Abstract
Antineutrophil cytoplasmic autoantibody (ANCA) causes vascular injury that leads to small-vessel vasculitis. Patients with ANCA aberrantly express neutrophil granule–encoding genes, including 2 that encode autoantigens: proteinase 3 (PR3) and myeloperoxidase (MPO). To uncover a potential transcriptional regulatory mechanism for PR3 and MPO disrupted in patients with ANCA vasculitis, we examined the PR3 and MPO loci in neutrophils from ANCA patients and healthy control individuals for epigenetic modifications associated with gene silencing. We found that levels of the chromatin modification H3K27me3, which is associated with gene silencing, were depleted at PR3 and MPO loci in ANCA patients compared with healthy controls. Interestingly, in both patients and controls, DNA was unmethylated at a CpG island in PR3, whereas in healthy controls, DNA was methylated at a CpG island in MPO. Consistent with decreased levels of H3K27me3, JMJD3, the demethylase specific for H3K27me3, was preferentially expressed in ANCA patients versus healthy controls. In addition, we describe a mechanism for recruiting the H3K27 methyltransferase enhancer of zeste homolog 2 (EZH2) to PR3 and MPO loci mediated by RUNX3. RUNX3 message was decreased in patients compared with healthy controls, and may also be under epigenetic control. DNA methylation was increased at the RUNX3 promoter in ANCA patients. These data indicate that epigenetic modifications associated with gene silencing are perturbed at ANCA autoantigen–encoding genes, potentially contributing to inappropriate expression of PR3 and MPO in ANCA patients.
Introduction
Systemic small-vessel vasculitis is characterized by microvascular inflammation, tissue necrosis, and circulating antineutrophil cytoplasmic autoantibodies (ANCAs). Clinical and experimental evidence indicates that ANCAs cause vascular injury by activating neutrophils (1–5). Neutrophils are the primary mediators of inflammation in ANCA vasculitis, because depletion of neutrophils protects against vascular lesions (6). Activated neutrophils have increased adherence and transmigration to the vascular endothelium, where they produce reactive oxygen species and release granule constituents, including proteolytic enzymes (7). These oxygen radicals and proteases activate the alternative complement pathway, in an animal and in vitro model, which amplifies neutrophil mediated inflammation (8).
The major ANCA autoantigens proteinase 3 (PR3) and myeloperoxidase (MPO) are neutrophil granule proteins (9). Neutrophil granules are classified by their intragranular proteins and determined by the stage of neutrophil development at which the granule proteins are produced (10). PR3 and MPO are predominantly expressed during the myeloblast and promyelocyte stage of neutrophil development (11), and their protein products sort into azurophil (primary) granules. PR3 and MPO are aberrantly expressed in mature neutrophils of ANCA patients, in contrast to their normally silenced state in mature neutrophils of healthy controls (12, 13). Inappropriate expression of PR3 and MPO may alter the availability of these antigens by targeting these proteins to granules that are more readily exocytosed. The regulation of neutrophil gene expression becomes critical to the etiology of ANCA vasculitis.
Transcriptional profiling of neutrophils from different diseases reveals unique transcriptional signatures that correspond to diseases, and changes in neutrophil gene expression occur upon in vitro stimulation, which indicates that neutrophils can modulate gene expression depending on external stimuli (14–17). These and other observations depict the neutrophil not as a terminally differentiated, transcriptionally silent cell, but as a cell poised to respond at the transcriptional level. A consequence of transcriptionally dynamic mature neutrophils is that proper silencing mechanisms must be in place to ensure that genes silenced during myelopoiesis remain silenced. Using the aberrant expression of PR3 and MPO in ANCA vasculitis patients as a model, we tested whether epigenetic gene silencing processes occur in neutrophils and whether aberrant PR3 and MPO expression result from disrupted epigenetic silencing.
Results
Histone methylation of PR3 and MPO genes.
