Abstract
Despite their evolutionary significance, little is known about the adaptation dynamics of genomically rewired cells in evolution. We have confronted yeast cells carrying a rewired regulatory circuit with a severe and unforeseen challenge. The essential HIS3 gene from the histidine biosynthesis pathway was placed under the exclusive regulation of the galactose utilization system. Glucose containing medium strongly represses the GAL genes including HIS3 and these rewired cells are required to operate this essential gene. We show here that although there were no adapted cells prior to the encounter with glucose, a large fraction of cells adapted to grow in this medium and this adaptation was stably inherited. The adaptation relied on individual cells that switched into an adapted state and, thus, the adaptation was due to a response of many individual cells to the change in environment and not due to selection of rare advantageous phenotypes. The adaptation of numerous individual cells by heritable phenotypic switching in response to a challenge extends the common evolutionary framework and attests to the adaptive potential of regulatory circuits.
Organisms and cells exhibit significant adaptation capabilities to diverse environmental conditions as evident by the realized biodiversity. The Darwinian natural selection framework states the order of steps in the evolutionary process that leads to diversification. First, heritable phenotypic variability exists in the population and later, upon a change in conditions, the environment might impose selection on certain phenotypes that will change their frequency from one generation to the next. The neo-Darwinian view extends this paradigm by maintaining that underlying the heritable phenotypic diversity are genes and genetic variation, which can be ascribed to neutral and advantageous mutations that occur rarely, spontaneously at random locations, and independently of any selection processes imposed by the environmental conditions. Since then, many studies demonstrated the importance of genetic variability that confers fitness advantage for the emergence of novel functional elements in a given selective environment (Luria and Delbruck, 1943; Paquin and Adams, 1983; Lenski and Travisano, 1994; Drake et al., 1998; Marini et al., 1999; Elena and Lenski, 2003; Fong et al., 2005; Maharjan et al., 2006;Perfeito et al., 2007).
In many evolutionary significant cases, phenotypes evolve not due to the emergence of a new protein or modified protein functionality but rather due to novelty in gene regulation (King and Wilson, 1975). Regulatory modes can be flexible and diverse and, indeed, comparative studies have delineated cases in which novel phenotypes that emerged due to a modification of gene regulation had created a new functional context for an existing gene (Carroll et al., 2001; Wilkins, 2002; Alonso and Wilkins, 2005; Carroll, 2005; Davidson, 2006; Wray, 2007; Tuch et al., 2008a, 2008b). The large variability in regulatory circuits we witness today was shaped by many past regulatory challenges that were successfully resolved in evolution. Nevertheless, as large as this observed variability is, it represents only a fraction of the vast combinatorial space of possible regulatory modes. Thus, it is reasonable to hypothesize that existing and rare genetic variation cannot provide an immediate advantageous solution for every possible novel regulatory challenge (Gerhart and Kirschner, 1997; West-Eberhard, 2003) and, thus, alternative mechanisms for adaptation should be considered.
Most of our knowledge in biology is based on studies of and comparisons among evolutionary end points, namely, current life forms. Little information exists on the dynamics of processes that lead to functional biological novelties and the intermediate states of evolving forms (West-Eberhard, 2003). Traditionally, studies in experimental evolution focused mostly on the evolved organism, characterizing its higher fitness and the underlying advantageous mutations that were selected (Paquin and Adams, 1983; Elena and Lenski, 2003). However, detecting an alternative adaptation process requires careful monitoring of the adaptation process itself and the dynamic trajectory of the whole population rather than merely studying the end point, which is only the long term outcome of the process. Moreover, studying the adaptation to a novel challenge that was never before encountered by the cells along their evolutionary history bares higher chances to detect an alternative process compared to studying a repeated challenge for which cells might already have a “predesigned” solution.
We have explored a possible alternative adaptation process by placing cells with a novel rewired genome in a challenging environment. In a genomic rewiring event, an existing gene becomes linked to a foreign (i.e., previously functionally unrelated) regulatory system. Such events are well documented by comparative genomic studies and were reported to drive significantly the evolution of gene regulation [(Carroll et al., 2001; Wilkins, 2002; Davidson, 2006) also, see the discussions in Alonso and Wilkins (2005); Stern and Orgogozo (2008); Erwin and Davidson (2009)]. In our experiments, a strain of the yeast Saccharomyces cerevisiae was engineered to recruit the gene HIS3, an essential enzyme from the histidine biosynthesis pathway, exclusively to the GAL system that is responsible for galactose utilization (Stolovicki et al., 2006). The arbitrary regulatory linkage between these two evolutionary conserved and highly specific modules was stressful, challenging and created incompatibilities in gene expression (Stolovicki et al., 2006). The cells had not exercised regulation of HIS3 by carbon sources availability along their history, so a substantial adaptive response was required for them to survive in a medium lacking histidine. Recently, we have shown that a cell population carrying this GAL-HIS3 rewired genome can rapidly adapt to grow competitively in histidine-lacking glucose medium despite the strong repression of the GAL system and HIS3 (Stolovicki et al., 2006; Stern et al., 2007). Once established, the adaptation have been propagated stably for hundreds of generations (Stolovicki et al., 2006).
Our previous study (Stolovicki et al., 2006) was focused on the population average adaptation dynamics, which did not allow us to follow the population structure during this process. Thus, it was impossible to conclude whether the observed adaptation phenomenon was due to the selection of a special subpopulation or other processes. To understand how the population achieved this rapid adaptation, in this paper, we monitored by colony counts and microscopy measurements the growth behavior of individual cells upon their first encounter with the challenging environment. We show that in spite of the overall population growth upon the first encounter with liquid glucose medium, there was no observable fraction of adapted cells, neither prior to the first exposure nor during the initial growth phase of the population. Remarkably, the potential of cells to adapt declined sharply during the first few generations of exposure to glucose, contrary to the expected behavior of an exponentially growing population. Plating samples from the growing population confirmed that numerous individual cells could eventually grow on glucose similar to wild-type cells but this capacity was gained only later on the plates and not during the initial growth in liquid glucose. Cells that gained the growth capacity on glucose were truly adapted since they could propagate this capacity to their descendants for hundreds of generations. Since we show that the adaptive state was gained only after the exposure to glucose and by numerous cells simultaneously, this heritable adaptation must have been induced in individual cells by this environment. This study, therefore, details a process that is different from the fundamental common view of adaptation. Here adaptation seems not to rely on random and rare genetic variability that accumulated independently from the selection agent.
