Abstract
Peroxisome proliferator-activated receptor (PPAR)α, β (also known as δ), and γ function as sensors for fatty acids and fatty acid derivatives and control important metabolic pathways involved in the maintenance of energy balance. PPARs also regulate other diverse biological processes such as development, differentiation, inflammation, and neoplasia. In the nucleus, PPARs exist as heterodimers with retinoid X receptor-α bound to DNA with corepressor molecules. Upon ligand activation, PPARs undergo conformational changes that facilitate the dissociation of corepressor molecules and invoke a spatiotemporally orchestrated recruitment of transcription cofactors including coactivators and coactivator-associated proteins. While a given nuclear receptor regulates the expression of a prescribed set of target genes, coactivators are likely to influence the functioning of many regulators and thus affect the transcription of many genes. Evidence suggests that some of the coactivators such as PPAR-binding protein (PBP/PPARBP), thyroid hormone receptor-associated protein 220 (TRAP220), and mediator complex subunit 1 (MED1) may exert a broader influence on the functions of several nuclear receptors and their target genes. Investigations into the role of coactivators in the function of PPARs should strengthen our understanding of the complexities of metabolic diseases associated with energy metabolism.
1. Introduction
The foundation for the discovery and designation of the PPAR subfamily of nuclear receptors in the early 1990s is the cumulative work over the preceding 25 years with peroxisome proliferators, a group of structurally diverse chemicals that induce characteristic and predictable pleiotropic responses including the transcriptional activation of genes involved in the fatty acid oxidation [1–6]. The PPAR subfamily consists of three members namely PPARα (NR1C1), PPARβ (also known as δ) (NR1C2), and PPARγ (NR1C3) with a high degree of sequence conservation across the species [1, 2, 7–9]. All three PPARs in the human and mouse are encoded by separate genes that are on different chromosomes [9]. PPARγ has two isoforms, PPARγ1, and an N-terminal 30 amino acid extended form PPARγ2, both encoded by the same gene using two distinct promoters and alternate splicing [10, 11]. All three members of PPAR subfamily function as sensors for fatty acids and fatty acid derivatives and control metabolic pathways involved in energy homeostasis [12, 13]. PPARs display high levels of homologies at the protein level, but exhibit distinct and noninterchangeable functional roles in mammalian energy metabolism [9]. PPARα is expressed in tissues with high fatty acid oxidation activities, which include liver, kidney, small intestine, heart, and skeletal muscle, consistent with its predominant functional role in regulating lipid catabolism. In the liver, PPARα is the master regulator of mitochondrial, peroxisomal, and microsomal fatty acid oxidation systems where it is activated by synthetic peroxisome proliferators and in addition senses the influx of fatty acids during fasting to upregulate the fatty acid burning capacity [14]. PPARα also plays a role in lipoprotein synthesis, inflammatory responses and the development of cancer in the rodent liver [15–19]. PPARβ is ubiquitously expressed with relatively higher levels found in brain, adipose tissue, and skin [20]. Activation of PPARβ also induces expression of genes required for fatty acid oxidation and energy dissipation in skeletal muscle and adipose tissue which in turn lead to improved lipid profiles and reduced adiposity [21]. In the liver, PPARβ can be activated by plasma free fatty acids influxed during fasting conditions [22]. PPARγ which is expressed at a relatively high level in adipose tissue serves as an essential regulator for adipocyte differentiation and promotes lipid/energy storage in mature adipocytes by increasing the expression of several key genes in this pathway [23]. These two important functions of PPARγ, namely adipogenesis and fat storage in adipocytes account for the insulin sensitizing effects of the anti-diabetic thiazolidinediones [24]. In summary, PPARα and PPARβ participate in energy burning, whereas PPARγ is critical in regulating adipocyte differentiation and energy storage by adipocytes [11, 25, 26].
2. Transcriptional Regulation of PPARs
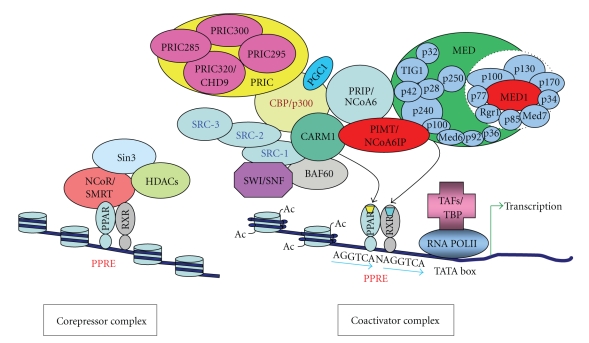
PPARs are ligand-activated transcription factors similar to other members of the nuclear hormone receptor superfamily [7, 8]. PPARs are nuclear in location, where they remain heterodimerized with the 9-cis retinoic acid receptor, RXRα (NR2B) [13] and bind to the upstream cis-acting regulatory regions termed as peroxisome proliferator response element (PPRE) of target genes [9, 27]. The canonical PPRE consists of two direct repeats AGGTCA separated by a single nucleotide so-called DR-1 element [28]. The two half-sites are distinguishable by their 5′ and 3′ positioning on the DR1 element whereby the DNA binding domain of PPAR binds 5′ half-site while RXR binds to the 3′ half-site [29, 30]. In addition to core DR-1 sequence, PPRE element contains an additional AAACT motif at the 5′ upstream region [30]. The hinge region of PPAR forms extensive interaction with the upstream AAACT element [30]. In the absence of ligand, the unliganded PPAR-RXR heterodimer remains bound to the nuclear receptor corepressor (NCoR) and silencing mediator of retinoid and thyroid hormone receptor (SMRT), two well-characterized corepressors (Figure 1) that are mostly present in the corepressor complex [31]. Both NCoR and SMRT directly interact with the Sin3 complex to form a multisubunit repressor complex [32]. SMRT functions as a platform protein and facilitates the recruitment of histone deacetylases (HDACs) to the DNA promoters bound by specific interacting transcription factors [32]. Another corepressor, Receptor Interacting Protein 140 (RIP140) also known as NRIP1 (nuclear receptor interacting protein 1) directly recruits HDAC and represses the activity of various nuclear receptor including PPARs by competing with their coactivators [33–35]. In the absence of ligand activation of nuclear receptor, the corepressor protein complex is known to subdue target gene transcription by causing the deacetylation of histones [31].
Figure 1.
A schematic representation of ligand-dependent recruitment of coactivators for PPAR-regulated target gene transcription. In the absence of ligand, the PPAR-RXR heterodimer recruits corepressors, which in turn, assemble additional components of a repressor complex including histone deacetylase (HDAC). When ligand (yellow trapezium representing PPAR ligand, and blue trapezium representing 9-cis-retinoic acid as RXR ligand) binds, conformational changes in PPAR-RXR induce dissociation of corepressor complex. Active transcriptional complex assembles with coactivator proteins either sequentially or preassembled subcomplex modules. PPAR binds to peroxisome proliferator response element (PPRE) and assemble coactivator complexes that acetylate (SRCs, p300) or methylate (CARM1) nucleosomes for chromatin remodeling. Mediator components [36, 37] contact PPARs and facilitate the recruitment of the basal transcription machinery (TATA-box-binding protein [TBP]/TBP-associated factors [TAFs]) to form linkage with RNA polymerase II for transcription of specific target genes.
The nuclear receptor-regulated transcriptional activation of target genes depends on the binding of a cognate ligand to the receptor (activator). For example, activation of PPARα-RXR heterodimer by a PPARα ligand triggers conformational changes in the receptor which releases the corepressor complex and recruits cofactor complexes to the promoter region of target genes (Figure 1) to initiate transcription [38, 39]. Transcription coactivators increase gene transcription via the acetylation of histones and through the recruitment and stabilization of the transcriptional complexes, mainly the Mediator complex which interacts directly with activator proteins and pol II [40–42]. During the past 15 years, more than 300 cofactors (coactivators, coregulators, corepressors etc.) have been identified but the in vivo physiological regulatory functions of many of these molecules in receptor/gene/cell-specific transcription remain to be explored [43]. This paper summarizes the current state of knowledge about the roles of coactivators and coactivator associated proteins (Table 1), with special emphasis on p160/SRC family members and PPAR-binding protein (PBP/PPARBP)/thyroid hormone receptor-associated protein 220 (TRAP220)/mediator complex subunit 1 (Med1), in the functioning of PPARs.
Table 1.
Some known coactivator and coactivator associated proteins that regulate PPAR function.
| Coactivator proteins | Enzyme activity | Function | References |
|---|---|---|---|
| SRC-1/NCoA-1 | HAT | Histone acetylation | [51, 52] |
| SRC-2/TIF2/GRIP1 | HAT | Histone acetylation | [53] |
| SRC3/pCIP/AIB1 | HAT | Histone acetylation | [54, 55] |
| CBP/p300 | HAT | Histone acetylation followed by recruitment of p160/SRCs | [56] |
| MED1/TRAP220/PBP | None | Anchor for Mediator complex | [57, 58] |
| PGC-1α | None | Recruit coactivator with HAT activities | [59] |
| PGC-1β/PERC | None | Recruit coactivator with HAT activities | [60] |
| PRIP/NCoA6 | None | Recruit ASC complex | [61, 62] |
| PRIC285 | Helicase | Chromatin remodeling by histone displacement and nucleosomal sliding | [63] |
| PRIC320/CHD9 | Helicase | Chromatin remodeling by histone displacement and nucleosomal sliding | [64] |
| SWI/SNF | ATPase | ATP dependent mobilization of nucleosome | [65, 66] |
| BAF60a/SMARCD1 | None | Recruit SWI/SNF complex | [67, 68] |
| BAF60c/SMARCD3 | None | Recruit SWI/SNF complex | [69] |
|
| |||
| Coactivator-associated proteins | |||
|
| |||
| PIMT/NCoA6IP | Methyltransferase | Methylation of caps of snRNAs and snoRNAs | [70] |
| CARM1/PRMT4 | Methyltransferase | Potentiate SRCs by methylation of Histone H3 | [71] |
| CoAA | None | RNA splicing | [72] |
3. Coactivators for PPAR Function
Transcriptional activation of PPAR-regulated genes is enhanced by coactivators. Most coactivators possess one or more LXXLL motifs (L: leucine and X: any amino acid) some of which may make contact with a coactivator-binding groove in the ligand-binding domain of nuclear receptor [44]. The assembly or preassembled coactivator complexes facilitate the liganded PPAR to achieve transcriptional activation of specific target genes in a tissue/cell-specific manner [45, 46]. Once coactivators are recruited to a liganded nuclear receptor they remodel chromatin structure by the intrinsic histone acetyltransferase (HAT) or methyl transferase activities [46]. In order to achieve this, steroid receptor coactivator (p160/SRC) family of proteins, which possess HAT activity, are recruited to the activation function 2 (AF-2) domain of the nuclear receptor and complex with the universal coactivator cyclic-AMP responsive element binding protein (CREB)-binding protein (CBP) and its homologue p300 [47–49]. CBP and p300 also exhibit potent HAT activity [50].
The second category of coactivators, with no known enzymatic functions, participates in the formation of a well-known multisubunit protein complex, variously called TRAP/DRIP/ARC/Mediator complex, consisting of 15–30 proteins [36, 37, 41, 57, 73–77]. Mediator complex, which is anchored by PBP (PPARBP)/TRAP220/DRIP205/Med1 facilitates interaction with poll II of the basal transcription machinery [41, 73, 75]. Members of this Mediator complex appear to be devoid of intrinsic enzymatic activities [46], but play an important role in connecting CBP/p300 bound coactivators with pol II containing preinitation complex [76]. Disruption of CBP/p300 and Med1 genes in the mouse results in embryonic lethality around E11.5 days, indicating that deletion of these pivotal anchoring coactivators affects the integrity of the cofactor complexes, thus altering the function of many nuclear receptors and most likely of other transcription factors [77–79].
A number of other coactivators and coactivator-associated proteins that possess enzymatic activities like methyltransferase (CARM1) [71], helicase (PRIC285, p68) [63, 80], and ATP dependent chromatin remodeling properties (PRIC320, SWI/SNF) [63, 80] or those without any enzymatic activities such as PPARγ coactivator-1α (PGC-1α), PGC-1β, and BAFs [81, 82] have been identified in the active PPAR transcriptional complex, referred to as PRIC (PPARα-receptor interacting cofactor) complex [64, 80]. Some other important coactivators such as PRIP (peroxisome proliferator-activated receptor-interacting protein)/ASC2/AIB3/RAP250/NCoA6 [61, 83–85] and PIMT (PRIP-interacting protein with methyltransferase domain/NCoA6IP) [70] are also identified, which serve as linkers between the initial HAT complex of CBP/p300 and p160 coactivators and the downstream mediator complex [76]. CCPG (constitutive coactivator of PPARγ) is identified as a novel coactivator for PPARγ [86]. As mentioned above, sequential or combinatorial recruitment of various multisubunit coactivator proteins on the liganded nuclear receptor leads to the establishment of a stable preinitiation complex with multiple possible configurations on to the target gene promoter.
