Abstract
To gain insights on cellular mechanisms regulating actin polymerization, we used the Virtual Cell to model FRAP and chromophore assisted laser inactivation (CALI) experiments on EGFP-capping protein (EGFP-CP). Modeling the FRAP kinetics demonstrated that the in vivo rate for the dissociation of CP from actin filaments is much faster (~0.1 s−1) than that measured in vitro (0.01–0.0004 s−1). The CALI simulation revealed that in order to induce sustainable changes in cell morphology after CP inactivation, the cells should exhibit anti-capping ability. We included the VASP protein as the anti-capping agent in the modeling scheme. The model predicts that VASP affinity for barbed ends has a cooperative dependence on the concentration of VASP-barbed end complexes. This dependence produces a positive feedback that stabilizes the complexes and allows sustained growth at clustered filament tips. We analyzed the range of laser intensities that are sufficient to induce changes in cell morphology. This analysis demonstrates that FRAP experiments with EGFP-CP can be performed safely without changes in cell morphology, because, the intensity of the photobleaching beam is not high enough to produce the critical concentration of free barbed ends that will induce filament growth before diffusional replacement of EGFP-CP occurs.
Keywords: ENA/VASP, cooperativity, fluorescent protein, filopodia, Virtual Cell
INTRODUCTION
Numerous motile processes in cell and developmental biology are driven by actin polymerization and depend on rapid actin filament assembly and disassembly at specific cellular locations. Thus, the precise spatial and temporal control over actin dynamics is vitally important for cell function. Actin polymerization is tightly regulated by various actin binding proteins, which nucleate, promote, stabilize, or sever actin filaments (Chesarone and Goode 2009; Chhabra and Higgs 2007; Cooper and Sept 2008; Le Clainche and Carlier 2008; Pollard 2007). The extent of filament elongation in vivo is limited by the presence of high-affinity barbed end capping proteins but this inhibition is antagonized by anti-cappers, among them the proteins from the Ena/VASP family (Bear et al. 2002; Paul and Pollard 2009). Capping protein β binds the barbed ends of actin filaments preventing continued polymerization and rapid growth from this end (Hug et al. 1995; Wear and Cooper 2004) and this maintains a short, highly-branched lamellipodial actin network as well as a pool of monomeric G-actin, an arrangement more suitable forproductive protrusion (Pollard and Borisy 2003). The presenceof Ena/VASP proteins at the leading edge of cells antagonizes CPs at barbed ends of actin filaments, enabling actin polymerization to continue, generating long, unbranched actin filaments (Barzik et al. 2005; Bear et al. 2002; Pasic et al. 2008; Trichet et al. 2008)
The mechanism and regulation of actin polymerization have been studied extensively in vitro for many years but now there is increasing interest to compare and contrast this body of information with that obtained in vivo. Recently, several microscopic techniques, such as speckle microscopy (Ponti et al. 2004); ICS, image correlation microscopy (Digman et al. 2005), FRAP, fluorescence recovery after photobleaching (Roy et al. 2002), and CALI, chromophore assisted laser inactivation (Jacobson et al. 2008) have begun to yield important qualitative and quantitative information on the processes that promote and regulate actin polymerization in living cells. Interpretation and analysis of the spatiotemporal data obtained from these techniques requires mathematical analysis and quantitative modeling.
FRAP is a technique to measure translational mobility in membranes and the cytoplasm by first photobleaching the fluorescence emitted from a labeled component from a small region of the cell and subsequently measuring the recovery of fluorescence into the previously bleached region; the kinetics of recovery are related to the transport process that dominates the recovery process. CALI, in particular, has attracted attention because of its potential to effect nearly instantaneous loss-of-function and thereby complement more conventional genetic manipulations. In this method, target proteins are inactivated by reactive photoproducts such as reactive oxygen species generated by intense irradiation of chromophores that are immediately adjacent to the protein. Because FRAP also uses a bright flash of light to bleach fluorophores, the question naturally arises as to how much CALI occurs during a typical FRAP experiment. Indeed, the motivation for this study was to understand why one could perform a FRAP measurement on EGFP-capping protein (EGFP-CP) in vivo without obtaining the protrusive phenotype observed in CALI (Vitriol et al. 2007). To achieve both our original goal and an expanded set of objectives that arose during this study, we employed the Virtual Cell platform and a mathematical description embodying the dendritic nucleation model for actin polymerization (Ditlev et al. 2009) to simulate FRAP and CALI experiments on EGFP-CP (Vitriol et al. 2007). By comparing simulation results directly to data obtained from FRAP experiments, we show that the rate constant for the dissociation of CPs from barbed ends must be much larger than the values reported for in vitro measurements. Our results are consistent with recent experimental results (Iwasa and Mullins 2007; Miyoshi et al. 2006).
Simulation of CALI experiments on knockdown-rescue EGFP-CP cells revealed that anti-capping activity is required to induce sustainable changes in cell morphology after CP inactivation. This prompted us to explicitly include VASP protein as an anti-capping agent in the model. Interestingly, the model predicts that VASP affinity for barbed ends must be cooperatively dependent on concentration of VASP-barbed ends complexes (VASP_BE). Thus, locally high concentration of uncapped barbed ends, such as those produced by CALI, is able to create large number of VASP-BE complexes in close proximity to each other with the consequence that the VASP dissociation rate from barbed ends is decreased, possibly due to VASP clustering. This positive feedback stabilizes VASP, antagonizes recapping and leads to rapid filament elongation. Finally, by analyzing the range of laser intensities that produce morphological changes in CALI of EGFP-CP, we address the issue of whether intensities typically used for FRAP experiments produce significant collateral CALI.
METHODS
The Virtual Cell modeling and simulation software was used for this work (http://vcell.org). The Virtual Cell provides a formal framework for modeling biochemical, electrophysiological, and transport phenomena while considering the subcellular localization of the molecules that take part in them. This localization can take the form of a three-dimensional (3D) arbitrarily shaped cell, where the molecular species might be heterogeneously distributed. Each 2D or 3D model explicitly considers the diffusion of the molecules within the geometry, as well as their reactions. The spatiotemporal changes to the concentrations of molecular species are governed by a mass conservation law: the rate of concentration change of a molecular species inside a volume element is caused by all the reactions that affect this species and diffusion fluxes coming in and out of the element. (Slepchenko et al. 2003). The mass and charge conservation laws, which form the physical basis in the Virtual Cell, are mathematically expressed by a system of differential equations
In this study, we employ a biochemical and mathematical description embodying the dendritic nucleation model for actin polymerization on Virtual Cell platform. The biochemistry and polymer physical chemistry underlying the Actin Dendritic Nucleation model has been thoroughly described (Ditlev et al. 2009) and it includes:
activation of Arp2/3 at the cell membrane by a nucleation promoting factor (e.g. NWASP);
nucleation and branching on preexisting F-actin by activated Arp2/3 and 2 actin monomers (including profilin-bound G-actin)(Beltzner and Pollard 2008; Carlsson et al. 2004);
dissociation of Arp2/3 branches in the cytoplasm (Mahaffy and Pollard 2006);
interconversions between the ATP, ADP and ADP·Pi forms of both monomeric G-actin and filamentous F-actin (Blanchoin and Pollard 2002; Fujiwara et al. 2007; Kinosian et al. 2000; Melki et al. 1996);
assembly and disassembly of these 3 nucleotide-bound monomer forms to each of the 3 forms of barbed and pointed ends (i.e. 18 unique pairs of forward and reverse reactions) (Kinosian et al. 2002; Pollard and Cooper 1986);
acceleration of nucleotide exchange on G-actin by profilin (Blanchoin and Pollard 1998; Godschmidt-Clermont et al. 1991; Kinosian et al. 2000);
addition of G-actin associated with profilin to barbed ends (Godschmidt-Clermont et al. 1991; Kang et al. 1999; Kinosian et al. 2002);
capping of the 3 forms of barbed ends (Kuhn and Pollard 2007; Schafer et al. 1996; Wear and Cooper 2004);
length-dependent annealing and fragmentation of actin filaments (Andrianantoandro et al. 2001; Sept et al. 1999);
buffering of G-actin by thymosin-β4 (Carlier et al. 1993; Kang et al. 1999; Nachmias 1993);
severing and accelerated disassembly of actin filaments by ADF/cofilin (Andrianantoandro and Pollard 2006; Blanchoin and Pollard 1998; Blanchoin and Pollard 1999; Blanchoin et al. 2000; Cai et al. 2007; Cao et al. 2006; Carlsson 2006; Ressad et al. 1999; Ressad et al. 1998).