RESULTS
Adaptation of cells on Glu-his plates
The adaptation capability of rewired cells by a colony growth assay on plates was demonstrated in Stolovicki et al. (2006). In this study, we have expanded this analysis to examine the variability of this process. Naive rewired cells, which were not exposed to glucose before, were sampled from a single colony and batch cultured in histidine-lacking galactose-based (Gal-his) medium. A sample of cells from this culture was washed and placed on histidine-lacking glucose (Glu-his) plates to test for adaptation and on rich medium plates as a viability control. On rich medium plates, which contained histidine, rewired cells grew as wild-type cells and formed mature colonies within 2–3 days. In contrast, on Glu-his plates, no colonies were observed until day 6 post-plating and only after additional incubation, colonies started to form and gradually accumulated until the maximal number of colonies was counted at day 20 post-plating. Importantly, as was shown before in numerous studies that used the deletion of HIS3 as an oxotrophic marker, also in our experiments cells without any copy of HIS3 could not grow on Glu-his plates even after longer incubation times. Common to the experiment described in Stolovicki et al. (2006) and to the 11 experiments we report here was that cells during their incubation in the repressive Glu-his environment gradually became capable of producing histidine, propagate and form mature colonies. On average, the fraction of cells that could grow mature colonies on Glu-his was about 50% of the total number of viable cells plated, as estimated from colony numbers on rich medium plates [Figs. 1a, 1b, 1e]. Moreover, this fraction turned out to be variable among experiments that started from different colonies and similar variability was found also among replicates of the same experiment that were drawn from a single starting culture [Fig. 1e].
Figure 1. Adaptation and propagation on plates.
From a culture of naive cells in galactose, an equal amount of cells were plated on Glu-his (a) and rich medium (b) plates. Images were captured after 12 days (a) and 48 h (b) of incubation, respectively. Note the high proportion of cells that adapted and formed colonies on glucose as well as the variable sizes of the colonies. Cells from a single adapted colony on the adaptation plate (a) were taken and equal amounts were further plated on Glu-his (c) and rich medium (d) plates. Both images [(c) and (d)] were taken at 48 h post-plating. Note the similar growth and uniform colony size of adapted cells on both medium types. Several adaptation experiments on Glu-his plates [as in panels (a) and (b)], each in replicates, were carried out and the proportion of adaptation was determined as the colony number on Glu-his divided by that on rich medium (e). Note the overall high proportion of adaptation (average of 0.5) and the variation, which was similar among replicates and among experiments.
This large and variable fraction of mature colonies on Glu-his plates was unexpected and raised the concern that the cell growth was a mere transient physiological response accommodating the stressful condition. However, this was not the case as cells from these colonies retained their growth capability over hundreds of generations on Glu-his both in liquid medium as well as on plates. Furthermore, cells from colonies that adapted on Glu-his formed mature colonies already within 2–3 days upon respreading on new Glu-his plates [Figs. 1c, 1d], showing growth rates similar to that on the rich medium plates. Thus, the adapted phenotype is inherited and based on this normal growth on Glu-his, the adaptive phenotype becomes easily distinguishable from the naive one. Furthermore, this study corroborated previous experiments (Stolovicki et al., 2006) and confirmed that the growth phenotype of adapted cells was stably inherited and did not require a repeatable adaptation period after switching back and forth between Gal-his and Glu-his environments. Thus, the growth of ∼50% of the cells into mature colonies on glucose plates was a genuine adaptation process that was stably inherited across generations, allowing the survival of the genome-rewired strain in this challenging environment.
The population growth dynamics during adaptation
In the context of evolution, the term adaptation is often used to describe the change in the population average phenotype over generations as a result of selection against or in favor of certain variants. The large fraction of cells that could lead to adapted colonies on the plates was surprising and to understand how such a large fraction of cells adapted, the process of the population adaptation was studied by adding the time dimension; measuring the population growth dynamics and analyzing the behavior of individual cells from the onset of exposure to glucose to the final adapted state. In previous studies, the growth dynamics of the population upon exposure to Glu-his was studied in a chemostat (Novick and Szilard, 1950) in which large populations of rewired cells were grown in Gal-his and switched to Glu-his medium under controlled and otherwise stable environmental conditions. Our chemostat setup allowed us to monitor online the cell density [optical density (OD)] and automatically sample the population at a high temporal resolution for further analyses of the cells [see examples in Stolovicki et al. (2006) and Stern et al. (2007)]. In this study, repeated chemostat experiments were carried out under identical conditions and the reproducible patterns of the population growth dynamics pointed out to a few robust features (Fig. 2). In particular, phase I of all populations, immediately following the switch into Glu-his medium, was characterized by a prolonged exponential increase in cell density lasting for about eight generations after which, in phase II, the cultures exhibited an exponential collapse. The increased density during phase I was probably due to the elevated growth rate that is typical of yeast cells when switched from galactose-based to glucose-based medium since the latter is a more efficient carbon source. For rewired cells however, this was an unexpected behavior because of the strong repression of HIS3 that was previously observed (Stolovicki et al., 2006). We still do not fully understand this robust phase but it is fair to assume that the prolonged exponential growth relies on existing resources, either reservoirs of histidine, HIS3 mRNA or His3p from the previous period in galactose, allowing a limited number of cell divisions. We leave this interesting issue for future work and concentrate in this paper on the growth dynamics of the population and its individual cells that eventually lead to adaptation.
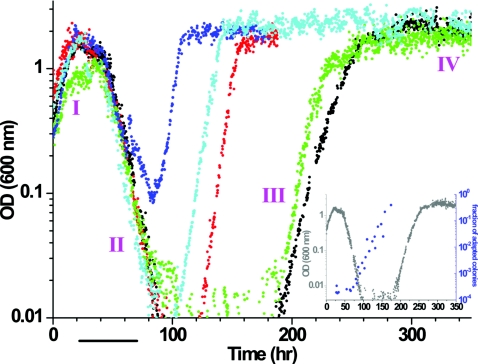
Figure 2. Adaptation dynamics of chemostat populations.
Cell density (OD at 600 nm) as a logarithmic function of time for repeated chemostat experiments conducted under the same nominal conditions. As a comparison, the light and dark blue curves corresponds to chemostat experiments described in (Stolovicki et al., 2006). The histidine-lacking medium was switched from galactose to glucose as a sole carbon source at t=0, leaving all other nutrients the same. A steady-state typical of galactose metabolism was established prior to the switch into glucose. Bar—ten generations and the population growth phases are denoted in purple (I-IV). Inset: The fraction of adapted colonies during phase II (blue dots) on the background of the population density (gray) showing that the population became fully adapted by the end of phase II.
