Abstract
Cell adhesion molecules (CAMs) mediate cell attachment and stress transfer through extracellular domains. Here we forcibly unfold the Ig domains of a prototypical Ig superfamily CAM that contains intradomain disulfide bonds. The Ig domains of all such CAMs have conformations homologous to cadherin extracellular domains, titin Ig-type domains, and fibronectin type-III (FNIII) domains. Atomic force microscopy has been used to extend the five Ig domains of Mel-CAM (melanoma CAM)—a protein that is overexpressed in metastatic melanomas—under conditions where the disulfide bonds were either left intact or disrupted through reduction. Under physiological conditions where intradomain disulfide bonds are intact, partial unfolding was observed at forces far smaller than those reported previously for either titin's Ig-type domains or tenascin's FNIII domains. This partial unfolding under low force may be an important mechanism for imparting elasticity to cell–cell contacts, as well as a regulatory mechanism for adhesive interactions. Under reducing conditions, Mel-CAM's Ig domains were found to fully unfold through a partially folded state and at slightly higher forces. The results suggest that, in divergent evolution of all such domains, stabilization imparted by disulfide bonds relaxes requirements for strong, noncovalent, folded-state interactions.
Keywords: AFM
Cell adhesion molecules (CAMs) play pivotal and ubiquitous roles in maintaining tissue organization, in facilitating organogenesis, and in transmembrane signaling. They are divided into at least four major superfamilies: integrin, selectin, cadherin, and Ig. With more than one hundred members of the Ig superfamily already identified (1), Mel-CAM (melanoma CAM—also known as MCAM, MUC18, and CD146) appears prototypical in having a number of extracellular Ig domains linked in tandem (Table 1). Mel-CAM is also of specific interest: Its expression correlates with the metastatic potential of melanomas, and it is also found on normal endothelium among several other tissue types (2–5). More generally however, the evolved contributions of Ig domain structures to CAM function, and in particular to receptor and cell membrane stresses, are not completely clear. It is well known that most cells express multiple CAMs and often exhibit dynamic interactions with adjacent cells, but regulation of these alternative adhesion systems is not well understood. It is nonetheless clear that mechanical stresses are imposed on the membranes of cells and their associated CAMs as cells migrate, divide, and attach to other cells and extracellular matrix.
Table 1.
Structural arrangement of extracellular Mel-CAM domains with two variable (V) and three constant type-2 (C2) domains
| Domain | No. aa outside Cys | Native Lc (nm) | No. aa total | Reduced Lc (nm) |
|---|---|---|---|---|
| V1 | 49 | 18.1 | 116 | 42.8 |
| V2 | 49 | 18.1 | 110 | 40.6 |
| C21 | 46 | 17.0 | 93 | 34.3 |
| C22 | 46 | 17.0 | 87 | 32.1 |
| C23 | 51 | 18.8 | 97 | 35.8 |
| Average | 48.2 | 17.8 | 100.6 | 37.1 |
The C23 domain is the C-terminal domain in the recombinant protein and is adjacent to a plasma membrane alpha-helix in the full-length protein. The number of amino acids (aa) not contained within the disulfide loop of each domain are extensible under oxidizing conditions and are listed in the fourth column. Contour lengths (Lc) have been calculated with a unit peptide length of 0.37 nm.
The Ig domains of CAMs share a tertiary structure in common with cadherin domains as well as Ig-like domains of nonadhesion proteins. The Ig domains of titin, an elastomeric protein of muscle, and the fibronectin type-III (FNIII) domains of tenascin, an extracellular matrix component, are especially notable homologs in that both types of domains have been shown to unfold when forcibly extended one molecule at a time by using an Atomic Force Microscope (AFM) (6, 7). The Ig domains of CAMs are distinctive, however; a disulfide bond is almost universally found to span the domain core (1) that is contained within seven beta strands (or nine for V-type domains) in a sandwich-type arrangement (Fig. 1a). The central disulfide bridge typically defines a loop harboring about 50%, on average, of each domain (Table 1). All-atom computer simulations of forced unfolding have already suggested significant differences in forces of unfolding for different beta-sandwich domains (8); and although partially unfolded intermediates of titin have previously been related to some key hydrogen bonds (9), the role of more stable internal disulfide bonds has not been probed. The importance of such internal disulfide bridges to Ig domain mechanics and stability (Fig. 1a) is put to experimental test here by forcibly extending Mel-CAM constructs with an AFM. This extension is done in the presence or absence of the reducing agent DTT.
