Abstract
A recent study used a forward genetic approach to identify a new gene whose protein product controls erythrocyte iron recycling mediated through macrophages in the spleen. Initially the investigators found a genetic region on chromosome 9 accounting for one third of the variation in spleen iron level in mice. Additional approaches to narrow the genomic region identified the gene Mon1a, which codes for a protein that acts as a novel regulator of spleen iron release. Cell-based studies showed that Mon1a is necessary for vesicular trafficking of proteins, including the iron export protein ferroportin, to the macrophage cell membrane. The forward genetics approach, which has currently only been used sparingly by the nutrition research community, offers a powerful and unbiased approach to identifying genes important in nutritional metabolism.
Keywords: Mon1a, quantitative trait loci, spleen iron, ferroportin
Introduction
Iron homeostasis is a complex process that has long been studied, but despite many recent advances there are many gaps in our understanding of how iron metabolism is regulated. Dysregulation of iron metabolism can result in disease states characterized by iron overload (genetic hemochromatosis) or depletion (iron deficiency anemia) and it has also been linked to the development of cancer and neurodegenerative disorders.1,2 Iron balance is controlled primarily at the level of intestinal absorption; there is no route for iron loss other than blood loss. Because of this, another critical point controlling iron homeostasis is within the reticuloendothelial system.1,3 Two-thirds of total body iron is incorporated into hemoglobin within erythrocytes.4 As erythrocytes die, macrophages within the spleen phagocytize the senescent cells as the organ filters the blood. This results in the release and recycling of iron from heme. Iron recycling by macrophage erythrophagocytosis is responsible for providing the 24 mg of iron needed each day for the production of new red blood cells.3 Due to the large amount of iron that passes through them, macrophages in the spleen can also be a major producer of the iron storage protein ferritin.1,2 Although this clearly shows that macrophage-mediated iron regulation is a critical component of iron homeostasis, there are significant gaps in our understanding of how iron recycling is accomplished in these cells. A recent report by Wang et al.5 applied a forward genetics approach whereby variation in spleen iron was used to identify genomic variation controlling for the trait to identify another critical protein in the macrophage iron story.
Identification of the Mon1a Gene by Quantitative Trait Locus (QTL) Analysis as a Gene Associated with Spleen Iron Content
Wang et al. initially examined 9 inbred strains of mice and found an 8-fold range in spleen iron concentration. This indicates there is a genetic difference in spleen iron accumulation that could be exploited to discover new mechanism controlling the process. To identify the genetic contributors, Wang et al. crossed mice with a low spleen iron phenotype (C57BL/10J or “B10”) and a high spleen iron phenotype (SWR/J or “SWR”) to produce a F1 population (with one gene allele coming from each parental line) that was then intercrossed (i.e. brother-sister pairings) to make an “F2” population or backcrossed to the SWR parental line to make a “N2” population. Quantitative trait loci (QTL) analysis was conducted on these populations; QTL analysis is a statistical approach used to correlate natural genetic variation in the genome (assessed using a panel of genetic markers) with a biological feature – in this case spleen iron accumulation. Using this approach, a highly significant locus controlling spleen iron content was found on chromosome 9 between 45 Mb and 115 Mb. This locus accounted for one third of the variation in spleen iron levels seen between B6 and SWR mice. The locus at chromosome 9 had been reported previously in another F2 population from a cross between C57BL/6J and SWR mice.6 Unfortunately, the region identified in that earlier study was too large and contained too many candidate genes to definitively determine the genetic variant on chromosome 9 controlling spleen iron accumulation. Wang et al. extended this earlier observation by narrowing the region on chromosome 9 to a locus containing just 16 genes by comparing congenic lines of mice (i.e. mouse lines with small sections of the chromosome 9 QTL region from the SWR genome transferred on to the B10 genetic background). Using this experimental approach with congenic mouse lines, they showed that the known genetic variation in the coding region of the Mon1a gene was associated with the ability of 7 inbred mouse lines to accumulate iron in the spleen. Specifically, they found the 374th amino acid residue, an asparagine (N374), is replaced by serine in the Mon1a gene in the B10 genome and that this replacement is conserved in other lines with low spleen iron levels. The N374 residue is also evolutionarily conserved in other species and so variation in this site is likely to have important functional consequences.