Following the initial growth in phase I, the population collapsed and the cell density decreased exponentially, indicating that regardless of the resources allowing the initial growth, those were limited and the cells eventually faced a severe growth challenge during phase II. Further in time, the population reached a turning point in which the cell density started to increase, indicating that most cells were growing faster than the chemostat’s dilution rate (phase III), followed by a stable growth and constant cell density stage reflecting a population in a steady-state (phase IV).
Based on the repeatable growth dynamics in chemostat experiments, it was clear that the population was not adapted upon the exposure to Glu-his but it became fully adapted by the time it reached phase IV. To understand better the adaptive state of the population in between these time limits, the phenotype of individual cells was studied by sampling at consecutive time points during the chemostat phases and spreading these cells on Glu-his plates. This analysis indicated the fraction of adapted cells was minimal at the end of phase I but it increased exponentially during phase II and became a 100% by the end of this phase (Fig. 2, inset). This analysis explained why the cell density increased exponentially during phase III, which turned out to serve as an adjustment period in which the population that was composed of 100% adapted cells fulfilled its maximal growth potential in Glu-his. But more importantly, new time limits were placed on the adaptation process that is now shown to be completed by the end of phase II. Repeated chemostat experiment under precisely the same conditions revealed a variable adaptation time that ranged between 10 and 30 generations (Fig. 2). This adaptation time is surprisingly shorter than those usually encountered in laboratory experiments involving the fixation of rare mutations in large populations of microorganisms, which were on the order of hundreds to thousands of generations (Luria and Delbruck, 1943; Paquin and Adams, 1983; Cairns et al., 1988; Lenski and Travisano, 1994; Drake et al., 1998; Marini et al., 1999; Elena and Lenski, 2003; Fong et al., 2005; Roth et al., 2006; Perfeito et al., 2007).
The time-dependent adaptation potential in glucose
Realizing the short timeframe during which the population achieved its fully adapted state pointed out to the significance of the first generations in Glu-his and in particular to the population structure during phase I. Thus, we sampled the population during this particular phase at a high temporal resolution and tested the adaptive state of individual cells from these samples on Glu-his plates. On rich medium control plates, the cells grew at a normal rate, forming uniform colonies after 2–3 days and exhibiting similar viability counts at all time points. On Glu-his plates, however, the growth behavior of these cells was completely different. Similar to the plate experiments described before, no visible colonies appeared before day 6 post-plating and additional colonies per plate accumulated with incubation time until a maximum of visible colonies was reached at about 20 days post-plating. Thus, adapted cells, those that would form mature colonies within 2–3 days, were not detected in any of the phase I time points. Mature colonies that were detected as of day 6 post-plating, each initiated from a single cell, turned out to be fully adapted since cells from these colonies could form uniform new colonies within 2–3 days upon replating on Glu-his plates. Adapted colonies were also capable of propagating stably in liquid Glu-his medium. Thus, the analysis of cells from phase I of the chemostat recapitulates the plate experiments in terms of the adaptation dynamics of individual cells.
Notably, however, this experiment showed that despite the initial exponential growth of the chemostat population, there was no observable fraction of adapted cells when the population first encountered the challenging environment or during the entire period of phase I. None of the ∼105 cells sampled at each time point from phase I of the chemostat experiment could normally grow on Glu-his plates as expected from an adapted cell. Instead, numerous individual cells in the chemostat culture clearly carried the potential to become adapted as they eventually, after a long and variable incubation time, grew colonies on Glu-his plates. These results indicated that the adaptive phenotype, which could be faithfully inherited to future generations, was developed independently in numerous cells following the exposure to the challenging environment. Clearly, it was not a property that cells carried with them from the Gal-his growth period, as could have been suspected from the short adaptation time in the chemostat. Thus, adaptation in our experiments was a property of individual cells rather than a property of the population and the process that led each cell to the adapted state was induced by the challenging environment.
In fact, further findings corroborated this striking result. In a conventional adaptation process, the fraction of adapted individuals is expected to increase with time in the selective condition or else it would have been difficult for rare advantageous mutations to become established. Surprisingly, in this case the cells from consecutive time points in Glu-his exhibited the opposite trend. The further into the challenging environment of phase I, the fewer the cells that were capable of forming adapted colonies on Glu-his plates (Fig. 3). The largest fraction of the population that could form adapted colonies (about 50%) was sampled at the onset of phase I while this adaptation capacity declined sharply with time. At the end of phase I, only ∼1% of the cells that were plated could form visible adapted colonies. Notably, the decline in the adaptive potential over time argues against the existence of an advantageous subpopulation during phase I and supports the notion that adaptation was achieved by cells only after the transition into the challenging environment.
Figure 3. Population characteristics upon exposure to glucose.
The maximal fraction of adapted colonies (dots) along phase I of the chemostat (switching to glucose at t=0). The colonies initiated from individual cells that were sampled from the chemostat and grown on Glu-his plates for 20 days. Note the logarithmic scale. The chemostat population density was plotted in the background (gray line).
To validate and further study the adaptation process of naive cells, we switched 15 parallel populations from Gal-his to Glu-his in batch cultures, sampled cells along their growth, and measured their adaptation rate on Glu-his plates. Upon switching to Glu-his, all populations grew exponentially for about seven generations, resembling phase I of the chemostat (Supplementary Material Fig. S1a ). Again here, in none of the replicate experiments did we notice adapted cells in plated samples. Like in the chemostat experiment, visible colonies appeared on Glu-his plates from day 6 and up to 20 days post-plating and the maximal fraction of adapted colonies on Glu-his plates reached above 50% for samples from early time points and declined exponentially with the exposure time to glucose (Supplementary Material Fig. S1b). Thus, plate, chemostat and batch culture experiments were all in agreement about the high adaptation potential of naive cells and about the variable time it took individuals to achieve the adapted state. These two points stress on one hand, the wide accessibility of this adaptation process to the cells but emphasize on the other hand, that achieving the adapted state is a process that each cell undergoes individually. Both the chemostat and the batch culture experiments in which the dynamics of the adaptation process could be measured, also confirmed that the potential of cells to develop adaptation declines with time along the first seven to eight generations in the challenging environment, indicating the critical dependency between the cellular status and the time in glucose.
