Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) is a molecular DNA damage sensor that catalyzes the synthesis of the complex biopolymer poly(ADP-ribose) [PAR] under consumption of NAD+. PAR engages in fundamental cellular processes such as DNA metabolism and transcription, and interacts non-covalently with specific binding proteins involved in DNA repair and regulation of chromatin structure. A factor implicated in DNA repair and chromatin organization is the DEK oncoprotein, an abundant and conserved constituent of metazoan chromatin, and the only member of its protein class. We have recently demonstrated that DEK, under stress conditions, is covalently modified with PAR by PARP-1, leading to a partial release of DEK into the cytoplasm. Additionally, we have also observed a non-covalent interaction between DEK and PAR, which we detail in the present work. Using sequence alignment, we identify three functional PAR-binding sites in the DEK primary sequence and confirm their functionality in PAR binding studies. Furthermore, we show that the non-covalent binding to DEK is dependent on PAR chain length as revealed by an overlay blot technique and PAR EMSA. Intriguingly, DEK promotes the formation of a defined complex with a 54mer PAR (KD=6 × 10−8 M), whereas no specific interaction is detected with a short PAR chain (18mer). In stark contrast to covalent poly(ADP-ribosyl)ation of DEK, the non-covalent interaction does not affect the overall ability of DEK to bind to DNA. Instead the non-covalent interaction interferes with subsequent DNA-dependent multimerization activities of DEK, as seen in South-Western, EMSA, topology and aggregation assays. In particular, non-covalent attachment of PAR to DEK promotes the formation of DEK-DEK complexes by competing with DNA binding. This was seen by the reduced affinity of PAR-bound DEK for DNA templates in solution. Taken together, our findings deepen the molecular understanding of the DEK-PAR interplay and support the existence of a cellular “PAR code” represented by PAR chain length.
Keywords: Chromatin, Post-translational Modification, DNA-repair, Oncogene, Poly(ADP-ribose) Polymerase
Poly(ADP-ribosyl)ation is a dramatic posttranslational modification of proteins carried out by the superfamily of poly(ADP-ribose) polymerases (PARPs) (1, 2). PARP-1 is the best understood member of this class of enzymes and is responsible for about 90% of cellular poly(ADP-ribose) (PAR) formation after DNA damage (3). PARP-1 is crucial for the maintenance of genomic stability and plays an important role during DNA repair, in particular base excision repair (BER) (4–12). Binding to DNA strand breaks activates PARP-1, which catalyzes the transfer of ADP-ribose moieties onto acceptor proteins under the consumption of NAD+. The PAR which is thus formed is a highly complex biopolymer and was shown to interact in a non-covalent fashion with various proteins involved in DNA damage checkpoint control and repair, and most likely also influences other biological processes (6). In turn, hydrolysis of PAR by the enzyme poly(ADP-ribose) glycohydrolase (PARG) also critically influences genomic stability and cellular survival (13, 14). PAR binding is mediated by a consensus motif, which has been identified in crucial domains of many proteins and may therefore interfere with their respective functions (15). Lately, a zinc-finger motif was described by Ahel and co-workers that displays specific PAR binding activity and is present in some DNA repair-associated proteins (16). We could recently show that the well known PAR-binding protein p53, a tumor suppressor protein with functions in double strand break repair (17), exhibits a high binding affinity to PAR, with a KD in the low nanomolar range (18). Non-covalent interaction between PAR and p53 has been demonstrated to inhibit both the sequence- and non-sequence-specific DNA-binding of p53 in a PAR-dependent manner (19). Importantly, several BER proteins harbor the PAR consensus motif, e.g. XRCC1, DNA ligase III and DNA polymerase ε, underscoring the role of PAR in the spatio-temporal organization of BER (15, 20, 21). Very recently, interplay of the protein kinase ATM, an early DNA damage sensor, and PAR has been described, indicating that rapid and transient PAR formation may directly or indirectly activate the ATM signaling pathway (22).
Recently, we have provided evidence that DEK, an abundant non-histone chromosomal factor (23), is a PARP-1 substrate implicated in the repair of DNA strand breaks, assigning DEK a function in PAR-dependent maintenance of genomic stability (24). Human DEK was initially discovered in a chimeric fusion protein with the nucleoporin CAN/NUP214 in a subset of patients with acute myeloid leukemia (AML) (25). Overexpression of DEK mRNA and DEK protein has subsequently been identified in a growing number of aggressive human tumors (26–29). Furthermore, high levels of DEK support cell immortalization and inhibit both senescence and apoptosis (30, 31), and DEK overexepression itself was shown recently to be sufficient for HRAS-driven epithelial hyperplasia induction and epithelial transformation, classifying it as a bona fide oncogene (32, 33). We further demonstrated that DEK is a true oncoprotein and a potential target for chemotherapy in malignant melanoma (28). Interestingly, DEK is also associated with several autoimmune disorders such as juvenile rheumatoid arthritis and systemic lupus erythematosus, thereby representing a major auto-antigen (34 ).
The amino acid (aa) sequence of human DEK (375 aa) harbors a nuclear localization sequence (NLS) and four highly acidic stretches, shown to have inhibitor of histone acetyltransferase activity (INHAT) in vitro (35). DEK harbors two structurally and functionally distinct DNA-binding domains (36–40). The central domain, spanning aa 87–187, represents an unprecedented form of the DNA-binding motif SAP-box, which consists of two SAP-folds, with each of these folds being able to independently interact with dsDNA and to stimulate DNA-dependent protein-protein-interactions (36, 41, 42). This domain mediates the characteristic DNA-binding and folding feature of DEK, i.e., a predominantly structure-specific (four-way-junction, supercoiled, distorted DNA) DNA-binding activity, resulting in a general compaction of chromatin as well as DNA templates and the introduction of positive superhelical turns into closed circular DNA and chromatin templates in vitro (36, 38, 39, 42, 43). Interestingly, a SAP domain has also been identified via a PSI-BLAST search in plant PARP-1 (44). Similar to human DEK, PARP-1 is also capable of binding to unusual DNA structures such as cruciforms and loops (45, 46). A second DNA-binding domain, located in the C-terminal region of DEK (aa 250–350), represents a winged-helix motif found in transcription factors of the E2F-family, and upon phosphorylation stimulates self-association of DEK molecules and/or other yet to be elucidated chromatin-associated factors (36, 37, 47).
DEK has been linked to a variety of intracellular activities, as it has been implicated in DNA replication (43), RNA processing (48), as a positive (28, 49, 50) or negative (31, 51–54) regulator of transcription, and as participating in DNA double strand repair (24). DEK is a target of a plethora of post-translational modifications, and these most likely regulate or integrate its various intra- and extracellular activities. In fact, phosphorylation (24, 37, 55), acetylation (56) and poly(ADP-ribosyl)ation (24, 52) were shown to affect DEK’s sub-cellular and sub-nuclear distribution and interaction partners.
Strikingly, DEK shares several functional similarities with HMGB1 (high mobility group B protein 1) (40), which is a known acceptor for covalent poly(ADP-ribosyl)ation (57). Moreover, both PARP-1 and DEK have been detected in a HeLa cell chromatin fraction enriched for the variant histone macroH2A, and were found to co-reside on chromatin fragments released by micrococcal nuclease, underscoring their close connection (24, 58). Furthermore, we showed that DEK is not only a target for covalent poly(ADP-ribosyl)ation, but also binds PAR in a non-covalent fashion (24). We have demonstrated that the binding affinity of PAR to specific proteins is very high and that this interaction is very selective with respect to PAR chain length and the binding partner (18). Here, we identify specific PAR binding motifs within the DEK primary sequence and quantify this non-covalent interaction as a function of chain length. Moreover, we address the question of whether the non-covalent binding of PAR to DEK affects the well-described functional properties of DEK.
MATERIAL AND METHODS
Materials
Human PARP-1 was expressed in Sf9 insect cells and purified as described (18). His-tagged full-length DEK as well as DEK fragments were also produced in insect cells and purified as previously described (36, 37). It is important to note that recombinant DEK purified from the baculovirus system is highly phosphorylated. Throughout this manuscript, recombinant His-DEK was either used in its phosphorylated form (Figure 1–3), or in its dephosphorylated form (after treatment with λ-phosphatase), for DNA binding studies (Fig. 4–6). Phosphorylation moderately reduced the overall PAR-binding capacity of DEK, however, did not interfere with observed preferential binding selectivity of DEK for long-chained PAR polymer (see Fig. 4A, and Supplementary Fig. S2). The DEK fragment 68–226 was expressed and purified from E. Coli as previously described (42), and is therefore un-phosphorylated. (24, 37(59). DEK peptides 195–222, 158–181, and 99–119 were custom-synthesized by Coring System Diagnostix GmbH, and peptides 329–352 and 314–334 were synthesized using an Fmoc/tBu based solid phase method. Affinity-purified polyclonal DEK antibodies were used as reported (24, 37). Mouse monoclonal antibody 10H, that recognized PAR, was purified from culture supernatant of 10H hybridoma cells (59).
