SUMMARY
Synchrotron x-ray protein footprinting is used to study structural changes upon formation of the ClpA hexamer. Comparative solvent accessibilities between ClpA monomer and ClpA hexamer samples are in agreement throughout most of the sequence with calculations based on two previously proposed hexameric models. The data differ substantially from the proposed models in two parts of the structure: the D1 sensor 1 domain and the D2 loop region. The results suggest that these two regions can access alternate conformations in which their solvent protection is greater than in the structural models based on crystallographic data. In combination with previously reported structural data, the footprinting data provide support for a revised model in which the D2 loop contacts the D1 sensor 1 domain in the ATP-bound form of the complex. These data provide the first direct experimental support for the nucleotide-dependent D2 loop conformational change previously proposed to mediate substrate translocation.
INTRODUCTION
ATP-dependent proteases are responsible for a variety of essential cellular regulatory functions, the most notable of which are the dissolution of protein aggregates and the degradation of unwanted proteins; both of these processes are required for cell growth, mediation of stress responses, and protein quality control. Typically, the unfolding and degradation of protein substrates proceeds via ATP-dependent translocation of these substrates from the ATPase component through a narrow pore into the degradation chamber of the protease component. Understanding the conformational changes that enable ATP-dependent proteases to manipulate their substrates is a useful first step in designing agents to modulate bacterial physiology by activating (Brotz-Oesterhelt et al., 2005) or inhibiting these molecular machines; however, the relevant conformational changes are not yet well-defined. In this study, we investigate the arrangement of protein domains in the pre-hydrolytic state of the energy-dependent protease ClpA.
ClpAP is a roughly cylindrical E. coli ATP-dependent protease complex comprised of a double-ringed tetradecameric ClpP protease core flanked by a hexameric ClpA ring on one or both ends (Kessel et al., 1995). The ClpA chaperone is required for substrate recognition, unfolding and translocation of the unfolded substrate to the ClpP active sites. Natural substrates for ClpAP include the plasmid P1 replication intiator RepA, the heme biosynthetic enzyme HemA and a number of carbon starvation proteins (Wickner et al., 1994); (Wang et al., 1999); (Damerau and St John, 1993). In addition, ClpAP is able to degrade proteins with one of two types of identifying ‘tags.’ The first type of marker is the identity of the N-terminal amino acid. N-terminal arginine, lysine, leucine, phenylalanine, tyrosine and tryptophan all target a protein for N-end rule degradation (Tobias et al., 1991). The second type of tag is the 11-amino acid ssrA sequence (AANDENYALAA), the addition of which can target stably folded proteins to ClpAP for ATP-dependent degradation, including tagged lambda repressor and tagged GFP (Gottesman et al., 1998) (Weber-Ban et al., 1999).
ClpA undergoes self-assembly from the monomeric into the active hexameric form in the presence of ATP or the non-hydrolyzable analog ATPγS. Hexamerization is required for both substrate binding to ClpA and for the formation of the ClpAP complex (Maurizi et al., 1998), (Hoskins et al., 1998); the ATPγS-bound form of the ClpA hexamer is competent to activate ClpP for the degradation of large peptide substrates (~30 residues), although ATP hydrolysis is required for efficient proteolysis of protein substrates (Thompson et al., 1994). ATP hydrolysis is required for ClpA-catalyzed unfolding of large proteins and for translocation of all substrates except small peptides. Protein substrates typically require several rounds of ATP hydrolysis for complete conversion into peptide products; the ClpAP complex remains associated throughout this process and through several rounds of degradation (Singh et al., 1999).
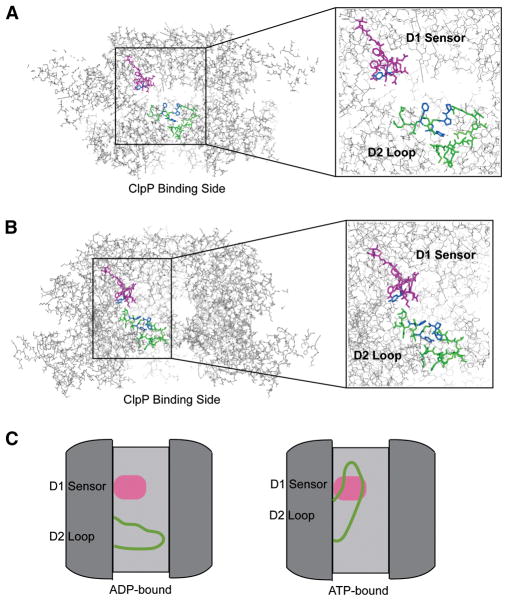
A 3-D reconstruction at a resolution of 29 Å via cryo-electron microscopy (Beuron et al., 1998) indicates that the macromolecular ClpAP complex forms three compartments: the digestion chamber inside ClpP, a small compartment between ClpA and ClpP, and a chamber inside of ClpA. More detailed structural information is available for the individual components of the structure in the form of crystal structures of the ClpP tetradecamer (Wang et al., 1997) (Bewley et al., 2006) (Szyk and Maurizi, 2006) and of the ClpA monomer (Guo et al., 2002). In addition, based on the ClpA monomeric crystal structure, two ClpA hexameric models have been published (Figure 1A and 1B). The first model (Guo et al., 2002) was constructed using crystal structures of the hexameric forms of NSF-D2 and HslU as templates. The second model (Hinnerwisch et al., 2005) was constructed using the p97 hexameric crystal structure as a template. With the exception of the ClpA N-domain, which is missing from the second model, the two models are qualitatively very similar to one another; each includes a central pore and is consistent with the structure inferred from electron microscopy. However, little direct, high-resolution structural information is available concerning the ClpA hexamer structure, nor have these models been tested against experimental data.
Figure 1.