Cell division capacity determines the chances of adaptation
Cells developed their adaptive state after exposure to Glu-his both in liquid and solid media experiments. Measuring the dynamics of the population in liquid medium experiments indicated that while the population grew exponentially, its individuals lost their capability to become adapted. Thus, to understand the nature of the adaptation process, we had to explain the counterintuitive observation that less and less cells adapted with time in Glu-his. The answer, we hypothesized, could be found by following the response of individual cells after switched into glucose medium. The same plates that were used to estimate the adaptation capacity were also analyzed by microscopy imaging to follow the growth of individual colonies. Samples from different time points of the liquid culture were extracted and placed on Glu-his plates. Each single cell on the plate served as a seed for a growing colony. These colonies were imaged repeatedly during the long incubation period of 20 days. Two types of colonies were observed after this long incubation time. The first were adapted colonies in a size that was visible to the naked eye and, thus containing about 106 cells. As previously described, some of these adapted colonies appeared about 6 days post-plating and others joined in continuously until the maximal fraction of adapted colonies was reached at about 20 days (Supplementary Material Fig. S2). The variability in formation time of adapted colonies on plates indicated that adaptation was independently gained by each cell lineage. The second type of colonies was microcolonies that could be observed only under the microscope and remained as such for the entire incubation period of around 20 days (Supplementary Material Fig. S3). Microcolonies had variable sizes, from single to few hundred cells, suggesting that the cells from which they originated had a variable, yet limited, division capacity compared to adapted cells. Such a discrepancy between colonies that proliferated to a visible size and ones that remained aborted at small sizes could be clearly observed at later stages of the incubation period (Supplementary Material Fig. S3) but essentially could be detected already during the first days after plating even before the adapted colonies were large enough to be counted by eye [Fig. 4a].
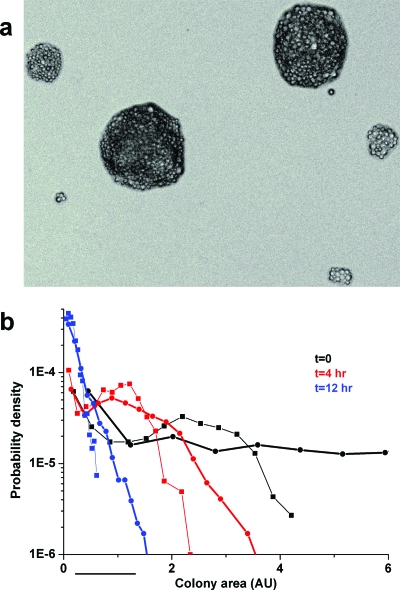
Figure 4. Distribution of colony sizes.
(a) A representative microscopy image of colonies grown on a Glu-his plate after a few days. These colonies were formed by seed cells sampled from the liquid culture at the onset of transition to Glu-his. Note the coexistence of a wide range of colony sizes down even to a single cell. Scale of image ∼320 μm. (b) The distributions of colonies area (in arbitrary units) based on numerous microscopy images, such as in panel (a), including a few hundred colonies each. Curves at different colors are for different times after the transition of the batch culture into Glu-his (black, t=0; red, t=4 h; and blue, t=12 h). Each two curves that have the same color but different symbols are from the same plates but were measured earlier (46–57 h; squares) and later (∼120 hrs; dots) after plating. The curves are normalized to unit area and presented as probability density for comparison (x-axis divided by104). The bar denotes the approximate distance on the x-axis of a colony with ∼70 cells.
To quantify this variability, the sizes of colonies grown from seed cells extracted at different time points of the batch cultures were measured by microscopy a few days post-plating. Similar to the exponential decay in the fraction of adapted colonies, also, the average colony area decreased with the exposure time to glucose (Supplementary Material Fig. S1b). Thus, the decline in the adaptation capability of cells with the exposure time to glucose has its signature also on the average and distribution of colony sizes already a few days post-plating. Not only that, on average, cells sampled immediately after exposure to Glu-his formed larger microcolonies but also their size distribution was broader compared to cells sampled later from the batch culture [Fig. 4b]. In addition, with time on Glu-his plates, for each time point in batch culture, the size distribution tail of microcolonies shifted toward larger colonies that will eventually become adapted. However, a wide range of smaller microcolonies that will not adapt remained even after 20 days. Similar microscopy results were obtained for cells harvested from phase I of the chemostat (Supplementary Material Fig. S4). Thus, the population structure is such that cells, on average formed smaller microcolonies and succeeded less to adapt, the longer they experienced the challenging environment.
The presence of matured colonies alongside with aborted microcolonies is not likely to be due to differences in growth rates. Tiny microcolonies were found even after 20 days, a time by which the number of adapted colonies already reached its maximal value (Supplementary Material Fig. S2). Moreover, similar growth rates in batch cultures were measured for adapted and naive cells at least for a few generations following their initial exposure to Glu-his (Fig. 5). Thus, the limited number of cell divisions that could be carried out by each lineage was the reason for the wide size variability of microcolonies. Advancing deeper into phase I caused more cells to arrest division after fewer generations, explaining the collapse of the chemostat population in phase II. Therefore, what determined the growth dynamics of individual cells and, thus, that of the population, was the exposure time to the new and challenging Glu-his environment. However, cells and their lineages could actually escape the fate of division arrest by switching into the adapted state and forming mature colonies that could be further propagated. Therefore, the essence of this adaptation was switching into a cell state that allows regular stable metabolism including histidine production and, thus, unlimited propagation. Here again, variable cell division capacity, as a parameter that determines the chances for adaptation, corroborates the notion that this adaptation was developed in individual lineages as a response to the challenging environment and was not a rare property carried by a few cells through the Gal-his to Glu-his medium switch.
Figure 5. Growth curves of parallel populations in batch cultures.
Adapted (red) versus naive (nonadapted, black) cells that were switched from Gal-his to Glu-his and as a control adapted cells (blue) that were propagated just in Glu-his. All strains exhibited similar growth rates in the Glu-his environment. The different lines are measurements of different clones averaged over three replicates, showing less than 7% variability in their growth rates.
