Abstract
The focus of this paper is the hotdog-fold thioesterase THEM2 from human (hTHEM2; Swiss-Prot entry Q9NPJ3). In an earlier communication (Cheng, Z., Song, F., Shan, X., Wei, Z., Wang, Y., Dunaway-Mariano, D., and Gong, W. (2006) Crystal structure of human thioesterase superfamily member 2, Biochem Biophys Res Commun 349, 172-177.) we reported the apo crystal structure of hTHEM2. Herein, we report the results of an extensive hTHEM2 substrate screen, the structure determination of hTHEM2 complexed with the inert substrate analog undecan-2-one-CoA (in which O=C-CH2-S substitutes for O=C-S) and the kinetic analysis of active site mutants. The work described in this paper represents the first reported structure-function based analysis of a human hotdog-fold thioesterase. The catalytic mechanism proposed involves the Asp65/Ser83 assisted attack of a water molecule at the Gly57/Asn50 polarized thioester C=O and the Asn50 assisted departure of the thiolate leaving group. Thioesterase activity was observed with acyl-CoAs but not with the human acyl-ACP or with an acyl-Cys peptide. The medium-to-long chain fatty acyl-CoAs displayed the smallest Km values. The substrate specificity profile was analyzed within the context of the liganded enzyme to define the structural determinants of substrate recognition. Based on the results of this structure-function analysis we hypothesize that the physiological role of hHTEM2 involves catalysis of the hydrolysis of cytosolic medium-to-long chain acyl-CoA thioesters.
Keywords: human, thioesterase, acyl-coenzyme A, X-ray structure, hotdog fold enzyme superfamily, general base catalysis, thioester hydrolysis, lipid metabolism, fatty acid, catalytic mechanism, catalytic scaffold, structure-function, biological function, substrate specificity, substrate range, THEM2, hTHEM2
The focus of this paper is the single domain acyl-CoA hotdog fold thioesterase THEM2 from human (hTHEM2; Swiss-Prot entry Q9NPJ3). hTHEM2 occurrence is most predominant in the kidney (1, 2) and although its physiological function is not known, studies have shown that hTHEM2 is regulated by the hepatocyte nuclear factor 4alpha (3), which in turn might play an important role in renal cell carcinoma (4). In this investigation we use a structure-function based approach to characterize hTHEM2 as a catalyst for thioester hydrolysis.
The hTHEM2 three-dimensional structure and thioesterase activity are traits of the hotdog-fold thioesterase family. The structural unit of this family is a monomer that consists of a five or six-stranded β-sheet that wraps around a long α-helix, reminiscent of a hotdog. The association of two subunits generates an elongated β-sheet, along which the two α-helices run anti-parallel to one another. Each dimer contains two substrate-binding sites, located at the respective ends of the subunit interface. Two clades are distinguished by “back-to-back” (sheet-to-sheet) (prototype 4-hydroxybenzoyl-CoA (4-HBA-CoA) thioesterase from Arthrobacter sp strain SU (5); Figure 1A) vs. “face-to-face” (helix-to-helix) (prototype 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp strain CBS3(6); Figure 1B) dimer-dimer association.
Figure 1.
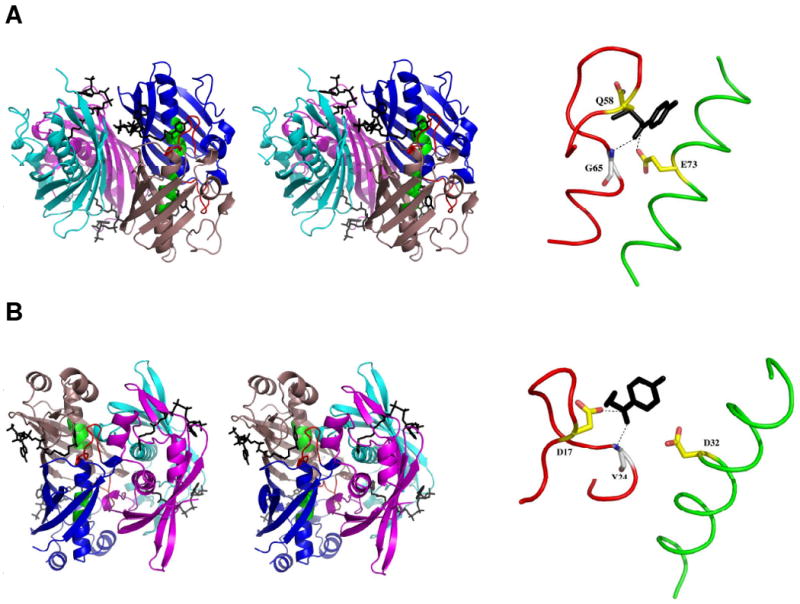
(A) The Pymol generated stereoview of the back-to-back tetramer of Arthrobacter 4-HBA-CoA thioesterase (5) (left) and the dissected catalytic scaffold (right). (B) The Pymol generated stereoview of the front-to-front tetramer of Pseudomonas 4-HBA-CoA thioesterase (6) (left) and the dissected catalytic scaffold (right). In both figures the subunits are colored individually to show the dimer units (blue and salmon; cyan and magenta). The 4-hydroxy-phenyacyl ligands are shown in black. The catalytic scaffold of the blue (protomer I) and salmon (protomer II) colored dimer is shown in red (the loop leading to the N-terminus of the polarizing α-helix) and green (the midsection of the polarizing α-helix). The catalytic scaffolds are shown in dissection to left in order to highlight the alignment of the catalytic carboxylate residue (yellow carbon atoms, red oxygen atoms) with the C=O carbon atom of the trunctated ligand (black) and the backbone amide NH of the helix N-terminal residue (white carbon atoms; blue nitrogen and red oxygen atoms) with the ligand C=O oxygen atom.
The catalytic scaffold of the hotdog-fold thioesterase is formed by the N-terminus of the elongated α-helix and its connecting loop from one subunit (protomer I), and the center segment of the elongated α-helix on the opposing subunit (protomer II). In Figure 1, these two segments are colored red and green, respectively. The backbone amide NH of the N-terminal residue of the α-helix in protomer I engages in hydrogen-bond interaction with the substrate thioester C=O, thereby orienting and polarizing the C=O for nucleophilic attack (see Figure 1). In the Pseudomonas 4-HBA-CoA thioesterase clade the catalytic carboxylate residue is positioned on this same segment (Figure 1B) whereas in the Arthrobacter 4-HBA-CoA thioesterase clade it is positioned on the α-helix of protomer II (Figure 1A).
In an earlier communication (7) we reported the apo crystal structure of hTHEM2, which revealed that it is a member of the Arthrobacter 4-HBA-CoA thioesterase clade. Limited activity screening showed that hTHEM2 possesses acyl-CoA thioesterase activity (6), however the identity of its physiological substrate and the mechanism by which it catalyzes the hydrolysis of this substrate remained to be defined. Herein, we report the findings from a structure-function analysis which integrates the results obtained from an extensive hTHEM2 substrate screen, structure determination of hTHEM2 complexed with the inert substrate analog undecan-2-one-CoA (in which O=C-CH2-S substitutes for O=C-S) and kinetic analysis of hTHEM2 site-directed mutants.
Experimental
Commercial Materials
All restriction enzymes and T4 DNA ligase were purchased from Invitrogen (Carlsbad, CA). Pfu Turbo DNA polymerase was purchased from Strategene (Cedar Creek, TX). Oligonucleotide primers were custom-synthesized by Invitrogen. Competent Escherichia coli cells (BL21-DE3) were obtained from Strategene and the pET22b vector was purchased from QIAGEN (Valencio, CA). DNA sequence determination was performed by the DNA Sequencing Facility of the University of New Mexico. Acetyl-CoA, 2-butenoyl-CoA, n-propionyl-CoA, hexanoyl-CoA, methylmalnoyl-CoA, ß-methylcrotonyl-CoA, tiglyl-CoA, linoleoyl-CoA, n-butyryl-CoA, ß-hydroxybutyryl-CoA, n-octanoyl-CoA, n-palmitoyl-CoA, glutaryl-CoA, malonyl-CoA, crotonyl-CoA, DL-3-hydroxy-3-methylglutaryl-CoA, palmitoleoyl-CoA, myristoyl-CoA, decanoyl-CoA, oleoyl-CoA, lauroyl-CoA, stearoyl-CoA, phenylacetyl-CoA and arachidonoyl-CoA were purchased from Sigma-Aldrich (St. Louis, MO). The tripeptide NAc-Gly-Ala-Cys-S-palmitoyl was custom synthesized by EZBiolab (Westfield, IN). All other chemicals were purchased from Sigma-Aldrich except where noted.
