Abstract
The evolution of biocrystallography from the pioneers’ time to the present era of global biology is presented in relation to the development of methodological and instrumental advances for molecular sample preparation and structure elucidation over the last 6 decades. The interdisciplinarity of the field that generated cross-fertilization between physics- and biology-focused themes is emphasized. In particular, strategies to circumvent the main bottlenecks of biocrystallography are discussed. They concern (i) the way macromolecular targets are selected, designed, and characterized, (ii) crystallogenesis and how to deal with physical and biological parameters that impact crystallization for growing and optimizing crystals, and (iii) the methods for crystal analysis and 3D structure determination. Milestones that have marked the history of biocrystallography illustrate the discussion. Finally, the future of the field is envisaged. Wide gaps of the structural space need to be filed and membrane proteins as well as intrinsically unstructured proteins still constitute challenging targets. Solving supramolecular assemblies of increasing complexity, developing a “4D biology” for decrypting the kinematic changes in macromolecular structures in action, integrating these structural data in the whole cell organization, and deciphering biomedical implications will represent the new frontiers.
Contemporary biocrystallography is interdisciplinary, by essence, in combining biology with physics, chemistry, and engineering. The field has always been knowledge driven by the need to visualize and to comprehend the molecules that underlie the basic life processes. This explains why its history paralleled the highlights of biological research and has regularly been distinguished by the Nobel Committee. Thus, in 2009 Venki Ramashrisknan, Thomas Steitz, and Ada Yonath shared the Chemistry Nobel Prize for their contribution to the determination of the crystal structure of the ribosome, the macromolecular machine that fabricates proteins (see comments by Carter, 2009; Nierhaus, 2009). Because of this tribute, this essay will highlight data on ribosomes and partners of the protein synthesis machinery that contributed to the development of modern crystallography.
The field was also methodology and technology driven. The first examples from the early ages concern the implementation of appropriate methods to solve structures (Arnold et al., 2010). Continuous developments have later been focused on the improvement of protein expression and purification (Christendat et al., 2000; Koehn and Hunt, 2009) as well as of crystallization (Sauter et al., 2010) and diffraction data collection and processing (Arnold et al., 2010). In this respect, the novel generations of synchrotron sources, of 3D graphics, and computing facilities for solving, building, and refining structures were essential. The field benefited also from protein and nucleic acid sequencing and synthesis technologies that provided the material to be crystallized and the chemicalinformation to be fitted to the electron density maps.
Over the years with the increasing number of solved crystal structures, biocrystallography reached the mature age and transformed into structural biology. Stimulated by the wealth of data originating from genomic programs, a new branch called structural genomics or structural proteomics emerged in the mid-1990s. It was based on a systematic high-throughput approach aimed to rapidly determine the ensemble of structures coded by selected genomes or belonging to specific biological functions or pathways (Terwilliger et al., 2009). In parallel, the interest in understanding the architecture, functioning, and dynamics of large supramolecular assemblies, as well as ultrahigh-resolution of essential structures increased. As a result, new bottlenecks and challenges appeared while the questions addressed in biocrystallography gained in complexity.
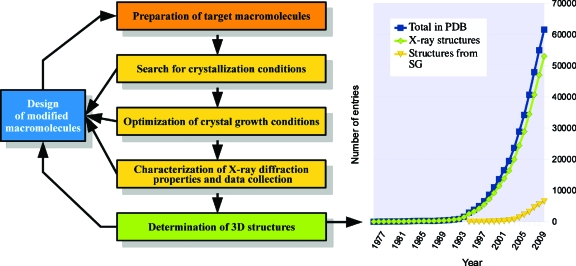
Figure 1 outlines the five steps that have to be mastered in order to determine a 3D structure. They first concern the choice of the most appropriate target macromolecule, its cloning, expression, purification, and assessment of purity in terms of chemical and conformational homogeneity. Although mainly dependent on biology methodologies, this step also requires bioinformatics and structure analysis tools to select a native target or to design variants. Likewise, the next three steps dealing with crystallization and crystal characterization definitely make an extensive use of interdisciplinary approaches (Sauter et al., 2010). They cover (i) the search of initial crystallization conditions by trial-and-error strategies using sparse matrix screening or rational-guided diagnostics, (ii) the optimization of crystal quality by seeding, phase diagram exploration, or more advanced approaches such as growth in diffusive media or in the presence of additives among them natural ligands or inhibitors, and (iii) the assessment of the diffraction properties of crystals such as resolution, mosaicity, and isotropy. Note that at this stage, crystals also constitute interesting objects to investigate physics related issues—crystal perfection studies by X-ray topography, rheology and other mechanical aspects, impurity inclusion, crystal surface poisoning, and crystal engineering—as well as in crystallo enzymology. These four initial steps are the main scope of biocrystallogenesis; the field that has been developed since the late 1980s to rationalize the preparation of well-diffracting crystals (McPherson and Giegé, 2007). The ultimate step, that is the determination and the analysis of the 3D structure, benefited as well from constant methodological and instrumental innovations (Arnold et al., 2010). However, despite all the gained expertise, a biocrystallographic project can be stuck at each of these steps and overcoming the bottlenecks often requires inventiveness and efforts. This essay will discuss these different aspects from the viewpoints of past, present, and future.
Figure 1. Biocrystallography, the multidisciplinary route to the 3D vision of biological processes.
(Left) main steps to a 3D crystal structure. (Right) contribution of biocrystallography to the PDB. The plot illustrates the growth of the PDB content since 1975 (blue curve) and shows that the biocrystallography community is by far the strongest contributor (green curve) with X-ray structures solved by structural genomics consortia already reaching 10% of the total (yellow curve). These data were extracted from the PDB http://www.rcsb.org/pdb/
HISTORICAL BACKGROUND—FROM SMALL TO LARGE
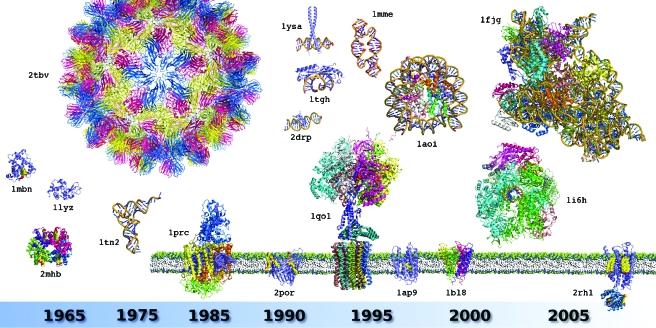
Biocrystallography started in the mid-1930s when it was realized that X-ray diffraction patterns recorded from macromolecular crystals (Bernal and Crowfoot, 1934) contain structural information that can be translated in atomic models of the crystalline macromolecules (Kendrew et al., 1958). The first bottleneck was the lack of suitable methods for structure solving, in particular to overcome the phase problem. As soon as these methods were developed and the first structures solved, a dozen of structure determinations followed, comprising small proteins and enzymes as well as nucleic acid fragments (Fig. 2). They wonderfully confirmed the models of α-helices and β-sheets in proteins (Pauling and Corey, 1951) and of the DNA double-helix (Watson and Crick, 1953). Over the years, biocrystallography targets gained in size and complexity, covering soluble proteins of increasing size, small RNAs, pieces of DNA and their complexes with proteins, spherical viruses with high intrinsic symmetry, membrane proteins, and nucleoprotein complexes to culminate nowadays with assemblies as intricate as the bacterial ribosome, a ∼2.3 MDa particle comprising three RNAs and ∼50 proteins, which structure was solved in various forms without or with combinations of bound tRNA, mRNA, and antibiotics substrates. This diversity is depicted in Fig. 2 by a series of emblematic milestone structures (see also Supplementary Material Table S1).
Figure 2. Milestone structures that recapitulate the history of biocrystallography.
The figure illustrates the diversity of 3D structures solved and the evolution of their complexity since the birth of the field in the late 1950s. A star ( *) indicates crystal structures linked to a Nobel Prize award. The proposed selection is displayed in chronological rank and includes: sperm whale myoglobin (PDB identifier: 1mbn*), horse hemoglobin (2mhb*), hen egg white lysozyme (1lyz), Saccharomyces cerevisiae tRNAPhe (1tn2), icosahedral Tomato Bushy Stunt Virus (2tbv), Rhodopseudomonas viridis photosynthetic reaction center (1prc*), Rhodobacter capsulatus porin (2por), human TATA binding protein in complex with TATA box DNA (1tgh), S. cerevisiae GCN4 leucine zipper (1ysa), Drosophila melanogaster Tramtrack zinc finger domain complexed with its DNA target (2drp), bovine ATP synthase (1qo1*), synthetic construct of a hammerhead ribozyme (1mme), Xenopus laevis nucleosome with synthetic DNA construct (1aoi), Halobacterium salinarum bacteriorhodopsin (1ap9), Streptomyces lividans K+ channel (1bl8*), S. cerevisiae RNA polymerase II (1i6h*), Thermus thermophilus 30S ribosomal subunit (1fjg*), and a human G Protein Coupled Receptor or GPCR (2rh1). Ligands, cofactors, and additives are shown in CPK form. For details and other milestones, see Supplementary Material Table S1. All structures are displayed at the same scale using PyMol (Delano Scientific—http://www.pymol.org). Membrane proteins are shown with their trans membrane region emphasized in a schematized membrane (notice the lysozyme module fused to the intracellular part of the GPCR structure, see text for details).
