Timp3-RT | M | AATTCCTCTGCTCAGGCTCCTGTT | F | 60 | 30 | 429 |
| AGCATGGTCATTTCCTCGGGTTCT | R | |||||
| Rprm-RT | M | TATTCGCCACGCTGCAGACCTAAT | F | 60 | 30 | 359 |
| AAACCGTGCAGACAGGAGCTAAGA | R | |||||
| Smpd3-RT | M | ACATCGATTCTCCCACCAACACCT | F | 60 | 30 | 227 |
| AATTCGCACAATGCAGCTGTCCTC | R | |||||
| Atp1b2-RT | M | TGCTTCTGGAATGTCCTTGACCCT | F | 60 | 30 | 368 |
| TGGGTGGATGAGACCATTTCCTGT | R | |||||
| Dusp2-RT | M | TGGTCCTGTGGAAATCTTGCCCTA | F | 60 | 31 | 307 |
| AATCAGGTATGCCAGGCAGATGGT | R | |||||
| FoxJ1-RT | M | AAGGTAACTTTGACTGGGAGGCCA | F | 60 | 30 | 225 |
| AGGATGTGCCCAAGAAGGTCTCAT | R |
Table II.
Methylation-Specific PCR (MSP) Primers
| Primer Designation | Human/Mouse | Primer Sequence (5′ ➝ 3′) | Orientation (Forw/Rev) | Annealing Temp (°C) | Thermo- cycles | Product Size (bp) |
|---|---|---|---|---|---|---|
| Timp3-MSP-M | M | CGGAGGGGGCGGATTTC | F | 60 | 40 | 84 |
| CACGAATCCGCGAAACCAAC | R | |||||
| Timp3-MSP-U | M | TGGAGGGGGTGGATTTT | F | 55 | 40 | 184 |
| ACACAAATCCACAAAACCAAC | R | |||||
| Rprm-MSP-M | M | GTCGGGTATTTGTTCGTTTCGC | F | 58 | 40 | 96 |
| ACTAAATTCATCACGAAAACAAACGAAAACCG | R | |||||
| Rprm-MSP-U | M | GTTAGTGTTTTGTTGGGTATTTGTTTGTTTTGTG | F | 58 | 40 | 96 |
| ACACTAAATTCATCACAAAAACAAACAAAAACCA | R | |||||
| Smpd3-MSP-M | M | GGGTTTTTTAGAGACGGTTTTCGC | F | 59 | 40 | 90 |
| GCTCCAAATACTAAATCCGAAAAAAACTCCG | R | |||||
| Smpd3-MSP-U | M | TGATTGGGGTTTTTTAGAGATGGTTTTTGTG | F | 58 | 40 | 90 |
| CCACTCCAAATACTAAATCCAAAAAAAACTCCA | R | |||||
| Atp1b2-MSP-M | M | CGTGGAGGGTTTTAGCGC | F | 58 | 40 | 74 |
| AAACACGAAACGAATCCCGACG | R | |||||
| Aatp1b2-MSP-U | M | GTGTGTGGAGGGTTTTAGTGTG | F | 57.5 | 40 | 74 |
| AACCATCTTAAAAACACAAAACAAATCCCAACA | R | |||||
| Dusp2-MSP-M | M | GTTCGTTTTTGGTTTTTTGTCGGAGTTAC | F | 58.5 | 40 | 102 |
| AAAACAACGACGAAAATACCGCG | R | |||||
| Dusp2-MSP-U | M | TGTTGTTTGTTTTTGGTTTTTTGTTGGAGTTATG | F | 58 | 40 | 102 |
| ACAAACAAACAAAAACAACAACAAAAATACCACA | R | |||||
| FoxJ1-MSP-M | M | GGTTTTTTTAGCGGTTAAATTCGACGC | F | 59 | 40 | 86 |
| GCTAAATCTCACGACGAATACTCG | R | |||||
| FoxJ1-MSP-U | M | GGTTGGGTTTTTTTAGTGGTTAAATTTGATGTG | F | 58 | 40 | 86 |
| CCCACACTAAATCTCACAACAAATACTCA | R | |||||
| SMPD3-MSP-M | H | TTAGATTTGCGGTCGGGAGTTC | F | 58.5 | 40 | 100 |
| AACACTAACCTCTAACGACGAACG | R | |||||
| SMPD3-MSP-U | H | TTGTTGTTAGATTTGTGGTTGGGAGTTTG | F | 58.5 | 40 | 100 |
| AAATTCAACAACACTAACCTCTAACAACAAACA | R | |||||
| ATP1B2-MSP-M | H | GGCGATCGCGGGTTTC | F | 59 | 40 | 116 |
| CCGATAATAACTCAACCGCGAAAATAAAACG | R | |||||
| ATP1B2-MSP-U | H | GTATAAAGATTGTGTTTGGTGATTGTGGGTTTTG | F | 58.5 | 40 | 116 |
| ACACCAATAATAACTCAACCACAAAAATAAAACA | R | |||||
| DUSP2-MSP-M | H | TTATCGTTACGAGAGTTCGGGAC | F | 58 | 40 | 119 |
| CGCAACAACGTACCCAACG | R | |||||
| DUSP2-MSP-U | H | GGTAGTGTTATTGTTATGAGAGTTTGGGATG | F | 58.5 | 40 | 119 |
| AAATCCCACAACAACATACCCAACA | R | |||||
| FOXJ1-MSP-M | H | TTTTTAGAACGAACGGATTTATAGC | F | 59 | 40 | 193 |
| ACACTTACCACTCTCCCTAACGAC | R | |||||
| FOXJ1-MSP-U | H | TTTAGAATGAATGGATTTATAGTGG | F | 58 | 40 | 193 |
| AACACTTACCACTCTCCCTAACAAC | R | |||||
| Ecad-MSP-M | M | GGTTTAATTCGGTTTTGTTCGATCGTATTC | F | 58 | 40 | 75 |
| GAACTCCCATAACGAACCCG | R | |||||
| Ecad-MSP-U | M | GTTTGGTTTAATTTGGTTTTGTTTGATTGTATTTG | F | 58 | 40 | 75 |
| ACCAAACTCCCATAACAAACCCA | R | |||||
| RassF1-MSP-M | M | GGGCGGTCGTTGGAATTTC | F | 58.5 | 40 | 99 |
| CCCTCCGAATAACTCGCTATAAAAAAACG | R | |||||
| RassF1-MSP-U | M | AGGGGTGGTTGTTGGAATTTTG | F | 57.5 | 40 | 99 |
| CAAAACCCTCCAAATAACTCACTATAAAAAAACA | R | |||||
| Gstp1-MSP-M | M | GAATTCGTATTTAGTAGGTAGTGAGTTCGTTTC | F | 58 | 40 | 106 |
| CGATACAAAAAACGCCAATCGTAAACG | R | |||||
| Gstp1-MSP-U | M | TGAATTTGTATTTAGTAGGTAGTGAGTTTGTTTTG | F | 58 | 40 | 106 |
| AACTACCCAATACAAAAAACACCAATCATAAACA | R | |||||
| Rarβ-MSP-M | M | GATTTTAAGTTTTTTTTTTTAGGCGACGTTC | F | 56.5 | 40 | 89 |
| GAACAAAAAACGTCTAAACACCG | R | |||||
| Rarβ-MSP-U | M | ATTGATTTTAAGTTTTTTTTTTTAGGTGATGTTTG | F | 56.5 | 40 | 89 |
| CCCAAACAAAAAACATCTAAACACCA | R | |||||
| Dapk1-MSP-M | M | TTCGGGTCGCGGAGTC | F | 59 | 40 | 110 |
| CAATTCCTCGCCGATATCGTAATAATCG | R | |||||
| Dapk1-MSP-U | M | GATGGTTTGGGTTGTGGAGTTG | F | 57.5 | 40 | 110 |
| CTACCCAATTCCTCACCAATATCATAATAATCA | R | |||||
| Wif1-MSP-M | M | GGGTATCGCGGTTTCGTTTTC | F | 58.5 | 40 | 96 |
| CTACTAACAAAAAACGAACACTAAAAACGACCG | R | |||||
| Wif1-MSP-U | M | GGGTTTATTGGGTATTGTGGTTTTGTTTTTG | F | 58 | 40 | 96 |
| ACTACTAACAAAAAACAAACACTAAAAACAACCA | R |
Table III.
Pyrosequencing Primers (Amplification and Sequencing)
| Primer Designation | Human/Mouse | Primer Sequence (5′ ➝ 3′) | Orientation (Forw/Rev) | Annealing Temp (°C) | Thermo- cycles | Product Size (bp) |
|---|---|---|---|---|---|---|
| Rprm-PYRO | M | TTGTAGTTAAGGAAGAAGATGTTAAAAATTAT | F | 50.5 | 50 | 344 |
| AACCTAAAACTTTAAAAAAAAAACTAAAAAAC | R | |||||
| GTTTGGGTGTAGTAG | SEQ* | |||||
| YGTATYGYGYGTTTTAGYGTTTYGTTAT | SEQ to analyze** | |||||
| Timp3-PYRO | M | GATGAATTTTTGTTTAGGTTT | F | 50 | 50 | 195 |
| AAAATCCAAAACTACAAAATAAT | R | |||||
| TTTTTATAAGGATTTGAA | SEQ* | |||||
| YGGTTYGGTYGGTTTYGTTTTGYGTTTYGYGTTTTTYGGTTTYGTT | SEQ to analyze** | |||||
| ATP1B2-PYRO | H | GGTGGGAAGGGGGTGGGTATTT | F | 56 | 50 | 123 |
| CAACCAAAAATAAAAAACCCCCCAAA | R | |||||
| GGGGTGGGTATTTTT | SEQ* | |||||
| YGYGTTYGGYGATYGYGGGTTTTYGTATT | SEQ to analyze** | |||||
| DUSP2-PYRO | H | GAAGTTTTTAGGTTAGTTTAGATTG | F | 53 | 50 | 242 |
| CCTCCAACCCCATAACCACC | R | |||||
| GTTTTTGTATGGTGTTT | SEQ* | |||||
| YGGAGGTYGGTTTAYGGGTATTTAATTYGGGTYGTYGYGGA | SEQ to analyze** | |||||
| FOXJ1-PYRO | H | TTTTAGTTTTAGTTTTTAATTTTAGGTTT | F | 52 | 50 | 290 |
| ACACTTACCACTCTCCCTAAC | R | |||||
| GTTTTAGGAGTAGTTGA | SEQ* | |||||
| YGTTTTAAAGYGTAYGGGATAGGAAGGGGGTGGGGGYGA | SEQ to analyze** | |||||
| SMPD3-PYRO | H | GGATGTGGTTTTGAAGGAGAGATTT | F | 53.5 | 50 | 305 |
| GAATTCAACAACACTAACCTCTA | R | |||||
| GGGTTGTGAGGAG | SEQ* | |||||
| TYGGATTYGAGAGTYGAGAGTYGTYGTYGTTGYGGTYGTYGTT | SEQ to analyze** |
SEQ: Sequencing primer
SEQ to analyze: Gene-specific sequence analyzed by pyrosequencing (Y: Potential methylation site)
Table IV. Microarray Data.
Shown are the 984 genes found to be ≥5-fold upregulated in 5-azadC treated EMT6 cells, genes subject to further study are indicated in magenta.
| Primary Sequence Name | Sequence Description | Accession # | Fold Change | P-value |
|---|---|---|---|---|
| 6720406K03 | hypothetical protein 6720406K03 | AK032705 | 63.47921 | 2.69E-07 |
| Gcg | Glucagon, mRNA (cDNA clone MGC:14058 IMAGE:4218718) | AK050538 | 63.45748 | 2.73E-07 |
| 2900027M19Rik | Transmembrane protein 28 (Tmem28), mRNA | AK045452 | 63.31421 | 2.98E-07 |
| D830044D21Rik | RIKEN cDNA D830044D21 gene (D830044D21Rik), mRNA | NM_177210 | 61.82942 | 2.46E-07 |
| Pkd1l2 | Polycystic kidney disease 1 like 2 (Pkd1l2), mRNA | NM_029686 | 60.27456 | 2.10E-44 |
| Tspan33 | Tetraspanin 33 (Tspan33), mRNA | NM_146173 | 58.68657 | 8.71E-33 |
| 4930442J19Rik | Adult male testis cDNA, RIKEN full-length enriched library, clone:4930442J19 product: unclassifiable, full insert sequence | AK015363 | 56.56114 | 9.32E-08 |
| 2610024A01Rik | RIKEN cDNA 2610024A01 gene (2610024A01Rik), mRNA | BC059048 | 49.58956 | 4.61E-08 |
| Fgf11 | Fibroblast growth factor 11 (Fgf11), mRNA | BC066859 | 48.44208 | 8.99E-26 |
| AF366264 | CDNA sequence AF366264 (AF366264), mRNA | NM_153093 | 47.83025 | 3.85E-08 |
| Ung2 | PREDICTED: similar to uracil-DNA glycosylase 2 [Mus musculus], mRNA sequence | AK053890 | 47.79745 | 3.05E-17 |
| 6430548M08Rik | RIKEN cDNA 6430548M08 gene, mRNA (cDNA clone MGC:38801 IMAGE:5359842) | AK122299 | 44.75741 | 1.10E-15 |
| Serpini1 | Serine (or cysteine) peptidase inhibitor, clade I, member 1, mRNA (cDNA clone MGC:6413 IMAGE:3588856) | NM_009250 | 44.68072 | 1.38E-15 |
| Dpep1 | Dipeptidase 1 (renal), mRNA (cDNA clone MGC:6318 IMAGE:2812088) | NM_007876 | 44.65215 | 2.73E-29 |
| Cabc1 | Chaperone, ABC1 activity of bc1 complex like (S. pombe), mRNA (cDNA clone MGC:31744 IMAGE:4922892) | NM_023341 | 41.85896 | 2.83E-24 |
| 1190002H23Rik | RIKEN cDNA 1190002H23 gene (1190002H23Rik), mRNA | NM_025427 | 39.94254 | 9.82E-29 |
| 1600029D21Rik | RIKEN cDNA 1600029D21 gene (1600029D21Rik), mRNA | NM_029639 | 39.51588 | 1.83E-22 |
| 4930522H14Rik | RIKEN cDNA 4930522H14 gene (4930522H14Rik), mRNA | NM_026291 | 39.45048 | 5.43E-08 |
| TC982356 | Unknown | 39.22191 | 3.98E-43 | |
| Xist | Xist (X inactive specific transcript) mRNA for open reading frame | X59289 | 37.43628 | 1.34E-23 |
| Dusp2 | Dual specificity phosphatase 2 (Dusp2), mRNA | NM_010090 | 37.08016 | 1.15E-17 |
| BB133117 | Transcribed locus | BB133117 | 37.05725 | 2.84E-13 |
| Nlk | Nemo like kinase (Nlk), mRNA | AK031461 | 35.36866 | 1.11E-08 |
| 6430548M08Rik | RIKEN cDNA 6430548M08 gene, mRNA (cDNA clone MGC:38801 IMAGE:5359842) | NM_172286 | 33.78417 | 6.01E-14 |
| BC011467 | CDNA sequence BC011467, mRNA (cDNA clone MGC:37865 IMAGE:5100525) | BC025823 | 32.62732 | 6.73E-17 |
| LOC545748 | similar to hypothetical protein LOC280487 | AK031095 | 32.16875 | 3.93E-17 |
| 9630025C22 | hypothetical protein 9630025C22 | NM_177597 | 30.76949 | 9.72E-12 |
| Hspa1a | Heat shock protein (hsp68) mRNA, clone MHS213 | NM_010479 | 30.68889 | 6.14E-11 |
| Timp3 | Tissue inhibitor of metalloproteinase 3, mRNA (cDNA clone MGC:25359 IMAGE:4239015) | NM_011595 | 30.407 | 3.63E-12 |
| H2-DMb1 | Histocompatibility 2, class II, locus Mb1, mRNA (cDNA clone MGC:5741 IMAGE:3486844) | NM_010387 | 30.20498 | 1.12E-44 |
| Arrdc4 | Arrestin domain containing 4 (Arrdc4), mRNA | NM_025549 | 30.05997 | 1.22E-14 |
| 9330117B14 | Mus musculus adult male diencephalon cDNA, RIKEN full-length enriched library, clone:9330117B14 product: unclassifiable, full insert sequence. | AK033923 | 29.13712 | 8.04E-09 |
| Sema4f | Sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain (Sema4f), mRNA | NM_011350 | 29.0931 | 1.55E-14 |
| Fzd9 | Mus musculus 16 days embryo head cDNA, RIKEN full-length enriched library, clone:C130077B15 product: frizzled homolog 9 (Drosophila), full insert sequence. | AK021164 | 28.87872 | 2.71E-19 |
| Zfpn1a4 | Zinc finger protein, subfamily 1A, 4 (Eos) (Zfpn1a4), mRNA | AK041640 | 27.41483 | 4.91E-20 |
| Osbp2 | Oxysterol binding protein 2 (Osbp2), mRNA | AK049816 | 27.39835 | 3.84E-24 |
| A930035J23Rik | PREDICTED: Mus musculus RIKEN cDNA A930035J23 gene (A930035J23Rik), mRNA | AK041893 | 27.09363 | 3.60E-28 |
| Tmem25 | Transmembrane protein 25 (Tmem25), mRNA | NM_027865 | 26.96004 | 2.00E-25 |
| Selenbp1 | Selenium binding protein 1, mRNA (cDNA clone MGC:18519 IMAGE:4209393) | NM_009150 | 26.94885 | 3.24E-18 |
| Tfpi | Tissue factor pathway inhibitor, mRNA (cDNA clone MGC:37332 IMAGE:4975683) | NM_011576 | 26.6853 | 9.47E-30 |
| Klk16 | Kallikrein 16, mRNA (cDNA clone MGC:18522 IMAGE:4165443) | NM_008454 | 26.63172 | 8.21E-09 |
| Cd248 | CD248 antigen, endosialin (Cd248), mRNA | NM_054042 | 26.56997 | 7.43E-19 |
| Pde2a | Phosphodiesterase 2A, cGMP-stimulated, mRNA (cDNA clone IMAGE:4949471) | BC006845 | 26.40924 | 2.97E-19 |
| Agxt2l1 | Alanine-glyoxylate aminotransferase 2-like 1 (Agxt2l1), mRNA | AK030395 | 25.35919 | 8.75E-09 |
| Slc30a1 | Solute carrier family 30 (zinc transporter), member 1 (Slc30a1), mRNA | AK048209 | 25.18293 | 7.34E-19 |
| 8030431J09Rik | Ankyrin repeat and BTB (POZ) domain containing 2, mRNA (cDNA clone MGC:62845 IMAGE:6493840) | AK033113 | 24.99684 | 1.12E-15 |
| 1700030G11Rik | PREDICTED: RIKEN cDNA 1700030G11 [Mus musculus], mRNA sequence | AK006546 | 24.99246 | 0.00001 |
| Mlf1 | Myeloid leukemia factor 1, mRNA (cDNA clone MGC:41163 IMAGE:1495721) | NM_010801 | 24.45573 | 7.37E-15 |
| A_51_P306031 | Unknown | 23.98088 | 4.47E-25 | |
| Chi3l3 | Chitinase 3-like 3 (Chi3l3), mRNA | NM_009892 | 23.96213 | 1.01E-38 |
| Dbccr1 | BRINP mRNA for BMP/retinoic acid-inducible neural-specific protein (BRINP) | NM_019967 | 23.93755 | 3.66E-17 |
| B230362M20Rik | RIKEN cDNA B230362M20 gene (B230362M20Rik), mRNA | NM_177273 | 23.77677 | 1.66E-08 |
| 9030221C07Rik | Aromatic-preferring amino acid transporter (Arpat) | NM_177802 | 23.37548 | 0.00001 |
| Sipa1l2 | Signal-induced proliferation-associated 1 like 2, mRNA (cDNA clone IMAGE:30360538) | AK122501 | 23.25885 | 4.47E-36 |
| Gm364 | PREDICTED: similar to hypothetical protein [Mus musculus], mRNA sequence | AV280269 | 23.15239 | 6.48E-38 |
| Selenbp2 | Selenium binding protein 2, mRNA (cDNA clone MGC:35927 IMAGE:5096831) | NM_019414 | 23.06403 | 1.19E-14 |
| Lin7b | Lin 7 homolog b (C. elegans), mRNA (cDNA clone MGC:18338 IMAGE:4164883) | NM_011698 | 22.83365 | 6.72E-27 |
| Tacstd1 | Tumor-associated calcium signal transducer 1, mRNA (cDNA clone MGC:11680 IMAGE:3711069) | NM_008532 | 22.5851 | 4.03E-33 |
| Eml5 | Echinoderm microtubule associated protein like 5, mRNA (cDNA clone IMAGE:4953665) | BC027154 | 22.21267 | 2.27E-28 |
| Fank1 | Fibronectin type 3 and ankyrin repeat domains 1 (Fank1), mRNA | NM_025850 | 22.05824 | 3.68E-22 |
| 1700001J03Rik | RIKEN cDNA 1700001J03 gene (1700001J03Rik), mRNA | BC048528 | 21.14908 | 3.13E-26 |
| NAP059666-1 | Unknown | 21.09742 | 3.79E-06 | |
| Klf2 | Kruppel-like factor 2 (lung) (Klf2), mRNA | NM_008452 | 20.99121 | 2.75E-09 |
| AK054415 | Mus musculus 2 days pregnant adult female ovary cDNA, RIKEN full-length enriched library, clone:E330023G08 product: phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide, full insert sequence. | AK054415 | 20.9248 | 0.00003 |
| D330017J20Rik | MKIAA1170 protein | AK122459 | 20.91998 | 8.39E-35 |
| NAP108451-1 | Unknown | 20.55861 | 0.00005 | |
| Gp2 | Glycoprotein 2 (zymogen granule membrane) (Gp2), mRNA | NM_025989 | 20.50423 | 3.80E-08 |
| Ush2a | PREDICTED: Mus musculus similar to usherin isoform B (LOC545397), mRNA | AK044721 | 20.08695 | 2.99E-08 |
| Abca3 | Mus musculus B6-derived CD11 +ve dendritic cells cDNA, RIKEN full-length enriched library, clone:F730004H09 product: ATP-binding cassette, sub-family A (ABC1), member 3, full insert sequence. | AK089316 | 19.91308 | 2.21E-08 |
| Sned1 | Secreted nidogen domain protein (Snep gene), splice form SNEP | NM_172463 | 19.73947 | 1.44E-21 |
| 4930488E11Rik | RIKEN cDNA 4930488E11 gene, mRNA (cDNA clone MGC:106799 IMAGE:3373135) | NM_207267 | 19.66398 | 1.42E-30 |
| Etv5 | Ets variant gene 5, mRNA (cDNA clone MGC:28414 IMAGE:4036564) | NM_023794 | 19.61583 | 5.13E-27 |
| Disp2 | Dispatched homolog 2 (Drosophila) (Disp2), mRNA | NM_170593 | 19.59815 | 5.48E-08 |
| ENSMUST00000064479 | Unknown | 19.4577 | 8.91E-11 | |
| TC952297 | AF302077 neprilysin-like peptidase gamma {Mus musculus}, partial (4%) [TC952297] | 19.41319 | 8.90E-30 | |
| Igbp1 | Mus musculus 12 days embryo male wolffian duct includes surrounding region cDNA, RIKEN full-length enriched library, clone:6720407F13 product: immunoglobulin (CD79A) binding protein 1, full insert sequence. | AK032709 | 19.30429 | 2.63E-08 |
| TC1019179 | Unknown | 19.29024 | 1.03E-22 | |
| Cd248 | CD248 antigen, endosialin (Cd248), mRNA | NM_054042 | 19.23851 | 4.63E-24 |
| Arhgap29 | Rho GTPase activating protein 29 (Arhgap29), mRNA | NM_172525 | 19.18384 | 3.71E-10 |
| E430004F17Rik | RIKEN cDNA E430004F17 gene (E430004F17Rik), mRNA | NM_183199 | 19.05738 | 5.93E-16 |
| Stard10 | START domain containing 10 (Stard10), mRNA | NM_019990 | 18.91585 | 1.20E-12 |
| Fn3k | Fructosamine 3 kinase (Fn3k), mRNA | NM_022014 | 18.91504 | 1.03E-24 |
| Pfn2 | Profilin 2, mRNA (cDNA clone MGC:35930 IMAGE:5059568) | NM_019410 | 18.78823 | 1.79E-18 |
| Ptgfr | Prostaglandin F receptor (Ptgfr), mRNA | AK086563 | 18.75204 | 3.09E-08 |
| Man1a | Mannosidase 1, alpha, mRNA (cDNA clone MGC:18448 IMAGE:4223319) | NM_008548 | 18.7403 | 3.14E-08 |
| Vwf | Von Willebrand factor (Vwf) mRNA, exon 1 and 5′UTR | AK044921 | 18.63142 | 5.82E-08 |
| Kif27 | Kinesin family member 27 (Kif27), mRNA | NM_175214 | 18.39237 | 2.38E-42 |