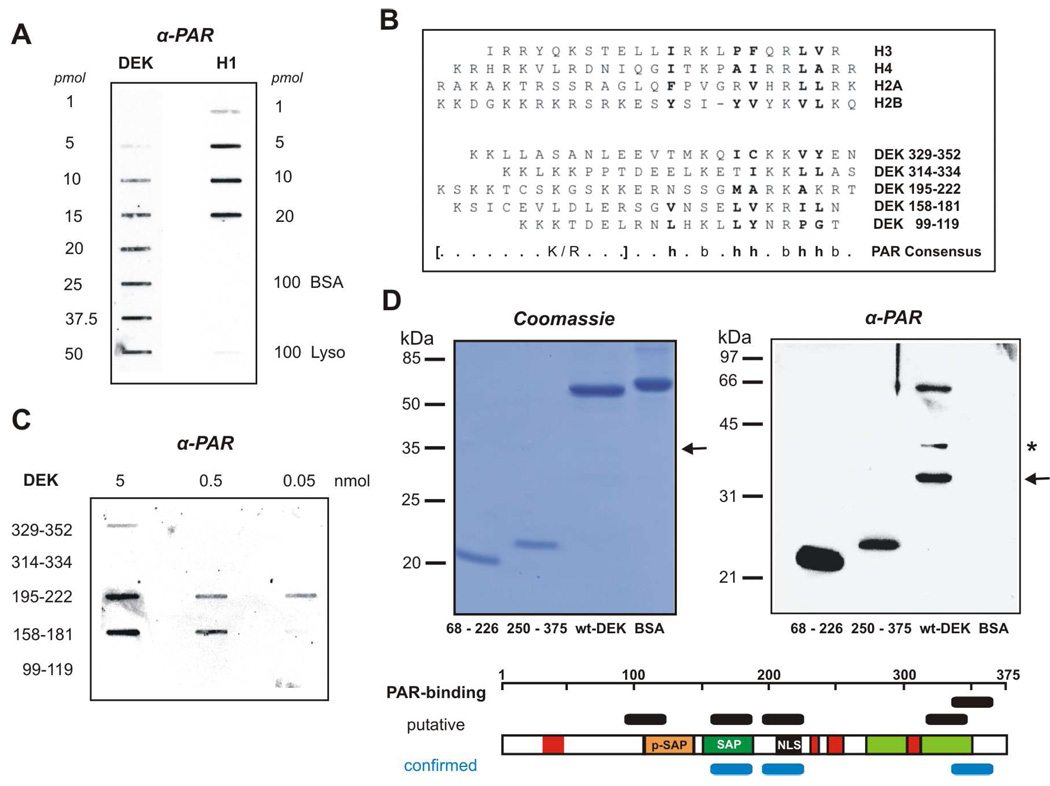
Figure 1. Mapping of PAR-interaction modules in the DEK protein.
(A) DEK specifically interacts with PAR. Increasing amounts of recombinant DEK and histone H1 (positive control) as well as BSA, and lysozyme in excess (negative controls) were slot-blotted onto a nitrocellulose membrane. Following incubation with PAR (1 µM) the membrane was treated with high-salt washes to disrupt nonspecific binding. Bound polymer was immuno-detected using specific PAR antibodies (10H). (B) Alignment of the PAR binding consensus sequence with the DEK amino acid sequence. Top: PAR binding motifs identified in different histones according to Pleschke et al. (15). Bottom: Putative PAR binding motifs in DEK found by alignment according to the consensus sequence: h: hydrophobic, b: basic amino acids. (C) PAR binding to DEK peptides. Decreasing amounts of DEK peptides synthesized according to the alignment were slot-blotted onto a nitrocellulose membrane. After incubation with biotinylated PAR (50 nM) the membrane was treated with high-salt washes to disrupt nonspecific binding. Bound polymer was visualized using streptavidin-POD. (D) PAR binding analysis using His-tagged DEK fragments. The respective fragments (aa 68–226; aa 250–375; aa 1–375) and BSA (BSA) as negative control were subjected to 15% SDS-PAGE and visualized by Coomassie staining (100 pmol, left panel), or analyzed using a PAR overlay blot (15 pmol, right panel). After semi-dry blotting and incubation with free biotinylated PAR (0.4 µM) the bound polymer was visualized as described in (A). The described 35 kDa breakdown product of wt-DEK is denoted by an arrow. An additional intermediate breakdown product, capable of PAR binding, is indicated by an asterisk. The scheme summarizes the putative (black) and experimentally confirmed (blue) PAR-binding domains in the DEK amino acid sequence (red boxes: acidic domains; orange: pseudo-SAP box; green: SAP-box, black: nuclear localization sequence; light-green: second DNA binding/ multimerization domain).
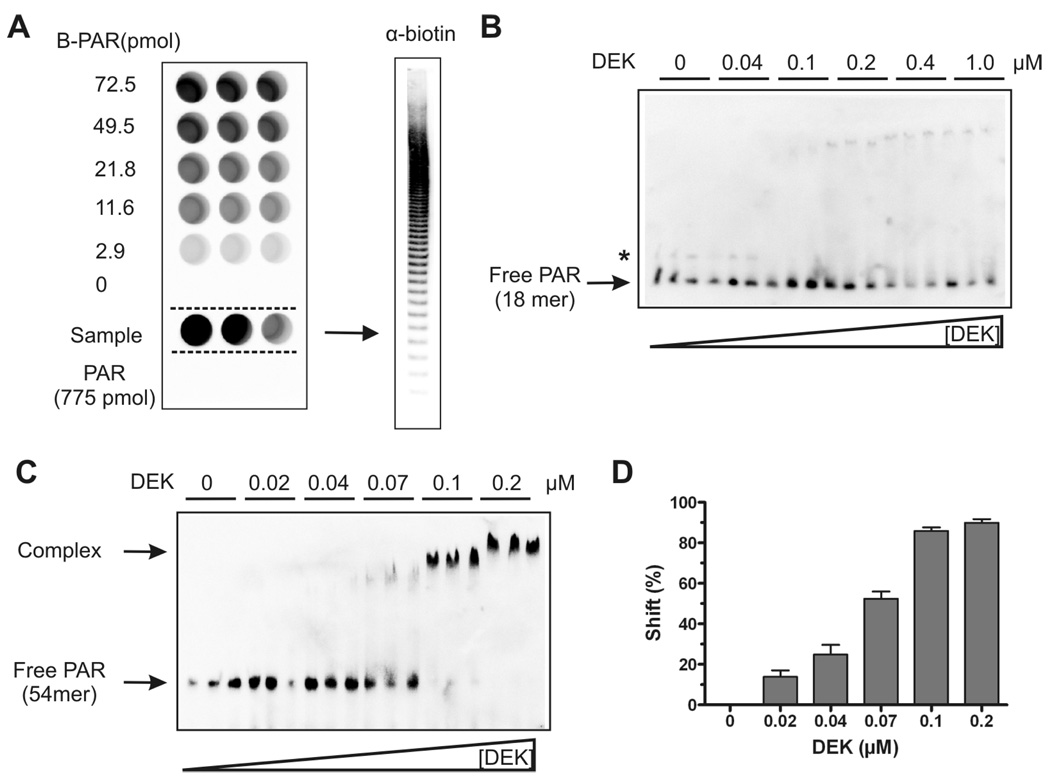
Figure 3. Specific End-labeling of PAR chains and PAR EMSA.
(A) Characterization of terminal biotin labeling by neutravidin-ELISA. Biotinylated PAR was captured in the neutravidin-coated wells (left panel) and detected using PAR-specific antibodies (10H). B-PAR: biotinylated PAR standard; end-labeled PAR sample using biocytin hydrazide (171/86/17 pmol from left to right). The end-labeled PAR sample was also analyzed by a native 20% PAGE followed by semi-dry transfer on a nylon membrane. Biotinylated PAR chains were visualized by streptavidin-POD.
(B) and (C) Interaction of size-fractionated PAR and DEK in solution. Biotin-labeled PAR of defined chain length was incubated with increasing concentrations of DEK and subjected to native PAGE followed by semi-dry blotting. Bound and free polymer was detected using streptavidin-POD. Free and complexed PAR is indicated by an arrow. In (B), a minor cross-contamination with PAR of longer chain length is indicated by an asterisk. (B) Binding of DEK to a short PAR 18mer (250 fmol/lane). (C) Binding of DEK to long PAR chains (54mer, 125 fmol/lane). (D) Quantitative evaluation of DEK gel shifts. Shift (%) was calculated as follows: signal intensity complexed PAR/(complexed + free PAR). Data are expressed as mean + SEM of triplicates from two independent experiments.
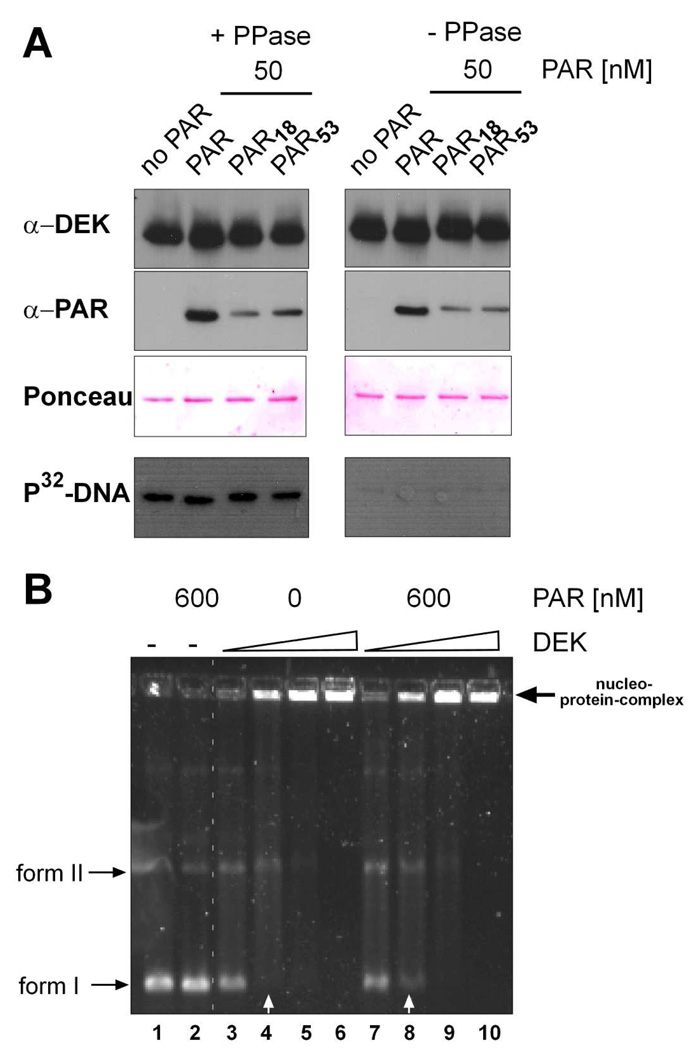
Figure 4. Non-covalent binding of PAR does not affect overall DNA-binding of DEK.