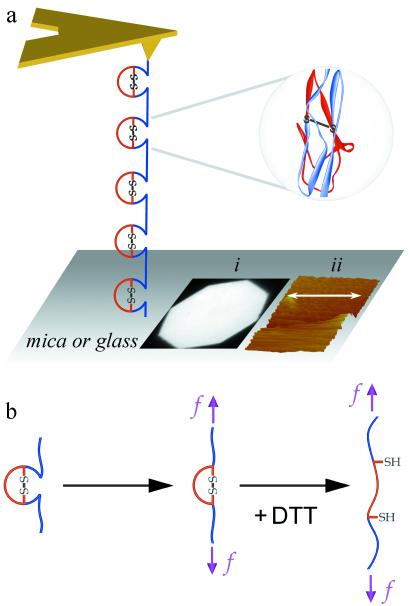
Figure 1.
Experimental AFM set-up and proposed reduction-coupled mechanism for forced unfolding of a multidomain Ig-CAM. (a) A single surface-to-tip adsorbed protein is shown with intact intradomain disulfide bonds. In separate experiments, the disulfide bonds are chemically reduced by DTT present in the surrounding medium. A prototypical ribbon structure of an Ig-CAM domain shows, in red, the disulfide-delimited cores. Protein is adsorbed to substrates of freshly cleaved mica or amino-silanized glass and nonadsorbed excess is washed away before AFM experiments. The use of mica or glass substrates here and elsewhere (13), rather than gold substrates, avoids artifactual formation of thio-gold bonds. Inset (i) shows fluorescently labeled Mel-CAM homogeneously adsorbed to mica at an estimated surface density of less than ≈10 protein molecules per 100 nm2. Inset (ii) shows a tapping-mode AFM image subsequent to contact-mode scraping over half the domain (arrow indicates direction); the image is consistent with no more than a monolayer of Mel-CAM molecules (1–2 nm thick) adsorbed to the surface. DTT does not affect the extent of fluorescent-Mel-CAM adsorption, and 1 mM DTT treatments of tenascin's FNIII domains (6) also indicate that DTT does not significantly alter the unfolding forces of Ig-like domains lacking intradomain disulfides. (b) Proposed unfolding pathway for a single domain under force. Under physiological conditions, only the first step can occur. In the presence of DTT, however, the partially unfolded domain exposes the disulfide bond to DTT so that, after reduction, the S–S-secluded region unfolds.
Materials and Methods
Protein Preparation.
The extracellular portion of Mel-CAM was expressed recombinantly and purified as described (10). For AFM experiments, protein was dialyzed into PBS and stored on ice. Immediately before use, any protein aggregates were removed by centrifugation at 166,000 g for 1 h at 2°C. Separate ultracentrifugation studies show Mel-CAM exists only as a monomer in solution. Fifty microliters of protein at 0.1 mg/ml was then allowed to adsorb for 15 min at room temperature onto a substrate of either freshly cleaved mica or amino-silanized glass coverslip. This was then rinsed with PBS and placed without drying under the head of the AFM with all measurements carried out in PBS. Lower protein concentrations generated minimal AFM results. For experiments with DTT, protein was incubated before adsorption with 1 mM DTT for 15 min; and the PBS used both for rinsing away unadsorbed protein and for measurements also contained 1 mM DTT. This tremendous molar excess of DTT relative to Mel-CAM translates to 1 DTT molecule per 10 nm3 of solution.
Dynamic Force Spectroscopy.
Two AFMs were used with similar results: (i) A Nanoscope IIIa Multimode AFM (Digital Instruments, Santa Barbara, CA) equipped with a liquid cell and (ii) an Epi-Force Probe from Asylum Research (Santa Barbara, CA). Sharpened silicon nitride (SiN3) cantilevers (Park Scientific, Sunnyvale, CA) with a nominal spring constant of kC = 10 pN/nm were commonly used, with some experiments performed by using 30 pN/nm cantilevers. kC was measured for each cantilever by manufacturer-supplied methods based on either forced resonance (11) or thermal fluctuations (12, 13). Agreement between measurements and the nominal values was invariably better than 30%. Initial calibration of the Epi-Force Probe by using titin adsorbed to glass showed good agreement with literature values for average force and extension. As needed, vertical displacements were calibrated to ± 2 nm with a VLSI Standard having an 18.0 nm step height (Model STS2–180P, VLSI Standard, San Jose, CA). AFM measurements were done at room temperature (≈23°C). For the reported results, tens of thousands of extension curves were analyzed with the aid of a custom visual analysis program; 100% of the data is presented.