| Sh2d4b | SH2 domain containing 4B (Sh2d4b), mRNA | NM_177816 | 18.37578 | 5.13E-08 |
| Sh2d1b | SH2 domain protein 1B (Sh2d1b), mRNA | NM_012009 | 18.35538 | 3.50E-08 |
| Arhgap29 | Rho GTPase activating protein 29 (Arhgap29), mRNA | NM_172525 | 17.9961 | 1.04E-10 |
| AI450469 | Transcribed locus | AI450469 | 17.7114 | 4.97E-28 |
| 4930504O13Rik | RIKEN cDNA 4930504O13 gene (4930504O13Rik), mRNA | NM_207527 | 17.60137 | 7.87E-08 |
| Anxa6 | Mus musculus 10, 11 days embryo whole body cDNA, RIKEN full-length enriched library, clone:2810407D22 product: annexin A6, full insert sequence. | AK013026 | 17.41486 | 1.20E-08 |
| Mical2 | MKIAA0750 protein | NM_177282 | 17.22894 | 1.87E-11 |
| Lrrc15 | Leucine rich repeat containing 15 (Lrrc15), mRNA | BC050245 | 17.22617 | 1.28E-27 |
| NM_133237 | Mus musculus cDNA sequence AB023957 (AB023957), mRNA [NM_133237] | NM_133237 | 17.09971 | 2.44E-31 |
| Myl2 | Myosin, light polypeptide 2, regulatory, cardiac, slow (Myl2), mRNA | NM_010861 | 17.05658 | 5.16E-06 |
| Trib2 | Tribbles homolog 2 (Drosophila), mRNA (cDNA clone MGC:32449 IMAGE:5043179) | NM_144551 | 16.9309 | 1.31E-20 |
| A930037G23Rik | RIKEN cDNA A930037G23 gene, mRNA (cDNA clone MGC:67253 IMAGE:6403474) | NM_178787 | 16.70077 | 1.01E-25 |
| Derl3 | Der1-like domain family, member 3 (Derl3), mRNA | NM_024440 | 16.70039 | 2.27E-08 |
| Rasl11b | RAS-like, family 11, member B, mRNA (cDNA clone MGC:7064 IMAGE:3156762) | AK004534 | 16.64886 | 4.78E-26 |
| A130052D22 | Mus musculus hypothetical protein A130052D22 (A130052D22), mRNA [NM_183306] | NM_183306 | 16.27097 | 3.94E-29 |
| Soat1 | Sterol O-acyltransferase 1, mRNA (cDNA clone IMAGE:4007277) | BC025091 | 16.22614 | 2.66E-44 |
| Adam30 | A disintegrin and metallopeptidase domain 30 (Adam30), mRNA | AK077058 | 16.15904 | 0.00015 |
| Abtb2 | Ankyrin repeat and BTB (POZ) domain containing 2, mRNA (cDNA clone MGC:62845 IMAGE:6493840) | NM_178890 | 16.12632 | 2.87E-27 |
| Dock4 | MKIAA0716 protein | AK085106 | 15.90782 | 4.44E-37 |
| 6720468P15Rik | 0 day neonate thymus cDNA, RIKEN full-length enriched library, clone:A430057M21 product: unclassifiable, full insert sequence | AK078497 | 15.87182 | 4.79E-06 |
| Cxcl4 | Chemokine (C-X-C motif) ligand 4 (Cxcl4), mRNA | NM_019932 | 15.81666 | 7.84E-36 |
| Otub2 | OTU domain, ubiquitin aldehyde binding 2 (Otub2), mRNA | NM_026580 | 15.80523 | 1.53E-15 |
| Thyn1 | Thymocyte protein mThy28 | AK037947 | 15.5622 | 1.23E-07 |
| E130308A19Rik | RIKEN cDNA E130308A19 gene (E130308A19Rik), transcript variant 1, mRNA | AK038359 | 15.44643 | 1.28E-07 |
| 0610042C05Rik | RIKEN cDNA 0610042C05 gene, mRNA (cDNA clone MGC:37398 IMAGE:4977394) | AK076350 | 15.44004 | 4.96E-07 |
| Chn2 | Beta chimaerin (bch gene) | NM_023543 | 15.41819 | 2.69E-07 |
| Rab3b | RAB3B, member RAS oncogene family (Rab3b), mRNA | NM_023537 | 15.2858 | 2.35E-15 |
| Anxa6 | Annexin A6, mRNA (cDNA clone MGC:6574 IMAGE:3482035) | NM_013472 | 15.14969 | 1.10E-13 |
| Nedd9 | Neural precursor cell expressed, developmentally down-regulated gene 9, mRNA (cDNA clone IMAGE:3499250) | NM_017464 | 15.10892 | 1.79E-15 |
| Dazl | Deleted in azoospermia-like (Dazl), mRNA | NM_010021 | 15.03384 | 3.11E-43 |
| Glcci1 | Glucocorticoid induced transcript 1 (Glcci1), transcript variant 1, mRNA | AK042221 | 14.97325 | 2.00E-07 |
| Tfdp2 | Transcription factor Dp 2, mRNA (cDNA clone IMAGE:30604420) | NM_178667 | 14.96471 | 2.55E-17 |
| Cacna1g | Calcium channel, voltage-dependent, T type, alpha 1G subunit (Cacna1g), mRNA | NM_009783 | 14.95897 | 1.24E-16 |
| Ifna4 | Interferon alpha family, gene 4 (Ifna4), mRNA | NM_010504 | 14.95108 | 1.76E-07 |
| Notch3 | Notch gene homolog 3 (Drosophila) (Notch3), mRNA | NM_008716 | 14.90218 | 1.13E-28 |
| Pacsin1 | Protein kinase C and casein kinase substrate in neurons 1, mRNA (cDNA clone MGC:25285 IMAGE:4527708) | NM_011861 | 14.88882 | 1.74E-11 |
| Atp1b2 | ATPase, Na+/K+ transporting, beta 2 polypeptide (Atp1b2), mRNA | NM_013415 | 14.7351 | 1.07E-22 |
| Habp2 | HYALURONIC ACID BINDING PROTEIN 2; HYADURONIC ACID-BINDING PROTEIN 2. [Source:RefSeq;Acc:NM_146101] [ENSMUST00000026060] | 14.65301 | 2.18E-06 | |
| Fabp7 | Fatty acid binding protein 7, brain (Fabp7), mRNA | NM_021272 | 14.34666 | 4.16E-06 |
| D14Ertd171e | DNA segment, Chr 14, ERATO Doi 171, expressed, mRNA (cDNA clone MGC:59433 IMAGE:6331997) | AK039674 | 14.3364 | 2.78E-07 |
| Oas1d | 2′-5′olygoadenylate synthetase 1D (Oas1d) | NM_133893 | 14.18881 | 1.11E-14 |
| Nid1 | Nidogen 1 (Nid1), mRNA | NM_010917 | 14.08903 | 5.04E-23 |
| Mcf2l | Mcf.2 transforming sequence-like (Mcf2l), mRNA | NM_178076 | 14.07378 | 6.06E-26 |
| Hbb-b1 | Hemoglobin, beta adult major chain, mRNA (cDNA clone MGC:40691 IMAGE:3988455) | NM_008220 | 14.02514 | 4.51E-07 |
| Prg4 | PREDICTED: proteoglycan 4 [Mus musculus], mRNA sequence | AB034730 | 14.01659 | 1.28E-07 |
| 4930524B15Rik | RIKEN cDNA 4930524B15 gene (4930524B15Rik), mRNA | NM_026262 | 13.99617 | 8.54E-07 |
| 4833416E15Rik | 0 day neonate head cDNA, RIKEN full-length enriched library, clone:4833416E15 product: unclassifiable, full insert sequence | AK014707 | 13.9541 | 1.99E-43 |
| Zfp422-rs1 | zinc finger protein 422, related sequence 1 | AK083028 | 13.94195 | 3.70E-34 |
| AI607043 | Adult male small intestine cDNA, RIKEN full-length enriched library, clone:2010310C07 product: unclassifiable, full insert sequence | AI607043 | 13.90419 | 1.08E-12 |
| Dnajc5b | DnaJ (Hsp40) homolog, subfamily C, member 5 beta (Dnajc5b), mRNA | NM_025489 | 13.89499 | 0.00001 |
| Slc2a6 | Solute carrier family 2 (facilitated glucose transporter), member 6 (Slc2a6), mRNA | NM_172659 | 13.84051 | 3.99E-43 |
| Drctnnb1a | down-regulated by Ctnnb1, a | NM_053090 | 13.83104 | 1.14E-34 |
| Sdc4 | Syndecan 4, mRNA (cDNA clone MGC:11456 IMAGE:3154160) | NM_011521 | 13.79767 | 7.42E-23 |
| Ell3 | Elongation factor RNA polymerase II-like 3 (Ell3), mRNA | NM_145973 | 13.77959 | 5.40E-23 |
| Stard10 | START domain containing 10 (Stard10), mRNA | NM_019990 | 13.74577 | 6.06E-19 |
| Mapk8ip1 | JNK interacting protein-1b (JIP-1b) | AK053819 | 13.62189 | 1.54E-12 |
| 1110051M20Rik | RIKEN cDNA 1110051M20 gene, transcript variant 2, mRNA (cDNA clone MGC:103036 IMAGE:6440075) | AK053282 | 13.62105 | 1.43E-18 |
| Ppfia4 | MKIAA0897 protein | XM_129443 | 13.46761 | 1.10E-27 |
| Itga6 | Integrin alpha 6 (Itga6), mRNA | BC024571 | 13.44888 | 1.15E-11 |
| Nfatc2 | Mus musculus 16 days embryo head cDNA, RIKEN full-length enriched library, clone:C130082H17 product: nuclear factor of activated T-cells, cytoplasmic 2, full insert | AK081853 | 13.39808 | 9.81E-45 |
| 4933401N24Rik | RIKEN cDNA 4933401N24 gene (4933401N24Rik), mRNA | NM_175367 | 13.39483 | 1.76E-41 |
| Mpp7 | Membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7), mRNA (cDNA clone IMAGE:6532780) | AK036956 | 13.36708 | 1.64E-28 |
| Acpp | Acid phosphatase, prostate (Acpp), transcript variant 2, mRNA | NM_207668 | 13.35389 | 1.35E-17 |
| AK038010 | 16 days neonate thymus cDNA, RIKEN full-length enriched library, clone:A130071G05 product: unclassifiable, full insert sequence | AK038010 | 13.30008 | 5.17E-07 |
| XM_156408 | Mus musculus similar to RIKEN cDNA 2610524H06 gene (LOC239727), mRNA [XM_156408] | XM_156408 | 13.28002 | 4.58E-14 |
| Itga6 | Integrin alpha 6 (Itga6), mRNA | NM_008397 | 13.26801 | 5.17E-12 |
| 1300013J15Rik | RIKEN cDNA 1300013J15 gene, mRNA (cDNA clone MGC:28678 IMAGE:4238749) | NM_026183 | 13.1909 | 1.26E-20 |
| Hsd17b13 | Hydroxysteroid (17-beta) dehydrogenase 13, mRNA (cDNA clone MGC:30360 IMAGE:5132342) | AK050378 | 13.159 | 8.50E-07 |
| Ankrd13d | Ankyrin repeat domain 13 family, member D (Ankrd13d), mRNA | NM_026720 | 13.07501 | 1.65E-18 |
| Sh3tc1 | SH3 domain and tetratricopeptide repeats 1, mRNA (cDNA clone IMAGE:3490511) | NM_194344 | 13.05323 | 1.40E-45 |
| Gas6 | Growth arrest specific 6, mRNA (cDNA clone MGC:6124 IMAGE:3592398) | NM_019521 | 13.04704 | 3.30E-34 |
| Fank1 | Fibronectin type 3 and ankyrin repeat domains 1 (Fank1), mRNA | NM_025850 | 13.03388 | 2.66E-34 |
| 6430590A10Rik | Mus musculus RIKEN cDNA 6430590A10 gene (6430590A10Rik), mRNA [NM_177017] | NM_177017 | 12.97531 | 0.00001 |
| Nck2 | Non-catalytic region of tyrosine kinase adaptor protein 2, mRNA (cDNA clone MGC:7093 IMAGE:3157491) | AK042129 | 12.94784 | 4.72E-07 |
| 1700123K08Rik | RIKEN cDNA 1700123K08 gene (1700123K08Rik), mRNA | NM_029693 | 12.92483 | 2.79E-11 |
| Jag1 | Jagged1 (Jag1) | NM_013822 | 12.89675 | 1.67E-18 |
| Cacna1g | Calcium channel, voltage-dependent, T type, alpha 1G subunit (Cacna1g), mRNA | NM_009783 | 12.88716 | 9.48E-20 |
| 2410003J06Rik | RIKEN cDNA 2410003J06 gene, mRNA (cDNA clone MGC:60956 IMAGE:30007513) | AK010362 | 12.79956 | 2.82E-16 |
| Pgbd5 | PiggyBac transposable element derived 5, mRNA (cDNA clone MGC:106643 IMAGE:30606749) | AK013562 | 12.79654 | 1.93E-30 |
| Nfe2l3 | Nuclear factor, erythroid derived 2, like 3, mRNA (cDNA clone MGC:5702 IMAGE:3485182) | NM_010903 | 12.78367 | 8.28E-39 |
| D330012F22Rik | Mus musculus RIKEN cDNA D330012F22 gene (D330012F22Rik), mRNA. | NM_178752 | 12.76423 | 3.55E-12 |
| Odz3 | ODZ3 (Odz3) | NM_011857 | 12.75018 | 6.34E-06 |
| Plat | Plasminogen activator, tissue, mRNA (cDNA clone MGC:18508 IMAGE:4038404) | NM_008872 | 12.73118 | 5.16E-23 |
| Hist1h1e | Histone 1, H1e (Hist1h1e), mRNA | AK047852 | 12.71911 | 1.20E-35 |
| Atp6v0e2 | ATPase, H+ transporting, lysosomal, V0 subunit E isoform 2 (Atp6v0e2), mRNA | NM_133764 | 12.6221 | 7.20E-18 |
| Capn5 | Calpain 5, mRNA (cDNA clone MGC:25253 IMAGE:4458488) | NM_007602 | 12.51361 | 5.38E-23 |
| Fzd4 | Frizzled homolog 4 (Drosophila), mRNA (cDNA clone MGC:18403 IMAGE:4238940) | NM_008055 | 12.51026 | 5.20E-28 |
| Nppb | Natriuretic peptide precursor type B (Nppb), mRNA | NM_008726 | 12.3018 | 2.94E-13 |
| 4631402N15Rik | RIKEN cDNA 4631402N15 gene | AK028622 | 12.293 | 1.87E-06 |
| Hey1 | Hairy-related transcription factor 1 (Hrt1) | NM_010423 | 12.27919 | 9.21E-35 |
| Cldn19 | Claudin 19 (Cldn19), mRNA | NM_153105 | 12.26932 | 2.86E-06 |
| Olfr488 | Olfactory receptor 488 (Olfr488), mRNA | NM_146732 | 12.23274 | 1.87E-06 |
| 4930402F06Rik | PREDICTED: hypothetical protein LOC74854 [Mus musculus], mRNA sequence | XM_130219 | 12.21738 | 2.16E-06 |
| Aldh6a1 | Aldehyde dehydrogenase family 6, subfamily A1, mRNA (cDNA clone MGC:28587 IMAGE:4212084) | NM_134042 | 12.14788 | 1.68E-44 |
| Olfr512 | Olfactory receptor 512 (Olfr512), mRNA | NM_146724 | 12.01476 | 1.44E-06 |
| Baiap2l1 | BAI1-associated protein 2-like 1 (Baiap2l1), mRNA | NM_025833 | 11.99713 | 2.71E-21 |
| Gcnt1 | Glucosaminyl (N-acetyl) transferase 1, core 2, mRNA (cDNA clone MGC:11452 IMAGE:3154671) | NM_010265 | 11.98347 | 1.35E-11 |
| C330006P03Rik | Homer homolog 1 (Drosophila) (Homer1), transcript variant L, mRNA | AK049142 | 11.98031 | 1.54E-10 |
| Wfdc12 | WAP four-disulfide core domain 12, mRNA (cDNA clone MGC:32134 IMAGE:4923469) | NM_138684 | 11.9789 | 4.85E-23 |
| Als2cr15 | Amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 15, mRNA (cDNA clone MGC:90771 IMAGE:30355976) | NM_027407 | 11.97403 | 2.12E-20 |
| Phtf2 | Putative homeodomain transcription factor 2, mRNA (cDNA clone MGC:60851 IMAGE:30050876) | AK035536 | 11.97298 | 4.35E-26 |
| Itga4 | Mus musculus 16 days neonate thymus cDNA, RIKEN full-length enriched library, clone:A130050E18 product: integrin alpha 4, full insert sequence. | AK037794 | 11.95943 | 1.52E-06 |
| Col5a3 | Asp6 mRNA for adipocyte-specific protein 6 | NM_016919 | 11.95215 | 2.56E-07 |
| Egr1 | Early growth response 1 (Egr1), mRNA | NM_007913 | 11.93462 | 4.12E-43 |
| Muc2 | Mucin 2, mRNA (cDNA clone MGC:37529 IMAGE:4986546) | AK008250 | 11.89613 | 5.08E-08 |
| Drctnnb1a | down-regulated by Ctnnb1, a | NM_053090 | 11.8877 | 1.99E-36 |
| Ctss | Cathepsin S, mRNA (cDNA clone MGC:6643 IMAGE:3495719) | NM_021281 | 11.87519 | 2.68E-24 |
| D7Ertd413e | Mus musculus DNA segment, Chr 7, ERATO Doi 413, expressed (D7Ertd413e), mRNA [NM_177468] | NM_177468 | 11.87163 | 2.22E-07 |
| Fcmd | Fukuyama type congenital muscular dystrophy homolog (human) (Fcmd), mRNA | NM_139309 | 11.84098 | 2.11E-06 |
| Rab26 | RAB26, member RAS oncogene family, mRNA (cDNA clone MGC:107516 IMAGE:6436242) | AK043912 | 11.75247 | 1.64E-12 |
| Gm520 | Mus musculus similar to hypothetical protein FLJ35880 (LOC245027), mRNA [XM_147082] | XM_147082 | 11.75046 | 4.73E-11 |
| Rasgef1a | PREDICTED: RasGEF domain family, member 1A [Mus musculus], mRNA sequence | AK018120 | 11.73011 | 8.35E-29 |
| Chrna10 | cholinergic receptor, nicotinic, alpha polypeptide 10 | AK033068 | 11.71933 | 6.66E-15 |
| Ptpn13 | Protein tyrosine phosphatase, non-receptor type 13 (Ptpn13), mRNA | AK033818 | 11.71558 | 4.43E-33 |
| BC039632 | CDNA sequence BC039632, mRNA (cDNA clone IMAGE:5124409) | BC039632 | 11.63456 | 4.31E-15 |
| 4632419I22Rik | Mus musculus 0 day neonate skin cDNA, RIKEN full-length enriched library, clone:4632419I22 product: unclassifiable, full insert sequence. | AK014588 | 11.6079 | 3.26E-06 |
| Rab3il1 | RAB3A interacting protein (rabin3)-like 1 (Rab3il1), mRNA | NM_144538 | 11.55758 | 7.46E-24 |
| Rapgef2 | Rap guanine nucleotide exchange factor (GEF) 2, mRNA (cDNA clone IMAGE:30545886) | BM247313 | 11.55429 | 3.38E-06 |
| Cpeb4 | Cytoplasmic polyadenylation element binding protein 4 (Cpeb4), mRNA | AK079421 | 11.54925 | 5.71E-19 |
| Ctgf | Connective tissue growth factor, mRNA (cDNA clone MGC:8122 IMAGE:3589136) | NM_010217 | 11.54686 | 1.93E-08 |
| Wnt6 | Wingless-related MMTV integration site 6 (Wnt6), mRNA | NM_009526 | 11.37961 | 1.23E-17 |
| Atp7a | ATPase, Cu++ transporting, alpha polypeptide (Atp7a), mRNA | AK033254 | 11.34701 | 1.47E-28 |
| Efemp1 | Epidermal growth factor-containing fibulin-like extracellular matrix protein 1 (Efemp1), mRNA | NM_146015 | 11.34476 | 3.71E-35 |
| Cyp2s1 | Cytochrome P450, family 2, subfamily s, polypeptide 1, mRNA (cDNA clone IMAGE:4986756) | NM_028775 | 11.3342 | 8.41E-25 |
| Plat | Plasminogen activator, tissue, mRNA (cDNA clone MGC:18508 IMAGE:4038404) | NM_008872 | 11.28647 | 2.45E-19 |
| Hmg20a | High mobility group 20A, mRNA (cDNA clone MGC:12002 IMAGE:3602239) | AK032790 | 11.27672 | 1.40E-45 |
| NAP002592-003 | Unknown | 11.26244 | 2.03E-08 | |
| Ppl | Periplakin (Ppl), mRNA | NM_008909 | 11.23955 | 1.47E-09 |
| TC976091 | AY129963 GPI-gamma 4; GPIgamma4 {Mus musculus}, partial (72%) [TC976091] | 11.21971 | 2.49E-24 | |
| BC067047 | PREDICTED: PREX1 protein [Mus musculus], mRNA sequence | AK090301 | 11.18218 | 2.25E-21 |
| Tekt1 | Tektin 1 (Tekt1), mRNA | NM_011569 | 11.17168 | 2.41E-26 |
| Atrnl1 | Attractin like 1, mRNA (cDNA clone IMAGE:4221668) | AK050882 | 11.16275 | 1.82E-14 |
| 4930470F04Rik | RAB11 family interacting protein 2 (class I) (Rab11fip2), mRNA | AK015532 | 11.14061 | 0.00748 |
| Svep1 | Sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1, mRNA (cDNA clone IMAGE:3582135) | AK035333 | 11.11646 | 3.62E-06 |
| AK052909 | 16 days neonate heart cDNA, RIKEN full-length enriched library, clone:D830036I24 product: weakly similar to CYTOCHROME C OXIDASE POLYPEPTIDE VA, MITOCHONDRIAL PRECURSOR (EC 1.9.3.1) [Rattus norvegicus], full insert sequence | AK052909 | 11.00683 | 1.38E-15 |
| Lrp4 | MKIAA0816 protein | AK032360 | 11.00157 | 7.64E-06 |
| Khk | Ketohexokinase, mRNA (cDNA clone MGC:18722 IMAGE:4235560) | NM_008439 | 10.99577 | 1.01E-23 |
| Mpp7 | Membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7), mRNA (cDNA clone IMAGE:6532780) | BC049662 | 10.97808 | 2.54E-27 |
| Muc3 | Mucin 3, intestinal, mRNA (cDNA clone IMAGE:4989899) | BC058768 | 10.94339 | 2.55E-18 |
| 3-Mar | Membrane-associated ring finger (C3HC4) 3 (March3), mRNA | AK085528 | 10.91169 | 6.53E-19 |