The individual mechanisms and their parameters were based on published data, as detailed in the “Table of Sources” provided in the supplementary material (Ditlev et al. 2009). and within a BioModel entitled “Actin Dendritic Nucleation” that is accessible in the Virtual Cell database under username “les”.
The model was formulated as a continuum model consisting of systems of partial differential equations. This necessitated the formulation of approaches that could consolidate the infinite number of possible states of actin to a reduced set of variables. Thus, the concentrations of actin in the different nucleotide forms were expressed in terms of the concentration of actin subunits. The hydrolysis of ATP to ADPPi and the dissociation of the ADPPi forms to ADP are explicitly included in the reaction network for all the actin species. The model independently keeps track of the concentration of barbed and pointed ends in each of the nucleotide-bound forms; these depend on both the hydrolysis and dissociation reactions of the nucleotide as well as the addition or dissociation of the various nucleotide forms of monomers. The total concentration of subunits in actin filaments is given by the sum of all the state variables corresponding to different forms of F-actin, including all forms of free and capped ends, Arp2/3-bound and cofilin-bound subunits. The concentration of filaments can be calculated from the sum of all the pointed ends, i.e., in all three nucleotide forms and both free and Arp2/3-capped in branches. The average filament length, in number of subunits, is given by the total concentration of F-actin divided by the concentration of filaments.
Parameters
For all simulations in this paper we used the following total concentrations of molecular species: [profilin]=10 μM, [cofilin]=5 μM, [thymosin β4]=100μM, [Arp2/3]=1μM and a total actin concentration=200 μM. All proteins within the cytoplasm were assumed to have a diffusion coefficient of 5 μm2/s, while for actin filaments, the diffusion coefficient was assumed to be 0.01 μm2/s. The diffusion coefficient, concentration and actin binding and dissociation rate constants for the CP were varied in the simulations.
In this study, the original Actin Dendritic Nucleation model was modified to include mechanisms for anticapping activity by Ena/VASP and for simulation of FRAP and CALI of EGFP-CP. The modified computational models for FRAP and CALI simulations are available as the BioModels entitled “CALI”, “FRAP” “Filopodia formation” in the Virtual Cell database under username “marynka”.
Modifications of the model
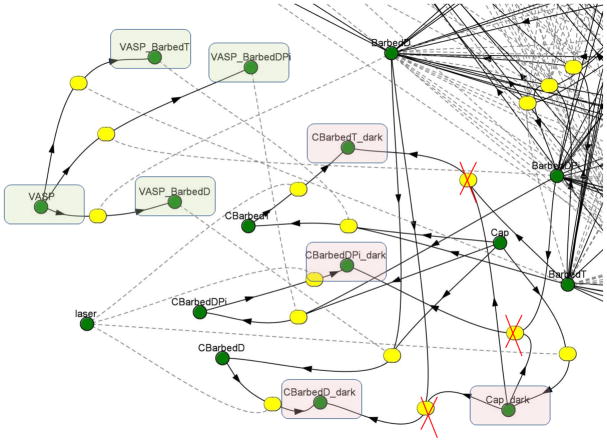
To simulate the photobleaching during FRAP or CALI computational experiments several new “species” (“laser”, “Cap_dark”) were added to the reaction scheme as shown in Fig. 1. During the bleaching phase, the new species “laser” transforms EGFP-CP, both free molecules and those bound to three barbed ends forms (CBarbedT, CBarbedDPi, CBarbedD corresponding to the 3 nucleotide-bound forms, respectively: ATP, ADP-Pi, or ADP), into “dark” forms of the same molecules (Cap_dark, CBarbedT_dark, CBarbedDPi_dark, CBarbedD_dark). All “dark” forms of CP were assumed to have the same diffusion coefficient as unbleached EGFP-CP. To model the FRAP experiments, the dark form of EGFP-CP was assumed to participate in all the chemical reactions that unbleached EGFP-CP does. In the CALI simulations, laser inactivated EGFP-CP can not bind to barbed ends, so kon for inactive EGFP-CP (species Cap_dark) is equal to zero.
FIGURE 1.
The part of initial reaction scheme on Vcell platform, which was used to simulate FRAP and CALI experiment, green circles denote the species which participate in simulation; yellow circles denote the reaction nodes. During the irradiation, the laser transforms Capping protein both free (Cap) and all forms bound to the Barbed ends (CBarbedD, CBarbedDPi, CBarbedT) to the “dark” form (pink rectangle). In FRAP simulation the photobleached, “dark” form, is still active and able to participate in all reactions. For CALI experiments the “dark” form is inactive and the crossed reaction edges show what reaction is only one way reaction for CALI simulation. The new VASP species can participate in binding reaction equally with each type of free Barbed end (green rectangle). The “on” reaction rate is a constant. The “off” reaction rate is a variable that depends on the total concentration of VASP_Barbed end complexes. A higher concentration of these complexes reduces the dissociation rate constant for VASP from barbed ends. A VASP_Barbed end complex is assumed to be equivalent to a free Barbed end for the actin polymerization reactions.
The VASP molecule (Fig. 1, green rectangle) was included in four different forms, the free VASP molecule and complexed with the three nucleotide-bound barbed ends (VASP_CBarbedT, VASP_CBarbedDPi, and VASP_CBarbedD). VASP-barbed end complexes were assumed to be able to participate in all the biochemical reactions that free barbed ends do but were not able to bind capping protein. The VASP binding and dissociation rate constants are discussed in Results.
Cell geometry
FRAP and CALI experiments were performed on fibroblasts adherent to glass substrates and fully spread (Vitriol et al. 2007). For the CALI experiments, the diameter of TEM00 Gaussian profile laser beam (width at 1/e2 of peak intensity) was 23.4 μm and the ratio of the irradiated area to the entire cell surface was between 1/6 and ¼. To approximate the ratio of the laser beam diameter to the cell size, the cell radius in the model was set to 25 μm. To model cell geometry in the numerical simulations, we modified the analytical geometry employed in the actin polymerization model (Ditlev et al. 2009). The computational domain consisted of a cell with radius R=25 μm, a height (H) in the center of 4μm and thickness at the edge of the lamellipodium of 0.3μm (Fig. 1A). To decrease the computational time, the model equations for the FRAP simulations were solved only in half of the cell, because the irradiated area during the FRAP (R=2.5 μm) was substantially smaller than the entire cell body. The modified computational models for FRAP and CALI simulations are available as the BioModels entitled “CALI”, “FRAP” “Filopodia formation” in the Virtual Cell database under username “marynka”.
Simulations
The resulting system of partial differential algebraic equations was solved using the finite volume solver available in Virtual Cell (Moraru et al. 2008; Novak et al. 2007; Schaff et al. 1997; Schaff et al. 2001; Slepchenko et al. 2000), using a regular rectangular grid of 150 × 75 × 12 elements and 0.002 s time step for FRAP simulations and 150 × 150 × 36 elements and 0.02 s time step for CALI simulations. The accuracy of the simulations was checked by refining the mesh and using shorter time steps. The computations were carried out on the computer cluster of Center for Cell Analysis and Modeling (CCAM).