Hexameric models with D1 sensor and D2 loop highlighted in magenta and green, respectively. A) A cut-away side view of Hexamer 1, the ClpA hexameric model published by Maurizi and coworkers (Guo et al., 2002), on the left, with a zoom of the pore region on the right. The D1 sensor of chain F and the D2 loop of chain A are highlighted, with probe residues Y324, P537, P538, Y540, and F543 shown in blue. B) A cut-away side view of Hexamer 2, the ClpA hexameric model published by Horwich and coworkers (Hinnerwisch et al., 2005), on the left, with a zoom of the pore region on the right. The D1 sensor and D2 loop from chain A and the same probe residues as in A are highlighted. C) Proposed movement of the D2 loop upon ATP hydrolysis with the D1 sensor and D2 loop colored as in A and B. The D2 loop is in the “down” conformation in the ADP-bound post-hydrolytic state. The D2 loop moves to the “up” conformation upon rebinding of ATP (the pre-hydrolytic state).
The details of the hexameric model are mechanistically important because large conformational changes in ClpA have been proposed to mediate unfolding of protein substrates and their translocation through the ClpAP complex (Hinnerwisch et al., 2005). Recent results with photoreactive substrates indicate that ClpA’s D2 loop contacts the substrate in the course of translocation. Furthermore, mutations in a region of the protein immediately adjacent to the D2 loop allow substrate binding but not degradation. Based on these results, a mechanism was proposed in which motions of the D2 loop mediate movement of the substrate through the ClpA pore during unfolding and translocation (Hinnerwisch et al., 2005). In the proposed mechanism, the D2 loop binds the substrate in an “up” conformation (Figure 1C). Upon ATP hydrolysis, the D2 loop is proposed to drag the substrate through the central pore of the ClpA hexamer toward the face of the complex that binds ClpP. Once the D2 loop and the substrate are in this “down” conformation, the substrate can be released.
Other functional data are also consistent with the hypothesis that the D2 loop binds substrate tightly in a pre-hydrolytic conformation and releases it in a post-hydrolytic conformation. When ClpA is bound to the poorly-hydrolyzable nucleotide analogue ATPγS, it binds peptide substrates with high affinity (Piszczek et al., 2005). Single-molecule fluorescence experiments indicate that ClpA can assume both high-affinity and low-affinity conformations, with high-affinity peptide binding favored in the presence of ATPγS and low-affinity binding favored in the presence of hydrolyzable ATP (Farbman et al., 2007).
Existing structural data can be interpreted in terms of a post-hydrolytic “down” conformation, but direct evidence for a pre-hydrolytic “up” conformation has not previously been reported. The hexameric structural models currently in use are derived from a ClpA monomer structure containing bound ADP (Guo et al., 2002). These models place the D2 loop and nearby residues close to the ClpP-binding face of ClpA: i.e., in a “down” conformation. These residues are observable in the structure, suggesting that the “down” conformation represents a reasonably stable and well-ordered state of ClpA. Observation of the proposed alternate “up” conformation in the pre-hydrolytic state would provide significant evidence in favor of the proposed mechanism of translocation by D2 loop motions.
To test the hypothesis that the pre-hydrolytic form of the ClpA hexamer places the D2 loop in the “up” conformation, we used synchrotron protein footprinting to investigate the solvent accessibilities of domains in ClpA. Protein footprinting probes the solvent accessibility of side chains in the macromolecule, allowing protein interaction sites and conformational changes occurring upon complex formation to be mapped to specific areas of the protein. Synchrotron protein footprinting works via the generation of large quantities of hydroxyl radicals by direct irradiation of a protein solution over milliseconds. These radicals oxidatively modify protein side chains (Xu and Chance, 2005) (Xu and Chance, 2007) when they react with solvent-exposed protein regions, while areas of the protein not exposed to solvent are protected from these modifications. Protease digestion of the sample followed by liquid-chromatography-coupled mass spectrometry (LCMS) and tandem mass spectrometry (MSMS) allows identification of affected regions and often the specific residue that is modified. Changes in modification rate with the addition of complex components identify interprotein interactions and conformational changes.
In this study, we report the results of synchrotron protein footprinting experiments on both disassembled ClpA subunits and the ClpA hexamer bound to the poorly-hydrolyzable nucleotide analogue ATPγS. The footprinting data are most consistent with an ATPγS-bound ClpA structural model in which the D2 loop and associated residues contact the D1 sensor 1 domain, a region within one of ClpA’s Walker A ATPase sites. This conformation would position the D2 loop at the end of the ClpA hexamer closest to the substrate entry site, rather than at the end closest to the ClpP binding site. The footprinting data thus provide new evidence for a pre-hydrolytic “up” conformation of the ClpA D2 loop. These results provide the first structural support for a previously proposed mechanism in which translocation by ClpA is mediated by movements of the D2 loop.
RESULTS
Synchrotron x-ray footprinting was carried out under two sets of conditions: in the absence of nucleotide, where ClpA is disassembled, and in the presence of ATPγS, where it is hexameric (Maurizi et al., 1998), (Hoskins et al., 1998). MSMS-verifiable coverage of the ClpA tryptic digest encompassed 78% of the protein (Figure 2). For those peptides not observed in the mass spectrum, no information can be concluded. Ten tryptic peptides were identified in the ClpA monomer samples with modification confirmed by MSMS; seven of these also displayed modification in hexamer samples. Relative modification rates are listed in Table 1, and the locations of the peptides in the monomer crystal structure and in the hexamer models are shown in Figure 3. Although the same concentration of protein (2 μM) was used for both ClpA monomer and hexamer experiments, 1 mM ATPγS was required for the formation of the hexamers. This ATP analogue provides a significant quenching effect (factor of 4.35) on the dose received by the hexamer samples due to scavenging of the hydroxyl radicals; all hexamer rates stated are therefore normalized for this effect (Figure S1).
Figure 2.