| Baiap2l1 | BAI1-associated protein 2-like 1 (Baiap2l1), mRNA | NM_025833 | 10.90945 | 1.29E-08 |
| Ptger2 | Prostaglandin E receptor 2 (subtype EP2), mRNA (cDNA clone MGC:6041 IMAGE:3487087) | NM_008964 | 10.83013 | 1.65E-07 |
| Tnfaip6 | Tumor necrosis factor alpha induced protein 6, mRNA (cDNA clone MGC:13914 IMAGE:4208738) | AK078295 | 10.7698 | 2.30E-17 |
| AK037893 | Adult male hypothalamus cDNA, RIKEN full-length enriched library, clone:A230098A08 product: hypothetical protein, full insert sequence | AK037893 | 10.7616 | 3.07E-15 |
| Ppic | Peptidylprolyl isomerase C, mRNA (cDNA clone MGC:31015 IMAGE:5026674) | NM_008908 | 10.73102 | 1.49E-26 |
| Kcnj1 | Potassium inwardly-rectifying channel, subfamily J, member 1, mRNA (cDNA clone MGC:18350 IMAGE:4241262) | NM_019659 | 10.65826 | 0.00005 |
| Zfp423 | Early B-cell factor-associated zinc finger protein (Ebfaz) | NM_033327 | 10.59541 | 6.09E-06 |
| Rora | RAR-related orphan receptor alpha, mRNA (cDNA clone MGC:5892 IMAGE:3592667) | AK087905 | 10.59469 | 9.75E-10 |
| AK050947 | 9 days embryo whole body cDNA, RIKEN full-length enriched library, clone:D030042L12 product: unclassifiable, full insert sequence | AK050947 | 10.59118 | 5.88E-06 |
| AK076715 | Mus musculus adult male testis cDNA, RIKEN full-length enriched library, clone:4930420A11 product: unclassifiable, full insert sequence. | AK076715 | 10.58991 | 2.55E-06 |
| Ttn | Titin, mRNA (cDNA clone IMAGE:5149168) | AK009965 | 10.56327 | 0.00001 |
| AK038867 | Adult male hypothalamus cDNA, RIKEN full-length enriched library, clone:A230069K02 product: hypothetical protein, full insert sequence | AK038867 | 10.55157 | 0.00001 |
| Ifrd1 | Interferon-related developmental regulator 1 (Ifrd1), mRNA | AK078888 | 10.54876 | 2.35E-12 |
| Bmp8a | Bone morphogenetic protein 8a (Bmp8a), mRNA | NM_007558 | 10.50666 | 1.72E-12 |
| NP_TR6JSE50FPA | olfactory receptor NP_TR6JSE50FPA | NM_203509 | 10.50266 | 6.41E-23 |
| TC1084975 | AE003651 CG13260-PA {Drosophila melanogaster}, partial (10%) [TC1084975] | 10.49094 | 2.76E-07 | |
| Tnfaip6 | Tumor necrosis factor alpha induced protein 6, mRNA (cDNA clone MGC:13914 IMAGE:4208738) | NM_009398 | 10.44943 | 2.17E-09 |
| LOC432449 | hypothetical gene supported by AK038984 | AK038984 | 10.44738 | 2.83E-07 |
| 2900041A09Rik | RIKEN cDNA 2900041A09 gene (2900041A09Rik), mRNA | NM_182839 | 10.44139 | 8.16E-28 |
| 1600015H20Rik | RIKEN cDNA 1600015H20 gene, mRNA (cDNA clone IMAGE:3592288) | NM_024237 | 10.43001 | 4.20E-45 |
| 4930412F09Rik | Mus musculus adult male testis cDNA, RIKEN full-length enriched library, clone:4930412F09 product: unclassifiable, full insert sequence. | AK015119 | 10.4231 | 6.86E-26 |
| 1700108L22Rik | RIKEN cDNA 1700108L22 gene, mRNA (cDNA clone MGC:38253 IMAGE:5324328) | AK037099 | 10.41494 | 4.96E-24 |
| Tjp3 | Tight junction protein 3, mRNA (cDNA clone MGC:11960 IMAGE:3600848) | NM_013769 | 10.41438 | 5.26E-21 |
| NAP053018-1 | Unknown | 10.38831 | 1.26E-44 | |
| Ibrdc3 | IBR domain containing 3, mRNA (cDNA clone IMAGE:5254400) | AK015966 | 10.37732 | 9.05E-35 |
| Slco4a1 | Solute carrier organic anion transporter family, member 4a1 (Slco4a1), mRNA | NM_148933 | 10.35873 | 7.30E-26 |
| E430024C06Rik | Mus musculus 2 days neonate thymus thymic cells cDNA, RIKEN full-length enriched library, clone:E430003N15 product: unclassifiable, full insert sequence. | AK088091 | 10.34123 | 3.10E-23 |
| 1500026H17Rik | Adult male corpora quadrigemina cDNA, RIKEN full-length enriched library, clone:B230317N08 product: unclassifiable, full insert sequence | AK005299 | 10.33636 | 4.98E-16 |
| 3110057O12Rik | RIKEN cDNA 3110057O12 gene, mRNA (cDNA clone IMAGE:4007705) | NM_026622 | 10.33092 | 1.71E-11 |
| Ptprm | Protein tyrosine phosphatase, receptor type, M (Ptprm), mRNA | AK047905 | 10.30778 | 8.35E-06 |
| Hemt1 | 17 days embryo kidney cDNA, RIKEN full-length enriched library, clone:I920059J21 product:11 beta-hydroxylase homolog [Mus sp], full insert sequence | AK078619 | 10.30203 | 7.99E-06 |
| Stau1 | Staufen (RNA binding protein) homolog 1 (Drosophila), mRNA (cDNA clone MGC:13708 IMAGE:4188394) | AK030890 | 10.2707 | 2.31E-16 |
| Hist1h1d | Histone 1, H1d (Hist1h1d), mRNA | NM_145713 | 10.25591 | 3.12E-13 |
| D16H22S680E | DNA segment, Chr 16, human D22S680E, expressed (D16H22S680E), mRNA | AK047383 | 10.23081 | 1.29E-32 |
| Rgl1 | Ral guanine nucleotide dissociation stimulator,-like 1, mRNA (cDNA clone MGC:18430 IMAGE:4241244) | AK032340 | 10.22274 | 1.40E-45 |
| 8430422M09Rik | Mus musculus RIKEN cDNA 8430422M09 gene (8430422M09Rik), mRNA [XM_148115] | XM_148115 | 10.21893 | 0.00001 |
| Kcnab3 | Potassium voltage-gated channel, shaker-related subfamily, beta member 3, mRNA (cDNA clone MGC:27635 IMAGE:4507029) | NM_010599 | 10.2158 | 1.74E-06 |
| Slc25a21 | Solute carrier family 25 (mitochondrial oxodicarboxylate carrier), member 21, mRNA (cDNA clone MGC:67702 IMAGE:5042653) | NM_172577 | 10.21333 | 5.59E-14 |
| Col20a1 | Collagen, type XX, alpha 1, mRNA (cDNA clone IMAGE:5366843) | AK006759 | 10.16397 | 3.65E-12 |
| Hrbl | HIV-1 Rev binding protein-like (Hrbl), transcript variant 1, mRNA | AK037434 | 10.14872 | 3.47E-17 |
| NM_029423 | Mus musculus RIKEN cDNA 4833414G05 gene (4833414G05Rik), mRNA [NM_029423] | NM_029423 | 10.1245 | 3.34E-13 |
| Kcnma1 | Large conductance calcium activated potassium BK channel STREX-1 variant (Slo) | AK052354 | 10.12088 | 7.76E-06 |
| Hivep3 | Clone 5′RACE1 ZAS3 (Krc) mRNA, 5′ UTR | AK087330 | 10.11146 | 8.57E-37 |
| Isl2 | Insulin related protein 2 (islet 2) (Isl2), mRNA | NM_027397 | 10.06446 | 6.70E-06 |
| A930034L06Rik | RIKEN cDNA A930034L06 gene (A930034L06Rik), mRNA | NM_175692 | 10.0482 | 3.73E-14 |
| Rhbg | Rhesus blood group-associated B glycoprotein, mRNA (cDNA clone MGC:35874 IMAGE:4218889) | NM_021375 | 10.00581 | 2.67E-18 |
| Grtp1 | GH regulated TBC protein 1, mRNA (cDNA clone MGC:27905 IMAGE:3500563) | NM_025768 | 9.93642 | 5.59E-28 |
| Pfkfb2 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2, mRNA (cDNA clone MGC:25723 IMAGE:3979400) | AK016729 | 9.92957 | 3.07E-35 |
| BC049806 | CDNA sequence BC049806, mRNA (cDNA clone MGC:76460 IMAGE:30431671) | NM_172513 | 9.90803 | 1.62E-19 |
| Cdc14a | CDC14 cell division cycle 14 homolog A (S. cerevisiae), mRNA (cDNA clone MGC:100044 IMAGE:30356962) | AK048250 | 9.87444 | 0.0038 |
| Hist1h4i | Mus musculus histone 2, H4, mRNA (cDNA clone IMAGE:4205460), partial cds. | BC019757 | 9.86807 | 1.47E-19 |
| Mcam | Melanoma cell adhesion molecule, mRNA (cDNA clone MGC:35823 IMAGE:5372497) | NM_023061 | 9.84222 | 1.58E-07 |
| Mfi2 | Antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 (Mfi2), mRNA | NM_013900 | 9.83413 | 4.36E-21 |
| Dock4 | MKIAA0716 protein | BC051125 | 9.81909 | 2.29E-11 |
| Xlr3b | X-linked lymphocyte-regulated 3B (Xlr3b), mRNA | NM_011727 | 9.78323 | 6.44E-14 |
| NAP069339-1 | Unknown | 9.77056 | 2.40E-32 | |
| 2610018I05Rik | Mus musculus RIKEN cDNA 2610018I05 gene (2610018I05Rik), mRNA [NM_183279] | NM_183279 | 9.77006 | 1.01E-17 |
| Nrarp | Notch-regulated ankyrin repeat protein (Nrarp), mRNA | NM_025980 | 9.74784 | 4.38E-07 |
| Eml5 | Mus musculus echinoderm microtubule associated protein like 5, mRNA (cDNA clone MGC:40838 IMAGE:5368750), complete cds. | BC030361 | 9.71342 | 1.76E-27 |
| Slc26a4 | Pendrin (Pds) | NM_011867 | 9.7024 | 2.38E-37 |
| Sdfr1 | Stromal cell derived factor receptor 1 (Sdfr1), mRNA | AK039616 | 9.69602 | 1.98E-06 |
| A430105I05Rik | B6-derived CD11 +ve dendritic cells cDNA, RIKEN full-length enriched library, clone:F730031B04 product: Serine-threonine kinase (Fragment) homolog [Rattus norvegicus], full insert sequence | AK040531 | 9.69534 | 3.56E-18 |
| Dgat2 | Diacylglycerol O-acyltransferase 2 (Dgat2), mRNA | NM_026384 | 9.69276 | 2.73E-35 |
| W29301 | 2 days pregnant adult female oviduct cDNA, RIKEN full-length enriched library, clone:E230029K24 product: hypothetical protein, full insert sequence | W29301 | 9.68389 | 1.54E-44 |
| Gng8 | Guanine nucleotide binding protein (G protein), gamma 8 subunit (Gng8), mRNA | NM_010320 | 9.68159 | 4.39E-19 |
| Ppfibp2 | Protein tyrosine phosphatase, receptor-type, F interacting protein, binding protein 2, mRNA (cDNA clone IMAGE:3488375) | NM_008905 | 9.67203 | 0.00004 |
| Aplp2 | Amyloid beta (A4) precursor-like protein 2, mRNA (cDNA clone IMAGE:4504805) | NM_009691 | 9.54457 | 8.29E-26 |
| 4921524J17Rik | RIKEN cDNA 4921524J17 gene (4921524J17Rik), mRNA | AK006764 | 9.52196 | 0.00001 |
| Nppb | Natriuretic peptide precursor type B (Nppb), mRNA | NM_008726 | 9.5214 | 2.01E-10 |
| AK082626 | 0 day neonate cerebellum cDNA, RIKEN full-length enriched library, clone:C230071E14 product: unclassifiable, full insert sequence | AK082626 | 9.49805 | 0.00002 |
| Ccpg1 | Cell cycle progression 1, mRNA (cDNA clone MGC:12151 IMAGE:3711012) | NM_028181 | 9.49574 | 1.25E-22 |
| 4933401N24Rik | RIKEN cDNA 4933401N24 gene (4933401N24Rik), mRNA | NM_175367 | 9.48097 | 1.88E-15 |
| Etv5 | Ets variant gene 5, mRNA (cDNA clone MGC:28414 IMAGE:4036564) | NM_023794 | 9.46181 | 1.65E-37 |
| Upb1 | Ureidopropionase, beta, mRNA (cDNA clone IMAGE:4974180) | NM_133995 | 9.45902 | 7.68E-06 |
| AK041503 | PREDICTED: Mus musculus hypothetical protein LOC (LOC545502), mRNA | AK041503 | 9.45271 | 0.00022 |
| Arc | Activity regulated cytoskeletal-associated protein, mRNA (cDNA clone MGC:27890 IMAGE:3498057) | NM_018790 | 9.4495 | 6.20E-31 |
| 4931406B18Rik | RIKEN cDNA 4931406B18 gene (4931406B18Rik), mRNA | NM_028737 | 9.44613 | 0.00004 |
| Atg12 | Apg12 | NM_026217 | 9.43954 | 2.67E-16 |
| Zswim5 | Zinc finger, SWIM domain containing 5 (Zswim5), mRNA | AK046815 | 9.43427 | 0.00003 |
| Pacsin3 | Protein kinase C and casein kinase II substrate 3 (Pacsin3) | AK014830 | 9.42759 | 4.14E-13 |
| AW125688 | Expressed sequence AW125688 (AW125688), mRNA | NM_001001885 | 9.37585 | 5.66E-26 |
| Odz2 | ODZ2 (Odz2) | AK083775 | 9.36205 | 0.00002 |
| Smpd3 | Sphingomyelin phosphodiesterase 3, neutral (Smpd3), mRNA | NM_021491 | 9.34228 | 2.30E-12 |
| Ceacam2 | CEA-related cell adhesion molecule 2, mRNA (cDNA clone MGC:18435 IMAGE:4241449) | NM_007543 | 9.33621 | 5.14E-12 |
| Mmp10 | Matrix metallopeptidase 10 (Mmp10), mRNA | NM_019471 | 9.28633 | 1.13E-07 |
| BC013712 | CDNA sequence BC013712, mRNA (cDNA clone IMAGE:3155889) | BC013712 | 9.27602 | 1.41E-15 |
| Cfh | Complement component factor h (Cfh), mRNA | NM_009888 | 9.21081 | 1.00E-18 |
| H2-Oa | Histocompatibility 2, O region alpha locus (H2-Oa), mRNA | NM_008206 | 9.20769 | 2.53E-16 |
| Cacng5 | Calcium channel, voltage-dependent, gamma subunit 5 (Cacng5), mRNA | NM_080644 | 9.20551 | 4.94E-09 |
| Slc28a2 | Purine-selective Na+ nucleoside cotransporter (Cnt2) | NM_172980 | 9.20316 | 2.97E-09 |
| AK089607 | Mus musculus activated spleen cDNA, RIKEN full-length enriched library, clone:F830004H11 product: unclassifiable, full insert sequence. | AK089607 | 9.1922 | 0.00004 |
| Gcnt1 | Glucosaminyl (N-acetyl) transferase 1, core 2, mRNA (cDNA clone MGC:11452 IMAGE:3154671) | NM_010265 | 9.17498 | 8.19E-10 |
| Serpinb1a | EIA (Serpinb1) | NM_025429 | 9.17307 | 6.72E-18 |
| Ccbl2 | Cysteine conjugate-beta lyase 2 (Ccbl2), mRNA | NM_173763 | 9.15601 | 3.33E-11 |
| Nsccn1 | Mus musculus non-selective cation channel 1 (Nsccn1), mRNA [NM_010940] | NM_010940 | 9.15204 | 0.0001 |
| Hrc | Histidine rich calcium binding protein (Hrc), mRNA | NM_010473 | 9.13879 | 6.63E-24 |
| H2-DMb2 | Histocompatibility 2, class II, locus Mb1, mRNA (cDNA clone MGC:5741 IMAGE:3486844) | NM_010388 | 9.10407 | 1.15E-27 |
| Rgl1 | Ral guanine nucleotide dissociation stimulator,-like 1, mRNA (cDNA clone MGC:18430 IMAGE:4241244) | NM_016846 | 9.0778 | 1.59E-41 |
| Dapk2 | Death-associated kinase 2, mRNA (cDNA clone MGC:13742 IMAGE:4016911) | NM_010019 | 9.06365 | 4.58E-16 |
| Lmtk2 | Lemur tyrosine kinase 2, mRNA (cDNA clone IMAGE:6407188) | AK039738 | 9.05942 | 1.27E-38 |
| Cfh | Complement component factor h, mRNA (cDNA clone IMAGE:5132971) | M29010 | 8.96985 | 2.80E-06 |
| Spry1 | Sprouty homolog 1 (Drosophila), mRNA (cDNA clone MGC:18307 IMAGE:3672353) | NM_011896 | 8.96343 | 8.83E-21 |
| Gm693 | PREDICTED: similar to novel protein [Mus musculus], mRNA sequence | XM_204104 | 8.95937 | 1.91E-21 |
| Ryr1 | RyR1 mRNA for skeletal muscle ryanodine receptor | AK042482 | 8.94761 | 2.14E-13 |
| 8430437N05Rik | DACH protein (Dach) | AK018473 | 8.93946 | 0.00004 |
| Arl5c | ADP-ribosylation factor-like 12 (Arl12), mRNA | NM_207231 | 8.93395 | 2.10E-19 |
| Rasip1 | Ras interacting protein 1, mRNA (cDNA clone IMAGE:1245387) | AK033276 | 8.93241 | 7.99E-10 |
| Adam25 | Testase 2 (Adam-25) | NM_011781 | 8.91583 | 0.00004 |
| 4930579G22Rik | RIKEN cDNA 4930579G22 gene (4930579G22Rik), mRNA | NM_026916 | 8.91564 | 1.40E-14 |
| NAP062857-1 | Unknown | 8.89087 | 0.0001 | |
| Pbx3 | Pre B-cell leukemia transcription factor 3 (Pbx3), mRNA | AK052017 | 8.85804 | 0.00004 |
| Kcnip1 | A-type potassium channel modulatory protein 1.2 (Kchip1.2) | NM_027398 | 8.84607 | 0.00004 |
| Pard6b | Par-6 (partitioning defective 6) homolog beta (C. elegans), mRNA (cDNA clone MGC:35871 IMAGE:2655658) | NM_021409 | 8.83071 | 3.85E-15 |
| Flrt3 | Strain C57BL/6xCBA fibronectin leucine rich transmembrane protein 3 (Flrt3) | NM_178382 | 8.81882 | 2.05E-14 |
| AK032440 | Adult male olfactory brain cDNA, RIKEN full-length enriched library, clone:6430547C23 product: unclassifiable, full insert sequence | AK032440 | 8.80642 | 0.00004 |
| Cpn1 | Carboxypeptidase N, polypeptide 1, mRNA (cDNA clone MGC:25883 IMAGE:4211169) | NM_030703 | 8.79262 | 7.85E-18 |
| 2810405K02Rik | RIKEN cDNA 2810405K02 gene, mRNA (cDNA clone MGC:40622 IMAGE:5401948) | NM_025582 | 8.78415 | 0.00006 |
| 9230110K08Rik | Adult male epididymis cDNA, RIKEN full-length enriched library, clone:9230110K08 product: hypothetical HMG-I and HMG-Y DNA-binding domain (A+T-hook) containing protein, full insert sequence | AK020331 | 8.77293 | 4.72E-29 |
| Igsf1 | Immunoglobulin superfamily, member 1, transcript variant 2, mRNA (cDNA clone IMAGE:30249729) | NM_183336 | 8.75732 | 0.00008 |
| Ndrg4 | N-myc downstream regulated gene 4 (Ndrg4), mRNA | NM_145602 | 8.75653 | 6.57E-36 |
| Mst1r | Macrophage stimulating 1 receptor (c-met-related tyrosine kinase) (Mst1r), mRNA | NM_009074 | 8.71853 | 6.63E-18 |
| Cdon | Cell adhesion molecule-related/down-regulated by oncogenes (Cdon), mRNA | NM_021339 | 8.71613 | 1.34E-16 |
| Serpine1 | Serine (or cysteine) peptidase inhibitor, clade E, member 1 (Serpine1), mRNA | NM_008871 | 8.66415 | 7.69E-24 |
| Atg12 | Apg12 | NM_026217 | 8.66365 | 5.05E-14 |
| AK051813 | Mus musculus 12 days embryo eyeball cDNA, RIKEN full-length enriched library, clone:D230004F15 product: unclassifiable, full insert sequence. | AK051813 | 8.6557 | 6.88E-09 |
| F830010H11Rik | Mus musculus activated spleen cDNA, RIKEN full-length enriched library, clone:F830010H11 product: unclassifiable, full insert sequence. | AK089716 | 8.65219 | 0.00002 |
| Clu | Clusterin (Clu), mRNA | NM_013492 | 8.64934 | 3.24E-12 |
| Akap12 | A kinase (PRKA) anchor protein (gravin) 12 (Akap12), mRNA | NM_031185 | 8.6406 | 1.71E-27 |
| Pacsin1 | Protein kinase C and casein kinase substrate in neurons 1, mRNA (cDNA clone MGC:25285 IMAGE:4527708) | NM_178365 | 8.62899 | 5.37E-08 |
| 4930449I04Rik | Adult male testis cDNA, RIKEN full-length enriched library, clone:4930449I04 product: hypothetical protein, full insert sequence | AK015432 | 8.6266 | 3.24E-33 |
| Ap3m2 | Adaptor-related protein complex 3, mu 2 subunit, mRNA (cDNA clone MGC:40858 IMAGE:5369288) | NM_029505 | 8.62306 | 1.02E-10 |
| Olfr1383 | Olfactory receptor 1383 (Olfr1383), mRNA | NM_207574 | 8.60579 | 1.28E-24 |
| Lip1 | Lysosomal acid lipase 1 (Lip1), mRNA | AK083099 | 8.59815 | 2.87E-09 |