4. p160/SRC Family of Coactivators with HAT Activity
p160/SRC family of coactivators consists of three members, namely, SRC-1/NCoA-1, SRC-2/TIF2 (transcriptional intermediary factor 2)/GRIP1 (glucocorticoid receptor interacting protein 1), and SRC-3/pCIP (CBP-interacting protein)/RAC3 (receptor-associated coactivator-3)/ACTR/AIB1 (amplified in breast cancer-1)/TRAM-1(thyroid hormone receptor activator molecule 1) [51–55]. These proteins are required for mediating the transcriptional function of nuclear receptors and other transcription factors in a ligand-dependent manner [87, 88]. All three p160/SRC family members contain bHLH and PAS domains, which are involved in protein-protein interactions. These coactivators also contain three LXXLL motifs, that mediate recognition of, and binding to AF-2 region of a variety of nuclear receptors [89]. They possess HAT activity and are part of the first multiprotein coactivator complex with CBP/p300 on DNA bound-liganded nuclear receptors and participate in the acetylation of histones and remodel chromatin structure to unravel DNA for transcription [76, 88]. SRC-1 interacts with many nuclear receptors including PPARγ and PPARα, and the X-ray crystal structure of SRC-1 and the liganded PPARγ complex revealed that binding is between highly conserved glutamate and lysine residues in the PPARγ ligand binding domain and the backbone atoms of the LXXLL helices of SRC-1 [29]. Protein-protein interactions between PPARα and SRC-1 and SRC-3 have been documented and these interactions appears to be ligand independent [52, 54]. p160/SRC family members exhibit strong sequence homology and somewhat similar functions under in vitro transactivation conditions. But gene knockout mouse models have provided valuable insights into the in vivo functional properties of these molecules [75, 88]. These mouse models include SRC-1−/−, SRC-2−/− and SRC-3-/ single gene disruptions and SRC-1−/−/SRC-2−/− and SRC-1−/−/SRC-3−/− double nulls [50, 90–95].
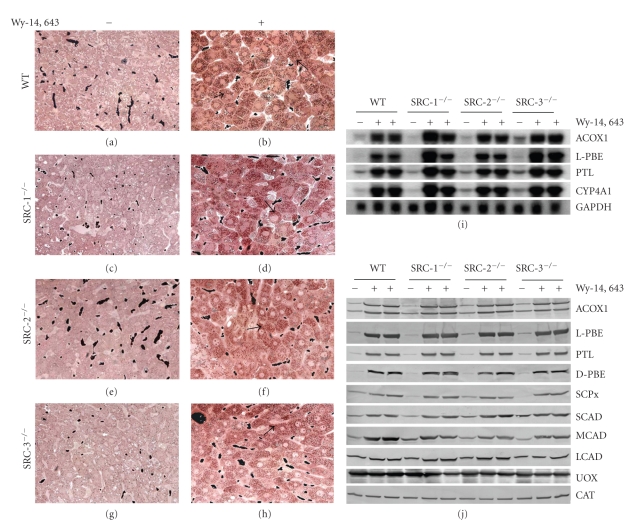
Mice lacking SRC-1 were generated to delineate its role in estrogen receptor, progesterone receptor, and PPARα signaling [90, 92]. SRC-1 null mice are viable and fertile but show somewhat subdued response to sex hormonal stimuli after orchiectomy or ovariectomy [90, 96]. However, SRC-1−/− mice when challenged with PPARα ligands, such as Wy-14,643 or ciprofibrate, display the characteristic robust pleiotropic responses, including hepatomegaly, hepatic peroxisome proliferation and PPARα-target gene activation [92]. These responses appear essentially similar to those exhibited by SRC-1+/+ littermates indicating that SRC-1 is not essential for PPARα signaling in liver [92]. Likewise, as shown here, studies with SRC-2 and SRC-3 null mice also revealed that PPARα target gene activation in liver is not dependent on these coactivators. Hepatic responses of SRC-1, SRC-2 and SRC-3 null mice following Wy-14,643 administration appear similar to those of wild-type mice treated with a PPARα ligand (Figure 2). Histological evaluation of liver sections, processed to visualize peroxisomal catalase, show similar increases in the number of peroxisomes in hepatic parenchymal cells of wild-type and SRC null mice treated with a peroxisome proliferator (Figures 2(a)–2(h)). To further investigate the influence of SRC family on PPARα function, we evaluated the changes in fatty acid-metabolizing enzymes in the liver of SRCs null and wild-type mice by Northern and Western blot analyses (Figures 2(i) and 2(j)). Northern blot analysis of total liver RNA shows similar basal levels of peroxisomal fatty acyl-CoA oxidase 1 (ACOX1), enoyl-CoA hydratase/L-3hydroxyacyl-CoA dehydrogenase bifunctional protein (L-PBE), peroxisomal 3-ketoacyl-CoA thiolase (PTL), and microsomal cytochrome P450 fatty acid ω-hydroxylase CYP4A1 in the livers of both wild-type and SRC-1, -2 and -3 null mice (Figure 2(i)). Massive increases in hepatic mRNA levels of these enzymes were noted in all SRC null mice treated with a PPARα ligand (Figure 2(i)). The increases appear similar to that noted in the livers of Wy-14,643 treated wild-type mice (Figure 2(i)). Western blot analysis reveals increases in the content of fatty acid oxidation enzyme proteins in liver of intact and SRC null mice (Figure 2(j)). The expression levels of hepatic peroxisomal fatty acid β-oxidation enzymes ACOX1, L-PBE, D-PBE, PTL, and SCPx (sterol carrier protein x) were increased significantly after Wy-14,643 administration in both wild-type and SRC-1, -2, and -3 null mice (Figure 2(j)). Furthermore, no significant differences in the magnitude of increases are observed in hepatic mitochondrial enzymes short-chain acyl-CoA dehydrogenase (SCAD), medium-chain acyl-CoA dehydrogenase (MCAD), and long-chain acyl-CoA dehydrogenase (LCAD) among wild-type and SRCs null treated mice (Figure 2(j)). Together, these data indicate that no member of p160/SRC family of coactivators (SRC-1, SRC-2 and SRC-3) is required for PPARα-mediated transcriptional activation in vivo [90].
Figure 2.
(a–h) Peroxisome proliferation in liver cells of wild-type (WT), SRC-1−/− SRC-2−/− and SRC-3−/− mice treated with Wy-14,643 for 4 days. Liver sections were processed for the cytochemical localization of peroxisomal catalase by using alkaline 3′,3′-diaminobenzidine substrate. Control diet (upper panels; a, c, e, g). Wy-14,643 diet (lower panels; b, d, f, h). Peroxisomes appear as brown dots (arrows) distributed throughout the cytoplasm in these 0.5 μm thick sections. All mice, wild-type and SRC nulls displayed extensive peroxisome proliferation after treatment with Wy-14, 643 indicating that these coactivators are not required for PPARα-regulated pleiotropic responses including fatty acid oxidation. (i) Northern blot analysis to confirm changes in mRNA expression of peroxisomal and microsomal fatty acid metabolizing enzymes in wild-type and SRC nulls after 4-days treatment with PPARα ligand Wy-14, 643. All genes are regulated by PPARα. Fatty acyl-CoA oxidase-1 (ACOX1), peroxisomal enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase bifunctional enzyme (L-PBE), and peroxisomal thiolase (PTL) represent peroxisomal β-oxidation system while CYP4A1 is involved in microsomal ω-oxidation of fatty acids. GAPDH is used as an indicator of RNA loading. (j) Western blot analysis from the above-mentioned livers was used to verify the degree of expression of peroxisomal and mitochondrial fatty acid metabolizing enzymes in wild-type and SRC knockout mice. Liver homogenates (20 μg) from each group of mice were run on 4–20% SDS PAGE gel and immunoblotted using antibodies for peroxisomal (ACOX1, L-PBE, PTL, D-PBE, and SCPx) and mitochondrial (SCAD, MCAD, and VLCAD) fatty acid metabolizing enzymes. No difference in the induction was observed between SRC nulls and wild-type for the β-oxidation enzymes.
Although the single gene-knockout mice have demonstrated that loss of individual members of p160/SRC family of coactivators is redundant for PPARα function in liver, it remains to be ascertained if deletion of two or all three of these coactivators affects the PPARα signaling. SRC-1 is required for the assembly of a complex that includes CBP/p300 for enhancing the coactivator function of PPARγ coactivator-1 (PGC-1) [95]. It appears that the docking of PGC-1 to PPARγ stimulates an apparent conformational change in PGC-1 that permits binding of SRC-1 and CBP/p300, resulting in a large increase in transcriptional activity, and this transcriptional enhancement function of PGC-1 fails to manifest in SRC-1 null cells [97]. SRC-1 null mice exhibit partial impairment of PPARγ function with decreased PGC-1 regulated thermogenic activity in brown adipose tissue [95]. The lower energy expenditure in SRC-1 null mice predisposes to higher sensitivity to obesity upon high fat feeding [88, 95, 98]. Fatty acid oxidation in brown fat is also decreased due to partial impairment of PPARγ/PPARα function in the absence of SRC-1.
SRC-2 has been implicated in a broader range of physiological processes including reproduction, mammary morphogenesis, uterine function and energy metabolism by affecting the regulation of adaptive thermogenesis [99]. SRC-2 null mice are viable, but the fertility of both sexes is impaired [94, 100]. These mice are resistant to diet-induced obesity and displayed enhanced adaptive thermogenesis [95]. In white adipose tissue, disruption of SRC-2 increases leptin expression as well as that of genes involved in lipolysis. Additionally, these mice manifest a decreased potential for fatty acid storage [95, 101]. Disruption of SRC-2 gene in the mouse reduces PPARγ function in white adipose tissue resulting in a lesser degree of fat accumulation [76, 95]. This enhances the function and development of brown adipose tissue, leading to increased levels of uncoupling protein 1, PGC 1α and ACOX1, promoting energy expenditure [76, 87, 95]. Gene knockout studies have demonstrated that SRC-3 is required for normal growth, puberty and mammary gland development [93]. SRC-3 null mice also show reduced body weight and adipose tissue mass with a significant decrease in PPARγ expression. At molecular level, SRC-3 interacts with the transcription factor CAAT/enchancer-binding protein(C/EBP) to control gene expression through PPARγ. These results imply that SRC-3 exerts a key role in adipocyte differentiation in vitro and in vivo, and that this regulation of adipogenesis is upstream of PPARγ [101, 102]. SRC-3 gene is often amplified or overexpressed in many types of cancers [93, 94].
The redundancy of p160/SRC family of coactivators in the energy balance or expenditure function of PPARs noted in SRC single-knockout mice suggests the existence of possible cooperative effects among the three members of the SRC family. To illuminate the physiological functions affected by double deletion of these coactivators, mice deficient in both SRC-1 and SRC-2 [100], or deficient in both SRC-1 and SRC-3 [50] have been generated. Most SRC-1/SRC-2 double null mice die at birth, generally before the weaning stage [100]. SRC-2−/− mice are protected against obesity and display enhanced adaptive thermogenesis, whereas SRC-1−/− mice are prone to obesity due to reduced energy expenditure. Together, these two members of SRC family control energy balance between white and brown adipose tissues through regulating PPARγ activity [101].
Most SRC-1 and SRC-3 double null mice also die before birth and surviving combined-knockout mice are lean and resistant to high-fat diet induced obesity [50]. These mice exhibit a developmental arrest in interscapular brown adipose tissue and defective thermogenesis due to a deficiency in the regulation of selective PPARγ target genes involved in adipogenesis and mitochondrial uncoupling. It is of interest that these double null mice consume more food because of lower leptin levels, but remain lean mostly due to a higher basal metabolic rate and enhanced physical activity [50]. Taken together, SRC-1 and SRC-3 play critical roles in energy balance by mediating both energy intake and energy expenditure [75].
To investigate the changes in gene expression profiles, microarray analysis has been used with the RNA from livers of SRC-1, SRC-2, and SRC-3 single null animals [98]. The overall pattern of altered hepatic gene expression in the SRC-1 null mice was one of upregulation as compared to wild-type mice. SRC-2 null mice appeared an overall downregulation compared to wild-type mice. In SRC-3 deficient mice, a minimal change of gene expression in liver was observed. All these data suggest that changes in gene expression for each SRC member show specific and nonoverlapping expression patterns and that the three members of SRC family play a key role in coregulating energy homeostasis and obesity [76, 98]. It is clear from experiments in mice that all three members of the SRC family contain both redundant and distinct functions and that each individual SRC contains the capacity to regulate different biological functions [53].
5. CBP/p300 with HAT Activity
CBP and p300, generally referred to as CBP/p300, are universal coactivators that link transcriptional activators to the basal transcription apparatus and provide a scaffold to integrate multiple cofactors. CBP was first identified as a protein that binds CREB (cAMP response element-binding protein), whereas p300 was cloned as an adenovirus oncoprotein E1A-associated protein [56]. Subsequent studies established that CBP and p300 are significantly related and that human CBP resembles the human p300 more closely [103, 104]. These proteins are well conserved amongst mammals and homologs of CBP/p300 have been found in Drosophila, Caenorhabditis elegans, and Arabidopsis thaliana [105]. No CBP/p300 homologs are found in prokaryotes or yeast [104]. CBP/p300 proteins possess HAT activity and the HAT domain has the ability to recruit other proteins such as p160/SRC family members with HAT activity to further enhance the acetylation potential of the coactivator complex to remodel chromatin structure for efficient gene transcription [76, 106–108]. CBP/p300 proteins share several conserved regions, which constitute most of their known functional domains such as bromodomain [109], cysteine/histidine-rich domains, a KIX domain to which transcription factor CREB binds, glutamine- and proline-rich domain, receptor-interacting domain, and SRC-1 interaction domain [108]. The C-terminal glutamine-rich domain of CBP/p300 forms contacts with other coactivators, most notably those involved in nuclear hormone-receptor signaling pathways. The complexity and breadth of CBP/p300 interactions attest to the unique involvement of CBP/p300 in the transcriptional control as universal and versatile cointegrators.