A model of adaptation containing two competing cellular processes
Based on the experimental results presented above we formulated a model of the population dynamics. In a previous population average model (Stolovicki et al., 2006), we showed that a two-population structure, allowing switching between adapted and nonadapted states with a fitness advantage of the adapted state, is inconsistent with the observed chemostat dynamics if a large fraction of adapted cells already exists at the onset of transition to glucose. Here, we take into account the fact that adapted cells do not in fact have a fitness advantage and model the two competing cellular processes underlying the population dynamics: a phenotypic switch from the limited growth capacity on Glu-his either to an arrested state or to an adapted state with an unlimited division capacity. The rewired cells could initially proliferate in glucose medium at a rate similar to adapted cells but for only a limited number of generations after which their division was arrested unless they switched into a stable adapted state allowing them to maintain proliferation. Switching of a cell, from a naive to an adapted or an arrested state, occurred over a wide time range and was assumed to be a random event. This event can either be described as a probability per unit time in simulations following single cells (Monte-Carlo simulations) or as a rate constant for numerical integration of the average population dynamic equations in a chemostat setting. Importantly, long exposure time of naive cells to Glu-his medium reduced their chance to adapt and, thus, the interesting ingredient of this model was the utilization of transition rates (or probabilities) that depend strongly on the integrated exposure time to Glu-his and, thus on the history of the cell. The probability of a cell to maintain its divisions was assumed to be an exponentially decreasing function of its past exposure time to Glu-his. Similarly, its probability to adapt was assumed to be an exponentially increasing function of that time (inset of Supplementary Material Fig. S5). These processes competed as their rates of occurrence depended on the history of exposure to glucose. The model was computed in two forms; in a chemostat-like setting in which the two subpopulations grew in a limiting resource environment (requiring in addition to the equations describing the dynamics of the two subpopulations (naive or adapted), a third equation for describing the dynamics of the limiting resource), and for colonies on a plate with no limiting resource. Solving the plate model numerically showed that a seed-cell forming a larger microcolony before all its daughter cells ceased dividing, would have a significantly larger overall probability (probability per cell multiplied by the number of cells per colony) to yield an adapted colony. The model reproduced the experimentally observed colony size distributions on plates (Fig. 6), and the fast decay of the maximal fraction of adapted colonies with the exposure time to glucose in chemostat and batch culture experiments (Fig. 6 inset). Similarly, the chemostat-like model reproduced the main features of the population growth dynamics (Supplementary Material Fig. S5).
Figure 6. Simulated distributions of colony size.
Monte-Carlo simulations of colonies that were initiated from single cells and proliferated under two opposing processes: a decaying probability to continually divide and an increasing probability to acquire adaptation and unlimited divisions, both as an exponential function of exposure time to Glu-his (see Materials and methods section for details). The different curves are for different values of the parameter tg, controlling these processes and representing the exposure time to Glu-his of the seed cells (see Materials and methods section). The colony sizes are measured in a number of cells in a colony. The curves are normalized to unit area and presented as probability density for comparison. Inset: the maximal fraction of adapted simulated colonies (i.e., colonies containing at least one adapted cell and, thus, can keep cell division indefinitely) in a log scale, as a function of the exposure time to Glu-his of their seed cells.
DISCUSSION
By studying cells with a rewired genome we demonstrated an alternative adaptation process with the following characteristics. (1) The fraction of naive cells that adapted was extremely high and variable with an average of about 50%. (2) This adaptation was genuine since the phenotype of adapted cells was distinct from that of naive cells and since this phenotype was stably inherited. (3) The adaptation of the population occurred rapidly within 10–30 generations, probably due to the large fraction of cells that adapted. (4) The adaptation of the population relied on individual cells that independently developed the adaptive phenotype as a response to the challenging environment. Undoubtedly, these characteristics signify an adaptation phenomenon that does not conform to the common view of the evolutionary process and, thus, extends this framework and our understanding of how new phenotypes evolve.
Adaptation is commonly viewed as a change in phenotype and gene frequencies in response to a change in environmental conditions. Here the adaptation was carried out in the familiar glucose-containing environment and the challenge was to satisfy the internal cellular requirements, imposed by the frustrating environmental conditions due to the exclusive linkage of HIS3 to the repressed GAL system. The dynamics of the population in phases I and II of the chemostat were somewhat unexpected given the observed behavior of its individuals. During phase I the population grew exponentially, when its individuals progressively lost their adaptation capability while during phase II the population collapsed while many of its individuals gained their adaptation. In fact, there are several indications that upon exposure to the Glu-his medium, naive cells had a very similar metabolism to that of adapted cells despite the internal challenge imposed by the genome rewiring. Naive cells, which were not exposed to glucose before, grew at a well defined single rate after the first encounter with the Glu-his liquid medium as evident from optical density and plate counts measurements (Fig. 2 and Supplementary Material Fig. S1a). Moreover, similar growth rates were measured for naive and adapted cells in batch cultures, for the former when moved from Gal-his to Glu-his and for the latter when propagated in fresh Glu-his medium (Fig. 5). In agreement, the chemostat measurements showed similar growth exponents in phases I and III, which contained naive and fully adapted cells, respectively, (Fig. 2). Furthermore, the maximal cell density reached by nonadapted cells in phase I of the chemostat was similar to that of fully adapted cells at the glucose steady-state (phase IV). The density of cells in the chemostat is a sensitive function of the average cellular metabolic state, which determines the capacity of the population to maintain a stable growth rate. Both similar densities and similar growth rates, thus, indicate that cells in phases I and IV have similar metabolic states. Furthermore, the similarity between the initial growth rate of naive cells and the propagation rate of adapted cells lends further support to the lack of an advantageous subpopulation during phase I. Therefore, given the familiar glucose-containing environment and the similarity in metabolism, it is noteworthy that the adaptation phenomenon observed in our experiments was not so much about acquisition of a new metabolism but rather about reorganization and stabilization of a familiar growth capacity under the constraints of the genome rewiring event and the regulatory challenge it presented in Glu-his. This is precisely the challenge posed by genome rewiring events in evolution; stabilization of familiar protein functions in new contexts. Thus, here we used the term adaptation, which is usually utilized to describe an increase in fitness of the population average by means of selection, to rather describe adjustments in regulation that enable single cells to stabilize a familiar metabolic state. These adjustments, as we have shown, can be rapidly gained by individual cells and stably propagated for many generations and, thus, should be considered an adaptation that might have a significant role in evolution of regulatory systems.
