Abstract
Aging affects all known organisms and has been studied extensively. Yet, the underlying mechanisms are insufficiently understood, possibly due to the multiscale complexity involved in this process: the aging of multicellular organisms depends on the aging of their cells, which depends on molecular events occurring in each cell. However, the aging of unicellular populations seeded in new niches and the aging of metazoans are surprisingly similar, indicating that the multiscale aspects of aging may have been conserved since the beginnings of cellular life on Earth. This underlines the importance of aging research in unicellular organisms such as a recent study by Lorenz et al., [(2009) Proc. Natl. Acad. Sci. U.S.A. 106, 1145–1150]. In their paper, the authors combine computational network identification with extensive experimentation and literature mining to discover and validate numerous regulatory interactions among ten genes involved in the cellular response to glucose starvation. Since low levels of glucose (calorie restriction) have been known to extend the longevity of various eukaryotes, the authors test the effect of Snf1 kinase overexpression on chronological aging and discover that this key regulator of glucose repression and two of its newly discovered synergistic repressors significantly affect the chronological lifespan of baker’s yeast.
AGING ACROSS THE KINGDOMS OF LIFE
“Forever young, I want to be forever young”—so goes a well-known song, evoking the ancient human aspiration for youth and eternal life. Yet, from the moment we are born, our bodies are subject to aging and every passing day takes us closer to the inevitable conclusion of human life. And, of course, we are not the only species affected by aging. Animals, plants, and even our farther unicellular-eukaryotic relatives (such as the baker’s yeast) undergo age-related changes starting with birth, continuing through adulthood and ultimately leading to death. More surprisingly, recent discoveries indicate that aging affects even bacteria (Stewart et al., 2005), which were previously assumed to be immortal, symmetrically dividing machines capable of indefinite growth limited only by nutrient availability.
Is there an evolutionary benefit of aging or is it just an unavoidable byproduct of being alive? Its apparent universality across all domains of life may support both alternatives. Clearly, aging confers nongenetic diversity to biological populations, which aids population survival and growth in lethal stress (Avery, 2006; Blake et al., 2006), provided that some surviving individuals can reproduce. On the other hand, aging is inevitable due to the error-prone nature of molecular synthesis, degradation, and signaling on which all organisms rely to cope with a changing environment (Orgel, 1963). While some of the errors can be corrected, the molecular repair machinery is itself subject to damage, creating a vicious circle of accelerated error accumulation that culminates in death. To circumvent this perilous scenario, organisms undergo asymmetric reproduction, where most of the accumulated damage segregates into the parent while the offspring gets a fresh start as a single cell with relatively error-free molecular contents (Danchin, 2009).
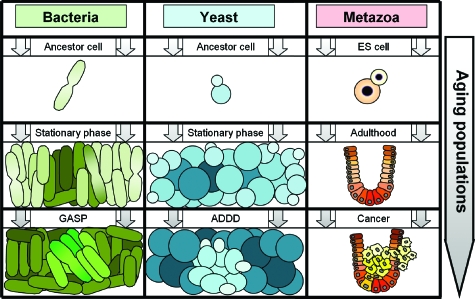
The lifespan of single cells can be characterized either in terms of their replicative ability or as their time of survival in a nonreplicative (senescent) state. Accordingly, replicative aging and lifespan (Steinkraus et al., 2008) refer to the ability of a mother cell to divide and the number of divisions it can undergo before senescence while chronological aging and lifespan (Fabrizio and Longo, 2003) describe the cell’s survival ability in a nondividing state such as cell cycle arrest or nutrient deprivation. While the dependence of animal aging on the organism’s individual cells is less well understood (Patil et al., 2005), it may be insightful to compare microbial growth in a new niche to an animal’s lifespan starting at conception (Fig. 1). Both processes start with a single cell that divides and gives rise to an exponentially growing population of young cells that predominantly undergo replicative aging (by definition, terminally differentiated metazoan cells have reached their maximum replicative lifespan). As the microbial and animal cell populations expand and age, they both reach a state corresponding to the microbial stationary phase and animal adulthood, where cell population growth slows and eventually stops due to space or nutrient limitations or intercellular growth inhibition (Aoki et al., 2005; Stark et al., 1998). Importantly, a nongrowing population may consist of dividing cells as long as cell division and cell death are at equilibrium. Finally, cell counts drop as the balance is tipped toward cell death and the population becomes enriched in senescent cells undergoing chronological aging. The amount of molecular damage is high and the chance of mutations increases at this stage, resulting in genetic diversity and clonal expansion of mutant subpopulations, called “growth advantage in stationary phase” (GASP) for bacteria (Finkel, 2006), “age-dependent dedifferentiation” (ADDD) for yeast (Madia et al., 2007), and cancer for metazoa (Skinner and Turker, 2005). In contrast to the prevailing view (Wolpert and Szathmáry, 2002), these similarities between the aging of very distant species suggest a universal scenario, where the genome controls cell population-level phenotypes that are passed to the next population through a single-cell bottleneck (Fig. 1). Undoubtedly, the molecular mechanisms driving microbial population dynamics in isolated niches and animal aging can be very different. Still, considering the possibility that multicellular organisms evolved from unicellular populations that gradually acquired skills of complex intercellular communication (Bassler and Losick, 2006) leading to task allocation through differentiation (Wahl, 2002), it is intriguing to ask what we can learn from one process while studying the other. This underlines the importance of a recent interdisciplinary effort by (Lorenz et al., 2009) to map the complex regulatory interactions among a handful of genes affecting longevity in response to calorie restriction in baker’s yeast (Saccharomyces cerevisiae).