In the early time, a second bottleneck appeared rapidly, namely, how to grow “good” crystals of “biologically hot” macromolecules. In this respect, two methodological breakthroughs were essential. The first one that paralleled the development of X-ray methods occurred in the late 1960s and was the implementation of micromethods allowing crystallization trials in assays of 10–50 μl. This allowed solving structures with sample quantities decreasing from more than 100 mg down to less than 1 mg, nowadays, in most favorable cases (Supplementary Material Table S2). The second breakthrough came in the early 1990s with the development of screening kits to rapidly explore crystallization parameter-spaces (Jancarik and Kim, 1991) together with the generalized use of biotechnological tools for sample preparation and the availability of novel computing and synchrotron facilities.
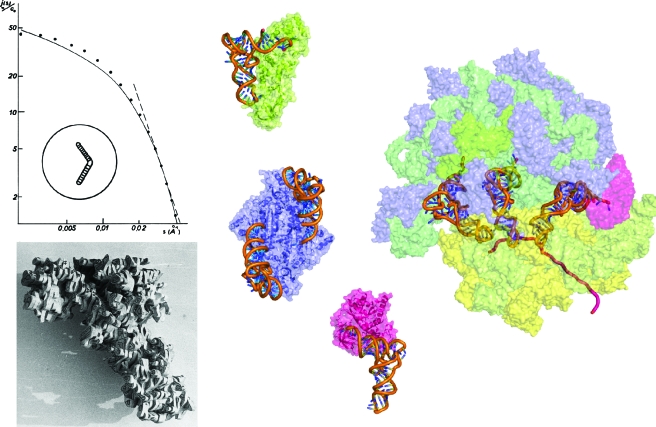
During its rather short history, biocrystallography had a tremendous impact on biology. The study of transfer RNAs (tRNAs) in the context of protein synthesis (Fig. 3) illustrates well how the field has evolved and transformed the structural view biologists had on major macromolecular actors of life. The tRNA story started in the 1960s when small angle X-ray scattering (SAXS) studies on bulk E. coli tRNA revealed an overall L-shaped envelope for these molecules. During the next decade, several crystal structures of free tRNAs revealed their internal atomic anatomy and another 10-year period was necessary to unravel the architecture of tRNA∕protein complexes. The next breakthrough arose at the turn of the millennium when crystallography opened new routes to visualize tRNAs on the ribosome and to understand their behavior during protein synthesis. Thus mechanistic anticipations from the past could be explicitly demonstrated (Schmeing and Ramakrishnan, 2009), such as the allosteric three-site model (Wilson and Nierhaus, 2006) with the tRNA “pas-de-trois” on the ribosome surface. Looking at tRNA molecules under different functional states (Giegé, 2008) also highlighted how crystallography can reveal the structural plasticity of biomacromolecules, a property of pivotal importance for understanding function. As will be discussed below, the tRNA story further illustrates how progress in methods and technologies were the driving force to move from low- to high-resolution and from low- to high-structural complexity (Fig. 3).
Figure 3. From the single macromolecule to its integration in supramolecular biological systems: the example of transfer RNAs.
(Left) SAXS curve from which a boomerang-shaped model of tRNA was deduced (Witz, 1964; Witz, 2003) and a balsawood model built after the X-ray analysis of yeast tRNAAsp at 3 Å resolution (Moras et al., 1980). (Middle) first X-ray structures of tRNAs in complex with proteins from the translation machinery: E. coli tRNAGln in interaction with monomeric class Ib GlnRS (in green) (Rould et al., 1989), S. cerevisiae tRNAAsp in interaction with its cognate dimeric class IIb AspRS (in blue) (Ruff et al., 1991), and phenylalanyl-tRNAPhe in interaction with bacterial elongation factor EF-Tu (in pink) (Nissen et al., 1995). (Right) view of the E. coli ribosome in translation with the three tRNA molecules bound to A, P, and E sites obtained by fitting an atomic model (derived from X-ray structures) in a 6.7 Å resolution cryo-EM map (Villa et al., 2009). Ribosomal proteins from 30 S and 50 S subunits are shown in yellow and blue, respectively, and 23 S, 5 S, and 16 S RNAs in dark, medium, and light green, respectively. The mRNA is symbolized by a violet ribbon and the tRNA in the A site is bound to EF-Tu (pink). Models in the middle and on the right are shown at the same scale.
DEFINING THE APPROPRIATE MACROMOLECULAR TARGET
In structural biology, success is often determined by the appropriate choice of the target macromolecule and different strategies were envisaged depending on the biological question. On the one hand, when the aim is to solve a milestone structure from an essential biological process and∕or from a given biochemical family—transcription or translation, nucleic acids or membrane proteins—the taxonomic origin of the macromolecule is less critical than its crystallizability. In such a situation the biological origin becomes a variable in crystallization. On the other hand, when the aim is to solve structures from a given organism—for pharmacological reasons or proteome establishment—crystallizability may be a real bottleneck and a great deal of effort can be necessary to produce a target amenable to crystallization. It is not our purpose to discuss the many solutions reported in literature to circumvent these concerns. Instead we will illustrate the subject by examples relevant to structural investigations on the translation machinery and on membrane proteins that have been leading in many respects to major developments in biocrystallogenesis.
Regarding the choice of the taxonomic origin of the target one has to remember that many organisms are adapted to extreme life conditions, notably with a temperature of up to 110 °C, pressures of up to 100 MPa, and high radiation levels or salt concentrations. To do so, they have evolved macromolecules, which are stable under such conditions. The pivotal finding that triggered the rush toward extremophiles was the good crystallizability of the tyrosyl-tRNA synthetase (TyrRS), a member of the aminoacyl-tRNA synthetase (aaRS) family, isolated from heat-loving Bacillus stearothermophilus (Reid et al., 1973). The many structures in the protein data bank (PDB) from extremophiles confirm the idea of the relative ease to crystallize their macromolecular components (Liebl, 2004). The concept is particularly true for the aaRS family, where ∼60% of the 3D structures stem from extremophiles (Giegé et al., 2008). Likewise, the known ribosome structures stem from three different extremophiles—T. thermophilus (Cate et al., 1999; Clemons et al., 1999; Tocilj et al., 1999),Haloarcula marismortui (Ban et al., 1999; Gluehmann et al., 2001), and Deinococcus radiodurans (Davidovich et al., 2007)—and were solved as the result of years of intensive and innovative worldwide research efforts (e.g., Moore and Steitz, 2003; Noller, 1991; Wilson and Nierhaus, 2006; Yonath et al., 1998; Yusupov et al., 1991). Note that the opportunity to obtain structures of the same biological entity originating from different taxa changes the traditional way to approach molecular evolution that can now be addressed by 3D structure instead of by 1D sequence analysis. Important applications have already emerged from studies on aminoacyl-tRNA synthetases (O’Donoghue and Luthey-Schulten, 2003) and it can be anticipated that evolutionary biology will be deeply impacted by the 3D vision of protein structures (Abad-Zapatero, 2009).
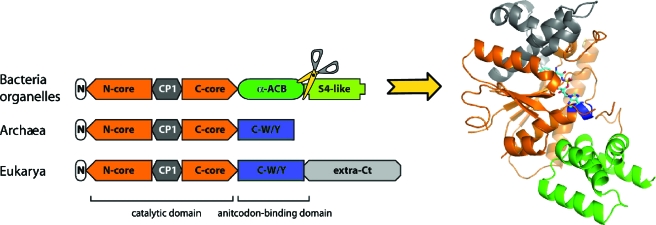
Another key observation is that many proteins and most nucleic acids have multidomain architectures leading to intrinsic structural flexibility, which is a priori not favorable for crystallization. Similarly, membrane proteins often have intracellular and extracellular hydrophilic and mobile domains that are susceptible to prevent their crystallization. For all these reasons, the use of molecular engineering—for removing appendices, inserting domains that constrain flexible regions, or producing isolated domains—was of great benefit to make such structures more compact and stable. A typical example comes from the TyrRS family (Fig. 4). These proteins have a modular architecture overall conserved across evolution with a N-terminal catalytic domain comprising a Rossmann-fold and a dimerization interface (CP1 insertion) and a C-terminal domain of variable architecture that binds tRNA anticodon. In the first crystal structure of a TyrRS, which was from the Bacteria B. stearothermophilus, this domain was not seen in the electron density map because of mobility (Brick et al., 1989). It was later observed that it is also mobile in eukaryal TyrRSs. Therefore in most crystallized TyrRSs, this tail was resected, in particular, in human mitochondrial TyrRS, where the full-length enzyme did not yield crystals suitable for structure determination (Bonnefond et al., 2007a). Likewise, structure determination of yeast AspRS (apo form) was only possible with a truncated protein lacking its flexible 70 residue-long N-terminal extension (Sauter et al., 2000). Other examples concern membrane proteins (Supplementary Material Table S1), notably the K+ channel from Streptomyces lividans and the human β2-adrenergetic G protein coupled receptor (GPCR) protein (Fig. 2). The first one needed two types of engineering for structure determination, namely, the resection of its C-terminal extension and the growth of crystals of a mutant with a single amino acid change that diffracted better than crystals grown from the wild-type protein (Doyle et al., 1998). As to the second example, a mobile intracellular domain prevented crystallization of the receptor. Here, the difficulty was circumvented by stabilizing the GPCR structure by the insertion of a T4 lysozyme molecule in the flexible region (Rosenbaum et al., 2007).