Synthetic Substrates and Inhibitors
3-Hydroxyphenylacetyl-CoA, 3,4-dihydroxyphenylacetyl-CoA, 3,5-dihroxyphenacetyl-CoA, 4-hydroxyphenylacetyl-CoA, 3-hydroxybenzoyl-CoA, 4-chlorobenzoyl-CoA, 4-hydroxybenzoyl-CoA, 4-hydroxybenzyl-CoA, 4-hydroxyphenylacyl-CoA, phenylacetylmethyl-CoA were synthesized as previously reported. (6, 8-11)
Undecan-2-one-CoA was synthesized from bromoundecan-2-one (12) and CoA as follows. Bromoundecan-2-one (108 mg) was dissolved in ethanol (3 ml) and benzene (400 μl) and then added dropwise to a stirred solution of 68 mg of coenzyme A lithium salt in 3.5 ml deionized H2O. The mixture was stirred for 16 h while maintaining the pH between 7 and 8 by the addition of 1.0 M LiOH in deionized water. The mixture was extracted three times with ethyl acetate. The aqueous fraction was concentrated by lyophilization and then chromatographed on a Sephadex-G15 (Amersham Pharmacia) column with deionized water serving as the eluant. The column factions were analyzed by reversed phase HPLC, and the desired fractions were combined to give pure undecan-2-one in 13 % yield. MW calculated: 935.2670; observed:[M-] 934.2667. 1H-NMR (D2O solvent) ppm: 0.86 (s, 3H), 0.89 (t, 3H), 0.99 (s, 3H), 1.27 (m, 14H), 1.58 (m, 2H), 2.68 (m, 3H), 3.24 (q, 1H), 3.38 (t, 2H), 3.57 (m, 3H), 3.64 (m, 1H), 3.91 (m, 1H), 4.09 (s, 1H), 4.31 (s, 1H), 4.66 (s, 1H), 6.28 (d, 1H), 8.47 (s, 1H), 8.72 (s, 1H).
Holo-acyl carrier proteins (ACP) were synthesized by reacting the apo human cytosolic ACP with acyl-CoAs catalyzed by the human phosphopantetheinyl transferase (PPTase). Human cytosolic ACP fragment and PPTase plasmids were a kind gift from Dr. Stuart Smith, Children's Hospital Oakland Research Institute, Oakland, CA. The two recombinant proteins were purified as described earlier (13, 14). The reaction mixture contained 60 μM apo-ACP, 150 μM acyl-CoA and 2 μM PPTase in 20 ml of 20 mM Tris/1mM MgCl2 (pH 7.0; 25 °C) for 3 h. The concentrated acyl-ACP was prepared using a PALL 10K centrifuge A device and the structure was verified by ES-MS. Oleoyl-ACP calculated: 13673, observed [M+] 13673, lauroyl-ACP calculated: 13597, observed [M+] 13598, octanoyl-ACP calculated: 13534, observed [M+] 13534, palmitoyl-ACP calculated: 13646, observed: [M+] 13647, palmitoeyl-ACP calculated: 13644, observed: [M+] 13645, myristoyl-ACP calculated: 13618, observed: [M+] 13619, steroyl-ACP calculated: 13678, observed: [M+] 13679.
Preparation of recombinant wild-type and mutant hTHEM
The C-terminal His6-tagged wild-type hTHEM2 (Swiss-Prot entry Q9NPJ3) was prepared as previously described (7). Site directed mutagenesis was carried out using a PCR-based strategy with the WT-hTHEM2/pET-22b(+) plasmid as template, commercial primers (Invitrogen), the PCR kit supplied by Stratagene, and the Techgene thermal cycler manufactured by TECHNE (Princeton, NJ). The PCR products were used to transform competent E. coli BL21(DE3) cells and the plasmid was prepared using a QIAprep Spin Miniprep Kit (Qiagen). The sequence of the mutated gene was confirmed by DNA sequencing carried out by the Health Sciences Center of the University of New Mexico. The hTHEM2 mutants were purified to homogeneity (as shown by SDS–PAGE analysis) in a yield of ∼ 4 mg protein/gm wet cells by the same procedure (7) used to purify the wild-type enzyme.
Enzymatic assays
Steady-state kinetic methods were used to determine the kcat and Km for wild-type and mutant hTHEM2. The thioesterase activity was monitored at 412 nm (Δε = 13.6 mM-1 cm-1) by coupling the reaction of 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB) with the CoA liberated from the acyl-CoA substrate. All reactions were carried out in 50 mM K+HEPES containing 1 mM DTNB at pH 7.5 and 25 °C.
The initial velocity data, measured as a function of substrate concentration, were analyzed using Eq. (1) and the computer program KinetAsyst (IntelliKinetics, PA).
| (Eq.1) |
V, initial velocity; Vmax, maximum velocity; [S], substrate concentration; and Km, Michaelis constant. The kcat was calculated from Vmax/[E] where [E] is the total enzyme concentration determined using the Bradford method (15).
The inhibition constant Ki for competitive inhibition was obtained by fitting the initial velocities of reactions of 3-hydroxyphenyacetyl-CoA (at varied concentration) in the absence or presence of inhibitor (at 1 × Ki and 2 × Ki concentration) to equation 2,
| (Eq.2) |
Where [I] is the concentration of the inhibitor and Ki is the inhibition constant.
The hTHEM2 catalyzed reactions of the acyl-ACPs were monitored by ES-MS (palmitoeyl-ACP and myristoyl-ACP) or by the DTNB based spectrophotometric assay. Reaction solutions monitored at 412 nm initially contained 2 μM hHTEM2, 5-100 μM acyl-ACP, 1 mM DTNB in 50 mM K+HEPES at pH 7.5 and 25 °C. Reaction solutions analyzed by ES-MS following incubation for 60 min initially contained 10 μM hHTEM2 and 80 μM palmitoeyl-ACP or myristoyl-ACP in 50 mM K+HEPES at pH 7.5 and 25 °C. The palmitoeyl-ACP ([M+] 13645 Da) or myristoyl-ACP ([M+] 13619 Da) signal observed in the control solution (lacking hHTEM2) were also present the spectra measured for reaction solutions. The signal for the product ACP on the other hand, was not observed.
The hTHEM2 catalyzed reaction of the tripeptide NAc-Gly-Ala-Cys-S-palmitoyl was monitored at 412 nm for 60 min using a reaction solution initially containing 10 μM hTHEM2 and 100 μM NAc-Gly-Ala-Cys-S-palmitoyl in 5 mM K+HEPES at pH 7.5 and 25 °C. In addition, ES-MS mass spec was used to analyze the reaction mixture initially containing 10 μM hHTEM2 and 100 μM NAc-Gly-Ala-Cys-S-palmitoyl in 5 mM K+HEPES at pH 7.5 and 25 °C following a 60 min incubation period. The ([M+Na+] = 551 and [M+K+] = 567 Da) signals observed for the acylated peptide in the control reaction (lacking hHTEM2) were also observed in the spectrum of the reaction solution, however the signals ([M+Na+] = 312 and [M+K+] = 328 Da) for the deacylated peptide were not observed.
hTHEM2 Crystallization and Structure Determination
The hTHEM2(undecan-2-one-CoA) complex was crystallized by using the hanging drop vapor diffusion method. The drops were set up by mixing 1 μl of solution containing 10 mg/ml hTHEM2, 2 mM undecan-2-one-CoA in 20 mM Tris (pH 7.5), 50 mM NaCl and 10 mM DTT and 1 μl of solution containing 15% PEG3350, 0.1 M ammonium citrate, dibasic, and 5-10% glycerol. A cloudy mixture initially formed, however after 1 wk crystals having the typical dimensions of 0.3 × 0.3 × 0.3 mm3 were observed. The crystals were removed using nylon cryo-loops and immediately flash-frozen at 100K in a stream of nitrogen gas (the glycerol served as the cryoprotectant). A 1.7 Å data set was collected on a FR-E+ Cu rotating-anode generator (Rigaku, Japan), equipped with a R-AXIS IV++ imaging-plate detector. The data set was processed with HKL2000 (16). In order to determine the structure of hTHEM2(undecan-2-one-CoA) complex, molecular replacement was performed by MOLREP (17), using the tetrameric hTHEM2 structure (PDB accession ID 2F0X) as the search model. After manual adjustment in Coot (18), the structure was refined using isotropic B-factor refinement in the REFMAC5 (19) to 1.7 Å with Rwork of 19.8% and Rfree of 23.6%. The statistics of data reduction and structure refinement are summarized in Table 1. The stereochemical quality of the final model was examined by PROCHECK (20). The existence of 7 chloride ions was confirmed by an anomalous difference Fourier map constructed using the anomalous difference signal and phases calculated from protein only. The densities corresponding to the chloride ions and undecan-2-one-CoA sulfur atoms were observed in the map, as these two atoms have similar anomalous signals at the wavelength of Cu K-alpha radiation (f″-Cl = 0.7e, f″-S = 0.6e). The fo-fc omit map was constructed using phases calculated based on the hTHEM2(undecan-2-one-CoA) complex structure, in the absence of undecan-2-one ligand M, chloride ion, water ligand, using FFT (21). The decanoly-CoA ligand was modeled in place of the undecan-2-one ligand M and chloride ion by using Coot (18).
Table 1.
Statistics of data reduction and structure refinement.
| Data collection statistics | |
| Space group | P212121 |
| Unit cell (Å) | a=91.0, b=110.6, c=119.8 |
| Resolution range (Å) | 50.00-1.7 (1.76-1.7)a |
| No. of unique reflections | 134034 |
| Average redundancy | 4.5 (4.2) |
| Completeness | 99.1(99.2) |
| Rmerge(%)b | 5.0(50.2) |
| I/σ (I) | 23.1(2.7) |
| Refinement statistics | |
| Rcryst/ Rfree (%)c | 19.8/23.6 |
| R.m.s.d Bond length (Å)d | 0.015 |
| R.m.s.d Bond angle (°) | 2.0 |
| Number of protein residues | 1098 |
| Number of H2O molecules | 863 |
| Number of PEG fragments | 4 |
| Number of Cl- | 7 |
| Average temperature factor (Å2) | |
| Protein main-chain atoms | 25.0 |
| Protein side-chains atoms | 26.9 |
| Inhibitor atoms | 43.0 |
| PEG stoms | 66.0 |
| Cl ions | 31.0 |
| Water molecules | 40.6 |
Data for the highest resolution bin is in parentheses.