Tryptic peptide coverage of ClpA. Color coding of amino acids: Black: MSMS identified peptides, Blue: peptides with modification, Red: specifically identified modified residues, Grey: not identifiable in spectra. Red boxes indicate areas of nucleotide interaction, green boxes indicate residues implicated in substrate binding and translocation, and the black box indicates the ‘IGL’ sequence required for binding to ClpP.
Table 1.
Rate constants for the modification of monomeric and hexameric ClpA
| Peptide | Sequence (MS/MS Verified Oxidation sites in Blue) | Oxidation Rate | Monomer/Hexamer Ratio | |
|---|---|---|---|---|
| Monomer | Hexamer | |||
| 77–86 | DTQPTLSFQR | 0.3±0.03 | - | - |
| 144–168 | KDEPTQSSDPGSQPNSEEQAGGEER | 1.7±0.10 | (1.3±0.09) | (1.3) |
| 169–181 | TENFTTNLNQLAR | 0.43±0.096 | - | - |
| 221–230 | TAIAEGLAWR | 0.6±0.04 | (0.8±0.65) | (0.7) |
| 318–333 | VIGSTTYQEFSNIFEK | 1.7±0.42 | (0.2±0.17) | (9.8) |
| 458–465 | MLVFGQDK | 16.9±0.51 | (8.2±0.82) | (2.0) |
| 519–527 | FDMSEYMER | 113.8±14.93 | (0) | (∞) |
| 533–555 | LIGAPPGYVGFDQGGLLTDAVIK | 6.0±0.72 | (0.5±0.27) | (10.9) |
| 616–633 | SIGLIHQDNSTDAMEEIK | 30.5±2.29 | (9.6±0.37) | (3.2) |
| 740–751 | NELTYGFQSAQK | 0.6±0.11 | - | - |
Verified modification sites are in bold and underlined.
Numbers in parentheses are normalized for ATPγS quenching (factor of 4.35).
Figure 3.
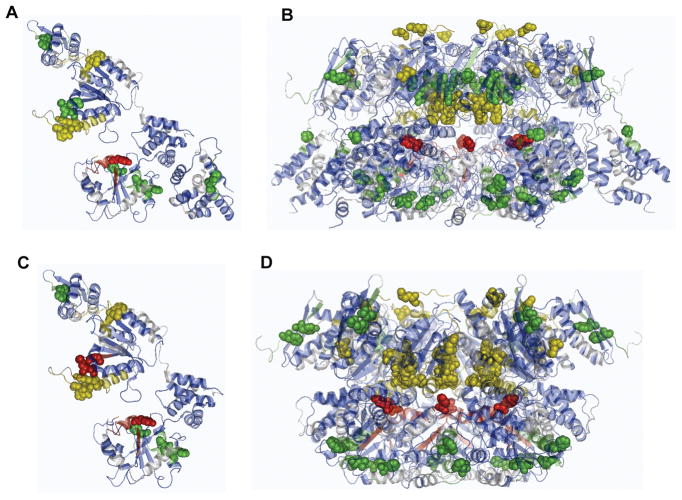
Comparison of experimental ClpA monomer/hexamer modification rate ratios and expected solvent accessibilities. Grey indicates a lack of coverage via MSMS. Blue indicates peptides for which only the unmodified peptide was identified via MSMS. Data/model comparison color coding: Green, relative agreement; Yellow, modest differences; Red, major disagreement. Specific modified residues are shown as spheres. A) ClpA monomer crystal structure 1KSF (Guo et al., 2002). B) Hexamer 1 model (Guo et al., 2002). C) ClpA monomer crystal structure 1KSF with the N-domain removed for comparison to D), Hexamer 2 model (Hinnerwisch et al., 2005).
The peptides analyzed in this experiment cover a variety of regions of the protein thought to be important for function. In each region, the experimental protection factors can be compared with predictions based on solvent accessibility analysis of the published monomeric structure or the two reported hexameric models (Tables 1, 2). For most residues, the agreement is reasonably good, but significant deviations are observed in a number of areas (Figure 3). Two of the anomalous regions, the D2 loop (corresponding to peptide 533–555) and the D1 sensor 1 (corresponding to peptide 318–333), are particularly noteworthy in terms of the magnitude of the effects observed and the possible mechanistic implications; however, it is important to discuss the entire set of footprinting observations in order to place the D2 loop and D1 sensor 1 results in the proper context.
Table 2.
Calculated peptide solvent accessibility values for structural models of ClpA
| Peptide | Domain | Solvent Accessibility (Ǻ2) | Monomer/Hexamer1 Ratio | Monomer/Hexamer2 Ratio | Monomer/FP Model Ratio | ||||
|---|---|---|---|---|---|---|---|---|---|
| Monomera | MonoΔNb | Hexamer1c | Hexamer2d | FPe | |||||
| 77–86 | N | 651.3 | - | 713.4 | - | - | 0.91 | - | - |
| AA168 | N-D1 loop | 194.7 | 194.7 | 73.6 | 194.3 | 194.4 | 2.65 | 1.00 | 1.00 |
| 169–181 | D1 | 890.0 | 890.0 | 744.2 | 789.9 | 789.8 | 1.20 | 1.13 | 1.13 |
| 221–230 | D1 near WA | 271.0 | 297.9 | 271.7 | 298.5 | 298.5 | 1.00 | 1.00 | 1.00 |
| 318–333 | D1 sensor | 711.2 | 711.2 | 676.2 | 521.2 | 307.3 | 1.05 | 1.36 | 2.31 |
| 458–465 | D2 nucl. int. | 470.5 | 470.5 | 511.6 | 488.7 | 488.8 | 0.92 | 0.96 | 0.96 |
| 519–527 | D2 | 527.7 | 527.7 | 218.4 | 401.4 | 484.7 | 2.42 | 1.31 | 1.09 |
| 533–555 | D2 loop | 1318.3 | 1318.3 | 855.0 | 702.7 | 591.9 | 1.54 | 1.88 | 2.23 |
| 616–633 | D2 | 828.8 | 828.8 | 773.5 | 867.8 | 869.3 | 1.07 | 0.96 | 0.95 |
| 740–751 | D2 C-term | 852.2 | 852.2 | 902.4 | 849.5 | 849.7 | 0.94 | 1.00 | 1.00 |
1KSF(Guo et al., 2002);
1KSF without N-domain;
footprinting model.