Figure 4. Multidomain organization of macromolecular structures and crystallizability: the case of the TyrRS family.
(Left) schematic structural alignment of TyrRS monomers highlighting the variation in domain composition during evolution. (Left) 3D fold of a resected monomer of dimeric human mitochondrial TyrRS represented with the same color code. In the case of this mitochondrial TyrRS orthologous to bacterial TyrRSs, the full-length form only produced poorly diffracting crystals (Bonnefond et al., 2007b). Based on this observation, the S4-like domain (mimicking ribosomal protein S4) was removed and the engineered catalytically active construct actually led to the 3D structure (Bonnefond et al., 2007a). This bacterial-type C-terminal domain has never been captured in the X-ray structure of an isolated TyrRS, either because it remains mobile in the crystals or because the protein cannot be crystallized in the presence of this appendix.
Structural plasticity of biomacromolecules, although detrimental for crystallization, is essential for function. Thus addition of ligands or of any type of small molecules able to restrain the conformational space of the macromolecule can help its crystallization. This has proven particularly useful for proteins such as aaRSs, where addition of small substrate derivatives or of tRNA allowed crystallization or led to crystals of improved diffraction properties (Giegé et al., 2008). On the other hand, crystallizing complexes containing macromolecular ligands is a way to explore the conformational space of these ligands. Again, this is well illustrated with tRNAs that show a large repertoire of conformations when interacting, for example, with maturation enzymes, aaRSs, elongation factor, or the ribosome (Giegé, 2008).
THE QUEST FOR THE HOLY GRAIL!
Obtaining a good crystal is a mandatory but not easy step. Indeed biological crystallization for long remained poorly understood because of its multiparametric nature. It is now becoming unraveled as a result of interdisciplinary research efforts (Chernov, 2003; McPherson and Giegé, 2007; Sauter et al., 2010). Whatever the type of molecule, crystallization includes four steps: prenucleation, nucleation, growth, and cessation of growth. The whole process can be conveniently visualized in a phase diagram (Fig. 5) that contains an undersaturated region, where macromolecules are soluble and a supersaturated region, where they crystallize, both regions being delimited by the solubility curve (Asherie, 2004; Sauter et al., 1999). The wisdom of the crystal grower will be to find the best and quickest way to explore the parameter space.
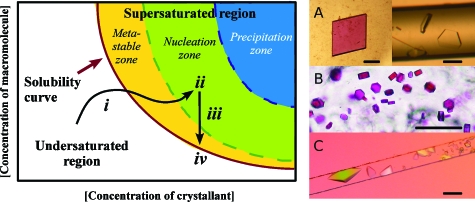
Figure 5. From a better understanding of the crystallization process to the design of advanced strategies of crystallogenesis.
(Left) typical phase diagram illustrating the evolution of a crystallization system as a function of macromolecular and crystallant concentrations. The successive steps leading to the production of crystals are (i) the generation of a supersaturated state (prenucleation), (ii) the formation of a stable nucleus (nucleation), (iii) the growth of the crystals, and (iv) the cessation of growth when the system comes back to equilibrium on the solubility curve. Every crystallization method will present a characteristic trajectory and a different way of exploring the phase diagram (Sauter et al., 2010). The crystal grower can use this knowledge to optimize initial hit(s) by moving the system inside the desired domain. (Right) crystallization at microliter- to nanoliter-scale using nonconventional methods in convection-free environments: (a) crystals of a bacterial AspRS (left) and of an archaeal Holliday junction cutting enzyme (right) grown in agarose gel by vapor diffusion or by counterdiffusion in a capillary, respectively, (Biertümpfel et al., 2005; Zhu et al., 2001). (b) Crystals of bacteriorhodopsin grown in a lipidic cubic phase (courtesy of Prof. Martin Caffrey; see Caffrey, 2008). (c) Crystallization of thaumatin by counterdiffusion in a microfluidic channel of 100×100 μm2 section (Dhouib et al., 2009). Note the shower of microcrystals at the entrance of the channel (at the right), where supersaturation is high, and the large monocrystal at the opposite end, where supersaturation is lower. Scale bars correspond to 100 μm.
Toward rational biocrystallization
Crystallization usually starts by a blind or a semirational screening approach. When a first “hit” is found, refining the parameters that affect crystallization and exploring the most important ones—purity and homogeneity of samples, nature and concentration of crystallants, pH, ionic strength, and temperature—can be conducted more rationally. Note that the meaning of “purity” and “homogeneity” in biocrystallization goes beyond their usual chemical definition, and also refers to physicochemical aspects. Thus, for crystallization one should be seeking for conformational purity that depends on both solvent conditions and structural features of the macromolecules. For that purpose, dynamic light scattering methods became useful tools for solubility and crystallizability diagnostics (Mikol et al., 1990; Wilson, 2003). This led to the recent development of dedicated instruments allowing measurements on samples in the microliter-scale.
Optimizing the production of macromolecular crystals relies presently on robust experimental data arising from studies on the nucleation and crystal growth behaviors of a panel of model proteins. Thus the supersaturated region, which is out of thermodynamic equilibrium in the phase diagram (Fig. 5), contains three kinetically-dependent zones, where crystallizability differs radically. Rapid separation of macromolecules from solution in an amorphous or microcrystalline state occurs at extreme supersaturation in the precipitation zone. In contrast, good crystals can be obtained at lower supersaturation in both nucleation and metastable zones. Lowest supersaturation defines the metastable zone, where nucleation cannot occur spontaneously, and thus is suitable for seeding (D’Arcy et al., 2007). In the nucleation zone, where nucleation occurs spontaneously, number of nuclei, growth rate, and growth mechanisms depend on precise solution conditions that should be tuned appropriately.
Nucleation can be homogeneous in the bulk of the solution, but in most of the cases, it is heterogeneous and occurs on solid surfaces such as the walls of the crystallization chamber or dust particles and other impurities. The means that minimize heterogeneous nucleation and thus favor reproducibility of experiments have been found (Sauter et al., 2010).
Growth of macromolecular crystals can occur by two mechanisms that highly depend on supersaturation. While crystals grow by screw dislocation—by a helical path propagating around a lattice defect—at low supersaturation, they predominantly grow by 2D island formation—from 2D clusters∕nuclei that form randomly on flat crystal faces—at higher supersaturation. High-quality crystals are obtained at lowest supersaturation and under constant growth regime but these are not easy to obtain in practice since crystal growth is accompanied by a decrease in supersaturation in the mother liquor that could trigger a modification of the growth regime. Atomic force microscopy revealed this effect during tRNAPhe crystallization (Ng et al., 1997). Perturbation of growth regime, likely accounts for nonreproducibility of diffraction properties, can also result from impurity incorporation in growing crystals. Such poisoning is favored when the impurity has resemblance with the crystallizing macromolecule. Therefore, the macromolecule itself can be the worst contaminant due to conformational heterogeneity or partially fragmented isoforms.
Crystallization improvements in current practice
All methods used in biocrystallization aim to bring the macromolecule to an appropriate state of supersaturation (Sauter et al., 2010). Although structural biologists favor vapor phase equilibrium techniques, batch, dialysis, and free-interface diffusion methods are alternatives. One shall recall that besides physical and chemical variables, the crystallization method itself and the geometry of the setup also affect crystallization. As mentioned above, crystal growth seldom occurs at constant protein concentration, thus introducing changes in supersaturation and, hence, possible changes in the growth regime. Crystallization at constant macromolecule concentration could be achieved in liquid circulation cells but is not obvious to implement in practice.
Batch crystallization was the method of choice in the pioneers’ time and remains the simplest since it just requires mixing macromolecules and crystallants until supersaturation is reached. It is the first crystallization method that was automated in a microdroplet version under oil (Chayen et al., 1990) and more recently was further miniaturized (D’Arcy et al., 2003). Dialysis permits easy variation in parameters but is less adapted for small sample volumes and screening procedures. In contrast, crystallization by vapor diffusion, which was invented for the production of tRNA crystals (Hampel et al., 1968), is very handy and has rapidly become the favored method in most laboratories. It is practiced in a variety of forms, mainly in microliter-size sitting drops. In free-interface and counterdiffusion methods, equilibration occurs by direct diffusion of the crystallant into the macromolecule solution (García-Ruiz and Moreno, 1994). Both methods require minimal convection and, therefore, experiments are conducted in capillaries. The advantage of counterdiffusion is that a wide range of supersaturation conditions can be tested in a single experiment and that all steps from crystallization to structure determination can be performed in situ without any crystal handling (Gavira et al., 2002).
Advanced crystallization strategies
Over the past decade, new strategies have been developed either to screen physical variables or to give emphasis to peculiar growth media and to take advantage of novel biotechnological tools for stabilizing macromolecular conformations by chaperone macromolecules. These methods were shown to be efficient for both ab initio search of crystallization conditions and optimization procedures for improving crystal quality. As an illustration, Fig. 5 displays crystals grown by three unconventional methods taking advantage of gelled media, cubic mesophases, or microfluidic counterdiffusion channels. On the other hand and in view of high-throughput structural genomics projects, automated instruments and entirely integrated systems have been developed to accelerate crystallization and optimization procedures (Newman et al., 2008).