| Ero1lb | ERO1-like beta (S. cerevisiae) (Ero1lb), mRNA | NM_026184 | 8.59729 | 5.41E-29 |
| Zfp509 | Mus musculus 2 days pregnant adult female ovary cDNA, RIKEN full-length enriched library, clone:E330031I18 product: weakly similar to ZINC FINGER PROTEIN ZFP-93 (ZINC FINGER PROTEIN HZF6) (FRAGMENT) [Homo sapiens], full insert sequence. | AK054488 | 8.5938 | 1.99E-28 |
| Fgd4 | FYVE, RhoGEF and PH domain containing 4 (Fgd4), transcript variant beta, mRNA | AK043656 | 8.53643 | 5.81E-08 |
| Il1rap | Interleukin 1 receptor accessory protein (Il1rap), transcript variant 2, mRNA | AK045686 | 8.52846 | 5.61E-45 |
| 1700109H08Rik | RIKEN cDNA 1700109H08 gene, mRNA (cDNA clone MGC:118394 IMAGE:30920581) | AK007150 | 8.52551 | 1.98E-13 |
| Ptger4 | Mus musculus 12 days embryo spinal ganglion cDNA, RIKEN full-length enriched library, clone:D130004L12 product: prostaglandin E receptor 4 (subtype EP4), full insert sequence. | AK051137 | 8.52297 | 6.25E-26 |
| 4933408J17Rik | Small nuclear ribonucleoprotein polypeptide F, mRNA (cDNA clone MGC:118343 IMAGE:30917013) | AK016739 | 8.52005 | 0.00012 |
| Jrkl | 0 day neonate thymus cDNA, RIKEN full-length enriched library, clone:A430098H07 product: hypothetical protein, full insert sequence | AK079877 | 8.51387 | 4.41E-18 |
| Cxcl9 | Chemokine (C-X-C motif) ligand 9, mRNA (cDNA clone MGC:6179 IMAGE:3257716) | NM_008599 | 8.50345 | 1.39E-17 |
| Cfh | Complement component factor h (Cfh), mRNA | NM_009888 | 8.49969 | 1.56E-11 |
| AK042806 | Mus musculus 7 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:A730026C13 product: unclassifiable, full insert sequence. | AK042806 | 8.4994 | 1.23E-08 |
| Gdpd5 | Glycerophosphodiester phosphodiesterase domain containing 5, mRNA (cDNA clone MGC:37148 IMAGE:4953174) | NM_201352 | 8.48545 | 5.38E-09 |
| Tgm4 | Experimental autoimmune prostatitis antigen 1 (Eapa1) | NM_177911 | 8.48033 | 0.00012 |
| Btbd5 | BTB (POZ) domain containing 5, mRNA (cDNA clone MGC:47075 IMAGE:5347183) | NM_025707 | 8.46778 | 1.71E-15 |
| Rtn4r | Reticulon 4 receptor (Rtn4r), mRNA | NM_022982 | 8.45249 | 7.05E-28 |
| Ibrdc3 | IBR domain containing 3, mRNA (cDNA clone IMAGE:5254400) | AK040902 | 8.4511 | 6.53E-22 |
| Aldh1l2 | Aldehyde dehydrogenase 1 family, member L2 (Aldh1l2), mRNA | AK085074 | 8.41092 | 2.26E-14 |
| LOC433886 | hypothetical gene supported by AK049058; BC025881 | BC025881 | 8.39834 | 0.00022 |
| Kiss1 | Golgi transport 1 homolog A (S. cerevisiae) (Golt1a), mRNA | NM_178260 | 8.39812 | 0.00014 |
| Egr3 | Early growth response 3 (Egr3), mRNA | AK078299 | 8.39771 | 1.35E-12 |
| NAP112027-1 | Unknown | 8.38015 | 1.69E-11 | |
| Myh3 | PREDICTED: myosin, heavy polypeptide 3, skeletal muscle, embryonic [Mus musculus], mRNA sequence | M11154 | 8.3727 | 4.98E-25 |
| Vil1 | Villin 1, mRNA (cDNA clone MGC:18506 IMAGE:4236751) | NM_009509 | 8.36524 | 5.30E-09 |
| Aplp2 | Amyloid beta (A4) precursor-like protein 2, mRNA (cDNA clone IMAGE:4504805) | NM_009691 | 8.36361 | 8.30E-43 |
| Gats | Opposite strand transcription unit to Stag3 (Gats), mRNA | BC026208 | 8.35244 | 8.31E-10 |
| Olfr1052 | Olfactory receptor 1052 (Olfr1052), mRNA | NM_147010 | 8.34985 | 0.00013 |
| Akap13 | PREDICTED: hypothetical protein LOC75547 [Mus musculus], mRNA sequence | AK006382 | 8.31218 | 1.41E-14 |
| Nanos1 | Nanos homolog 1 (Drosophila), mRNA (cDNA clone MGC:102288 IMAGE:6849859) | BC056473 | 8.30066 | 1.94E-13 |
| Eif2c3 | Eukaryotic translation initiation factor 2C, 3 (Eif2c3), mRNA | AK087577 | 8.26887 | 1.33E-22 |
| AK086157 | Mus musculus 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930008M17 product: unclassifiable, full insert sequence. | AK086157 | 8.26126 | 0.00012 |
| Art3 | ART3 mono(ADP-ribosyl)transferase (art3 gene), splice variant 1 | NM_181728 | 8.2485 | 9.93E-16 |
| TC1085358 | AF179867 STE20-like kinase {Homo sapiens}, partial (56%) [TC1085358] | 8.24529 | 1.54E-22 | |
| Eif2c3 | Eukaryotic translation initiation factor 2C, 3 (Eif2c3), mRNA | BC060127 | 8.21917 | 1.00E-29 |
| Cd52 | CD52 antigen, mRNA (cDNA clone MGC:40993 IMAGE:1396480) | NM_013706 | 8.21899 | 0.00035 |
| Slc25a29 | Solute carrier family 25 (mitochondrial carrier, palmitoylcarnitine transporter), member 29, mRNA (cDNA clone MGC:7958 IMAGE:3584570) | NM_181328 | 8.20668 | 2.73E-17 |
| Kif13b | MKIAA0639 protein | AK031370 | 8.20576 | 2.43E-10 |
| Itpka | Inositol 1,4,5-trisphosphate 3-kinase A (Itpka), mRNA | NM_146125 | 8.20068 | 2.20E-07 |
| Efhd1 | EF hand domain containing 1, mRNA (cDNA clone MGC:28727 IMAGE:4459208) | NM_028889 | 8.18839 | 4.79E-24 |
| Ccpg1 | Cell cycle progression 1, mRNA (cDNA clone MGC:12151 IMAGE:3711012) | BC006717 | 8.15286 | 2.83E-33 |
| 1810020D17Rik | RIKEN cDNA 1810020D17 gene, mRNA (cDNA clone MGC:36240 IMAGE:5027461) | AK031619 | 8.13287 | 0.00011 |
| Ripk3 | Receptor-interacting serine-threonine kinase 3, mRNA (cDNA clone MGC:35896 IMAGE:3590770) | NM_019955 | 8.12366 | 1.65E-15 |
| Zdhhc15 | Zinc finger, DHHC domain containing 15 (Zdhhc15), mRNA | BU610239 | 8.10685 | 0.00016 |
| Scgb1c1 | PREDICTED: similar to RYD5 [Mus musculus], mRNA sequence | XM_355976 | 8.10518 | 1.20E-23 |
| XM_356367 | Mus musculus similar to cyclin-dependent kinase-like 5; serine/threonine kinase 9 (LOC382253), mRNA [XM_356367] | XM_356367 | 8.10486 | 1.44E-12 |
| Rab3il1 | Mus musculus RAB3A interacting protein (rabin3)-like 1 (Rab3il1), mRNA. | NM_178881 | 8.09444 | 1.48E-10 |
| Egfl5 | MKIAA0818 protein | NM_172694 | 8.09398 | 1.04E-08 |
| Csnd | Mus musculus 10 days lactation, adult female mammary gland cDNA, RIKEN full-length enriched library, clone:D730003B11 product: casein delta, full insert sequence. | AK052789 | 8.09332 | 0.00016 |
| Fkbp6 | FK506 binding protein 6 (Fkbp6), mRNA | NM_033571 | 8.09194 | 5.51E-33 |
| Dll4 | Dll-4 mRNA for Delta-4 | NM_019454 | 8.08534 | 8.83E-21 |
| Sema4g | Semaphorin subclass 4 member G (sema4g) | NM_011976 | 8.0833 | 5.15E-11 |
| Rccd1 | Protein regulator of cytokinesis 1 (Prc1), mRNA | AK047578 | 8.0818 | 4.76E-07 |
| Cul4a | Cullin 4A (Cul4a), mRNA | AK084008 | 8.07649 | 0.00002 |
| Tec | Cytoplasmic tyrosine kinase, Dscr28C related (Drosophila), mRNA (cDNA clone MGC:46873 IMAGE:5029430) | NM_013689 | 8.06 | 3.75E-18 |
| Entpd2 | Ectonucleoside triphosphate diphosphohydrolase 2, mRNA (cDNA clone MGC:5806 IMAGE:3582550) | NM_009849 | 8.05603 | 1.54E-20 |
| Krtap16-4 | Keratin associated protein 16-4 (Krtap16-4), mRNA | NM_130873 | 8.05589 | 0.00004 |
| Aqp11 | Aquaporin 11, mRNA (cDNA clone IMAGE:5126527) | NM_175105 | 8.05418 | 1.48E-42 |
| Cd36 | CD36 antigen, mRNA (cDNA clone MGC:6068 IMAGE:3481681) | L23108 | 8.05303 | 7.36E-26 |
| 1700001G17Rik | RIKEN cDNA 1700001G17 gene (1700001G17Rik), mRNA | NM_026204 | 8.04819 | 1.22E-14 |
| Pgr | Progesterone receptor (Pgr), mRNA | AK036862 | 8.04787 | 0.00003 |
| Maf | c-maf=c-Maf protein {proto-oncogene} [mice, BALB/c, cerebellum, mRNA, 2736 nt]. | S74567 | 8.03437 | 0.00007 |
| Sdc4 | Syndecan 4, mRNA (cDNA clone MGC:11456 IMAGE:3154160) | NM_011521 | 8.0206 | 1.74E-10 |
| Aytl1 | Hypothetical protein A330042H22 (A330042H22), mRNA | NM_173014 | 8.01654 | 7.00E-11 |
| 2310047O13Rik | RIKEN cDNA 2310047O13 gene, mRNA (cDNA clone MGC:28605 IMAGE:4217391) | AK049694 | 8.01005 | 1.88E-15 |
| Trp73 | P73 alpha protein (P73 gene) | AK014503 | 8.00474 | 0.00017 |
| Tagln3 | Transgelin 3 (Tagln3), mRNA | NM_019754 | 8.00047 | 0.00012 |
| NAP052665-1 | Unknown | 7.99834 | 3.51E-13 | |
| Cd40 | CD40 antigen (Cd40), transcript variant 5, mRNA | NM_170701 | 7.98914 | 2.13E-26 |
| 4732465E10Rik | MKIAA0716 protein | AK028870 | 7.98626 | 1.12E-30 |
| 1700109F18Rik | Homeo box D4 (Hoxd4), mRNA | AK006878 | 7.96332 | 0.00015 |
| Slc25a29 | Solute carrier family 25 (mitochondrial carrier, palmitoylcarnitine transporter), member 29, mRNA (cDNA clone MGC:7958 IMAGE:3584570) | NM_181328 | 7.95316 | 5.52E-28 |
| Ebi3 | Epstein-Barr virus induced gene 3, mRNA (cDNA clone MGC:11457 IMAGE:3154957) | NM_015766 | 7.94831 | 1.93E-15 |
| D330017J20Rik | Mus musculus RIKEN cDNA D330017J20 gene (D330017J20Rik), mRNA [NM_177204] | NM_177204 | 7.9325 | 1.86E-28 |
| A930037G23Rik | RIKEN cDNA A930037G23 gene, mRNA (cDNA clone MGC:67253 IMAGE:6403474) | AK044382 | 7.92998 | 0.00002 |
| Wdr47 | WD repeat domain 47 (Wdr47), mRNA | NM_181400 | 7.92668 | 7.41E-15 |
| Ppl | Periplakin (Ppl), mRNA | BC057152 | 7.88627 | 4.33E-13 |
| 3110021A11Rik | Rab6 interacting protein 2 (Rab6ip2), transcript variant 1, mRNA | AK052063 | 7.84546 | 0.00001 |
| Dmn | MKIAA0353 protein | NM_201639 | 7.82763 | 3.07E-17 |
| Olfr541 | Olfactory receptor 541 (Olfr541), mRNA | NM_146962 | 7.82426 | 0.0002 |
| Lgr4 | PREDICTED: G protein-coupled receptor 48 [Mus musculus], mRNA sequence | AK044357 | 7.82294 | 1.53E-17 |
| Star | Steroidogenic acute regulatory protein (Star), mRNA | NM_011485 | 7.81952 | 6.22E-16 |
| Plekha7 | Pleckstrin homology domain containing, family A member 7, mRNA (cDNA clone MGC:90927 IMAGE:3602481) | NM_172743 | 7.81937 | 5.45E-12 |
| D330028D13Rik | RIKEN cDNA D330028D13 gene, mRNA (cDNA clone MGC:67688 IMAGE:4487036) | NM_172727 | 7.81506 | 3.42E-10 |
| Rprm | Reprimo, TP53 dependent G2 arrest mediator candidate, mRNA (cDNA clone MGC:41066 IMAGE:1434823) | NM_023396 | 7.7925 | 1.35E-15 |
| Lcn12 | Lipocalin 12 (Lcn12), mRNA | AK020307 | 7.78453 | 0.00011 |
| Spire1 | Spire homolog 1 (Drosophila), mRNA (cDNA clone MGC:106458 IMAGE:30535423) | AK053505 | 7.77423 | 9.18E-16 |
| Glt25d2 | Glycosyltransferase 25 domain containing 2, mRNA (cDNA clone MGC:92944 IMAGE:5706069) | NM_177756 | 7.7719 | 0.00026 |
| Ddit3 | DNA-damage inducible transcript 3, mRNA (cDNA clone MGC:18641 IMAGE:4012433) | NM_007837 | 7.77126 | 3.83E-06 |
| Gab3 | Growth factor receptor bound protein 2-associated protein 3 (Gab3), mRNA | NM_181584 | 7.75833 | 1.55E-13 |
| 1110031B11Rik | RIKEN cDNA 1110031B11 gene (1110031B11Rik), mRNA | NM_026811 | 7.73455 | 0.00024 |
| Tcf7 | Transcription factor 7, T-cell specific (Tcf7), mRNA | NM_009331 | 7.7187 | 0.00008 |
| Lcn5 | Lipocalin 5 (Lcn5), mRNA | NM_007947 | 7.70424 | 0.00016 |
| BB064041 | Activated spleen cDNA, RIKEN full-length enriched library, clone:F830206M21 product: unclassifiable, full insert sequence | BB064041 | 7.6678 | 1.96E-26 |
| Tagln | Transgelin, mRNA (cDNA clone MGC:6045 IMAGE:3600413) | NM_011526 | 7.64204 | 1.12E-11 |
| Tktl1 | Transketolase-like 1 (Tktl1), mRNA | NM_031379 | 7.63328 | 0.00021 |
| Mlph | Melanophilin (Mlph), mRNA | NM_053015 | 7.6104 | 0.00021 |
| Armc9 | RIKEN cDNA 4930438O05 gene (4930438O05Rik), mRNA | NM_030184 | 7.59315 | 8.43E-21 |
| 9330102E08Rik | PREDICTED: hypothetical protein XP_489154 [Mus musculus], mRNA sequence | AK033854 | 7.58577 | 7.54E-14 |
| Cbara1 | Calcium binding atopy-related autoantigen 1 (Cbara1), mRNA | AK029884 | 7.57763 | 9.77E-14 |
| Hisppd2a | 4 days neonate male adipose cDNA, RIKEN full-length enriched library, clone:B430315C20 product: hypothetical Histidine acid phosphatase containing protein, full insert sequence | AK046696 | 7.56076 | 1.42E-12 |
| TC1052172 | Unknown | 7.55807 | 1.37E-10 | |
| Dcun1d4 | Expressed sequence AI836376, mRNA (cDNA clone IMAGE:3995042) | NM_178896 | 7.55542 | 4.88E-10 |
| Ralgps1 | Ral GEF with PH domain and SH3 binding motif 1, mRNA (cDNA clone MGC:99991 IMAGE:30536315) | AK129123 | 7.52544 | 1.74E-06 |
| Flrt3 | Strain C57BL/6xCBA fibronectin leucine rich transmembrane protein 3 (Flrt3) | NM_178382 | 7.51845 | 1.29E-33 |
| Efnb2 | Ephrin B2 (Efnb2), mRNA | NM_010111 | 7.50936 | 8.34E-13 |
| Mum1l1 | Melanoma associated antigen (mutated) 1-like 1, mRNA (cDNA clone IMAGE:30619674) | NM_175541 | 7.50527 | 4.98E-06 |
| Zfp185 | Zinc finger protein 185, mRNA (cDNA clone MGC:49126 IMAGE:4191053) | NM_009549 | 7.50102 | 5.10E-06 |
| Epb4.1l5 | Erythrocyte protein band 4.1-like 5 (Epb4.1l5), mRNA | AK079301 | 7.49371 | 1.30E-11 |
| Tnfaip8l2 | Tumor necrosis factor, alpha-induced protein 8-like 2 (Tnfaip8l2), mRNA | NM_027206 | 7.48745 | 3.98E-30 |
| Rad9b | RAD9 homolog B (S. cerevisiae) (Rad9b), mRNA | NM_144912 | 7.47671 | 3.20E-11 |
| Irf8 | Interferon consensus sequence binding protein 1, mRNA (cDNA clone MGC:6194 IMAGE:3487214) | NM_008320 | 7.47332 | 6.42E-32 |
| 9430085L16Rik | Early B-cell factor 2 (Ebf2), mRNA | AK020505 | 7.46734 | 0.00033 |
| 9630050P21Rik | 16 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:9630050P21 product: unclassifiable, full insert sequence | AK036274 | 7.46671 | 1.10E-07 |
| Gpr155 | G protein-coupled receptor 155, mRNA (cDNA clone IMAGE:4167684) | BC026382 | 7.45823 | 3.04E-31 |
| Habp4 | Hyaluronic acid binding protein 4 (Habp4), mRNA | NM_019986 | 7.45096 | 7.37E-17 |
| 9830169C18Rik | RIKEN cDNA 9830169C18 gene, mRNA (cDNA clone MGC:69959 IMAGE:6509090) | NM_172581 | 7.4493 | 6.68E-15 |
| Lrp1b | Low density lipoprotein-related protein 1B (deleted in tumors) (Lrp1b), mRNA | AK034373 | 7.43347 | 0.00006 |
| Btg2 | B-cell translocation gene 2, anti-proliferative (Btg2), mRNA | NM_007570 | 7.43345 | 2.06E-10 |
| 2410066E13Rik | RIKEN cDNA 2410066E13 gene (2410066E13Rik), mRNA | NM_026629 | 7.43071 | 6.08E-15 |
| Got1l1 | RIKEN cDNA 1700083M11 gene (1700083M11Rik), mRNA | AK006984 | 7.42792 | 8.13E-06 |
| Mell1 | Neprilysin-like metallopeptidase 1 (Nl1) | NM_013783 | 7.41912 | 4.75E-11 |
| Pde6c | Phosphodiesterase 6C, cGMP specific, cone, alpha prime, mRNA (cDNA clone MGC:38745 IMAGE:5358611) | NM_033614 | 7.40919 | 0.00024 |
| 3-Mar | Membrane-associated ring finger (C3HC4) 3 (March3), mRNA | AK036622 | 7.38467 | 0.00002 |
| TC1013578 | AF110520 NG28 {Mus musculus}, partial (30%) [TC1013578] | 7.37995 | 2.09E-06 | |
| 2310046K01Rik | RIKEN cDNA 2310046K01 gene (2310046K01Rik), mRNA | NM_027172 | 7.37995 | 6.69E-29 |
| Ldhc | Lactate dehydrogenase 3, C chain, sperm specific (Ldh3), mRNA | NM_013580 | 7.36571 | 3.03E-20 |
| Mgst3 | Microsomal glutathione S-transferase 3, mRNA (cDNA clone MGC:35738 IMAGE:5372427) | NM_025569 | 7.36371 | 7.31E-12 |
| AK084877 | 0 day neonate eyeball cDNA, RIKEN full-length enriched library, clone:E130014I08 product: unclassifiable, full insert sequence | AK084877 | 7.35577 | 7.22E-23 |
| 4732467B22 | hypothetical protein 4732467B22 | NM_177822 | 7.33593 | 6.02E-12 |
| Spib | Spi-B transcription factor (Spi-1/PU.1 related) (Spib), mRNA | U87620 | 7.32567 | 1.70E-08 |
| Aldh1a1 | Ahd-2=acetaldehyde dehydrogenase [mice, BALB/c, livers, mRNA Partial, 1632 nt] | NM_013467 | 7.32059 | 1.68E-29 |
| BB128963 | Expressed sequence BB128963, mRNA (cDNA clone IMAGE:3964696) | AK088715 | 7.29587 | 3.02E-21 |
| Samd8 | Sterile alpha motif domain containing 8 (Samd8), mRNA | NM_026283 | 7.28944 | 2.04E-13 |
| Dnmt3b | DNA cytosine methyltransferase 3b6 (Dnmt3b6) mRNA, complete cds; alternatively spliced | NM_010068 | 7.28075 | 1.55E-06 |
| Txnip | Thioredoxin interacting protein, mRNA (cDNA clone MGC:25534 IMAGE:3591421) | AK004653 | 7.27867 | 6.51E-09 |
| D930040F23Rik | Mus musculus adult male olfactory brain cDNA, RIKEN full-length enriched library, clone:6430407I07 product: unclassifiable, full insert sequence. | AK032178 | 7.26988 | 3.49E-09 |
| Wdr47 | WD repeat domain 47 (Wdr47), mRNA | NM_181400 | 7.26652 | 1.73E-06 |
| Pla2g5 | Phospholipase A2, group V, mRNA (cDNA clone MGC:31537 IMAGE:4502326) | NM_011110 | 7.25852 | 7.82E-12 |
| Tex101 | Testis expressed gene 101 (Tex101), mRNA | NM_019981 | 7.22574 | 4.77E-20 |
| Gprk5 | G protein-coupled receptor kinase 5 (GRK5) | AK079806 | 7.217 | 0.00001 |
| Cry2 | Cryptochrome 2 (photolyase-like), mRNA (cDNA clone IMAGE:3593948) | NM_009963 | 7.21232 | 4.87E-34 |
| Epb4.1l3 | Protein 4.1B (Epb4.1l3) | AK086340 | 7.20588 | 0.00037 |
| Cpeb4 | Cytoplasmic polyadenylation element binding protein 4 (Cpeb4), mRNA | AY313775 | 7.18189 | 1.32E-08 |