Italics indicate missing or disparate residues; Amino acid 168 is Ala in Hexamer1 and Arg in the monomer, the Hexamer2 model and the FP model. Aside from amino acid 168, peptide 144–168 is absent from the models. Residues 616–623 are also missing from the models; the accessibilities are calculated using only residues 624–633.
The peptide comprised of amino acids 458–465 contains the VFGQD sequence considered to be responsible for nucleotide binding in the D2 AAA+ domain (Guo et al., 2002). While peptide side chain solvation calculations using either of the previously published hexameric models predict that this region of the protein becomes more accessible upon hexamer formation, the footprinting experiments indicate that it is slightly less accessible in hexameric form. However, because the modified region is thought to participate in nucleotide binding, a process required for hexamer formation, the small increase in protection observed may be attributable to the space taken up by the nucleotide and localized conformational changes associated with the binding of the small molecule.
The IGL triplet is found within peptide 616–629 at positions 617–619. This loop is known to be essential for binding to ClpP (Kim et al., 2001) (Singh et al., 2001) (Joshi et al., 2004) and is predicted to remain on the surface upon hexamer formation, implying a small decrease in solvent accessibility. The data shows significant protection (>3-fold), indicating a conformational change that buries the probe side chains. It is important to note that residues 616–623, which contain the region of interest, are missing from the ClpA crystal structure (and the model), and thus are not included in the accessibility calculations. Also, the probe residues are H621 and M629, and while these are protected in the hexamer, this may not translate into protection of the IGL triplet, which is expected to remain accessible in order to bind to ClpP.
Peptide 144–168 is almost entirely unobservable in the crystal structure of the ClpA monomer (only residue 168 provided resolvable electron density), indicating that there is a high level of flexibility in this segment. The model places both ends of this fragment at the hexamer surface facing away from ClpP. This peptide shows small but significant protection from modification upon hexamer formation, possibly indicating an increased ordering of the strand as it makes contacts along the surface.
Peptide 77–86 is within the N-domain of the protein, a region thought to be fairly flexible. The rate of modification of this peptide is quite low, even in the monomer. Although no modification was evident in hexamer samples, the quenching effect of the ATPγS may have placed any fragments beyond the detection limits of the technique. Peptide 169–181, found within the D1 domain, exhibits a similar pattern of modification. Peptide 221–230 is proximal to the AAA+ Walker A nucleotide-binding consensus sequence motif in the D1 domain. This region of ClpA shows no difference in modification rate between the monomeric form of ClpA, with no nucleotide bound, and the hexameric form, with nucleotide present. Solvent accessibility calculations are consistent with these observations.
Peptide 519–527, a region immediately N-terminal to the D2 loop, exhibits significant protection upon formation of the hexamer. This region is highly oxidized in the monomer, with a modification rate of >100 s−1; multiple residues are observed to be oxidized in this peptide in the MSMS spectra. Even unexposed monomer samples showed a small amount of oxidation within this peptide. However, hexameric samples showed no oxidation above this background level, even upon greater exposure times, indicating complete burial of the probe residues as a function of complex formation. This is in reasonable agreement with the structural models on the residue level (Table S1), which predict that hexamerization will lead to a significant decrease in solvent-accessibility for the M525 probe residue that accounts for the majority of the oxidized product. On the peptide level, the solvent-accessible area in the hexamer is predicted to be relatively small (~300 Å2) compared to that of the monomer (~500Å2), but would nevertheless predict a measurable rate of modification for these residues.
The hexameric models’ predictions and the experimental data differ most substantially in two regions of ClpA (Figure 3). One is the β-sheet D1 sensor 1 sequence (referred to herein as the D1 sensor), contained within peptide 318–333. This peptide becomes highly protected upon hexamer formation (~10-fold). Its modification rate is at the lower limit of detection when ClpA is in the hexameric form, while the rate is easily measurable for the ClpA monomer (Table 1). This is in contrast to calculations based on the models, which predict only a very minor decrease in overall solvent accessibility.
The oxidation behavior of peptide 533–555 also deviates from the predicted solvent accessibilities provided by the models. This peptide contains the majority of the D2 loop and all of the amino acid residues within the D2 domain shown to affect substrate binding and/or ClpA function in mutational analyses (Hinnerwisch et al., 2005). The GYVG sequence within this peptide (including one of the oxidized residues, Y540) is highly conserved. It is expected to have particular functional significance based on the observation that the Y540A mutant is able to bind ssrA-linked substrates, but ClpAP proteolytic activity is inhibited (Hinnerwisch et al., 2005). Four different residues are oxidized in this segment during exposure, but the signals are overlapping in the chromatogram, so the individual residue oxidation patterns are not distinguishable. Formation of the ClpA hexamer significantly reduces the solvent accessibility of these amino acids (~10-fold decrease in modification rate). As observed for the D1 sensor, the modification rate for residues 533–555 is close to the lower limit of detection in the hexamer form, in contrast to the substantial modification rate observed for the monomer (Table 1). The modest decrease in solvent accessibility predicted for this region is unlikely to account for this level of protection, particularly considering the significant solvent-accessible area (~800Å) predicted to remain in both hexamer models (Table 2).