CBP and p300 directly interact with the ligand-binding domain of several nuclear receptors including PPARs [110]. C-terminal of PPARα that corresponds to AF-2 domain (residues 448–468) is required for the interaction with N-terminal region of p300 spanning aa 39–117, and also the N-terminal fragment of CBP encompassing aa 1–115 in a ligand dependent manner. Fragments of both p300 and CBP that interact with PPARα contain one LXXLL motif [110]. Interaction of CBP and p300 with the ligand-binding domain of PPARα or PPARβ was demonstrated in human intestine-like Caco2 cell line [111]. Induction of conformational change and transactivation potential of PPARβ was considerably lower than that of PPARα in response to arachidonic acid, as well as other polyunsaturated fatty acids [112]. Arachidonic acid enhanced binding of p300 to PPARα but not to PPARβ. Additionally, arachidonic acid induced in vitro binding of both PPARα-RXRα and PPARβ-RXRα heterodimers to several PPREs [113, 114]. CBP, which is highly expressed in brown fat, also coactivates PPARα-dependent regulation of the UCP-1 gene promoter in the HepG2 cells in the presence of PPARα ligand, Wy-14,643 [115]. Presence of CBP in PRIC complex that interacts with full-length PPARα in the presence of ciprofibrate and leukotriene B4 also substantiate the coactivator status of CBP [63].
p300 interacts with the N- and C-terminal regions of PPARγ in a ligand-independent and -dependent manner, respectively [116]. Leu-311, Asn-312, and Thr-316 in helix 4 of PPARγ ligand-binding domain are involved in PPARγ binding with CBP [117]. Deletion of A/B-domain of PPARγ compromises recruitment or stabilization of CBP- and p300-containing cofactor complexes on a subset of target genes involved in lipid storage [118]. Both PPARγ and CBP are expressed in preadipocytes and differentiated adipocytes suggesting that CBP serves as a physiologically relevant coactivator for PPARγ signal transduction [119]. p300 also transactivates PPARγ in the presence of natural ligand 15-deoxy-Δ12, 14-prostaglandin J2, but troglitazone, a synthetic PPARγ ligand, failed to induce PPARγ interactions with p300. CBP/p300 also increases the transcriptional activity of PPARγ through PGC-1 which stimulates an apparent conformational change in PGC-1 that permits binding of SRC-1 and CBP/p300 resulting transactivation of target genes [91]. Recruitment of CBP/p300 and PGC-1α was shown recently on PPARγ/RXRα heterodimer bound to the promoter of UCP-1 gene after the activation of β-adrenergic receptor in Jhdm2a knockout mice [120]. Jhdm2a is a H3K9-specific demethylase that directly regulates PPARα and Ucp1 expression [120]. PPARγ recruits CBP to the aP2 gene promoter as evidence by chromatin immunoprecipitation and in vitro immunoprecipitation assay in the MEFs induced for adipogenic differentiation [121]. Recently, it has been shown that beraprost, a synthetic analogue of prostacyclin serves as a ligand for PPARβ that enhances transcriptional activation of p21/p27 by increasing CBP nuclear translocation, which contributes to the vasoprotective action in rat aortic smooth muscle cells [122].
6. PBP (PPARBP)/TRAP220/DRIP205/MED1
PPAR-binding protein (PBP/PPARBP) was first cloned through yeast two-hybrid system using Gal4-PPARγ as bait to screen a mouse liver cDNA library and identified as a nuclear receptor coactivator with 2 LXXLL motifs [58]. Subsequently, PBP was shown as a critical component of TRAP/DRIP/ARC/Mediator complex [57, 73, 74, 123, 124] and it is variously referred to as PBP (PPARBP)/TRAP220/DRIP205/Med1 subunit of Mediator complex [41]. The Mediator complex was first discovered in yeast and was shown to be essential for pol II dependent transcription [41]. The mammalian Mediator complex consists of ~31 subunits and PBP/TRAP220/Med1 is the prominent member of this complex [73, 75]. Med1 binds to several nuclear receptors such as PPARα, RARα, TRβ1, RXR, VDR, FXR, ERα and GR via two conserved LXXLL motifs in a ligand-dependent manner [58, 74, 121, 123–125], Med1 also interacts with a variety of other transcriptional factors, including tumor suppressor p53, five GATA family members, p300, PGC-1 and C/EBPβ [46, 79, 126–129]. These interactions imply a major role for Med1 in nuclear receptor mediated cellular proliferation, differentiation and homeostatic regulation [76, 130].
Med1 serves as an anchor for the Mediator complex and facilitates the linkage between HAT containing CBP/p300 and p160/SRC protein complex and pol II basal transcription machinery in regulating transcription [41, 46]. Phosphorylation of Med1 by mitogen-activated protein kinase- extracellular signal-regulated kinase (MAPK-ERK) promotes its association with Mediator [131, 132]. Med1 is widely expressed in many tissues of adult mice, including brain, heart, lung, liver, kidney, adipose tissues, and the most prominent being the testis [76, 133]. Recently, it has been reported that Med1 is a target for miR-205. miR-205 interacts with a specific target in the 3′-UTR sequence of Med1 and silences its expression in human trophoblasts exposed to hypoxia [134].
Med1 ablation leads to embryonic lethality at midgestation, day 11.5 postcoitum (E11.5), which is attributed, in part, to defects in the development of placental vasculature, similar to those encountered in PPARγ [78, 135–137]. Embryonic development of the heart, eye, vasculature and the hematopoietic system is altered in Med1 null mice. This phenotype is similar to that in mice deficient in members of GATA, a family of transcription factors that modulates differentiation of adipocytes, megakaryocytes and erythrocytes [79].
As indicated above, Med1 was first identified as a PPARγ coactivator and it plays an important role in the PPARγ signaling pathway [58, 78, 136]. Med1 and PPARγ interaction requires two LXXLL nuclear receptor recognition motifs present in Med1 [138]. Med1 modestly increases the transcriptional activity of PPARγ, and a truncated form of Med1 (aa 487–735) acts as a dominant negative repressor [74, 123]. It has been shown that the deletion of 12 amino acids from the extreme carboxyl terminus of PPARγ results in the abolition of Med1-PPARγ interaction [58]. However, deletion of the PPARγ A/B-domain does not affect Med1 recruitment [118]. To study the role of Med1 in PPARγ-mediated adipogenesis in vitro, Med1+/+ and Med1−/− MEFs were isolated from E10.0 littermate embryos and infected with a retroviral vector driving PPARγ [139]. Disruption of TRAP220/Med1 in MEFs is refractory to PPARγ-stimulated adipogenesis but not MyoD-stimulated myogenesis [139]. Surprisingly however, a conserved N-terminal region of Med1 that lacks the LXXLL motifs but gets incorporated into Mediator fully supports PPARγ-stimulated adipogenesis [138]. A direct interaction between PPARγ and the mediator complex through Med1 is not essential for PPARγ-stimulated adipogenesis and for PPARγ target gene expression in cultured fibroblasts [138]. Furthermore, PPARγ target gene expression and recruitment of Mediator to a PPARγ response element on the aP2 promoter in undifferentiated MEFs do not require Med1 [138]. These findings imply that the presence of alternative mechanisms for Mediator recruitment, possibly through intermediate cofactors or other cofactors that are functionally redundant with Med1 [138].
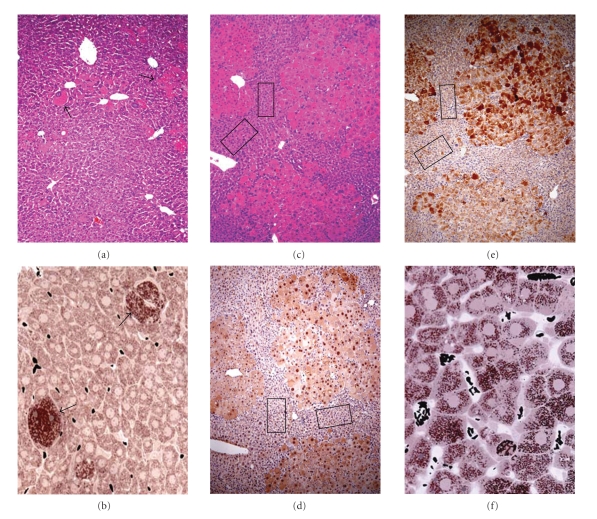
To further study the role of Med1 in specific tissues in vivo, mice carrying floxed Med1 alleles were generated for conditional null mutation [140, 141]. Conditional deletion of Med1 gene in liver results in the abrogation of PPARα ligand-induced pleiotropic effects, indicating that Med1 is essential for PPARα signaling and fatty acid oxidation [140]. Med1 deficiency in liver parenchymal cells results in the near abrogation of PPARα ligand-induced peroxisome proliferation, liver cell proliferation, and induction of PPARα-regulated genes. In contrast, scattered residual Med1+/+ hepatocytes that escape Cre-mediated excision of floxed alleles in Med1 liver nulls, show DNA synthesis and were markedly hypertrophic with peroxisome proliferation in response to PPARα ligands (Figures 3(a) and 3(b)). Med1−/− hepatocytes are refractory for PPARα ligand-induced peroxisome proliferation [140]. Moreover, Med1ΔLiv mice, chronically exposed to PPARα ligand Wy-14,643, show a striking proliferative response and clonal expansion of residual Med1+/+ hepatocytes (Figures 3(c) and 3(d)) but no proliferative expansion of Med1−/− hepatocytes occurs and these Med1 null hepatocyte appeared hypoplastic (boxed areas in Figures 3(c)–3(e)) as compared to hyperplastic large Med1+/+ hepatocytes.
Figure 3.
Effects of Med1 and PRIP deletion on PPARα ligand-induced pleiotropic responses in liver. Med1ΔLiv mice treated with Wy-14,643 (0.125% wt/wt) for 2 weeks (a, b) show an occasional hepatocyte that escaped Alb-Cre mediated deletion of Med1 floxed alleles. These Med1 positive hepatocytes respond to the peroxisome proliferative effects of PPARα-ligand (arrows indicate to Med1+/+ cells with numerous peroxisomes) but not the majority of Med1−/− hepatocytes. Chronic treatment of Med1ΔLiv mice with 0.02% Wy-14,643 for 5 months (c–e) results in clonal expansion of residual Med1fl/fl cells as demonstrated by H&E staining (c). In contrast, the adjacent hepatocyte lacking Med1 are generally smaller than normal hepatocytes (see boxed area in c). Immunohistochemical localization of Med1 reveals that expanding colonies of large hepatocytes are Med1 positive (nuclear Med1 in panel d). These cells also show abundant cytoplasmic expression of L-PBE, the second enzyme of the peroxisomal fatty acid β-oxidation system (panel e), whereas the smaller Med1 null hepatocytes (boxed areas) fail to show L-PBE induction (panel e). Disruption of coactivator PRIP in hepatocytes does not interfere with PPARα ligand-induced peroxisome proliferation as evidenced by the abundant catalase positive peroxisomes (brown dots) in all hepatocytes (panel f). Compare this panel (f) with panel (b) in which only an occasional Med1+/+ cell responds to PPARα ligand.
Surprisingly, the Med1 liver conditional null mice develop liver tumors on long-term exposure to PPARα ligand, but all tumors developing in Med1ΔLiv mice reveal Med1 expression and no tumors developed from Med1−/− hepatocytes [142]. These data suggest that Med1 plays a key role in PPARα ligand-induced liver tumor development and that cells deficient in Med1 do not give rise to tumors [142]. Furthermore, initiation by a genotoxic carcinogen diethylnitrosamine followed by phenobarbital promotion in Med1ΔLiv mice results in a failure of Med1 null hepatocytes to undergo proliferation. As in the case of Wy-14,643 treatment, all hepatocellular carcinomas developing in Med1ΔLiv mice are Med1 positive [143]. Liver tumors that develop in Med1ΔLiv mouse livers are transplantable in athymic nude mice and these maintain Med1fl/fl genotype. These observations imply that Med1 is essential for the development of hepatocellular carcinoma in the mouse [143]. The failure of Med1 null hepatocytes to develop liver tumors following PPARα ligand administration or after prolonged promotion with phenobarbital, which is an activator for nuclear receptor constitutive androstane receptor (CAR), implies that coactivator Med1 is a critical component of PPARα and CAR signaling and thus participates in the neoplastic process [142–144]. Med1 deficient livers fail to develop hepatic steatosis induced by glucorcorticoid receptor (GR) agonist [145] and also fail to develop hepatic steatosis when induced by PPARγ overexpression (unpublished data). In addition, using a conditional null mutation, it has been shown that Med1 is required for mammary gland development [146], and is also essential for the growth of Notch4-immortalized mammary cells by activating SOX10 expression [147]. Earlier studies have demonstrated the Med1 is either overexpressed or amplified in several breast carcinomas implying that Med1 plays a role in ER signaling and cancer [125, 148]. More recently, Med1 has been shown to play an important coregulatory role in prostate cancer cell proliferation and survival [149]. However, decrease of Med1 expression in human melanoma cells increases their tumorigenic phenotype and the reason for this discordancy is unclear [150].
In summary, using conditional knockout mice, it has been established that Med1 subunit is essential for the signaling of nuclear receptors PPARγ, PPARα, CAR and GR [139–145]. Evidence indicates that Med1-deficiency does not lead to the disintegration of the Mediator complex as originally speculated but it is possible that Mediator complex devoid of Med1 subunit may be impaired in its ability to recruit pol II to transduce the transcriptional signal [151].
7. PGC-1 Family in Coactivation of PPAR
PGC-1 family of coactivators, with three members, plays a critical role in the maintenance of mitochondrial function, thermogenesis and energy homeostasis [59]. The first member of the PGC-1 family was identified as a PPARγ-interacting protein from brown fat cDNA library using yeast two-hybrid screen and is now termed PGC-1α [59]. Thereafter, two related coactivators, PGC-1β (also termed PERC) and PGC-1-related coactivator (PRC) were discovered through searches of new data base entries [60, 152]. PGC-1α and PGC-1β share similar tissue distribution with highest levels of expression in brown fat, heart and slow-twitch skeletal muscle [59, 152], and their mRNA levels are induced significantly in the adult liver following fasting [152, 153]. Expression of PGC-1α mRNA is also elevated in brown fat after cold exposure, whereas PGC-1β does not respond [59, 152]. Less is known about the expression patterns and biological roles of PRC [60].