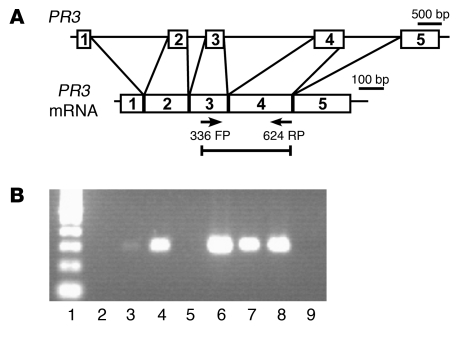
Previous studies demonstrated that PR3 and MPO transcripts are elevated in ANCA patients compared with healthy and disease controls (12, 13). This observation is consistent with failure to degrade PR3 and MPO message or active transcription in mature neutrophils. To test whether PR3 and MPO message results from active transcription of PR3 and MPO genes in ANCA disease patients, RNA immunoprecipitation was performed on isolated leukocytes with an antibody that recognizes the transcriptionally active form of RNA polymerase II. Immunoprecipitated RNA from 6 ANCA patients was analyzed by RT-PCR using primers that span intron 3. PR3 message was specifically and robustly amplified from 4 ANCA patients (Figure 1). Similarly, using primers that recognize MPO and span intron 7, we found 2 of 6 ANCA patients to be positive by Taqman (data not shown). In healthy controls, neither PR3 nor MPO message was amplified following immunoprecipitation with anti-RNA polymerase II antibody. These immunoprecipitation experiments indicated that PR3 and MPO were actively transcribed in ANCA patients (Figure 1). Evidence for active transcription of neutrophil granule genes suggests transcriptional silencing of PR3 and MPO is disrupted in neutrophils of ANCA patients. To test whether there is a defect in epigenetic gene silencing, we analyzed chromatin from neutrophils of ANCA disease patients and healthy controls for histone modifications associated with gene silencing.
Figure 1. PR3 gene is actively transcribed in ANCA patients.
(A) Schematic of PR3 gene and processed PR3 mRNA. Arrows mark the location of forward and reverse primers (FP and RP, respectively) used for RT-PCR analysis of RNA immunoprecipitated with anti-RNA polymerase II antibody. (B) Ethidium bromide–stained agarose gel showed RT-PCR product specific for PR3 mRNA present in 4 of 6 ANCA patients. Lane 1, 100-bp DNA ladder; lane 2, blank; lanes 3–8, ANCA patients; lane 9, water-only control.
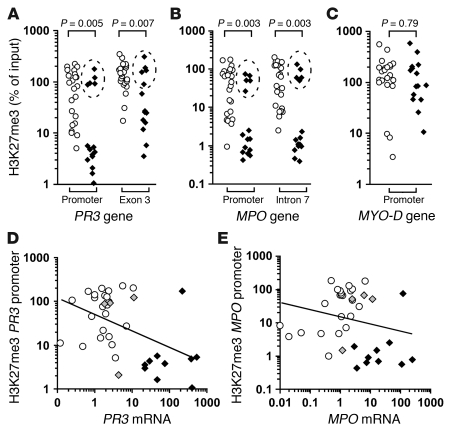
We used ChIP followed by quantitative real-time PCR to measure levels of trimethylated histone H3 at lysine 27 (H3K27me3) and dimethylated histone H3 at lysine 9 (H3K9me2) at PR3 and MPO in neutrophils from ANCA patients versus healthy controls. Both PR3 and MPO were depleted for the H3K27me3 modification in chromatin from ANCA patients compared with healthy controls (Figure 2, A and B). In contrast, no significant global differences were detected in H3K27me3 modifications between neutrophils from a patient with ANCA and those from a healthy control (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI40034DS1). The depletion of the H3K27me3 modification was detected at several locations across the gene body of both PR3 and MPO in neutrophils of ANCA patients compared with those of healthy controls (Figure 2, A and B, and data not shown). In addition to the average decrease in H3K27me3 in ANCA patients, we observed a bimodal distribution of H3K27me3 levels among ANCA patients. We observed that 5 patients had high H3K27me3 levels (Figure 2, A and B, dashed ellipses), similar to those detected in healthy controls, and had high H3K27me3 levels at all PR3 and MPO genomic regions tested (data not shown). Of these 5 patients, 4 were in remission, which suggests that the epigenetic state of PR3 and MPO correlates with disease state (Supplemental Table 1 and Figure 2, D and E). Moreover, 10 patients with ANCA had H3K27me3 levels — at both PR3 and MPO genes — well below the average measured among healthy controls; 9 of the 10 were categorized as having active ANCA vasculitis disease (Supplemental Table 1 and Figure 2, D and E). The bimodal distribution was absent at a transcriptionally silent gene, MYO-D (Figure 2C), which argues against more effective ChIP for H3K27me3 among some ANCA patients as an explanation for the distribution. These data suggest that H3K27me3 is associated with silencing PR3 and MPO in neutrophils from healthy controls and that neutrophils from ANCA patients are depleted for H3K27me3 at PR3 and MPO. Furthermore, although there were exceptions, the level of H3K27me3 inversely correlated with disease severity, scored by a physician as active or remission based on structured disease activity measure (Supplemental Table 1 and ref. 18).