For the majority of the modified peptides, the specific amino acid modified has been identified through tandem mass spectrometry. Despite the general agreement of the footprinting data with the proposed hexameric models when considering overall accessibility for most peptides, the predicted specific amino acid side chain accessibilities are significantly disparate from the data (Table S1). As the models are built from monomeric subunits with the goal to satisfy overall domain and general structural requirements observed in similar structures and consistent with cryoEM studies (Guo et al., 2002) (Beuron et al., 1998), they do not take into account changes at smaller scales that could cause these differences. Using constraints based on the protection maps created by the footprinting data, with the addition of the missing residues and information about their accessibility, the existing ClpA hexameric models can be refined to incorporate these changes.
DISCUSSION
Our footprinting results are generally consistent with the previous ClpA structural data and the previously proposed hexameric models. For the peptides analyzed, the regions containing very reactive residues that are predicted to be highly accessible in the hexameric models are generally reactive enough to produce detectable amounts of oxidation products. These regions include the nucleotide binding site and residues at the surface of the hexamer. Those residues for which modification is expected but not observed are predominantly found within the N-domain. This flexible region (Ishikawa et al., 2004) may assume a different conformation in solution than in the crystal structure. In addition, the residues that change dramatically in accessibility upon hexamer formation mostly lie along the central axis of the hexamer (Figure 3). This observation is consistent with the idea that the intersubunit contacts that define the ClpA central pore help to protect residues in this region from the solvent. Evidence from electron microscopy (Kessel et al., 1995), (Grimaud et al., 1998) (Beuron et al., 1998) puts a strong constraint on the global structures that can be considered for ClpA hexamers. The agreement of the footprinting data with the broad outlines of the hexameric model thus supports the utility of this technique in probing the ultrastructure of ClpA.
Previous work does not provide direct evidence for the positioning of individual domains within the hexamer, and it is on this issue that the footprinting data differ from previous models of the hexamer. There are two regions of the protein that react with hydroxyl radical at a moderate-to-high rate in disassembled ClpA subunits and at a low-to-undetectable rate in the ClpA hexamer: the D1 sensor and the D2 loop. These regions are more highly protected than expected based on solvent accessibility calculations of the hexameric models (Figure S2). There are two alternative explanations for this observation: either these two regions form a new contact in the ATPγS-bound hexamer, or they insert themselves into other regions of the hexamer that are protected from solvent.
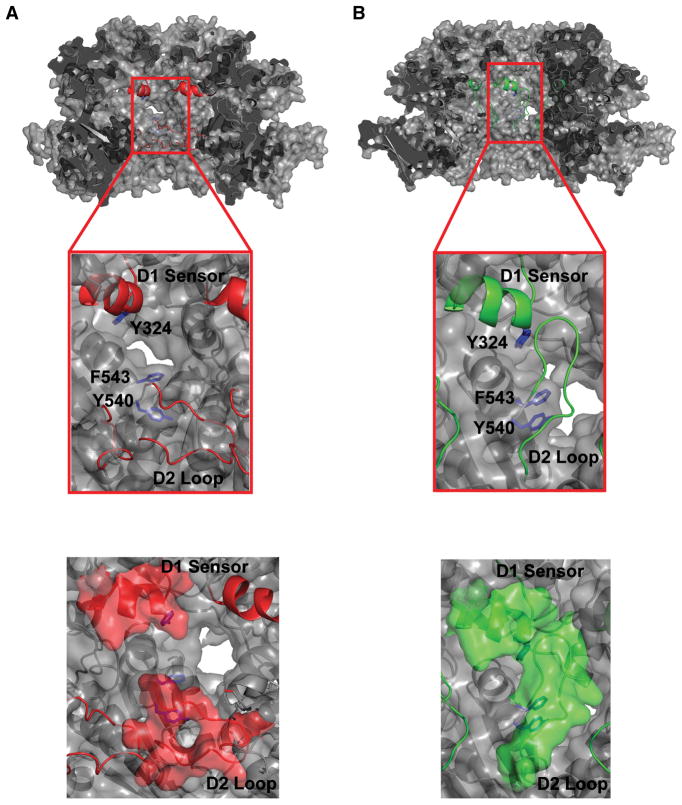
The most parsimonious explanation for the footprinting data is that the D1 sensor domain and the D2 loop contact each other in the ATPγS-bound hexamer. Based on the available crystallographic evidence, this interaction is feasible. A proposed structural model, shown in Figure 4B, was constructed in which the flexible D2 loop contacts the D1 sensor; this model has reasonable bond lengths and bond angles, and does not introduce unfavorable steric interactions. This interpretation of the structural data would be consistent with the proposed mechanism for translocation (Hinnerwisch et al., 2005), as it places the substrate-binding D2 loop in the proximity of the entrance to the ClpA pore when the complex is in the prehydrolytic state. The revised structural model in Figure 4B still does not account for the very high protection observed for residues 519-527 on the peptide level. However, the specific solvent accessibility of the reactive sulfur of M525 is reduced from 12.3 Å2 in the monomeric structure to 1.7 Å2 in the revised hexameric model and that of M521 is 0 Å2 for both monomer and hexamer structures. At the resolution provided by the current experiments and analysis, it is not possible to rule out a subtle effect of local dynamics as the source of the anomalously high oxidation rate observed for this peptide in the ClpA monomer (Zheng et al., 2008).
Figure 4.
Cross section of the ClpA hexamer illustrating the pore region. A) depicts the Hexamer2 model (Hinnerwisch et al., 2005) and B) the FP model. The sequences containing the D1 sensor 1 region (318–333) and the D2 loop (526–538) peptides are highlighted and labeled. Coloring is the same as in Figure 3 (Green, relative agreement with footprinting results; Red, major disagreement with footprinting results). In A) the loop is in the “down” position and does not protect residues Y324, Y540, or F543 (blue) from solvent access in the pore. In B) the loop is in the “up” position where it is able to protect Y324, Y540, and F543 from solvent. The FP model (B) provides better agreement with the footprinting results. Bottom figures depict the D1 Sensor 1 and D2 Loop regions in spacefill form.