In addition to PPARγ, PGC-1α also coactivates a variety of other nuclear receptors, including PPARα [154], PPARβ [21], TRβ [59], ERα [59], GR [155], FXR [156], LXR [157], HNF4 [152] and RAR, but not RXRα [59]. Cotransfection experiments in cells show that PGC-1α increases the PPARα-mediated transcriptional activity and that AF2-LXXLL interaction is necessary for the coactivation of PPARα by PGC-1α [154]. Furthermore, overexpression of PPARα and PGC-1α in 3T3L1 cells cooperatively induces the expression of mitochondrial fatty acid β-oxidation enzyme system genes and increases cellular palmitate oxidation rates [154]. PPARα-driven mitochondrial biogenic response reveals that expression of PGC-1α is activated in wild-type mice but not in PPARα-deficient mice [158]. PGC-1α promotes expression of mammalian tribbles homolog TRB-3 through PPARα and knockdown of hepatic TRB-3 expression improves glucose tolerance, whereas hepatic overexpression of TRB-3 reversed the insulin-sensitive phenotype of PGC-1-deficient mice [159].
PGC-1α utilizes a domain rich in proline residues to bind to a region that overlaps the DNA binding and hinge region of PPARγ [59]. The strong interaction of PGC-1α and PPARγ is mediated through both hydrophobic and specific polar interactions. Mutations within the context of the full-length PGC-1α indicate that the first PGC-1α motif is necessary and sufficient for PGC-1α to coactivate PPARγ in the presence or absence of rosiglitazone [160]. Thiazolidinediones and rexinoids induce PGC-1α gene expression in brown and white adipocytes by a PPARγ-dependent pathway. This is attributed to the presence of a PPARγ-responsive element in the distal region of the PGC-1α gene promoter that binds PPARγ/RXR heterodimers [161]. The interaction between PGC-1α and PPARβ depends on the LXXLL motif in PGC-1α (aa 142–146) and this interaction is enhanced in the presence of PPARβ agonist GW501516 [21]. Tetradecylthioacetic acid, a pan-PPAR ligand, induces hepatic fatty acid oxidation in PPARα −/− mice possibly through PGC-1α mediated PPARβ coactivation [162]. Pharmacological activation of PPARβ induces fatty acid oxidation and this depends upon PGC-1α as the induction is completely abolished in the absence of both PGC-1α and PGC-1β [163]. To study the role of PGC-1α in vivo, PGC-1α knockout mice have been generated [164, 165], which show normal embryonic development, suggesting that PGC-1α driven coactivation is not critical for PPARγ and PPARβ functions in the maintenance of placental adequacy [164, 165]. Interestingly, PGC-1α −/− mice survive with modestly blunted postnatal cardiac growth, suggesting that PGC-1α is essential for the maintenance of maximal, efficient cardiac mitochondrial fatty acid oxidation, ATP synthesis, and myocardial lipid homeostasis [165, 166].
Hepatic PGC-1β overexpression results in the attenuation of changes induced by Wy-14,643, a PPARα ligand [167]. PGC-1β poorly activates the expression of gluconeogenic genes in hepatocytes or liver in vivo. The reduced ability of PGC-1β to induce gluconeogenic genes is due, in part, to its inability to physically associate with and coactivate HNF4α and FOXO1 [153]. PGC-1β null mice are viable and fertile and show no overt phenotype. However, PGC-1β deficient mice display an altered expression in a large number of nuclear-encoded genes governing mitochondrial functions in multiple tissues including heart, skeletal muscle, brain, brown adipose tissue, and liver [168]. PGC-1α null mice appeared hyperactive in comparison to somewhat sluggish PGC-1β null mice. When acutely exposed to cold, these mice develop abnormal hypothermia and morbidity [168]. Furthermore, high-fat feeding induced hepatic steatosis and increases serum triglyceride and cholesterol levels in the mutant mice [168].
8. PRIP/NCoA6
Nuclear receptor coactivator PRIP (PPAR-interacting protein) [168], also referred to as activating signal cointegrator-2(ASC-2) [83]/nuclear receptor activating protein 250 (RAP250) [61]/nuclear receptor coregulator (NRC) [84]/thyroid hormone receptor (TR)-binding protein (TRBP) [85], was cloned by different groups using yeast two-hybrid screens with a nuclear receptor as bait. PRIP (NCoA6) was identified as a ligand-dependent nuclear receptor-interacting protein. PRIP forms large steady-state complex of approximately 2MDa (ASC-2 complex [ASCOM] with retinoblastoma-binding protein RBQ-3, α/β-tubulins and subset of Trithorax-related proteins [169]. PRIP, like MED1 gene is also amplified and overexpressed in breast, colon and lung cancers [170, 171]. PRIP and PRIP-interacting protein with methyltransferase domain (PIMT/NCoA6IP) appears to serve as a linker between CBP/p300-anchored and Mediator complexes. PRIP contains two LXXLL motifs, one at the N-terminal region (aa 892 to 896) plays a pivotal role for ligand-dependent interactions with a wide spectrum of nuclear receptors including PPARs, TRs, RXRs, ERs, GR, VDR and RARs [64, 81, 83] and the second LXXLL located at the C-terminal does not bind with PPARs, RAR, RXRs, and GR but does bind with LXRα and LXRβ [85, 172–174]. These two LXXLLs interact with LXRs and other nuclear receptors and regulate insulin secretion and maintain β-cell function [175].
PRIP acts as a strong coactivator for PPARγ, and a truncated form of PRIP (aa 786–1132) acts as a dominant-negative repressor [62]. It is worth noting that PRIP is also detected in the transcriptionally active PPARα interacting cofactor (PRIC) complex isolated from rat liver [63]. PRIP protein expression has been detected in many tissues, especially in the reproductive organs such as testis, prostate and ovary [76, 176]. In the testis and ovary, PRIP immunostaining shows intense staining of the nuclei of Sertoli and follicular granulosa cells respectively [177]. Mice with disrupted PRIP/RAP250/NRC/AIB3 gene die at embryonic stage E11.5 and E12.5 days. PRIP mutant embryo mortality has been attributed to placental dysfunction including the failure of labyrinthine development, the dilation of maternal blood sinuses, the massive erythrophagocytosis by trophoblastic cells, alteration in trophoblast population and the formation of fewer blood vessels in extra-embryonic membrane covering the embryo [76, 85, 170, 173, 178, 179]. In addition, developmental abnormalities in heart, liver, and the nervous system have been noted [85, 173, 178]. Interestingly, MEFs derived from PRIP/NRC null embryos display growth retardation and apoptosis [176]. Further studies with heterozygous PRIP/NRC+/- show a spontaneous wound healing deficiency, suggesting that PRIP/NRC is important in maintaining integrity during wound healing [176, 180]. Haploid inactivation of PRIP/AIB3 in AIB3+/-/PyMT bitransgenic mice cause inhibition of cell proliferation mediated by PPARγ/RXR [181].
PRIP null MEFs are also resistant to PPARγ stimulated adipogenesis [121]. This defect occurs because of apparent disruption of the linkage between the CBP/p300 anchored subunit complex and the MED1-dependent mediator complex [121]. In order to investigate the physiological role of PRIP in vivo, conditional knockout mice have been generated [182]. Conditional PRIP null mutation in the mouse mammary gland results in defective mammopoiesis, similar to that encountered in Med1 deficient mammary glands [146, 168]. To further understand the function of PRIP in mammary gland tumorigenesis, a mammary tumor cell line with the PRIPloxP/loxP genotype was established and disruption of the PRIP gene in these cells has been shown to abrogate their tumorigenic potential. PRIP deficiency substantially reduced the expression of FOS gene [171]. Liver-specific disruption of the PRIP gene fails to affect the induction of PPARα-regulated pleiotropic responses, including hepatomegaly, hepatic peroxisome proliferation (Figure 3(f)), and induction of genes involved in the fatty acid oxidation systems [182]. These results are dissimilar to those encountered with liver specific MED1 gene disruption [140], indicating that PRIP is not essential for PPARα target gene activation in liver [182].
9. PRIC285
PRIC complex isolated from the rat liver nuclear extract using full-length GST-PPARα fusion protein in the presence of a PPARα ligand comprises of ~25 subunits [63]. A protein complex similar to this was also obtained using PPARα ligand (ciprofibrate)-affinity matrix [64]. MALDI-TOF analysis of the components of PRIC complex showed identities of many already known genes identified in yeast two-hybrid screens known to be involved in transcriptional regulation and some novel proteins [63]. This complex includes CBP/p300, p160/SRC-1, MED1, PRIP, PIMT and novel coactivators designated as PRIC285, PRIC295, and PRIC320 based on estimated molecular size (see [63, 64], unpublished data). PRIC285 is a component in the PRIC complex isolated using ciprofibrate or LTB4 as ligand in GST PPARα pull down system [63]. Subsequently, a longer isoform has been cloned using human PPARγ as bait in yeast two-hybrid screen and has been referred to as PPARγ-DBD-interacting protein 1 (PDIP1)-α [183]. PRIC285 is expressed in multiple human tissues such as skeletal muscle, colon, spleen, liver, kidney, heart, lung, pancreas, small intestine, thymus, prostate, ovary, peripheral blood, and placenta [63]. PRIC285 has been detected in several human cancer lines such as HeLa, colorectal adenocarcinoma SW480, melanoma, HepG2, medulloblastoma HTB185, and DU145 (prostate) [63, 183].
The human PRIC285 gene, which spans ~16.1 kb, is located on chromosome 20 at position 20q13.33 and it encodes a protein of 2080 amino acid with an estimated molecular mass of 285 kDa [63]. PRIC285 contains five LXXLL signature motifs at aa 506–510; 549–553; 604–608; 1443–1447; 1660–1664 [183]. It appears that none of these LXXLL motifs of PRIC285 is needed for interaction with PPARs as demonstrated by mutating LXXLL motifs [183]. PRIC285 binds to the DBD-hinge (DBD-H) of the PPARs through its C-terminal region mapped at aa 1675–1823 [183]. Comparison of the amino acid sequences flanking core LXXLL motifs in PRIC285 with those identified in other coactivators revealed that this configuration did not fit well with the proposed alignment rules [184]. Other than LXXLL signature motifs human PRIC285 also displays amino acid sequence homologous to RnaseB (RNB) and UvrD/REP motifs, a superfamily I DNA helicase [63, 183]. It is found to be transcriptional coactivator for the PPARα, PPARδ/β and PPARγ in transfected cells. Cotransfection of PPARα and PRIC285 into HEK293 cells stimulates transcription of PPRE-TK-Luc gene in the presence of ciprofibrate, a PPARα specific ligand. This interaction between PRIC285 and PPARα was also shown by the colocalization in nucleus of 293 cells [63]. Transactivation of PPARα by PRIC285 also occurs after treatment with a different PPARα ligand, fenofibrate [183]. Human PRIC285 has been shown to enhance PPARγ-mediated transactivation of DR1 reporter gene using the synthetic PPARγ ligand troglitazone. PRIC285 also coactivates PPARβ in the transactivation assay where CV-1 cells were treated with PPARβ ligand cyclic prostaglandin [183].
To assess the biological significance of PRIC285, we have generated whole-body gene knockout mice using two-loxP and two-frt system and characterized them for PPARα-mediated transcriptional activation in vivo [185]. Mice homozygous for PRIC285 mutation (PRIC285−/−) are apparently healthy and fertile and show no consistent phenotypic differences when compared to their wild-type floxed littermates (PRIC285fl/fl). When challenged with PPARα ligands, such as Wy-14,643 or ciprofibrate, no differences were observed in the magnitude of pleiotropic responses, which include hepatomegaly, peroxisome proliferation in hepatocytes, and increased levels of PPARα target genes such as peroxisomal and mitochondrial β-oxidation enzymes [185]. The role of PRIC285 in PPARγ mediated adipogenesis in the liver has been examined using PRIC285 null mice. Adenovirally driven PPARγ gene when injected through tail vein induced hepatic steatosis in both PRIC285 null and wild type floxed littermates to delineate the role of the coactivator PRIC285 in hepatic steatosis. No discernible differences in the PPARγ-mediated hepatic adipogenic steatosis in the normal and mutant PRIC285 mouse liver has been noted [L. Bai, unpublished]. These results may point to a functional redundancy of PRIC285 in the general transcriptional machinery as far as PPARα and PPARγ are concerned. The discordance between in vitro and in vivo results of PRIC285 function reflects the complexity and redundancy in that loss of a single component of a multisubunit protein complex could be compensated in vivo by other members of this mega complex. Nonetheless, it would be a challenge for the immediate future to assess the role of PRIC285 in the signaling of other nuclear receptors in vivo using the whole-body or conditional deletion.