| Rasgef1b | RasGEF domain family, member 1B (Rasgef1b), transcript variant 1, mRNA | NM_181318 | 7.17597 | 4.25E-16 |
| Atrnl1 | Attractin like 1, mRNA (cDNA clone IMAGE:4221668) | AK048968 | 7.15363 | 4.73E-28 |
| Ablim1 | Actin-binding LIM protein 1, mRNA (cDNA clone IMAGE:5323557) | AK031453 | 7.15289 | 6.91E-24 |
| D930005D10Rik | MKIAA1849 protein | NM_178702 | 7.13213 | 5.89E-24 |
| Rgl1 | Ral guanine nucleotide dissociation stimulator,-like 1, mRNA (cDNA clone MGC:18430 IMAGE:4241244) | NM_016846 | 7.12829 | 2.93E-40 |
| Olfr295 | Olfactory receptor 295 (Olfr295), mRNA | NM_146851 | 7.12443 | 2.88E-06 |
| Ccpg1 | Cell cycle progression 1, mRNA (cDNA clone MGC:12151 IMAGE:3711012) | BC006717 | 7.12363 | 1.51E-24 |
| Zbtb10 | PREDICTED: zinc finger and BTB domain containing 10 [Mus musculus], mRNA sequence | AK083443 | 7.11228 | 0.00061 |
| Icosl | Icos ligand, mRNA (cDNA clone MGC:35971 IMAGE:4217333) | NM_015790 | 7.1024 | 0.0004 |
| Mycn | Neuroblastoma myc-related oncogene 1, mRNA (cDNA clone MGC:6240 IMAGE:3495446) | NM_008709 | 7.10045 | 0.0004 |
| D030074K08Rik | YY1 associated factor 2 (Yaf2), mRNA | AK083749 | 7.09956 | 0.00016 |
| Slc6a18 | Solute carrier family 6 (neurotransmitter transporter), member 18, mRNA (cDNA clone MGC:18346 IMAGE:4235659) | AK085473 | 7.09686 | 0.00038 |
| Tex12 | Testis protein TEX12 (Tex12) | NM_025687 | 7.09649 | 1.01E-10 |
| Usp49 | Ubiquitin specific peptidase 49 (Usp49), mRNA | NM_198421 | 7.09535 | 1.27E-14 |
| Magi3 | Membrane associated guanylate kinase, WW and PDZ domain containing 3 (Magi3), mRNA | NM_133853 | 7.08507 | 8.81E-28 |
| Mgl1 | Macrophage galactose N-acetyl-galactosamine specific lectin 1 (Mgl1), mRNA | NM_010796 | 7.07263 | 8.28E-25 |
| Pdlim5 | PDZ and LIM domain 5, mRNA (cDNA clone MGC:46824 IMAGE:4457868) | NM_019809 | 7.05824 | 9.20E-15 |
| A630005I04Rik | RIKEN cDNA A630005I04 gene (A630005I04Rik), mRNA | AK088332 | 7.03002 | 2.01E-26 |
| AK089990 | RCB-0559 K-1. F1 cDNA, RIKEN full-length enriched library, clone:G430046J05 product: unclassifiable, full insert sequence | AK089990 | 7.0234 | 9.15E-19 |
| Dmn | MKIAA0353 protein | NM_201639 | 7.0219 | 2.13E-11 |
| Fabp4 | Fatty acid binding protein 4, adipocyte (Fabp4), mRNA | NM_024406 | 7.02153 | 1.00E-18 |
| 4930463G05Rik | PREDICTED: Mus musculus RIKEN cDNA 4930463G05 gene (4930463G05Rik), mRNA | AK015217 | 7.02125 | 1.22E-42 |
| Rshl3 | PREDICTED: similar to radial spoke head-like 1 [Mus musculus], mRNA sequence | AK038980 | 7.01713 | 0.00039 |
| Smpd3 | Sphingomyelin phosphodiesterase 3, neutral (Smpd3), mRNA | NM_021491 | 7.00037 | 1.98E-09 |
| AI931714 | PREDICTED: hypothetical protein LOC102182 [Mus musculus], mRNA sequence | AK045296 | 7.00021 | 4.36E-14 |
| Diras1 | DIRAS family, GTP-binding RAS-like 1 (Diras1), mRNA | NM_145217 | 6.99843 | 8.46E-28 |
| Olfr460 | Olfactory receptor 460 (Olfr460), mRNA | NM_146383 | 6.98029 | 0.00004 |
| Lphn3 | Latrophilin 3, mRNA (cDNA clone IMAGE:30549106) | AK053210 | 6.97894 | 0.00044 |
| 1700026L06Rik | RIKEN cDNA 1700026L06 gene (1700026L06Rik), mRNA | BC055108 | 6.97247 | 2.83E-27 |
| Kremen1 | Kringle containing transmembrane protein 1 (Kremen1), mRNA | AK081707 | 6.95532 | 2.39E-24 |
| B3gnt4 | Beta1,3 N-acetylglucosaminyltransferase-4 (B3gnt4) | NM_198611 | 6.94475 | 2.02E-12 |
| Hcn3 | Hyperpolarization-activated, cyclic nucleotide-gated K+ 3 (Hcn3), mRNA | NM_008227 | 6.93488 | 8.78E-08 |
| Plxdc1 | Plexin domain containing 1 (Plxdc1), mRNA | NM_028199 | 6.93463 | 8.74E-18 |
| Meig1 | Meiosis expressed gene 1 (Meig1), mRNA | NM_008579 | 6.92131 | 9.92E-14 |
| Cdx1 | Caudal type homeo box 1, mRNA (cDNA clone MGC:28644 IMAGE:4224352) | NM_009880 | 6.91978 | 0.00044 |
| Zp3 | Zona pellucida glycoprotein 3 (Zp3), mRNA | NM_011776 | 6.91854 | 8.16E-36 |
| 6530409C15Rik | PREDICTED: hypothetical protein LOC76224 [Mus musculus], mRNA sequence | AK018330 | 6.91722 | 0.00041 |
| 4930518F03Rik | Mus musculus RIKEN cDNA 4930518F03 gene (4930518F03Rik), mRNA [NM_177191] | NM_177191 | 6.91301 | 0.00073 |
| Mmp2 | Matrix metallopeptidase 2 (Mmp2), mRNA | BF147716 | 6.91006 | 1.84E-11 |
| Herc5 | Iduronato sulfatase | AK037442 | 6.90618 | 7.13E-07 |
| Neud4 | Clone m62 zinc finger protein neuro-d4 (neuro-d4) | NM_013874 | 6.90322 | 1.87E-11 |
| Slc16a10 | Solute carrier family 16 (monocarboxylic acid transporters), member 10, mRNA (cDNA clone MGC:60866 IMAGE:30046388) | AK011813 | 6.89975 | 2.58E-10 |
| TC947784 | SNAG_HUMAN Gamma-soluble NSF attachment protein (SNAP-gamma) (N-ethylmaleimide- sensitive factor attachment protein, gamma). [Human] {Homo sapiens}, partial (13%) [TC947784] | 6.88434 | 0.00048 | |
| Tmem64 | Transmembrane protein 64, mRNA (cDNA clone MGC:40684 IMAGE:3661112) | AK035894 | 6.87747 | 0.00049 |
| AK043237 | Mus musculus 7 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:A730074B07 product: unclassifiable, full insert sequence. | AK043237 | 6.86471 | 0.00065 |
| Gm172 | PREDICTED: similar to hypothetical protein [Mus musculus], mRNA sequence | AV254918 | 6.86109 | 7.31E-13 |
| Apobec2 | Apolipoprotein B editing complex 2, mRNA (cDNA clone MGC:41184 IMAGE:1433092) | NM_009694 | 6.85954 | 0.0007 |
| Bcl2l11 | BCL2-like 11 (apoptosis facilitator), transcript variant 1, mRNA (cDNA clone MGC:28730 IMAGE:4459720) | BC058175 | 6.85608 | 1.80E-13 |
| Gcnt1 | Glucosaminyl (N-acetyl) transferase 1, core 2, mRNA (cDNA clone MGC:11452 IMAGE:3154671) | NM_173442 | 6.84626 | 6.17E-10 |
| Fcgr3 | Fc gamma receptor III (Fcgr3) mRNA, Fcgr3-b allele | NM_010188 | 6.844 | 1.64E-15 |
| Zdhhc6 | Zinc finger, DHHC domain containing 6, mRNA (cDNA clone MGC:30394 IMAGE:4459242) | NM_025883 | 6.84083 | 0.00001 |
| Pbx4 | Pre-B-cell leukemia transcription factor 4, mRNA (cDNA clone MGC:40990 IMAGE:1263363) | NM_030555 | 6.83768 | 6.20E-23 |
| Eif2c3 | Eukaryotic translation initiation factor 2C, 3 (Eif2c3), mRNA | BC060127 | 6.80015 | 7.50E-12 |
| 1810057P16Rik | RIKEN cDNA 1810057P16 gene (1810057P16Rik), mRNA | AK075801 | 6.77942 | 5.18E-12 |
| Osbpl8 | Oxysterol binding protein-like 8 (Osbpl8), transcript variant 2, mRNA | AK036477 | 6.77762 | 1.16E-17 |
| Lysmd4 | RIKEN cDNA 4930506D23 gene, mRNA (cDNA clone MGC:65434 IMAGE:4504057) | AK039294 | 6.75923 | 2.83E-18 |
| Fsd1 | Fibronectin type 3 and SPRY domain-containing protein (Fsd1), mRNA | NM_183178 | 6.74462 | 0.0004 |
| Lcmt1 | Leucine carboxyl methyltransferase 1, mRNA (cDNA clone IMAGE:3595687) | NM_025304 | 6.72952 | 3.34E-35 |
| Bach2 | BTB and CNC homology 2 (Bach2), mRNA | NM_007521 | 6.72785 | 5.54E-12 |
| Kif17 | Kinesin family member 17 (Kif17), mRNA | NM_010623 | 6.70819 | 8.92E-14 |
| LOC195281 | similar to Proton-associated sugar transporter A (PAST-A) (Deleted in neuroblastoma 5 protein) (DNb-5) | XM_111577 | 6.70613 | 4.14E-29 |
| D330012F22Rik | RIKEN cDNA D330012F22 gene, mRNA (cDNA clone MGC:47349 IMAGE:4458984) | AK084531 | 6.70519 | 3.43E-07 |
| Arhgef7 | Rho guanine nucleotide exchange factor (GEF7) (Arhgef7), mRNA | AK129064 | 6.70342 | 1.94E-21 |
| D930046H04Rik | Mus musculus RIKEN cDNA D930046H04 gene (D930046H04Rik), mRNA [NM_176934] | NM_176934 | 6.70196 | 0.00005 |
| Tmem24 | Transmembrane protein 24, mRNA (cDNA clone IMAGE:5360163) | AK004920 | 6.68313 | 1.32E-19 |
| Cyp46a1 | Cytochrome P450, family 46, subfamily a, polypeptide 1, mRNA (cDNA clone MGC:18311 IMAGE:4195579) | NM_010010 | 6.67773 | 6.83E-22 |
| Usp22 | MKIAA1063 protein | AK086502 | 6.67222 | 1.15E-09 |
| 9530059J11Rik | Mus musculus adult male urinary bladder cDNA, RIKEN full-length enriched library, clone:9530059J11 product: weakly similar to 12 DAYS EMBRYO HEAD CDNA, RIKEN FULL-LENGTH ENRICHED LIBRARY, CLONE:3010005E14, FULL INSERT SEQUENCE [Mus musculus], full insert sequence. | AK020615 | 6.66825 | 1.21E-08 |
| Mgst3 | Microsomal glutathione S-transferase 3, mRNA (cDNA clone MGC:35738 IMAGE:5372427) | AK008211 | 6.65726 | 9.29E-09 |
| 9330107J05Rik | PREDICTED: hypothetical protein LOC73884 [Mus musculus], mRNA sequence | AK047625 | 6.65622 | 0.00061 |
| Grb7 | Growth factor receptor bound protein 7, mRNA (cDNA clone MGC:5653 IMAGE:3583896) | NM_010346 | 6.65154 | 6.22E-12 |
| Pros1 | Protein S (alpha) (Pros1), mRNA | NM_011173 | 6.6501 | 9.72E-31 |
| Usf1 | M.musculus USF1 (non-coding exon 1). | X95315 | 6.64978 | 1.88E-09 |
| Grin3b | Glutamate receptor, ionotropic, NMDA3B (Grin3b), mRNA | NM_130455 | 6.64959 | 3.38E-22 |
| A730016F12Rik | Mus musculus RIKEN cDNA A730016F12 gene (A730016F12Rik), transcript variant 2, mRNA [NM_175079] | NM_175079 | 6.64561 | 1.08E-35 |
| Casz1 | MKIAA3026 protein | AK010559 | 6.63118 | 9.68E-36 |
| Nckap1l | NCK associated protein 1 like (Nckap1l), mRNA | NM_153505 | 6.62986 | 7.63E-07 |
| AI324046 | Immunoglobulin heavy chain 6 (heavy chain of IgM), mRNA (cDNA clone MGC:18788 IMAGE:4189350) | NM_198640 | 6.62565 | 0.00096 |
| 9530001N24Rik | Peroxiredoxin 1 (Prdx1), mRNA | AK020533 | 6.60763 | 2.92E-11 |
| 4732496G21Rik | RIKEN cDNA 4732496G21 gene (4732496G21Rik), mRNA | AK035094 | 6.59781 | 0.00384 |
| Evpl | Envoplakin, mRNA (cDNA clone IMAGE:3985477) | AK007353 | 6.59541 | 1.14E-40 |
| Postn | Periostin, osteoblast specific factor, mRNA (cDNA clone MGC:25368 IMAGE:4457222) | NM_015784 | 6.59457 | 2.74E-12 |
| Lyzs | Lysozyme, mRNA (cDNA clone IMAGE:2655292) | NM_017372 | 6.58874 | 0.00173 |
| Apol2 | PREDICTED: hypothetical protein XP_139463 [Mus musculus], mRNA sequence | AK036408 | 6.56783 | 8.71E-25 |
| Paqr3 | Progestin and adipoQ receptor family member III (Paqr3), mRNA | NM_198422 | 6.56779 | 5.81E-14 |
| 4833421E05Rik | RIKEN cDNA 4833421E05 gene (4833421E05Rik), mRNA | AK014743 | 6.56384 | 7.26E-13 |
| 4930519L02Rik | PREDICTED: hypothetical protein LOC75085 [Mus musculus], mRNA sequence | AK015845 | 6.55752 | 0.00113 |
| Hrbl | HIV-1 Rev binding protein-like (Hrbl), transcript variant 1, mRNA | NM_178162 | 6.55743 | 8.29E-31 |
| Bai2 | Brain-specific angiogenesis inhibitor 2, mRNA (cDNA clone MGC:66520 IMAGE:5705854) | NM_173071 | 6.5429 | 2.54E-10 |
| Map2k6 | MAP Kinase Kinase | NM_011943 | 6.53525 | 2.77E-11 |
| Ttll1 | Mus musculus tubulin tyrosine ligase-like 1 (Ttll1), mRNA. | NM_178869 | 6.52155 | 6.57E-30 |
| Hand2 | BHLH transcription factor dHand | NM_010402 | 6.51319 | 0.00064 |
| Cpeb4 | Cytoplasmic polyadenylation element binding protein 4 (Cpeb4), mRNA | AK173229 | 6.50992 | 7.59E-11 |
| C920005C14Rik | RIKEN cDNA C920005C14 gene (C920005C14Rik), mRNA | NM_177391 | 6.49949 | 0.0012 |
| Fbp2 | Fructose bisphosphatase 2, mRNA (cDNA clone MGC:14060 IMAGE:4205515) | NM_007994 | 6.49091 | 1.69E-19 |
| AK042798 | Mus musculus 7 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:A730025J02 product: unclassifiable, full insert sequence. | AK042798 | 6.47491 | 0.00001 |
| D930005D10Rik | MKIAA1849 protein | NM_178702 | 6.4664 | 3.36E-17 |
| Irf6 | Interferon regulatory factor 6, mRNA (cDNA clone MGC:5918 IMAGE:3592582) | NM_016851 | 6.46367 | 8.31E-13 |
| A530064D06Rik | RIKEN cDNA A530064D06 gene (A530064D06Rik), mRNA | NM_178796 | 6.46325 | 0.00093 |
| Fbxl16 | PREDICTED: similar to spinal cord injury and regeneration related protein 1 [Mus musculus], mRNA sequence | XM_128530 | 6.46071 | 5.26E-11 |
| 1700013F07Rik | PREDICTED: hypothetical protein LOC75504 [Mus musculus], mRNA sequence | AK005946 | 6.44623 | 1.24E-23 |
| Cdh15 | Cadherin 15 (Cdh15), mRNA | NM_007662 | 6.44407 | 1.52E-22 |
| AK033995 | Mus musculus adult male diencephalon cDNA, RIKEN full-length enriched library, clone:9330137C05 product: unclassifiable, full insert sequence. | AK033995 | 6.44198 | 2.74E-11 |
| Slc36a2 | Solute carrier family 36 (proton/amino acid symporter), member 2 (Slc36a2), mRNA | NM_153170 | 6.43572 | 4.47E-18 |
| Agt | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8), mRNA (cDNA clone MGC:19077 IMAGE:4194190) | NM_007428 | 6.43479 | 0.00014 |
| 4921513E08Rik | RIKEN cDNA 4921513E08 gene (4921513E08Rik), mRNA | NM_025725 | 6.42815 | 1.00E-30 |
| Hey2 | Basic helix-loop-helix factor 1 (CHF1) | AK048343 | 6.425 | 0.0015 |
| Krtap6-1 | Keratin associated protein 6-1 (Krtap6-1), mRNA | D86421 | 6.42422 | 0.0014 |
| AK046930 | 10 days neonate cerebellum cDNA, RIKEN full-length enriched library, clone:B930004K07 product: histocompatibility 2, T region locus 18, full insert sequence | AK046930 | 6.42302 | 4.29E-12 |
| Rtf1 | Gene trap locus 7, mRNA (cDNA clone IMAGE:6740526) | XM_283757 | 6.42082 | 3.63E-18 |
| Rnf122 | Ring finger protein 122, mRNA (cDNA clone MGC:36976 IMAGE:4947596) | NM_175136 | 6.40147 | 0.00007 |
| 5730508B09Rik | RIKEN cDNA 5730508B09 gene (5730508B09Rik), mRNA | AK017758 | 6.39187 | 2.05E-39 |
| D230014K01Rik | RIKEN cDNA D230014K01 gene (D230014K01Rik), mRNA | NM_172573 | 6.38975 | 1.69E-06 |
| Spdef | SAM pointed domain containing ets transcription factor, mRNA (cDNA clone MGC:13763 IMAGE:4218741) | NM_013891 | 6.37865 | 8.43E-08 |
| Adc | Arginine decarboxylase (Adc), mRNA | NM_172875 | 6.37788 | 6.03E-36 |
| Adora2b | Adenosine A2b receptor (Adora2b), mRNA | NM_007413 | 6.37729 | 8.30E-10 |
| TC949982 | AF465729 cyclin fold protein 1 variant b {Homo sapiens}, partial (83%) [TC949982] | 6.36781 | 5.12E-29 | |
| Ccdc37 | RIKEN cDNA C230069K22 gene, mRNA (cDNA clone MGC:90884 IMAGE:5693354) | NM_173775 | 6.36741 | 4.66E-17 |
| Epas1 | Endothelial PAS domain protein 1 (Epas1), mRNA | AK087208 | 6.35043 | 1.62E-18 |
| 9630020I17Rik | Adult male aorta and vein cDNA, RIKEN full-length enriched library, clone:A530095L03 product: unclassifiable, full insert sequence | AK079318 | 6.34776 | 3.78E-06 |
| ENSMUST00000055324 | OLFACTORY RECEPTOR GA_X6K02T2PSCP-1637650–1636712 (FRAGMENT). [Source:SPTREMBL;Acc:Q7TRL0] [ENSMUST00000055324] | 6.3429 | 0.00019 | |
| AI646023 | Expressed sequence AI646023 (AI646023), mRNA | NM_198860 | 6.3422 | 1.37E-12 |
| Ptpn12 | Protein tyrosine phosphatase, non-receptor type 12 (Ptpn12), mRNA | AK087609 | 6.32927 | 0.00114 |
| Wdr40b | WD repeat domain 40B, mRNA (cDNA clone MGC:76797 IMAGE:30431243) | NM_178739 | 6.32111 | 0.00162 |
| Rap2ip | Rap2 interacting protein (Rap2ip), mRNA | AK007019 | 6.3115 | 4.38E-17 |
| 6230427J02Rik | RIKEN cDNA 6230427J02 gene (6230427J02Rik), mRNA | AK020090 | 6.30412 | 4.72E-38 |
| 5730420B22Rik | RIKEN cDNA 5730420B22 gene, mRNA (cDNA clone IMAGE:5356411) | NM_172597 | 6.29909 | 2.99E-13 |
| Zhx1 | Zinc fingers and homeoboxes protein 1 (Zhx1), mRNA | AK051410 | 6.29396 | 3.18E-08 |
| Lmtk2 | Lemur tyrosine kinase 2, mRNA (cDNA clone IMAGE:6407188) | BC058653 | 6.2919 | 1.25E-23 |
| 2410003J06Rik | RIKEN cDNA 2410003J06 gene, mRNA (cDNA clone MGC:60956 IMAGE:30007513) | AK010362 | 6.2909 | 1.27E-08 |
| Dgat2 | Diacylglycerol O-acyltransferase 2 (Dgat2), mRNA | NM_026384 | 6.28587 | 3.72E-19 |
| Rab3d | RAB3D, member RAS oncogene family, mRNA (cDNA clone MGC:6707 IMAGE:3584902) | NM_031874 | 6.28515 | 3.96E-08 |
| 2610031L17Rik | RIKEN cDNA 2610031L17 gene, mRNA (cDNA clone MGC:38351 IMAGE:5344332) | AK043897 | 6.2836 | 0.00003 |
| Jmjd2d | Hypothetical protein 4932416A15 (4932416A15), mRNA | NM_173433 | 6.27488 | 2.19E-07 |
| 1700023A16Rik | PREDICTED: hypothetical protein LOC69371 [Mus musculus], mRNA sequence | AK006257 | 6.27378 | 2.95E-13 |
| Tmcc3 | Transmembrane and coiled coil domains 3, mRNA (cDNA clone MGC:30586 IMAGE:3660220) | NM_172051 | 6.27294 | 9.85E-06 |
| Clec2e | C-type lectin domain family 2, member e (Clec2e), mRNA | NM_153506 | 6.27057 | 0.0012 |
| H2-DMa | Histocompatibility 2, class II, locus DMa, mRNA (cDNA clone MGC:5743 IMAGE:3591389) | NM_010386 | 6.26974 | 1.31E-33 |
| Habp4 | Hyaluronic acid binding protein 4 (Habp4), mRNA | NM_019986 | 6.26891 | 6.44E-11 |
| LOC434018 | hypothetical gene supported by AK029757 | AK029757 | 6.2591 | 0.0016 |