Figure 1. Connection between the aging of single cells and cell populations for bacteria, yeast and metazoa.
Bacteria and yeast are seeded from spores and grow in a new niche. Cells primarily undergo replicative aging in the beginning (top row) while chronological aging dominates toward the end of each population’s lifespan (bottom row). Cell members of old populations (bottom row) have increasing tendency to acquire mutations that confer growth advantage due to genomic instability. Abbreviations: ES cell is embryonic stem cell, GASP is growth advantage in stationary phase, and ADDD is age-dependent dedifferentiation.
LONGEVITY AND CALORIE RESTRICTION IN YEAST
The replicative lifespan of yeast cells was first measured half a century ago by counting the number of buds that individual mother cells produced (Mortimer and Johnston, 1959). By contrast, studies of chronological aging were initiated relatively recently (Longo et al., 1996) and involved monitoring post-diauxic cell viability in synthetic dextrose-complete (SDC) medium or water (Fabrizio and Longo, 2003). Since these pioneering studies, significant effort has been spent discovering the molecular-genetic factors and the environmental conditions that influence replicative and chronological aging in yeast. The relative ease of genetic manipulation and the vast amount of biological knowledge available for this fungus have been further complemented by the surprising conservation of pathways and conditions that affect aging from yeast to higher eukaryotes. Consequently, the molecular biology of aging is probably best (but still insufficiently) understood for yeast and the accumulating information is invaluable for studying longevity in higher organisms.
The molecular factors affecting replicative lifespan have been reviewed recently (Guarente and Kenyon, 2000; Steinkraus et al., 2008) and will not be discussed in detail here. In a nutshell, three important factors have been found to promote replicative aging: extrachromosomal ribosomal circles, oxidative damage and protein aggregation. The level and effect of these (and other) factors can be modulated by silent information regulator (Sir) proteins, especially Sir2; by proteins involved in glucose repression, including the Snf1 kinase; and by protein kinase A pathways, among others. On the other hand, chronological aging is affected by general stress response proteins, such as Msn2 and Msn4, superoxide dismutase (Sod) proteins, as well as the Rasand Sch9 pathways (Fabrizio and Longo, 2003). Surprisingly, the effect of Sir2 on chronological lifespan seems to be opposite to its effect on replicative lifespan (Fabrizio et al., 2005). However, all of these seemingly disparate lifespan-modulating pathways share one important property: they are related to calorie restriction, the primary and most studied environmental condition that prolongs both replicative and chronological lifespan in every organism that has been tested, from yeast to mammals (Steinkraus et al., 2008).
Calorie restriction consists of limiting the concentration of glucose, the sugar that yeast prefers to ferment, in the growth medium. As glucose levels drop, yeast undergo a temporary growth arrest (diauxie) before switching to respiratory growth. However, once devoid of glucose, yeast mother cells produce more buds before they become senescent. Moreover, they survive three times longer in a nonreplicative state in sugar-free water compared to SDC medium (Fabrizio and Longo, 2003).