10. PRIC320/CReMM/CHD9: ATP Dependent Chromatin Remodeling Activity
PRIC320 was identified in high molecular weight protein complex isolated using ciprofibrate coupled AH-sepharose affinity pulldown from the rat liver nuclear extract [64]. PRIC320 is also known as chromodomain helicase DNA binding protein 9 (CHD9)/Chromatin Related Mesenchymal Modulator (CReMM). It is a member of the CHD (chromodomain-helicase-DNA-binding) family of proteins that interacts with nucleosomes and plays a role in chromatin remodeling to modulate transcription [64, 186–188]. Members of the CHD family of enzymes belong to the SWI/SNF2 (SWItch/Sucrose Nonfermentable) superfamily of ATP-dependent chromatin remodelers. PRIC320 displays two tandem N-terminal chromodomains (aa 692–752; 774–826) that function as interaction surfaces for a variety of chromatin components. It also contains SNF2-like ATPase/DEAD-like helicase domain (aa 879–1028) located in the central region of the protein structure [189, 190]. A C-terminal cluster of domains such as paired BRK (Brahma and Kismet; aa 2483–2532; 2557–2601) domains, a SANT-like domain, and a DNA-binding domain are also present in PRIC320 [64, 191, 192]. The SNF2-like ATPase/DEAD-like helicase domain contains a conserved set of amino acid motifs that has been found in proteins involved in many of cellular processes including chromatin assembly, transcription regulation, DNA repair, DNA replication, development and differentiation [69, 193]. PRIC320 contains five LXXLL signature motifs that mediate interaction with nuclear receptors [64].
Two isoforms of PRIC320 designated as PRIC320-1 and PRIC320-2 that encode aa 2882 and 1995, respectively, have been identified in the human [64]. PRIC320-1 with an estimated molecular weight of 320 kDa contains all five LXXLL motifs located at aa 868–872; 1036–1040; 2031–2035; 2706–2710; and 2778–2782, whereas PRIC320-2 with an estimated molecular weight of 240 kDa contains distal four LXXLL motifs. The gene encoding human PRIC320 is mapped to long arm of chromosome at 16q12.2 (Ensembl; www.ensembl.org). PRIC320 transcript is present in various human tissues though at very low levels. PRIC320 mRNA has been detected in cancer cell lines such as HL-60, HeLa, Burkitt's lymphoma Raji and colorectal and lung carcinoma [64]. Cancer cell lines such as HL-60, HeLa cells, are shown to express two isoforms of mRNA 11.5 kb and 10.5 kb corresponding to PRIC320-1 and PRIC320-2. In mice, expression of PRIC320 is higher in brain, followed by heart, kidney, and skeletal muscle [64].
PRIC320 interacts with PPARα and functions as a coactivator in vitro [64]. Full length PPARα fused to GST interacts with both PRIC320 isoforms in a ligand dependent manner whereas interaction with PPARγ appeared minimal [64]. This selectivity for PPARs indicates a differential role of PRIC320 in the regulation of downstream target genes. The recognition in PRIC320/CHD9 of chromatin remodeling function and nuclear receptor coactivator function is suggestive of the multiple roles played by these nuclear receptor cofactors.
11. SWI/SNF: ATP-Dependent Chromatin Remodeling Complex
The SWI/SNF (mating type switch/Sucrose Nonfermenting) families of chromatin remodeling complexes mobilize nucleosomes and function as master regulators of transcription factor function. Although the precise mechanisms by which SWI/SNF modifies chromatin structure remains unclear, this process involves a conformational change of nucleosome and chromatin-remodeling in an ATP-dependent manner [65, 66]. SWI/SNF complex contains one or two possible ATPases, BRM (Brahma) or BRG1 (Brahma-Related gene 1) [194]. Chromatin remodelling represents an important step in adipocyte differentiation. C/EBPα, which is known to interact with the pol II-associated general transcription factors TBP/TFIIB, also interacts with BRM of the human SWI/SNF complex [195]. PPARγ depends on a specific BRG1-containing SWI/SNF complex to activate adipogenesis under in vitro conditions [196]. SWI/SNF complex and TFIIH are recruited on the promoter of PPARγ to transactivate PPARγ [197]. The docking of SWI/SNF complex on PPARγ promoter occurs through the subunit BAF60c (BRG1/Brm-associated factor subunit c) [69]. Recently, an interaction between SWI/SNF complexes and PPARα was demonstrated through BAF60a. SWI/SNF also plays a role in the regulation of the hepatic lipid metabolism through the fatty acid oxidation [67].
12. BAF(s) Family
The BAF (BRG1/Brm-associated factor) family represents the accessory subunits of SWI/SNF complexes that act as the connection between transcription factors and SWI/SNF complexes. Several BAFs been identified which include BAF250, BAF170, BAF155, BAF60, BAF57 and BAF53a [198–200]. Recent studies have implicated the BAF60 family members, including BAF60a, BAF60b and BAF60c, in mediating the interaction between the SWI/SNF complexes and target transcription factors. BAF60a or SMARCD1 (SWI/SNF related, matrix associated, actin-dependent regulator of chromatin subfamily d, member 1), a protein of 60 kDa, is known to be the connection between SWI/SNF and GR [68]. More recently, BAF60a was identified as a molecular link between SWI/SNF complexes and hepatic lipid metabolism. Adenoviral expression of BAF60a has been shown to stimulate fatty acid β-oxidation in primary hepatocytes culture to ameliorate hepatic steatosis in vivo. PGC-1α mediates the recruitment of BAF60a to the PPRE and enhances the transcription of PPARα regulated fatty acid oxidation system genes [67]. BAF60a is considered as a regulator of hepatic lipid metabolism. BAF60c or SMARCD3 (SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily d, member 3), which is also a 60 kDa protein, binds to several nuclear receptors, including PPARγ, ERα, and RORα [69]. Recently, a new regulator of PPARγ, the subunit BAF60c2 has been identified in a yeast two-hybrid screen of a human adipose tissue cDNA library. BAF60c2 represents a new isoform of BAF60c, which allows the recruitment of SWI/SNF to the nuclear receptor. Two isoforms BAF60c1 and BAF60c2 are localized primarily in the nucleus and are expressed in a wide variety of tissues. BAF60c proteins interact in a ligand-independent manner with PPARγ and enhance its transcriptional activity [69].
13. PIMT
PIMT (NCoA6IP, TGS1) was first isolated as a PRIP (NCoA6)-interacting protein [70]. PIMT is an RNA methyltransferase and was cloned from a human liver cDNA library using PRIP as bait in yeast two-hybrid assay. Human PIMT protein contains 852 amino acids. It has a methyltransferase motif at the C- terminus and an RNA-binding domain at the N-terminus [70]. PIMT is an evolutionarily conserved protein found in C. elegans, Arabidopsis thaliana, and yeast [70]. PIMT serves as a linker between multiprotein complexes anchored by CBP/p300 and PBP/MED1. PIMT enhances Med1mediated transcriptional activity of the PPARγ which was increased by PRIP [70, 79, 201]. Consistent with its RNA methyltransferase function, PIMT homologue in yeast known as trimethylguanosine synthase1 (TGS1), plays a role in the formation of the 2, 2, 7-trimethylguanosine (m3G) 5′-cap structure of snRNAs and small snoRNAs [202]. In Drosophila it is designated as DTL (Drosophila Tat-like) where it is important in development [203]. PIMT is localized predominantly to the nucleus. It is expressed in most adult tissues and in all embryonic stages in the mouse [76]. Inhibition of PIMT by siRNA in HeLa cells results in G2/M phase arrest [204].
In order to investigate the biological functions of this gene in mammalian development and growth, PIMT gene knockout mice have been generated [205]. Heterozygous (PIMT+/-) mice grow normally and are indistinguishable from their wild-type (PIMT+/+) littermates. Disruption of both PIMT alleles results in early embryonic lethality due to apoptosis and decreased proliferative potential of the blastocyst cells [205]. PIMT deficient embryos die shortly after implantation and then resorbed. PIMTfl/fl MEFs treated with adenovirus expressing Cre showed defective wound healing and G2 phase arrest of cell cycle. These results suggest that PIMT is important for early embryonic development of mice [205].
14. CARM1
Protein methylation is involved in regulating protein-protein interactions that affect key cellular events, including regulation of transcription [206]. Proteins can be methylated irreversibly on the side-chain nitrogens of the amino acids arginine, lysine, and histidine in a reaction with S-adenosylmethionine (AdoMet) [207]. Coactivator-associated arginine methyltransferase (CARM1)/protein arginine Nmethyltransferase 4 (PRMT4) is identified as a binding partner of SRC-2/GRIP1 (glucocorticoid receptor-interacting protein 1) [71]. Recently, CARM1 shown to promote adipocyte differentiation by coactivating PPARγ using cDNA microarray and serial analysis of gene expression (SAGE) [208]. CARM1 also stimulates transcriptional activation by nuclear receptors in combination with the p160 family of coactivators [71]. The p160/SRC coactivators recruit CBP/p300 and CARM1 via two activation domains, AD1 and AD2 [71]. AD1 binds CBP or p300, whereas AD2 has been shown to activate transcription through the recruitment of the arginine methyltransferase CARM1. The ternary complex of p160-CARM1-CBP/p300 functions synergistically to enhance transcriptional activation by nuclear receptors. CARM1 efficiently methylates three arginine residues (R714, R742 and R768) spanning aa 685–774 of CBP which are also conserved in p300 to transactivate SRC-2/GRIP-1 [209]. Other than these three methylated arginine residues, CARM1 also methylates KIX domain of CBP/p300 to block the interaction with KID domain of CREB (Cyclic AMP response element binding protein) [210]. Methylation is an irreversible process but peptidyl deiminase 4 removes methylated arginine from the p300 (Arg-2142) which is localized in the p160-binding domain to inhibit the bimolecular interactions between p300 and GRIP1. The functional significance of the methylation and demethylimination of the arginine residue of p300 may be a key mechanism in p300/CBP-p160-CARM1 coactivator synergy. Methylation of p300/CBP by CARM1 promotes a conformational change that allows the p300-p160 interaction in the complex and facilitates additional steps in transcriptional activation [211].
Although CARM1 methylates CBP/p300 to enhance protein-protein interaction in the activated nuclear receptor complex, it also methylates histone tail, preferentially H3 which in turn relaxes chromatin to generate a docking site for coactivators and other transcriptional factors on the promoter of the target genes [71, 212, 213]. Methylation of the Q-rich domain of SRC-3 through CARM1 has an antagonizing activity on ER-mediated transcriptional activation [214]. During estrogen signaling methylation of SRC-3/AIB1 by CARM1 attenuates the transcriptional response by dissociating SRC-3/CARM1 coactivator complex from the ER receptor and thus completing a dynamic equilibrium of receptor-mediated coactivator assembly and disassembly at the promoter [214]. Mice deficient in CARM1 die at the perinatal stage emphasizing that CARM1 is crucial during late embryonic development or immediately after birth [215]. Methylation of CBP/p300 was shown to be abolished in the CARM1 knockout embryos and cells. Thus, it appears that CARM1 mediated methylation is needed for interaction between p/160 family of proteins and CBP/p300 to maintain general transcript integrity.
15. Coactivator Activator (CoAA) with RNA Splicing Activity
The Coactivator activator (CoAA) was first identified as a protein associated with PRIP/TRBP (thyroid hormone receptor-binding protein) in a yeast two-hybrid screen [72]. CoAA functions as a general activator of transcription for several nuclear receptors and stimulates transcription through its interaction with the C-terminal of PRIP/TRBP [72]. CoAA interacts with both PRIP/TRBP and p300 in vitro. The PRIP-interaction domain on CoAA protein is localized at the central region, which is encoded by exon 2 of CoAA gene [72]. In addition, CoAA potently coactivates transcription mediated by multiple hormone-response elements and acts synergistically with PRIP/TRBP and CBP. Thus, CoAA appears to function as a coactivator associated protein. Apart from participating in PRIP/TRBPmediated transcription, CoAA also regulates alternative splicing in a promoter-dependent manner [72]. The N-terminal region of the CoAA protein contains two RNA recognition motifs (RRMs) at amino acid position 3–68 and 81–144. Both RRMs are composed of two conserved ribonucleoprotein (RNP) consensus motifs that regulate posttranslational RNA splicing [216]. Coactivator modulator (CoAM), a splice variant is generated as a result of alternative splicing of exon 2 of the CoAA. CoAM, which lacks a PRIP/TRBP-interacting domain, represses both PRIP/TRBP and CBP action suggesting that CoAM may modulate endogenous CoAA function [72]. In conclusion, CoAA and PIMT, both capable of interacting with PRIP/TRBP/NCoA6 appear to function as regulators of RNA processing.
16. Conclusion
During the past 15 years, using yeast 2 hybrid screening [51, 52, 58, 59, 84], affinity pulldown of nuclear extracts via covalently bound ligand to the sepharose matrix [63], GST-receptor pulldown [64] and proteomic approaches [217], over 300 nuclear receptor transciption cofactors have been identified. Transcriptional control is a multistep process, a fact reflected in the diversity of the coregulators, and their intrinsic enzyme activities. These coregulators are possibly organized into stable, preformed multiprotein complexes, the modular character of which may facilitate the efficient assembly of functionally diverse complexes by a liganded nuclear receptor. In addition, the modular character of these complexes provides the potential for different activators to assemble diverse configurations of regulatory complexes at their cognate cis-acting elements. Emerging genomic and proteomic approaches promise to advance the characterization of coactivator proteins and their physiological functions. It should be worth noting that of the many cofactors, about 165 coactivators have been implicated to date in various human diseases [218]. Gene knockout mouse models have clearly established that Med1 is necessary for PPARα and PPARγ functions and that SRC-1, SRC-2, and SRC-3 are redundant for PPARα function. It is anticipated that further studies of nuclear receptor coregulators and their complexes will yield significant insights into the basis of the complexity of signaling by PPARs and their ligands.
Acknowledgment
This paper was supported by NIH Grant no. DK083163 (J. K. Reddy).