Rmerge = Σ|Ii - Im|/ΣIi, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
Rcryst = Σ‖Fobs| - |Fcalc‖/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factors.
Rfree = ΣT‖Fobs| - |Fcalc‖/ΣT|Fobs|, where T is a test data set of about 10% of the total reflections randomly chosen and set aside prior to refinement.
R.m.s.d = Root-mean-square deviations
The atomic coordinates of hTHEM2(undecan-2-one-CoA) complex have been deposited into the Protein Data Bank (PDB accession code 35FO).
Results and Discussion
Overall structure of the hTHEM2(undecan-2-one-CoA) complex
Global
The structure of this complex was solved and refined to 1.7 Å resolution (Table 1). The C-terminus (residue 140 and the His6 tag) is disordered in each subunit. Within one asymmetric unit, there are 2 tetramers of hTHEM2 (Figure 2A, subunits are labeled A-H). Eight undecan-2-one-CoA ligands (colored blue and labeled I-P in Figure 2A) are bound to the respective substrate-binding sites. Seven chloride ions (colored green in Figure 2A) are bound to seven of the eight subunits. For the eighth subunit, a water molecule is observed in place of the chloride ion (colored magenta in Figure 2A). The chloride ions were identified by the anomalous difference Fourier map (see Figure SI1; Supporting Information). Two subunits of each tetramer are bound with a PEG molecule (colored red in Figure 2A). Approximately five oxyethylene units of the PEG ligand are observed in a surface depression near the rear exit of the undecan-2-one-CoA binding site. The PEG and chloride ligands were acquired from the crystallization solution that contained 15% PEG3350 in the reservoir solution and 50 mM NaCl in the protein solution. The effect of NaCl on the kcat and Km of hHTEM2 catalyzed hydrolysis of 3-hydroxyphenylacetyl-CoA was measured over a NaCl concentration range of 0 to 100 mM. The results (Table SI1; Supporting information) showed that neither kinetic constant is altered. Likewise, there is no difference in the undecan-2-one-CoA inhibition constant Ki measured in the absence of NaCl (Ki = 3.0 ± 0.2 μM) and in the presence of 100 mM NaCl (Ki = 3.3 ± 0.3 μM) (Table 2). We therefore conclude that the presence of the chloride ion in the active site is not pertinent to the catalytic function of the enzyme or to the binding affinity of the undecan-2-one-CoA ligand. On the other hand, as will be described in the following section, the incidental chloride ion assumes the position in the active site that we anticipate would otherwise be occupied by the oxygen atom of the undecan-2-one O=C-CH2-S unit. The presence of the chloride ion might be, at least in part, responsible for the orientation of the O=C-CH2-S unit observed for undecan-2-one-CoA ligands J-P.
Figure 2.
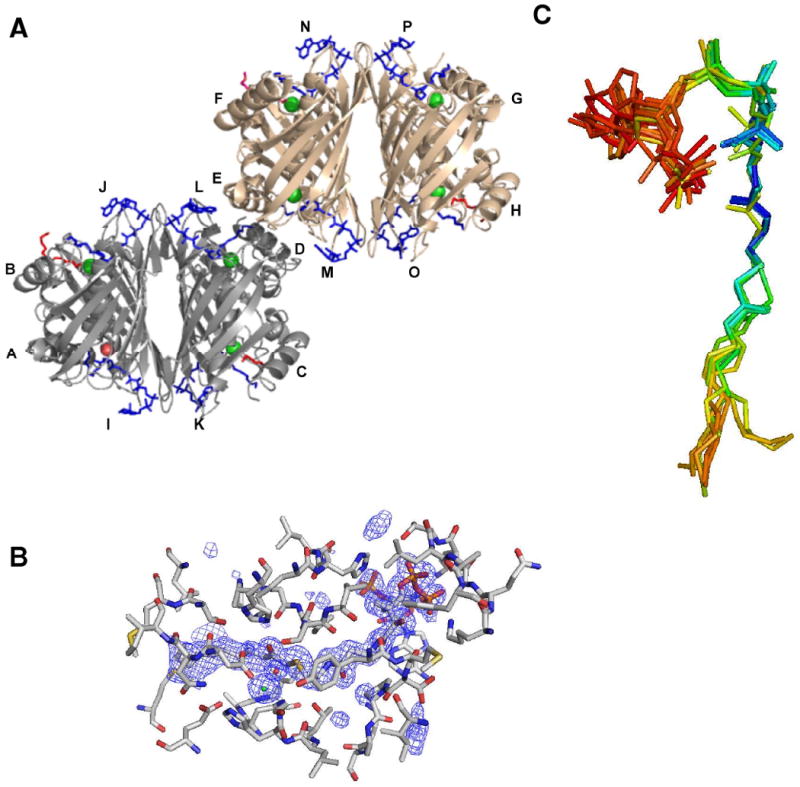
(A). The asymmetric unit of hTHEM2 bound with undecan-2-one-CoA defining two tetramers colored gray (subunits A-D) and brown (subunits H-K) bound with undecan-2-one-CoA ligands (I-L and M-P, respectively) colored blue. The seven chloride ions are represented as green colored spheres and the water molecule that replaces the chloride ion in one subunit is represented as a red colored sphere. The PEG ligands are colored red. (B) The fo-fc electron density map of undecan-2-one-CoA ligand M. The densities of the map (shown in blue) are contoured at the level of 2 sigma. The undecan-2-one-CoA carbon atoms are colored cyan, the hTHEM2 carbon atoms are colored gray, and all oxygen, nitrogen and sulfur atoms are colored red, blue and yellow respectively. (C). The superposition of the eight undecan-2-one-CoA ligands. The atoms of these ligands are colored according to isotropic B values in order to illustrate the change in ordered structure as one proceeds from the nucleotide unit (red to orange to yellow: B factors 60 to 50 to 40) to the pantetheine arm to the O=C-CH2S unit (green to blue: B factors 30 to 20) and progressively through the hydrocarbon tail. The average B value for an amino acid side chain is 27 (Table 1).
Table 2.
Steady-state constants for hTHEM2 wild-type catalyzed hydrolysis of acyl-CoA thioesters in 50 mM K+HEPES (pH 7.5 and 25 °C) and 1 mM DTNB. The error limits shown reflect the fit to a data set and do not represent the standard deviation calculated from repetitive determination.