(A) PAR-overlay. 400 ng dephosphorylated (+ PPase, left panel) or phosphorylated (− PPase, right panel) His-DEK per lane was blotted to a nitrocellulose membrane, cut into strips and incubated with 50 nM of un-fractiontated PAR, short- PAR (18mer), or long-chained PAR (53mer) overnight (see also Supplementary Fig. S2). Strips were subsequently probed with DEK (α-DEK) or PAR-specific antibodies (α-PAR, 10H). As a loading control, Ponceau S staining is shown. Lowest panel: South-Western analysis: after blocking and renaturating, the individual strips were incubated with radioactively labeled SV40 DNA prior to detection of binding by autoradiography. (B) EMSA. 30 ng SV40 DNA were incubated with increasing amounts (100, 200, 400 or 800 ng) of either untreated or PAR-treated DEK, analyzed on a 0.6% agarose gel, and visualized by SYBRGold staining. Form I: supercoiled DNA; form II: relaxed DNA. The arrow indicates DEK-specific large nucleo-protein complexes, which remain in the wells of the gel due to their high molecular weight. White arrows point to quantitative differences in the presence of form I DNA in samples with or without prior PAR treatment.
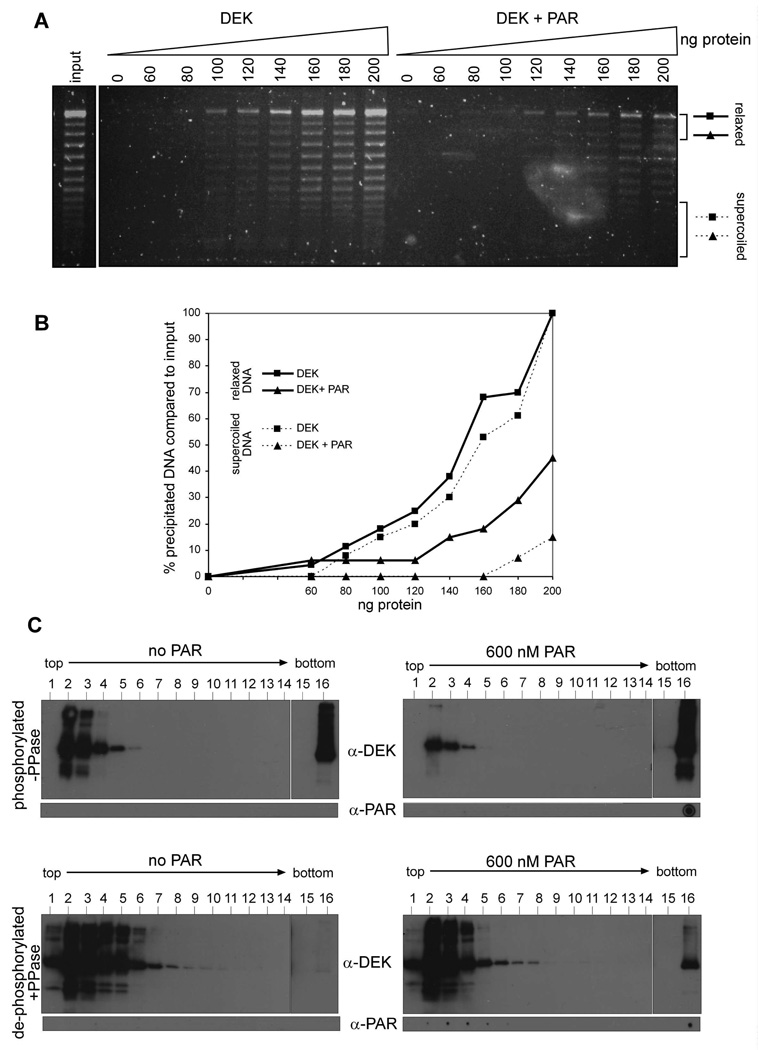
Figure 6. PAR attenuates DEK’s affinity for highly supercoiled DNA templates by competitively stimulating the self-association activity of DEK molecules.
(A) DEK aggregation assay: 0, 60, 80, 100, 120, 140, 160, 180 or 200 ng of His-DEK were incubated with 20 ng of partially relaxed SV40 DNA in the presence or absence of 600 nM PAR in a total volume of 30 µl. After 20 min incubation at RT, samples were centrifuged for 15 min and pellets (i.e., precipitated material) were analyzed by 0.8% agarose gel electrophoresis and SYBRGold staining.
(B) DEK-dependent precipitation of relaxed DNA topoisomeres (indicated by solid lines), or strongly superhelical topoisomers (dashed lines) was further analyzed by densitometry and is expressed as percentage of precipitated DNA compared to input. (C) Sucrose density gradient analysis. Phosphorylated (−PPase) or dephosphorylated (+PPase) His-DEK was incubated in the presence (right panels) or absence (left panels) of 600 nM unfractionated PAR, and subjected to sedimentation analysis on 5–40 % sucrose density gradients. Individual fractions of the gradients were either analyzed by SDS-PAGE and immunoblotting for DEK (α-DEK), or by dot-blot using PAR-specific antibodies (α-PAR).
In vitro PAR synthesis and purification
PAR was synthesized essentially as described (18). Briefly, 150 nM human PARP-1 was incubated with 1 mM NAD+, 1 mM DTT, 60 µg/ml histone H1, 60 µg/ml histone type IIa and 50 µg/ml octameric “activator” oligonucleotide GGAATTCC in a total volume of 20 ml containing 1x reaction buffer (100 mM TrisHCl pH 7.8, 10 mM MgCl2). After 15 min the PAR produced was precipitated by addition of 20 ml 20% TCA and the pellet was washed several times with ice-cold 99.8% ethanol. PAR was further purified according to Malanga et al. (60), and finally precipitated with ethanol overnight.
Terminal biotinylation and neutravidin-ELISA
PAR was dissolved in sodium acetate buffer pH 5.5 containing 4 mM biocytin hydrazide and biotinylated under reductive amination conditions for 8 h as described (18). Following dialysis and ethanol precipitation, PAR concentration was determined using UV-absorbance at 258 nm (61). Successful terminal labeling of PAR chains was checked using a neutravidin-ELISA. Biotin-labeled PAR samples diluted in 50 mM NaHCO3 pH 7.5 were transferred to ELISA plates and incubated for 1 hour at room temperature. Captured biotinylated PAR was detected with primary antibody 10H (4 µg/ml) in conjunction with a secondary peroxidase-conjugated anti-mouse IgG (DakoCytomation, 1:2,000) and visualized in a Tecan GeniosPlus ELISA reader. A biotinylated PAR standard was synthesized in the presence of NAD+ and 6-biotin-17-NAD+ (ratio 15:1) resulting in the incorporation of biotinylated ADP-ribose units into the polymer (62). An excess of unlabeled PAR served as a specificity control to rule out non-specific PAR binding. In addition, terminal biotin-labeling of PAR chains was assessed after electrophoretic separation and semi-dry blotting by incubation with streptavidin-POD (1:2,000).
HPLC fractionation of biotinylated PAR
Separation of biotinylated PAR was carried out on a Shimadzu LC-8A HPLC system equipped with a semi-preparative DNA Pac PA100 column (DIONEX). PAR was eluted according to chain length using a multistep NaCl gradient in 25 mM Tris-HCl pH 9.0 and collected manually after UV-detection at 258 nm (18, 63). Biotin-labeled ADP-ribose polymers were characterized on modified sequencing gels (64) and visualized using GELCODE Color silver stain (Pierce) as described (60).
PAR overlay blot
Increasing amounts of recombinant DEK (1 – 50 pmol), histone H1 (1 – 20 pmol) as well as BSA and lysozyme (100 pmol each) were vacuum-aspirated onto a nitrocellulose membrane (Amersham Biosciences) using a slot-blot manifold (Schleicher & Schuell). To map the binding of PAR to specific DEK domains, His-tagged DEK fragments (15 pmol for PAR overlay blot; 100 pmol for Coomassie staining) were separated by 15% SDS-PAGE and stained by Coomassie or transferred onto a nitrocellulose membrane by semi-dry blotting. Membranes were incubated with Tris-buffered saline -Tween 20 (TBS-T) containing 1 nmol (0.4 µM) of purified PAR for 1 h. Membranes were subsequently washed in TBS-T - 1 M NaCl and blocked with 5% (w/v) skim milk powder in TBS-T. Detection of bound PAR was performed using 10H antibodies and secondary antibodies (goat α-mouse/HRP). Bands were visualized in the FujiLAS1000 device using enhanced chemiluminescence and blots were evaluated with AIDA software (Raytest).
Equimolar amounts of synthetic DEK peptides were dissolved in PBS and slot-blotted with the indicated concentrations onto a nitrocellulose membrane. After incubation with biotinylated PAR (50 nM in TBS-T), the membrane was washed three times with 1 M NaCl in TBS-T and blocked 1 h in 5% skim milk powder in TBS-T. Subsequently, PAR binding was detected after incubation with streptavidin-POD (1:5,000) in TBS-T for 1 h employing enhanced chemiluminescence.
To assess the binding of separated PAR chains to immobilized DEK, 15 pmol of recombinant protein was transferred onto nitrocellulose membranes using a slot-blot apparatus. The membranes were cut into strips and incubated with 500 pmol (0.25 µM) of the respective ADP-ribose fraction in TBS-T at 4 °C overnight. After high-stringency wash steps in TBS-T - 1 M NaCl, strips were blocked and bound polymer was detected using 10H antibodies as described above.