References
- 1.Reddy JK, Krishnakantha TP. Hepatic peroxisome proliferation: induction by two novel compounds structurally unrelated to clofibrate. Science. 1975;190(4216):787–789. doi: 10.1126/science.1198095. [DOI] [PubMed] [Google Scholar]
- 2.Reddy JK, Rao MS, Moody DE. Hepatocellular carcinomas in acatalasemic mice treated with Nafenopin, a hypolipidemic peroxisome proliferator. Cancer Research. 1976;36(4):1211–1217. [PubMed] [Google Scholar]
- 3.Reddy JK, Azarnoff DL, Hignite CE. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature. 1980;283(5745):397–398. doi: 10.1038/283397a0. [DOI] [PubMed] [Google Scholar]
- 4.Reddy JK, Goel SK, Nemali MR. Transcriptional regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(6):1747–1751. doi: 10.1073/pnas.83.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347(6294):645–650. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 6.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68(5):879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 7.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocrine Reviews. 1999;20(5):649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 8.Michalik L, Auwerx J, Berger JP, et al. International union of pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacological Reviews. 2006;58(4):726–741. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- 9.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126(4):789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, Qi C, Korenberg JR, et al. Structural organization of mouse peroxisome proliferator-activated receptor γ (mPPARγ) gene: alternative promoter use and different splicing yield two mPPARγ isoforms. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(17):7921–7925. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lazar MA. Becoming fat. Genes and Development. 2002;16(1):1–5. doi: 10.1101/gad.964002. [DOI] [PubMed] [Google Scholar]
- 12.Krey G, Braissant O, L’Horset F, et al. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Molecular Endocrinology. 1997;11(6):779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 13.Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the x-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto T, Cook WS, Qi C, Yeldandi AV, Reddy JK, Rao MS. Defect in peroxisome proliferator-activated receptor α-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. The Journal of Biological Chemistry. 2000;275(37):28918–28928. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- 15.Lee SS-T, Pineau T, Drago J, et al. Targeted disruption of the α isoform of the peroxisome proliferator- activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Molecular and Cellular Biology. 1995;15(6):3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukherjee R, Locke KT, Miao B, et al. Novel peroxisome proliferator-activated receptor α agonists lower low-density lipoprotein and triglycerides, raise high-density lipoprotein, and synergistically increase cholesterol excretion with a liver X receptor agonist. Journal of Pharmacology and Experimental Therapeutics. 2008;327(3):716–726. doi: 10.1124/jpet.108.143271. [DOI] [PubMed] [Google Scholar]
- 17.Mei C-L, He P, Cheng B, Liu W, Wang Y-F, Wan J-J. Chlamydia pneumoniae induces macrophage-derived foam cell formation via PPAR α and PPAR γ-dependent pathways. Cell Biology International. 2009;33(3):301–308. doi: 10.1016/j.cellbi.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 18.Crisafulli C, Cuzzocrea S. The role of endogenous and exogenous ligands for the peroxisome proliferator-activated receptor alpha (PPAR-α) in the regulation of inflammation in macrophages. Shock. 2009;32(1):62–73. doi: 10.1097/shk.0b013e31818bbad6. [DOI] [PubMed] [Google Scholar]
- 19.Pyper SR, Viswakarma N, Yu S, Reddy JK. PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nuclear Receptor Signalling. 2010;16(8, article e002) doi: 10.1621/nrs.08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peters JM, Lee SST, Li W, et al. Growths, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor β(δ) Molecular and Cellular Biology. 2000;20(14):5119–5128. doi: 10.1128/mcb.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y-X, Lee C-H, Tiep S, et al. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113(2):159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 22.Sanderson LM, Degenhardt T, Koppen A, et al. Peroxisome proliferator-activated receptor β/δ (PPARβ/δ) but not PPARα serves as a plasma free fatty acid sensor in liver. Molecular and Cellular Biology. 2009;29(23):6257–6267. doi: 10.1128/MCB.00370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosen ED, Spiegelman BM. PPARγ: a nuclear regulator of metabolism, differentiation, and cell growth. The Journal of Biological Chemistry. 2001;276(41):37731–37734. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- 24.Yamauchi T, Kamon J, Waki H, et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. The Journal of Biological Chemistry. 2001;276(44):41245–41254. doi: 10.1074/jbc.M103241200. [DOI] [PubMed] [Google Scholar]
- 25.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79(7):1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 26.Kliewer SA, Xu HE, Lambert MH, Willson TM. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Progress in Hormone Research. 2001;56:239–263. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 27.Yamauchi T, Kamon J, Waki H, et al. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor γ (PPARγ) deficiency and PPARγ agonist improve insulin resistance. The Journal of Biological Chemistry. 2001;276(44):41245–41254. doi: 10.1074/jbc.M103241200. [DOI] [PubMed] [Google Scholar]
- 28.Varanasi U, Chu R, Huang Q, Castellon R, Yeldandi AV, Reddy JK. Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. The Journal of Biological Chemistry. 1996;271(4):2147–2155. doi: 10.1074/jbc.271.4.2147. [DOI] [PubMed] [Google Scholar]
- 29.Direnzo J, Söderström M, Kurokawa R, et al. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. Molecular and Cellular Biology. 1997;17(4):2166–2176. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandra V, Huang P, Hamuro Y, et al. Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karagianni P, Wong J. HDAC3: taking the SMRT-N-CoRrect road to repression. Oncogene. 2007;26(37):5439–5449. doi: 10.1038/sj.onc.1210612. [DOI] [PubMed] [Google Scholar]
- 32.Nagy L, Kao H-Y, Chakravarti D, et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89(3):373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 33.Wei L-N, Hu X, Chandra D, Seto E, Farooqui M. Receptor-interacting protein 140 directly recruits histone deacetylases for gene silencing. The Journal of Biological Chemistry. 2000;275(52):40782–40787. doi: 10.1074/jbc.M004821200. [DOI] [PubMed] [Google Scholar]
- 34.Miyata KS, McCaw SE, Meertens LM, Patel HV, Rachubinski RA, Capone JP. Receptor-interacting protein 140 interacts with and inhibits transactivation by, peroxisome proliferator-activated receptor α and liver- X-receptor α . Molecular and Cellular Endocrinology. 1998;146(1-2):69–76. doi: 10.1016/s0303-7207(98)00196-8. [DOI] [PubMed] [Google Scholar]
- 35.Debevec D, Christian M, Morganstein D, et al. Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor α . Molecular Endocrinology. 2007;21(7):1581–1592. doi: 10.1210/me.2007-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang YK, Guermah M, Yuan C-X, Roeder RG. The TRAP/mediator coactivator complex interacts directly with estrogen receptors α and β through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(5):2642–2647. doi: 10.1073/pnas.261715899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conaway JW, Florens L, Sato S, et al. The mammalian Mediator complex. FEBS Letters. 2005;579(4):904–908. doi: 10.1016/j.febslet.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 38.Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, Leid M. Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor α interacting protein. The Journal of Biological Chemistry. 1999;274(22):15901–15907. doi: 10.1074/jbc.274.22.15901. [DOI] [PubMed] [Google Scholar]
- 39.Xu HE, Lambert MH, Montana VG, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377(6548):454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 41.Kornberg RD. The molecular basis of eukaryotic transcription. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(32):12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tetel MJ, Auger AP, Charlier TD. Who’s in charge? Nuclear receptor coactivator and corepressor function in brain and behavior. Frontiers in Neuroendocrinology. 2009;30(3):328–342. doi: 10.1016/j.yfrne.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Malley BW, Kumar R. Nuclear receptor coregulators in cancer biology. Cancer Research. 2009;69(21):8217–8222. doi: 10.1158/0008-5472.CAN-09-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 45.McKenna NJ, O’Malley BW. Minireview: nuclear receptor coactivators—an update. Endocrinology. 2002;143(7):2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 46.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Letters. 2005;579(4):909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 47.Kwok RPS, Lundblad JR, Chrivia JC, et al. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370(6486):223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 48.Chekravarti D, LaMorte VJ, Nelson MC, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383(6595):99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 49.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes and Development. 2000;14(13):1553–1577. [PubMed] [Google Scholar]
- 50.Wang Z, Qi C, Krones A, et al. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metabolism. 2006;3(2):111–122. doi: 10.1016/j.cmet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Onate SA, Tsai SY, Tsai M-J, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270(5240):1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 52.Zhu Y, Qi C, Calandra C, Rao MS, Reddy JK. Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator-activated receptor gamma. Gene Expression. 1996;6(3):185–195. [PMC free article] [PubMed] [Google Scholar]
- 53.Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(10):4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Gomes PJ, Chen JD. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(16):8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277(5328):965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 56.Chrivia JC, Kwok RPS, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365(6449):855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 57.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends in Biochemical Sciences. 2005;30(5):256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 58.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. The Journal of Biological Chemistry. 1997;272(41):25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- 59.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 60.Lin J, Puigserver P, Donovan J, Tarr P, Spiegelman BM. Peroxisome proliferator-activated receptor γ coactivator 1β (PGC-1β), a novel PGC-1-related transcription coactivator associated with host cell factor. The Journal of Biological Chemistry. 2002;277(3):1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 61.Caira F, Antonson P, Pelto-Huikko M, Treuter E, Gustafsson J-Å. Cloning and characterization of RAP250, a novel nuclear receptor coactivator. The Journal of Biological Chemistry. 2000;275(8):5308–5317. doi: 10.1074/jbc.275.8.5308. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Y, Kan L, Qi C, et al. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. The Journal of Biological Chemistry. 2000;275(18):13510–13516. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- 63.Surapureddi S, Yu S, Bu H, et al. Identification of a transcriptionally active peroxisome proliferator-activated receptor α-interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11836–11841. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Surapureddi S, Viswakarma N, Yu S, Guo D, Rao MS, Reddy JK. PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex. Biochemical and Biophysical Research Communications. 2006;343(2):535–543. doi: 10.1016/j.bbrc.2006.02.160. [DOI] [PubMed] [Google Scholar]
- 65.Becker PB, Hörz W. Atp-dependent nucleosome remodeling. Annual Review of Biochemistry. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 66.Neely KE, Workman JL. The complexity of chromatin remodeling and its links to cancer. Biochimica et Biophysica Acta. 2002;1603(1):19–29. doi: 10.1016/s0304-419x(02)00067-7. [DOI] [PubMed] [Google Scholar]
- 67.Li S, Liu C, Li N, et al. Genome-wide coactivation analysis of PGC-1α identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metabolism. 2008;8(2):105–117. doi: 10.1016/j.cmet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hsiao P-W, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Molecular and Cellular Biology. 2003;23(17):6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Debril M-B, Gelman L, Fayard E, Annicotte J-S, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. The Journal of Biological Chemistry. 2004;279(16):16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 70.Zhu Y, Qi C, Cao W-Q, Yeldandi AV, Rao MS, Reddy JK. Cloning and characterization of PIMT, a protein with a methyltransferase domain, which interacts with and enhances nuclear receptor coactivator PRIP function. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10380–10385. doi: 10.1073/pnas.181347498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen D, Ma H, Hong H, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284(5423):2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 72.Iwasaki T, Chin WW, Ko L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM) The Journal of Biological Chemistry. 2001;276(36):33375–33383. doi: 10.1074/jbc.M101517200. [DOI] [PubMed] [Google Scholar]
- 73.Yuan C-X, Ito M, Fondell JD, Fu Z-Y, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(14):7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rachez C, Lemon BD, Suldan Z, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398(6730):824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 75.Reddy JK, Guo D, Jia Y, Yu S, Rao MS. Nuclear receptor transcriptional coactivators in development and metabolism. Advances in Developmental Biology. 2006;16:389–420. [Google Scholar]
- 76.Yu S, Reddy JK. Transcription coactivators for peroxisome proliferator-activated receptors. Biochimica et Biophysica Acta. 2007;1771(8):936–951. doi: 10.1016/j.bbalip.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 77.Yao T-P, Oh SP, Fuchs M, et al. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93(3):361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 78.Zhu Y, Qi C, Jia Y, Nye JS, Rao MS, Reddy JK. Deletion of PBP/PARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. The Journal of Biological Chemistry. 2000;275(20):14779–14782. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- 79.Crawford SE, Qi C, Misra P, et al. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. The Journal of Biological Chemistry. 2002;277(5):3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 80.Clark EL, Coulson A, Dalgliesh C, et al. The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Research. 2008;68(19):7938–7946. doi: 10.1158/0008-5472.CAN-08-0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370(6489):477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 82.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119(2):157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 83.Lee S-K, Anzick SL, Choi J-E, et al. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. The Journal of Biological Chemistry. 1999;274(48):34283–34293. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- 84.Mahajan MA, Samuels HH. A new family of nuclear receptor coregulators that integrate nuclear receptor signaling through CREB-binding protein. Molecular and Cellular Biology. 2000;20(14):5048–5063. doi: 10.1128/mcb.20.14.5048-5063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu Y-J, Crawford SE, Stellmach V, et al. Coactivator PRIP, the peroxisome proliferator-activated receptor-interacting protein, is a modulator of placental, cardiac, hepatic, and embryonic development. The Journal of Biological Chemistry. 2003;278(3):1986–1990. doi: 10.1074/jbc.C200634200. [DOI] [PubMed] [Google Scholar]