No doubt that the fraction of cells, which adapted on the plates, either in solid or in liquid medium experiments, was unprecedentedly high and, thus, stands in contrast to previous evolutionary experiments that found much lower rates (one in 105–109 cells) at which adaptive phenotypes rose under stressful conditions (Paquin and Adams, 1983; Lenski and Travisano, 1994; Storchová et al., 1998; Elena and Lenski, 2003; Heidenreich, 2007). In previous studies adaptive phenotypes were observed in rates that matched the rates of genetic mutations and, indeed, in several such studies, mutations were found to be the causative agents of the new phenotypes. Here, the rate at which adapted phenotypes rose does not match the known rates of mutations. However, if mutations, indeed, provided the mechanism for adaptation of every other cell in our experiments then they should have arisen by one of the following processes. Either random mutations occurred at the conventional low rate and rare advantageous ones propagated in the population prior to the first exposure to glucose, explaining the high fraction of adapted cells on plates and the rapid adaptation of the chemostat population. Or, cells acquired adaptive mutations (mutations directed at advantageous positions) at a very high rate after the exposure to glucose. Based on the plate growth patterns of all experiment types, the galactose populations do not seem to contain a detectable fraction of cells with advantageous phenotypes. Moreover, most cells lost their capacity to adapt with exposure time to glucose, yet, the overall adaptation process was rapid. Therefore, the first option in which upon the exposure to glucose, the population already contained a substantial fraction of advantageous mutated genotypes that facilitated the rapid adaptation is inconsistent with our results. On the other hand, adaptive mutations might arise as a response to stressful environments and allow such a widespread adaptation of individuals and the rapid adaptation of the whole population. However, up until now, in spite of major efforts to identify such and although some cases are still under debate, there is no conclusive evidence in the literature for the existence of adaptive mutations (Cairns et al., 1988; Cairns and Foster, 1991; Steele and Jinks-Robertson, 1992; Hall, 1998a, 1998b; Rosenberg, 2001; Roth et al., 2006). Even if adaptive mutations were in consensus, in previous studies revertants in response to stressful conditions and induced adaptive mutations were reported to occur only in a minute fraction (∼10−6) of the population (Storchová et al., 1998; Heidenreich, 2007) and not in 50% of the cells such as in our experiments. Other phenomena with some resemblance to what we described here were also studied. Phenotypic switching cases in which the phenotype was unstable and changed according to the environment were studies (Hallet, 2001; Balaban et al., 2004; Levin and Rozen, 2006). In other classes of phenotypic switching, it was suggested that chaperons act as capacitors of phenotypic variation that expose hidden genetic variation in response to environmental changes (Rutherford and Lindquist, 1998; Queitsch et al., 2002). While the adaptation occurring in our experiments shares some features with the above phenomena, it fundamentally differs from them in others, mainly in the rate in which the adaptive phenotype rose and in the conclusion that the inherited phenotype was induced by the environment and not selected by it. We do not know yet what the actual mechanism of adaptation is and whether genetic mutations take part in this evolutionary process. However, since a stable advantageous change in phenotype was observed, the two more probable explanations for the high rate of adaptation are adaptive mutations or mechanisms other than mutations, which can be referred to as epigenetic processes (Lachmann and Jablonka, 1996; Jablonka and Lamb, 2005; Allis et al., 2007; Jablonka and Raz, 2009).
The results presented here place the environment and the history of the population as significant factors in determining the potential of cells to adapt as evident in the dynamics of the adaptation process. Pending on their exposure time to glucose, seed cells, or their descendents by individually switching their phenotype had a chance to stabilize an adaptive metabolic state and avoid arresting. The phenotypic features of the population dynamics were faithfully captured by an apparently simple model involving two competing processes, causing a switch from the naive initial state either to an arrested state or to an adapted one that enabled an unlimited cell division capacity. The phenotypic switching event depends upon the time spent in glucose, namely, on the history of the cell. The process that leads to cell arrest in our experiments is presumably different from the one observed during starvation since it is imposed only on some of the cells and because it facilitates the generation of an adapted state in others. This observation might be supported by our previous work showing that along phase I there has been a genomewide arbitrary transcriptional response involving a large fraction of the genome and having a very small overlap with known stress responses (Stern et al., 2007). Such a dramatic transcriptional response might significantly affect the physiology in ways that on the one hand will lead to cell arrest but on the other will open up new opportunities for adaptation.
Thus, our experiments prove the existence of a cellular mechanism enabling an inherited cellular adaptation that was induced by an unforeseen challenge in many cells simultaneously. The regulatory nature of the challenge, the abundance of adapted cells, and the rapid dynamics make cellular plasticity supported by epigenetic processes a reasonable candidate to mediate the induced adaptation process and its stable inheritance (Lachmann and Jablonka, 1996; Jablonka and Lamb, 2005; Ahmed and Brickner, 2007; Allis et al., 2007; Brickner et al., 2007; Jablonka and Raz, 2009). Since unforeseen challenges are bound to be encountered by every organism, such accessible and rapid adaptation based on cell plasticity could enable cells to overcome immediate severe challenges without relying on preexisting solutions until further and maybe rare advantageous genotypes could be realized and fixed in the population (West-Eberhard, 2003). The implications of such a mechanism are far reaching in diverse areas of biology; from behavior of single cells and tissues in health and disease to the dynamics of populations in nature. In particular, such adaptation capability supports the significant potential of genomic rewiring in evolution of biological networks and life itself.
MATERIALS AND METHODS
Strain and growth conditions
Experiments were carried out with the haploid yeast strain YPH499 [Mata, ura3–52, lys2–801, ade2–101,trp1-Δ63, his3Δ200, leu2Δ1] carrying the plasmid pESC-LEU (Stratagene) containing the pGAL1-pGAL10 divergent promoter with HIS3 under pGAL1 (Stolovicki et al., 2006). The deletion his3Δ200 removed the entire HIS3 coding region plus the upstream promoter region. Cells were grown in a custom made chemostat (Stolovicki et al., 2006) in synthetic dropout medium lacking histidine and leucine with the appropriate amino acid supplement and 2% of either high grade pure galactose or pure glucose as a sole carbon source as in (Stolovicki et al., 2006). Histidine and leucine-lacking galactose or glucose medium are referred throughout the text as Gal-his or Glu-his, respectively. Growth in the chemostat was limited by the concentration of the amino acid supplement. The chemostat contains ∼130 ml (109–1010 cells at steady-state). The generation time equals chemostat dilution time x ln 2, which was about 5 h. An online measurement system was used to measure the OD (at 600 nm) of cells in the chemostat (Stolovicki et al., 2006).
For the experiments in batch cultures, cells were first grown in a Gal-his medium to midexponential phase then washed and switched to Glu-his medium. Parallel populations were incubated and shaken in a 96 well plate (LabNet plate shaker at 30°C). Samples of cells from each population were plated on agar rich medium and colony counts were used to measure growth rates. In addition, the growth rates of adapted and nonadapted cells were measured in a Tecan Infinite-200 plate reader with shaking and incubation at 30°C.