An alternative explanation for the footprinting data is that both the D2 loop and the D1 sensor become buried in different solvent-protected regions of the ATPγS-bound hexamer structure. If so, the D2 loop and the D1 sensor would be more solvent-protected than predicted by the original hexameric model, but they would not be in contact with each other. However, it would be difficult to accommodate this possibility without also including large, global changes in the hexameric model. The D2 loop might be able to insert itself into the gap between the D1 and D2 domains on the equator of the complex, but large motions of the D1 sensor would not appear to be possible without a large rearrangement of the structure. Such a rearrangement might be possible, but would be without precedent in existing structural studies of both the hexamer and the monomer. Furthermore, such a rearrangement would likely perturb the solvent accessibility in other regions in addition to the D1 sensor domain; such perturbations are not observed.
When both the footprinting results and previous structural studies are taken into account, the structural model in which the D2 loop contacts the D1 sensor is thus most consistent with the data. It accounts for large increases in protection for these regions that would be difficult to explain without postulating unprecedented rearrangements of the tertiary structure. Small-scale rearrangements of local structure/chemical environment could account for the effects of hexamerization on residues 519–527 (i.e., the almost complete protection observed for a region predicted to be incompletely buried). It is worth noting that this region is adjacent to the D2 loop. Movement of the D2 loop might cause local structural changes in this region that would account for the protection data, however, the resolution of our data does not allow us to speculate about the nature of such rearrangements. With more sophisticated computational modeling (Diemand and Lupas, 2006) (Zheng et al., 2008), it may be possible to explain the protection observed in this region.
The larger, more easily interpretable effects observed for the D2 loop proper and the D1 sensor provide useful information about state-dependent conformational changes in ClpA. The revised structural model (Figure 4B) is in agreement with a previously proposed mechanism for translocation (Hinnerwisch et al., 2005), in which the D2 loop resides near the entry of the central pore before nucleotide hydrolysis (Figure 1C). The previously reported hexameric models might be viewed as a good description of the ADP-bound state of the hexamer. These models are in fact derived from monomer structures that contain bound ADP (Guo et al., 2002) (Hinnerwisch et al., 2005). With that assumption, the two hexamer models would represent the two key intermediates in the proposed translocation mechanism: the pre-hydrolytic state with the D2 loop “up” and the post-hydrolytic state with the D2 loop “down” (Figure 1C).
In the future, efforts will be made to use transient state and/or single-molecule kinetic experiments to detect the proposed conformational change. Stopped-flow synchrotron footprinting studies have previously been carried out (Shcherbakova et al., 2004); (Sclavi et al., 1998), and adapting them to the ClpA system appears to be feasible. These time-resolved footprinting experiments would allow determination of whether the proposed conformational change occurs and is kinetically competent for translocation. Single-molecule fluorescence experiments have also been carried out on ClpA (Farbman et al., 2007), and might be adapted to allow observation of the proposed conformational change (e.g., by detection of FRET between fluorophores attached to the D2 loop and D1 sensor).
The results of the current study also suggest that synchrotron x-ray footprinting will be generally useful for the study of domain motions in other proteolytic machines and chaperones. Like the Clp proteases, archaeal and mammalian proteasomes must translocate protein substrates through a central pore to present them to protease active sites. The conformational changes involved in translocation by these proteolytic complexes are still incompletely understood; synchrotron footprinting techniques may be useful in addressing the question of which protein domains mediate substrate translocation in these systems and other molecular machines.
EXPERIMENTAL PROCEDURES
Protein purification
ClpA (with a M169T mutation that enhances solubility and increases levels of full-length protein expression (Seol et al., 1994)) was purified in the Licht lab as previously described (Choi and Licht, 2005), (Maurizi et al., 1994). After purification and before storage, buffer exchange into RXN buffer (50 mM sodium cacodylate, pH 7.0, 400 mM KCl, and 20 mM MgCl2) was performed using a PD10 column (GE Healthcare) according to the manufacturer’s instructions. The ATP hydrolysis rate of the enzyme in this buffer system is 70–80% of the rate observed using standard HEPES buffer conditions (50mM HEPES, pH 7.5, 300mM KCl, 20mM MgCl2, 10% glycerol, 0.1% nonylphenylpolyethylene glycol). The enzyme stock solution was aliquoted, flash frozen in liquid nitrogen, and shipped overnight in dry ice to Brookhaven National Laboratory (Upton, NY), where it was immediately stored at −80°C.