What happens when glucose becomes scarce? Similar to other cell types (Balázsi et al., 2005, 2008), yeast have a hierarchically organized starvation-specialized system (Balázsi and Oltvai, 2005) that consists of sensory input nodes coupled to downstream signal-processing transit nodes ensuring that the appropriate transcriptional response is mounted at the output. The Snf1 kinase, together with its protein interaction partners and its downstream signaling targets [Figs. 2A, 2B] are crucial components of this hierarchical starvation-response system (Hedbacker and Carlson, 2008). In the absence of glucose, Snf1 becomes phosphorylated, associates with a new protein interaction partner within the heterotrimeric SNF1 complex and translocates to the nucleus, where it activates stress-induced transcription factors including Cat8 and Sip4, and relieves repression by Mig1, the major transcription factor involved in glucose repression (Hedbacker and Carlson, 2008).
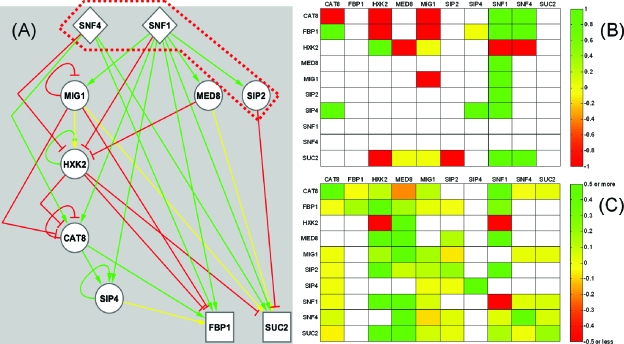
Figure 2. Glucose repression network identification using the NIR algorithm.
(A) Known regulatory interactions among the ten genes involved in glucose repression. The genes are arranged in a three-tier hierarchy according to their role in signal transduction, and are classified as either input nodes (diamonds), transit nodes (circles) or output nodes (squares). The colors of the links indicate repression (red), activation (green) or dual∕incompletely characterized regulatory interaction (yellow). The dark red dashed line indicates a repressory form of the SNF1 protein complex that prevents Snf1 translocation to the nucleus in the presence of glucose. (B) Known regulatory interactions represented as a matrix. The colors indicate the same type of regulatory relationships as on the left. (C) Matrix representation of the reconstructed network. Many new interactions compared to panel (B) are identified.
COMPUTATIONAL DISCOVERY AND EXPERIMENTAL VALIDATION OF NEW REGULATORY CONNECTIONS AFFECTING CHRONOLOGICAL LIFESPAN IN YEAST
Network inference is a difficult but necessary intermediate step in the systems biology workflow that converts genome-scale data into biological discovery and practical applications (Veiga et al., 2010). Once constructed and validated, the molecular interaction network facilitates testable hypothesis generation by summarizing known interactions among the genes of interest. For example, the discovery of new regulatory interactions among genes that influence lifespan by calorie restriction could improve the treatment of aging diseases (such as Werner’s syndrome) or could lead to improved cancer prevention strategies.
The “network identification by multiple regression” (NIR) algorithm (Gardner et al., 2003) infers regulatory interactions from gene expression measurements following individual perturbations of each gene’s expression. The method is based on three assumptions: (i) the gene network is at steady-state; (ii) the perturbations are small; and (iii) the network is sparse (the number of regulatory inputs to each gene is smaller than a threshold k). Assumptions (i) and (ii) imply that the network’s response to each perturbation is linear and the system Ax=u describes the effect of perturbations, where the N×N matrices A, x, and −u represent the network, the gene expression levels in response to each perturbation and the perturbations, respectively. Assumption (iii) ensures that this system is over-determined and A can be reliably estimated by multiple linear regressions from gene expression measurements in individual yeast strains overexpressing each of the N genes.
Lorenz et al., (2009) applied a variant of NIR (Gardner et al., 2003) to infer regulatory relationships among ten glucose repression genes (CAT8, FBP1, HXK2, MED8, MIG1, SIP2, SIP4, SNF1, SNF4, and SUC2), some of which also affect lifespan. First, they established ten overexpression strains, each carrying a doxycycline-inducible, integrated extra copy of a glucose repression gene, and one control strain carrying Green Fluorescent Protein (GFP) under the control of the same promoter. Next, to obtain perturbation data for NIR, they performed 12 replicate real-time quantitative RT-PCR (qRT-PCR) measurements comparing the expression of each glucose repression gene in the overexpression strains to the control strain. The inferred network contained a large number of interactions that had not been previously documented in the literature [Fig. 2C].