Results and Discussion
An important initial perspective of the AFM results that follow is provided by solution measurements of disulfide bond reduction with and without chaotropic agents. First, under native conditions, the disulfide bridges in Mel-CAM's Ig domains are found to be intact, as expected, but also unaffected by either moderate (1 mM DTT) or strong (5 mM TCEP) reducing conditions as assessed by cysteine alkylation (Table 2). Under strong reducing plus denaturing conditions, however, Mel-CAM's cysteines are quantitatively alkylated. The results are indicative of obligate unfolding before disulfide reduction and suggest a multistep pathway for AFM-forced unfolding when DTT is in the surrounding solution (Fig. 1b). Furthermore, hydrodynamic radius measurements made by HPLC gel filtration separately show that if Mel-CAM is denatured without reduction, its domains will properly refold with high fidelity. In contrast, denaturation with reduction leads to misfolding of Mel-CAM with formation of large aggregates and intermolecular disulfide bonds. Also, alkylation of reduced cysteines in the unfolded state severely interferes with proper refolding (data not shown). Taken together, these solution measurements demonstrate that a disulfide bridge in an Ig-CAM domain plays a critical role in facilitating proper and stable folding.
Table 2.
Number of carboxymethyl-cysteines per protein after exposure to reducing agents (plus or minus chaotropic agents) followed by iodoacetamide, which alkylates free cysteines
| Mel-CAM | Modified Cys per molecule |
|---|---|
| +1 mM DTT/room temperature | 0.0 |
| +5 mM TCEP/room temperature | 0.0 |
| +10 mM DTT/Guanadine/37°C | 9.2 |
| +5 mM TCEP/Guanadine/37°C | 9.5 |
Because there are ten cysteines in Mel-CAM, the maximum possible number of modified cys is 10.0. 0.0 indicates no disulfide bonds were reduced within the measurement precision of ± 0.6 amino acids. TCEP is a significantly more potent reducing agent than DTT (23). The data show that any significant reduction of Mel-CAM's disulfide bonds requires at least partial unfolding of its Ig domains.
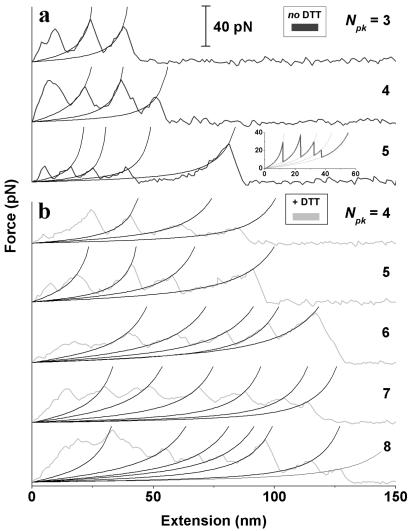
AFM-imposed extension of Mel-CAM, under both physiological and reducing conditions (Fig. 2 a and b, respectively), leads to a sawtooth pattern of unfolding peaks similar in form to cited results for titin and tenascin. In the physiological, oxidized state, the interval between Mel-CAM force peaks appears between 10 and 20 nm and is significantly shorter than the peak-to-peak spacing reported for titin and tenascin. In contrast, with DTT added (Mel-CAM+DTT), the distances between force peaks appear predominantly in the 20–30 nm range. This range is largely the same as that reported for titin and tenascin, and it also corresponds to a large fraction of any given Mel-CAM domain's full contour length (Table 1). However, the heights of the force peaks with either Mel-CAM or Mel-CAM+DTT are invariably much smaller than the 150–300 pN forces reported for either titin or FNIII domains under similar rates of extension (0.01–10 nm/msec) (6, 7, 14). Fits of the exponentially increasing portions of the force–extension curves are fit well by the entropic–elastic worm-like chain (WLC) model (15) and correspond to extension of an unfolded domain up to the point where a neighboring (folded) domain unfolds. Monte Carlo (MC) simulations of elastically coupled two-level systems (16) capture the basic sawtooth pattern of unfolding (Fig. 2a Inset) with suitably chosen parameters elaborated below for both partial and full unfolding.
Figure 2.