The ways physical variables affect crystallization are manifold (Sauter et al., 2010). Thus gravity influences fluid properties and movement of molecules, pressure and temperature alter conformation of macromolecules, magnetic fields orient crystals, and electric fields can reduce nucleation rates. Likewise gelled and microfluidic environments reduce convection and thus favor crystallization while cubic gel-like mesophases provide conditions to crystallize hydrophobic proteins. Convection and sedimentation always take place in current procedures and severely influence crystallization. In the absence of gravity, theory predicts regular crystal growth under diffusive regime that should enhance crystal quality. Such considerations have justified space-crystallization programs and have contributed to a deeper understanding of protein crystallization (Kundrot et al., 2001). However, due to experimental limitation, crystallization in weightlessness is not a panacea and ways to mimic its beneficial effects on earth were searched. Because convection depends on viscosity, gels represent a convection-free environment and thus a good media to improve crystal quality (Lorber et al., 2009). As was anticipated, crystals grown in gels are often of superior quality than controls grown from solutions. They can easily be removed from their soft environment and set up for X-ray analysis. Microfluidic devices also provide a diffusive environment due to their small size. The first microfluidic applications in biocrystallization were a free-interface system (Hansen et al., 2002) followed by a “batch in nanodroplets” chip (Zheng et al., 2003) suitable for high-throughput screening. The absence of convection in microfluidic channels makes microsystems very appealing for counterdiffusion experiments. When made of appropriate polymer material, counterdiffusion chips allow a direct on-chip characterization of the crystals by X-ray diffraction without any further sample handling (Dhouib et al., 2009; Ng et al., 2008). Finally, it was conjectured that suppression of convection could be achieved under hypergravity or when magnetic or electric fields are applied. Although macromolecule crystal growth under such conditions is not widespread and the underlying physics not completely validated, these approaches can be useful in special cases, for instance, to reduce the number of nucleation sites (Sauter et al., 2010).
Temperature and pressure are two thermodynamic parameters that can trigger nucleation and sustain protein crystal growth (Rosenberger et al., 1993; Suzuki et al., 2002) but were hardly exploited although temperature-induced crystallization often occurs consequently of accidental temperature variation in the laboratory. Dedicated crystallization systems have been designed for temperature-induced crystallization that find application, e.g., in the growth of large crystals for neutron crystallography (Budayova-Spano et al., 2007). Pressure-induced crystallization is trickier and requires other specialized equipments (Suzuki et al., 2002). Interestingly, cowpea mosaic virus crystals compressed at 330 MPa in a diamond anvil cell demonstrated pressure-induced ordering of the crystals, lower ADPs, and a larger number of ordered water molecules (Girard et al., 2005).
In a more biological perspective, use of “crystallization helper” chaperones is becoming useful to crystallize recalcitrant proteins (Koide, 2009) or RNA fragments (Ye et al., 2008). A typical example of the chaperone strategy is the structure determination of Escherichia coli tRNACys from crystals, where the tRNA was sequestered by elongation factor (Nissen et al., 1999). First tested with antibody fragment chaperones, it was rejuvenated with the DARPin technology based on the natural ankyrin repeat protein fold with randomized surface residue positions allowing specific binding to virtually any target protein (Sennhauser and Grütter, 2008).
BETTER AND FASTER METHODS FOR STRUCTURE DETERMINATION
When the pioneers of biocrystallography showed that biological samples could potentially produce high-resolution diffraction patterns (Bernal and Crowfoot, 1934; Perutz, 1985), structure determination was still very empirical and tedious. The first breakthrough came with the development of a robust phasing method based on the introduction of heavy atoms in the crystals, a procedure called multiple isomorphous replacement (MIR). This approach led to the atomic models of myoglobin and hemoglobin in the early 1960s (Kendrew et al., 1960; Perutz et al., 1968). It is still in use in various forms nowadays and was the key for phasing diffraction data from ribosome crystals soaked with large heavy-atom clusters (Ban et al., 1998; Yonath et al., 1998). Of course, the concomitant boom of computing systems was also pivotal. Following the first success stories and the increase in the number of biological systems investigated, the major difficulty became soon, and still remains, the availability of the biomolecule and the difficulty to produce crystals of adequate quality.
The 1990s brought a radical change in the practice of biocrystallography. The first major development was crystal cryocooling at around 100 K in a stream of nitrogen gas, a method known for long in chemistry and introduced in biology with ribosome crystals (Yonath et al., 1998). It is presently systematized in biocrystallography to slow down radiation damages and to increase the lifetime of samples during X-ray analysis (Garman, 2003). Further, post-crystallization methods to enhance crystal quality—dehydration, annealing, soaking, and other treatments—became popular and cured many “poor” crystals (Heras and Martin, 2005). Here as well, observations on crystals of proteins from the translation machinery—a GlnRS and EF-Tu—where among the first that opened the field (Rould et al., 1991; Schick and Jurnak, 1994). Second, the access to strong synchrotron X-ray light sources increased sharply with the building of new third generation facilities worldwide. The availability of intense and tunable radiations facilitated the development a new phasing method, the multiwavelength anomalous dispersion (MAD) (Hendrickson, 1991), which simplifies the original MIR approach. It eliminates the necessity of preparing several crystal derivatives and associated isomorphism distortion since the entire structure determination can be performed on a single cryocooled crystal including an anomalous scatterer. The combination of all these methods was instrumental to the explosion of 3D data in the PDB that occurred in the mid-1990s (Fig. 1) and here again the ribosome adventure is a striking illustration of their impact in structural biology (Abrahams and Ban, 2003; Gluehmann et al., 2001; Yonath et al., 1998).
At the end of the 1990s, the structural biology community invented the concept of structural genomics and jumped into the post-genomics era. The effective implementation of structural genomics implied to deal with hundreds of targets in a massively parallel manner in order to achieve high-throughput at each stage of a project (Terwilliger et al., 2009) and these developments found also applications at small scale in academic laboratories. They include the production and fast purification of tagged molecules, the use of automated data collection protocols on cryocooled samples (Arzt et al., 2005), and of automated pipelines for X-ray structure solution and refinement (Adams et al., 2009) exploiting the incorporation of selenomethionine in proteins for MAD or single autonomous dispersion (SAD) phasing (Joachimiak, 2009). On the other hand, the increase in the 3D repertoire with ∼50% of new folds in the PDB provided by structural genomics consortia, is rejuvenating the effectiveness of molecular replacement methods as alternate phasing and refinement tools.
Other breakthroughs stem from methodological advances in single particle cryoelectron microscopy (cryo-EM) and were applied in the ribosome field for imaging various ribosomes (Becker et al., 2009; Frank, 2009; Spahn and Penczek, 2009), including minimalist mitochondrial ribosomes (Sharma et al., 2009). When cryo-EM was combined with X-ray crystalloghraphy, molecular dynamics and modeling, new biological questions could be addressed such as uncovering high-resolution snapshots of functional ribosomes during initiation (Simonetti et al., 2009) or elongation (Villa et al., 2009; Fig. 3) of protein synthesis. They were also essential for structure determination of other large assemblies such as viruses with asymmetric properties (Mueller et al., 2007; Rossmann et al., 2007; Steven and Baumeister, 2008).
The recent progress in terms of synchrotron and data collection facilities will certainly help to tackle new appealing biological systems. Of particular interest are the microfocused beams that allow analysis of crystals with a size down to a few microns (Moukhametzianov et al., 2008; Schneider, 2008). Thus, a 2 Å crystal structure of both recombinant and infectious silkworm cypovirus polyhedra could be determined using crystals of 5–12 μm, the smallest crystals yet used for de novo X-ray protein structure determination (Coulibaly et al., 2007). Likewise, the development of a new generation of ultrafast and sensitive X-ray detectors (Kraft et al., 2009) enables the exploitation of radiation-sensitive or weakly diffracting samples, and gives the possibility to analyze crystals in their growth environment—either in microplates or microfluidic devices (Dhouib et al., 2009; Jacquamet et al., 2004)—avoiding potentially detrimental handling.
Today, structure determination of a new target can generally be carried out in a few months, where it may have taken years if not decades, in the past. The situation is well depicted by the metaphor of the “flying crystallographer,” rushing from one synchrotron facility to the next, solving and refining his new structures on the way back to the laboratory, if not directly on the beamline while collecting the data. However, this apparent ease, which might reflect a majority of cases, should not mask that every single project is unique and that the starting point will always remain a good, well-diffracting crystal. Further, it should not be forgotten that entire regions∕areas of the “3D-space” underlying the tree of life remain essentially unexplored such as that of native metazoan proteins with post-translational modifications and without resected motifs.