PAR sequence alignment
Putative PAR-binding sequences in DEK were aligned with the consensus PAR binding motif as reported previously (15).
Avidin-affinity purification of biotinylated PAR
End-biotinylated separated PAR chains were affinity-purified using SoftLink™ Soft Release Avidin Resin (Promega) before being used in EMSA studies. Briefly, biotinylated polymer was diluted to 2 ml in bind & wash (BW) buffer comprising 50 mM Tris-HCl pH 8.0, 50 mM NaCl and loaded onto the avidin column. After collection of the flow-through, the column was rinsed with 6 ml BW buffer, and bound biotinylated PAR chains were gently eluted in 1 ml steps using a solution of 5 mM D(+)-biotin. The purification process was analyzed by native 20% PAGE and end-labeled PAR was detected using streptavidin-POD and enhanced chemiluminescence. Concentration of affinity-purified PAR was assessed by native PAGE using a biotinylated 49-mer DNA oligonucleotide (Invitrogen) as the standard.
PAR EMSA
To characterize PAR-protein complexes in solution, we have recently developed a PAR EMSA (18). Varying amounts of recombinant His-DEK were incubated in an appropriate volume of 10 mM Tris-HCl pH 7.4, 1 mM EDTA for 10 min at 25 °C before affinity-purified PAR of defined chain length (18mer and 54mer, respectively) was added. Complex formation was allowed to proceed for 20 min at 25 °C to reach equilibrium. Subsequently, the reaction mixture was supplemented with 10x loading dye resulting in a final volume of 25 µl. The samples were subjected to a native 5% PAGE for 2.5 h at 160 V to separate free and bound ADP-ribose polymer. Thereafter, samples were blotted onto a nylon membrane by semidry transfer at 20 V for 50 min followed by a heat-fixation at 90 °C for 1 hour. After blocking with 2% BSA in TBS-T for 1 hour, biotinylated ADP-ribose chains were detected by incubation with streptavidin-POD (1:2,000 in blocking solution) for 1 hour. Blots were visualized using a FujiLAS 1000 imager, and quantification was performed using AIDA Software. Shift (%) was determined as follows: signal intensity complexed PAR / signal intensity (complexed + free PAR). The data obtained were analyzed using GraphPad Prism 4 software, and KD values were calculated by using a sigmoidal dose-response curve with variable slope. Curves were fitted using non-linear regression (see Supplementary Figure S5).
EMSA, topology, aggregation assay and South-Western analyses with recombinant His-DEK
EMSA and topology assays were carried out as described (24, 36). Recombinant His-DEK, purified from insect cells, was dephosphorylated with λ-phosphatase (New England Biolabs) prior to utilization. His-DEK was incubated with either un-fractionated PAR or PAR of indicated chain length and concentrations as specified for the individual experiment for 20 min at RT prior to the start of the reaction. Individual samples were further supplemented with indicated PAR concentrations throughout the reactions. South-Western analyses were performed to investigate DNA-binding of DEK upon non-covalent PAR incubation and were carried out as described recently (37). Briefly, indicated amounts of His-DEK were separated on a 10% SDS-PAGE and blotted to a nitrocellulose membrane. Individual lanes were cut into strips and incubated overnight in TBS-T with PAR at indicated concentrations in a total volume of 2 ml. Following three wash-steps with TBS-T containing 1 M NaCl, the strips were blocked with 5% skim milk powder in 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM DTT and 1 mM EDTA for 1 hour at RT. As a probe, HinfI-digested, end-labeled SV40 DNA was used. After 1 hour incubation, the individual strips were washed collectively five times with 10 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM DTT and 1 mM EDTA and subjected to autoradiography followed by immunoblotting. Aggregation assays, a measure for DNA-dependent multimerization of SAP-box proteins, were performed as reported previously (24), using PAR incubation procedures as described above. Two-dimensional DNA topology assays were carried out to identify the orientation of supercoils introduced and were essentially performed as described (42).
Sucrose density gradient analysis
Sedimentation analysis of DEK was carried out as described (36, 37). Briefly, 1500 ng (31 pmol) of recombinant His-DEK was either treated with λ-phosphatase (+PPase) or left untreated (−PPase). Both DEK preparations were subsequently incubated with 600 nM unfractionated PAR (in a total volume of 100 µl), or were incbubated without PAR, for 30 min at RT, and layered on top of a 5–40 % sucrose density gradient (150 mM NaCl, 20 mM Tris-Cl, pH 7.5, 1 mM EDTA, 1 mM MgCl2, 10 mM sodiumbisulfite). After centrifugation for 12 h at 36,000 rpm (SW-41), the gradient was fractionated from the top in fractions containing 700 µl. 100 µl of each fraction was spotted onto a nitrocellulose membrane using a dot-blot apparatus, and further subjected to immunoblotting using PAR-specific antibodies (10H). The remaining 600 µl were processed for SDS-PAGE and further analyzed by immunoblotting using DEK-specific antibodies.
RESULTS
Mapping of DEK domains involved in non-covalent PAR interaction
We have recently shown that DEK is covalently modified with PAR during apoptosis or genotoxic stress, and our initial data suggested that DEK is also capable of interacting in a non-covalent fashion with free PAR polymers (24). To detail the specificity of the latter interaction, we used a slot-blot approach. Increasing amounts of recombinant DEK and histone H1, a well-known PAR-binding protein, were transferred onto a nitrocellulose membrane followed by PAR incubation and high-salt washes. Immobilized DEK interacted with PAR in a concentration-dependent manner (Fig. 1A). Histone H1, as expected, showed even stronger binding, whereas the negative controls BSA and lysozyme, which were supplied in excess, did not bind. To identify potential PAR interaction sites, the DEK peptide sequence was aligned with the PAR consensus binding motif (15). Five putative PAR binding sites were identified (Fig. 1B and D, lower panel). Interestingly, two motifs (aa 99–119; aa 158–181) are located in the major DNA binding domain (aa 87–187) of DEK and display high sequence homology to the established PAR-binding consensus motif (Fig. 1B, PAR consensus). Another candidate binding site is located C-terminally adjacent to the major DNA-binding domain (aa 195–222). Two motifs (aa 314–334; aa 329–352) were found in the C-terminal part of DEK, overlapping with the second DNA-binding or multimerization domain. Prompted by the sequence alignment, we synthesized corresponding DEK peptides to test for functional PAR binding using overlay slot-blots (Fig. 1C). Strikingly, DEK peptide 195–222 showed a strong PAR signal in a concentration-dependent manner starting at 50 pmol PAR. A slightly weaker signal was detected using peptide 158–181, and rather weak binding was observed for the C-terminal peptide 329–352. The peptides 99–119 and 314–334 failed to bind, even at high PAR concentrations. To further verify our peptide-derived results, we subjected selected recombinant DEK fragments, either produced in bacteria (aa 68–226) or insect cells (aa 250–375, aa 1–375) to the same approach (Fig. 1D). The DEK fragment 68–226, which comprises the two PAR-binding sites revealed by the peptide approach, produced a very strong signal (Fig. 1D, 68–226, right panel). The second DEK fragment 250–375 (Fig. 1D, 250–375, right panel) showed a lower affinity for PAR comparable to that of wt-DEK (Fig. 1D, 1–375). Wt-DEK and the fragment 250–375 were purified from insect cells, and were therefore phosphorylated (see also Material and Methods). We further compared PAR binding affinities of phosphorylated and dephosphorylated DEK, and found only a slightly reduced overall PAR-binding capacity for phosphorylated DEK (see Fig. 4 A, and Supplementary Fig. S2). Another prominent band at about 35 kDa was observed in the wt-DEK preparation, which corresponds to a previously described N-terminal degradation product (65), and was visible in the protein staining of the individual protein preparations (Fig. 1D, arrow). Additionally, an intermediate degradation product of unknown nature, present in the wt-DEK preparation, also bound efficiently to PAR (Fig. 1D, asterisk). BSA failed to react with PAR (Fig. 1D, BSA), underscoring the selectivity of this assay. Taken together, we have identified three functional PAR-binding sites in DEK by means of a peptide approach and confirmed the interaction using recombinant DEK fragments spanning the sites of interest.