Representative force–extension curves for Mel-CAM, under either physiological (a) or reducing conditions (+DTT; b). Various numbers of unfolding peaks (Npk) are shown; fewer Npk are observed when Mel-CAM is oxidized. The peak-to-peak spacing also appears shorter with intact S–S. The exponential increase in force, for peaks beyond the first that may or may not be initial tip or chain desorption from the substrate, are fitted with the worm-like chain (WLC) equation (15): f(x) = (kBT/p)[(x/Lc) + 0.25/(1 − x/Lc)2 − 0.25]. A differential contour length between peaks averaged ΔLc = 16.5 nm for intact disulfides and ΔLc = 26.0 nm for reduced samples. For both oxidized and reduced samples, the persistence length that characterizes the minimal flexible length of an unfolded domain was determined to average P = 0.3–0.5 nm, consistent with reports of others (7). Dynamic MC simulations of unfolding, shown as Inset for oxidized Mel-CAM, were performed as prescribed by Rief et al. (14) by using a force-free, unfolding rate constant of kuo = 3 × 10−3 s−1 and a transition state length of xu = 1.6 nm (see Figure 6). Various contour lengths were taken from Table 1. For reference, titin Ig domain unfolding at much higher forces has previously been fit with kuo = 2.8 − 3.3 × 10−5 s−1 and xu = 0.25 − 0.3 nm (7, 14, 17)
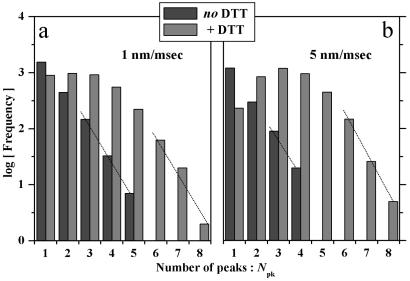
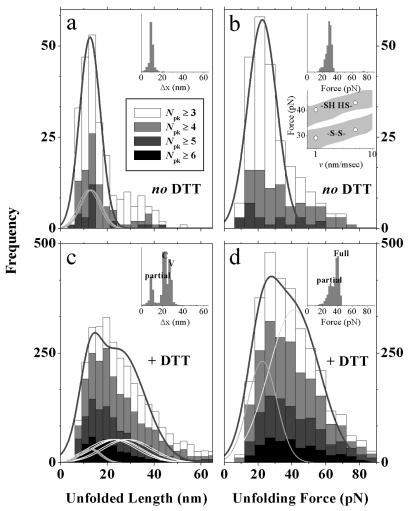
The number of visually counted force peaks, Npk, is a maximum of either 4 or 5 under oxidizing conditions (Fig. 3). The maximum depends on the substrate velocity v, but Npk appears consistent with Mel-CAM's pentadomain structure and, per domain, a two-state unfolding process. The first and last peaks could nonetheless represent desorption rather than unfolding (7). In addition, thermal noise on the force scale of Fig. 2 is significant as estimated from the root–product (kBT kC)1/2 ≈ 6 pN, where kC ≈ 10 pN/nm is the cantilever spring constant, kB is Boltzmann's constant, and T ≈ 296 K is the temperature. Strongly asymmetric peaks are nevertheless discernible above the random noise (Fig. 2), and so Npk would seem to reflect which domain(s) the randomly applied AFM tip adsorbs to and thus how many domains are subject to extension. Under reducing conditions, the finding that Npk often exceeds 5 but is never more than 7 or 8 suggests some amount of partial unfolding. A fuller analysis of the high-Npk tails of the distributions (see Fig. 3 legend) tentatively suggests 20–25% more ways of unfolding under reducing conditions. Such an estimate will prove reinforced by analyses below of distributions for both unfolding lengths and forces. Furthermore, histograms of total length (Fig. 4) are well bounded by single-chain contour length limits (Table 1). This appears consistent with the lack of any multi-meric Mel-CAM aggregates (see Materials and Methods).
Figure 3.
Frequency distributions for the number of peaks observed under oxidizing or reducing conditions where the imposed extension rate was either 1 nm/msec (a) or 5 nm/msec (b). Exponential decays at the higher values of Npk were fit with P(Npk) ∝ mNpk, where m may be interpreted as the number of ways that the AFM tip, after randomly contacting one domain in a chain, can extend and unfold a subset of all of the multiple domains between tip and substrate (before desorption). Extension of single chains is assumed to predominate under the adsorption conditions used; furthermore, chains are known from separate ultracentrifugation experiments to exist only as monomers in solution. For the intact disulfide bonds, mS-S | 1Hz = 4.6 and mS-S | 5Hz = 4.5, signifying, perhaps, that each of four or five domains has an equal chance of partially unfolding or not in a two-state process before domain detachment. Desorption, however, also seems likely to be accelerated at higher Npk because protein–tip or protein–substrate interactions are at a minimum in this limit. For the reduced disulfide bonds, sequential partial unfolding (see Figure 1b), in addition to full unfolding, appears implicated by higher values of mSH | 1Hz = 5.6 and mSH | 5Hz = 5.5.
Figure 4.