| Substrate | kcat (s-1) | Km(μM) | kcat/Km (M-1 s-1) |
|---|---|---|---|
| Short Chain Fatty Acyl-CoAs | |||
| n-Decanoyl-CoA | (1.6± 0.1) × 10-2 | 4.9 ± 0.6 | 3.3 × 103 |
| n-Octanoyl-CoA | (4.7 ± 0.1) × 10-3 | (2.6 ± 0.3) × 101 | 1.8× 102 |
| n-Hexanoyl-CoA | (2.2 ± 0.1) × 10-2 | (7.0 ± 0.6) × 101 | 3.1 × 102 |
| n-Butyryl–CoA | (4.7± 0.5) × 10-2 | (6.0 ± 0.1) × 102 | 7.8 × 101 |
| 2-Butenoyl-CoA | (1.4 ± 0.5) × 10-2 | (2.2 ± 0.3) × 102 | 6.4 × 101 |
| β-Methylcrotonyl-CoA | (2.3± 0.1) × 10-2 | (1.2 ± 0.1) × 102 | 1.9 × 102 |
| Crotonyl-CoA | (7.6 ± 0.2) × 10-3 | (1.8 ± 0.1) × 102 | 4.7 × 102 |
| Tiglyl-CoA | (2.0± 0.2) × 10-2 | (2.5 ± 0.5) × 102 | 8.2 × 101 |
| n-Propionyl-CoA | (3.6 ± 0.3) × 10-2 | (1.9 ± 0.4) × 102 | 1.9 × 102 |
| Acetyl-CoA | (5.4 ± 0.2) × 10-2 | (2.9 ± 0.8) × 102 | 1.8 × 102 |
| Medium-Long Chain Fatty Acyl-CoAs | |||
| Lauroyl-CoA | (1.9 ± 0.1) × 10-2 | 9.9 ± 0.8 | 1.9 × 103 |
| Myristoyl-CoA (25 °C) | (2.0± 0.1) × 10-2 | 5 ± 1 | 4.0 × 103 |
| (37 °C) | (7.0 ± 0.1) × 10-2 | 9 ± 2 | 7.7 × 103 |
| n-Palmitoyl-CoA | (4.4 ± 0.1) × 10-3 | (1.6 ± 0.1) × 101 | 2.8 × 102 |
| Palmitoleoyl-CoA | (1.7± 0.1) × 10-2 | 9 ± 2 | 1.9 × 103 |
| Stearoyl-CoA | (1.1 ± 0.1) × 10-2 | (2.0 ± 0.4) × 101 | 5.5 × 102 |
| Oleoyl-CoA | (1.1 ± 0.1) × 10-2 | 9 ± 1 | 1.3 × 103 |
| Linoleoyl-CoA | (1.7± 0.1) × 10-2 | (3.1 ± 0.4) × 102 | 5.4 × 101 |
| Arachidonoyl-CoA | (2.2 ± 0.1) × 10-2 | (2.0 ± 0.2) × 101 | 1.1 × 103 |
| Aromatic Acyl-CoAs | |||
| 3-Hydroxyphenylacetyl-CoA | 1.4 ± 0.1 | (4.1 ± 0.4) × 101 | 3.5 × 104 |
| 3,4-Dihydroxyphenylacetyl-CoA | (1.9 ± 0.1) × 10-1 | (1.0 ± 0.1) × 101 | 1.8 × 104 |
| 3,5-Dihydroxyphenylacetyl-CoA | (1.9 ± 0.1) × 10-1 | (2.5 ± 0.3) × 101 | 7.9 × 103 |
| 4-Hydroxyphenylacetyl-CoA | (1.4 ± 0.1) × 10-1 | (4.9 ± 0.4) × 101 | 2.9 × 103 |
| Phenylacetyl-CoA | (8.4 ± 0.6) × 10-2 | (2.4 ± 0.3) × 102 | 3.5 × 102 |
| 3-Hydroxybenzoyl-CoA | (1.1 ± 0.1) × 10-2 | (3.2 ± 0.2) × 101 | 3.5 × 102 |
| 4-Hydroxybenzoyl-CoA | No activity | - | - |
| 4-Chlorobenzoyl-CoA | (3.2 ± 0.2) × 10-3 | (1.3 ± 0.2) × 102 | 2.5 × 101 |
| Polar Aliphatic Acyl-CoAs | |||
| β-Hydroxybutyryl-CoA | (1.8 ± 0.1) × 10-1 | (1.1 ± 0.2) × 102 | 1.6 × 103 |
| Glutaryl-CoA | (5.0 ± 0.2) × 10-2 | (6.7 ± 0.5) × 102 | 7.5 × 102 |
| 3-Hydroxy-3-methylglutaryl-CoA | (7.9 ± 0.2) × 10-2 | (2.9 ± 0.2) × 103 | 2.7 × 101 |
| Malonyl-CoA | (6.6 ± 0.6) × 10-2 | (2.8 ± 0.8) × 102 | 2.4× 102 |
| Methylmalonyl-CoA | (8.3± 0.1) × 10-2 | (2.1 ± 0.1) × 102 | 3.9 × 102 |
| Fatty acyl-ACPS | |||
| Lauroyl-ACP | No activity | ||
| Myristoyl-ACP | No activity | ||
| Palmitoleoyl-ACP | No activity | ||
| Octanoyl-ACP | No activity | ||
| Oleoyl-ACP | No activity | ||
| Acylated Cys Peptide | |||
| AC-GAC(Palmitoylated)-NH2 | No activity | ||
| Inhibitors | Ki(μM) | ||
| Undecan-2-one-CoA 0 mM NaCl | 3.0 ± 0.2 | ||
| 100 mM Nacl | 3.3 ± 0.3 | ||
| Phenylacetylmethyl-CoA | 500 ±100 | ||
| DesulfoCoA | 9 ± 1 | ||
The two hTHEM2 tetramers of the asymmetric unit are identical, and are formed by the back-to-back (i.e., sheet-to-sheet) association of dimer 1 (formed by subunits A and B or E and F) and dimer 2 (formed by subunits C and D or G and H) (Figure 2A). The subunit organization observed for the apo hTHEM2 (7) and undecan-2-one-CoA bound hTHEM2 are identical as is the subunit fold (RMSD of 0.603 for the alpha carbon atoms of the superimposed apo and liganded tetramers) (Figure SI2; Supporting Information).
The undecan-2-one-CoA ligands
The electron density map of undecan-2-one-CoA ligand M bound at the interface of subunits D and E is presented in Figure 2B. Of the eight undecan-2-one-CoA ligands, only ligand I (bound at the interface of subunits A and B) shows poor electron density. An overlay of the eight undecan-2-one-CoA ligands is shown in Figure 2C. The atoms of each ligand are colored according to their isotropic B factors to illustrate that the atoms of the pantetheine arm and the O=C-CH2S unit are highly ordered, whereas the atoms of nucleotide and decanyl units are significantly less ordered than the atoms of the surrounding amino acid side chains.
The substrate binding site
The undecan-2-one-CoA (Ki = 3 μM; Table 2) ligands mark the locations of the four substrate-binding sites of each tetramer (Figure 2A). A comparison of the liganded hTHEM2 tetramer to that of the Arthrobacter 4-HBA-CoA thioesterase tetramer bound with the inert substrate analog 4-hydroxyphenacyl-CoA (5) reveals the conservation of the substrate binding mode between these two thioesterases (see Figures 3A and 1A). Although of the same clade, hTHEM2 does not share significant sequence identity with the Arthrobacter 4-HBA-CoA thioesterase.
Figure 3.
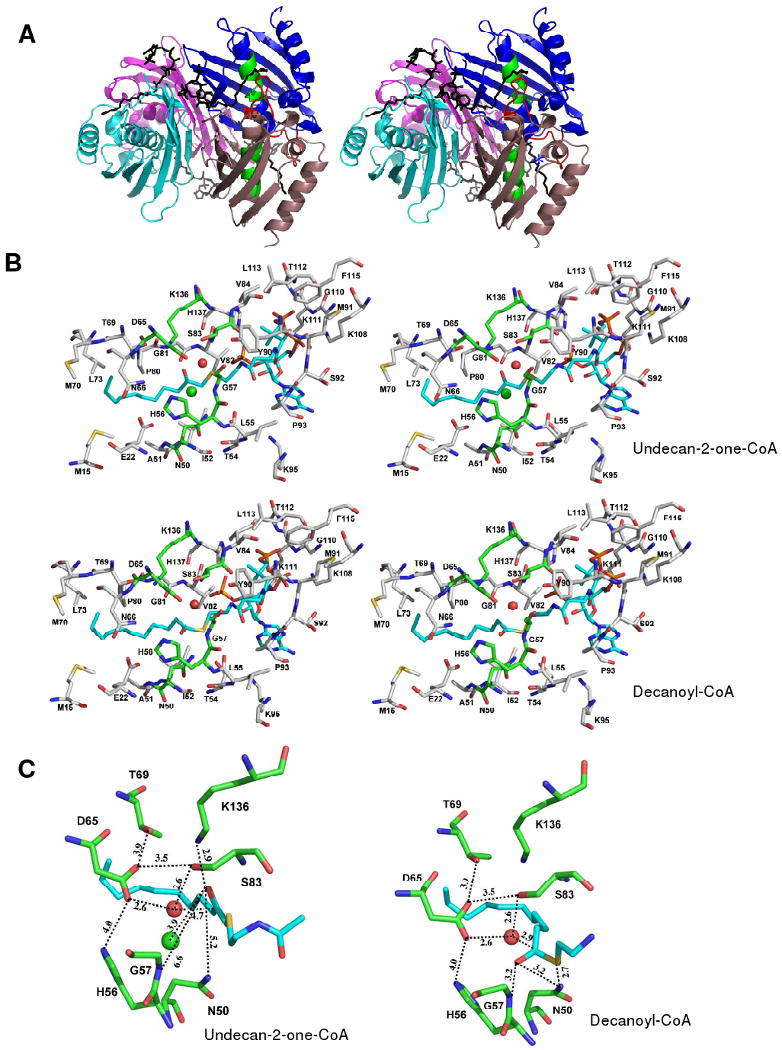
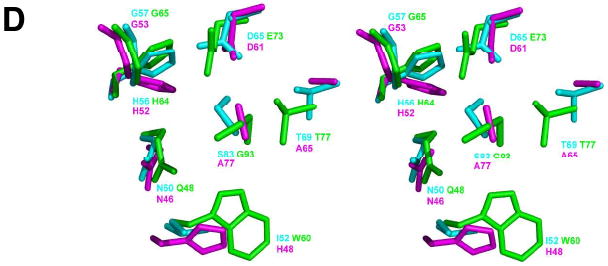
(A) The Pymol generated stereoview of the back-to-back tetramer of hTHEM2 bound with undecan-2-one-CoA for illustration of subunits as functional protomers and the location of the catalytic scaffold (as described in the Introduction section and illustrated for the prototypes of the two hotdog-fold thioesterase clades in Figure 1). The subunits are colored individually to show the dimer units (blue and salmon; cyan and magenta). The undecan-2-one-CoA ligands are shown in black. The catalytic scaffold of the blue (protomer I) and salmon (protomer II) colored dimer is shown in red (the loop leading to the N-terminus of the polarizing helix) and green (the midsection of the polarizing helix). The location of the protomer III loop (residues 108-113) that interacts with the CoA pyrophosphate moiety of the undecan-2-one-CoA ligand bound of the opposing dimer is identified by * (B) The Pymol generated stereoview of the hHTEM2 active site showing the amino acid residues that immediately surround the undecan-2-one-CoA ligand M. The ligand carbon atoms are colored cyan, the carbon atoms of the amino acid residues of the catalytic site are colored green, and all other carbon atoms are colored gray. The oxygen atoms are colored red, nitrogen atoms blue and sulfur atoms yellow. (C) The Pymol generated stereoview of the zoom-in of hHTEM2 active site showing the amino acid residues that immediately surround the undecan-2-one-CoA ligand M (left) and the modeled decanoyl-CoA ligand (right). The coloring scheme used is the same as that described in (B). (D) The Pymol generated stereoview of the superimposed active site residues of hHTEM2 (cyan), Arthrobacter 4-HBA-CoA thioesterase (16) (green), and the E. coli PaaI (23) (magenta). The figure was created by first super-positioning the three structures using the N-terminus of the polarizing helix to set the structural alignment.