DEK exhibits a high binding affinity for long PAR chains
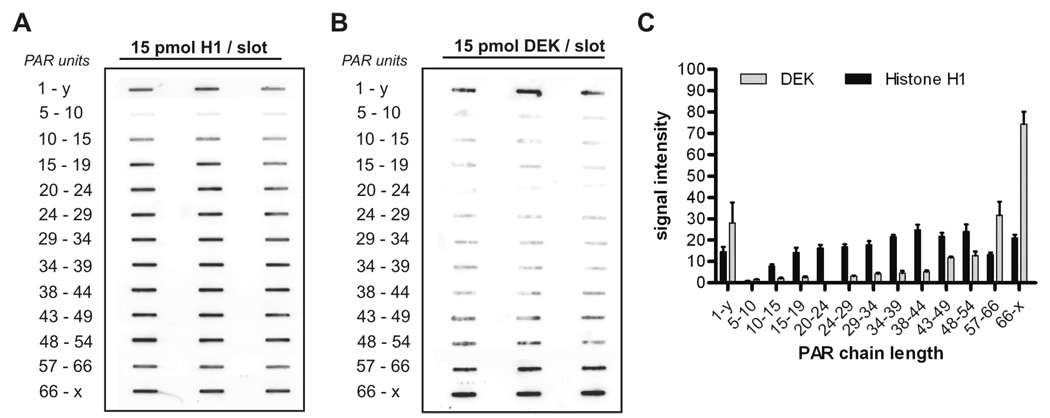
Having identified the regions for non-covalent PAR binding in the DEK molecule, we next investigated whether DEK exhibits any selectivity with respect to PAR chain length. Therefore, PAR was fractionated according to chain length by high resolution HPLC as described recently (18), and the individual fractions subsequently used in PAR overlay studies with purified recombinant DEK (Fig. 2). For simplicity, five adjacent HPLC fractions were pooled, resulting in samples comprising approximately five distinct PAR chain lengths, i.e. 5–10, 11–15 etc. This was further verified by modified sequencing gel electrophoresis followed by silver staining (data not shown) of the pooled fractions. First, histone H1, known to exhibit chain-dependent PAR binding, was analyzed using the fractionated PAR preparations, and clearly demonstrated chain length-dependent non-covalent interaction (Fig. 2A). Oligo(ADP-ribose) molecules ranging from 5–10 units displayed weak affinity for histone H1, whereas longer polymers (starting with 15–19 units), or un-fractionated PAR (1-y), were tightly bound to the immobilized protein. The identical PAR preparations were then used to study binding to recombinant full-length DEK (Fig. 2B). Binding of short PAR chains containing up to 39 units was very weak, with longer PAR chains, ranging from 34–54 units, showing slightly enhanced binding to DEK. In contrast, PAR containing more than 57 ADP-ribose units, or un-fractionated PAR (1-y), showed an extremely high affinity for DEK, producing the strongest PAR signals. Results obtained with DEK and histone H1 were further analyzed by densitometry, and signal intensities confirm a clear preference of DEK for long PAR chains that exceeds that of histone H1 (Fig. 2C).
Figure 2. Interaction of fractionated PAR and immobilized DEK: DEK preferentially binds long chains of PAR.
For each binding assay, 15 pmol of full-length DEK or histone H1 were vacuum aspirated on a nitrocellulose membrane and cut into strips, followed by incubation with 500 pmol of the respective PAR fraction (0.25 µM, 1-y denotes unfractionated PAR). Each reaction was run in triplicate. Nonspecific binding was abrogated by high-salt washes and bound PAR chains were detected using PAR-specific antibodies (10H). (A) Binding of fractionated PAR to immobilized histone H1. (B) Binding of fractionated PAR to immobilized DEK. (C) Densitometric evaluation of slot blots. Signal intensity is indicated in arbitrary units. The bars represent mean + SEM of triplicates.
To further validate this result, a PAR-specific EMSA approach using biotin labeled polymers of defined sizes was carried out. First, specific end-labeling of PAR was achieved by using a carbonyl-reactive biotin analogue, termed biocytin hydrazide, and was monitored by a neutravidin-ELISA, which allows for the immobilization of biotinylated PAR chains. As a reference, a biotinylated standard (B-PAR) was synthesized in the presence of NAD+ and 6-biotin-17-NAD+, leading to the incorporation of biotinylated ADP-ribose units into the growing polymer chain. As little as 2.9 pmol of this standard was readily detectable (Fig. 3A, left panel). Three different dilutions (171, 86 and 17 pmol) of end-biotinylated PAR samples were also applied and revealed biotin-coupled PAR in a concentration-dependent manner, whereas a 10-fold excess of unlabeled PAR displayed virtually no signal (data not shown). Additionally, efficient biotin-labeling of these PAR polymers was confirmed by native PAGE, and detection using streptavidin-POD (Fig. 3A, right panel). End-labeled PAR was then separated by anion exchange HPLC according to chain length as described in Material and Methods. Two PAR fractions (18mer and 54mer PAR) were selected, affinity-purified using a monomeric avidin column, and purification was monitored as described in Supplementary Figure S1. The concentration of the collected affinity-purified 18mer and 54mer PAR samples was determined in comparison with a commercially available biotinylated 49mer oligonucleotide (Fig. S1).
The two biotin-labeled size-fractionated PAR samples were then used to determine the binding to recombinant DEK (Fig. 3B). Short 18mer PAR (250 fmol) was incubated in triplicate in the presence or absence of increasing amounts of recombinant DEK (0 to 1 µM) under EMSA conditions. DEK failed to mediate the formation of a specific complex with the 18mer PAR. A faint, slowly migrating complex is visible, starting with 0.1 µM DEK, and is due to minor cross-contaminations with longer PAR chains that were introduced during the affinity-purification procedure (Fig. 3B, asterisk). Furthermore, high DEK concentrations led to a reduction of the signal intensity of free 18mer PAR, most likely caused by low-affinity binding to DEK. This was confirmed by prolonged exposure of the blot, which revealed a non-specific smear at higher DEK concentrations (data not shown). In contrast, DEK at a concentration of 70 nM bound about 50% of the 54mer PAR, and produced a defined, clearly identifiable, complex at concentrations starting with 100 nM DEK (Fig. 3C). A concentration of 200 nM DEK also promoted the formation of a defined complex, which displayed a slightly reduced electrophoretic mobility as compared to incubation with 100 nM DEK. The signals were further quantitated densitometrically and revealed a strict dependency of PAR complex formation on DEK concentration (Fig. 3D). The EMSA data obtained were subsequently used to calculate the KD-value for the interaction of DEK with long PAR chains (see also Supplementary Figure S5). The KD-value was in the low nM range, indicative of high binding affinity (Tab. 1). It is noteworthy that DEK exhibits higher affinity for long ADP-ribose chains as compared to p53 and XPA. Similar to XPA, DEK was not capable of producing a defined complex with short ADP-ribose molecules (18).
Table 1.
Comparison of KD-values obtained by EMSA
| Protein | 16/18 mer PAR KD [M] |
55/54 mer PAR KD [M] |
|---|---|---|
| XPA | No binding | 3.2 × 10−7 ± 7.7 10−9 |
| p53 | 2.5 × 10−7 ± 3.8 × 10−8 |
1.3 × 10−7 ± 4.2 × 10−9 |
| DEK | No binding | 6.1 × 10−8 ± 5.2 × 10−9 |
Non-covalent PAR modification competitively interferes with DNA in the formation of DEK multimers
We have recently reported that covalent poly(ADP-ribosyl)ation of DEK catalyzed by recombinant PARP-1 results in impaired general DNA binding, and, as a consequence, in the loss of DEK’s supercoiling activity (24). In addition, we observed that the presence of covalently bound PAR to DEK disrupts DEK’s ability to bind DNA via the SAP-box-triggered “mass-binding” mechanism, as measured by impaired formation of large nucleo-protein complexes in aggregation assays (42). As noted above, the recombinant His-DEK used in these experiments (Fig. 1–3) was expressed in insect cells, and purified in its phosphorylated form. As phosphorylation abolishes DEK’s DNA binding activity (36, 37), we had to dephosphorylate His-DEK before studying the impact of PAR on DEK’s DNA-binding activity (see also Material and Methods) (36, 24, 36). Consequently, we first tested if the phosphorylation status of DEK influences its ability to bind to unfractionated, short-chain or long-chain PAR polymers. Using PAR overlay-blots as described above, we observed a moderately reduced overall PAR-binding capacity in phosphorylated DEK samples. However, phosphorylation did not influence the preferential binding selectivity for long-chained PAR polymer. This binding selectivity for long PAR chains was equally visible in both DEK species (Fig. 4A, α-PAR and Supplementary Figure S2), thus validating the use of dephosphorylated DEK for further experiments.
First, we wanted to asses if PAR of various chain lengths alters the DNA binding properties of DEK. To discriminate between the direct interaction of DEK with DNA and its subsequent DNA-dependent multimerization activities, we initially employed immobilized DEK in a South-Western approach. Interestingly, dephosphorylated DEK (+ PPase) bound to P32-labeled DNA, regardless of prior incubation with un-fractionated (PAR), or PAR chains comprising 18 (PAR18) or 53 (PAR53) units (Fig. 4A, left panel), even at concentrations of up to 600 nM PAR (Supplementary Figure S3). As expected, phosphorylated DEK did not bind to DNA (24, 36, 37), but it remained capable of interacting with PAR (Fig. 4A, right panel).
Next, we assessed the influence of PAR on DEK’s DNA binding in solution by standard EMSA techniques as previously described (36, 37), now taking DEK’s multimerization activity into consideration. Dephosphorylated His-DEK was incubated in the absence (Fig. 4B, lane 1, and lanes 3–6), or in the presence of increasing concentrations of PAR (0, 100, 200, 400 [data not shown], or 600 nM, Fig 4B, lanes 7–10), and subjected to native agarose gel electrophoresis (Fig 4B). As expected, DEK readily bound to DNA, with a preference for supercoiled DNA at lower DEK concentrations (39) (compare ratio between form I (supercoiled) to form II (relaxed) in lane 4) and formed very large nucleo-protein complexes at higher DEK concentrations (e.g. lane 6, arrow). The latter effect is due to its DNA-dependent multimerization activity and ability to promote the formation of inter and intra molecular interactions between DNA strands (38, 39, 42, 43). Pretreatment of DEK with un-fractionated PAR (600 nM) did not influence the overall DNA-binding activity, but appeared to slightly weaken the preference for supercoiled DNA forms (compare lane 4 to lane 8, white arrows). PAR itself had no influence on the electrophoretic mobility of the utilized DNA (Fig. 4B, lane 2).