Network inference is a computationally intensive method of varying reliability that depends on the amount of data used for reconstruction, as well as measurement noise. To evaluate the performance of NIR and other computational network inference methods, they should be tested to infer relatively well-known, validated “gold standard” networks assembled by literature mining or genome-scale interaction mapping experiments. After extensive mining of glucose repression literature, the authors assembled such a reference network based on more than 50 journal articles. A simplified version of this network [Fig. 2A] can be organized into a three-layer hierarchy considering each node’s in- and out-degree connectivity (Balázsi et al., 2005, 2008). Specifically, input nodes have no regulators (in-degree=0), output nodes have no regulated targets (out-degree=0) while transit nodes have nonzero in- and out-degrees. Interestingly, most transit nodes regulate their own expression and Hxk2, the predominant glucose kinase enzyme catalyzing the first step of glycolysis, emerges as a highly regulated core integrator with many regulatory targets. Comparing the known and inferred networks, the authors found that the sensitivity (true positive rate) and precision (positive predictive value) of the NIR algorithm were 65% and 22%, respectively. These values are remarkably high compared to those of genome-scale network reconstruction efforts in Escherichia coli (Faith et al., 2007), as expected considering the small size of the glucose repression network and the accessibility of greater experimental detail. Importantly, these benchmarks do not consider the type of regulation (repression versus activation), which is reasonable considering that gene regulation is environment- and context-dependent.
Since NIR predicted many new regulatory interactions [Figs. 2B, 2C], the authors performed additional experiments to validate these new links. After deleting all but one essential gene (MED8), they compared the betagalactosidase activity for each deletion strain carrying different target promoter-lacZ fusion constructs to the corresponding wild-type strain carrying the same construct. Moreover, they performed ChIP-qPCR experiments to identify Med8 and Hxk2 binding locations upstream and downstream from all target promoters. These experiments confirmed the role of Snf1, Hxk2, and Med8 as key regulators in the glucose repression network, in addition to validating the majority of novel interactions predicted by the NIR algorithm. Whereas Snf1 partners with Snf4 to post-translationally regulate gene expression, Hxk2 and Med8 (a Mediator complex subunit bridging transcriptional activators and the general transcription machinery) bind in various combinations to target promoters and regulate target gene transcription. The dual function of Hxk2 as key transcription factor and major glucose kinase in the first step of glycolysis underlines its role as a core integrator in the glucose repression network.
Besides its role in the glucose repression network, Snf1 is involved in response to multiple forms of environmental stress, including starvation for a variety of nutrients, toxic cation stress, alkaline pH, and heat shock (Hedbacker and Carlson, 2008). Since stress response genes are known to affect chronological lifespan (Fabrizio and Longo, 2003), the authors tested the effect of Snf1 overexpression on post-diauxic survival in SDC medium and found significantly reduced longevity for the overexpression mutant, as well as for double-deletion mutants of the newly discovered synergistic SNF1 repressors Mig1, and Hxk2.
CONCLUSIONS AND OUTLOOK
While numerous studies have focused on genome-scale network reconstruction, Lorenz, Cantor and Collins showed that even small networks that had been partially mapped “one molecule at a time” can offer many surprises (Lorenz et al., 2009). Using an unsupervised approach (the NIR algorithm), they uncovered numerous unknown regulatory interactions, and discovered new contributors to chronologic aging. Their work generates new questions and opens up new avenues of research.
For example, it would be interesting to test how the synergistic regulators of SNF1 affect replicative (as opposed to chronological) lifespan. The same question could be asked about other genes and regulators in the network-in particular, how would Med8 overexpression affect lifespan? Moreover, the key regulatory nodes identified in the yeast aging network are functionally conserved in higher eukaryotes, including components of the SNF1 complex (mammalian Adenosine Monophosphate-Activated Protein Kinase), Hxk2 (human glucokinase), and the Mediator complex. Do the complex regulatory interactions between these proteins translate to other species, and can they shed new insight into the regulation of longevity in animals, including humans?
In the future it will be important to study cell population-level aspects of longevity, such as its variability across the cell population. While this can be easily accomplished for replicative aging, which is measured at the single cell level (by monitoring bud formation), it would also be interesting to determine the effect of various growth conditions on the intercellular variation in chronological lifespan. This requires monitoring single cells for a week or longer, which may soon become possible with the help of clever microfluidics and automated fluorescence microscopy (Bennett et al., 2008) since chronological lifespan is measured in a nonreplicative state, where cell lineage tracking is not necessary.