The undecan-2-one-CoA ligand is bound at the interface formed by three adjacent hTHEM2 subunits, two (protomers I and II) of which belong to the same dimer and the third (protomer III) to the opposing dimer (Figure 3A and Figure SI3; Supporting information). The catalytic scaffold consists of the connecting loop and N-terminal residue of the α-helix of protomer I (colored red in Figure 3A) and the midsection of the α-helix of protomer II (colored green in Figure 3A). The scaffold is very similar to that observed in the Arthrobacter 4-HBA-CoA thioesterase (5) (see Figure 1A).
The adenine 3′-monophosphate-5′-pyrophosphate unit binds to the protein surface at the entrance of the active site, whereas the pantetheine arm and acyl tail snake through a channel formed at the interface of protomers I and II. The residues that surround the undecan-2-one-CoA ligand M are illustrated in Figure 3B. The zoom-in snap shot of the catalytic site (Figure 3C) reveals that O=CCH2S unit is “flipped” away from the N-terminal residue (Gly47) of the protomer I α-helix N-terminus, and consequently the formation of a hydrogen bond between the O=C and the Gly57 backbone amide NH is not possible. The chloride ion, which occupies the site that we would anticipate to be occupied by the O=C for hydrogen bond formation with the Gly57 backbone amide NH, might promote this particular binding mode. The undecan-2-one-CoA ligand I is bound to the one active site that does not contain the chloride ion (Figure 2A). The O=C-CH2S unit of this ligand appears to be rotated in such a way as to bring the oxygen atom in closer contact with the Gly57 backbone amide NH (Figure SI4).
The substrate model
Whereas the ligand O=C-CH2S is misaligned with the N-terminal residue (G57) of the protomer I polarizing α-helix, it is aligned with the Lys136 of protomer II (Figure 3C). Because Ala replacement of Lys136 resulted in a relatively small perturbation in catalytic efficiency (kcat reduced 4-fold and Km increased 3-fold; Table 3) we contend that the orientation of the O=C-CH2S is an artifact of crystallization. In order to obtain a “virtual” picture of the catalytic site in the enzyme-substrate complex, the decanoyl-CoA ligand was modeled in place of the undeac-2-one-CoA ligand M and the incidental chloride ion. The resulting model (see Figure 3B and 3C) shows the substrate thioester C=O to be within hydrogen bond distance of the Gly57 backbone amide and the Asn50 side chain, and the thioester sulfur atom to be within hydrogen bond distance of the Asn50 side chain. Otherwise, the substrate ligand conformation is identical to that of undecan-2-one-CoA ligand M (Figure 3B). An overlay of the undecan-2-one-CoA ligand M and the modeled decanoyl-CoA ligand is shown in Figure SI5 of Supporting Information.
Table 3.
Steady-state constants for hTHEM2 wild-type and mutants catalyzed hydrolysis of 3-hydroxyphenylacetyl-CoA in 50 mM K+HEPES (pH 7.5 and 25 °C) and 1 mM DTNB. The error limits shown reflect the fit to a data set and do not represent the standard deviation calculated from repetitive determination.
| Enzyme | kcat (s-1) | Km (μM) | kcat/Km (M-1s-1) |
|---|---|---|---|
| WT | 1.4 ± 0.1 | (4.1 ± 0.4) × 101 | 3.5 × 104 |
| D65A | <1.0× 10-5 | - | - |
| D65N | (3.6 ± 0.1) × 10-1 | (3.6 ± 0.3) × 102 | 1.0 × 103 |
| D65E | (1.6 ± 0.1) × 10-1 | (2.3 ± 0.3) × 102 | 6.7 × 102 |
| H56A | (6.2 ± 2) × 10-1 | (2.6 ± 0.2) × 101 | 2.4 × 104 |
| S83A | (1.5 ± 0.1) × 10-1 | (3.1 ± 0.5) × 102 | 5.8 × 102 |
| T69A | (6.6 ± 0.4) × 10-1 | (1.6 ± 0.2) × 102 | 4.1 × 103 |
| H134A | (1.6 ± 0.1) × 10-1 | (6.2 ± 0.4) × 102 | 1.7 × 102 |
| N50A | (7.0 ± 1) × 10-3 | (3.0± 0.5) × 102 | 2.3 × 101 |
| K136A | (3.2 ± 0.1) × 10-1 | (1.2 ± 0.1) × 102 | 2.6 × 103 |
| H137A | (8.3 ± 0.8) × 10-1 | (9.0 ± 1.5) × 102 | 9.2 × 102 |
The hHTEM2 Catalytic Mechanism
The active site residues of special interest include Asp65, Gly57, Asn50, Ser83, His56 and Thr69. Each residue is conserved among the hTHEM2 eukaryotic sequence homologs having at least 42 % sequence identity with hTHEM2 (the sequence alignment is shown Figure SI6; Supporting Information). The counter parts to these residues in the like-clade members Arthrobacter 4-HBA-CoA thioesterase (5) and E. coli phenylacetyl-CoA thioesterase (PaaI) (22) are shown in Figure 3D. Gly57 is conserved as Gly65 in Arthrobacter 4-HBA-CoA thioesterase and as Gly53 in E. coli PaaI. The hTHEM2 Asp65 and Asn50 are conserved as Asp61 and Asn46 in the E. coli PaaI and are conservatively substituted with Glu73 and Gln48 in the Arthrobacter 4-HBA-CoA thioesterase. His56 is conserved as His64 in Arthrobacter 4-HBA-CoA thioesterase and as His52 in E. coli PaaI. Residues that are not conserved are Ser83, which is replaced by Gly93 in the Arthrobacter 4-HBA-CoA thioesterase and by Ala77 in E. coli PaaI and Thr69, which is conserved in Arthrobacter 4-HBA-CoA thioesterase as Thr71 but replaced by Ala65 in E. coli PaaI.
The Gly53 of Arthrobacter 4-HBA-CoA thioesterase orients and polarizes the substrate thioester C=O, whereas the Glu73 functions in nucleophilic catalysis, the Thr71 helps orient the water molecule that hydrolyzes the mixed anhydride intermediate, the Gln48 activates the CoA leaving group and the His52 participates in a hydrogen bond network which includes the substrate ring C(4)OH substituent (5, 23-24). Ala replacement of Arthrobacter 4-HBA-CoA thioesterase Thr71 resulted in a 3-fold reduction in kcat and no change in Km. The T69A hTHEM2 mutant kcat is reduced 2-fold and the Km is increased 4-fold (Table 3). The Thr69 of hTHEM2 forms a “long” hydrogen bond with the Asp65 carboxylate as does the His56. Ala replacement of the hTHEM2 His56 reduces the kcat value 2-fold and increases the Km value 6-fold (Table 3). In comparison, Ala replacement of His64 has a dramatic effect on the Arthrobacter 4-HBA-CoA thioesterase catalytic efficiency as indicated by a 4000-fold reduction in the kcat value and a 500-fold increase in the Km value. Ala replacement of the Arthrobacter 4-HBA-CoA thioesterase Gln58 reduces the kcat value 53-fold without having a significant impact on the Km value. Ala replacement of the hHTEM2 Asn50 has a more pronounced effect: the kcat value is reduced 200-fold and the Km value is increased 10-fold (Table 3).
The greatest deviation in the proposed mechanisms of hHTEM2 and Arthrobacter 4-HBA-CoA thioesterase catalysis is the roles of the respective catalytic carboxylate residues Asp65 and Glu73. Conservative replacement of the Glu73 with Gln results in a 2,000-fold reduction in the kcat value, consistent with its suggested role in nucleophilic catalysis (5, 23-24). Whereas the substitution of the hTHEM2 Asp65 with Ala is not tolerated, conservative replacement with Asn or Glu results in relatively small reductions in the hTHEM2 turnover rate (the kcat values of the D65N2 and D65E mutants are reduced only 3-fold and 10-fold, respectively) (Table 3).3 This finding is consistent with a less demanding role for Asp65 wherein it collaborates with Ser83 to orient and activate the water nucleophile. The replacement of the hHTEM2 Ser83 with Ala reduces the kcat value 10-fold and increases the Km value 10-fold.