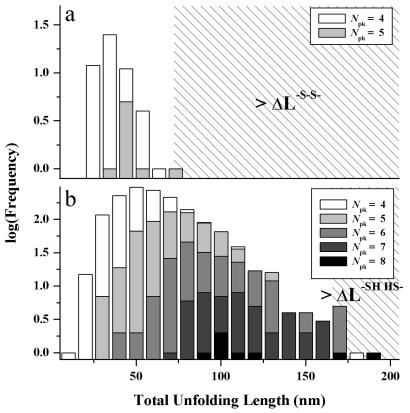
Histograms of full unfolding lengths for Mel-CAM under oxidizing (a) and reducing (b) conditions. Unfolding lengths summed between all identified unfolding peaks were cumulated for extension curves containing four and five peaks for the oxidized conditions and curves containing four, five, six, seven, and eight peaks for the reduced conditions. The right-most hatched regions correspond to the theoretical full unfolding lengths of the protein in either oxidized or reduced states, as given by the respective contour lengths minus a maximum of 4 nm per domain for the folded state. Consistent with extension of monomeric chains, < 0.25% of the data collected exceeds the theoretical limits for a single chain.
Histograms of unfolding lengths for oxidized and reduced Mel-CAM (Fig. 5 a and c, respectively) compare well with MC simulations based on either partial or full unfolding (Insets). However, experimental noise significantly broadens the experimental peaks. Fits of peak-to-peak length (lpk-pk) distributions were therefore separately performed with weighted sums of Gaussians (17) suitably constrained in their means by contour length proportionalities (Table 1). For oxidized Mel-CAM (Fig. 5a), the results differed very little from the overall mean but did entail fitting with
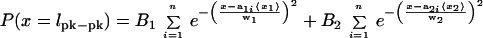 |
1 |
where n = 5 for the five domains, B2 ≡ 0, and B1, w1, 〈x1〉 are fit-optimized variables with the a1i specified relative to domain V1 (i.e., a11 = 49/49, a12 = 49/49, a13 = 46/49, a14 = 46/49, a15 = 51/49; see the number of amino acids in Table 1). The fit yields a set of proportional means {a11〈x1〉, a12〈x1〉, . . . } ranging from 12–13.6 nm that are all less than the mean increment in contour length of 16.5 nm (Fig. 2). The set of unfolding lengths also compares well with expected values of 17.0–18.1 nm (last column of Table 1) even when a folded domain size of about 3–4 nm is subtracted from these expected values (7). The set of small average extensions thus appears consistent with partial unfolding of Mel-CAM domains.
Figure 5.
Histograms of unfolding lengths and unfolding forces for Mel-CAM under oxidizing (a and b) and reducing (c and d) conditions. Extension curves with three or more, four or more, etc., peaks are cumulated separately, but only for those intervals beyond the first interval (l12) because the initial peak may well reflect chain desorption. By including the interval between the next-to-last and the last peak, a significant percentage (< 20%) of the lpk-pk are in excess of 25 nm, as apparent in the Npk = 5 extension curve in Fig. 2a. However, the distributions of unfolding force do not include the first or last peaks. The Upper Insets show corresponding histograms from MC simulations: Both partial unfolding and full domain unfolding simulations were separately carried out by using data in Table 1 and the parameters listed in the legends of Figs. 2 and 6. In c and d, simulated distributions for full and partial unfolding were scaled to reflect experimental proportions; and the measured distributions were fitted with Gaussians to account for measurement noise as described in the text. The Lower Inset in b shows the log-rate dependence for the unfolding force under oxidizing and reducing conditions. The experimental data points (circles) fall well within one standard deviation of the average disruption force calculated by simulation at various substrate velocities and indicated by the two gray bands.
Under reducing conditions, in contrast, an overall mean of 28 nm for lpk-pk is suggestive of full domain unfolding when compared with the average, reduced contour length of 37.1 nm (Table 1). However, a strong asymmetry in DTT-treated Mel-CAM's distribution, together with the earlier suggestion of partial unfolding under reducing conditions (Fig. 3), suggest a fit composed of both partial and full unfolding (i.e., B2 ≠ 0 in Eq. 1). This introduces additional a2i given by ratios of the full contour lengths relative, once more, to the domain V1. Holding fixed the w1 and 〈x1〉 determined in the fit of Fig. 5a, the relative proportion of full to partial unfolding, B2w2/(B1w1 + B2w2) = 0.80, indicates that full-chain unfolding predominates under reducing conditions. In addition, the second set of means {a21〈x2〉, a22〈x2〉, . . . } range from 21.8–29 nm and are again significantly less than the full contour lengths in Table 1 (i.e., 32.1–42.8 nm even when adjusted down by the folded domain size of 3–4 nm). Domain unfolding transitions thus occur with relative ease and well before full chain extension.