RESULTS
Steady state distribution of actin filaments
It is known that cell adhesion and spreading involves complex and dynamic rearrangements of the actin cytoskeleton (Dobereiner et al. 2004) that are coordinated and controlled by the Arp2/3 complex (Lai et al. 2008). The nucleation function of the Arp2/3 complex is activated by a member of the Wiskott-Aldrich syndrome family protein, N-WASP (Anton et al. 2007; Chesarone and Goode 2009) located on the membrane
Initially simulations were run for a sufficiently long time to achieve a steady state and to generate a distribution of actin filaments that is similar to those observed in spread cells. In this case, high concentrations of short filaments reside near the edge of lamellipodium while longer filaments are present toward the center of the cell. To produce this filament distribution, the concentration of N-WASP is taken to be small (5 to 20 molecules/μm2) on the majority of the plasma membrane but greatly increases (25 to 200 molecules/μm2) in regions within 2.5 μm of the cell edge. A similar pattern of N-WASP distribution was reported in (Sukumvanich et al. 2004). For each parameter set, the model initially was simulated for 200 s, which was sufficient to allow all concentrations to come to steady state. The steady state concentration profiles were used later as the initial conditions for the FRAP or CALI simulations.
Fig. 2 demonstrates the cell geometry (Fig. 2a) and initial conditions for simulation. Fig. 2b shows the concentration of N-WASP at the beginning of the simulation while Fig. 2c presents the distribution of active Arp2/3, which has been recruited to the membrane by N-WASP, after a 200 s simulation. An example of the filament length distributions at equilibrium is given in Fig. 2d and e. The differences in the average filament lengths between the lamellipodium (~540 nm, or 200 actin monomers) and the cell center (~710 nm, or 300 actin monomers) is clearly visible.
FIGURE 2.
Initial conditions for simulations. (a) X-Z depiction of the cell geometry, used for simulations (cell dimension R= 25μm). (b) Concentration of N-WASP at the beginning of simulation. (c) Equilibrated distribution of active Arp2/3 after a 200 s simulation. (d,e) Equilibrated actin filament network prior to performing FRAP and CALI simulation. # of subunits refers to the number of G actin subunits in individual filaments. Initial conditions: N-WASP=20 molecules/μm2 around the cell and additional 50 molecules/μm2 near the edges (denoted in the legends as N-WASP=20+50); [CP]= 1 μM; simulation time = 200s; (f,g)) Plot of filament lengths (f) and filament concentrations (g) along the radius of the cell after equilibration with different concentrations of CP (μM) and N-WASP (mol/μm2). Black dashed lines on (d,e) shows where the number of filaments and their concentrations were measured.
To investigate how the amounts of N-WASP and CP influence the actin filament distribution and the concentrations of free/capped barbed ends, we performed additional simulations with different total amounts of N-WASP and CP (Fig. 2f and g). The results show that, indeed, increasing the CP concentration produces shorter filaments with higher concentration. Simulations with a tenfold increase in the abundance of N-WASP (50 molecules/μm2 over the entire plasma membrane and an additional 200 molecules/μm2 near the edges) also decreased the filament length, but the effect was weaker than that of the CP.
Simulation of CP FRAP kinetics
Before attempting to model the CALI data, we used the model to fit FRAP data and estimate the values of key model parameters. The recovery kinetics of EGFP-CP following photobleaching depend on several factors: (1) the total concentrations of EGFP-CP and barbed ends that bind CPs; (2) rate constants for EGFP-CP binding to and dissociating from barbed ends; and (3) diffusion coefficients for EGFP-CP and actin filaments in the cytosol. We investigated how each of these parameters affects the recovery curve. We assumed that all endogenous CPs are replaced by EGFP-CP which exhibit identical chemical properties as endogenous CP. Furthermore, we assumed that transport and binding properties of bleached EGFP-CP were identical to those of unbleached EGFP-CP.
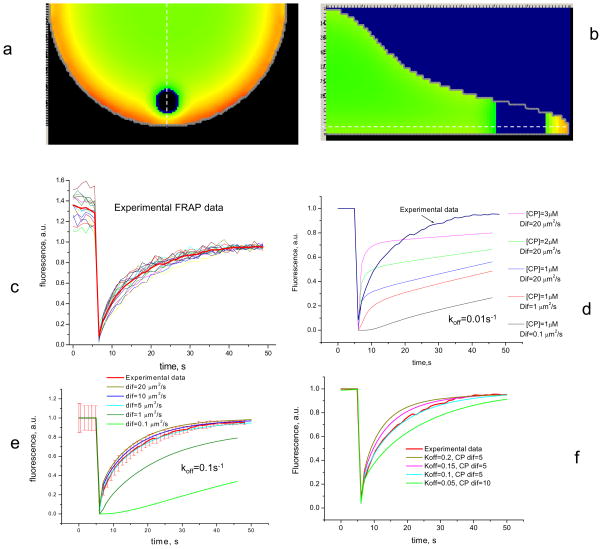
The experimental FRAP measurements were performed on an Olympus FV1000 confocal microscope in the tornado scanning mode, which allows generation of a circular bleaching spot within a short time interval (t=200 ms). To mimic the experimental setup in the simulation, we uniformly ’bleached’, in 200 ms, a 5 μm diameter cylinder through the cytoplasm positioned so that its center was located 5 μm from the cell edge. The parameter characterizing the laser intensity (I) has the dimension of s−1 similar to the rate constant for a zero-order reaction. This parameter was adjusted to reduce fluorescence in the bleached region to 2% of the initial fluorescence, similar to that measured in the experiments.
In our simulation the fluorescence intensity is proportional to total concentration (bound and free) of unbleached EGFP-CP, and, during the recovery phase, the change in fluorescence was monitored within the entire bleached region. Figs. 2a and b show the distribution of fluorescence within the cell and the result of laser photobleaching of EGFP-CP in side and top views, respectively.
The fluorescence in the simulation recovers nearly completely, with the failure to reach 100% recovery due to the ~ 1% of the total EGFP-CP that was bleached. In experimental FRAP measurements, there is generally an immobile fraction that leads to less than 100% recovery (Fig. S1 in the Supplementary Material). We estimated from analyzing each separate data curve that recovery was nearly complete (95%) between 42 s and 48 s after photobleaching. Using this estimation, the immobile part of experimental fluorescence recovery was extracted and experimental FRAP data was normalized by dividing each time point in the recovery curve by the 100% fluorescence recovery value for that curve (Fig. 3c). This procedure did not change the value of the half-time for fluorescence recovery after bleaching. After normalization, the immobile fraction for each curve can be estimated by the comparing the fluorescence value before the photobleaching to that after recovery is deemed complete.
FIGURE 3.
Simulation of FRAP experiments. (a,b) Fluorescence distribution immediately after photobleaching applied to an equilibrated system. Laser beam diameter = 5μm; time of photobleaching = 200 ms. White dashed line on (a) shows where the cross-sectional profile in (b) is taken. White dashed line in (b) shows the depth at which the 2 dimensional image in (a) is taken. (c) Experimental data for FRAP of EGFP-CP (N=10). Data are normalized by the mean value of intensity between 42 and 48s of recovery, considering this value as 95% of recovery. (d,f) Simulation results of a FRAP experiment. (d) With CP koff = 0.01 s−1, traces show poor fit with any combination of diffusion coefficient and CP concentration. (e) Simulation for koff = 0.1 s−1 with different diffusion coefficients for CP. Red line shows experimental results with standard deviations. (f) Variation of koff (= 0.1, 0.15, 0.2 s−1) and CP diffusion coefficient of 5 and 10 μm2/s Traces show the best fit with experiment for CP koff = 0.1 s−1 and diffusion rate between 5–10 μm2/s. A larger diffusion coefficient with a smaller koff does not produce good agreement with experimental FRAP data.