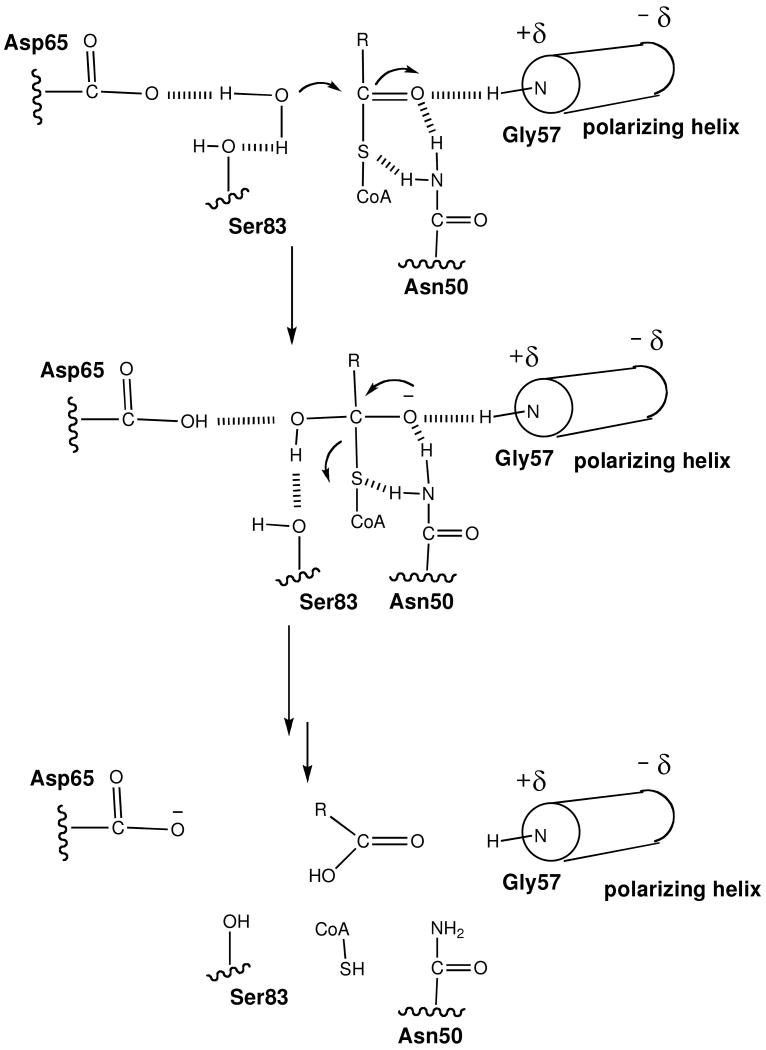
The catalytic mechanism that we propose for hTHEM2 catalysis is depicted in Figure 4. The reaction pathway is shown as a two-step sequence, however we do not dismiss the possibility that the acyl transfer is concerted. The key components of this mechanism are the orientation and polarization of the thioester C=O and the water nucleophile. The N-terminus residue of the polarizing α-helix of protomer I Gly57 is suggested collaborate with Asn50, also of protomer I, in binding and polarizing the substrate thioester C=O. The Asp65 and Ser83, both of protomer II, are believed to collaborate in the orientation and activation of the water nucleophile. Asn50 might facilitate the departure of the thiolate leaving group. The proposed mechanism is different from, and much less elaborate than, the general base catalysis mechanism proposed for the T. thermophilus PaaI (25). This enzyme is reported to employ an asymmetric substrate-induced fit mechanism. We found no structural evidence for substrate-induced fit operative in hTHEM2 (vide supra).
Figure 4.
A cartoon created in Chemdraw that depicts the proposed hTHEM2 catalytic mechanism. Although the reaction is shown to involve two chemical steps, we do not rule out the possibility of a concerted mechanism.
Structure-function Analysis of the hHTEM2 Acyl Binding Site
Substrate Screen
The acyl-CoAs listed (by structural category) in Table 2 were tested as substrates in order to define the hTHEM2 substrate range. Using the substrate kcat/Km value as the measure of substrate activity the results show that the medium-to-long chain fatty acyl-CoAs are preferred over short chain and very long chain fatty acyl-CoAs. With the exception of β-hydroxybutyryl-CoA, the small polar aliphatic acyl-CoAs are inferior substrates. Among the aromatic acyl-CoAs tested hydroxyphenylacetyl-CoA (but not phenylacetyl-CoA) is a good substrate. Because the kcat values measured for all substrates (excluding 3-hydroxyphenylacetyl-CoA) are low, it is reasonable to assume that the substrate binding step is at thermodynamic equilibrium and therefore that the Km value approximates the Kd value. Indeed, the Km values measured for decanoyl-CoA and phenylacetyl-CoA agree with the Ki values measured for undecan-2-one-CoA and phenylacetylmethyl-CoA, respectively (Table 2). The Km values reported in Table 2 range from 5 μM to 3 mM. The very short chain (nonpolar as well as polar) acyl-CoAs bind less tightly than do the medium to long chain fatty acyl-CoAs, which suggests that desolvation of the acyl chain contributes to favorable binding energy. The tight binding observed for the medium-to-long chain fatty acyl-CoAs is suggestive of structure based targeting of a class of molecules rather than the targeting of a specific metabolite.
Structure of the Acyl Binding Site
The hTHEM2 acyl binding site, formed by Ile52, Asn50, Ala51, Thr54, Leu55 and His56 of protomer I and by Met15, Glu22, Asn66, Thr69, Met70, Leu73, Pro80, Gly81 and Val 82 of protomer II, is a long, largely hydrophobic, opened-ended channel (see Figure 3B and Figure 5A). This accounts for why the medium-to-long chain fatty acyl-CoAs bind tightly. On the other hand, we are left to rationalize the observation that the turnover rates of the medium-to-long chain fatty acyl-CoAs are ∼100-fold slower than that measured for 3-hydroxyphenylacetyl-CoA, which we note is not a known human metabolite. The 170-fold tighter binding of undecan-2-one-CoA vs phenylacetylmethyl-CoA (Table 2) indicates that the linear aliphatic chain offers more favorable binding energy than does the aromatic ring and the kcat measured phenylacetyl-CoA is comparable to those of the medium-to-long chain fatty acyl-CoAs.
Figure 5.
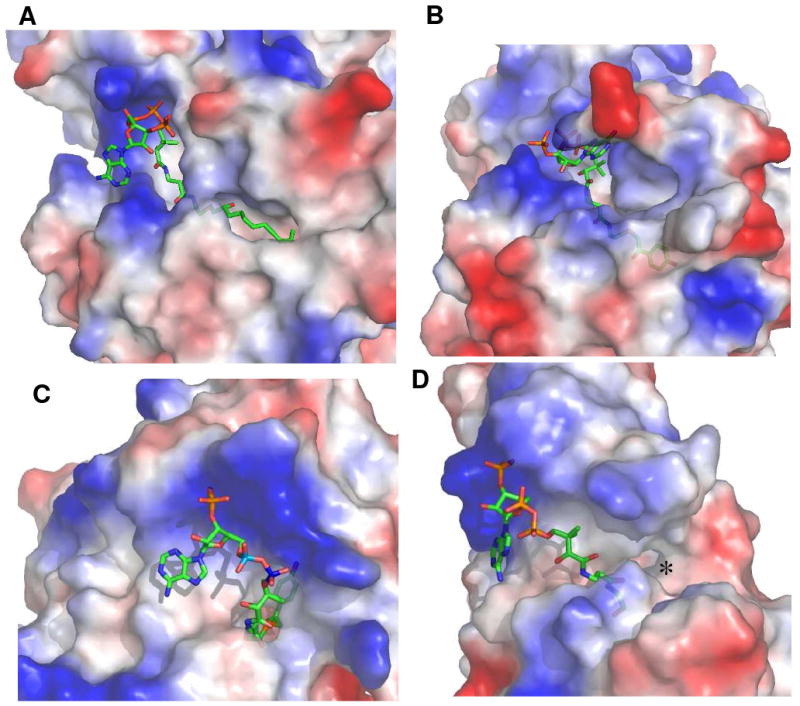
The Grasp generated representations of the steric and electrostatic topological features of the acyl-CoA binding sites of (A) hHTEM2 bound with undecan-2-one-CoA, (B) Arthrobacter 4-HBA-CoA thioesterase bound with 4-hydroxyphenacyl-CoA (16), (C) Pseudomonas 4-HBA-CoA thioesterase bound with 4-hydroxyphenacyl-CoA (17), and (D) H. influenza YciA bound bound with CoA (19). The electron poor surface is represented in blue color and the electron rich surface is represented in red color.
In order to explore why 3-hydroxyphenylacetyl-CoA is a superior substrate, a model of the hTHEM2(3-hydroxyphenylacetyl-CoA) complex (not shown) was generated which shows the potential for hydrogen bond interaction between the channel residue Glu22 and the substrate ring C(3)OH. The 10-fold higher turnover rate observed for the β-hydroxybutyryl-CoA compared to butyryl-CoA (Table 2) might also be attributed to interaction of the hydroxyl group with Glu22. It is conceivable that the hydroxyl groups of these two substrates help orient the thioester C=O in the active site and in this manner enhances the turnover rate. Given this, a β-hydroxy medium-to-long chain fatty acyl-CoA might prove to be a more active in vitro substrate for hTHEM2 than is the corresponding medium-to-long chain fatty acyl-CoA, however this has not been tested.4
Structure-function Analysis of the hHTEM2 CoA Binding Site
In this section we use structure-function analysis to address the question of whether hTHEM2 is specialized to distinguish between an acyl-CoA thioester, an acylated acyl carrier protein (acyl-ACP) and a protein acylated at a Cys residue.