Figure 2. PR3 and MPO genes are depleted for H3K27me3 in neutrophils of ANCA vasculitis patients.
(A–C) Quantitative ChIP analysis for H3K27me3 enrichment on chromatin isolated from neutrophils of healthy controls (white circles; n = 23) and ANCA vasculitis patients (black diamonds; n = 15) is shown for PR3 (A), MPO (B), and MYO-D (C). Levels of H3K27me3 immunoprecipitated chromatin are reported as percent of input DNA. 4 of 5 ANCA patients were clinically in remission (dashed ellipse); and 9 of the remaining 10 had active disease (Supplemental Table 1 and Supplemental Figure 3). (D) Pearson correlation analysis of logarithmically transformed H3K27me3 levels at PR3 promoter versus logarithmically transformed PR3 expression levels among ANCA patients (black diamonds, active; gray diamonds, remission) and healthy controls (white circles) showed a modest inverse correlation (r = –0.494; P = 0.0019). (E) Pearson correlation analysis of logarithmically transformed H3K27me3 levels at MPO promoter versus logarithmically transformed MPO expression levels among ANCA patients and healthy controls (symbols as in D) trended toward an inverse correlation, but was not statistically significant (r = –0.189; P = 0.264). Only 4 ANCA patients in remission are shown because expression data for PR3 and MPO were not obtained for ANCA patient no. 1 (Supplemental Table 1).
Since the expression of PR3 and MPO directly correlated with disease severity (Supplemental Figure 3, A and B, and ref. 12), and the level of H3K27me3 was inversely correlated with disease severity, we predicted that PR3 and MPO expression inversely correlates with H3K27me3 levels. In fact, calculating correlations with the logarithmically transformed values for H3K27me3 enrichment (Figure 2, A and B) and the logarithmically transformed values for expression revealed a statistically significant inverse correlation between H3K27me3 levels at the PR3 promoter and expression of PR3 and a slight inverse correlation between H3K27me3 levels at the MPO promoter and expression of MPO (Figure 2, D and E). For both PR3 and MPO, relatively low H3K27me3 levels clustered with high expression levels in patients, whereas relatively high H3K27me3 levels clustered with low expression levels in healthy controls (Figure 2, D and E).
The lack of more robust inverse correlations could be a consequence of measuring expression and histone methylation on a heterogenous population of cells. A population of neutrophils with a relatively small number of cells strongly expressing PR3 or MPO would have a large number of transcriptionally inactive neutrophils with H3K27me3 levels similar to those of healthy controls. This situation would result in a sample with relatively high levels of both H3K27me3 and gene expression. Alternatively, a stronger inverse correlation would be confounded by the absence of positive regulators, such as the lack of transcription factors responsible for PR3 or MPO expression in mature neutrophils in samples with low H3K27me3 levels. Also, consistent with our observation that not all patients were actively transcribing PR3 and MPO (Figure 1), patients with low H3K27me3 levels at PR3 and MPO may have not acquired signals for active transcription. For example, some genes on the inactive X chromosome remain silent in the absence of H3K27me3 because they have not acquired histone modifications (such as acetylation) associated with transcriptionally active genes (19). Silencing of PR3 and MPO genes may also be achieved by other mechanisms.