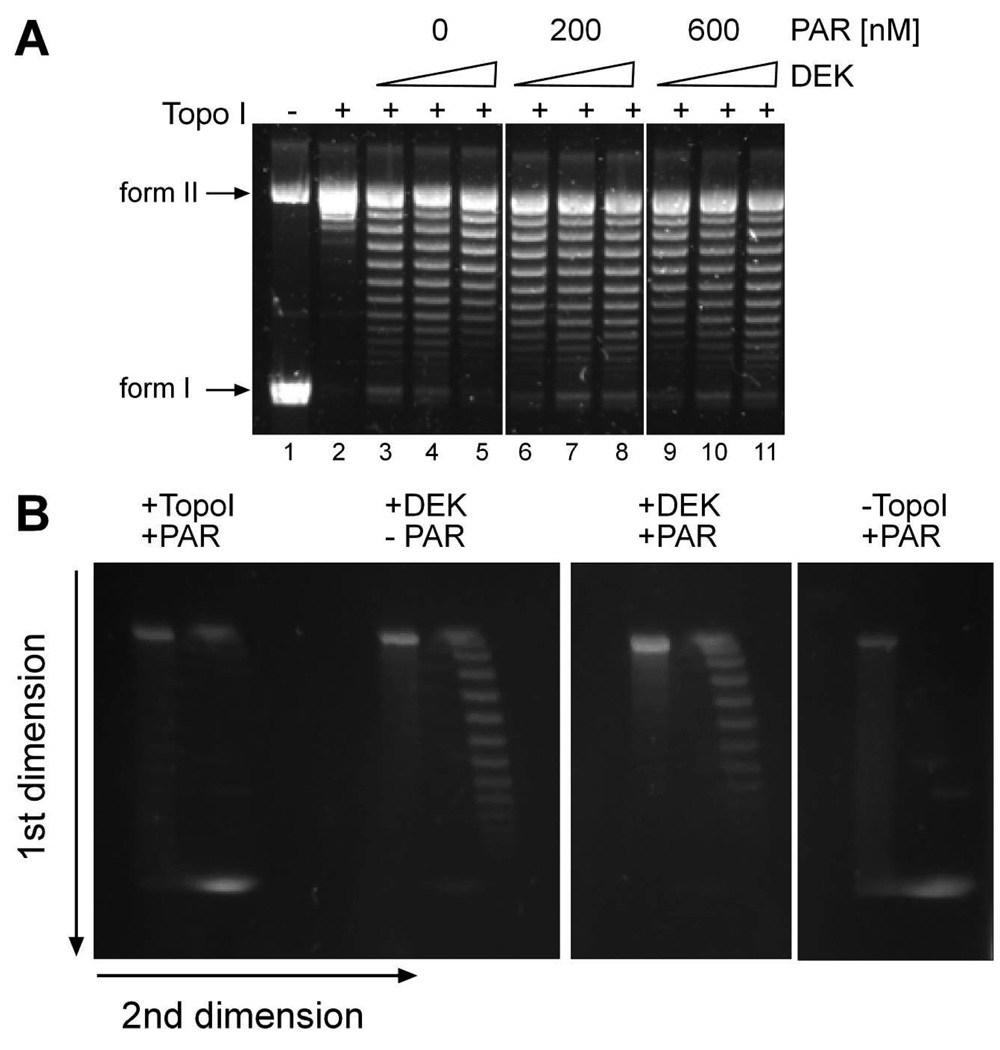
We next wished to assess the impact of PAR binding on the ability of DEK to introduce positive supercoils into DNA templates. Therefore, we employed topology assays using negatively supercoiled Simian Virus 40 (SV40) DNA as a template (38). In order to visualize DEK-induced topological changes to DNA, topoisomerase I (Topo I) is present in this assay. Control experiments excluded the influence of PAR on topoisomerase I function (data not shown, and Fig. 5B, + Topo I, +PAR), as Topo I harbors three non-covalent PAR binding sites (66). As shown in figure 5A, SV40 DNA is fully converted from its supercoiled (form I) into the relaxed form (form II) in the presence of topoisomerase I (lane 2). Addition of increasing amounts of DEK resulted in well-defined, constrained DNA topoisomers (Fig. 5A, lanes 3–5), reaching saturation at 800 ng DEK (lane 5), indicated by a reduced intensity of supercoils in the lower part of the gel (compare lane 4 to lane 5) (38). Pre-incubation of DEK with different concentrations of unfractionated PAR (Fig. 5A, 200 nM (lanes 6–8), or 600 nM PAR (lanes 9–11)), slightly affected this supercoilong activity, as no saturation was observed at the highest DEK concentration used (Fig. 5 A, compare lanes 5, 8 and 11, and see Supplementary Figure S4).
Figure 5. DEK’s supercoiling activity is moderately influenced by non-covalent PAR interaction.
(A) DNA topology assay. 70 ng of SV40 DNA were incubated with 200, 400 or 800 ng of His-DEK in the presence of 200 or 600 nM PAR and topoisomerase I. The samples were then deproteinized by Proteinase K treatment, the DNA was purified and analyzed by 0.8 % agarose gel electrophoresis followed by SYBRGold staining. Form I: supercoiled DNA; form II: relaxed DNA (B) 2D-topology assay. DNA topology reaction was performed as in (A) and the deproteinized and purified DNA was separated by standard agarose gel electrophoresis in the first dimension (from top to bottom). Subsequently, the gel was incubated with 0.25 mg/ml chloroquine for 2 hours, rotated by 90° and, after a run in the second dimension (from left to right), DNA was visualized by SYBRGold staining.
Full-length DEK induces positive supercoiling (38), however, Bohm et al. (42) have identified two domains in DEK (aa 68–87 and 187–208) that, upon deletion of either or both, result in a switch to negative DNA winding activity of DEK. Negative supercoils, as opposed to positive supercoils, favor local unwinding of the DNA double helix, thus facilitating processes such as transcription, DNA replication or repair. Strikingly, the PAR binding motif with the strongest affinity identified (aa 195–222, Fig 1) overlaps with one of the switch domains. Non-covalent attachment of PAR could therefore modulate the function of this domain, possibly resulting in negative supercoiling activity of DEK. To test for this hypothesis, we performed 2D gel agarose electrophoresis (Fig 5B). As expected, incubation of SV40 DNA with DEK, followed by subsequent incubation with chloroquine in the second dimension, produced DNA topoisomers that migrate in a clock-wise direction (positive supercoils, Fig. 5B, +DEK, -PAR). This positive supercoiling activity of DEK was, however, not affected by the presence of PAR (Fig. 5B, +DEK, +PAR). In summary, non-covalent interaction between PAR and DEK produces a subtle, negative effect on DEK’s DNA binding activity. This negative effect becomes visible in solution assays (EMSA and topology assays). We considered that this may be evidence of a role for PAR in modulating DEK’s self-interacting ability.
Therefore, we next employed an aggregation assay that allows us to measure this activity. We used this method, as described (42), to test if PAR modifies affinities of DEK for DNA molecules of variable superhelicity. Partially relaxed SV40 DNA (comprising SV40 DNA topoisomers ranging from 25 (form I, highly supercoiled) to 0 supercoils (form II, relaxed) was incubated with increasing concentrations of dephosphorylated DEK, either in the presence or absence of 600 nM PAR, followed by centrifugation of the formed nucleo-protein complexes. The recovered pellet fractions (corresponding to bound DNA present in large nucleo-protein complexes) were then analyzed by gel electrophoresis. As shown before, full-length DEK binds efficiently to a wide range of superhelical DNA forms using this assay (Fig. 6A and B, DEK). In the presence of PAR, binding affinity of DEK for DNA topoisomers of high superhelicity was strongly reduced (Fig. 6A and B, DEK + PAR). Furthermore, less DNA was precipitated overall. The decreased precipitation suggested weaker DNA binding, and interference of PAR with DEK’s DNA-dependent multimerization activity. To further support this interpretation, we performed density gradients as reported previously (36, 37). Recombinant His-DEK, either phosphorylated (Fig. 6C, -PPase) or dephosphorylated (Fig. 6C, +PPAse), was incubated in the presence or absence of 600 nM unfractionated PAR, and subjected to sedimentation analysis. Surprisingly, PAR strongly augmented the formation of large DEK-multimers beyond the phosphorylation-triggered multimerization activity (Fig. 6B, top panels, -PPase, α-DEK) (36). This is underscored by the exclusive presence of PAR in the bottom fraction (Fig. 6B, top panels, α-PAR). Importantly, reduced abundance of DEK molecules in the top fractions coincided with a PAR-dependent accumulation. These large PAR-bound DEK complexes in the bottom fraction of the gradient were also observed with dephosphorylated DEK, however to a lesser extent (Fig. 6B, lower panels, +PPase, α-DEK, α-PAR).
Taken together, the strong non-covalent interaction of PAR and DEK does not impair DEK’s DNA binding per se; however, PAR seems to be able to replace DNA in catalyzing the formation of high-molecular DEK complexes.
Discussion
In the present study, we have dissected the non-covalent interaction of PAR, a complex biopolymer formed in response to genotoxic stress, and the oncoprotein DEK, and studied its effect on DEK function. Based on our previous observation that DEK is capable of binding non-covalently to PAR, we set out to map the PAR-binding sites using a sequence alignment according to Pleschke et al. (15) in conjunction with PAR binding assays. This resulted in the identification of three functional PAR-binding sites within DEK’s primary sequence, similar to the situation in the tumor suppressor protein p53 (19). The binding affinity of DEK for PAR was slightly weaker compared to that of histone H1, which is among the strongest PAR binders, as judged by a slot blot assay. Using size-fractionated PAR we then showed that DEK preferably binds to longer PAR chains, which was also found for XPA (18), but differs from histone H1. The observed binding specificity was confirmed using a more sophisticated EMSA approach with size-fractionated, end-biotinylated PAR of defined chain length. Long PAR molecules (54mer) produced a specific complex with DEK in a concentration-dependent manner allowing for the determination of the KD-value (6 × 10−8 M), which is in the low nM range. This binding affinity is even higher than those determined previously for p53 and XPA (18). Intriguingly, DEK was not capable of forming a defined complex with very short ADP-ribose molecules, which supports earlier observations that binding occurs as a function of chain length (18, 67, 68). The biological role of PAR chain length was previously underscored by Yu and colleagues, who demonstrated that PAR, depending on its chain length, can function as a death signal, leading to caspase-independent cell death (69, 70).