Adaptation assays on test plates
Approximately 2 ml of cells were harvested from the chemostat at precise time points, washed and diluted to OD=1. A hundred microliters of each cell suspension was diluted 1,000-fold and spread on agar Glu-his plate to monitor the fraction of adapted cells and in parallel on rich medium (YPD) plates as viability controls. The medium composition of the plates was similar to that of the chemostat except for the addition of 2% agar. The plates were incubated at 30°C for 20 days and colonies were counted every day. The colony counts were normalized by the counts from the rich medium plates. The same plating procedure was used for the batch culture experiments. To study the growth dynamics of individual cells, agar plates were imaged under bright field microscopy (Zeiss Axiovert 135). Colony area distributions were measured using IMAGEPRO software.
Cell growth model
All computations were performed using MATLAB. The model computes the dynamics of two subpopulations; a naive one supporting a limited number of cell divisions ending eventually in cell arrest, and the other in the adapted state supporting indefinite divisions. The growth rates were assumed the same for these two subpopulations. Arrested cells were not allowed to switch back to the growing phase and the adapted state was inherited to daughter cells. A similar model was computed in two different settings: (a) for cells growing in a chemostat and for which, in addition to the equations describing the dynamics of the two subpopulations a third equation, describing the dynamics of a limiting resource was added and (b) for colony growth on plates. For the latter, the equations have been solved by Monte-Carlo simulations. In each time step a decision whether to divide (if above a threshold cell mass) was made by comparing to a constant probability. For the two resulting cells after division, the cell state was switched into arrested or adapted according to probabilities computed as a function of integrated time in glucose for the cell lineage (see the probability functions below). Otherwise, the cell continues to grow and divide again.
The population density in a chemostat as a function of time was computed by integrating the following chemostat equations:
where x1,2 are the fractions of nonadapted and adapted subpopulations, respectively, μ is their maximal growth rate, and a is the Michaelis–Menten (half saturation) constant (both growth parameters assumed to be the same for the two subpopulations). The naive cells of population 1 can switch to the arrested state at rate fs or to the adapted state at ratefa, which transfers cells from population 1 to population 2.b, c, d, and e are constants of these transition rates and τs and τa are the response time constants (time units are normalized). S is the concentration of limiting nutrient in the chemostat. The units are normalized so that S, a, and x1,2 are measured in units of the feeding concentration of the limiting nutrient and the yield is normalized to one. Time is measured in units of the chemostat dilution time [see Smith and Waltman (1995) for details]. Supplementary Material Fig. S5 was produced with the following parameters: μ=1.8, a=0.2, b=3, c=3, d=1, e=125, τs=18, and τa=25. The equations have been integrated from the initial conditions: x1(t=0)=0.1, x2(t=0)=0, and S=1.
For cells growing in colonies (Fig. 6), the rate functions are interpreted as probabilities per unit time to switch phenotype with the following probability density functions: Pac=0.02 exp(tcell∕τs) and Pad=0.06∕{3+exp(−tcell∕τa)}, where Pac is the probability density to remain active (keep dividing), Pad is the probability density to switch into the adaptive state, and tcell=t+tg is the running time including the past exposure to glucose tg in the liquid medium. τs=τa=1 are the time constants related to the switch to arrested and adapted states, respectively. The times are in units of the integration time, dt=0.1. The models have been computed for a wide range of parameters without a qualitative change in results.
ACKNOWLEDGMENTS
The authors wish to acknowledge N. Brenner, K. Keren, S. Marom, Y. Soen, J. Dean, L. Steinmetz, G. Yvert, and G. Simchen for the fruitful discussions and critical review of the manuscript. We thank the reviewers for their useful comments that helped improving the manuscript. This research was supported by grants from the Israeli Science Foundation (L.D. and E.B.), the Yeshaya Horowitz Association through the Center of Complexity Science (E.S and E.B), the German-Israeli Foundation, and the U.S.-Israeli Binational Foundation for E.B.
References
- Ahmed, S, and Brickner, J H (2007). “Regulation and epigenetic control of transcription at the nuclear periphery.” Trends Genet. 23, 396–402. 10.1016/j.tig.2007.05.009 [DOI] [PubMed] [Google Scholar]
- Allis, C D, Jenuwein, T, and Reinberg, D (eds.) (2007). Epigenetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Alonso, C R, and Wilkins, A S (2005). “The molecular elements that underlie developmental evolution.” Nat. Rev. Genet. 6, 709–715. 10.1038/nrg1676 [DOI] [PubMed] [Google Scholar]
- Balaban, N Q, Merrin, J, Chait, R, Kowalik, L, and Leibler, S (2004). “Bacterial persistence as a phenotypic switch.” Science 305, 1622–1625. 10.1126/science.1099390 [DOI] [PubMed] [Google Scholar]
- Brickner, D G, Cajigas, I, Fondufe-Mittendorf, Y, Ahmed, S, Lee, P-C, Widom, J, and Brickner, J H (2007). “H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state.” PLoS Biol. 5, e81. 10.1371/journal.pbio.0050081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, J, and Foster, P L (1991). “Adaptive reversion of a frameshift mutation in escherichia coli.” Genetics 128, 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns, J, Overbaugh, J, and Miller, S (1988). “The origin of mutants.” Nature (London) 335, 142–145. 10.1038/335142a0 [DOI] [PubMed] [Google Scholar]
- Carroll, S B (2005). “Evolution at two levels: on genes and form.” PLoS Biol. 3, e245. 10.1371/journal.pbio.0030245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, S B, Grenier, J K, and Weatherbee, S D (2001). From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design, Blackwell Science, Malden, MA. [Google Scholar]
- Davidson, E H (2006). The Regulatory Genome, Elsevier, Amsterdam. [Google Scholar]