Network reconstruction could also benefit from more precise gene perturbations. Current approaches of gene expression control (including the doxycycline-inducible system used in this study) suffer from two drawbacks (Nevozhay et al., 2009): nonlinear dose-response and cell-cell variability (biological noise). The increasingly detailed understanding of gene regulation at the single cell level (Becskei et al., 2005; Blake et al., 2003; Rosenfeld et al., 2005) will certainly improve gene expression control. For example, the recently described “linearizer” synthetic gene circuit (Nevozhay et al., 2009) eliminates both drawbacks of current gene expression control, and makes it possible to tune gene expression very precisely, obtaining nearly identical levels of expression that are linearly proportional to the extracellular inducer in every member of the cell population.
ACKNOWLEDGMENTS
The author would like to thank the National Institutes of Health Director’s New Innovator Award Program (Grant No. 1DP2 OD006481-01), the Institutional Research Grant Program sponsored by the Office of Research Administration at the M. D. Anderson Cancer Center, and the TB PAN-NET consortium sponsored by the European Commission’s Seventh Framework Programme for Research (FP7) for research support. The author would also like to thank James J. Collins, Jordan Gutterman, Dmitry Nevozhay, and David R. Lorenz for comments.
REFERENCES
- Aoki, S K, Pamma, R, Hernday, A D, Bickham, J E, Braaten, B A, and Low, D A (2005). “Contact-dependent inhibition of growth in Escherichia coli.” Science 309, 1245–1248. 10.1126/science.1115109 [DOI] [PubMed] [Google Scholar]
- Avery, S V (2006). “Microbial cell individuality and the underlying sources of heterogeneity.” Nat. Rev. Microbiol. 4, 577–587. 10.1038/nrmicro1460 [DOI] [PubMed] [Google Scholar]
- Balázsi, G, Barabási, A L, and Oltvai, Z N (2005). “Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli.” Proc. Natl. Acad. Sci. U.S.A. 102, 7841–7846. 10.1073/pnas.0500365102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázsi, G, Heath, A P, Shi, L, and Gennaro, M L (2008). “The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest.” Mol. Syst. Biol. 4, 225. 10.1038/msb.2008.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balázsi, G, and Oltvai, Z N (2005). “Sensing your surroundings: how transcription-regulatory networks of the cell discern environmental signals.” Sci. STKE 2005, pe20. 10.1126/stke.2822005pe20 [DOI] [PubMed] [Google Scholar]
- Bassler, B L, and Losick, R (2006). “Bacterially speaking.” Cell 125, 237–246. 10.1016/j.cell.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Becskei, A, Kaufmann, B B, and van Oudenaarden, A (2005). “Contributions of low molecule number and chromosomal positioning to stochastic gene expression.” Nat. Genet. 37, 937–944. 10.1038/ng1616 [DOI] [PubMed] [Google Scholar]
- Bennett, M R, Pang, W L, Ostroff, N A, Baumgartner, B L, Nayak, S, Tsimring, L S, and Hasty, J (2008). “Metabolic gene regulation in a dynamically changing environment.” Nature (London) 454, 1119–1122. 10.1038/nature07211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, W J, Balázsi, G, Kohanski, M A, Isaacs, F J, Murphy, K F, Kuang, Y, Cantor, C R, Walt, D R, and Collins, J J (2006). “Phenotypic consequences of promoter-mediated transcriptional noise.” Mol. Cell 24, 853–865. 10.1016/j.molcel.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Blake, W J, KÆrn, M, Cantor, C R, and Collins, J J (2003). “Noise in eukaryotic gene expression.” Nature (London) 422, 633–637. 10.1038/nature01546 [DOI] [PubMed] [Google Scholar]
- Danchin, A (2009). “Natural selection and immortality.” Biogerontology 10, 503–516. 10.1007/s10522-008-9171-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio, P, Gattazzo, C, Battistella, L, Wei, M, Cheng, C, McGrew, K, and Longo, V D (2005). “Sir2 blocks extreme life-span extension.” Cell 123, 655–667. 10.1016/j.cell.2005.08.042 [DOI] [PubMed] [Google Scholar]
- Fabrizio, P, and Longo, V D (2003). “The chronological life span of Saccharomyces cerevisiae.” Aging Cell 2, 73–81. 10.1046/j.1474-9728.2003.00033.x [DOI] [PubMed] [Google Scholar]