Initially, all simulations were performed using kon=10 μM−1s−1 and koff=0.01 s−1, respectively as values for the association and dissociation rate constants for EGFP-CP to and from the barbed ends respectively (Schafer et al. 1996). However, simulations with these rate constants derived from in vitro measurements, failed to reproduce the experimental results even when other parameters important for the recovery kinetics were varied widely. In particular, the CP diffusion coefficient was varied between 0.1 to 20 μm2/s, the CP concentration was varied from 0.5 to 4 μM, and the concentration of barbed ends was varied from 0.5 to 2 μM (Fig. 3d). By contrast, good fit between the simulations and experimental data was obtained when the uncapping rate was increased by one order of magnitude (koff = 0.1 s−1) (Fig. 3e and f). This requirement for a large value of the rate constant for uncapping is supported by the results of single speckle microscopy experiments performed recently by two independent research groups. Iwasa & Mullins showed that the average lifetime for CP on barbed ends is ~15 s (koff~0.05 s−1) (Iwasa and Mullins 2007), and Watanabe’s group (Miyoshi et al. 2006) measured an unexpectedly fast dissociation rate for CPs at the leading edge (koff~0.58 s−1).
Simulations with various diffusion coefficients for CP and a dissociation rate constant equal to 0.1 s−1 (Fig. 3e) show that a diffusion coefficient between 5 and 15 μm2/s produce results that fit the experimental data within two standard deviations. Fixing the diffusion coefficient for CP at 5μm2/s and varying the dissociation rate constant between 0.05 s−1 and 0.2 s−1 revealed that the best fit was obtained for koff=0.1 s−1 (Fig. 3f).
Simulation of CALI experiments performed on capping proteins
Our first approach to model the effect of CALI was to use the same scheme as for the FRAP simulations, but with one significant difference: Laser inactivated EGFP-CP can not bind to barbed ends, so kon for inactive EGFP-CP (species Cap_dark in Vcell scheme) is equal to zero (Fig. 1). The dissociation rate, koff, for inactive (photodamaged) EGFP-CP was varied for different simulations.
To compare the simulations to experimental results, we assumed that an increase in filament length corresponds to the dorsal ridges observed experimentally in fibroblasts (Vitriol et al. 2007). This assumption is justified by the additional f-actin associated with these structures (Vitriol et al. 2007). For quantification, the number of ridges in concentric rings centered on the optical axis was counted using an image processing algorithm that detects edges (Vitriol et al. 2007) and plotted versus time. This experimental time dependence was compared qualitatively with simulated filament length in corresponding areas.
The time of simulated laser inactivation was chosen to equal the experimental irradiation time of 100 ms. The model parameter that corresponds to laser intensity was chosen to produce ≥ 95% inactivation of CP on the optical axis. The laser beam has a Gaussian intensity profile (Fig. 3a) characterized by a 1/e2 diameter where the intensity drops to 1/e2 of the maximum value on the optical axis of the microscope; this diameter was 23.4 μm. The laser optical axis was located 10 μm from the cell edge.
We ran several CALI simulations using different parameter sets. Fig. 4c shows the time course of average F-actin length inside a circle of radius 6 um centered on the optical axis. As seen, these simulations produced the expected elongation of F-actin inside the irradiated zone, but the time course of this increase differed significantly from experimental observations (Fig. 4b). The elongation phase was too rapid and reached maximum within 15– 20 s. By contrast, the experiments exhibited slower kinetics, reaching a maximum between 100–120 s. Also, the experiments showed that new actin-filled ridges after CALI diminished slowly and many of them persisted for the entire observation period whereas the simulations produced a rapid diminution of the long filaments. An attempt to slow down the elongation phase by decreasing the dissociation rate for CALI damaged EGFP-CP (Fig. 4c, (black)) from 1 s−1 to 0.1 s−1 did not substantially slow the growth phase but simply produced a smaller increase in F-actin.
FIGURE 4.
Comparison between experimental growth of protrusive ridges and V-cell modeling of the F-actin increase after CALI. (a) Pseudocolored image shows the distribution of laser intensity and 6,10,15 μm distance from optical axis; (b) Experimental results of protrusion development, analyzed by an edge detection method from (Vitriol et al. 2007); (black), protrusions within a 6μm circle centered on the optical axis; (red), protrusions in an annulus from 6–10 μm from optical axis; (green), protrusions in an annulus from 10–15 μm from optical axis; (c) F-actin elongation after laser inactivation of CP inside 6 μm circle. Simulation scheme doesn’t include anti-capping agent. Plots were computed for two different unbinding rates for photo deactivated CPs (koff=0.1 s−1, blue, koff==1 s−1, red). (d) The growth of actin filaments as a function of time and distance from the optical axis for simulation that includes VASP as an anti-capper. Color scheme is the same as for (b). (e) Pseudocolored images show the position of increased filament growth at different times after CALI. Point indicates the center of laser beam.
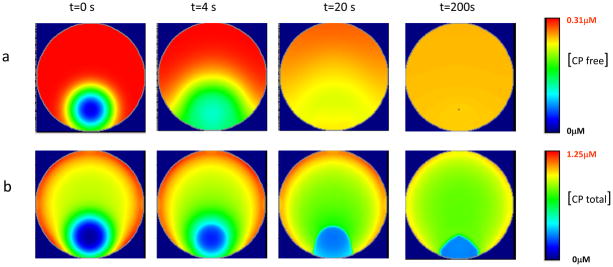
These results can be explained by the fast diffusion of free CPs. The concentration of free undamaged EGFP-CP inside the irradiated area is restored to nearly 80% of normal level within the first 20–40 s (Fig. 5a) and the rapid binding of CPs to filaments quickly inhibits further F-actin elongation. We analyzed the fluorescence recovery after photobleaching inside an area with a 12μm radius. The computational results predict that the half-time for fluorescence recovery (corresponding to the sum of free and bound EGFP-CP) according to our model should be around 50s (Fig. S2) which contradicts the experimental results where substantial recovery of fluorescence inside the CALI’ed region was observed only after 150 s following irradiation (Vitriol et al. 2007).
FIGURE 5.
Time development for the replacement of active CP after CALI in the irradiated region. (a) Kinetics for the repopulation of free undamaged CP inside the irradiated region. Rapid diffusion of free CP results in a nearly uniform distribution with a slightly lower concentration within the first 50 s compared to the bulk of the cytoplasm. (b) Kinetics of the fluorescence recovery after CALI, which reflects the total concentration of free CP and that bound to Barbed ends. Note that there is no fast fluorescence recovery after irradiation because barbed ends which were capped prior to CALI are replaced by VASP-Barbed end complexes and that leaves the irradiated area with less fluorescence.
The comparison between computational and experimental data suggests that the cells have an additional mechanism to regulate capping dynamics that is absent in our model. To properly capture persistent filament elongation following CALI that lasts for hundreds of seconds, an anti-capping mechanism is required. We propose that ENA/VASP family of proteins are prime candidates for this role; these proteins have been shown to antagonize CP activity (Bear et al. 2002; Breitsprecher et al. 2008; Trichet et al. 2008). We introduced VASP protein into the Vcell modeling scheme and assumed that VASP molecules bind barbed ends independently of their ATP/ADP status. The VASP-Barbed end complex can participate in all reactions that free barbed ends can, but it cannot be capped by CP (Fig. 1).
For proper assignment of VASP modeling parameters, some important experimental evidence about VASP, CPs and their interactions with barbed ends needs to be considered. Thus far, the evidence is somewhat controversial and has been the topic of many recent investigations and discussions (Barzik et al. 2005; Bear and Gertler 2009; Bear et al. 2002; Pasic et al. 2008; Sechi and Wehland 2004; Trichet et al. 2008):
In vitro, all barbed ends are capped despite a 20–25 -fold molar excess of VASP (Breitsprecher et al. 2008; Pasic et al. 2008) which indicates that the affinity of CPs for barbed ends is a 2–3 orders of magnitude greater than the VASP affinity for barbed ends.