The disulfide-delimited, partial unfolding evident in the length distributions (Fig. 5a) occurs at a force of either ≈30 pN when averaged for the total distribution or 23 pN when fit with a single Gaussian (Fig. 5c; v = 1 nm/msec). Under reducing conditions, the unfolding force is shifted to a higher mean of 〈f2〉 = 41 pN, consistent with more work being done in full unfolding. This latter average was arrived at from a double-Gaussian fit analogous to Eq. 1 (n = 1, a1i = 1, x → f), but fixing 〈f1〉 at the oxidized value for partial unfolding. By this fitting, it was also determined that B2w2/(B1w1 + B2w2) = 0.73, which is essentially identical to the same ratio determined from the unfolding length distribution under reducing conditions (Fig. 5c) despite the noise-broadening apparent when compared with the “idealized” MC simulations (Fig. 5 Upper Insets). It is also important to recognize that broad distributions in Mel-CAM unfolding forces and intervals [like those of spectrin domains which unfold at similar forces (18)] reflect an effective domain softness. Nonetheless, for both oxidized and reduced Mel-CAM domains, the mean unfolding forces clearly increase with the rate of chain extension and are again well fit by the MC simulations (Fig. 5b Inset) for suitable values of kuo and xu (see Figs. 2 and 6).
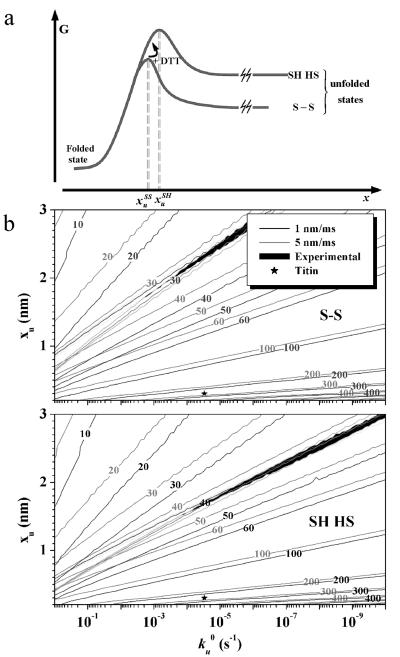
Figure 6.
Model and simulation results for unfolding of Mel-CAM. (a) Free energy profiles (in the absence of force) for unfolding of a domain. The kinetic barriers in these one-dimensional models are overcome in forced unfolding over a distance xu. DTT is assumed to effectively modulate the barrier height. In theory, xuSH ≤ xuSS. MC simulations in b and c follow the algorithm in ref. 16 and show isoforce curves for oxidized or reduced Mel-CAM extended at either 1 nm/ms (black lines) or 5 nm/ms (gray lines) for a broad range of the fitting parameters xu and kuo. Overlap between the simulations and rate-dependant measurements of peak forces (± 1 pN) are shown as black regions. Under either condition, the overlap region starts at xu ≈ 1.5 − 1.7 nm, indicating that xuSH ≈ xuSS. Parameters fit to titin unfolding (★) agree with previous fits to measured forces in the literature (7, 16) and suggest that suitable xuSS and xuSH for Mel-CAM are likely to be in the range 1.5–1.7 nm.
What seems most clear is that the forces for either full or partial unfolding of Mel-CAM's Ig domains are much smaller than those required to fully unfold titin's Ig domains or tenascin's homologous FNIII domains. The forces measured here are also small in comparison to the transition state forces revealed by molecular dynamics simulated unfolding of titin's Ig27 domain: two hydrogen bonds proximal to force-application were found disrupted at finit ≈ 50–100 pN (9).
MC simulations of domain unfolding introduced in Fig. 2 for oxidized Mel-CAM were expanded to simulate the effect of DTT by assuming that partial unfolding was an obligate but rapid step preceding reduction (Fig. 6a). A two-state process was thus deemed adequate for modeling the high force Gaussian of Fig. 5d. What emerges from simulation is a transition path-length of at least xu = 1.5–1.7 nm (see Fig. 6) for both oxidized and reduced Mel-CAM domains. A higher barrier height is found for reduction, however. A physical picture for a single domain, in the absence of DTT, would be that the AFM forces open some critical hydrogen bonds and hydrophobic interactions followed by complete unfolding up to the intact S–S bond. In the presence of DTT, the same critical pathway is followed, but DTT then very rapidly enters the partially unfolded state and reduces the disulfide, leading to the disruption of a few more hydrogen bonds and hydrophobic interactions that are critical for full-length unfolding.