| 2700038N03Rik | RIKEN cDNA 2700038N03 gene, mRNA (cDNA clone MGC:27927 IMAGE:3584484) | NM_027356 | 6.2369 | 1.38E-30 |
| 3526402J09Rik | RIKEN cDNA 3526402J09 gene, mRNA (cDNA clone IMAGE:30544810) | AK028312 | 6.23264 | 3.49E-06 |
| Dlgap1 | Discs, large (Drosophila) homolog-associated protein 1 (Dlgap1), transcript variant 2, mRNA | AK034215 | 6.21769 | 1.54E-06 |
| 9230116B18Rik | PREDICTED: hypothetical protein LOC78245 [Mus musculus], mRNA sequence | AK020348 | 6.21758 | 3.02E-09 |
| H2-Ke6 | H2-K region expressed gene 6 (H2-Ke6), mRNA | NM_013543 | 6.21346 | 6.39E-38 |
| Prdx1 | Peroxiredoxin 1 (Prdx1), mRNA | AK083642 | 6.21182 | 3.67E-17 |
| 4930550L24Rik | Mus musculus RIKEN cDNA 4930550L24 gene (4930550L24Rik), mRNA [NM_023774] | NM_023774 | 6.21139 | 6.89E-09 |
| Trim62 | Tripartite motif-containing 62 (Trim62), mRNA | NM_178110 | 6.20606 | 8.48E-07 |
| NAP044566-1 | Unknown | 6.20486 | 9.69E-13 | |
| Brsk2 | BR serine/threonine kinase 2 (Brsk2), transcript variant 2, mRNA | BC056498 | 6.18857 | 0.00023 |
| 9630019E01Rik | Mus musculus 12 days embryo spinal ganglion cDNA, RIKEN full-length enriched library, clone:D130045C05 product: unclassifiable, full insert sequence. | AK051384 | 6.17974 | 2.17E-15 |
| Itga6 | Mus musculus adult male corpora quadrigemina cDNA, RIKEN full-length enriched library, clone:B230112G01 product: integrin alpha 6, full insert sequence. | AK045391 | 6.17009 | 2.44E-07 |
| Acoxl | Acyl-Coenzyme A oxidase-like (Acoxl), mRNA | AV282483 | 6.16545 | 1.07E-07 |
| C030014L02 | hypothetical protein C030014L02 | AK047704 | 6.15908 | 0.00132 |
| 4921515A04Rik | Mus musculus RIKEN cDNA 4921515A04 gene (4921515A04Rik), mRNA [NM_172939] | NM_172939 | 6.15587 | 2.02E-16 |
| Frat1 | Frequently rearranged in advanced T-cell lymphomas (Frat1), mRNA | NM_008043 | 6.14419 | 9.07E-08 |
| D5Wsu178e | DNA segment, Chr 5, Wayne State University 178, expressed (D5Wsu178e), mRNA | AK046608 | 6.14236 | 5.29E-09 |
| AK012034 | Mus musculus 10 days embryo whole body cDNA, RIKEN full-length enriched library, clone:2610315N17 product: unclassifiable, full insert sequence. | AK012034 | 6.1156 | 2.54E-08 |
| Tax1bp1 | Tax1 (human T-cell leukemia virus type I) binding protein 1, mRNA (cDNA clone MGC:11692 IMAGE:3962810) | AK051030 | 6.11319 | 0.00526 |
| Syce1 | PREDICTED: Mus musculus RIKEN cDNA 4933406J07 gene (4933406J07Rik), mRNA | AK016694 | 6.1065 | 1.56E-15 |
| A430091O22Rik | RIKEN cDNA A430091O22 gene (A430091O22Rik), mRNA | AK039072 | 6.10372 | 0.00337 |
| Scrib | Scribbled homolog (Drosophila), mRNA (cDNA clone IMAGE:4459388) | AK084936 | 6.1006 | 8.84E-14 |
| Rhov | Ras homolog gene family, member V (Rhov), mRNA | NM_145530 | 6.0999 | 1.54E-23 |
| AK040776 | Adult male aorta and vein cDNA, RIKEN full-length enriched library, clone:A530024I17 product: unclassifiable, full insert sequence | AK040776 | 6.09753 | 1.78E-38 |
| D330014H01Rik | Mus musculus RIKEN cDNA D330014H01 gene (D330014H01Rik), mRNA [NM_177617] | NM_177617 | 6.08754 | 1.59E-14 |
| D0H4S114 | DNA segment, human D4S114 (D0H4S114), mRNA | NM_053078 | 6.08447 | 0.00133 |
| Herc1 | Hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1, mRNA (cDNA clone MGC:7618 IMAGE:3494753) | AK083444 | 6.08322 | 4.90E-24 |
| 1700065D16Rik | Protein kinase ATR (Atr) | AK030118 | 6.07886 | 0.00004 |
| Ppp1r13b | Protein phosphatase 1, regulatory (inhibitor) subunit 13B (Ppp1r13b), mRNA | NM_011625 | 6.0768 | 9.44E-25 |
| LOC213402 | hypothetical LOC213402 | XM_136978 | 6.07669 | 6.10E-19 |
| Capns2 | Hypothetical protein A330042H22 (A330042H22), mRNA | AK009171 | 6.0641 | 6.40E-07 |
| NM_027559 | Mus musculus RIKEN cDNA 4930463G05 gene (4930463G05Rik), mRNA [NM_027559] | NM_027559 | 6.05992 | 8.61E-10 |
| 2810427I04Rik | RIKEN cDNA 2810427I04 gene (2810427I04Rik), mRNA | NM_028146 | 6.05072 | 3.51E-08 |
| Mycbp | C-myc binding protein, mRNA (cDNA clone MGC:13820 IMAGE:3985166) | NM_019660 | 6.0454 | 3.53E-09 |
| D130076A03Rik | Mus musculus 12 days embryo spinal ganglion cDNA, RIKEN full-length enriched library, clone:D130076A03 product: unclassifiable, full insert sequence. | AK084007 | 6.04495 | 1.01E-16 |
| AA919669 | Adult male hippocampus cDNA, RIKEN full-length enriched library, clone:2900016J10 product: unclassifiable, full insert sequence | AA919669 | 6.04404 | 1.08E-19 |
| Ifrd1 | Interferon-related developmental regulator 1 (Ifrd1), mRNA | NM_013562 | 6.04347 | 2.10E-09 |
| Ddit4l | DNA-damage-inducible transcript 4-like, mRNA (cDNA clone MGC:47974 IMAGE:5254530) | NM_030143 | 6.04198 | 0.00184 |
| Gsdm1 | Gasdermin 1 (Gsdm1), mRNA | NM_021347 | 6.04098 | 2.79E-13 |
| 3110057O12Rik | RIKEN cDNA 3110057O12 gene, mRNA (cDNA clone IMAGE:4007705) | NM_026622 | 6.04095 | 1.90E-12 |
| Pdpk1 | Phosphoinositide-dependent protein kinase-1 beta (Pdk1beta) | NM_011062 | 6.03733 | 3.77E-31 |
| Zp3 | Zona pellucida glycoprotein 3 (Zp3), mRNA | NM_011776 | 6.02738 | 1.28E-12 |
| TC984987 | NDK6_HUMAN Nucleoside diphosphate kinase 6(NDK 6) (NDP kinase 6) (nm23-H6) (Inhibitor of p53-induced apoptosis-alpha) (IPIA-alpha). [Human] {Homo sapiens}, partial (14%) [TC984987] | 6.02678 | 0.00023 | |
| 1700011E04Rik | PREDICTED: Mus musculus RIKEN cDNA 1700011E04 gene (1700011E04Rik), mRNA | AK005860 | 6.02661 | 6.11E-06 |
| Sstr2 | Somatostatin receptor 2 (Sstr2), mRNA | NM_009217 | 6.0182 | 1.48E-11 |
| Pgm2l1 | Phosphoglucomutase 2-like 1, mRNA (cDNA clone MGC:29299 IMAGE:5004308) | NM_027629 | 6.01695 | 1.89E-15 |
| BE646931 | Transcribed locus | BE646931 | 6.00062 | 1.38E-15 |
| Ccdc37 | ZXD family zinc finger C, mRNA (cDNA clone MGC:7160 IMAGE:3256958) | AK021139 | 5.99479 | 5.23E-06 |
| Timd4 | T-cell immunoglobulin and mucin domain containing 4, mRNA (cDNA clone MGC:41117 IMAGE:1195144) | NM_178759 | 5.98633 | 0.00165 |
| Ap3m2 | Adaptor-related protein complex 3, mu 2 subunit, mRNA (cDNA clone MGC:40858 IMAGE:5369288) | NM_029505 | 5.97689 | 1.24E-08 |
| Htr2b | 5-hydroxytryptamine (serotonin) receptor 2B, mRNA (cDNA clone MGC:36132 IMAGE:5344328) | NM_008311 | 5.97448 | 0.00018 |
| Tfdp2 | DP-3=protein regulating cell cycle transcription factor DRTF1/E2F [mice, pl-2, F9 EC, mRNA, 1380 nt]. | S79780 | 5.97116 | 4.82E-07 |
| Rybp | RING1 and YY1 binding protein (Rybp), mRNA | NM_019743 | 5.97057 | 9.26E-38 |
| Sema3b | Sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B (Sema3b), mRNA | NM_009153 | 5.9703 | 2.43E-22 |
| Dcn | Decorin (Dcn), mRNA | NM_007833 | 5.96766 | 2.08E-06 |
| Lhx2 | LIM-homeodomain protein MLHX2 (Lhx2) | NM_010710 | 5.96324 | 1.93E-11 |
| Fbxo33 | F-box only protein 33, mRNA (cDNA clone IMAGE:3661715) | BC020022 | 5.96152 | 1.82E-17 |
| Cfh | Complement component factor h (Cfh), mRNA | M12660 | 5.95652 | 0.00006 |
| Psd2 | Pleckstrin and Sec7 domain containing 2 (Psd2), mRNA | NM_028707 | 5.95334 | 6.23E-14 |
| Ncoa1 | Nuclear receptor coactivator 1 (Ncoa1), mRNA | AA738860 | 5.94761 | 1.72E-25 |
| Robo2 | MKIAA1568 protein | BC055333 | 5.94552 | 0.00019 |
| AW125753 | Expressed sequence AW125753 (AW125753), mRNA | NM_029007 | 5.94357 | 1.90E-15 |
| Cfh | Complement component factor h (Cfh), mRNA | NM_009888 | 5.94161 | 6.84E-07 |
| 1700014D04Rik | PREDICTED: hypothetical protein LOC74224 [Mus musculus], mRNA sequence | AK077007 | 5.93902 | 9.40E-15 |
| 6-Sep | Mus musculus 2 days neonate thymus thymic cells cDNA, RIKEN full-length enriched library, clone:C920001C06 product: hypothetical protein, full insert sequence. | AK083302 | 5.93827 | 0.00283 |
| Pdpk1 | Phosphoinositide-dependent protein kinase-1 beta (Pdk1beta) | NM_011062 | 5.93442 | 2.75E-36 |
| BC048651 | CDNA sequence BC048651 (BC048651), mRNA | NM_207258 | 5.93022 | 1.19E-13 |
| Metrnl | Meteorin, glial cell differentiation regulator-like (Metrnl), mRNA | AK053718 | 5.9294 | 3.74E-21 |
| LOC229608 | Histone 2, H2bb (Hist2h2bb), mRNA | BC019122 | 5.92838 | 4.58E-08 |
| Arl5b | ADP-ribosylation factor-like 8 (Arl8), mRNA | NM_029466 | 5.92634 | 1.03E-09 |
| Olfr987 | Olfactory receptor 987 (Olfr987), mRNA | CB174485 | 5.92041 | 0.00266 |
| Malat1 | Adult male stomach cDNA, RIKEN full-length enriched library, clone:2210407K09 product: receptor (calcitonin) activity modifying protein 2, full insert sequence | AK020483 | 5.91958 | 6.65E-15 |
| Mall | CDNA sequence BC012256 (BC012256), mRNA | NM_145532 | 5.91692 | 8.22E-08 |
| Phtf2 | Putative homeodomain transcription factor 2, mRNA (cDNA clone MGC:60851 IMAGE:30050876) | NM_172992 | 5.91568 | 1.02E-30 |
| D330028D13Rik | RIKEN cDNA D330028D13 gene, mRNA (cDNA clone MGC:67688 IMAGE:4487036) | NM_172727 | 5.91051 | 1.54E-22 |
| Rhpn2 | Rhophilin, Rho GTPase binding protein 2 (Rhpn2), mRNA | NM_027897 | 5.90122 | 2.02E-22 |
| 3526402J09Rik | 8 days embryo whole body cDNA, RIKEN full-length enriched library, clone:5730590C14 product: unclassifiable, full insert sequence | AK019982 | 5.89752 | 1.80E-24 |
| Tigd5 | Tigger transposable element derived 5 (Tigd5), mRNA | NM_178646 | 5.89512 | 9.40E-16 |
| C230078M08Rik | RIKEN cDNA C230078M08 gene (C230078M08Rik), mRNA | NM_176995 | 5.88686 | 6.61E-06 |
| D930001I22Rik | RIKEN cDNA D930001I22 gene, mRNA (cDNA clone MGC:107602 IMAGE:6750721) | AK052945 | 5.86474 | 5.89E-43 |
| Tcf15 | Transcription factor 15, mRNA (cDNA clone MGC:41210 IMAGE:5143248) | NM_009328 | 5.86455 | 2.79E-14 |
| Wif1 | Wnt inhibitory factor 1, mRNA (cDNA clone MGC:13706 IMAGE:3984128) | NM_011915 | 5.85749 | 1.02E-06 |
| Egfl5 | MKIAA0818 protein | AK122378 | 5.85619 | 1.22E-10 |
| Fcer1g | Fc receptor, IgE, high affinity I, gamma polypeptide, mRNA (cDNA clone MGC:36077 IMAGE:5065647) | AI326608 | 5.85071 | 2.00E-10 |
| Mtap2 | Mus musculus microtubule-associated protein 2 (Mtap2), mRNA [NM_008632] | NM_008632 | 5.85052 | 5.98E-13 |
| Uros | Uroporphyrinogen III synthase (Uros), mRNA | NM_009479 | 5.84947 | 1.39E-29 |
| 3110098I04Rik | Transmembrane protein 49, mRNA (cDNA clone MGC:7590 IMAGE:3493738) | AK019443 | 5.84922 | 1.23E-18 |
| Snai3 | Snail-related zinc finger protein SMUC (Smuc) | NM_013914 | 5.84401 | 6.78E-12 |
| Arzc | Mus musculus hypothetical protein (Arzc) mRNA, complete cds. | AY344585 | 5.83844 | 1.81E-12 |
| BE987854 | Bone marrow macrophage cDNA, RIKEN full-length enriched library, clone:I830003K21 product: unclassifiable, full insert sequence | BE987854 | 5.8367 | 3.15E-09 |
| Npc1l1 | Mus musculus NPC1-like 1 (Npc1l1), mRNA. | XM_137497 | 5.83041 | 2.18E-14 |
| Etv1 | Ets variant gene 1, mRNA (cDNA clone MGC:6207 IMAGE:3257346) | NM_007960 | 5.82656 | 0.00003 |
| AK041974 | 3 days neonate thymus cDNA, RIKEN full-length enriched library, clone:A630050C01 product: unclassifiable, full insert sequence | AK041974 | 5.82616 | 0.00025 |
| Atp11b | ATPase, Class VI, type 11B (Atp11b), mRNA | AK041684 | 5.82607 | 3.61E-08 |
| Pgm2l1 | Phosphoglucomutase 2-like 1, mRNA (cDNA clone MGC:29299 IMAGE:5004308) | NM_027629 | 5.81964 | 0.0001 |
| Ppp1r13b | Protein phosphatase 1, regulatory (inhibitor) subunit 13B (Ppp1r13b), mRNA | NM_011625 | 5.81761 | 4.07E-12 |
| 2310068J10Rik | Adult male tongue cDNA, RIKEN full-length enriched library, clone:2310068J10 product: unclassifiable, full insert sequence | AK075917 | 5.81515 | 5.55E-09 |
| Nos3 | Nitric oxide synthase 3, endothelial cell (Nos3), mRNA | NM_008713 | 5.80909 | 1.30E-07 |
| Spsb1 | SplA/ryanodine receptor domain and SOCS box containing 1 (Spsb1), mRNA | NM_029035 | 5.80869 | 5.16E-13 |
| 8430416H19Rik | RIKEN cDNA 8430416H19 gene (8430416H19Rik), mRNA | NM_198636 | 5.80743 | 1.09E-11 |
| Gpr111 | PREDICTED: similar to G-protein coupled receptor 111 [Mus musculus], mRNA sequence | AY255579 | 5.79653 | 0.00225 |
| Pla2g4f | RIKEN cDNA 4732472I07 gene (4732472I07Rik), mRNA | BC039947 | 5.7812 | 5.59E-15 |
| Rfx2 | Regulatory factor X, 2 (influences HLA class II expression), mRNA (cDNA clone MGC:6105 IMAGE:3497910) | NM_009056 | 5.77832 | 1.13E-11 |
| Dpf3 | D4, zinc and double PHD fingers, family 3 (Dpf3), mRNA | NM_058212 | 5.77719 | 9.83E-15 |
| Zfp654 | Zinc finger protein 654, mRNA (cDNA clone IMAGE:5346461) | NM_028059 | 5.77371 | 1.78E-35 |
| Rnut1 | RNA, U transporter 1, mRNA (cDNA clone MGC:74414 IMAGE:30251661) | NM_178374 | 5.76909 | 7.92E-16 |
| Mpp7 | Membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7), mRNA (cDNA clone IMAGE:6532780) | AK017344 | 5.76872 | 3.39E-11 |
| Cx3cl1 | Mus musculus mRNA, complete cds, clone:1–53. | AB030188 | 5.75894 | 0.00037 |
| Map2k6 | Mus musculus 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930047J04 product: mitogen activated protein kinase kinase 6, full insert sequence. | AK086722 | 5.74548 | 2.97E-08 |
| Gsto2 | Glutathione S-transferase omega 2 (Gsto2), mRNA | NM_026619 | 5.73514 | 3.96E-07 |
| A030009H04Rik | RIKEN cDNA A030009H04 gene, mRNA (cDNA clone MGC:35887 IMAGE:5352453) | NM_020591 | 5.73374 | 2.63E-12 |
| Atox1 | ATX1 (antioxidant protein 1) homolog 1 (yeast), mRNA (cDNA clone MGC:41183 IMAGE:1328907) | AK089832 | 5.73109 | 2.04E-11 |
| Map2k6 | MAP Kinase Kinase | NM_011943 | 5.72903 | 0.00035 |
| Bmp2k | BMP2 inducible kinase (Bmp2k), mRNA | NM_080708 | 5.72563 | 2.28E-11 |
| St3gal5 | ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (St3gal5), mRNA | AI464789 | 5.72266 | 0.0001 |
| Rasd1 | RAS, dexamethasone-induced 1, mRNA (cDNA clone MGC:36188 IMAGE:4989312) | NM_009026 | 5.71641 | 9.16E-11 |
| C330001K17Rik | RIKEN cDNA C330001K17 gene (C330001K17Rik), mRNA | AK031175 | 5.70009 | 1.16E-08 |
| Olig2 | Olig2 bHLH protein | BC051967 | 5.69272 | 0.00135 |
| Zfp422-rs1 | zinc finger protein 422, related sequence 1 | NM_029952 | 5.68877 | 7.93E-14 |
| Ccni | Cyclin I, mRNA (cDNA clone MGC:5636 IMAGE:3583418) | AK083496 | 5.68864 | 0.00337 |
| AK028719 | 10 days neonate skin cDNA, RIKEN full-length enriched library, clone:4732444B01 product: unclassifiable, full insert sequence | AK028719 | 5.68121 | 0.00073 |
| 5730469D23Rik | RIKEN cDNA 5730469D23 gene, mRNA (cDNA clone MGC:66555 IMAGE:6825308) | AK017687 | 5.67016 | 6.96E-16 |
| Tspan1 | Tetraspan 1 (Tspan1), mRNA | NM_133681 | 5.66843 | 7.35E-08 |
| Ebf1 | Early B-cell factor 1 (Ebf1), mRNA | AK036716 | 5.66713 | 0.00329 |
| Susd2 | Sushi domain containing 2 (Susd2), mRNA | NM_027890 | 5.66242 | 2.96E-08 |
| XM_131928 | Mus musculus myeloid/lymphoid or mixed-lineage leukemia 3 (Mll3), mRNA [XM_131928] | XM_131928 | 5.65966 | 0.00033 |
| Trfr2 | Transferrin receptor 2, mRNA (cDNA clone MGC:18814 IMAGE:4196597) | NM_015799 | 5.65881 | 2.39E-07 |
| Tspyl4 | Mus musculus TSPY-like 4 (Tspyl4), mRNA. | NM_133745 | 5.65493 | 1.21E-11 |
| Hgf | Hepatocyte growth factor | AK082461 | 5.65072 | 0.00111 |
| Txnip | Thioredoxin interacting protein, mRNA (cDNA clone MGC:25534 IMAGE:3591421) | AK004653 | 5.6496 | 1.05E-06 |
| Angptl2 | Angiopoietin-like 2 (Angptl2), mRNA | AK035967 | 5.64678 | 9.32E-12 |
| Dos | Downstream of Stk11 (Dos), mRNA | AK035824 | 5.6364 | 2.23E-11 |
| Mars2 | Methionine-tRNA synthetase 2 (mitochondrial), mRNA (cDNA clone MGC:106216 IMAGE:3153138) | NM_175439 | 5.62867 | 1.35E-27 |
| Hamp1 | Prohepcidin (hepc1) | NM_032541 | 5.62597 | 1.61E-09 |
| BC049806 | CDNA sequence BC049806, mRNA (cDNA clone MGC:76460 IMAGE:30431671) | NM_172513 | 5.62361 | 6.88E-34 |
| Sbsn | suprabasin | NM_172205 | 5.62177 | 3.83E-18 |
| FHOS2 | formin-family protein FHOS2 | NM_175276 | 5.62016 | 0.00001 |
| Als2 | Amyotrophic lateral sclerosis 2 (juvenile) homolog (human), mRNA (cDNA clone MGC:27807 IMAGE:3257574) | NM_028717 | 5.61726 | 6.41E-30 |
| 5330420D20Rik | RIKEN cDNA 5330420D20 gene (5330420D20Rik), mRNA | NM_027442 | 5.61363 | 0.00005 |
| Gdap5 | Dishevelled associated activator of morphogenesis 1 (Daam1), mRNA | Y17854 | 5.61177 | 1.02E-26 |
| E030003N13Rik | Fibroblast growth factor receptor substrate 2, mRNA (cDNA clone MGC:64739 IMAGE:6830555) | AK051267 | 5.61074 | 0.00191 |