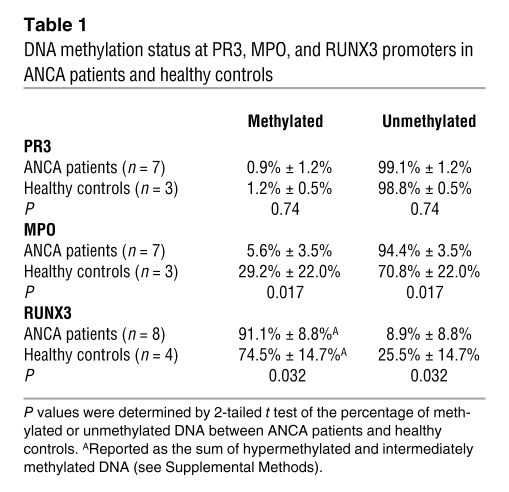
DNA methylation of CpG-rich regions is associated with gene silencing (20). Previous reports analyzing DNA methylation at PR3 and MPO paradoxically showed that CpG sites at PR3 and MPO are demethylated in normal mature granulocytes that no longer express PR3 and MPO (21, 22). We profiled DNA methylation status at CpG-rich regions within PR3 and MPO by treating genomic DNA with methylation-sensitive or methylation-dependent restriction enzymes. Following the restriction enzyme reaction, DNA was analyzed by real-time quantitative PCR using primers specific for the CpG islands in PR3 or MPO (23, 24). PR3 was unmethylated in both ANCA patients and healthy controls (Table 1 and Supplemental Figure 5). The unmethylated state of the PR3 CpG island in ANCA patients and healthy controls was confirmed by methyl-specific PCR after bisulfite treatment of genomic DNA (Supplemental Figure 5). For MPO, DNA methylation was measured at a CpG island spanning exons 2 and 3 of MPO, previously demonstrated to be methylated in the acute myeloid leukemia (AML) cell line OCI-AML2 and in some AML patients — both instances in which MPO is silenced (25). As expected, the DNA was almost completely unmethylated in ANCA patients; however, in healthy controls, about 30% of the DNA at this MPO CpG island was methylated (Table 1). Thus, epigenetic silencing of MPO by DNA methylation could also explain the failure to detect a stronger inverse correlation between H3K27me3 levels and MPO expression. This suggests that, although PR3 and MPO expression is highly correlated among ANCA patients (12), their transcriptional activity may be controlled by additional, independent pathways.
Table 1 .
DNA methylation status at PR3, MPO, and RUNX3 promoters in ANCA patients and healthy controls
In addition, we tested whether silencing mediated by H3K9me2 differed between ANCA patients and healthy controls. No significant differences were detected for H3K9me2, neither specifically at PR3 or MPO or globally, in neutrophils from ANCA patients compared with healthy controls (Supplemental Figure 2). One interpretation of this result is that the chromatin of mature neutrophils contains a low level of H3K9me2, which does not dramatically differ at PR3 and MPO between healthy controls and ANCA patients. This suggests that silencing of PR3 and MPO in normal neutrophils is not primarily mediated by H3K9me2.
Active demethylation or failure of methylation at H3K27.