Titration of cell lysates revealed that the molar ratios of VASP to CPs are approximately 5:1 with the concentration of VASP being 4±1μM (Breitsprecher et al. 2008; Laurent et al. 1999). The concentration of free CP in theintact cell was estimated to be approximately 1–2 μM. Thus, there appearedto be enough high affinity cappers to cap all the barbed ends in vivo (DiNubile et al. 1995). Indeed, a simple simulation shows that 5μM of VASP with a Kd=5μM together with 1μM of CP (Kd= 0.01 μM) results in 95% of the barbed ends being capped. Nevertheless, immediately after lysis with detergent, cells contained large number of barbed ends that are not capped and are able to bind pyrenyl-G-actin resulting in filament elongation (DiNubile et al. 1995).
Several investigations (Breitsprecher et al. 2008; Samarin et al. 2003) show that dense packing of VASP bound to beads significantly enhanced actin assembly even in the presence of CP, suggesting that if VASP molecules are located on the barbed ends in close proximity to each other, they are able to support processive filament elongation, possibly by protecting filaments from capping.
Applewhite & coworkers (Applewhite et al. 2007) performed a series of FRAP experiments which showed that EGFP-VASP exhibits rapid recovery in lamellipodia but almost no recovery in filopodia. This observation suggests that, in resting cells, VASP molecules in the lamella and lamellipodia have a high dissociation rate, in which case actin filaments grow only to short lengths before being capped. However, when barbed ends are concentrated, such as in filopodia, dissociation of VASP is much slower, allowing VASP to remain complexed with filaments for longer times, thereby protecting them from being capped and permitting filaments to elongate.
We modeled this effect using the following equation for the VASP-Barbed end dissociation rate constant, kdis:
| (1) |
where k0 is dissociation rate in the absence of VASP-Barbed end complexes, VASP_B is the concentration of VASP/Barbed end complexes, α is a scaling coefficient that determines the concentration of complex at which the dissociation rate is half its maximum, and n is a coefficient of cooperativity. This equation captures our suggestion that the dissociation of VASP from barbed ends decreases with increasing concentrations of VASP/Barbed end complexes in a cooperative fashion.
This equation was incorporated into the Vcell simulations of the EGFP-CP CALI experiment. Fig. 4d shows simulation results for the average filament length as a function of time derived in several regions (concentric circles R=6,10,15 μm) centered on the optical axis. The good qualitative agreement between the model and experimental results (Fig. 4b) supports the hypothesis that VASP binds cooperatively to barbed ends. In fact, a coefficient of cooperativity smaller than 4 did not produce a stable effect in time and a coefficient higher than 4 replaces all capping protein with anti-capper too quickly to match the experimental kinetics. These figures clearly indicate that where the intensity is highest at the center of the laser beam, filament growth is most pronounced corresponding to the strongest CALI effect.
Dynamics and position of morphological changes are similar in the model and experiments
By monitoring the dynamics of filament growth, we found that the position of stable morphological changes depends not only on the irradiation intensity but also on the distance from the cell edge. Shortly after inactivation, the majority of filament growth shifted toward the cell edge (Fig. 4e, t=40s). This effect occurs for two reasons. First, due to geometric considerations, the diffusional recovery of active CP is anisotropic, resulting in delayed replacement close to edge as compared to more central locations. Second, the higher concentration of free barbed ends near the edge requires more CP to halt growth, so depletion of this protein has a more pronounced effect near the cell edge. As a result, uncapped barbed ends at the cell edge collect more VASP molecules which increases the probability of VASP clustering and resultant stabilization at barbed ends, in turn protecting those barbed ends from being recapped when the concentration of active EGFP-CP is restored by diffusion.
The panels of Fig. 5a show a time course for the concentration of free active EGFP-CP as it recovers following CALI. The panels in Fig. 5b show a time course for the kinetics of fluorescence recovery, which takes into account the formation of new EGFP-CP-barbed end complexes. Note that free EGFP-CP quickly reaches a uniform distribution inside the cell due to the rapid diffusion of this protein. However, EGFP-CP bound to barbed ends exhibits a distinctive pattern with lower concentration close to the cell edge inside the irradiated region
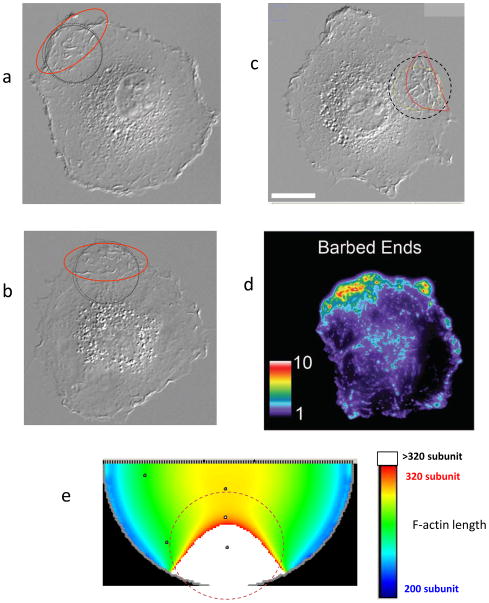
The simulations produce a number of satisfying similarities with the experimental results. Fig. 6(a–d) presents three different cells with developed dorsal ridges after CALI and pseudocolored fluorescent images of actin filament barbed-ends labeled with Alexa Fluor 568-conjugated G-actin at 3 min post-CALI (Vitriol et al. 2007). The computational panel (Fig. 6e) shows the increased filament length after CALI. The dashed circle on both panels marks the region where the laser beam is applied. Inspection of these figures reveals that the increase in filament length (corresponding to the protrusive ridges in the experiment) persists proximate to the cell edge longer than in the other parts of the cell.
FIGURE 6.
Similarities in the location of morphological changes development between the experimental (a–d) and computational (e) results. Dashed circle shows the 1/e2 width of applied laser beam. (a–c) Microscope images of three different cells after CALI. Red lines show the area of the most pronounced protrusive ridges development. Scale bar in c represents 20 μm. (d) A pseudocolored fluorescent image of actin filament barbed-ends labeled with Alexa Fluor 568-conjugated G-actin at 3 min post-CALI from (Vitriol et al. 2007). (e) Computational results of post-CALI showing where filament growth is most pronounced with white regions representing the longest filaments.
Simulation of FRAP and CALI experiments with various irradiation conditions
To determine how the development of the EGFP-CP CALI phenotype depends on irradiation conditions, we ran additional simulations with different laser intensities and beam radii. The typical FRAP bleaching power is between 0.5–30 mW, which is less than 5% of the CALI laser intensity used in our experiment (I=650 mW, measured on the specimen plane). A FRAP experiment with laser beam 1/e2 width = 23.4 μm and intensity equal to 5% of that used for CALI revealed that no appreciable changes in filament length occur after irradiation (Fig. 7a).
FIGURE 7.
Simulation of CALI experiments with various irradiation conditions. (a) Montage showing FRAP of EGFP-CP (center) with no appreciable CALI phenotype developing (DIC image before and after FRAP). Laser beam d=23.4μm. (b–d) Simulation with intensity equal to 5% of CALI intensity (I=1 s−1) showing that no appreciable increase in filament length occurs in the simulation. (e–g) Results of CALI with small radius of irradiation showing that the simulation does not produce CALI phenotype when the CALI beam has a diameter of 5 μm and intensity of 20 s−1. (h–k) CALI of CP without Knockdown/Rescue does not produce changes in the phenotype. The total concentration of CP is 1.5 μM of which ½ is EGFP-CP, laser intensity I=7 s−1. (B,E,H) show inactivation of EGFP-CP after CALI; (c,f,I,d,g,k) show the recovery of active CP (c,f,i), and average filament length (d,g,k) in the center of CALI’ed region
To simulate possible collateral damage in a FRAP experiment, we performed additional CALI simulations using Vcell with the same parameters as above, but with laser intensity only 5%–10% of that previously used in the simulations. Thus, during the CALI step, the laser beam inactivated only 10% of EGFP-CP on the optical axis, instead of the >95% as in previous simulations (Fig. 7b and c). The results show that no elongation of actin filaments developed after irradiation (Fig. 7d). Thus, our simulations also indicate that in the case of EGFP-CP, the FRAP experiments can be performed using different sizes of laser beam (5–25μm) without developing a CALI phenotype.