UNSOLVED ISSUES, HIGHER COMPLEXITY, AND 4D BIOLOGY
The questions addressed to biocrystallography have dramatically evolved since the first protein structures were solved. The fact that high quality X-ray diffraction data can be obtained from a single crystal of dimensions in the range of 20–50 μm has changed the objectives considerably. One shall recall that 40 years ago a structure analysis required many crystals in the mm size range. Since fewer and smaller crystals are now the rule—except for neutron diffraction (Budayova-Spano et al., 2007)—it is easier today to envisage more challenging projects dealing with membrane proteins, lipoproteins, intrinsically unstructured proteins, large RNAs, or nucleoprotein complexes or assemblies. Intrinsically unstructured proteins or proteins with disordered regions represent a real challenge. Such proteins are especially abundant in eukarya and remain poorly understood but may fold and play important roles upon binding to their cellular partners (Dyson and Wright, 2005; Fukuchi et al., 2009). Besides understanding their biology, it is anticipated that their study will provide clues to comprehend protein folding and protein dynamics. Membrane proteins constitute another tricky category due to their lipophylic nature and the difficulty to make them stable in solution and thus amenable to crystallization (Caffrey, 2008). The great deal of effort already invested to enlarge the repertoire of their 3D structures, certainly will be pursued, notably for applications since these proteins represent almost 50% of the promising pharmaceutical targets. In a wider perspective of applications, biocrystallography provides a powerful platform for the conception of new drugs with the possibility to screen for ligand binding in crystallo (Blundell et al., 2006) and can deliver valuable 3D data to fight against new threats such as emerging pathogenic viruses (Anand et al., 2003; Bollati et al., 2009). Crystallography alone, however, is not sufficient for applications such as ligand screening or drug design that may require complementary biophysical and computational techniques (Renaud and Delsuc, 2009).
Although crystallography gives access to static atomic snapshots of objects frozen in a crystal lattice, solving structures with and without ligands or exploiting different crystal forms generated during crystallization screening are powerful means to capture alternate functional states of biomacromolecules. Interestingly, packing plasticity does even exist in crystals diffracting to high-resolution that can sustain a high degree of disorder—up to 30–35%—in their packing (Touzé et al., 2007; Troffer-Charlier et al., 2007). This brings to the question of decrypting the kinematic changes in macromolecular structures in action, in other words the potential of a time-resolved crystallography with perspective of “4D biology,” where the fourth dimension would be the temporal component. Along these lines, the example of crystallographic snapshots obtained during the maturation of a tRNA, provided a “movie” of this enzymatic reaction and gave a robust support to the approach (Tomita et al., 2006). The more direct approach would be to capture transient 3D information in crystallo. This dream of crystallographers was already experimentally assayed in the late 1980s and gave electron density maps from millisecond diffraction data collected on Laue photographs (Hajdu et al., 1988). Recently sophisticated methodologies for time-resolved crystallography have been successfully validated with several model enzymes (Bourgeois and Royant, 2005). With the next generation of X-ray light sources, the coming era of single-molecule X-ray diffraction (Helliwell, 2004) and the development of new tools for analysis intensity changes in Laue diffraction experiments (Coppens et al., 2009), one can anticipate a flourishing future for time-resolved crystallography not only for enzymology but also to get a kinematic insight of macromolecular recognition processes.
Hybrid approaches combining X-ray diffraction with EM, NMR spectroscopy, biophysical, and computational methods will continue to improve and their importance in structural biology will undoubtedly increase (Steven and Baumeister, 2008). Thus, the association of correlative light microscopy and EM, electron or X-ray tomography with crystallography makes possible to apprehend a living cell at different scales, starting from its global organization and zooming down to capture macromolecular events at atomic resolution (Hoenger and McIntosh, 2009; McDermott et al., 2009; Plitzko et al., 2009). This old dream of biologists is now becoming a reality and promising results such as the visualization of the cadherin network bridging the extracellular space in the epidermal desmosome have already been obtained (Al-Amoudi et al., 2007). Interestingly, recent crystallographic work combined with cell biology and modeling also links cadherin biology with the angiostatic activity of a human aaRS, namely, TrpRS (Zhou et al., 2010), a step toward the integrative biology of these two classes of proteins. The perspectives are wide and it can be anticipated that structural biology in its perpetual evolution will become an integral component of integrative biology in a near future.
CONCLUSION
Since the first picture of myoglobin at 5 Å resolution (Kendrew et al., 1958) and the first use of a synchrotron to collect diffraction photographs on a virus crystal (Rosenbaum et al., 1971), immense progresses have been made in the precision of the structural data delivered by X-ray crystallography. The highest resolution for large soluble proteins has recently reached 0.66 Å for human aldolase reductase, a protein of 36 kDa (Podjarny et al., 2004), and 1.15 Å for a membrane protein, namely, a yeast aquaporin (Fischer et al., 2009). Neutron crystallography has progressed as well and provided a structure of bovine pancreatic RNase A at 1.7 Å resolution (Yagi et al., 2009). All along its history the field has been built on a strong interdisciplinary spirit that contributed to solve successive bottlenecks and helped to tackle biological questions of increasing complexity. As a result biocrystallography has become a must and continues to be at the frontiers of biological research. While structural knowledge remains sparse in many respects regarding membrane proteins or eukaryal proteomes, including the human proteome and related biomedical issues, supramolecular crystallography of large and even giant assemblies is just at the verge of a golden age.
As a concluding remark, let us note that biocrystallography transformed from a multi- to an interdisciplinary discipline with scientific fields a priori disconnected that progressively became tightly interconnected. This is well illustrated by the semantic and operational changes in the name of the discipline that transformed to structural biology with its experts coming from physics and chemistry progressively integrating biochemistry and molecular biology in their research practice. At the opposite an increasing number of groups with biochemistry and molecular biology background have adopted crystallography as a major investigation tool. We anticipate that the present structural biology will undoubtedly play a key role in the coming “mutations” toward integrated and global biology and will completely merge with these novel biodisciplines.
ACKNOWLEDGMENTS
We dedicate this essay to the memory of Dr. Warren L. Delano, creator of PYMOL, the powerful molecular visualization software used to prepare the illustrations. We thank Dr. Marat Yusupov for sharing with us his experience on the structural study of the ribosome. This work received support from the Centre National de la Recherche Scientifique (CNRS), the Université de Strasbourg, and the Agence Nationale de la Recherche (ANR) under Grant Nos. ANR-07-NANO-060 and ANR-09-BLAN-009-01.
REFERENCES
- Abad-Zapatero, C (2009). “A note of a protein crystallographer: the molecular structure of evolutionary theory.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 65, 1341–1349. 10.1107/S0907444909044370 [DOI] [PubMed] [Google Scholar]
- Abrahams, J P, and Ban, N (2003). “X-ray crystallographic structure determination of large asymmetric macromolecular assemblies.” Methods Enzymol. 374, 163–188. 10.1016/S0076-6879(03)74008-7 [DOI] [PubMed] [Google Scholar]
- Adams, P D, Afonine, P V, Grosse-Kunstleve, R W, Read, R J, Richardson, J S, Richardson, D C, and Terwilliger, T C (2009). “Recent developments in phasing and structure refinement for macromolecular crystallography.” Curr. Opin. Struct. Biol. 19, 566–572. 10.1016/j.sbi.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi, A, Diez, D C, Betts, M J, and Frangakis, A S (2007). “The molecular architecture of cadherins in native epidermal desmosomes.” Nature (London) , 450, 832–837. 10.1038/nature05994 [DOI] [PubMed] [Google Scholar]
- Anand, K, Ziebuhr, J, Wadhwani, P, Mesters, J R, and Hilgenfeld, R (2003). “Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs.” Science 300, 1763–1767. 10.1126/science.1085658 [DOI] [PubMed] [Google Scholar]