Synchrotron hydroxyl radical footprinting
Samples were thawed on ice and diluted to 2 μM ClpA in RXN buffer. Diluted samples were kept at 4°C prior to and during the experiments and used within 12 hours of thawing. All samples other than disassembled ClpA samples (referred to as “monomer,” although dimers and trimers might also be present (Hwang et al., 1987) (Maurizi, 1991) (Seol et al., 1994) were incubated in 1 mM ATPγS for five minutes prior to exposure. Exposure conditions were predetermined by following the dose-dependent degradation of the fluorescent compound Alexa 488 (Invitrogen, Carlsbad, CA) in the presence of RXN buffer (Gupta et al., 2007). The ratio of this degradation rate to that of a similar experiment also containing 1 mM ATPγS provided the normalization factor of 4.35 for comparison of monomer and hexamer data. Samples were exposed to a mirror-focused (Sullivan et al., 2008) synchrotron x-ray beam (7 mrad angle, focus value of 4.5) at the X28C beamline of the National Synchrotron Light Source at Brookhaven National Laboratory for 0–100 ms. The exposure time of the samples was controlled via flow rate through the flow cell in the KinTek (Austin, TX) stopped-flow apparatus (Gupta et al., 2007). Oxidation was quenched by the addition of methionine amide to a final concentration of 10 mM. Irradiated protein samples were digested with sequencing grade modified trypsin (Promega, Madison, WI) at an enzyme to protein ratio of 1:20 (w/w) at 37°C overnight. The digestion reaction was terminated by adding formic acid to a final concentration of 0.1%. The resulting peptides (1 pmol) were loaded onto a 300 μm ID × 5 mm C18, PepMap nano Reverse phase (RP) trapping column to pre-concentrate and wash away excess salts using a U 3000 nano HPLC (Dionex, Sunnyvale, CA). The loading flow rate was set to a 25 μl/min, with 0.1% formic acid (pH 2.9) as the loading solvent. Reverse phase separation was performed on a 75 μm ID × 15 cm C18, PepMap nano separation column using nano separation system U 3000 (Dionex). Peptide separation was accomplished using buffer A (100% water and 0.1% formic acid) and buffer B (20% water, 80% acetonitrile and 0.1% formic acid). Proteolytic peptide mixtures eluted from the column with a 2% per minute acetonitrile gradient were introduced into an LTQ FT mass spectrometer (ThermoFisher Scientific, Waltham, MA) equipped with a nano spray ion source and using a needle voltage of 2.2 kV. MS and tandem MS spectra were acquired in the positive ion mode, with the following acquisition cycle: a full scan recorded in the FT analyzer at resolution R=100000 followed by MSMS of the eight most intense peptide ions in the LTQ analyzer. MSMS spectra of the peptide mixtures were searched against an E. coli data base for modifications (oxidation) of the tryptic peptides from the ClpA protein using BioWorks 3.2 software (ThermoFisher Scientific). In addition, detected MSMS mass spectral data for modified peptides were manually interpreted and correlated with hypothetical MSMS spectra predicted for the proteolysis products of the ClpA protein with the aid of the ProteinProspector (UCSF, CA) algorithm. The detected total ion currents were utilized to determine the extent of oxidation by separate quantitation of the unmodified proteolytic peptides and their radiolytic products by dividing the peak area corresponding to the modified peptide by that of the total peptide (modified and unmodified) (Kiselar et al., 2002). Levels of modification versus exposure time were plotted and fitted with a single exponential (Xu et al., 2005) via chi-squared minimization to determine the rate constant.
Solvent accessibility calculations
Solvent accessible surface area was calculated using the ‘surface’ function in the CCP4 package (1994). The PDB structure 1KSF (Guo et al., 2002) was used for monomer calculations and the models of the ClpA hexamer used were obtained from the authors of Guo et al. (Guo et al., 2002) and Hinnerwisch et al. (Hinnerwisch et al., 2005).
Modeling of the D2 loop position
The proposed loop movement was modeled in COOT (Emsley and Cowtan, 2004), and the Regularize function was used to assist in modeling. The Regularize command minimizes the function S, defined as: S = Sbond + Sangle + Storsion + Splane, thus minimizing the difference between each term and the ideal value for that term (Emsley and Cowtan, 2004). All altered residues (526–540) remained in the Ramachandran-allowed regions.
Supplementary Material
Acknowledgments
Center for Synchrotron Biosciences, Center for Proteomics, Case Western Reserve University, is supported by the National Institute for Biomedical Imaging and Bioengineering under P41-EB-01979. The National Synchrotron Light Source at Brookhaven National Laboratory is supported by the Department of Energy under contract DE-AC02-98CH10886. Work in the Licht laboratory is supported in part by a Beckman Young Investigator Award. The authors would like to thank Drs. Janna Kiselar and Serguei Ilchenko, who performed FTMS for this work; Dr. Wuxian Shi, for assistance with solvent accessibility calculations and Dr. Sayan Gupta for footprinting advice; Mike Sullivan, John Toomey and Don Abel for expert technical support for the X28C beamline; and Bob Sauer and Tania Baker for the ClpA expression plasmid.
References
- 1.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 2.Beuron F, Maurizi MR, Belnap DM, Kocsis E, Booy FP, Kessel M, Steven AC. At sixes and sevens: Characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. Journal of Structural Biology. 1998;123:248–259. doi: 10.1006/jsbi.1998.4039. [DOI] [PubMed] [Google Scholar]
- 3.Bewley MC, Graziano V, Griffin K, Flanagan JM. The asymmetry in the mature amino-terminus of ClpP facilitates a local symmetry match in ClpAP and ClpXP complexes. Journal of Structural Biology. 2006;153:113–128. doi: 10.1016/j.jsb.2005.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brotz-Oesterhelt H, Beyer D, Kroll HP, Endermann R, Ladel C, Schroeder W, Hinzen B, Raddatz S, Paulsen H, Henninger K, et al. Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat Med. 2005;11:1082–1087. doi: 10.1038/nm1306. [DOI] [PubMed] [Google Scholar]
- 5.Choi KH, Licht S. Control of peptide product sizes by the energy-dependent protease ClpAP. Biochemistry. 2005;44:13921–13931. doi: 10.1021/bi0505060. [DOI] [PubMed] [Google Scholar]