| Zfp59 | Zinc finger protein 59 (Zfp59), mRNA | AK042484 | 5.60473 | 0.00217 |
| Rnd1 | Rho family GTPase 1, mRNA (cDNA clone MGC:58446 IMAGE:6535763) | BC048531 | 5.60436 | 2.78E-39 |
| Ypel4 | Ypel4 mRNA for yippee-like 4 | AK043577 | 5.60118 | 0.00005 |
| Hbld2 | HESB like domain containing 2 (Hbld2), mRNA | NM_026921 | 5.59811 | 1.03E-15 |
| Plekhk1 | Membrane-bound factor MBF1 (Mbf) | AY036116 | 5.59772 | 2.32E-19 |
| Asb11 | Mus musculus adult male colon cDNA, RIKEN full-length enriched library, clone:9030022F06 product: ankyrin repeat and SOCS box-containing protein 11, full insert sequence. | AK078851 | 5.59544 | 0.00382 |
| TC996139 | Unknown | 5.59089 | 1.46E-07 | |
| Bai2 | Brain-specific angiogenesis inhibitor 2, mRNA (cDNA clone MGC:66520 IMAGE:5705854) | NM_173071 | 5.57838 | 0.00002 |
| Rnf17 | Ring finger protein 17 (Rnf17), transcript variant 1, mRNA | NM_013894 | 5.57648 | 0.00408 |
| Scoc | Short coiled-coil protein, mRNA (cDNA clone MGC:28845 IMAGE:4506964) | NM_019708 | 5.57413 | 1.02E-21 |
| Lama3 | Laminin-5, alpha3B chain | X84014 | 5.56979 | 1.47E-30 |
| Bmp6 | Bone morphogenetic protein 6 (Bmp6), mRNA | NM_007556 | 5.5686 | 6.55E-09 |
| Dffa | DNA fragmentation factor, alpha subunit (Dffa), transcript variant 2, mRNA | AK051011 | 5.55806 | 2.27E-19 |
| Nfatc4 | Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4, mRNA (cDNA clone MGC:25538 IMAGE:3670623) | AK014164 | 5.55789 | 1.20E-09 |
| Klc3 | Kinesin light chain 3 (Klc3), mRNA | NM_146182 | 5.55558 | 2.85E-09 |
| Kif17 | Kinesin family member 17 (Kif17), mRNA | NM_010623 | 5.5494 | 0.00022 |
| Btbd10 | BTB (POZ) domain containing 10, mRNA (cDNA clone IMAGE:3484902) | AK052335 | 5.54822 | 4.06E-10 |
| Slc25a35 | Solute carrier family 25, member 35 (Slc25a35), mRNA | NM_028048 | 5.54317 | 9.37E-07 |
| AK076567 | Adult male testis cDNA, RIKEN full-length enriched library, clone:4921513I01 product: unclassifiable, full insert sequence | AK076567 | 5.54169 | 4.56E-17 |
| Kif17 | Kinesin family member 17 (Kif17), mRNA | NM_010623 | 5.53068 | 7.65E-13 |
| Trpt1 | TRNA phosphotransferase 1 (Trpt1), mRNA | NM_153597 | 5.52301 | 1.26E-44 |
| C030032O16Rik | Putative homeodomain transcription factor 1 (Phtf1), mRNA | AK021115 | 5.52287 | 0.00004 |
| NAP050738-1 | Unknown | 5.51793 | 0.00143 | |
| Slc40a1 | Solute carrier family 40 (iron-regulated transporter), member 1, mRNA (cDNA clone MGC:6489 IMAGE:2647365) | NM_016917 | 5.51526 | 5.26E-10 |
| Mll3 | Mixed-lineage leukemia 3 protein (Mll3) | XM_355579 | 5.51391 | 1.75E-19 |
| Ablim3 | Actin binding LIM protein family, member 3, mRNA (cDNA clone MGC:106450 IMAGE:30534236) | NM_198649 | 5.51387 | 4.79E-12 |
| Dmxl1 | Dmx-like 1, mRNA (cDNA clone MGC:28386 IMAGE:4022096) | AK082851 | 5.51375 | 9.31E-09 |
| Hivep3 | Clone 5′RACE1 ZAS3 (Krc) mRNA, 5′ UTR | AK013707 | 5.51275 | 3.18E-26 |
| Fhl2 | Four and a half LIM domains 2, mRNA (cDNA clone MGC:29060 IMAGE:5066565) | AK081770 | 5.51184 | 0.00028 |
| Arl5b | ADP-ribosylation factor-like 8 (Arl8), mRNA | NM_029466 | 5.50628 | 6.74E-06 |
| Usp53 | Ubiquitin specific peptidase 53, mRNA (cDNA clone IMAGE:4236151) | AK030317 | 5.5048 | 6.73E-16 |
| Eaf2 | Testosterone regulated apoptosis inducer and tumor suppressor (Traits) | NM_134111 | 5.49779 | 6.28E-07 |
| Lmbr1 | Limb region 1, mRNA (cDNA clone MGC:28737 IMAGE:4481088) | AK030631 | 5.49689 | 1.41E-08 |
| F730023N20 | hypothetical protein F730023N20 | NM_183158 | 5.48167 | 0.00469 |
| Vamp1 | Vesicle-associated membrane protein 1 (Vamp1), mRNA | NM_009496 | 5.47901 | 7.04E-22 |
| Abcc10 | ATP-binding cassette, sub-family C (CFTR/MRP), member 10 (Abcc10), transcript variant mrp7B, mRNA | NM_145140 | 5.47864 | 1.39E-10 |
| BG080473 | Transcribed locus | BG080473 | 5.47538 | 0.00524 |
| Ppp1r9a | Protein phosphatase 1, regulatory (inhibitor) subunit 9A, mRNA (cDNA clone MGC:59493 IMAGE:6331919) | NM_181595 | 5.47417 | 4.82E-07 |
| TC1012618 | RGS2_MOUSE Regulator of G-protein signaling 2 (RGS2). [Mouse] {Mus musculus}, partial (43%) [TC1012618] | 5.46503 | 6.76E-10 | |
| Smarcad1 | MRNA of enhancer-trap-locus 1 | AK088163 | 5.4628 | 3.56E-06 |
| Malat1 | Adult male stomach cDNA, RIKEN full-length enriched library, clone:2210407K09 product: receptor (calcitonin) activity modifying protein 2, full insert sequence | BC004722 | 5.46155 | 1.11E-09 |
| Btg2 | B-cell translocation gene 2, anti-proliferative (Btg2), mRNA | NM_007570 | 5.46006 | 1.64E-08 |
| Malat1 | Adult male stomach cDNA, RIKEN full-length enriched library, clone:2210407K09 product: receptor (calcitonin) activity modifying protein 2, full insert sequence | AK090111 | 5.45534 | 2.16E-21 |
| Kcnk7 | Dpkch3 mRNA for double-pore K channel 3 | AF110521 | 5.45419 | 0.00021 |
| XM_358570 | Mus musculus LOC381415 (LOC381415), mRNA [XM_358570] | XM_358570 | 5.4508 | 0.00001 |
| Tnrc15 | Trinucleotide repeat containing 15 (Tnrc15), mRNA | AK034426 | 5.44852 | 9.38E-14 |
| D16H22S680E | DNA segment, Chr 16, human D22S680E, expressed (D16H22S680E), mRNA | NM_138583 | 5.4478 | 6.43E-27 |
| Cxxc5 | CXXC finger 5 (Cxxc5), mRNA | NM_133687 | 5.44654 | 0.00002 |
| Pbx4 | Pre-B-cell leukemia transcription factor 4, mRNA (cDNA clone MGC:40990 IMAGE:1263363) | NM_030555 | 5.4461 | 4.76E-13 |
| Gm1418 | PREDICTED: similar to Ig kappa chain precursor V region - mouse [Mus musculus], mRNA sequence | J04610 | 5.44288 | 0.0003 |
| Slc25a2 | Mutant ornithine transporter 2 (Ornt2) | AK077159 | 5.43766 | 3.03E-14 |
| Hist3h2bb | Histone 3, H2bb (Hist3h2bb), mRNA | NM_206882 | 5.43583 | 1.97E-33 |
| Ralgps2 | Ral GEF with PH domain and SH3 binding motif 2 (Ralgps2), mRNA | NM_023884 | 5.43465 | 0.0001 |
| Lao1 | L-amino acid oxidase 1 (Lao1), mRNA | NM_133892 | 5.43168 | 0.00322 |
| Hist1h2bm | Histone 1, H2bm (Hist1h2bm), mRNA | NM_178200 | 5.42961 | 1.25E-36 |
| Zfp422-rs1 | zinc finger protein 422, related sequence 1 | AK020746 | 5.42824 | 3.08E-44 |
| Spire1 | Spire homolog 1 (Drosophila), mRNA (cDNA clone MGC:106458 IMAGE:30535423) | NM_176832 | 5.42537 | 9.41E-07 |
| Matn3 | Matrilin 3 (Matn3), mRNA | NM_010770 | 5.42402 | 0.00001 |
| Zfp422-rs1 | zinc finger protein 422, related sequence 1 | NM_029952 | 5.42321 | 2.97E-17 |
| Pcdh10 | OL-protocadherin isoform (Pcadh10) mRNA, complete cds; alternatively spliced | AK122503 | 5.42181 | 5.94E-28 |
| NAP042178-1 | Unknown | 5.41306 | 8.92E-21 | |
| Ranbp6 | RAN binding protein 6 (Ranbp6), mRNA | AK083089 | 5.41102 | 0.00015 |
| Myh3 | PREDICTED: myosin, heavy polypeptide 3, skeletal muscle, embryonic [Mus musculus], mRNA sequence | XM_354614 | 5.39493 | 3.95E-11 |
| Vill | Villin-like (Vill), mRNA | U72681 | 5.39006 | 1.44E-17 |
| Ephb3 | Eph receptor B3, mRNA (cDNA clone MGC:18409 IMAGE:3673003) | NM_010143 | 5.38235 | 1.15E-08 |
| Xpr1 | Xenotropic and polytropic murine retrovirus receptor (Xpr1) | NM_011273 | 5.38141 | 1.81E-13 |
| Bank1 | B-cell scaffold protein with ankyrin repeats 1 (Bank1), mRNA | AK041242 | 5.37887 | 0.00416 |
| TC1066872 | S3A1_HUMAN Splicing factor 3 subunit 1 (Spliceosome associated protein 114) (SAP 114) (SF3a120). [Human] {Homo sapiens}, partial (3%) [TC1066872] | 5.3751 | 0.00537 | |
| Wdr37 | MKIAA0982 protein | AK044740 | 5.36656 | 0.00008 |
| Mtap2 | Microtubule-associated protein 2, mRNA (cDNA clone MGC:63400 IMAGE:6412797) | BC052446 | 5.36496 | 7.87E-18 |
| AB112350 | NAPE-PLD mRNA for N-acyl-phosphatidylethanolamine-hydrolyzing phospholipase D | NM_178728 | 5.36332 | 3.14E-16 |
| Malat1 | Adult male stomach cDNA, RIKEN full-length enriched library, clone:2210407K09 product: receptor (calcitonin) activity modifying protein 2, full insert sequence | AK020134 | 5.36328 | 6.17E-11 |
| AI464131 | MKIAA1161 protein | BC036141 | 5.36291 | 4.74E-15 |
| H1f0 | H1 histone family, member 0, mRNA (cDNA clone MGC:19309 IMAGE:4166167) | NM_008197 | 5.35386 | 2.93E-07 |
| 4933404M02Rik | RIKEN cDNA 4933404M02 gene (4933404M02Rik), mRNA | AK017113 | 5.35281 | 0.00005 |
| Adarb1 | Adenosine deaminase, RNA-specific, B1 (Adarb1), transcript variant 1, mRNA | AF403109 | 5.3483 | 1.65E-15 |
| Tcf7 | Transcription factor 7, T-cell specific (Tcf7), mRNA | NM_009331 | 5.34121 | 1.40E-08 |
| Strbp | Spermatid perinuclear RNA binding protein, mRNA (cDNA clone MGC:25337 IMAGE:4502840) | AK081780 | 5.33831 | 0.00004 |
| Foxj1 | Forkhead Box j1 (Foxj1) | NM_008240 | 5.33683 | 3.52E-08 |
| Vill | Villin-like (Vill), mRNA | NM_011700 | 5.33523 | 2.17E-10 |
| A_51_P170529 | Unknown | 5.32731 | 0.00598 | |
| 2310047O13Rik | RIKEN cDNA 2310047O13 gene, mRNA (cDNA clone MGC:28605 IMAGE:4217391) | AK083925 | 5.32579 | 5.06E-32 |
| Ccdc28a | Coiled-coil domain containing 28A (Ccdc28a), mRNA | NM_144820 | 5.32149 | 2.42E-10 |
| NAP057224-1 | Unknown | 5.32046 | 8.05E-06 | |
| Arl5b | ADP-ribosylation factor-like 8 (Arl8), mRNA | NM_029466 | 5.31107 | 0.00004 |
| Zfp40 | Zinc finger protein 40 (Zfp40), mRNA | NM_009555 | 5.30822 | 4.36E-10 |
| Thnsl1 | Threonine synthase-like 1 (bacterial), mRNA (cDNA clone IMAGE:5037172) | NM_177588 | 5.30806 | 7.21E-20 |
| 4932411G14Rik | RIKEN cDNA 4932411G14 gene (4932411G14Rik), mRNA | NM_177711 | 5.30751 | 3.12E-11 |
| Map3k14 | Mitogen-activated protein kinase kinase kinase 14 (Map3k14), mRNA | NM_016896 | 5.30704 | 5.21E-18 |
| 1700110N18Rik | PREDICTED: hypothetical protein LOC73569 [Mus musculus], mRNA sequence | XM_283372 | 5.3057 | 1.79E-09 |
| ENSMUST00000050440 | Unknown | 5.30328 | 1.74E-27 | |
| Rapgef6 | Rap guanine nucleotide exchange factor (GEF) 6, mRNA (cDNA clone IMAGE:5321951) | AK040089 | 5.30322 | 2.62E-11 |
| Arhgap26 | MKIAA0621 protein | AK129176 | 5.30199 | 0.00026 |
| Wnt9a | Wingless-type MMTV integration site 9A (Wnt9a), mRNA | BC066165 | 5.3003 | 0.00003 |
| Hist1h2ba | Histone 1, H2ba (Hist1h2ba), mRNA | NM_175663 | 5.29806 | 2.18E-27 |
| Lincr | lung-inducible neuralized-related C3HC4 RING domain protein | NM_153408 | 5.29767 | 1.15E-07 |
| B230105J10 | hypothetical protein B230105J10 | AK045359 | 5.29449 | 1.35E-06 |
| TC984274 | Unknown | 5.29254 | 2.02E-17 | |
| 5730553K21 | 5′-AMP-ACTIVATED PROTEIN KINASE, BETA-2 SUBUNIT (AMPK BETA-2 CHAIN) homolog [Rattus norvegicus] [5730553K21] | 5.29004 | 4.53E-13 | |
| Nek1 | PREDICTED: Mus musculus NIMA (never in mitosis gene a)-related expressed kinase 1 (Nek1), mRNA | AK049625 | 5.28832 | 1.68E-17 |
| AK086484 | Mus musculus 15 days embryo head cDNA, RIKEN full-length enriched library, clone:D930031J16 product: microtubule-associated protein 2, full insert sequence. | AK086484 | 5.28708 | 1.64E-12 |
| Rb1 | Retinoblastoma 1 (Rb1), mRNA | NM_009029 | 5.27516 | 1.41E-07 |
| Olfml3 | Olfactomedin-like 3 (Olfml3), mRNA | NM_133859 | 5.26799 | 3.38E-15 |
| Etv3 | ETS-domain transcriptional repressor PE1 (PE1) | NM_012051 | 5.26705 | 2.17E-29 |
| Ptpn13 | Protein tyrosine phosphatase, non-receptor type 13 (Ptpn13), mRNA | NM_011204 | 5.26698 | 1.95E-21 |
| Tfdp2 | Transcription factor Dp 2, mRNA (cDNA clone IMAGE:30604420) | NM_178667 | 5.26621 | 2.69E-06 |
| TC970961 | AF169692 protocadherin-9 {Homo sapiens}, partial (9%) [TC970961] | 5.2653 | 5.85E-08 | |
| Plekhk1 | Membrane-bound factor MBF1 (Mbf) | AY036116 | 5.25979 | 2.22E-16 |
| Tmcc3 | Transmembrane and coiled coil domains 3, mRNA (cDNA clone MGC:30586 IMAGE:3660220) | AK083129 | 5.25708 | 2.98E-07 |
| Prph1 | Peripherin 1 (Prph1), mRNA | NM_013639 | 5.25389 | 4.55E-08 |
| Tbc1d1 | TBC1 domain family, member 1, mRNA (cDNA clone IMAGE:3500261) | NM_019636 | 5.2511 | 2.50E-15 |
| Map3k1 | MAP kinase kinase kinase 1 (Mekk1) | NM_011945 | 5.25036 | 0.00004 |
| 1810053B23Rik | 10 day old male pancreas cDNA, RIKEN full-length enriched library, clone:1810053B23 product: unclassifiable, full insert sequence | AK007854 | 5.24933 | 0.00479 |
| Ppp1r2 | Protein phosphatase 1, regulatory (inhibitor) subunit 2 (Ppp1r2), mRNA | AK037804 | 5.2492 | 8.39E-28 |
| Rnf24 | Ring finger protein 24, mRNA (cDNA clone MGC:106607 IMAGE:6406985) | NM_178607 | 5.24766 | 3.12E-31 |
| Bhlhb8 | Basic helix-loop-helix domain containing, class B, 8, mRNA (cDNA clone MGC:19046 IMAGE:4189225) | NM_010800 | 5.24479 | 8.19E-09 |
| A230105L22Rik | Cytochrome P450, family 4, subfamily f, polypeptide 18, mRNA (cDNA clone MGC:19144 IMAGE:4218398) | AK020714 | 5.24224 | 0.00468 |
| Herc1 | Hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1, mRNA (cDNA clone MGC:7618 IMAGE:3494753) | BC004027 | 5.23497 | 5.02E-14 |
| Upk3b | Uroplakin 3B, mRNA (cDNA clone IMAGE:30297822) | NM_175309 | 5.22777 | 0.00453 |
| Pja2 | Praja 2, RING-H2 motif containing (Pja2), transcript variant 2, mRNA | AK122282 | 5.22623 | 2.64E-30 |
| Usp2 | Ubiquitin specific peptidase 2, mRNA (cDNA clone MGC:27630 IMAGE:4506362) | NM_016808 | 5.22394 | 9.53E-08 |
| Pak3 | P21-activated kinase 3 (pak3 gene) | NM_008778 | 5.22033 | 7.00E-06 |
| 0610008C08Rik | RIKEN cDNA 0610008C08 gene (0610008C08Rik), mRNA | AK029224 | 5.21977 | 0.00125 |
| 6330416L11Rik | RIKEN cDNA 6330416L11 gene (6330416L11Rik), mRNA | NM_027518 | 5.21346 | 3.01E-07 |
| Homer1 | Homer homolog 1 (Drosophila) (Homer1), transcript variant L, mRNA | AK035139 | 5.20561 | 0.00057 |
| Armc8 | Armadillo repeat containing 8, mRNA (cDNA clone MGC:47965 IMAGE:1230321) | AK078280 | 5.20529 | 6.30E-35 |
| Lipg | Lipase, endothelial, mRNA (cDNA clone MGC:13719 IMAGE:3981856) | NM_010720 | 5.20391 | 0.00001 |
| Dnahc2 | Axonemal dynein heavy chain (partial, ID mdhc5) | Z83813 | 5.20226 | 0.00081 |
| Cotl1 | Coactosin-like 1 (Dictyostelium) (Cotl1), mRNA | NM_028071 | 5.20139 | 3.70E-07 |
| 2700050L05Rik | RIKEN cDNA 2700050L05 gene (2700050L05Rik), transcript variant 1, mRNA | NM_145995 | 5.19216 | 5.70E-15 |
| Rab11fip4 | MKIAA1821 protein | AK122559 | 5.19174 | 0.00128 |
| Eml5 | Echinoderm microtubule associated protein like 5, mRNA (cDNA clone IMAGE:4953665) | AK047762 | 5.19141 | 0.00003 |
| Ptpn13 | Protein tyrosine phosphatase, non-receptor type 13 (Ptpn13), mRNA | NM_011204 | 5.19002 | 1.54E-26 |
| 3-Mar | Membrane-associated ring finger (C3HC4) 3 (March3), mRNA | NM_177115 | 5.18876 | 3.33E-22 |
| Synpo2 | Adult male urinary bladder cDNA, RIKEN full-length enriched library, clone:9530006G20 product: unclassifiable, full insert sequence | AK035258 | 5.18793 | 9.93E-06 |
| Zfp36 | Zinc finger protein 36, mRNA (cDNA clone MGC:29129 IMAGE:5053664) | NM_011756 | 5.18672 | 4.78E-06 |
| Epb4.1l5 | Erythrocyte protein band 4.1-like 5 (Epb4.1l5), mRNA | NM_145506 | 5.18616 | 1.68E-43 |
| Pip5k3 | Phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (Pip5k3), mRNA | NM_011086 | 5.18443 | 1.80E-10 |
| Tec | Cytoplasmic tyrosine kinase, Dscr28C related (Drosophila), mRNA (cDNA clone MGC:46873 IMAGE:5029430) | NM_013689 | 5.1779 | 7.22E-11 |
| C130065N10Rik | PREDICTED: hypothetical protein XP_488897 [Mus musculus], mRNA sequence | AK048435 | 5.17772 | 1.71E-43 |
| Pctk2 | PCTAIRE-motif protein kinase 2 (Pctk2), mRNA | NM_146239 | 5.17294 | 1.41E-12 |
| A130019P10Rik | Eukaryotic translation initiation factor 4E binding protein 2, mRNA (cDNA clone MGC:13950 IMAGE:4218820) | AK037456 | 5.17158 | 7.10E-27 |
| C920006C10Rik | RIKEN cDNA C920006C10 gene (C920006C10Rik), mRNA | AK052176 | 5.17083 | 1.37E-21 |
| Camkk1 | Calcium/calmodulin-dependent protein kinase kinase 1, alpha, mRNA (cDNA clone MGC:27706 IMAGE:4924656) | NM_018883 | 5.17034 | 0.0001 |
| Tusc2 | Tumor suppressor candidate 2 (Tusc2), mRNA | NM_019742 | 5.16901 | 1.18E-25 |
| AK048541 | Mus musculus 16 days embryo head cDNA, RIKEN full-length enriched library, clone:C130071D07 product: ATPase, Cu++ transporting, alpha polypeptide, full insert sequence. | AK048541 | 5.16848 | 1.68E-13 |
| Golph3 | Mus musculus 10 days neonate skin cDNA, RIKEN full-length enriched library, clone:4733401N08 product: golgi phosphoprotein 3, full insert sequence. | AK014644 | 5.16802 | 3.09E-33 |