- Arnold, E, Himmel, D M, and Rossmann, M, eds. (2010). Crystallography of Biological Macromolecules, 2nd Ed., Wiley, Chichester. [Google Scholar]
- Arzt, S, et al. (2005). “Automation of macromolecular crystallography beamlines.” Prog. Biophys. Mol. Biol. 89, 124–152. 10.1016/j.pbiomolbio.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Asherie, N (2004). “Protein crystallization and phase diagrams.” Methods 34, 266–272. 10.1016/j.ymeth.2004.03.028 [DOI] [PubMed] [Google Scholar]
- Ban, N, Freeborn, B, Nissen, P, Penczek, P, Grassucci, R A, Sweet, R, Frank, J, Moore, P B, and Steitz, T A (1998). “A 9 Å resolution x-ray crystallographic map of the large ribosomal subunit.” Cell 93, 1105–1115. 10.1016/S0092-8674(00)81455-5 [DOI] [PubMed] [Google Scholar]
- Ban, N, Nissen, P, Hansen, J, Capel, M, Moore, P B, and Steitz, T A (1999). “Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit.” Nature (London) 400, 841–847. 10.1038/23641 [DOI] [PubMed] [Google Scholar]
- Becker, T, et al. (2009). “Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome.” Science 326, 1369–1373. 10.1126/science.1178535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal, J D, and Crowfoot, D (1934). “X-ray photographs of crystalline pepsin.” Nature (London) 133, 794–795. 10.1038/133794b0 [DOI] [Google Scholar]
- Biertümpfel, C, Basquin, J, Birkenbihl, R P, Suck, D, and Sauter, C (2005). “Characterization of crystals of the Hjc resolvase from Archaeoglobus fulgidus grown in gel by counter-diffusion.” Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 61, 684–687. 10.1107/S1744309105018269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell, T L, Sibanda, B L, Montalvao, R W, Brewerton, S, Chelliah, V, Worth, C L, Harmer, N J, Davies, O, and Burke, D (2006). “Structural biology and bioinformatics in drug design: opportunities and challenges for target identification and lead discovery.” Philos. Trans. R. Soc. London, Ser. B 361, 413–423. 10.1098/rstb.2005.1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati, M, et al. (2009). “Structure and functionality in flavivirus ns-proteins: perspectives for drug design.” Antiviral Res. (in press). [DOI] [PMC free article] [PubMed]
- Bonnefond, L, Frugier, M, Touzé, E, Lorber, B, Florentz, C, Giegé, R, Rudinger-Thirion, J, and Sauter, C (2007b). “Tyrosyl-tRNA synthetase: the first crystallization of a human mitochondrial aminoacyl-tRNA synthetase.” Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 63, 338–341. 10.1107/S1744309107012481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefond, L, Frugier, M, Touzé, E, Lorber, B, Florentz, C, Giegé, R, Sauter, C, and Rudinger-Thirion, J (2007a). “Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features.” Structure (London) 15, 1505–1516. 10.1016/j.str.2007.09.018 [DOI] [PubMed] [Google Scholar]
- Bourgeois, D, and Royant, A (2005). “Advances in kinetic protein crystallography.” Curr. Opin. Struct. Biol. 15, 538–547. 10.1016/j.sbi.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Brick, P, Bhat, T N, and Blow, D M (1989). “Structure of tyrosyl-tRNA synthetase refined at 2.3 Å resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate.” J. Mol. Biol. 208, 83–98. 10.1016/0022-2836(89)90090-9 [DOI] [PubMed] [Google Scholar]
- Budayova-Spano, M, Dauvergne, F, Audiffren, M, Bactivelane, T, and Cusack, S (2007). “A methodology and an instrument for the temperature-controlled optimization of crystal growth.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 63, 339–347. 10.1107/S0907444906054230 [DOI] [PubMed] [Google Scholar]
- Caffrey, M (2008). “On the mechanism of membrane protein crystallization in lipidic mesophases.” Cryst. Growth Des. 8, 4244–4254. 10.1021/cg800693r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, C W, Jr. (2009). “E pluribus tres: the Nobel Prize in chemistry.” Structure (London) 17(12), 1588–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J H, Yusupov, M M, Yusupova, G Z, Earnest, T N, and Noller, H F (1999). “X-ray crystal structures of 70S ribosome functional complexes.” Science 285, 2095–2104. 10.1126/science.285.5436.2095 [DOI] [PubMed] [Google Scholar]
- Chayen, N E, Shaw Stewart, P D, Maeder, D L, and Blow, D M (1990). “An automated system for microbatch protein crystallisation and screening.” J. Appl. Crystallogr. 23, 297–302. 10.1107/S0021889890003260 [DOI] [Google Scholar]
- Chernov, A A (2003). “Protein crystals and their growth.” J. Struct. Biol. 142, 3–21. 10.1016/S1047-8477(03)00034-0 [DOI] [PubMed] [Google Scholar]
- Christendat, D, Yee, A, Dharamsi, A, Kluger, Y, Gerstein, M, Arrowsmith, C H, and Edwards, A M (2000). “Structural proteomics: prospects for high throughput sample preparation.” Prog. Biophys. Mol. Biol. 73, 339–345. 10.1016/S0079-6107(00)00010-9 [DOI] [PubMed] [Google Scholar]
- Clemons, W M, Jr., May, J LC, Wimberly, B T, McCutcheon, J P, Capel, M S, and Ramakrishnan, V (1999). “Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution.” Nature (London) 400, 833–840. 10.1038/23631 [DOI] [PubMed] [Google Scholar]
- Coppens, P, et al. (2009). “The RATIO method for time-resolved Laue crystallography.” J. Synchrotron Radiat. 16, 226–230. 10.1107/S0909049508040892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly, F, Chiu, E, Ikeda, K, Gutmann, S, Haebel, P W, Schulze-Briese, C, Mori, H, and Metcalf, P (2007). “The molecular organization of cypovirus polyhedra.” Nature (London) 446, 97–101. 10.1038/nature05628 [DOI] [PubMed] [Google Scholar]
- D’Arcy, A, Mac Sweeney, A, Stihle, M, and Haber, A (2003). “The advantages of using a modified microbatch method for rapid screening of protein crystallization conditions.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 59, 396–399. 10.1107/S0907444902022011 [DOI] [PubMed] [Google Scholar]
- D’Arcy, A, Villard, F, and Marsh, M (2007). “An automated microseed matrix-screening method for protein crystallization.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 63, 550–554. 10.1107/S0907444907007652 [DOI] [PubMed] [Google Scholar]
- Davidovich, C, Bashan, A, Auerbach-Nevo, T, Yaggie, R D, Gontarek, R R, and Yonath, A (2007). “Induced-fit tightens pleuromutilins binding to ribosomes and remote interactions enable their selectivity.” Proc. Natl. Acad. Sci. U.S.A. 104, 4291–4296. 10.1073/pnas.0700041104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhouib, K, et al. (2009). “Microfluidic chips for the crystallization of biomacromolecules by counter-diffusion and on-chip crystal x-ray analysis.” Lab Chip 9, 1412–1421. 10.1039/b819362b [DOI] [PubMed] [Google Scholar]
- Doyle, D A, Morais Cabral, J, Pfuetzner, R A, Kuo, A, Gulbis, J M, Cohen, S L, Chait, B T, and MacKinnon, R (1998). “The structure of the potassium channel: molecular basis of K+ conduction and selectivity.” Science 280, 69–77. 10.1126/science.280.5360.69 [DOI] [PubMed] [Google Scholar]
- Dyson, H J, and Wright, P E (2005). “Intrinsically unstructured proteins and their functions.” Nat. Rev. Mol. Cell Biol. 6, 197–208. 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- Fischer, G, Kosinska-Eriksson, U, Aponte-Santamaria, C, Palmgren, M, Geijer, C, Hedfalk, K, Hohmann, S, de Groot, B L, Neutze, R, and Lindkvist-Petersson, K (2009). “Crystal structure of a yeast aquaporin at 1.15 angstrom reveals a novel gating mechanism.” PLoS Biol. 7, e1000130. 10.1371/journal.pbio.1000130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, J (2009). “Single-particle reconstruction of biological macromolecules in electron microscopy–30 years.” Q. Rev. Biophys. 42, 139–158. 10.1017/S0033583509990059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi, S, Homma, K, Minezaki, Y, Gojobori, T, and Nishikawa, K (2009). “Development of an accurate classification system of proteins into structured and unstructured regions that uncovers novel structural domains: its application to human transcription factors.” BMC Struct. Biol. 9, 26. 10.1186/1472-6807-9-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Ruiz, J M, and Moreno, A (1994). “Investigations on protein crystal growth by the gel acupuncture method.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 50, 484–490. 10.1107/S0907444993014350 [DOI] [PubMed] [Google Scholar]
- Garman, E (2003). “‘Cool’ crystals: macromolecular cryocrystallography and radiation damage.” Curr. Opin. Struct. Biol. 13, 545–551. 10.1016/j.sbi.2003.09.013 [DOI] [PubMed] [Google Scholar]
- Gavira, J A, Toh, D, Lopez-Jaramillo, J, Garcia-Ruiz, J-M, and Ng, J D (2002). “Ab initio crystallographic structure determination of insulin from protein to electron density without crystal handling.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 58, 1147–1154. 10.1107/S0907444902006959 [DOI] [PubMed] [Google Scholar]
- Giegé, R (2008). “Toward a more complete view of tRNA biology.” Nat. Struct. Mol. Biol. 15, 1007–1014. 10.1038/nsmb.1498 [DOI] [PubMed] [Google Scholar]
- Giegé, R, Touzé, E, Lorber, B, Théobald-Dietrich, A, and Sauter, C (2008). “Crystallogenesis trends of free and liganded aminoacyl-tRNA synthetases.” Cryst. Growth Des. 8, 4297–4306. 10.1021/cg8007766 [DOI] [Google Scholar]