Substrate Screen
The ACP of interest is the ACP domain of the human cytosol fatty acid synthase (FAS). The ACP domain has been prepared as a separate protein unit and charged to form the acyl-ACP using the human pantetheinephosphate transferase in conjunction with the corresponding acyl-CoA (13, 14). We prepared the fatty acyl-ACPs listed in Table 2 and tested them as substrates for hTHEM2 (see Material and Methods for details). Despite the high enzyme concentration and the extended incubation times used, the expected hydrolysis products were not observed. This finding indicates that hTHEM2 does not recognize fatty acyl-ACPs derived from the engineered (FAS) ACP domain as substrates.
For the purpose of probing the possibility that hTHEM2 might target a palmitoylated protein (6) in vivo the peptide Ac-GAC(palmitoylated)-NH2 1 was tested as substrate. Significantly, no activity was found.
These results, together with the substrate activities of the acyl-CoAs reported in the previous section, suggest that hTHEM2 requires CoA as the substrate thiol unit, which in turn demands that hTHEM2 has a CoA binding site. We tested this prediction by measuring the inhibition constant of the CoA analog desulfoCoA, in which the CH2SH is replaced with CH3. Because the activity assay is based on measuring CoA formation using DTNB, CoA itself could not be tested. The desulfoCoA was shown to be tight binding competitive inhibitor of hTHEM2 catalysis (Ki = 9 μM; Table 2).
The CoA Binding Site
Next, we examined the binding interactions with the CoA unit as observed from the structure of the hTHEM2(undecan-2-one-CoA) complex. A stereoview of the hTHEM2 CoA binding site residues is shown in Figure 3B and the topological and electrostatic features of the binding site are shown in Figure 5A. The CoA 3′-phosphate is within hydrogen bond distance (3.0 Å) of the protomer II His137 side chain. Ala replacement of this residue has a minimal effect on the kcat, however the Km is increased 460-fold (Table 3). His137 is conserved among vertebrate homologs (mammal, fish, frog), although in more distant homologs (tree, insect, nematode) it is replaced with Tyr or Ala (Figure SI6; Supporting Information). Direct binding interactions involving the hairpin loop (residues 108-113) of protomer III (see Figure 2A) include Lys108 ion pair formation (3.2 Å) and Lys111 backbone amide NH hydrogen bond formation (2.6 Å) with the 5′-α-phosphate (both Lys residues are conservatively replaced with Arg in some homologs) and Leu113 (3.1 Å; not conserved) and Thr112 (3.0 Å; not conserved) backbone amide NH hydrogen bond formation with the 5′-β-phosphate.
The isotropic B values which are high for the atoms of the undecan-2-one-CoA nucleotide unit, low for the atoms of the pantetheine unit and high again for the atoms of the acyl tail (Figure 2C) indicate that the pantetheine unit of the acyl-CoA substrate plays a key role in hTHEM2 substrate recognition. The hTHEM2 channel residues that engage pantetheine unit in hydrogen bond interaction include the backbone amide C=Os of residues Ser83 (3.1 Å to N(9)H; protomer I; stringently conserved), Tyr90 (3.1 Å, N(5)H; protomer II; conservatively replaced with Phe), and Met91 (2.9 Å, C(3)OH; protomer II; conservatively replaced with Leu). In addition, the pantetheine unit C(8)=O interacts with several water molecules that are in turn bound by protomer II residues Ser92 (conservatively replaced with Thr in some sequences), Leu55 (stringently conserved) and Thr54 (not conserved). Packing of the hydrocarbon moieties of the pantetheine unit with the hydrocarbon moieties of channel residues Ile52, Le55, Pro93, Lys108, Val82, Tyr112, Tyr90, and Leu113 (Figure 3A) is also strongly suggestive of electrostatic complementation between the binding site and the ligand.
The pantetheine unit can bridge the distance between the entrance to the active site (where the nucleotide is positioned) and the short tunnel in which the thioester C=O is positioned for reaction. In contrast, the Cys unit of the acylated peptide is not long enough to bridge this distance and therefore the inactivity of this thioester is easily accounted for. The FAS acyl-ACPs on the other hand do fulfill the distance requirement, yet the acyl-ACPs derived from the engineered ACP domain tested did not show substrate activity. This suggests that the topological and/or electrostatic surface of the ACP is not compatible with that of the hTHEM2. However, we cannot rule out the possibility that the acyl-ACP of the holo FAS is substrate active.
hTHEM2 Divergence of Structure and Function From its Bacterial Counterparts
The liganded structure of hTHEM2 provides the opportunity to compare its structural determinants of substrate recognition with those of its characterized bacterial counterparts. In Figure 5 the topological and electrostatic features that define the landscape of the hTHEM2 substrate-binding site are represented along with those of three bacterial hotdog-fold thioesterases. The Arthrobacter 4-HBA-CoA thioesterase (5) and the high activity, broad substrate range, cytosolic thioesterase YciA (26, 27) from H. influenza belong to the same clade as hTHEM2, whereas the Pseudomonas 4-HBA-CoA thioesterase (6, 28) belongs to the other clade. All three bacterial thioesterases catalyze the hydrolysis of their physiological acyl-CoA substrates with high catalytic efficiency (kcat/Km > 1 × 106 M-1s-1) (26, 29-30). Structure determinations (5-6, 27) have shown that for each of these liganded bacterial thioesterases, as well as the liganded hTHEM2, the pantetheine unit of the CoA snakes through a narrow channel that leads to a short tunnel (where the thioester C=O binds) that in turn opens to the acyl-binding site (Figure 5). The CoA nucleotide binds to the protein surface (at the door to the channel), which can be best described as a canyon that is highly electropositive in the vicinity of the CoA nucleotide phosphoryl groups. Like hTHEM2, all three bacterial thioesterases bind CoA tightly and this tight binding appears to derive from favorable interaction (desolvation of the hydrophobic units and hydrogen bond formation with the polar units) with the pantetheine unit and from the electrostatic forces at protein surface that act on the nucleotide phosphate groups (27, 28).
The enclosed substrate binding pockets of the Arthrobacter and Pseudomonas 4-HBA-CoA thioesterases (Figure 5B and 5C) allow these enzymes to restrict their substrate range and thereby avoid unwanted hydrolysis of acyl-CoA metabolites. Owing to their “open” acyl-binding sites, the bacterial thioesterase YciA (26, 27) (Figure 5A) and hTHEM2 (Figure 5D) are less discriminatory. YciA is subject to strong CoA feedback inhibition (Kd ≪ 1 μM) but hTHEM2 is not.
hTHEM2 Function
Human hotdog-fold thioesterases have been linked to diseases that range from cancer, to inflammatory disease, to obesity (31-32). Although most of these enzymes are known to catalyze thioester hydrolysis in fatty acyl-CoAs, the connection between this activity and the physiology of the healthy vs. diseased human is elusive. On the cellular level, the biochemical role that a particular fatty acyl-CoA thioesterase might perform will depend in part on its location. hTHEM2 is cytosolic and therefore the fatty acyl-CoAs that it might encounter originate from fatty acids acquired by import across the plasma membrane or via biosynthesis on the cytoplasmic FAS. What is the purpose of hTHEM2 catalysis: to decrease the level fatty acyl-CoA, to recover the fatty acid or to recover the CoA? We do not yet know the answer to this question.
The combination of slow catalytic turnover coupled with tight substrate binding observed for hTHEM2 might the mechanism by which this thioesterase selects its medium-to-long chain fatty acyl-CoA substrates from the cytoplasmic metabolite pool. Alternatively, hTHEM2 might be activated by a protein partner. A recent report suggests that the phosphatidylcholine transfer protein (StarD2) binds to hTHEM2 and increases its thioesterase activity (2), yet we note that the 4-fold increase is seemingly insignificant, that the physical basis for formation of a complex between these two proteins is not obvious from inspection of their individual structures, and that a possible connection between the biochemical activities of these two proteins has not been made.
Conclusion
The work reported in this paper represents the first structure-function based analysis of a human hotdog-fold thioesterase. The mechanism of hydrolysis proposed involves the Asp65/Ser83 assisted attack of a water molecule at the Gly57/Asn50 polarized thioester C=O and the Asn50 assisted departure of the thiolate leaving group. The substrate specificity profile was analyzed within the context of the liganded enzyme and the ultimate conclusion that we reach is that hHTEM2 is, by structural design and by cellular location, set to target cytoplasmic medium-to-long chain acyl-CoA thioesters for hydrolysis. The most challenging objective, determination of the physiological function of hTHEM2, remains to be achieved.
Supplementary Material
Acknowledgments
The authors thank Dr. Stuart Smith of the Children's Hospital Oakland Research Institute, Oakland, CA for the kind gift of plamids encoding the Human cytosolic ACP fragment and the PPTase and thank Dr. Karen Allen of Boston University for a critical read of the manuscript and for helpful suggestions.
Abbreviations used are
- hTHEM2
human thioesterase superfamily member 2
- 4-HBA-CoA
4-hydroxybenzoyl-CoA
- DTNB
5,5′-dithio-bis(2-nitrobenzoic acid)
- HPLC
high performance liquid chromatography
- ES-MS
electro spray mass spectroscopy
- K+HEPES
potassium salt of N-2-hydroxyethylpiperzine-N′-ethanesulfonic acid
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- ACP
acyl carrier protein
- PaaI
phenylacetyl-CoA thioesterase
Footnotes
This work was supported by grants to W.G. and H. X. from the National Natural Science Foundation of China (Grant Nos. 30600115, 10490193, and 30721003), the Ministry of Science and Technology of China (Grant Nos.2006AA02A316, 2006CB910903, 2007CB914304 and 2009CB825501) and the Chinese Academy of Sciences (Grant No. KSCX2-YW-R-61) and by NIH grant GM28688 to D. D. M.