It is well known that non-covalent PAR binding may modulate the function of its target proteins. On the one hand, PAR can positively regulate the enzymatic properties of its binding partner, e.g. an increased DNA joining activity of DNA ligase III (20) or activation of ATM kinase (71). On the other hand, PAR also negatively affects the function of other interacting proteins, e.g. PAR-mediated inhibition of the DNA-binding activity of p53 (19). This prompted us to study the effect of non-covalent PAR-binding on DEK functions. Surprisingly, concentrations up to 600 nM PAR did not influence the DNA-binding of immobilized DEK, although an increase in negative charge might be expected to cause electrostatic repulsion and subsequent dissociation of DEK from DNA. Indeed, DEK can simultaneously bind PAR and DNA, as revealed by our South-Western blot experiments. This is in contrast to the covalent modification of DEK with PAR, which strongly inhibits its DNA-binding activity (24). By further dissecting DEK’s DNA-binding activities, we found subtle, yet reproducible, effects of non-covalent PAR modifications on DEK’s DNA binding activity in solution. PAR interfered weakly, yet measurably, with DEK’s supercoiling activity. This occurred without causing a switch in the orientation of DNA winding. In contrast, we have previously shown that covalently modified DEK completely lost its ability to alter DNA topology (24). Importantly, we observed that PAR competitively suppressed DEK-DNA aggregation and, by promoting the self-interaction of DEK molecules, reduced the affinity of DEK for supercoiled DNA forms. Interestingly, this effect on DEK’s multimerization was particularly pronounced using phosphorylated DEK, but was also clearly evident in dephosphorylated DEK samples. Therefore, we speculate that non-covalent PAR attachment to DEK may have two possible functions in the cell: 1) further augment self-interaction of phosphorylated DEK molecules, which have already detached from DNA, 2) competitively counteract DNA binding, by favoring PAR-dependent formation of DEK multimeres.
This may represent a mechanism of how non-covalently attached PAR impinges on DEK’s spreading along chromatin fibers in a competitive manner. This modulation could play a role during DNA repair, as PAR-bound DEK might dissociates from supercoiled DNA at the site of a DNA lesion to allow repair and/or redistributes to adjacent sites of chromatin, where it may be stored in large complexes until the site of damage is cleared.
A growing body of evidence has linked the oncoprotein DEK to the DNA-damage sensor PARP-1 and its product PAR, suggesting a functional interplay in PAR-dependent maintenance of genomic integrity and chromatin remodeling. Consistent with our findings described here and elsewhere (24), in silico analysis based on a refined consensus sequence as reported by Pleschke et al. (15) has also confirmed DEK as part of the PAR-interaction network (72). Very recently, DEK was found in ecdysone-induced puffs, i.e. a local loosening of polytene chromatin in Drosophila (73). Interestingly PARP-1 is required for the formation of normal-sized puffs, and elevated PAR levels were detected in ecdysone-dependent puffs (74). The chromatin relationship is further supported by the finding that both DEK and PARP-1 are present together in a chromatin fraction enriched for the macrodomain-containing histone mH2A1.1 (58). Timinszky and colleagues recently reported that DEK, PARP-1 and mH2A1.1 are physically associated in HeLa cells and showed the PAR-mediated recruitment of mH2A1.1 to sites of laser-induced DNA damage, resulting in the modulation of chromatin structure (75). Therefore, it is tempting to speculate that PARP-1, DEK and mH2A1.1 act in concert to spatially modulate chromatin structure and that this functional interplay is mediated by PAR, which binds to DEK and mH2A1.1. This could also explain the rather limited effect of PAR binding on DEK function in vitro described here, since PAR may be rather involved in the local recruitment of DEK to sites of DNA lesions, as shown for the scaffold protein XRCC-1 (21, 76), or may induce its redistribution to surrounding chromatin regions.
In summary, our findings reveal DEK as a potent PAR-binding protein that shows differential binding to PAR as a function of chain length. This strong non-covalent interaction does not affect DNA-binding of DEK per se, however, may modulate functions that depend on the formation of larger DEK-DEK complexes. This is exemplified here by the competitive PAR-dependent modulation of DEK’s selectivity for supercoiled DNA, which could be critical during DNA repair processes. Our present study contributes to the molecular understanding of the DEK-PAR interplay and further supports the existence of a cellular “PAR code”.
Supplementary Material
Acknowledgements
The authors thank Prof. A. Marx and Dr. R. Kranaster, Konstanz, Germany for kindly providing the HPLC facility and for support with the large-scale HPLC fractionation of PAR; Prof. M. Przybylski, Konstanz, Germany, for his advice and support with the peptide synthesis; and Prof. M. Miwa and Prof. T. Sugimura, Tokyo, Japan, for kindly providing 10H hybridoma cells.
Abbreviations
- PAR
poly(ADP-ribose)
- PARP-1
poly(ADP-ribose) polymerase-1
- BER
base excision repair
- aa
amino acid
- Topo I
topoisomerase I
- EMSA
electrophoretic mobility shift assay
- PARG
poly(ADP-ribose) glycohydrolase
Footnotes
This work was supported by grants from the German Research Foundation (FOR 434 to A.B. and MA2385/2-3 to E.F.M). F.K. was supported by a William D. Robinson Fellowship from the Arthritis Foundation/Michigan Chapter and is a recipient of an Arthritis Foundation Postdoctoral Fellowship. Work in the laboratory of D.M.M. was supported by NIH grants R01-AI062248 and R01-AI087128 and by a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
Supplementary Material
Avidin chromatography of end-biotinylated 18mer, or 54mer PAR (Figure S1), PAR-overlay blot using 100 nM PAR (Figure S2), South-Western analysis (Figure S3), magnified depiction of Fig. 5A (Figure S4), and a graph depicting the determination of the KD value (Figure S5). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Burkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ Febs J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 2.Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 3.Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- 4.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 5.Simbulan-Rosenthal CM, Haddad BR, Rosenthal DS, Weaver Z, Coleman A, Luo R, Young HM, Wang ZQ, Ried T, Smulson ME. Chromosomal aberrations in PARP(−/−) mice: genome stabilization in immortalized cells by reintroduction of poly(ADP-ribose) polymerase cDNA. Proc Natl Acad Sci U S A. 1999;96:13191–13196. doi: 10.1073/pnas.96.23.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 7.Kupper JH, Muller M, Burkle A. Trans-dominant inhibition of poly(ADP-ribosyl)ation potentiates carcinogen induced gene amplification in SV40-transformed Chinese hamster cells. Cancer Res. 1996;56:2715–2717. [PubMed] [Google Scholar]
- 8.Meyer R, Muller M, Beneke S, Kupper JH, Burkle A. Negative regulation of alkylation-induced sister-chromatid exchange by poly(ADP-ribose) polymerase-1 activity. Int J Cancer. 2000;88:351–355. doi: 10.1002/1097-0215(20001101)88:3<351::aid-ijc5>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 9.Berger F, Lau C, Ziegler M. Regulation of poly(ADP-ribose) polymerase 1 activity by the phosphorylation state of the nuclear NAD biosynthetic enzyme NMN adenylyl transferase 1. Proc Natl Acad Sci U S A. 2007;104:3765–3770. doi: 10.1073/pnas.0609211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 11.Flohr C, Burkle A, Radicella JP, Epe B. Poly(ADP-ribosyl)ation accelerates DNA repair in a pathway dependent on Cockayne syndrome B protein. Nucleic Acids Res. 2003;31:5332–5337. doi: 10.1093/nar/gkg715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitchell J, Smith GC, Curtin NJ. Poly(ADP-Ribose) polymerase-1 and DNA-dependent protein kinase have equivalent roles in double strand break repair following ionizing radiation. Int J Radiat Oncol Biol Phys. 2009;75:1520–1527. doi: 10.1016/j.ijrobp.2009.07.1722. [DOI] [PubMed] [Google Scholar]
- 13.Ame JC, Fouquerel E, Gauthier LR, Biard D, Boussin FD, Dantzer F, de Murcia G, Schreiber V. Radiation-induced mitotic catastrophe in PARG-deficient cells. J Cell Sci. 2009;122:1990–2002. doi: 10.1242/jcs.039115. [DOI] [PubMed] [Google Scholar]
- 14.Niere M, Kernstock S, Koch-Nolte F, Ziegler M. Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol. 2008;28:814–824. doi: 10.1128/MCB.01766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 16.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 17.Gatz SA, Wiesmuller L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 18.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J Biol Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 20.Leppard JB, Dong Z, Mackey ZB, Tomkinson AE. Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair. Mol Cell Biol. 2003;23:5919–5927. doi: 10.1128/MCB.23.16.5919-5927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haince JF, Kozlov S, Dawson VL, Dawson TM, Hendzel MJ, Lavin MF, Poirier GG. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J Biol Chem. 2007;282:16441–16453. doi: 10.1074/jbc.M608406200. [DOI] [PubMed] [Google Scholar]