A small radius of irradiation does not produce the CALI phenotype
The experimental results reported earlier in (Vitriol et al. 2007) demonstrated that when cells were irradiated with a smaller beam size with high laser intensity (5 μm; 6.1 mW/μm2), no increase in dorsal surface protrusive ridges occurred. In that study it was suggested that the CALI effects require inactivating a large enough fraction of the EGFP-CP pool to avoid rapid recovery of active CP through diffusion. Using Vcell, we tested this prediction. In the simulations, a beam diameter of 5 μm was used and the laser intensity was varied from I=20 s−1 to I=100 s−1, which is consistent with the experimental conditions. The dissociation rate constant was taken as koff=0.1 s−1 for active CP and koff=0.2 s−1 for inactivated CP. The pseudocolor image of active CP concentration immediately following CALI is shown in Fig. 7 E. The recovery of active EGFP-CP in the center of the irradiated area and kinetics of filament length are presented in Figs. 6f and g. The results of this simulation demonstrate that filament length does not increase after a high intensity irradiation in a small area.
The dissociation rate of photodamaged EGFP-CP is another parameter that can affect the development of morphological change after CALI. More rapid EGFP-CP dissociation from filament tips after CALI will enhance the probability of VASP binding to vacant barbed ends before the diffusional replacement of active EGFP-CP occurs. Table S1 in Supporting Material presents results for different combinations of intensity and koff for inactive EGFP-CP, where ‘N’ means that the simulation produced no filament growth while ’Y’ means that substantial filament growth occurred. The table shows that employing a 5μm diameter of inactivation, with a laser intensity even 5–10 times higher than that normally used for wide beam CALI, does not produce increased filament growth unless the off rate for inactivated EGFP-CP is appreciably increased.
CALI without knockdown of endogenous capping protein
CALI experiments also were performed on a cell line in which both endogenous CP and EGFP-CP are present (Vitriol et al. 2007). To reproduce this condition in the model, we assumed that total concentration of CP in the cell is higher than was used in the previous simulations and equal to 1.5μM, with only half of this being EGFP-CP We ran simulations for 200s to allow the new system to reach steady state. Next we performed CALI with a laser intensity that was able to inactivate only 50% of available CP (Fig. 7h and i). The results of this simulation (Fig. 7k) demonstrate that under this condition CALI does not induce changes in filament length.
After VASP protein was introduced into the actin polymerization model, we repeated all our simulations for fluorescence recovery after photobleaching of EGFP-CP. The recovery kinetics were identical to those already reported for the FRAP experiments (data not shown), which showed that the VASP molecules did not interfere substantially with CPs binding to the barbed ends. This means that under resting conditions, the local concentration of free barbed ends is not high enough to create conditions for VASP_Barbed-end cooperative behavior. This result also supports several experimental observations (Pasic et al. 2008; Sechi and Wehland 2004) which demonstrated that in resting cells VASP molecules do not play significant role in actin dynamics; by contrast, VASP activity is required to regulate the rate of actin filament elongation and provide a mechanism to recruit barbed ends during assembly of specialized actin filament structures (Pasic et al. 2008; Sechi and Wehland 2004)
DISCUSSION
Virtual Cell simulations described in this paper, employing a detailed model of actin polymerization in the context of the dendritic nucleation scheme (Pollard 2007), provide insights into FRAP and CALI experiments involving CP and other actin binding proteins. The major results of computational modeling show that: (1) the CP dissociation rate constant must be around 0.1 s−1, which is much larger than measured in vitro; (2) in order to induce sustainable changes in cell morphology after CP inactivation, the cells require an anti-capping mechanism, which includes cooperativity with positive feedback; (3) at the low doses of irradiation that are employed in FRAP experiments, collateral CALI is not sufficient to produce significant alterations in morphology.
Most analytical equations developed for FRAP analysis must assume that the bleach spot is small relative to the size of the fluorescent compartment so that diffusional recovery is isotropic. Other limitations include the failure to account for the positioning of the bleached area within the 3D geometry of the cell and include the assumption of a homogenous prebleach distribution of the fluorophores (Carrero et al. 2003; Kang and Kenworthy 2008; Sprague et al. 2004; Tsibidis and Ripoll 2008). The complexity of the cell geometry and its position with respect to the bleaching beam as well as uneven fluorescent distributions in many cell experiments may violate the usual approximations. The numerical methods employed in this work permit the use of realistic geometries and heterogeneous distributions as well as complex biochemical mechanisms that may govern the FRAP kinetics observed experimentally. In general, the use of the Virtual Cell modeling platform permits facile incorporation of spatial and biochemical details in the analysis of FRAP and CALI data and this allows extraction of more accurate and important kinetic constants.
Comparison of the CALI simulations and the kinetics of morphological changes measured from actual CALI experiments showed that without an anti-capping agent the induced morphological changes are smaller and persist for a significantly shorter time than is experimentally observed. This was remedied in the simulation by including VASP as the anti-capping agent and resulted in simulations that reproduced the experimental kinetics. Furthermore, the area of dorsal ridge development, which is shifted from the center of irradiation toward the cell edge after CALI (Fig. 6), is also reproduced in the simulation This self-consistency provides an additional measure of confidence in the validity of our proposed biophysical mechanism of VASP action as incorporated in our system.
Finding the appropriate kinetic parameters for VASP modeling prompted us to look into the details of VASP binding kinetics. One hypothesis which reconciles the experimental work (Applewhite et al. 2007; Breitsprecher et al. 2008) is that when VASP binds to a single isolated filament, the rate of dissociation is determined primarily by dissociation from the membrane. FRAP kinetics for the EVH1 domain responsible for membrane binding are rapid with a half-time ~ 3 s (Applewhite et al. 2007). We hypothesize that when the concentration of free barbed ends is sufficiently high, there is an increased probability for binding many VASPs in close proximity to each other. In such cases, VASP molecules cluster with the consequence that the dissociation rate from barbed ends is decreased. Results of the parameter search demonstrate that the best fit to the experimental data is achieved with a coefficient of cooperativity characterizing the inverse dependence of the VASP-barbed end dissociation on the concentration of VASP-barbed ends complexes of around 4. Interestingly, this value is in agreement with experiments showing that tetramerization of VASP actively drives processive actin filament elongation (Bachmann et al. 1999; Bear and Gertler 2009; Breitsprecher et al. 2008).
Our model also suggest that recruitment of Ena/VASP proteins to the leading edge requires free barbed ends explaining why low to moderate doses of cytochalasin D (100–150 nM), which caps barbed ends, displaced all Ena/VASP proteins from the leading edge and filopodial tips (Sechi and Wehland 2004). The proposed mechanism also explains why Ena/VASP proteins are concentrated at the leading edge in proportion to the protrusion velocity (Rottner et al. 1999). As protrusion velocity and free barbed end density increase, greater levels of Ena/VASP proteins will bind the filaments creating a positive feedback effect which stabilizes Ena/VASP.
We performed a simulation to test if this mechanism could initiate the filopodia formation. Fig. 8 and Movie S1 in Supporting Material demonstrate the results for the situation where, after equilibration, additional molecules of active N-WASP were applied for a short time (5 s) to a limited membrane area (Fig. 8a). The simulation shows that the increase of the barbed end concentrations due to N-WASP activation on the leading edge, can very quickly switch the competition between CP and VASP toward VASP binding (Fig. 8c), leading to the increased actin polymerization. The pseudocolor images (Fig. 8d) of filament length distribution and the time plots of filament length and concentration (Fig. 8b) show that immediately after the stimulus, the filaments become shorter and denser with a higher concentration of free barbed ends. The excess free barbed end quickly exhausts the pool of available CP increasing the VASP binding. The growing concentration of VASP_Barbed end complexes creates a positive feedback via clustering of the complexes and results in filament elongation (Fig. 8d, t=100s) and possibly filopodia-like protrusions on the leading edge.