- Drake, J W, Charlesworth, B, Charlesworth, D, and Crow, J F (1998). “Rates of spontaneous mutation.” Genetics 148, 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena, S F, and Lenski, R E (2003). “Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation.” Nat. Rev. Genet. 4, 457–469. 10.1038/nrg1088 [DOI] [PubMed] [Google Scholar]
- Erwin, D H, and Davidson, E H (2009). “The evolution of hierarchical gene regulatory networks.” Nat. Rev. Genet. 10, 141–148. 10.1038/nrg2499 [DOI] [PubMed] [Google Scholar]
- Fong, S S, Joyce, A R, and Palsson, B O (2005). “Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states.” Genome Res. 15, 1365–1372. 10.1101/gr.3832305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart, J, and Kirschner, M (1997). Cells, Embryos, and Evolution, Blackwell Science, Malden, MA. [Google Scholar]
- Hall, B G (1998a). “Activation of the bgl operon by adaptive mutation.” Mol. Biol. Evol. 15, 1–5. [DOI] [PubMed] [Google Scholar]
- Hall, B G (1998b). “Adaptive mutagenesis: a process that generates almost exclusively beneficial mutations.” Genetica 102–103, 109–125. 10.1023/A:1017015815643 [DOI] [PubMed] [Google Scholar]
- Hallet, B (2001). “Playing Dr. Jekyll and Mr. Hyde: combined mechanisms of phase variation in bacteria.” Curr. Opin. Microbiol. 4, 570–581. 10.1016/S1369-5274(00)00253-8 [DOI] [PubMed] [Google Scholar]
- Heidenreich, E (2007). “Adaptive mutation in saccharomyces cerevisiae.” Crit. Rev. Biochem. Mol. Biol. 42, 285–311. 10.1080/10409230701507773 [DOI] [PubMed] [Google Scholar]
- Jablonka, E, and Lamb, M J (eds.) (2005). Evolution in Four Dimensions, MIT Press, Cambridge, MA. [Google Scholar]
- Jablonka, E, and Raz, G (2009). “Transgenerational epigenetic inheritance: prevalence, mechanisms and implications for the study of heredity and evolution.” Q. Rev. Biol. 84, 131–176. 10.1086/598822 [DOI] [PubMed] [Google Scholar]
- King, M C, and Wilson, A C (1975). “Evolution at two levels in humans and chimpanzees.” Science 188, 107–116. 10.1126/science.1090005 [DOI] [PubMed] [Google Scholar]
- Lachmann, M, and Jablonka, E (1996). “The inheritance of phenotypes: an adaptation to fluctuating environment.” J. Theor. Biol. 181, 1–9. 10.1006/jtbi.1996.0109 [DOI] [PubMed] [Google Scholar]
- Lenski, R E, and Travisano, M (1994). “Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations.” Proc. Natl. Acad. Sci. U.S.A. 91, 6808–6814. 10.1073/pnas.91.15.6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, B R, and Rozen, D E (2006). “Non-inherited antibiotic resistance.” Nat. Rev. Microbiol. 4, 556–562. 10.1038/nrmicro1445 [DOI] [PubMed] [Google Scholar]
- Luria, S E, and Delbruck, M (1943). “Mutations of bacteria from virus sensitivity to virus resistance.” Genetics 8, 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maharjan, R, Seeto, S, Notley-McRobb, L, and Ferenci, T (2006). “Clonal adaptive radiation in a constant environment.” Science 313, 514–517. 10.1126/science.1129865 [DOI] [PubMed] [Google Scholar]
- Marini, N, Matmati, N, and Morpurgo, G (1999). “Starvation in yeast increase non-adaptive mutation.” Curr. Genet. 35, 77–81. 10.1007/s002940050435 [DOI] [PubMed] [Google Scholar]
- Novick, A, and Szilard, L (1950). “Experiments with the chemostat on spontaneous mutations of bacteria.” Proc. Natl. Acad. Sci. U.S.A. 36, 708–719. 10.1073/pnas.36.12.708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin, C, and Adams, J (1983). “Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations.” Nature (London) 302, 495–500. 10.1038/302495a0 [DOI] [PubMed] [Google Scholar]
- Perfeito, L, Fernandes, L, Mota, C, and Gordo, I (2007). “Adaptive mutations in bacteria: high rate and small effects.” Science 317, 813–815. 10.1126/science.1142284 [DOI] [PubMed] [Google Scholar]
- Queitsch, C, Sangster, T A, and Lindquist, S (2002). “Hsp90 as a capacitor of phenotypic variation.” Nature (London) 417, 618–624. 10.1038/nature749 [DOI] [PubMed] [Google Scholar]
- Rosenberg, S M (2001). “Evolving responsively: adaptive mutation.” Nat. Rev. Genet. 2, 504–515. 10.1038/35080556 [DOI] [PubMed] [Google Scholar]
- Roth, J R, Kugelberg, E, Reams, A B, Kofoid, E, and Andersson, D I (2006). “Origin of mutations under selection: the adaptive mutation controversy.” Annu. Rev. Microbiol. 60, 477–501. 10.1146/annurev.micro.60.080805.142045 [DOI] [PubMed] [Google Scholar]
- Rutherford, S L, and Lindquist, S (1998). “Hsp90 as a capacitor for morphological evolution.” Nature (London) 396, 336–342. 10.1038/24550 [DOI] [PubMed] [Google Scholar]
- See supplementary material at http://dx.doi.org/10.2976/1.3353782 for Figs. S1–S5.
- Smith, H L, and Waltman, P (1995). The Theory of the Chemostat Dynamics of Microbial Competition, Cambridge University Press, NY. [Google Scholar]
- Steele, D F, and Jinks-Robertson, S (1992). “An examination of adaptive reversion in saccharomyces cerevisiae.” Genetics 132, 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D L, and Orgogozo, V (2008). “The loci of evolution: how predictable is genetic evolution?.” Evolution, (Lawrence, Kans.) 62, 2155–2177. 10.1111/j.1558-5646.2008.00450.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, S, Dror, T, Stolovicki, E, Brenner, N, and Braun, E (2007). “Transcriptional plasticity underlies cellular adaptation to novel challenge.” Mol. Syst. Biol. 3, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolovicki, E, Dror, T, Brenner, N, and Braun, E (2006). “Synthetic gene recruitment reveals adaptive reprogramming of gene regulation in yeast.” Genetics 173, 75–85. 10.1534/genetics.106.055442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová, Z, Gil, A PR, Janderova, B, and Vondrejs, V (1998). “The involvement of the RAD6 gene in starvation-induced reverse mutation in saccharomyces cerevisiae.” Mol. Gen. Genet. 258, 546–552. 10.1007/s004380050766 [DOI] [PubMed] [Google Scholar]
- Tuch, B B, Galgoczy, D J, Hernday, A D, Li, H, and Johnson, A D (2008a). “The evolution of combinatorial gene regulation in fungi.” PLoS Biol. 6, e38. 10.1371/journal.pbio.0060038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch, B B, Li, H L, and Johnson, A D (2008b). “Evolution of eukaryotic transcription circuits.” Science 319, 1797–1799. 10.1126/science.1152398 [DOI] [PubMed] [Google Scholar]
- West-Eberhard, M J (2003). Developmental Plasticity and Evolution, Oxford University Press, Oxford, U.K. [Google Scholar]
- Wilkins, A S, (2002) The Evolution of Developmental Pathways, Sinauer Associates, Sunderland, MA. [Google Scholar]
- Wray, G A (2007). “The evolutionary significance of cis-regulatory mutations.” Nat. Rev. Genet. 8, 206–216. 10.1038/nrg2063 [DOI] [PubMed] [Google Scholar]