- 6.Damerau K, St John AC. Role of Clp protease subunits in degradation of carbon starvation proteins in Escherichia coli. J Bacteriol. 1993;175:53–63. doi: 10.1128/jb.175.1.53-63.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diemand AV, Lupas AN. Modeling AAA+ ring complexes from monomeric structures. J Struct Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 9.Farbman ME, Gershenson A, Licht S. Single-molecule analysis of nucleotide-dependent substrate binding by the protein unfoldase ClpA. J Am Chem Soc. 2007;129:12378–12379. doi: 10.1021/ja074168x. [DOI] [PubMed] [Google Scholar]
- 10.Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes & Development. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimaud R, Kessel M, Beuron F, Steven AC, Maurizi MR. Enzymatic and structural similarities between the Escherichia coli ATP-dependent proteases, ClpXP and ClpAP. J Biol Chem. 1998;273:12476–12481. doi: 10.1074/jbc.273.20.12476. [DOI] [PubMed] [Google Scholar]
- 12.Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 13.Gupta S, Sullivan M, Toomey J, Kiselar J, Chance MR. The Beamline X28C of the Center for Synchrotron Biosciences: a national resource for biomolecular structure and dynamics experiments using synchrotron footprinting. J Synchrotron Radiat. 2007;14:233–243. doi: 10.1107/S0909049507013118. [DOI] [PubMed] [Google Scholar]
- 14.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Hoskins JR, Pak M, Maurizi MR, Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang BJ, Park WJ, Chung CH, Goldberg AL. Escherichia coli contains a soluble ATP-dependent protease (Ti) distinct from protease La. Proc Natl Acad Sci U S A. 1987;84:5550–5554. doi: 10.1073/pnas.84.16.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa T, Maurizi MR, Steven AC. The N-terminal substrate-binding domain of ClpA unfoldase is highly mobile and extends axially from the distal surface of ClpAP protease. J Struct Biol. 2004;146:180–188. doi: 10.1016/j.jsb.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 18.Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nature Structural & Molecular Biology. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- 19.Kessel M, Maurizi MR, Kim B, Kocsis E, Trus BL, Singh SK, Steven AC. Homology in Structural Organization between Escherichia-Coli Clpap Protease and the Eukaryotic 26s-Proteasome. Journal of Molecular Biology. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- 20.Kim YI, Levchenko I, Fraczkowska K, Woodruff RV, Sauer RT, Baker TA. Molecular determinants of complex formation between Clp/Hsp100 ATPases and the ClpP peptidase. Nature Structural Biology. 2001;8:230–233. doi: 10.1038/84967. [DOI] [PubMed] [Google Scholar]
- 21.Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Hydroxyl radical probe of protein surfaces using synchrotron X-ray radiolysis and mass spectrometry. International Journal of Radiation Biology. 2002;78:101–114. doi: 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
- 22.Maurizi MR. ATP-promoted interaction between Clp A and Clp P in activation of Clp protease from Escherichia coli. Biochem Soc Trans. 1991;19:719–723. doi: 10.1042/bst0190719. [DOI] [PubMed] [Google Scholar]
- 23.Maurizi MR, Singh SK, Thompson MW, Kessel M, Ginsburg A. Molecular properties of ClpAP protease of Escherichia coli: ATP-dependent association of ClpA and clpP. Biochemistry. 1998;37:7778–7786. doi: 10.1021/bi973093e. [DOI] [PubMed] [Google Scholar]
- 24.Maurizi MR, Thompson MW, Singh SK, Kim SH. Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods in Enzymology. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 25.Piszczek G, Rozycki J, Singh SK, Ginsburg A, Maurizi MR. The molecular chaperone, ClpA, has a single high affinity peptide binding site per hexamer. J Biol Chem. 2005;280:12221–12230. doi: 10.1074/jbc.M411733200. [DOI] [PubMed] [Google Scholar]
- 26.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 27.Seol JH, Yoo SJ, Kim KI, Kang MS, Ha DB, Chung CH. The 65-kDa protein derived from the internal translational initiation site of the clpA gene inhibits the ATP-dependent protease Ti in Escherichia coli. J Biol Chem. 1994;269:29468–29473. [PubMed] [Google Scholar]
- 28.Shcherbakova I, Gupta S, Chance MR, Brenowitz M. Monovalent ion-mediated folding of the Tetrahymena thermophila ribozyme. J Mol Biol. 2004;342:1431–1442. doi: 10.1016/j.jmb.2004.07.092. [DOI] [PubMed] [Google Scholar]
- 29.Singh SK, Guo F, Maurizi MR. ClpA and ClpP remain associated during multiple rounds of ATP-dependent protein degradation by ClpAP protease. Biochemistry. 1999;38:14906–14915. doi: 10.1021/bi991615f. [DOI] [PubMed] [Google Scholar]
- 30.Singh SK, Rozycki J, Ortega J, Ishikawa T, Lo J, Steven AC, Maurizi MR. Functional domains of the ClpA and ClpX molecular chaperones identified by limited proteolysis and deletion analysis. J Biol Chem. 2001;276:29420–29429. doi: 10.1074/jbc.M103489200. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan MR, Rekhi S, Bohon J, Gupta S, Abel D, Toomey J, Chance MR. Installation and testing of a focusing mirror at beamline X28C for high flux x-ray radiolysis of biological macromolecules. Review of Scientific Instruments. 2008;79 doi: 10.1063/1.2839027. in press. [DOI] [PubMed] [Google Scholar]
- 32.Szyk a, Maurizi MR. Crystal structure at 1.9 angstrom of E-coli ClpP with a peptide covalently bound at the active site. Journal of Structural Biology. 2006;156:165–174. doi: 10.1016/j.jsb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Thompson MW, Singh SK, Maurizi MR. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. Journal of Biological Chemistry. 1994;269:18209–18215. [PubMed] [Google Scholar]
- 34.Tobias JW, Shrader TE, Rocap G, Varshavsky A. The N-end rule in bacteria. Science. 1991;254:1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- 35.Wang JM, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 angstrom resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, Elliott M, Elliott T. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J Bacteriol. 1999;181:1211–1219. doi: 10.1128/jb.181.4.1211-1219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 38.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 40.Xu G, Kiselar J, He Q, Chance MR. Secondary reactions and strategies to improve quantitative protein footprinting. Anal Chem. 2005;77:3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- 41.Xu GZ, Chance MR. Radiolytic modification and reactivity of amino acid residues serving as structural probes for protein footprinting. Analytical Chemistry. 2005;77:4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- 42.Zheng X, Wintrode PL, Chance MR. Complementary structural mass spectrometry techniques reveal local dynamics in functionally important regions of a metastable serpin. Structure. 2008;16:38–51. doi: 10.1016/j.str.2007.10.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.