Although the xu estimated from simulation for Mel-CAM significantly exceeds the 0.25–0.3 nm reported for titin and tenascin domains, the lower unfolding force for Mel-CAM domains over a longer unfolding distance cumulates to a net work (= force × extension) that is very similar in magnitude to that for the unfolding of titin or tenascin domains. In particular, for full unfolding of Mel-CAM (at v = 1 nm/msec), the average force acting over the transition state length is given by the product 〈f2〉 xu = 41 pN × 1.6 nm ≈ 16 kBT (with T ≈ 296 K). This compares very well with the 13–14 kBT that can be estimated from the unfolding of both tenascin FNIII and titin Ig domains. A further consistency check on the simulations is provided by an equality between the difference in transition state energies and the difference in the work required to unfold (i.e., −ΔΔGu* ≈ ΔWork*, where ΔΔGu* = Δ kBT ln(kuo) and ΔWork* = 〈f2〉 xu2 − 〈f1〉 xu1. For Mel-CAM in the reduced versus oxidized states, ΔWork* ≈ (11 pN) × 1.6 nm = 4.3 kBT and ΔΔGu* = [ln(5 × 10−5) − ln(3 × 10−3)] kBT = −4.1 kBT. Very similar calculations can be made for Ig27 and Ig28 domains that differ in 〈f〉 by up to 100 pN (17), depending on the particular recombinant construct. Hence, whether an unfoldable, modular domain is beta–sheet or alpha–helix based, dozens of hydrogen bonds invariably surround an essentially hydrophobic core and so a common transition energy scale, if not force scale, for force-nucleated unfolding is very understandable.
Conclusion.
Mechanical extension of Mel-CAM, as a prototypical Ig-CAM, shows that “hyper” extension through unfolding occurs under unusually mild forces and is significantly constrained by the disulfide bonds characteristic of Ig-CAM domains. This subdomain unfolding under mild forces is likely to cushion stresses on cell–cell contacts (Fig. 7) by stretching the response out over greater lengths (19). A similar proposal for the adhesive function of tenascin (6, 20) has been argued to be realistic under physiological rates of stressing where molecular forces can be estimated to reach 10–100 pN.
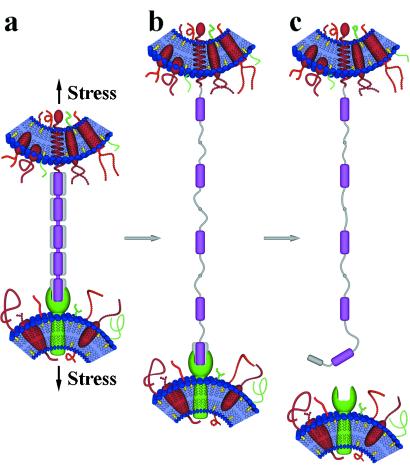
Figure 7.
Generic model of Ig-CAM domain unfolding in cell–cell contact sites. Under the stresses encountered during metastatic detachment, organogenesis, cell migration, etc. Additional extensibility and elasticity is provided by partial unfolding of Ig domains that bridge two cell membranes. (a) Model depicting heterotypic association between the N-terminal domain of a five-Ig domain CAM and a receptor on an opposing cell. (b) Under moderate stress, up to four domains could partially unfold without breaking cell–cell contact. (c) When tensile forces are sufficient to unfold domains directly involved in receptor associations, cell adhesion would be disrupted or at least moderated. The core regions delimited by the disulfides are purple. These regions may act as nuclei for domain refolding when tensile stresses are removed or reduced. Applying the model to Mel-CAM, which interacts with a different type of receptor but efficiency of unknown identity and at unknown site(s) on Mel-CAM, the clear suggestion is that molecular extensibility contributes to adhesive efficiency in metastases.
The present data are also readily extrapolated to suggest, generically, that forced unfolding of domains directly involved in receptor binding disrupts or modulates cell–cell attachments (Fig. 7c) and thereby sensitizes CAM signaling and protein associations to stress. Such mechanisms for modulating intermolecular interactions could well explain why selected CAM Ig domains (e.g., domains in ICAM-1, ICAM-2, and human CD2) have evolved a second disulfide bridge at or very near the N- or C-terminal loops: two disulfides would tend to more tightly regulate unfolding under tension. The model predicts that domains with two disulfide bonds play particularly critical roles in CAM function where resistance to tensile forces is required. The model further predicts that diversity among the many CAMs in the number of tandem domains (from just one to at least seven Ig domains) is a critical determinant of the “elastic reach” of these CAMs [i.e., the distance that two cell membranes can be displaced apart without breaking the cell–cell adhesive contact (Fig. 7)]. The folded subdomain protected by the disulfide bond thus limits the degree of molecular extension, but it is also likely to nucleate facile refolding, postextension, of the subdomain exterior to the disulfide. Detailed studies of Ig domain folding prove consistent by suggesting that cysteine contact and bridge formation is rate-limiting (21), although the role of less-conserved tryptophans and hydrophobic residues near disulfide bonds in Ig domains (22) could also prove crucial to nucleated refolding. Finally, whether reductases modulate disulfide bond formation in microenvironments around cells, and thereby influence adhesive functions such as metastatic potential or tissue architecture, is highly speculative. What does seem definitive, in light of prior AFM results with titin Ig and tenascin FNIII domains, is that the Ig-like fold shared with CAMs has evolved to respond in mechanically diverse ways.