- 86.Li D, Kang Q, Wang D-M. Constitutive coactivator of peroxisome proliferator-activated receptor (PPARγ), a novel coactivator of PPARγ that promotes adipogenesis. Molecular Endocrinology. 2007;21(10):2320–2333. doi: 10.1210/me.2006-0520. [DOI] [PubMed] [Google Scholar]
- 87.Zhou G, Hashimoto Y, Kwak I, Tsai SY, Tsai M-J. Role of the Steroid Receptor Coactivator SRC-3 in Cell Growth. Molecular and Cellular Biology. 2003;23(21):7742–7755. doi: 10.1128/MCB.23.21.7742-7755.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu J, Wu R-C, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nature Reviews Cancer. 2009;9(9):615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang H, Yi X, Sun X, et al. Differential gene regulation by the SRC family of coactivators. Genes and Development. 2004;18(14):1753–1765. doi: 10.1101/gad.1194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai M-J, O’Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279(5358):1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 91.Puigserver P, Adelmant G, Wu Z, et al. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 92.Qi C, Zhu Y, Pan J, et al. Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor α-regulated gene expression. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(4):1585–1590. doi: 10.1073/pnas.96.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O’Malley BW. The steroid receptor coactivator SRC-3 (P/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Molecular and Cellular Biology. 2002;22(16):5923–5937. doi: 10.1128/MCB.22.16.5923-5937.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Picard F, Géhin M, Annicotte J-S, et al. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111(7):931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- 96.Weiss RE, Xu J, Ning G, Pohlenz J, O’Malley BW, Refetoff S. Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO Journal. 1999;18(7):1900–1904. doi: 10.1093/emboj/18.7.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Puigserver P, Adelmant G, Wu Z, et al. Activation of PPARγ coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 98.Jeong J-W, Kwak I, Lee KY, et al. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Molecular Endocrinology. 2006;20(5):1138–1152. doi: 10.1210/me.2005-0407. [DOI] [PubMed] [Google Scholar]
- 99.Chopra AR, Louet J-F, Saha P, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322(5906):1395–1399. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mark M, Yoshida-Komiya H, Gehin M, et al. Partially redundant functions of SRC-1 and TIF2 in postnatal survival and male reproduction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4453–4458. doi: 10.1073/pnas.0400234101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Louet J-F, Coste A, Amazit L, et al. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(47):17868–17873. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Louet J-F, O’Malley BW. Coregulators in adipogenesis: what could we learn from the SRC (p160) coactivator family? Cell Cycle. 2007;6(20):2448–2452. doi: 10.4161/cc.6.20.4777. [DOI] [PubMed] [Google Scholar]
- 103.Eckner R, Ewen ME, Newsome D, et al. Molecular cloning and functional analysis of the adenovirus E1A- associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes and Development. 1994;8(8):869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 104.Chekravarti D, LaMorte VJ, Nelson MC, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383(6595):99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 105.Arany Z, Sellers WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77(6):799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 106.Smith CL, Oñate SA, Tsai M-J, O’Malley BW. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(17):8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Torchia J, Rose DW, Inostroza J, et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear- receptor function. Nature. 1997;387(6634):677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 108.Waters L, Yue B, Veverka V, et al. Structural diversity in p160/CREB-binding protein coactivator complexes. The Journal of Biological Chemistry. 2006;281(21):14787–14795. doi: 10.1074/jbc.M600237200. [DOI] [PubMed] [Google Scholar]
- 109.Polesskaya A, Naguibneva I, Duquet A, Bengal E, Robin P, Harel-Bellan A. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Molecular and Cellular Biology. 2001;21(16):5312–5320. doi: 10.1128/MCB.21.16.5312-5320.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dowell P, Ishmael JE, Avram D, Peterson VJ, Nevrivy DJ, Leid M. p300 functions as a coactivator for the peroxisome proliferator- activated receptor α . The Journal of Biological Chemistry. 1997;272(52):33435–33443. doi: 10.1074/jbc.272.52.33435. [DOI] [PubMed] [Google Scholar]
- 111.Mochizuki K, Suruga K, Sakaguchi N, Takase S, Goda T. Major intestinal coactivator p300 strongly activates peroxisome proliferator-activated receptor in intestinal cell line, Caco-2. Gene. 2002;291(1-2):271–277. doi: 10.1016/s0378-1119(02)00625-x. [DOI] [PubMed] [Google Scholar]
- 112.Mochizuki K, Suzuki T, Goda T. PPARα and PPARδ transactivity and p300 binding activity induced by arachidonic acid in colorectal cancer cell line Caco-2. Journal of Nutritional Science and Vitaminology. 2008;54(4):298–302. doi: 10.3177/jnsv.54.298. [DOI] [PubMed] [Google Scholar]
- 113.Dreyer C, Keller H, Mahfoudi A, Laudet V, Krey G, Wahli W. Positive regulation of the peroxisomal β-oxidation pathway by fatty acids through activation of peroxisome proliferator-activated receptors (PPAR) Biology of the Cell. 1993;77(1):67–76. doi: 10.1016/s0248-4900(05)80176-5. [DOI] [PubMed] [Google Scholar]
- 114.Kliewer SA, Sundseth SS, Jones SA, et al. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ . Proceedings of the National Academy of Sciences of the United States of America. 1997;94(9):4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Barberá MJ, Schlüter A, Pedraza N, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor α activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. The Journal of Biological Chemistry. 2001;276(2):1486–1493. doi: 10.1074/jbc.M006246200. [DOI] [PubMed] [Google Scholar]
- 116.Gelman L, Zhou G, Fajas L, Raspé E, Fruchart J-C, Auwerx J. p300 Interacts with the N- and C-terminal part of PPARγ2 in a ligand- independent and -dependent manner, respectively. The Journal of Biological Chemistry. 1999;274(12):7681–7688. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- 117.Chen S, Johnson BA, Li Y, et al. Both coactivator LXXLL motif-dependent and -independent interactions are required for peroxisome proliferator-activated receptor γ (PPARγ) function. The Journal of Biological Chemistry. 2000;275(6):3733–3736. doi: 10.1074/jbc.275.6.3733. [DOI] [PubMed] [Google Scholar]
- 118.Bugge A, Grøntved L, Aagaard MM, Borup R, Mandrup S. The PPARγ2 A/B-domain plays a gene-specific role in transactivation and cofactor recruitment. Molecular Endocrinology. 2009;23(6):794–808. doi: 10.1210/me.2008-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mizukami J, Taniguchi T. The antidiabetic agent thiazolidinedione stimulates the interaction between PPARγ and CBP. Biochemical and Biophysical Research Communications. 1997;240(1):61–64. doi: 10.1006/bbrc.1997.7602. [DOI] [PubMed] [Google Scholar]
- 120.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458(7239):757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Qi C, Surapureddi S, Zhu Y-J, et al. Transcriptional coactivator PRIP, the peroxisome proliferator-activated receptor γ (PPARγ)-interacting protein, is required for PPARγ-mediated adipogenesis. The Journal of Biological Chemistry. 2003;278(28):25281–25284. doi: 10.1074/jbc.C300175200. [DOI] [PubMed] [Google Scholar]
- 122.Sue Y-M, Chung C-P, Lin H, et al. PPARδ-mediated p21/p27 induction via increased CREB-binding protein nuclear translocation in beraprost-induced antiproliferation of murine aortic smooth muscle cells. American Journal of Physiology. 2009;297(2):C321–C329. doi: 10.1152/ajpcell.00069.2009. [DOI] [PubMed] [Google Scholar]
- 123.Näär AM, Beaurang PA, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398(6730):828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 124.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends in Biochemical Sciences. 2005;30(5):235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 125.Hittelman AB, Burakov D, Iñiguez-Lluhí JA, Freedman LP, Garabedian MJ. Differential regulation of glucocorticoid receptor transcriptional activation via AF-1-associated proteins. EMBO Journal. 1999;18(19):5380–5388. doi: 10.1093/emboj/18.19.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Drané P, Barel M, Balbo M, Frade R. Identification of RB18A, a 205 kDa new p53 regulatory protein which shares antigenic and functional properties with p53. Oncogene. 1997;15(25):3013–3024. doi: 10.1038/sj.onc.1201492. [DOI] [PubMed] [Google Scholar]
- 127.Lottin-Divoux S, Barel M, Frade R. RB18A enhances expression of mutant p53 protein in human cells. FEBS Letters. 2005;579(11):2323–2326. doi: 10.1016/j.febslet.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 128.Gordon DF, Tucker EA, Tundwal K, Hall H, Wood WM, Ridgway EC. MED220/thyroid receptor-associated protein 220 functions as a transcriptional coactivator with pit-1 and GATA-2 on the thyrotropin-β promoter in thyrotropes. Molecular Endocrinology. 2006;20(5):1073–1089. doi: 10.1210/me.2005-0115. [DOI] [PubMed] [Google Scholar]
- 129.Li H, Gade P, Nallar SC, et al. The Med1 subunit of transcriptional mediator plays a central role in regulating CCAAT/enhancer-binding protein-β-driven transcription in response to interferon-γ . The Journal of Biological Chemistry. 2008;283(19):13077–13086. doi: 10.1074/jbc.M800604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Belakavadi M, Fondell JD. Role of the Mediator complex in nuclear hormone receptor signaling. Reviews of Physiology, Biochemistry and Pharmacology. 2006;156:23–43. doi: 10.1007/s10254-005-0002-0. [DOI] [PubMed] [Google Scholar]
- 131.Misra P, Owuor ED, Li W, et al. Phosphorylation of transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP). Stimulation of transcriptional regulation by mitogen-activated protein kinase. The Journal of Biological Chemistry. 2002;277(50):48745–48754. doi: 10.1074/jbc.M208829200. [DOI] [PubMed] [Google Scholar]
- 132.Belakavadi M, Pandey PK, Vijayvargia R, Fondell JD. MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Molecular and Cellular Biology. 2008;28(12):3932–3942. doi: 10.1128/MCB.02191-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jain S, Pulikuri S, Zhu Y, et al. Differential expression of the peroxisome proliferator-activated receptor γ (PPARγ) and its coactivators steroid receptor coactivator-1 and PPAR-binding protein PBP in the brown fat, urinary bladder, colon, and breast of the mouse. American Journal of Pathology. 1998;153(2):349–354. doi: 10.1016/s0002-9440(10)65577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mouillet J-F, Chu T, Nelson DM, Mishima T, Sadovsky Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB Journal. 2010;24(6):2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Barak Y, Nelson MC, Ong ES, et al. PPARγ is required for placental, cardiac, and adipose tissue development. Molecular Cell. 1999;4(4):585–595. doi: 10.1016/s1097-2765(00)80209-9. [DOI] [PubMed] [Google Scholar]
- 136.Ito M, Yuan C-X, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Molecular Cell. 2000;5(4):683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- 137.Landles C, Chalk S, Steel JH, et al. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Molecular Endocrinology. 2003;17(12):2418–2435. doi: 10.1210/me.2003-0097. [DOI] [PubMed] [Google Scholar]
- 138.Ge K, Cho Y-W, Guo H, et al. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor γ-stimulated adipogenesis and target gene expression. Molecular and Cellular Biology. 2008;28(3):1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ge K, Guermah M, Yuan C-X, et al. Transcription coactivator TRAP220 is required for PPARγ2-stimulated adipogenesis. Nature. 2002;417(6888):563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 140.Jia Y, Qi C, Kashireddi P, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARα-regulated gene expression in liver. The Journal of Biological Chemistry. 2004;279(23):24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- 141.Chen W, Zhang X, Birsoy K, Roeder RG. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(22):10196–10201. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matsumoto K, Yu S, Jia Y, et al. Critical role for transcription coactivator Peroxisome Proliferator- activated Receptor (PPAR)-binding protein/TRAP220 in liver regeneration and PPARα ligand-induced liver tumor development. The Journal of Biological Chemistry. 2007;282(23):17053–17060. doi: 10.1074/jbc.M701956200. [DOI] [PubMed] [Google Scholar]
- 143.Matsumoto K, Huang J, Viswakarma N, et al. Transcription coactivator PBP/MED1-deficient hepatocytes are not susceptible to diethylnitrosamine-induced hepatocarcinogenesis in the mouse. Carcinogenesis. 2009;31(2):318–325. doi: 10.1093/carcin/bgp306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jia Y, Guo GL, Surapureddi S, et al. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(35):12531–12536. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jia Y, Viswakarma N, Fu T, et al. Conditional ablation of mediator subunit MED1 (MED1/PPARBP) gene in mouse liver attenuates glucocorticoid receptor agonist dexamethasone-induced hepatic steatosis. Gene Expression. 2009;14(5):291–306. doi: 10.3727/105221609788681213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jia Y, Qi C, Zhang Z, Zhu YT, Rao SM, Zhu Y-J. Peroxisome proliferator-activated receptor-binding protein null mutation results in defective mammary gland development. The Journal of Biological Chemistry. 2005;280(11):10766–10773. doi: 10.1074/jbc.M413331200. [DOI] [PubMed] [Google Scholar]
- 147.Zhu YT, Jia Y, Hu L, et al. Peroxisome-proliferator-activated receptor-binding protein (PBP) is essential for the growth of active Notch4-immortalized mammary epithelial cells by activating SOX10 expression. Biochemical Journal. 2010;425(2):435–444. doi: 10.1042/BJ20091237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kourtidis A, Jain R, Carkner RD, Eifert C, Brosnan MJ, Conklin DS. An RNA interference screen identifies metabolic regulators NR1D1 and PBP as novel survival factors for breast cancer cells with the ERBB2 signature. Cancer Research. 2010;70(5):1783–1792. doi: 10.1158/0008-5472.CAN-09-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vijayvargia R, May MS, Fondell JD. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Research. 2007;67(9):4034–4041. doi: 10.1158/0008-5472.CAN-06-3039. [DOI] [PubMed] [Google Scholar]