The atomic coordinates and structure factors of the hTHEM2(undecan-2-one-CoA) structure have been deposited in the Protein Data Bank (Accession code 35FO).
The modest reduction in the kcat value raised the question of whether the Asn65 had partially reverted to Asp65 during enzyme purification. However, because the because the Km value is significantly different from that of the wild-type enzyme we think that this is unlikely.
For the record, a general base catalysis mechanism has been proposed for the T. thermophilus PaaI (25). Asn replacement of the T. thermophilus PaaI (25) catalytic Asp48 resulted in a 160-fold reduction in catalytic efficiency. In contrast, Asn replacement of the catalytic Asp75 of the Azoarcus evansii PaaI reduced the kcat value 3,000-fold (22), which is comparable to the reduction in the kcat value observed for the Arthrobacter 4-HBA-CoA thioesterase E73Q and less than the > 10,000-fold reduction in the kcat value observed for Asn replacement of the Asp17 nucleophile of the Pseudomonas 4-HBA-CoA thioesterase (6). Asn replacement of the catalytic Asp16 in the Bacillus halodurans gentisyl-CoA thioesterase (BH1999) reduced the kcat value 230-fold (26).
To our knowledge β-hydroxy fatty acyl-CoAs are not formed in the cytoplasm.
Supporting Information. This section includes a table listing kcat and Km values measured hTHEM2 catalysis as a function of NaCl concentrations and for five graphics figures depicting hTHEM2 and one figure showing the alignment of hTHEM2 sequence homologs. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cheng Z, Bao S, Shan X, Xu H, Gong W. Human thioesterase superfamily member 2 (hTHEM2) is co-localized with beta-tubulin onto the microtubule. Biochem Biophys Res Commun. 2006;350:850–853. doi: 10.1016/j.bbrc.2006.09.105. [DOI] [PubMed] [Google Scholar]
- 2.Kanno K, Wu MK, Agate DS, Fanelli BJ, Wagle N, Scapa EF, Ukomadu C, Cohen DE. Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. J Biol Chem. 2007;282:30728–30736. doi: 10.1074/jbc.M703745200. [DOI] [PubMed] [Google Scholar]
- 3.Grigo K, Wirsing A, Lucas B, Klein-Hitpass L, Ryffel GU. HNF4 alpha orchestrates a set of 14 genes to down-regulate cell proliferation in kidney cells. Biol Chem. 2008;389:179–87. doi: 10.1515/BC.2008.011. [DOI] [PubMed] [Google Scholar]
- 4.Lucas B, Grigo K, Erdmann S, Lausen J, Klein-Hitpass L, Ryffel GU. HNF4alpha reduces proliferation of kidney cells and affects genes deregulated in renal cell carcinoma. Oncogene. 2005;24:6418–6431. doi: 10.1038/sj.onc.1208794. [DOI] [PubMed] [Google Scholar]
- 5.Thoden JB, Zhuang Z, Dunaway-Mariano D, Holden HM. The structure of 4-hydroxybenzoyl-CoA thioesterase from Arthrobacter sp. strain SU. J Biol Chem. 2003;278:43709–43716. doi: 10.1074/jbc.M308198200. [DOI] [PubMed] [Google Scholar]
- 6.Thoden JB, Holden HM, Zhuang Z, Dunaway-Mariano D. X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase. J Biol Chem. 2002;277:27468–27476. doi: 10.1074/jbc.M203904200. [DOI] [PubMed] [Google Scholar]
- 7.Cheng Z, Song F, Shan X, Wei Z, Wang Y, Dunaway-Mariano D, Gong W. Crystal structure of human thioesterase superfamily member 2. Biochem Biophys Res Commun. 2006;349:172–177. doi: 10.1016/j.bbrc.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D. Divergence of function in the hot dog fold enzyme superfamily: the bacterial thioesterase YciA. Biochemistry. 2008;47:2789–2796. doi: 10.1021/bi702334h. [DOI] [PubMed] [Google Scholar]
- 9.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J Biol Chem. 2006;281:11028–11038. doi: 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 10.Luo L, Taylor KL, Xiang H, Wei Y, Zhang W, Dunaway-Mariano D. Role of active site binding interactions in 4-chlorobenzoyl-coenzyme A dehalogenase catalysis. Biochemistry. 2001;40:15684–15692. doi: 10.1021/bi011536f. [DOI] [PubMed] [Google Scholar]
- 11.Merkel SM, Eberhard AE, Gibson J, Harwood CS. Involvement of coenzyme A thioesters in anaerobic metabolism of 4-hydroxybenzoate by Rhodopseudomonas palustris. J Bacteriol. 1989;171:1–7. doi: 10.1128/jb.171.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.B Michael W, S RAJ, W Lisa. Alkyne formation in the reaction of alpha-bromo ketones with arylsulfonylhydrazines. Aust J Chem. 1982;35:2529–2540. [Google Scholar]
- 13.Zhang L, Joshi AK, Smith S. Cloning, expression, characterization, and interaction of two components of a human mitochondrial fatty acid synthase. Malonyltransferase and acyl carrier protein. J Biol Chem. 2003;278:40067–40074. doi: 10.1074/jbc.M306121200. [DOI] [PubMed] [Google Scholar]
- 14.Bunkoczi G, Pasta S, Joshi A, Wu X, Kavanagh KL, Smith S, Oppermann U. Mechanism and substrate recognition of human holo ACP synthase. Chem Biol. 2007;14:1243–1253. doi: 10.1016/j.chembiol.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in osillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 17.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cyrst. 1997;30:1022–1025. [Google Scholar]
- 18.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 20.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cyrst. 1993;26:283–291. [Google Scholar]
- 21.Read RJ, S AJ. A phased translation function. J Appl Cryst. 1998;21:490–495. [Google Scholar]
- 22.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD. Structure, Function and Mechanism of the Phenylacetyl Pathway Hotdog-Fold Thioesterase PaaI. J Biol Chem. 2006;281:11028–11038. doi: 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang Z. Department of Chemistry and Chemical Biology PhD Thesis. University of New Mexico; Albuquerque NM: 2003. Structure-function relationship within 4-hydroxybenzoyl-CoA thioesterase enzyme; p. 239. [Google Scholar]
- 24.Song F. Department of Chemistry and Chemical Biology PhD Thesis. University of New Mexico; Albuquerque NM: 2005. Structure, function and mechanism of hotdog-fold enzyme superfamily thioesterases; pp. 38–81. [Google Scholar]
- 25.Kunishima N, Asada Y, Sugahara M, Ishijima J, Nodake Y, Miyano M, Kuramitsu S, Yokoyama S. A novel induced-fit reaction mechanism of asymmetric hot dog thioesterase PAAI. J Mol Biol. 2005;352:212–228. doi: 10.1016/j.jmb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhuang Z, Song F, Takami H, Dunaway-Mariano D. The BH 1999 protein of Bacillus halodurans C-125 is gentisyl-coenzyme A thioesterase. J Bacteriol. 2004;186:393–399. doi: 10.1128/JB.186.2.393-399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willis MA, Zhuang Z, Song F, Howard A, Dunaway-Mariano D, Herzberg O. Structure of YciA from Haemophilus influenzae (HI0827), a hexameric broad specificity acyl-coenzyme A thioesterase. Biochemistry. 2008;47:2797–2805. doi: 10.1021/bi702336d. [DOI] [PubMed] [Google Scholar]
- 28.Zhuang Z, Song F, Zhang W, Taylor K, Archambault A, Dunaway-Mariano D, Dong J, Carey PR. Kinetic, Raman, NMR, and site-directed mutagenesis studies of the Pseudomonas sp. strain CBS3 4-hydroxybenzoyl-CoA thioesterase active site. Biochemistry. 2002;41:11152–11160. doi: 10.1021/bi0262303. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang Z, Gartemann KH, Eichenlaub R, Dunaway-Mariano D. Characterization of the 4-hydroxybenzoyl-coenzyme A thioesterase from Arthrobacter sp. strain SU. Appl Environ Microbiol. 2003;69:2707–2711. doi: 10.1128/AEM.69.5.2707-2711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang KH, Liang PH, Beck W, Scholten JD, Dunaway-Mariano D. Isolation and characterization of the three polypeptide components of 4-chlorobenzoate dehalogenase from Pseudomonas sp. strain CBS-3. Biochemistry. 1992;31:5605–5610. doi: 10.1021/bi00139a025. [DOI] [PubMed] [Google Scholar]
- 31.King KL, Young ME, Kerner J, Huang H, O'Shea KM, Alexson SE, Hoppel CL, Stanley WC. Diabetes or peroxisome proliferator-activated receptor alpha agonist increases mitochondrial thioesterase I activity in heart. J Lipid Res. 2007;48:1511–1517. doi: 10.1194/jlr.M600364-JLR200. [DOI] [PubMed] [Google Scholar]
- 32.Knobbe CB, Reifenberger J, Blaschke B, Reifenberger G. Hypermethylation and transcriptional downregulation of the carboxyl-terminal modulator protein gene in glioblastomas. J Natl Cancer Inst. 2004;96:483–486. doi: 10.1093/jnci/djh064. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.