- Faith, J J, Hayete, B, Thaden, J T, Mogno, I, Wierzbowski, J, Cottarel, G, Kasif, S, Collins, J J, and Gardner, T S (2007). “Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles.” PLoS Biol. 5, e8. 10.1371/journal.pbio.0050008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, S E (2006). “Long-term survival during stationary phase: evolution and the GASP phenotype.” Nat. Rev. Microbiol. 4, 113–120. 10.1038/nrmicro1340 [DOI] [PubMed] [Google Scholar]
- Gardner, T S, di Bernardo, D, Lorenz, D, and Collins, J J (2003). “Inferring genetic networks and identifying compound mode of action via expression profiling.” Science 301, 102–105. 10.1126/science.1081900 [DOI] [PubMed] [Google Scholar]
- Guarente, L, and Kenyon, C (2000). “Genetic pathways that regulate ageing in model organisms.” Nature (London) 408, 255–262. 10.1038/35041700 [DOI] [PubMed] [Google Scholar]
- Hedbacker, K, and Carlson, M (2008). “SNF1/AMPK pathways in yeast.” Front. Biosci. 13, 2408–2420. 10.2741/2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V D, Gralla, E B, and Valentine, J S (1996). “Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: Mitochondrial production of toxic oxygen species in vivo.” J. Biol. Chem. 271, 12275–12280. 10.1074/jbc.271.21.12275 [DOI] [PubMed] [Google Scholar]
- Lorenz, D R, Cantor, C R, and Collins, J J (2009). “A network biology approach to aging in yeast.” Proc. Natl. Acad. Sci. U.S.A. 106, 1145–1150. 10.1073/pnas.0812551106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madia, F, Gattazzo, C, Fabrizio, P, and Longo, V D (2007). “A simple model system for age-dependent DNA damage and cancer.” Mech. Ageing Dev. 128, 45–49. 10.1016/j.mad.2006.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer, R K, and Johnston, J R (1959). “Life span of individual yeast cells.” Nature (London) 183, 1751–1752. 10.1038/1831751a0 [DOI] [PubMed] [Google Scholar]
- Nevozhay, D, Adams, R M, Murphy, K F, Josic, K, and Balázsi, G (2009). “Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression.” Proc. Natl. Acad. Sci. U.S.A. 106, 5123–5128. 10.1073/pnas.0809901106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel, L E (1963). “The maintenance of the accuracy of protein synthesis and its relevance to ageing.” Proc. Natl. Acad. Sci. U.S.A. 49, 517–521. 10.1073/pnas.49.4.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, C K, Mian, I S, and Campisi, J (2005). “The thorny path linking cellular senescence to organismal aging.” Mech. Ageing Dev. 126, 1040–1045. 10.1016/j.mad.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Rosenfeld, N, Young, J W, Alon, U, Swain, P S, and Elowitz, M B (2005). “Gene regulation at the single-cell level.” Science 307, 1962–1965. 10.1126/science.1106914 [DOI] [PubMed] [Google Scholar]
- Skinner, A M, and Turker, M S (2005). “Oxidative mutagenesis, mismatch repair, and aging.” Sci. SAGE KE 2005, re3. 10.1126/sageke.2005.9.re3 [DOI] [PubMed] [Google Scholar]
- Stark, G R, Kerr, I M, Williams, B R, Silverman, R H, and Schreiber, R D (1998). “How cells respond to interferons.” Annu. Rev. Biochem. 67, 227–264. 10.1146/annurev.biochem.67.1.227 [DOI] [PubMed] [Google Scholar]
- Steinkraus, K A, Kaeberlein, M, and Kennedy, B K (2008). “Replicative aging in yeast: the means to the end.” Annu. Rev. Cell Dev. Biol. 24, 29–54. 10.1146/annurev.cellbio.23.090506.123509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, E J, Madden, R, Paul, G, and Taddei, F (2005). “Aging and death in an organism that reproduces by morphologically symmetric division.” PLoS Biol. 3, e45. 10.1371/journal.pbio.0030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga, D F, Dutta, B, and Balázsi, G (2010). “Network inference and network response identification: moving genome-scale data to the next level of biological discovery.” Mol. Biosyst. 6, 469–480. 10.1039/b916989j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl, L M (2002). “The division of labor: genotypic versus phenotypic specialization.” Am. Nat. 160, 135–145. 10.1086/340601 [DOI] [PubMed] [Google Scholar]
- Wolpert, L, and Szathmáry, E (2002). “Multicellularity: evolution and the egg.” Nature (London) 420, 745. 10.1038/420745a [DOI] [PubMed] [Google Scholar]