FIGURE 8.
Hypothetical mechanism to develop filopodial protrusion after N-WASP activation. (a) The position of additional N_WASP application. Concentration of applied stimulus is 100 molecules/μm2. Time of stimulus application is 5 s. (b) Time dependence for normalized filament length and filament concentration in the center of the stimulus zone. (c) Time dependence of the relationship between concentrations of barbed ends complexed with CP (red) and with VASP (blue). (d) Pseudocolor images illustrate the time development of long filaments on the cell edge. Animated version of simulation are available as Movie S1.
Vitriol et al (Vitriol et al. 2007) also showed that a CALI induced dorsal ridge development ensues only when CP is totally replaced by EGFP CP. The simulation, which incorporated VASP as an anti-capper, also showed that no appreciable CALI phenotype could be obtained without the knockdown-rescue feature of this experiment (Fig. 7h–k). Without this, the morphological changes observed in the CALI experiments are not developed because the concentration of endogenous CP, unaffected by CALI irradiation, is sufficient to successfully compete with VASP molecules and replace the photodamaged CP so that significant increase in filament length does not occur.
Previously it also has been documented that, under conditions when a successful FRAP experiment was conducted, no CALI phenotype was detected (Vitriol et al. 2007). Our simulations demonstrated that a FRAP experiment with EGFP-CP does not produce sufficient collateral damage from CALI to effect changes in cell morphology; this is because the intensity of the photobleaching beam is not high enough to produce the critical concentration of free barbed ends that will induce filament growth before diffusional replacement of EGFP-CP occurs. This simulation is important because the successful application of CALI in cell biology highlights concerns about whether appreciable photodamage occurs during FRAP experiments. These results allow us to formulate a working hypothesis for the case where the quantum yield for photobleaching is significantly greater than that for CALI and the exchange between CALI’ed protein and the undamaged protein is rapid compared to the half-time for development of the biological phenotype. In such cases, bleaching can be accomplished with much lower intensity, meaning that only a minute fraction of the protein actually experiences the CALI effect. Because of the rapid exchange with the normal, undamaged protein, any damaged protein is quickly replaced by the normal protein before phenotypic effects can develop. In this case, FRAP experiments can be performed without danger of inducing an inactivation effect. However, photochemical effects in the cytosol are a complicated function of the particular fluorescent protein and the fusion protein itself, so that the independence of FRAP and CALI should be checked for each case.
Supplementary Material
Acknowledgments
This work was prompted by a suggestion from and initial experiments with Harold Erickson, Duke University. We also acknowledge many helpful discussions with James Bear, UNC-CH. This work was supported by U. S. Public Health Service grants from the National Institutes for General Medical Sciences grant RO1 GM078994 (TE), the Cell Migration Consortium (U54 GM064346 (KJ & LL), the National Center for Research Resources P41 RR013186 (LL) and the NIH Roadmap National Technology Centers for Networks and Pathways U54 RR022232 (LL).
References
- Andrianantoandro E, Blanchoin L, Sept D, McCammon JA, Pollard TD. Kinetic mechanism of end-to-end annealing of actin filaments. J Mol Biol. 2001;312(4):721–30. doi: 10.1006/jmbi.2001.5005. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24(1):13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Anton IM, Jones GE, Wandosell F, Geha R, Ramesh N. WASP-interacting protein (WIP): working in polymerisation and much more. Trends in Cell Biology. 2007;17(11):555–562. doi: 10.1016/j.tcb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima SI, Svitkina TM, Gertler FB, Borisy GG. Ena/VASP proteins have an anti-capping, independent function in filopodia formation. Molecular Biology of the Cell. 2007;18(7):2579–2591. doi: 10.1091/mbc.E06-11-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M. The EVH2 Domain of the Vasodilator-stimulated Phosphoprotein Mediates Tetramerization, F-actin Binding, and Actin Bundle Formation. Journal of Biological Chemistry. 1999;274(33):23549–23557. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. Journal of Biological Chemistry. 2005;280(31):28653–28662. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122(12):1947–1953. doi: 10.1242/jcs038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109(4):509–521. doi: 10.1016/s0092-8674(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem. 2008;283(11):7135–44. doi: 10.1074/jbc.M705894200. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J Biol Chem. 1998;273(39):25106–11. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem. 1999;274(22):15538–46. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41(2):597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Mullins RD. Interactions of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr Biol. 2000;10(20):1273–82. doi: 10.1016/s0960-9822(00)00749-1. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. Embo Journal. 2008;27(22):2943–2954. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B Coordinates Arp2/3 Complex and Cofilin Activities at the Leading Edge. Cell. 2007;128(5):915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361(2):257–67. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Jean C, Rieger KJ, Lenfant M, Pantaloni D. Modulation of the interaction between G-actin and thymosin beta 4 by the ATP/ADP ratio: possible implication in the regulation of actin dynamics. Proc Natl Acad Sci U S A. 1993;90(11):5034–8. doi: 10.1073/pnas.90.11.5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson AE. Stimulation of actin polymerization by filament severing. Biophys J. 2006;90(2):413–22. doi: 10.1529/biophysj.105.069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson AE, Wear MA, Cooper JA. End versus Side Branching by Arp2/3 Complex. Biophys J. 2004;86(2):1074–1081. doi: 10.1016/S0006-3495(04)74182-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero G, McDonald D, Crawford E, de Vries G, Hendzel MJ. Using FRAP and mathematical modeling to determine the in vivo kinetics of nuclear proteins. Methods. 2003;29(1):14–28. doi: 10.1016/s1046-2023(02)00288-8. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Current Opinion in Cell Biology. 2009;21(1):28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: Matching assembly factors with cellular structures. Nature Cell Biology. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. International Review of Cell and Molecular Biology. 2008;267:183. doi: 10.1016/S1937-6448(08)00604-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Sengupta P, Wiseman PW, Brown CM, Horwitz AR, Gratton E. Fluctuation correlation spectroscopy with a laser-scanning microscope: Exploiting the hidden time structure. Biophysical Journal. 2005;88(5):L33–L36. doi: 10.1529/biophysj.105.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNubile MJ, Cassimeris L, Joyce M, Zigmond SH. Actin filament barbed-end capping activity in neutrophil lysates: the role of capping protein-beta 2. Mol Biol Cell. 1995;6(12):1659–1671. doi: 10.1091/mbc.6.12.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Vacanti NM, Novak IL, Loew LM. An Open Model of Actin Dendritic Nucleation. Biophysical Journal. 2009;96(9):3529–3542. doi: 10.1016/j.bpj.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobereiner HG, Dubin-Thaler B, Giannone G, Xenias HS, Sheetz MP. Dynamic phase transitions in cell spreading. Physical Review Letters. 2004;93(10):108105/1–108105/4. doi: 10.1103/PhysRevLett.93.108105. [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. 2007:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godschmidt-Clermont PJ, Machesky LM, Doberstein SK, Pollard TD. Mechanism of the interaction of human platelet profilin with actin. Journal of Cell Biology. 1991;113:1081–1089. doi: 10.1083/jcb.113.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug C, Jay PY, Reddy I, McNally JG, Bridgman PC, Elson EL, Cooper JA. CAPPING PROTEIN-LEVELS INFLUENCE ACTIN ASSEMBLY AND CELL MOTILITY IN DICTYOSTELIUM. Cell. 1995;81(4):591–600. doi: 10.1016/0092-8674(95)90080-2. [DOI] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Current Biology. 2007;17(5):395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K, Rajfur Z, Vitriol E, Hahn K. Chromophore-assisted laser inactivation in cell biology. Trends in Cell Biology. 2008;18(9):443–450. doi: 10.1016/j.tcb.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang F, Purich DL, Southwick FS. Profilin promotes barbed-end actin filament assembly without lowering the critical concentration. J Biol Chem. 1999;274(52):36963–72. doi: 10.1074/jbc.274.52.36963. [DOI] [PubMed] [Google Scholar]