- 150.De La Croix Ndong J, Jean D, Rousselet N, Frade R. Down-regulation of the expression of RB18A/MED1, a cofactor of transcription, triggers strong tumorigenic phenotype of human melanoma cells. International Journal of Cancer. 2009;124(11):2597–2606. doi: 10.1002/ijc.24253. [DOI] [PubMed] [Google Scholar]
- 151.Zhang X, Krutchinsky A, Fukuda A, et al. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Molecular Cell. 2005;19(1):89–100. doi: 10.1016/j.molcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 152.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Molecular and Cellular Biology. 2001;21(11):3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lin J, Tarr PT, Yang R, et al. PGC-1β in the regulation of hepatic glucose and energy metabolism. The Journal of Biological Chemistry. 2003;278(33):30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 154.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor α in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Molecular and Cellular Biology. 2000;20(5):1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Knutti D, Kaul A, Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Molecular and Cellular Biology. 2000;20(7):2411–2422. doi: 10.1128/mcb.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes and Development. 2004;18(2):157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lin J, Yang R, Tarr PT, et al. Hyperlipidemic effects of dietary saturated fats mediated through PGC-1β coactivation of SREBP. Cell. 2005;120(2):261–273. doi: 10.1016/j.cell.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 158.Duncan JG, Fong JL, Medeiros DM, Finck BN, Kelly DP. Insulin-resistant heart exhibits a mitochondrial biogenic response driven by the peroxisome proliferator-activated receptor-α/PGC-1α gene regulatory pathway. Circulation. 2007;115(7):909–917. doi: 10.1161/CIRCULATIONAHA.106.662296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Koo S-H, Satoh H, Herzig S, et al. PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nature Medicine. 2004;10(5):530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 160.Li Y, Kovach A, Suino-Powell K, Martynowski D, Xu HE. Structural and biochemical basis for the binding selectivity of peroxisome proliferator-activated receptor γ to PGC-1α . The Journal of Biological Chemistry. 2008;283(27):19132–19139. doi: 10.1074/jbc.M802040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hondares E, Mora O, Yubero P, et al. Thiazolidinediones and rexinoids induce peroxisome proliferator-activated receptor-coactivator (PGC)-1α gene transcription: an autoregulatory loop controls PGC-1α expression in adipocytes via peroxisome proliferator-activated receptor-γ coactivation. Endocrinology. 2006;147(6):2829–2838. doi: 10.1210/en.2006-0070. [DOI] [PubMed] [Google Scholar]
- 162.Røst TH, Haugan Moi LL, Berge K, Staels B, Mellgren G, Berge RK. A pan-PPAR ligand induces hepatic fatty acid oxidation in PPARα −/− mice possibly through PGC-1 mediated PPARδ coactivation. Biochimica et Biophysica Acta. 2009;1791(11):1076–1083. doi: 10.1016/j.bbalip.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 163.Kleiner S, Nguyen-Tran V, Baré O, Huang X, Spiegelman B, Wu Z. PPARδ agonism activates fatty acid oxidation via PGC-1α but does not increase mitochondrial gene expression and function. The Journal of Biological Chemistry. 2009;284(28):18624–18633. doi: 10.1074/jbc.M109.008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Leone TC, Lehman JJ, Finck BN, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biology. 2005;3(4):p. e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Arany Z, He H, Lin J, et al. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metabolism. 2005;1(4):259–271. doi: 10.1016/j.cmet.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 166.Lehman JJ, Boudina S, Banke NH, et al. The transcriptional coactivator PGC-1α is essential for maximal and efficient cardiac mitochondrial fatty acid oxidation and lipid homeostasis. American Journal of Physiology. 2008;295(1):H185–H196. doi: 10.1152/ajpheart.00081.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lelliott CJ, Ljungberg A, Ahnmark A, et al. Hepatic PGC-1β overexpression induces combined hyperlipidemia and modulates the response to PPARα activation. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;27(12):2707–2713. doi: 10.1161/ATVBAHA.107.155739. [DOI] [PubMed] [Google Scholar]
- 168.Sonoda J, Mehl IR, Chong L-W, Nofsinger RR, Evans RM. PGC-1β controls mitochondrial metabolism to modulate circadian activity, adaptive thermogenesis, and hepatic steatosis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(12):5223–5228. doi: 10.1073/pnas.0611623104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Goo Y-H, Chang Sohn Y, Kim D-H, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Molecular and Cellular Biology. 2003;23(1):140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhu Y, Qi C, Jain S, et al. Amplication and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(19):10848–10853. doi: 10.1073/pnas.96.19.10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu YT, Hu L, Qi C, Zhu Y-J. PRIP promotes tumor formation through enhancing serum-responsive factor-mediated FOS expression. The Journal of Biological Chemistry. 2009;284(21):14485–14492. doi: 10.1074/jbc.M900935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim S-W, Park K, Kwak E, et al. Activating signal cointegrator 2 required for liver lipid metabolism mediated by liver X receptors in mice. Molecular and Cellular Biology. 2003;23(10):3583–3592. doi: 10.1128/MCB.23.10.3583-3592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kuang S-Q, Liao L, Zhang H, et al. Deletion of the cancer-amplified coactivator AIB3 results in defective placentation and embryonic lethality. The Journal of Biological Chemistry. 2002;277(47):45356–45360. doi: 10.1074/jbc.C200509200. [DOI] [PubMed] [Google Scholar]
- 174.Li Q, Chu M-J, Xu J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Molecular and Cellular Biology. 2007;27(23):8073–8086. doi: 10.1128/MCB.00451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yeom S-Y, Kim GH, Kim CH, et al. Regulation of insulin secretion and β-cell mass by activating signal cointegrator 2. Molecular and Cellular Biology. 2006;26(12):4553–4563. doi: 10.1128/MCB.01412-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mahajan MA, Samuels HH. Nuclear receptor coactivator/coregulator NCoA6(NRC) is a pleiotropic coregulator involved in transcription, cell survival, growth and development. Nuclear Receptor Signaling. 2008;6:p. e002. doi: 10.1621/nrs.06002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhang H, Liao L, Kuang S-Q, Xu J. Spatial distribution of the messenger ribonucleic acid and protein of the nuclear receptor coactivator, amplified in breast cancer-3, in mice. Endocrinology. 2003;144(4):1435–1443. doi: 10.1210/en.2002-0018. [DOI] [PubMed] [Google Scholar]
- 178.Antonson P, Schuster GU, Wang L, et al. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Molecular and Cellular Biology. 2003;23(4):1260–1268. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Mahajan MA, Samuels HH. Nuclear hormone receptor coregulator: role in hormone action, metabolism, growth, and development. Endocrine Reviews. 2005;26(4):583–597. doi: 10.1210/er.2004-0012. [DOI] [PubMed] [Google Scholar]
- 180.Lee S-K, Na S-Y, Jung S-Y, et al. Activating protein-1, nuclear factor-κB, and serum response factor as novel target molecules of the cancer-amplified transcription coactivator ASC-2. Molecular Endocrinology. 2000;14(6):915–925. doi: 10.1210/mend.14.6.0471. [DOI] [PubMed] [Google Scholar]
- 181.Zhang H, Kuang S-Q, Liao L, Zhou S, Xu J. Haploid inactivation of the amplified-in-breast cancer 3 coactivator reduces the inhibitory effect of peroxisome proliferator-activated receptor γ and retinoid X receptor on cell proliferation and accelerates polyoma middle-T antigen-induced mammary tumorigenesis in mice. Cancer Research. 2004;64(19):7169–7177. doi: 10.1158/0008-5472.CAN-04-1176. [DOI] [PubMed] [Google Scholar]
- 182.Sarkar J, Qi C, Guo D, et al. Transcription coactivator PRIP, the peroxisome proliferator-activated receptor (PPAR)-interacting protein, is redundant for the function of nuclear receptors PPARα and CAR, the constitutive androstane receptor, in mouse liver. Gene Expression. 2007;13(4-5):255–269. doi: 10.3727/000000006780666948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tomaru T, Satoh T, Yoshino S, et al. Isolation and characterization of a transcriptional cofactor and its novel isoform that bind the deoxyribonucleic acid-binding domain of peroxisome proliferator-activated receptor-γ . Endocrinology. 2006;147(1):377–388. doi: 10.1210/en.2005-0450. [DOI] [PubMed] [Google Scholar]
- 184.Savkur RS, Burris TP. The coactivator LXXLL nuclear receptor recognition motif. Journal of Peptide Research. 2004;63(3):207–212. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- 185.Viswakarma N, Matsumoto K, Jia Y, Rao MS, Reddy JK. Mice lacking transcription cofactor PRIC285 reveal attenuation of liver regeneration but are viable and develop normally. FASEB Journal. 2009;23:p. 117.6 . [Google Scholar]
- 186.Shur I, Socher R, Benayahu D. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. Journal of Cellular Physiology. 2006;207(2):374–378. doi: 10.1002/jcp.20586. [DOI] [PubMed] [Google Scholar]
- 187.Marfella CGA, Imbalzano AN. The Chd family of chromatin remodelers. Mutation Research. 2007;618(1-2):30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochemistry and Cell Biology. 2007;85(4):463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- 189.Delmas V, Stokes DG, Perry RP. A mammalian DNA-binding protein that contains a chromodomain and an SNF2/SW12-like helicase domain. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(6):2414–2418. doi: 10.1073/pnas.90.6.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Woodage T, Basrai MA, Baxevanis AD, Hieter P, Collins FS. Characterization of the CHD family of proteins. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(21):11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shur I, Benayahu D. Characterization and functional analysis of CReMM, a novel chromodomain helicase DNA-binding protein. Journal of Molecular Biology. 2005;352(3):646–655. doi: 10.1016/j.jmb.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 192.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Research. 1994;22(10):1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Tsukiyama T. The in vivo functions of ATP-dependent chromatin-remodelling factors. Nature Reviews Molecular Cell Biology. 2002;3(6):422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- 194.Peterson CL. Chromatin remodeling: nucleosomes bulging at the seams. Current Biology. 2002;12(7):R245–R247. doi: 10.1016/s0960-9822(02)00782-0. [DOI] [PubMed] [Google Scholar]
- 195.Pedersen TÅ, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPα TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes and Development. 2001;15(23):3208–3216. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 197.Salma N, Xiao H, Mueller E, Imbalzano AN. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor γ nuclear hormone receptor. Molecular and Cellular Biology. 2004;24(11):4651–4663. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang W, Côté J, Xue Y, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO Journal. 1996;15(19):5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 199.Nie Z, Xue Y, Yang D, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Molecular and Cellular Biology. 2000;20(23):8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Martens JA, Winston F. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Current Opinion in Genetics and Development. 2003;13(2):136–142. doi: 10.1016/s0959-437x(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 201.Misra P, Qi C, Yu S, et al. Interaction of PIMT with transcriptional coactivators CBP, p300, and PBP differential role in transcriptional regulation. The Journal of Biological Chemistry. 2002;277(22):20011–20019. doi: 10.1074/jbc.M201739200. [DOI] [PubMed] [Google Scholar]
- 202.Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonné R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Molecular Cell. 2002;9(4):891–901. doi: 10.1016/s1097-2765(02)00484-7. [DOI] [PubMed] [Google Scholar]
- 203.Komonyi O, Pápai G, Enunlu I, et al. DTL, the Drosophila homolog of PIMT/Tgs1 nuclear receptor coactivator-interacting protein/RNA methyltransferase, has an essential role in development. The Journal of Biological Chemistry. 2005;280(13):12397–12404. doi: 10.1074/jbc.M409251200. [DOI] [PubMed] [Google Scholar]
- 204.Enünlü I, Pápai G, Cserpán I, Udvardy A, Jeang K-T, Boros I. Different isoforms of PRIP-interacting protein with methyltransferase domain/trimethylguanosine synthase localizes to the cytoplasm and nucleus. Biochemical and Biophysical Research Communications. 2003;309(1):44–51. doi: 10.1016/s0006-291x(03)01514-6. [DOI] [PubMed] [Google Scholar]
- 205.Jia Y, Viswakarma N, Matsumoto K, Pyper SR, Rao MS, Reddy JK. Early embryonic lethality of mice with disrupted transcription cofactor PIMT/NCoA6IP gene. FASEB Journal. 2009;23:p. 739.6. [Google Scholar]
- 206.Stallcup MR, Kim JH, Teyssier C, Lee Y-H, Ma H, Chen D. The roles of protein-protein interactions and protein methylation in transcriptional activation by nuclear receptors and their coactivators. Journal of Steroid Biochemistry and Molecular Biology. 2003;85(2–5):139–145. doi: 10.1016/s0960-0760(03)00222-x. [DOI] [PubMed] [Google Scholar]
- 207.Aletta JM, Cimato TR, Ettinger MJ. Protein methylation: a signal event in post-translational modification. Trends in Biochemical Sciences. 1998;23(3):89–91. doi: 10.1016/s0968-0004(98)01185-2. [DOI] [PubMed] [Google Scholar]
- 208.Yadav N, Cheng D, Richard S, et al. CARM1 promotes adipocyte differentiation by coactivating PPARγ . EMBO Reports. 2008;9(2):193–198. doi: 10.1038/sj.embor.7401151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO Journal. 2002;21(20):5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Xu W, Chen H, Du K, et al. A transcriptional switch mediated by cofactor methylation. Science. 2001;294(5551):2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 211.Lee Y-H, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(10):3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Teyssier C, Chen D, Stallcup MR. Requirement for multiple domains of the protein arginine methyltransferase CARM1 in its transcriptional coactivator function. The Journal of Biological Chemistry. 2002;277(48):46066–46072. doi: 10.1074/jbc.M207623200. [DOI] [PubMed] [Google Scholar]
- 213.Bedford MT. Arginine methylation at a glance. Journal of Cell Science. 2007;120(24):4243–4246. doi: 10.1242/jcs.019885. [DOI] [PubMed] [Google Scholar]
- 214.Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Molecular and Cellular Biology. 2006;26(21):7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Yadav N, Lee J, Kim J, et al. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(11):6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Auboeuf D, Dowhan DH, Li X, et al. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Molecular and Cellular Biology. 2004;24(1):442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Jung SY, Malovannaya A, Wei J, O’Malley BW, Qin J. Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Molecular Endocrinology. 2005;19(10):2451–2465. doi: 10.1210/me.2004-0476. [DOI] [PubMed] [Google Scholar]
- 218.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocrine Reviews. 2007;28(5):575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]