Acknowledgments
We thank Prof. J. Fernandez for discussions of AFM methods, Prof. E. A. Evans for early discussions of the rate dependence in force spectroscopy measurements, and Dr. David Tees for helpful comments on the manuscript. We thank P. Dalhaimer for the custom visual analysis program, and R. Law for help in data analysis. This work was supported by National Institutes of Health and National Science Foundation grants (to D.E.D. and D.W.S.).
Abbreviations
- AFM
atomic force microscope (microscopy)
- CAM
cell adhesion molecule
- Mel-CAM
melanoma CAM
- FNIII
fibronectin type-III
- MC
Monte Carlo
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.031409698.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.031409698
References
- 1.Thiery J P. In: Guidebook to Extracellular Matrix, Anchor, and Adhesion Proteins. Kreis T, Vale R, editors. New York: Oxford Univ. Press; 2000. pp. 125–128. [Google Scholar]
- 2.Shih I M, Elder D E, Speicher D, Johnson J P, Herlyn M. Cancer Res. 1994;54:2514–2520. [PubMed] [Google Scholar]
- 3.Shih I M, Kurman R J. Lab Invest. 1996;75:377–388. [PubMed] [Google Scholar]
- 4.Shih I M, Hsu M Y, Palazzo J P, Herlyn M. Am J Pathol. 1997;151:745–751. [PMC free article] [PubMed] [Google Scholar]
- 5.Pickl W F, Majdic O, Fischer G F, Petzelbauer P, Fae I, Waclavicek M, Stockl J, Scheinecker C, Vidicki T, Aschauer H, et al. J Immunol. 1997;158:2107–2115. [PubMed] [Google Scholar]
- 6.Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 7.Rief M, Gautel M, Oesterhelt F, Fernandez J M, Gaub H E. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 8.Lu H, Schulten K. Proteins. 1999;35:453–463. [PubMed] [Google Scholar]
- 9.Marszalek P E, Lu H, Li H B, Carrion-Vazquez M, Oberhauser A F, Schulten K, Fernandez J M. Nature (London) 1999;402:100–103. doi: 10.1038/47083. [DOI] [PubMed] [Google Scholar]
- 10.Shih I M, Speicher D, Hsu M Y, Levine E, Herlyn M. Cancer Res. 1997;57:3835–3840. [PubMed] [Google Scholar]
- 11.Sader J E, Larson I, Mulvaney P, White L R. Rev Sci Instrum. 1995;66:3789–3798. [Google Scholar]
- 12.Walters D A, Cleveland J P, Thomson N H, Hansma P K, Wendman M A, Gurley G, Elings V. Rev Sci Instrum. 1996;67:3583–3590. [Google Scholar]
- 13.Hutter J L, Bechhoefer J. Rev Sci Instrum. 1993;64:1868–1873. [Google Scholar]
- 14.Li H, Oberhauser A F, Fowler S B, Clarke J, Fernandez J M. Proc Natl Acad Sci USA. 2000;97:6527–6531. doi: 10.1073/pnas.120048697. . (First Published May 23, 2000; 10.1073/pnas.120048697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marko J F, Siggia E D. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 16.Rief M, Fernandez J M, Gaub H E. Phys Rev Lett. 1998;81:4764–4767. [Google Scholar]
- 17.Carrion-Vazquez M, Marszalek P E, Oberhauser A F, Fernandez J M. Proc Natl Acad Sci USA. 1999;96:11288–11292. doi: 10.1073/pnas.96.20.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rief M, Pascual J, Saraste M, Gaub H E. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 19.Smith B L, Schaffer T E, Viani M, Thompson J B, Frederick N A, Kindt J, Belcher A, Stucky G D, Morse D E, Hansma P K. Nature (London) 1999;399:761–763. [Google Scholar]
- 20.Krammer A, Lu H, Isralewitz B, Schulten K, Vogel V. Proc Natl Acad Sci USA. 1999;96:1351–1356. doi: 10.1073/pnas.96.4.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorch M, Mason J M, Clarke A R, Parker M J. Biochemistry. 1999;38:1377–1385. doi: 10.1021/bi9817820. [DOI] [PubMed] [Google Scholar]
- 22.Ioerger T R, Du C, Linthicum D S. Mol Immunol. 1999;36:373–386. doi: 10.1016/s0161-5890(99)00032-2. [DOI] [PubMed] [Google Scholar]
- 23.Han J C, Han G Y. Anal Biochem. 1994;220:5–10. doi: 10.1006/abio.1994.1290. [DOI] [PubMed] [Google Scholar]