The Mon1a protein was not previously thought to have a role in the control of iron metabolism. However, based on their observation that Mon1a has a homolog in yeast that is responsible for vesicular trafficking, Wang et al. hypothesized that Mon1a participates in the trafficking of critical iron transport proteins to the membrane of macrophages.
Other research has identified the iron regulatory proteins that could be influenced by Mon1a during macrophage mediated iron recycling. After phagocytosis, iron is freed from heme by heme oxygenase 1 (HO1). After HO1 action, chelatable iron is freed into the phagosome and it is transported into the cell through the actions of Divalent Metal Transporter 1 (DMT1)7, which is present in the early phagosome, and Natural resistance-associated macrophage protein 1 (Nramp 1), a macrophage and neutrophil-specific protein that is active in the late phagosome.8 The iron transferred out of the phagosome into the macrophage cytosol by DMT1 and Nramp1 can be stored in the macrophage inside the protein ferritin, or it can be transported out of the cell through ferroportin, the only known iron exporter. Expression of ferroportin is strongly induced in macrophages by the presence of heme iron.4,9 However, when body iron levels are high, the peptide hormone hepcidin is released from the liver and acts on ferroportin, internalizing it and shutting down iron export, increasing macrophage iron stores, and causing iron accumulation in the spleen.10 Because of the important role that ferroportin plays in macrophage iron recycling, Wang et al., decided to focus on the impact that Mon1a has on this protein.
Mon1a Regulates Ferroportin Localization on the Macrophage Cell Membrane
Neither Mon1a nor ferroportin mRNA/protein level differed between primary macrophages isolated from B10 or SWR mice. Similarly, the liver levels of the ferroportin regulatory protein hepcidin where not different between the two mouse lines. This suggests that the genetic variation in Mon1a that controls spleen iron level is not modulating ferroportin gene transcription, mRNA stability, or protein stability, nor is it indirectly regulating the production of the ferroportin regulator protein hepcidin. In contrast, when ferroportin localization was examined by immunofluorescent staining under various iron loading conditions known to alter the distribution of ferroportin within macrophages, Wang et al. saw that macrophages isolated from B10 mice had significantly higher amounts of ferroportin localized on the cell membrane. This suggests that B10 macrophages have a greater ability to export iron and it is consistent with the reduced spleen iron accumulation seen in B10 mice. Similarly, although macrophages from both B10 and SWR mice responded to hepcidin treatment by internalizing ferroportin, more ferroportin remained on the cell membrane of B10 macrophages after hepcidin treatment. This demonstrates there is resistance to the normal signals that down-regulate iron export in B10 mice. To determine if Mon1a is directly involved in transporting ferroportin to the membrane, small-interfering RNA (siRNA) was used to knockdown Mon1a in primary macrophages. Consistent with the hypothesis, treatment of macrophages with Mon1a-siRNA, reduced ferroportin localization to the membrane, lowered iron export, and increased iron storage as ferritin. The authors noted that other studies have shown Mon1a has an essential role in the secretion of proteins involved in macrophage immune response. Wang et al. showed that IL-6 secretion from activated macrophages is reduced by reducing Mon1a levels with siRNA. This indicates that the role of Mon1a is not specific to iron metabolism but that iron metabolism uses a pre-existing vesicular trafficking network to regulate iron release. From this evidence Wang et al. concluded that the B10 allele of Mon1a confers a gain-of-function polymorphism that allows macrophages to more efficiently clear iron from the spleen, leading to lower spleen iron levels.
Conclusion
The study by Wang and colleagues has added a new protein, Mon1a, to our growing understanding of iron trafficking in the reticuloendothelial system (Figure 1). Despite the excellent detail and sheer scope of the Wang et al. study, it is still unclear how the sequence variation in the Mon1a gene is able to affect its function and alter ferroportin movement within cells. Future research will be necessary to determine the specific interactions between Mon1a and the vesicular transport system In addition, it is important to note that the variation in Mon1a accounts for just one third of the natural variation controlling spleen iron accumulation between B10 and SWR mice. Additional forward genetics studies are justified to discover the genomic regions accounting for the other 65% of the variation in spleen iron accumulation. The forward genetics approach has been utilized extensively by geneticists but sparingly by the nutrition research community. However, it has the great advantage of being unbiased; the phenotype (in this case spleen iron metabolism) drives the identification of genes that contain natural variation that influences iron metabolism. Similar approaches are likely to be fruitful for other key organs in iron metabolism (e.g. Wang et al. found a large range of iron accumulation in the livers of inbred mice) and for other mineral elements (e.g. similar to what has been done for mineral accumulation in plants11).