| Fa2h | Fatty acid 2-hydroxylase, mRNA (cDNA clone IMAGE:4983638) | NM_178086 | 5.16055 | 2.45E-15 |
| Sesn3 | Sestrin 3 (Sesn3), mRNA | NM_030261 | 5.15972 | 4.03E-09 |
| D030022P07Rik | RIKEN cDNA 4930451A13 gene, mRNA (cDNA clone MGC:25849 IMAGE:4194266) | AK045169 | 5.15849 | 2.37E-16 |
| Spock2 | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan 2 (Spock2), mRNA | NM_052994 | 5.15198 | 5.72E-12 |
| Elavl3 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C) (Elavl3), mRNA | NM_010487 | 5.14832 | 3.61E-06 |
| LOC620630 | similar to Contactin 5 precursor (Neural recognition molecule NB-2) | XM_146640 | 5.14694 | 0.0002 |
| Plxna3 | Plexin A3 (Plxna3), mRNA | NM_008883 | 5.14348 | 1.78E-10 |
| 2200001I15Rik | RIKEN cDNA 2200001I15 gene (2200001I15Rik), mRNA | NM_183278 | 5.12983 | 9.01E-11 |
| Stk38l | Serine/threonine kinase 38 like, mRNA (cDNA clone MGC:70096 IMAGE:30135051) | NM_172734 | 5.12867 | 1.65E-13 |
| NAP092202-001 | Unknown | 5.12151 | 9.54E-32 | |
| Dock4 | MKIAA0716 protein | AK049953 | 5.10798 | 0.0001 |
| Nek1 | Mus musculus 13 days embryo male testis cDNA, RIKEN full-length enriched library, clone:6030407P11 product: NIMA (never in mitosis gene a)-related expressed kinase 1, full insert sequence. | AK031330 | 5.1072 | 1.46E-15 |
| Rgs2 | Regulator of G-protein signaling 2, mRNA (cDNA clone MGC:36187 IMAGE:5365271) | NM_009061 | 5.10596 | 4.25E-10 |
| Mapk8ip1 | JNK interacting protein-1b (JIP-1b) | NM_011162 | 5.10563 | 1.85E-29 |
| Pard6b | Par-6 (partitioning defective 6) homolog beta (C. elegans), mRNA (cDNA clone MGC:35871 IMAGE:2655658) | BC025147 | 5.1056 | 2.16E-21 |
| H1f0 | H1 histone family, member 0, mRNA (cDNA clone MGC:19309 IMAGE:4166167) | NM_008197 | 5.10476 | 2.71E-06 |
| Dmp1 | Dentin matrix protein 1 (Dmp1), mRNA | NM_016779 | 5.10461 | 0.0003 |
| Usf1 | M.musculus USF1 (non-coding exon 1). | X95315 | 5.1015 | 0.00002 |
| 2810453I06Rik | RIKEN cDNA 2810453I06 gene (2810453I06Rik), mRNA | NM_026050 | 5.08664 | 2.86E-23 |
| Ndrg3 | N-myc downstream regulated gene 3, mRNA (cDNA clone MGC:21649 IMAGE:4500970) | NM_013865 | 5.08321 | 3.05E-26 |
| 2810013E07Rik | RIKEN cDNA 2810013E07 gene, mRNA (cDNA clone MGC:67555 IMAGE:6402150) | AK036255 | 5.08237 | 0.0059 |
| 6330406L22Rik | RIKEN cDNA 6330406L22 gene, mRNA (cDNA clone MGC:59510 IMAGE:6333971) | AK041380 | 5.07511 | 0.00009 |
| Tubgcp5 | Mus musculus 12 days embryo embryonic body between diaphragm region and neck cDNA, RIKEN full-length enriched library, clone:9430028E06 product: unclassifiable, full insert sequence. | AK034715 | 5.07129 | 0.00549 |
| AK037544 | Mus musculus 16 days neonate thymus cDNA, RIKEN full-length enriched library, clone:A130024K02 product: hypothetical Proteinase inhibititor I4, serpin containing protein, full insert sequence. | AK037544 | 5.07051 | 5.67E-11 |
| Creb3l3 | CAMP responsive element binding protein 3-like 3 (Creb3l3), mRNA | NM_145365 | 5.06499 | 2.30E-13 |
| Mfap3l | Microfibrillar-associated protein 3-like, mRNA (cDNA clone MGC:106286 IMAGE:5025281) | AK122332 | 5.0638 | 0.00006 |
| Ddah2 | Dimethylarginine dimethylaminohydrolase 2, mRNA (cDNA clone MGC:5866 IMAGE:3158175) | NM_016765 | 5.06334 | 6.63E-30 |
| Il1rap | Interleukin 1 receptor accessory protein (Il1rap), transcript variant 2, mRNA | NM_134103 | 5.06296 | 4.72E-21 |
| Cox7a1 | Cytochrome c oxidase, subunit VIIa 1 (Cox7a1), mRNA | NM_009944 | 5.06261 | 3.37E-27 |
| Unc13a | Glycosyltransferase 25 domain containing 1 (Glt25d1), mRNA | AK083196 | 5.06174 | 6.78E-08 |
| Mdm1 | Transformed mouse 3T3 cell double minute 1 (Mdm1), transcript variant 2, mRNA | NM_148922 | 5.0613 | 3.82E-13 |
| Flywch1 | FLYWCH-type zinc finger 1 (Flywch1), mRNA | BC025645 | 5.06097 | 8.55E-16 |
| 2410116G06Rik | RIKEN cDNA 2410116G06 gene (2410116G06Rik), mRNA | NM_026630 | 5.05623 | 7.14E-06 |
| AK087708 | Mus musculus 2 days pregnant adult female ovary cDNA, RIKEN full-length enriched library, clone:E330009M13 product: unclassifiable, full insert sequence. | AK087708 | 5.05545 | 0.00228 |
| Hist1h2bk | Histone 1, H2bk (Hist1h2bk), mRNA | NM_175665 | 5.0523 | 1.47E-33 |
| Dffa | DNA fragmentation factor, alpha subunit (Dffa), transcript variant 2, mRNA | NM_010044 | 5.05211 | 4.21E-08 |
| Gpr155 | G protein-coupled receptor 155, mRNA (cDNA clone IMAGE:4167684) | AY255560 | 5.05096 | 5.00E-13 |
| Cntn3 | Contactin 3 (Cntn3), mRNA | NM_008779 | 5.05051 | 8.40E-08 |
| Trpc6 | Transient receptor potential cation channel, subfamily C, member 6 (Trpc6), mRNA | NM_013838 | 5.04168 | 0.00005 |
| Xpr1 | Xenotropic and polytropic murine retrovirus receptor (Xpr1) | AK047436 | 5.0407 | 5.27E-29 |
| BC052046 | CDNA sequence BC052046 (BC052046), mRNA | NM_183177 | 5.03591 | 1.47E-06 |
| Scnn1g | Sodium channel, nonvoltage-gated 1 gamma, mRNA (cDNA clone MGC:29058 IMAGE:5042876) | NM_011326 | 5.0338 | 0.00021 |
| Rab11fip1 | MFLJ00294 protein | AK008412 | 5.03263 | 6.56E-16 |
| Zfpm1 | Zinc finger protein, multitype 1 (Zfpm1), mRNA | NM_009569 | 5.02713 | 4.89E-09 |
| Serpinb1c | Serine (or cysteine) peptidase inhibitor, clade B, member 1c (Serpinb1c), mRNA | NM_173051 | 5.02704 | 1.26E-06 |
| Ablim3 | Actin binding LIM protein family, member 3, mRNA (cDNA clone MGC:106450 IMAGE:30534236) | NM_198649 | 5.01635 | 0.00514 |
| Hic2 | Hypermethylated in cancer 2 protein (Hic2) | NM_178922 | 5.01624 | 5.56E-27 |
| Vkorc1 | Vitamin K epoxide reductase complex, subunit 1, mRNA (cDNA clone MGC:25747 IMAGE:3991412) | NM_178600 | 5.01352 | 1.91E-06 |
| 3-Mar | Membrane-associated ring finger (C3HC4) 3 (March3), mRNA | AK018160 | 5.01164 | 0.00268 |
| Kcnh1 | Potassium voltage-gated channel, subfamily H (eag-related), member 1 (Kcnh1), mRNA | NM_010600 | 5.00508 | 2.36E-08 |
| Rab37 | RAB37, member of RAS oncogene family (Rab37), mRNA | NM_021411 | 5.00277 | 1.08E-09 |
| Hist3h2a | Histone 3, H2a, mRNA (cDNA clone MGC:70265 IMAGE:6493838) | NM_178218 | 5.00243 | 0.00002 |
| Atp6v1b2 | ATPase, H+ transporting, V1 subunit B, isoform 2, mRNA (cDNA clone MGC:21587 IMAGE:4500843) | BB633273 | 5.00183 | 2.44E-07 |
Table V.
Confirmed/Putative TSGs identified in our screen that contain CpG islands in both human and mouse forms of the gene.
| Gene | CpG Island M/H |
RT-PCR* | MSP** M/H |
|---|---|---|---|
| Tspan33 | +/+ | + | −/− |
| Cabc1 | +/+ | + | −/− |
| Dusp2 | +/+ | + | +/+ |
| Nlk | +/+ | + | −/− |
| Timp3 | +/+ | + | +/+ |
| Dbccr1 | +/+ | − | −/− |
| Klf2 | +/+ | − | −/− |
| Dock4 | +/+ | − | −/− |
| Cacna1g | +/+ | + | −/− |
| Atp1b2 | +/+ | + | +/+ |
| Egr1 | +/+ | − | −/− |
| Smpd3 | +/+ | + | +/+ |
| Akap12 | +/+ | − | −/− |
| Rprm | +/+ | + | +/+ |
| Zfp185 | +/+ | − | −/− |
| Cry2 | +/+ | − | −/− |
| Scrib | +/+ | − | −/− |
| Btg2 | +/+ | − | −/− |
| Foxj1 | +/+ | + | +/+ |
| Sesn3 | +/+ | − | −/− |
| Kremen1 | +/+ | + | −/− |
| Bach2 | +/+ | + | −/− |
| Rb1 | +/+ | − | −/− |
| Ddit3 | +/+ | − | −/− |
| Irf6 | +/+ | − | −/− |
| Bcl2l11 | +/+ | − | −/− |
| Tusc2 | +/+ | ND | ND |
| Tfdp2 | +/+ | ND | ND |
| Ptpn13 | +/+ | ND | ND |
| Ephb3 | +/+ | ND | ND |
| Pcdh10 | +/+ | ND | ND |
| Eaf2 | +/+ | ND | ND |
| Sema3b | +/+ | ND | ND |
| Lrp1b | +/+ | ND | ND |
| Rad9b | +/+ | ND | ND |
| Cdh15 | +/+ | ND | ND |
| Dapk2 | +/+ | ND | ND |
M: mouse, H: human
ND: not done
For RT-PCR assays, + = increased gene expression in 5-azadC/TSA treated EMT6 cells
For MSP assays, + = gene methylation in any mouse mammary or human breast tumor line studied
Acknowledgments
The authors would like to thank Drs. Lingbao Ai and Wan-Ju Kim for technical assistance and advise during the completion of this study and Dr. Martha Campbell-Thompson for supplying breast tumors.
Supported, in part, by a grant from the NIH (R21 CA102220) to KDB.
References
- Ai L, Kim WJ, Kim TY, Fields CR, Massoll NA, Robertson KD, Brown KD. Epigenetic Silencing of the Tumor Suppressor Cystatin M Occurs during Breast Cancer Progression. Cancer Res. 2006;66:7899–7909. doi: 10.1158/0008-5472.CAN-06-0576. [DOI] [PubMed] [Google Scholar]
- Alesse E, Zazzeroni F, Angelucci A, Giannini G, Di Marcotullio L, Gulino A. The growth arrest and downregulation of c-myc transcription induced by ceramide are related events dependent on p21 induction, Rb underphosphorylation and E2F sequestering. Cell Death Differ. 1998;5:381–9. doi: 10.1038/sj.cdd.4400358. [DOI] [PubMed] [Google Scholar]
- Antonicek H, Persohn E, Schachner M. Biochemical and functional characterization of a novel neuron-glia adhesion molecule that is involved in neuronal migration. J Cell Biol. 1987;104:1587–95. doi: 10.1083/jcb.104.6.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bishop DT. BRCA1 and BRCA2 and breast cancer incidence: a review. Ann Oncol. 1999;10(Suppl 6):113–9. [PubMed] [Google Scholar]
- Chalmers IJ, Aubele M, Hartmann E, Braungart E, Werner M, Hofler H, Atkinson MJ. Mapping the chromosome 16 cadherin gene cluster to a minimal deleted region in ductal breast cancer. Cancer Genet Cytogenet. 2001;126:39–44. doi: 10.1016/s0165-4608(00)00376-9. [DOI] [PubMed] [Google Scholar]
- Chen F. Endogenous inhibitors of nuclear factor-kappaB, an opportunity for cancer control. Cancer Res. 2004;64:8135–8. doi: 10.1158/0008-5472.CAN-04-2096. [DOI] [PubMed] [Google Scholar]
- Chu Y, Solski PA, Khosravi-Far R, Der CJ, Kelly K. The mitogen-activated protein kinase phosphatases PAC1, MKP-1, and MKP-2 have unique substrate specificities and reduced activity in vivo toward the ERK2 sevenmaker mutation. J Biol Chem. 1996;271:6497–501. doi: 10.1074/jbc.271.11.6497. [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- Dobretsov M, Stimers JR. Neuronal function and alpha3 isoform of the Na/K-ATPase. Front Biosci. 2005;10:2373–96. doi: 10.2741/1704. [DOI] [PubMed] [Google Scholar]
- Dobrovic A, Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57:3347–50. [PubMed] [Google Scholar]
- Driouch K, Dorion-Bonnet F, Briffod M, Champeme MH, Longy M, Lidereau R. Loss of heterozygosity on chromosome arm 16q in breast cancer metastases. Genes Chromosomes Cancer. 1997;19:185–91. doi: 10.1002/(sici)1098-2264(199707)19:3<185::aid-gcc8>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–98. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, Bonilla F, Matias-Guiu X, Lerma E, Bussaglia E, Prat J, Harkes IC, Repasky EA, Gabrielson E, Schutte M, Baylin SB, Herman JG. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92:564–9. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR, Sukumar S. High frequency of hypermethylation at the 14–3-3 sigma locus leads to gene silencing in breast cancer. Proc Natl Acad Sci U S A. 2000;97:6049–54. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gloor S, Antonicek H, Sweadner KJ, Pagliusi S, Frank R, Moos M, Schachner M. The adhesion molecule on glia (AMOG) is a homologue of the beta subunit of the Na, K-ATPase. J Cell Biol. 1990;110:165–74. doi: 10.1083/jcb.110.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst CR, Nuovo GJ, Esteller M, Chew K, Baylin SB, Herman JG, Tlsty TD. Methylation of p16(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–601. [PubMed] [Google Scholar]
- Hong C, Maunakea A, Jun P, Bollen AW, Hodgson JG, Goldenberg DD, Weiss WA, Costello JF. Shared epigenetic mechanisms in human and mouse gliomas inactivate expression of the growth suppressor SLC5A8. Cancer Res. 2005;65:3617–23. doi: 10.1158/0008-5472.CAN-05-0048. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–500. [PubMed] [Google Scholar]
- Kim YH, Petko Z, Dzieciatkowski S, Lin L, Ghiassi M, Stain S, Chapman WC, Washington MK, Willis J, Markowitz SD, et al. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45:781–9. doi: 10.1002/gcc.20341. [DOI] [PubMed] [Google Scholar]
- Lin L, Spoor MS, Gerth AJ, Brody SL, Peng SL. Modulation of Th1 activation and inflammation by the NF-kappaB repressor Foxj1. Science. 2004;303:1017–20. doi: 10.1126/science.1093889. [DOI] [PubMed] [Google Scholar]
- Lu T, Stark GR. Cytokine overexpression and constitutive NFkappaB in cancer. Cell Cycle. 2004;3:1114–7. [PubMed] [Google Scholar]
- Luberto C, Hassler DF, Signorelli P, Okamoto Y, Sawai H, Boros E, Hazen-Martin DJ, Obeid LM, Hannun YA, Smith GK. Inhibition of tumor necrosis factor-induced cell death in MCF7 by a novel inhibitor of neutral sphingomyelinase. J Biol Chem. 2002;277:41128–39. doi: 10.1074/jbc.M206747200. [DOI] [PubMed] [Google Scholar]
- Nass SJ, Herman JG, Gabrielson E, Iversen PW, Parl FF, Davidson NE, Graff JR. Aberrant methylation of the estrogen receptor and E-cadherin 5′ CpG islands increases with malignant progression in human breast cancer. Cancer Res. 2000;60:4346–8. [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Senner V, Schmidtpeter S, Braune S, Puttmann S, Thanos S, Bartsch U, Schachner M, Paulus W. AMOG/beta2 and glioma invasion: does loss of AMOG make tumour cells run amok? Neuropathol Appl Neurobiol. 2003;29:370–7. doi: 10.1046/j.1365-2990.2003.00473.x. [DOI] [PubMed] [Google Scholar]
- Stoffel W, Jenke B, Holz B, Binczek E, Gunter RH, Knifka J, Koebke J, Niehoff A. Neutral sphingomyelinase (SMPD3) deficiency causes a novel form of chondrodysplasia and dwarfism that is rescued by Col2A1-driven smpd3 transgene expression. Am J Pathol. 2007;171:153–61. doi: 10.2353/ajpath.2007.061285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M, Timmerman MM, Brody LC, Tucker MA. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Engl J Med. 1997;336:1401–8. doi: 10.1056/NEJM199705153362001. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Suzuki M, Shigematsu H, Shivapurkar N, Echebiri C, Nomura M, Stastny V, Augustus M, Wu CW, Wistuba II, et al. Aberrant methylation of Reprimo in human malignancies. Int J Cancer. 2005;115:503–10. doi: 10.1002/ijc.20910. [DOI] [PubMed] [Google Scholar]
- Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003;35:152–6. doi: 10.2144/03351md02. [DOI] [PubMed] [Google Scholar]
- van den Boom J, Wolter M, Blaschke B, Knobbe CB, Reifenberger G. Identification of novel genes associated with astrocytoma progression using suppression subtractive hybridization and real-time reverse transcription-polymerase chain reaction. Int J Cancer. 2006;119:2330–8. doi: 10.1002/ijc.22108. [DOI] [PubMed] [Google Scholar]
- Vo QN, Kim WJ, Cvitanovic L, Boudreau DA, Ginzinger DG, Brown KD. The ATM gene is a target for epigenetic silencing in locally advanced breast cancer. Oncogene. 2004;23:9432–7. doi: 10.1038/sj.onc.1208092. [DOI] [PubMed] [Google Scholar]
- Weinberg RA. How cancer arises. Sci Am. 1996;275:62–70. doi: 10.1038/scientificamerican0996-62. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Chromatin remodeling: why it is important in cancer. Oncogene. 2001;20:2988–90. doi: 10.1038/sj.onc.1204322. [DOI] [PubMed] [Google Scholar]
- Wu P, Qi B, Zhu H, Zheng Y, Li F, Chen J. Suppression of staurosporine-mediated apoptosis in Hs578T breast cells through inhibition of neutral-sphingomyelinase by caveolin-1. Cancer Lett. 2007;256:64–72. doi: 10.1016/j.canlet.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Yin Y, Liu YX, Jin YJ, Hall EJ, Barrett JC. PAC1 phosphatase is a transcription target of p53 in signalling apoptosis and growth suppression. Nature. 2003;422:527–31. doi: 10.1038/nature01519. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, Raval A, Liu TH, Ding W, Mao C, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005a;37:265–74. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- Yu YP, Paranjpe S, Nelson J, Finkelstein S, Ren B, Kokkinakis D, Michalopoulos G, Luo JH. High throughput screening of methylation status of genes in prostate cancer using an oligonucleotide methylation array. Carcinogenesis. 2005b;26:471–9. doi: 10.1093/carcin/bgh310. [DOI] [PubMed] [Google Scholar]