- Girard, E, Kahn, R, Mezouar, M, Dhaussy, A C, Lin, T, Johnson, J E, and Fourme, R (2005). “The first crystal structure of a macromolecular assembly under high pressure: CpMV at 330 MPa.” Biophys. J. 88, 3562–3571. 10.1529/biophysj.104.058636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluehmann, M, et al. (2001). “Ribosomal crystallography: from poorly diffracting microcrystals to high-resolution structures.” Methods 25, 292–302. 10.1006/meth.2001.1241 [DOI] [PubMed] [Google Scholar]
- Hajdu, J, Acharya, K R, Stuart, D I, Barford, D, and Johnson, L (1988). “Catalysis in enzyme crystals.” Trends Biochem. Sci. 13, 104–109. 10.1016/0968-0004(88)90051-5 [DOI] [PubMed] [Google Scholar]
- Hampel, A, Labananskas, M, Conners, P G, Kirkegard, L, RajBhandary, U L, Sigler, P B, and Bock, R M (1968). “Single crystals of transfer RNA from formylmethionine and phenylalanine transfer RNA’s.” Science 162, 1384–1387. 10.1126/science.162.3860.1384 [DOI] [PubMed] [Google Scholar]
- Hansen, C L, Skordalakes, E, Berger, J M, and Quake, S R (2002). “A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion.” Proc. Natl. Acad. Sci. U.S.A. 99, 16531–16536. 10.1073/pnas.262485199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, J R (2004). “Overview and new developments in softer x-ray (2 Å<lambda<5 Å) protein crystallography.” J. Synchrotron Radiat. 11, 1–3. 10.1107/S0909049503024099 [DOI] [PubMed] [Google Scholar]
- Hendrickson, W A (1991). “Determination of macromolecular structures from anomalous diffraction of synchrotron radiation.” Science 254, 51–58. 10.1126/science.1925561 [DOI] [PubMed] [Google Scholar]
- Heras, B, and Martin, J (2005). “Post-crystallization treatments for improving diffraction quality of protein crystals.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 61, 1173–1180. 10.1107/S0907444905019451 [DOI] [PubMed] [Google Scholar]
- Hoenger, A, and McIntosh, J R (2009). “Probing the macromolecular organization of cells by electron tomography.” Curr. Opin. Cell Biol. 21, 89–96. 10.1016/j.ceb.2008.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquamet, L, et al. (2004). “Automated analysis of vapor diffusion crystallization drops with an x-ray beam.” Structure (London) 12, 1219–1225. 10.1016/j.str.2004.04.019 [DOI] [PubMed] [Google Scholar]
- Jancarik, J, and Kim, S-H (1991). “Sparse matrix sampling: a screening method for crystallization of proteins.” J. Appl. Crystallogr. 24, 409–411. 10.1107/S0021889891004430 [DOI] [Google Scholar]
- Joachimiak, A (2009). “High-throughput crystallography for structural genomics.” Curr. Opin. Struct. Biol. 19, 573–584. 10.1016/j.sbi.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrew, J, Brodo, G, Dintzis, H, Parrish, H, Wyckoff, H, and Phillips, D (1958). “A three-dimensional model of the myoglobin molecule obtained by x-ray analysis.” Nature (London) 181, 662–666. 10.1038/181662a0 [DOI] [PubMed] [Google Scholar]
- Kendrew, J C, Dickerson, R E, Strandberg, B E, Hart, R G, Davies, D R, Phillips, D C, and Shore, V C (1960). “Structure of myoglobin: a three-dimensional Fourier synthesis at 2 Å resolution.” Nature (London) 185, 422–427. 10.1038/185422a0 [DOI] [PubMed] [Google Scholar]
- Koehn, J, and Hunt, I (2009). “High-throughput protein production (HTPP): a review of enabling technologies to expedite protein production.” Methods Mol. Biol. 498, 1–18. 10.1007/978-1-59745-196-3_1 [DOI] [PubMed] [Google Scholar]
- Koide, S (2009). “Engineering of recombinant crystallization chaperones.” Curr. Opin. Struct. Biol. 19, 449–457. 10.1016/j.sbi.2009.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft, P, et al. (2009). “Performance of single-photon-counting PILATUS detector modules.” J. Synchrotron Radiat. 16, 368–375. 10.1107/S0909049509009911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrot, C E, Judge, R A, Pusey, M L, and Snell, E H (2001). “Microgravity and macromolecular crystallography.” Cryst. Growth Des. 1, 87–99. 10.1021/cg005511b [DOI] [Google Scholar]
- Liebl, W (2004). “Genomics taken to the extreme.” Nat. Biotechnol. 22, 524–525. 10.1038/nbt0504-524 [DOI] [PubMed] [Google Scholar]
- Lorber, B, Sauter, C, Théobald-Dietrich, A, Moreno, A, Schellenberger, P, Robert, M-C, Capelle, B, Sanglier, S, Potier, N, and Giegé, R (2009). “Crystal growth of proteins, nucleic acids, and viruses in gels.” Prog. Biophys. Mol. Biol. 101, 13–25. 10.1016/j.pbiomolbio.2009.12.002 [DOI] [PubMed] [Google Scholar]
- McDermott, G, Le Gros, M A, Knoechel, C G, Uchida, M, and Larabell, C A (2009). “Soft x-ray tomography and cryogenic light microscopy: the cool combination in cellular imaging.” Trends Cell Biol. 19, 587–595. 10.1016/j.tcb.2009.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, A, and Giegé, R (2007). “Crystallogenesis research for biology in the last two decades as seen from the international conferences on the crystallization of biological macromolecules.” Cryst. Growth Des. 7, 2126–2133. 10.1021/cg700683a [DOI] [Google Scholar]
- Mikol, V, Hirsch, E, and Giegé, R (1990). “Diagnostic of precipitant for biomacromolecule crystallization by quasi-elastic light-scattering.” J. Mol. Biol. 213, 187–195. 10.1016/S0022-2836(05)80130-5 [DOI] [PubMed] [Google Scholar]
- Moore, P, and Steitz, T A (2003). “The structural basis of large ribosomal subunit function.” Annu. Rev. Biochem. 72, 813–850. 10.1146/annurev.biochem.72.110601.135450 [DOI] [PubMed] [Google Scholar]
- Moras, D, Comarmond, M-B, Fischer, J, Weiss, R, Thierry, J-C, Ebel, J-P, and Giegé, R (1980). “Crystal structure of yeast tRNAAsp.” Nature (London) 288, 669–674. 10.1038/288669a0 [DOI] [PubMed] [Google Scholar]
- Moukhametzianov, R, Burghammer, M, Edwards, P C, Petitdemange, S, Popov, D, Fransen, M, McMullan, G, Schertler, G F, and Riekel, C (2008). “Protein crystallography with a micrometre-sized synchrotron-radiation beam.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 64, 158–166. 10.1107/S090744490705812X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, M, Jenni, S, and Ban, N (2007). “Strategies for crystallization and structure determination of very large macromolecular assemblies.” Curr. Opin. Struct. Biol. 17, 572–579. 10.1016/j.sbi.2007.09.004 [DOI] [PubMed] [Google Scholar]
- Newman, J, Pham, T M, and Peat, T S (2008). “Phoenito experiments: combining the strengths of commercial crystallization automation.” Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun., 64, 991–996. 10.1107/S1744309108029667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, J D, Clark, P J, Stevens, R C, and Kuhn, P (2008). “In situ x-ray analysis of protein crystals in low-birefringent and x-ray transmissive plastic microchannels.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 64, 189–197. 10.1107/S0907444907060064 [DOI] [PubMed] [Google Scholar]
- Ng, J D, Kuznetsov, Y G, Malkin, A J, Keith, G, Giegé, R, and McPherson, A (1997). “Visualization of RNA crystal growth by atomic force microscopy.” Nucleic Acids Res. 25, 2582–2588. 10.1093/nar/25.13.2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus, K H (2009). “Nobel Prize for the elucidation of ribosome structure and insight into the translation mechanism.” Angew. Chem., Int. Ed. Engl. 48, 9225–9228. 10.1002/anie.200905795 [DOI] [PubMed] [Google Scholar]
- Nissen, P, Kjeldgaard, M, Thirup, S, Polekhina, G, Reshetnikova, L, Clark, B F, and Nyborg, J (1995). “Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog.” Science 270, 1464–1472. 10.1126/science.270.5241.1464 [DOI] [PubMed] [Google Scholar]
- Nissen, P, Thirup, S, Kjeldgaard, M, and Nyborg, J (1999). “The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA.” Structure (London) 7, 143–156. 10.1016/S0969-2126(99)80021-5 [DOI] [PubMed] [Google Scholar]
- Noller, H F (1991). “Ribosomal RNA and translation.” Annu. Rev. Biochem. 60, 191–227. 10.1146/annurev.bi.60.070191.001203 [DOI] [PubMed] [Google Scholar]
- O’Donoghue, P, and Luthey-Schulten, Z (2003). “On the evolution of structure in aminoacyl-tRNA synthetases.” Microbiol. Mol. Biol. Rev. 67, 550–573. 10.1128/MMBR.67.4.550-573.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling, L, and Corey, R B (1951). “Configurations of polyptide chains with favored orientations around single bonds: two new pleeted sheets.” Proc. Natl. Acad. Sci. U.S.A. 37, 729–740. 10.1073/pnas.37.11.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutz, M F (1985). “Early days of protein crystallography.” Methods Enzymol. 114, 3–18. 10.1016/0076-6879(85)14003-6 [DOI] [PubMed] [Google Scholar]
- Perutz, M F, Miurhead, H, Cox, J M, Goaman, L C, Mathews, F S, McGandy, E L, and Webb, L E (1968). “Three-dimensional Fourier synthesis of horse oxyhaemoglobin at 2.8 Å resolution: (1) x-ray analysis.” Nature (London) 219, 29–32. 10.1038/219029a0 [DOI] [PubMed] [Google Scholar]
- Plitzko, J M, Rigort, A, and Leis, A (2009). “Correlative cryo-light microscopy and cryo-electron tomography: from cellular territories to molecular landscapes.” Curr. Opin. Biotechnol. 20, 83–89. 10.1016/j.copbio.2009.03.008 [DOI] [PubMed] [Google Scholar]