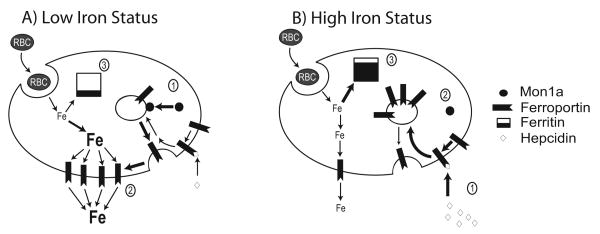
Figure 1. A model for the effects of iron status on the intracellular movement and cellular release of iron in the macrophage.
A) Low iron status increases macrophage iron export by modulating Mon1a association with vesicles. (1) Low iron increases association of Mon1a with membrane-bound vesicles containing ferroportin, (2) Increased Mon1a association leads to more ferroportin present on the membrane, (3) Iron from red blood cells (RBC) entering the macrophase is released into the cytosol and exported through ferroportin rather than stored within ferritin. B) High iron status decreases macrophage iron export by reducing Mon1a association with vesicles. (1) High iron induces hepatic production of hepcidin, a peptide hormone which is released into the serum where it binds to, and internalizes, ferroportin, 2) Reduced need for iron export decreases Mon1a association with membrane-bound vesicles containing ferroportin, thereby reducing ferroportin movement to the plasma membrane, 3) As iron export decreases iron storage in ferritin is increased.
References
- 1.Andrews NC. Forging a field: the golden age of iron biology. Blood. 2008;112:219–230. doi: 10.1182/blood-2007-12-077388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chua AC, Graham RM, Trinder D, Olynyk JK. The regulation of cellular iron metabolism. Crit Rev Clin Lab Sci. 2007;44:413–459. doi: 10.1080/10408360701428257. [DOI] [PubMed] [Google Scholar]
- 3.Knutson M, Wessling-Resnick M. Iron metabolism in the reticuloendothelial system. Crit Rev Biochem Mol Biol. 2003;38:61–88. doi: 10.1080/713609210. [DOI] [PubMed] [Google Scholar]
- 4.Knutson MD, Vafa MR, Haile DJ, Wessling-Resnick M. Iron loading and erythrophagocytosis increase ferroportin 1 (FPN1) expression in J774 macrophages. Blood. 2003;102:4191–4197. doi: 10.1182/blood-2003-04-1250. [DOI] [PubMed] [Google Scholar]
- 5.Wang F, Paradkar PN, Custodio AO, et al. Genetic variation in Mon1a affects protein trafficking and modifies macrophage iron loading in mice. Nat Genet. 2007;39:1025–1032. doi: 10.1038/ng2059. [DOI] [PubMed] [Google Scholar]
- 6.Grant GR, Robinson SW, Edwards RE, et al. Multiple polymorphic loci determine basal hepatic and splenic iron status in mice. Hepatology. 2006;44:174–185. doi: 10.1002/hep.21233. [DOI] [PubMed] [Google Scholar]
- 7.Tchernitchko D, Bourgeois M, Martin ME, Beaumont C. Expression of the two mRNA isoforms of the iron transporter Nramp2/DMTI in mice and function of the iron responsive element. Biochem J. 2002;363:449–455. doi: 10.1042/0264-6021:3630449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soe-Lin S, Sheftel AD, Wasyluk B, Ponka P. Nramp1 equips macrophages for efficient iron recycling. Exp Hematol. 2008;36:929–937. doi: 10.1016/j.exphem.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Delaby C, Pilard N, Puy H, Canonne-Hergaux F. Sequential regulation of ferroportin expression after erythrophagocytosis in murine macrophages: early mRNA induction by haem, followed by iron-dependent protein expression. Biochem J. 2008;411:123–131. doi: 10.1042/BJ20071474. [DOI] [PubMed] [Google Scholar]
- 10.Nemeth E, Tuttle MS, Powelson J, et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- 11.Salt DE, Baxter I, Lahner B. Ionomics and the study of the plant ionome. Annu Rev Plant Biol. 2008;59:709–733. doi: 10.1146/annurev.arplant.59.032607.092942. [DOI] [PubMed] [Google Scholar]