- Kang M, Kenworthy AK. A closed-form analytic expression for FRAP formula for the binding diffusion model. Biophysical Journal. 2008;95(2):L13–L15. doi: 10.1529/biophysj.108.135913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinosian HJ, Selden LA, Gershman LC, Estes JE. Interdependence of Profilin, Cation, and Nucleotide Binding to Vertebrate Non-Muscle Actin. Biochemistry. 2000;39(43):13176–13188. doi: 10.1021/bi001520+. [DOI] [PubMed] [Google Scholar]
- Kinosian HJ, Selden LA, Gershman LC, Estes JE. Actin Filament Barbed End Elongation with Nonmuscle MgATP-Actin and MgADP-Actin in the Presence of Profilin. Biochemistry. 2002;41(21):6734–6743. doi: 10.1021/bi016083t. [DOI] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. Single molecule kinetic analysis of actin filament capping: Polyphosphoinositides do not dissociate capping proteins. J Biol Chem. 2007:M705287200. doi: 10.1074/jbc.M705287200. [DOI] [PubMed] [Google Scholar]
- Lai FP, Szczodrak M, Block J, Faix J, Breitsprecher D, Mannherz HG, Stradal TE, Dunn GA, Small JV, Rottner K. Arp2/3 complex interactions and actin network turnover in lamellipodia. Embo Journal. 2008;27(7):982–992. doi: 10.1038/emboj.2008.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, Wehman A, Grobe L, Jockusch BM, Wehland J, Gertler FB, Carlier M-F. Role of Proteins of the Ena/VASP Family in Actin-based Motility of Listeria monocytogenes. J Cell Biol %R. 1999;144(6):1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Carlier MF. Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiological Reviews. 2008;88(2):489–513. doi: 10.1152/physrev.00021.2007. [DOI] [PubMed] [Google Scholar]
- Mahaffy RE, Pollard TD. Kinetics of the Formation and Dissociation of Actin Filament Branches Mediated by Arp2/3 Complex. Biophys J. 2006;91(9):3519–3528. doi: 10.1529/biophysj.106.080937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Fievez S, Carlier MF. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mercapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry. 1996;35(37):12038–45. doi: 10.1021/bi961325o. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Tsuji T, Higashida C, Hertzog M, Fujita A, Narumiya S, Scita G, Watanabe N. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. Journal of Cell Biology. 2006;175(6):947–955. doi: 10.1083/jcb.200604176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraru II, Schaff JC, Slepchenko BM, Blinov M, Morgan F, Lakshminarayana A, Gao F, Li Y, Loew LM. The virtual cell modeling and simulation software environment. IET Systems Biology. 2008;2:352–362. doi: 10.1049/iet-syb:20080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachmias VT. Small actin-binding proteins: the beta-thymosin family. Curr Opin Cell Biol. 1993;5(1):56–62. doi: 10.1016/s0955-0674(05)80008-0. [DOI] [PubMed] [Google Scholar]
- Novak IL, Gao F, Choi YS, Resasco D, Schaff JC, Slepchenko BM. Diffusion on a curved surface coupled to diffusion in the volume: Application to cell biology. Journal of Computational Physics. 2007;226(2):1271–1290. doi: 10.1016/j.jcp.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasic L, Kotova T, Schafer DA. Ena/VASP proteins capture actin filament barbed ends. Journal of Biological Chemistry. 2008;283(15):9814–9819. doi: 10.1074/jbc.M710475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the Mechanism of Processive Actin Filament Elongation by Formins. Cell Motility and the Cytoskeleton. 2009;66(8):606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annual Review of Biophysics and Biomolecular Structure. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112(4):453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annual Reviews of Biochemistry. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305(5691):1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Ressad F, Didry D, Egile C, Pantaloni D, Carlier MF. Control of actin filament length and turnover by actin depolymerizing factor (ADF/cofilin) in the presence of capping proteins and ARP2/3 complex. J Biol Chem. 1999;274(30):20970–6. doi: 10.1074/jbc.274.30.20970. [DOI] [PubMed] [Google Scholar]
- Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins. Comparison of plant and human ADFs and effect of phosphorylation. J Biol Chem. 1998;273(33):20894–902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J. VASP dynamics during lamellipodia protrusion. Nature Cell Biology. 1999;1(5):321–322. doi: 10.1038/13040. [DOI] [PubMed] [Google Scholar]
- Roy P, Rajfur Z, Pomorski P, Jacobson K. Microscope-based techniques to study cell adhesion and migration. Nature Cell Biology. 2002;4(4):E91–E96. doi: 10.1038/ncb0402-e91. [DOI] [PubMed] [Google Scholar]
- Samarin S, Romero S, Kocks C, Didry D, Pantaloni D, Carlier M-F. How VASP enhances actin-based motility. J Cell Biol. 2003;163(1):131–142. doi: 10.1083/jcb.2003.03.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D, Jennings P, Cooper J. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J Cell Biol. 1996;135(1):169–179. doi: 10.1083/jcb.135.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaff J, Fink CC, Slepchenko B, Carson JH, Loew LM. A general computational framework for modeling cellular structure and function. Biophysical Journal. 1997;73:1135–1146. doi: 10.1016/S0006-3495(97)78146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaff JC, Slepchenko BM, Choi Y, Wagner JM, Resasco D, Loew LM. Analysis of nonlinear dynamics on arbitrary geometries with the Virtual Cell. Chaos. 2001;11:115–131. doi: 10.1063/1.1350404. [DOI] [PubMed] [Google Scholar]
- Sechi AS, Wehland J. ENA/VASP proteins: Multifunctional regulators of actin cytoskeleton dynamics. Frontiers in Bioscience. 2004;9:1294–1310. doi: 10.2741/1324. [DOI] [PubMed] [Google Scholar]
- Sept D, Xu J, Pollard TD, McCammon JA. Annealing accounts for the length of actin filaments formed by spontaneous polymerization. Biophys J. 1999;77(6):2911–9. doi: 10.1016/s0006-3495(99)77124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slepchenko BM, Schaff JC, Choi YS. Numerical approach to fast reaction-diffusion systems: application to buffered calcium waves in bistable models. Journal of Computational Physics. 2000;162:186–218. [Google Scholar]
- Slepchenko BM, Schaff JC, Macara I, Loew LM. Quantitative cell biology with the Virtual Cell. Trends in Cell Biology. 2003;13(11):570–576. doi: 10.1016/j.tcb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophysical Journal. 2004;86(6):3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumvanich P, DesMarais V, Sarmiento CV, Wang Y, Ichetovkin I, Mouneimne G, Almo S, Condeelis J. Cellular localization of activated N-WASP using a conformation-sensitive antibody. Cell Motility and the Cytoskeleton. 2004;59(2):141–152. doi: 10.1002/cm.20030. [DOI] [PubMed] [Google Scholar]
- Trichet L, Sykes C, Plastino J. Relaxing the actin cytoskeleton for adhesion and movement with Ena/VASP. Journal of Cell Biology. 2008;181(1):19–25. doi: 10.1083/jcb.200710168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibidis GD, Ripoll J. Investigation of binding mechanisms of nuclear proteins using confocal scanning laser microscopy and FRAP. Journal of Theoretical Biology. 2008;253(4):755–768. doi: 10.1016/j.jtbi.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Vitriol EA, Uetrecht AC, Shen FM, Jacobson K, Bear JE. Enhanced EGFP-chromophore-assisted laser inactivation using deficient cells rescued with functional EGFP-fusion proteins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(16):6702–6707. doi: 10.1073/pnas.0701801104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends in Biochemical Sciences. 2004;29(8):418–428. doi: 10.1016/j.tibs.2004.06.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.