- 23.Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276:26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- 24.Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Burkle A, Markovitz DM, Ferrando-May E. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Lindern M, Poustka A, Lerach H, Grosveld G. The (6;9) chromosome translocation, associated with a specific subtype of acute nonlymphocytic leukemia, leads to aberrant transcription of a target gene on 9q34. Mol Cell Biol. 1990;10:4016–4026. doi: 10.1128/mcb.10.8.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, Muller H. DEK Expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5:1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 27.Grasemann C, Gratias S, Stephan H, Schuler A, Schramm A, Klein-Hitpass L, Rieder H, Schneider S, Kappes F, Eggert A, Lohmann DR. Gains and overexpression identify DEK and E2F3 as targets of chromosome 6p gains in retinoblastoma. Oncogene. 2005;24:6441–6449. doi: 10.1038/sj.onc.1208792. [DOI] [PubMed] [Google Scholar]
- 28.Khodadoust MS, Verhaegen M, Kappes F, Riveiro-Falkenbach E, Cigudosa JC, Kim DS, Chinnaiyan AM, Markovitz DM, Soengas MS. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 30.Wise-Draper TM, Allen HV, Thobe MN, Jones EE, Habash KB, Munger K, Wells SI. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J Virol. 2005;79:14309–14317. doi: 10.1128/JVI.79.22.14309-14317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA, Cripe TP, Grosveld GC, Wells SI. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wise-Draper TM, Morreale RJ, Morris TA, Mintz-Cole RA, Hoskins EE, Balsitis SJ, Husseinzadeh N, Witte DP, Wikenheiser-Brokamp KA, Lambert PF, Wells SI. DEK proto-oncogene expression interferes with the normal epithelial differentiation program. Am J Pathol. 2009;174:71–81. doi: 10.2353/ajpath.2009.080330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mor-Vaknin N, Punturieri A, Sitwala K, Faulkner N, Legendre M, Khodadoust MS, Kappes F, Ruth JH, Koch A, Glass D, Petruzzelli L, Adams BS, Markovitz DM. The DEK nuclear autoantigen is a secreted chemotactic factor. Mol Cell Biol. 2006;26:9484–9496. doi: 10.1128/MCB.01030-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko SI, Lee IS, Kim JY, Kim SM, Kim DW, Lee KS, Woo KM, Baek JH, Choo JK, Seo SB. Regulation of histone acetyltransferase activity of p300 and PCAF by proto-oncogene protein DEK. FEBS Lett. 2006;580:3217–3222. doi: 10.1016/j.febslet.2006.04.081. [DOI] [PubMed] [Google Scholar]
- 36.Kappes F, Scholten I, Richter N, Gruss C, Waldmann T. Functional domains of the ubiquitous chromatin protein DEK. Mol Cell Biol. 2004;24:6000–6010. doi: 10.1128/MCB.24.13.6000-6010.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kappes F, Damoc C, Knippers R, Przybylski M, Pinna LA, Gruss C. Phosphorylation by protein kinase CK2 changes the DNA binding properties of the human chromatin protein DEK. Mol Cell Biol. 2004;24:6011–6020. doi: 10.1128/MCB.24.13.6011-6020.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waldmann T, Eckerich C, Baack M, Gruss C. The ubiquitous chromatin protein DEK alters the structure of DNA by introducing positive supercoils. J Biol Chem. 2002;277:24988–24994. doi: 10.1074/jbc.M204045200. [DOI] [PubMed] [Google Scholar]
- 39.Waldmann T, Baack M, Richter N, Gruss C. Structure-specific binding of the proto-oncogene protein DEK to DNA. Nucleic Acids Res. 2003;31:7003–7010. doi: 10.1093/nar/gkg864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waldmann T, Scholten I, Kappes F, Hu HG, Knippers R. The DEK protein--an abundant and ubiquitous constituent of mammalian chromatin. Gene. 2004;343:1–9. doi: 10.1016/j.gene.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 41.Devany M, Kappes F, Chen KM, Markovitz DM, Matsuo H. Solution NMR structure of the N-terminal domain of the human DEK protein. Protein Sci. 2008;17:205–215. doi: 10.1110/ps.073244108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohm F, Kappes F, Scholten I, Richter N, Matsuo H, Knippers R, Waldmann T. The SAF-box domain of chromatin protein DEK. Nucleic Acids Res. 2005;33:1101–1110. doi: 10.1093/nar/gki258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 2000;14:1308–1312. [PMC free article] [PubMed] [Google Scholar]
- 44.Aravind L, Koonin EV. SAP - a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25:112–114. doi: 10.1016/s0968-0004(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 45.Gradwohl G, Mazen A, de Murcia G. Poly(ADP-ribose) polymerase forms loops with DNA. Biochem Biophys Res Commun. 1987;148:913–919. doi: 10.1016/s0006-291x(87)80219-x. [DOI] [PubMed] [Google Scholar]
- 46.Sastry SS, Kun E. The interaction of adenosine diphosphoribosyl transferase (ADPRT) with a cruciform DNA. Biochem Biophys Res Commun. 1990;167:842–847. doi: 10.1016/0006-291x(90)92102-6. [DOI] [PubMed] [Google Scholar]
- 47.Devany M, Kotharu NP, Matsuo H. Solution NMR structure of the C-terminal domain of the human protein DEK. Protein Sci. 2004;13:2252–2259. doi: 10.1110/ps.04797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3' splice site recognition by DEK. Science. 2006;312:1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 49.Cavellan E, Asp P, Percipalle P, Farrants AK. The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J Biol Chem. 2006;281:16264–16271. doi: 10.1074/jbc.M600233200. [DOI] [PubMed] [Google Scholar]
- 50.Campillos M, Garcia MA, Valdivieso F, Vazquez J. Transcriptional activation by AP-2alpha is modulated by the oncogene DEK. Nucleic Acids Res. 2003;31:1571–1575. doi: 10.1093/nar/gkg247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 52.Gamble MJ, Fisher RP. SET and PARP1 remove DEK from chromatin to permit access by the transcription machinery. Nat Struct Mol Biol. 2007;14:548–555. doi: 10.1038/nsmb1248. [DOI] [PubMed] [Google Scholar]
- 53.Fu GK, Grosveld G, Markovitz DM. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc Natl Acad Sci U S A. 1997;94:1811–1815. doi: 10.1073/pnas.94.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faulkner NE, Hilfinger JM, Markovitz DM. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J Biol Chem. 2001;276:25804–25812. doi: 10.1074/jbc.M006454200. [DOI] [PubMed] [Google Scholar]
- 55.Tabbert A, Kappes F, Knippers R, Kellermann J, Lottspeich F, Ferrando-May E. Hypophosphorylation of the architectural chromatin protein DEK in death-receptor-induced apoptosis revealed by the isotope coded protein label proteomic platform. Proteomics. 2006;6:5758–5772. doi: 10.1002/pmic.200600197. [DOI] [PubMed] [Google Scholar]
- 56.Cleary J, Sitwala KV, Khodadoust MS, Kwok RP, Mor-Vaknin N, Cebrat M, Cole PA, Markovitz DM. p300/CBP-associated factor drives DEK into interchromatin granule clusters. J Biol Chem. 2005;280:31760–31767. doi: 10.1074/jbc.M500884200. [DOI] [PubMed] [Google Scholar]
- 57.Tanuma S, Kawashima K, Endo H. Comparison of ADP-ribosylation of chromosomal proteins between intact and broken cells. Biochem Biophys Res Commun. 1985;127:896–902. doi: 10.1016/s0006-291x(85)80028-0. [DOI] [PubMed] [Google Scholar]
- 58.Ouararhni K, Hadj-Slimane R, Ait-Si-Ali S, Robin P, Mietton F, Harel-Bellan A, Dimitrov S, Hamiche A. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006;20:3324–3336. doi: 10.1101/gad.396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawamitsu H, Hoshino H, Okada H, Miwa M, Momoi H, Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- 60.Malanga M, Bachmann S, Panzeter PL, Zweifel B, Althaus FR. Poly(ADP-ribose) quantification at the femtomole level in mammalian cells. Anal Biochem. 1995;228:245–251. doi: 10.1006/abio.1995.1346. [DOI] [PubMed] [Google Scholar]
- 61.Shah GM, Poirier D, Duchaine C, Brochu G, Desnoyers S, Lagueux J, Verreault A, Hoflack JC, Kirkland JB, Poirier GG. Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal Biochem. 1995;227:1–13. doi: 10.1006/abio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 62.Cheung A, Zhang J. A scintillation proximity assay for poly(ADP-ribose) polymerase. Anal Biochem. 2000;282:24–28. doi: 10.1006/abio.2000.4604. [DOI] [PubMed] [Google Scholar]
- 63.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 64.Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 65.Sierakowska H, Williams KR, Szer IS, Szer W. The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis. Clin Exp Immunol. 1993;94:435–439. doi: 10.1111/j.1365-2249.1993.tb08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Malanga M, Althaus FR. Poly(ADP-ribose) reactivates stalled DNA topoisomerase I and Induces DNA strand break resealing. J Biol Chem. 2004;279:5244–5248. doi: 10.1074/jbc.C300437200. [DOI] [PubMed] [Google Scholar]
- 67.Althaus FR, Bachmann S, Hofferer L, Kleczkowska HE, Malanga M, Panzeter PL, Realini C, Zweifel B. Interactions of poly(ADP-ribose) with nuclear proteins. Biochimie. 1995;77:423–432. doi: 10.1016/0300-9084(96)88155-7. [DOI] [PubMed] [Google Scholar]
- 68.Panzeter PL, Realini CA, Althaus FR. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992;31:1379–1385. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 69.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 70.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goodarzi AA, Lees-Miller SP. Biochemical characterization of the ataxia-telangiectasia mutated (ATM) protein from human cells. DNA Repair (Amst) 2004;3:753–767. doi: 10.1016/j.dnarep.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 72.Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sawatsubashi S, Murata T, Lim J, Fujiki R, Ito S, Suzuki E, Tanabe M, Zhao Y, Kimura S, Fujiyama S, Ueda T, Umetsu D, Ito T, Takeyama K, Kato S. A histone chaperone, DEK, transcriptionally coactivates a nuclear receptor. Genes Dev. 24:159–170. doi: 10.1101/gad.1857410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tulin A, Spradling A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003;299:560–562. doi: 10.1126/science.1078764. [DOI] [PubMed] [Google Scholar]
- 75.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 76.El-Khamisy SF, Masutani M, Suzuki H, Caldecott KW. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 2003;31:5526–5533. doi: 10.1093/nar/gkg761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.