- Podjarny, A, Cachau, R E, Schneider, T, Van Zandt, M, and Joachimiak, A (2004). “Subatomic and atomic crystallographic studies of aldose reductase: implications for inhibitor binding.” Cell. Mol. Life Sci. 61, 763–773. 10.1007/s00018-003-3401-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, B R, Koch, G LE, Boulanger, Y, Hartley, B S, and Blow, D (1973). “Crystallization and preliminary x-ray diffraction studies on tyrosyl transfer RNA synthetase from Bacillus stearothermophilus.” J. Mol. Biol. 80, 199–200. 10.1016/0022-2836(73)90241-6 [DOI] [PubMed] [Google Scholar]
- Renaud, J-P, and Delsuc, M A (2009). “Biophysical techniques for ligand screening and drug design.” Curr. Opin. Pharmacol. 9, 622–628. 10.1016/j.coph.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Rosenbaum, D M, et al. (2007). “GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function.” Science 318, 1266–1273. 10.1126/science.1150609 [DOI] [PubMed] [Google Scholar]
- Rosenbaum, G, Holmes, K C, and Witz, J (1971). “Synchrotron radiation as a source for x-ray diffraction.” Nature (London) 230, 434–437. 10.1038/230434a0 [DOI] [Google Scholar]
- Rosenberger, F, Howard, S B, Sowers, J W, and Nyce, T A (1993). “Temperature dependence of protein solubility—determination and application to crystallization in x-ray capillaries.” J. Cryst. Growth 129, 1–12. 10.1016/0022-0248(93)90427-X [DOI] [Google Scholar]
- Rossmann, M G, et al. (2007). “From structure of the complex to understanding of the biology.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 63, 9–16. 10.1107/S0907444906047330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rould, M A, Perona, J J, Söll, D, and Steitz, T A (1989). “Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNAGln and ATP at 2.8 Å resolution.” Science 246, 1135–1142. 10.1126/science.2479982 [DOI] [PubMed] [Google Scholar]
- Rould, M A, Perona, J J, and Steitz, T A (1991). “Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase.” Nature (London) 352, 213–218. 10.1038/352213a0 [DOI] [PubMed] [Google Scholar]
- Ruff, M, Krishnaswamy, S, Boeglin, M, Poterszman, A, Mitschler, A, Podjarny, A, Rees, B, Thierry, J-C, and Moras, D (1991). “Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNAAsp.” Science 252, 1682–1689. 10.1126/science.2047877 [DOI] [PubMed] [Google Scholar]
- Sauter, C, Lorber, B, Kern, D, Cavarelli, J, Moras, D, and Giegé, R (1999). “Crystallogenesis studies on aspartyl-tRNA synthetase: use of phase diagram to improve crystal quality.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 55, 149–156. 10.1107/S0907444998010890 [DOI] [PubMed] [Google Scholar]
- Sauter, C, Lorber, B, McPherson, A, and Giegé, R (2010). “Crystallization. general methods.” Crystallography of Biological Macromolecules, Arnold, E, Himmel, D A, Rossmann, M, eds., 2nd Ed., Wiley, Chichester, Vol. F, in press. [Google Scholar]
- Sauter, C, Lorber, E, Cavarelli, J, Moras, D, and Giegé, R (2000). “The free yeast aspartyl-tRNA synthetase differs from the tRNAAsp-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain.” J. Mol. Biol. 299, 1313–1324. 10.1006/jmbi.2000.3791 [DOI] [PubMed] [Google Scholar]
- Schick, B, and Jurnak, F (1994). “Extension of the diffraction resolution of crystals.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 50, 563–568. 10.1107/S0907444994001976 [DOI] [PubMed] [Google Scholar]
- Schmeing, T M, and Ramakrishnan, V (2009). “What recent ribosome structures have revealed about the mechanism of translation.” Nature (London) 461, 1234–1242. 10.1038/nature08403 [DOI] [PubMed] [Google Scholar]
- Schneider, T R (2008). “Synchrotron radiation: micrometer-sized x-ray beams as fine tools for macromolecular crystallography.” HFSP J. 2, 302–306. 10.2976/1.2982661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennhauser, G, and Grütter, M G (2008). “Chaperone-assisted crystallography with DARPins.” Structure (London) 16, 1443–1453. 10.1016/j.str.2008.08.010 [DOI] [PubMed] [Google Scholar]
- Sharma, M R, Booth, T M, Simpson, L, Maslov, D M, and Agrawal, R K (2009). “Structure of a mitochondrial ribosome with minimal RNA.” Proc. Natl. Acad. Sci. U.S.A. 106, 9637–9642. 10.1073/pnas.0901631106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti, A, Marzi, S, Jenner, L, Myasnikov, A, Romby, P, Yusupova, G, Klaholz, B P, and Yusupov, M (2009). “A structural view of translation initiation in bacteria.” Cell. Mol. Life Sci. 66, 423–436. 10.1007/s00018-008-8416-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C MT, and Penczek, P A (2009). “Exploring conformational modes of macromolecular assemblies by multiparticle cryo-EM.” Curr. Opin. Struct. Biol. 19, 623–631. 10.1016/j.sbi.2009.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven, A C, and Baumeister, W (2008). “The future is hybrid.” J. Struct. Biol. 163, 186–195. 10.1016/j.jsb.2008.06.002 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.1063/1.3156311.
- Suzuki, Y, Sazaki, G, Miyashita, S, Sawada, T, Tamura, K, and Komatsu, H (2002). “Protein crystallization under high pressure.” Biochim. Biophys. Acta 1595(1–2), 345–356. 10.1016/S0167483801003557 [DOI] [PubMed] [Google Scholar]
- Terwilliger, T C, Stuart, D, and Yokoyama, S (2009). “Lessons from structural genomics.” Ann. Rev. Biophys. 38, 371–383. 10.1146/annurev.biophys.050708.133740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocilj, A, et al. (1999). “The small ribosomal subunit from Thermus thermophilus at 4.5 Å resolution: pattern fittings and the identification of a functional site.” Proc. Natl. Acad. Sci. U.S.A. 96, 14252–14257. 10.1073/pnas.96.25.14252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita, K, Ishitani, R, Fukai, S, and Nureki, O (2006). “Complete crystallographic analysis of the dynamics of CCA sequence addition.” Nature (London) 443, 956–960. 10.1038/nature05204 [DOI] [PubMed] [Google Scholar]
- Touzé, E, Lorber, B, Deniziak, M, Becker, H D, Kern, D, Giegé, R, and Sauter, C (2007). “Disorder can exist inside well-diffracting crystals.” Cryst. Growth Des. 7, 2195–2197. 10.1021/cg7007057 [DOI] [Google Scholar]
- Troffer-Charlier, N, Cura, V, Hassenboehler, P, Moras, D, and Cavarelli, J (2007). “Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains.” EMBO J. 26, 4391–4401. 10.1038/sj.emboj.7601855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, E, et al. (2009). “Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis.” Proc. Natl. Acad. Sci. U.S.A. 106, 1063–1068. 10.1073/pnas.0811370106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, J D, and Crick, F H (1953). “Molecular structure of nucleic acids.” Nature (London) 171, 737–738. 10.1038/171737a0 [DOI] [PubMed] [Google Scholar]
- Wilson, D N, and Nierhaus, K H (2006). “The e-site story: the importance of maintaining two tRNAs on the ribosome during protein synthesis.” Cell. Mol. Life Sci. 63, 2725–2737. 10.1007/s00018-006-6125-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W W (2003). “Light scattering as a diagnostic for protein crystal growth—a practical approach.” J. Struct. Biol. 142, 56–65. 10.1016/S1047-8477(03)00038-8 [DOI] [PubMed] [Google Scholar]
- Witz, J (1964). Etude de la structure de quelques polynucleotides en solution par diffusion centrale des rayons X, Université Louis Pasteur, Strasbourg. [Google Scholar]
- Witz, J (2003). “1964: the first model for the shape of a transfer RNA molecule. An account of an unpublished small-angle x-ray scattering study.” Biochimie 85, 1265–1268. 10.1016/j.biochi.2003.09.018 [DOI] [PubMed] [Google Scholar]
- Yagi, D, Yamada, T, Kurihara, K, Ohnishi, Y, Yamashita, M, Tamada, T, Tanaka, I, Kuroki, R, and Niimura, N (2009). “A neutron crystallographic analysis of phosphate-free ribonuclease A at 1.7 Å resolution.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 65, 892–899. 10.1107/S0907444909018885 [DOI] [PubMed] [Google Scholar]
- Ye, J D, Tereshko, V, Frederiksen, J K, Koide, A, Fellouse, F A, Sidhu, S S, Koide, S, Kossiakoff, A A, and Piccirilli, J A (2008). “Synthetic antibodies for specific recognition and crystallization of structured RNA.” Proc. Natl. Acad. Sci. U.S.A. 105, 82–87. 10.1073/pnas.0709082105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonath, A, et al. (1998). “Crystallographic studies on the ribosome, a large macromolecular assembly exhibiting severe nonisomorphism, extreme beam sensitivity and no internal symmetry.” Acta Crystallogr. 54, 945–955. 10.1107/S010876739800991X [DOI] [PubMed] [Google Scholar]
- Yusupov, M M, Garber, M B, Vasiliev, V D, and Spirin, A S (1991). “Thermus thermophilus ribosomes for crystallographic studies.” Biochimie 73, 887–897. 10.1016/0300-9084(91)90130-S [DOI] [PubMed] [Google Scholar]
- Zheng, B, Roach, L S, and Ismagilov, R F (2003). “Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets.” J. Am. Chem. Soc. 125, 11170–11171. 10.1021/ja037166v [DOI] [PubMed] [Google Scholar]
- Zhou, Q, et al. (2010). “Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality.” Nat. Struct. Mol. Biol. 17, 57–61. 10.1038/nsmb.1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D-W, Lorber, B, Sauter, C, Ng, J D, Bénas, P, Le Grimellec, C, and Giegé, R (2001). “Growth kinetics, diffraction properties and effect of agarose on the stability of a novel crystal form of Thermus thermophilus aspartyl-tRNA synthetase-1.” Acta Crystallogr., Sect. D: Biol. Crystallogr. 57, 552–558. 10.1107/S0907444901001743 [DOI] [PubMed] [Google Scholar]