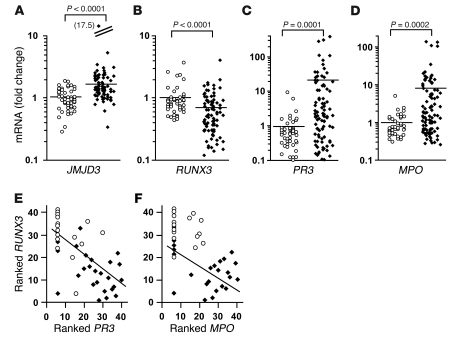
A potential explanation for the depleted levels of H3K27me3 at PR3 and MPO loci is that in ANCA patients, an active process preferentially removes methyl groups from H3K27. We measured mRNA for the H3K27me3 demethylase jumonji C domain–containing protein 3 (JMJD3; ref. 26). JMJD3 was expressed higher in leukocytes from ANCA patients than in those from healthy controls, as measured by quantitative RT-PCR (Figure 3A), and higher in ANCA patients with active disease than patients in remission (Supplemental Figure 3C). An alternative, but not mutually exclusive, explanation for depleted levels of H3K27me3 at PR3 and MPO loci in ANCA patients is that the process of active histone methylation at these sites is deficient. To explore this putative mechanism, we pursued potential transcriptional regulators that could recruit H3K27 methyltransferase activity.
Figure 3. Expression of autoantigen and potential regulators.
(A–D) Relative mRNA levels from total leukocytes of healthy controls (white circles; n = 49) and ANCA vasculitis patients (black diamonds; n = 97) were determined by quantitative RT-PCR with SYBR green (A and B) or Taqman (C and D). Mean fold change (horizontal bars) for patients versus controls was 1.77 ± 1.76 versus 1.00 ± 0.38 for JMJD3 (A), 0.67 ± 0.54 versus 1.00 ± 0.63 for RUNX3 (B), 19.77 ± 60.08 versus 1.00 ± 1.58 for PR3 (C), and 9.27 ± 24.85 versus 1.00 ± 0.78 for MPO (D). An outlier and its value (in parentheses) is shown for A. (E and F) Spearman ranked analysis of Affymetrix microarray data of healthy controls (white circles; n = 16) and ANCA patients (black diamonds; n = 25) showed an inverse correlation between RUNX3 mRNA and PR3 mRNA (P < 0.001; E) and between RUNX3 mRNA and MPO mRNA (P < 0.001; F).
RUNX3 expression inversely correlates with PR3 and MPO expression.
Analysis of Affymetrix expression arrays showed that expression of Runt-related transcription factor 3 (RUNX3) differed in leukocytes from ANCA patients compared with those from healthy controls (data not shown). RUNX3 has been implicated in gene regulation in a number of systems, including functioning as a tumor suppressor (27, 28), developing proprioceptive neurons within the dorsal root ganglia (29, 30), epigenetic silencing in CD8+ T cells (31, 32), TGF-β signaling in dendritic cells (33), and causing transcriptional repression in myeloid cells (34). In ANCA patients’ leukocytes, RUNX3 mRNA was significantly reduced compared with healthy controls, in contrast to the increased PR3 and MPO mRNA in the same cohorts (Figure 3, B–D). In addition, there was greater reduction in RUNX3 mRNA in ANCA patients with active disease compared with patients in remission (Supplemental Figure 3D).
Transcriptional repression of RUNX3 has previously been shown to be associated with DNA methylation at the RUNX3 promoter (35); therefore, we examined whether there are differences in DNA methylation of RUNX3 in ANCA patients compared with healthy controls. DNA methylation at the RUNX3 promoter was measured by real-time PCR on genomic DNA digested with methylation-sensitive or methylation-dependent restriction enzymes, as for PR3 and MPO (23, 24). There was a significantly greater percentage of methylated DNA at the RUNX3 promoter, and consequently a significantly lower percentage of unmethylated DNA at the RUNX3 promoter, in ANCA patients compared with healthy controls (Table 1). This suggests that reduced RUNX3 expression in ANCA patients might also be a consequence of aberrant epigenetic regulation.
Spearman rank analysis of expression levels for RUNX3 versus PR3 and MPO indicated a statistically significant inverse correlation between RUNX3 and PR3 mRNA levels and between RUNX3 and MPO mRNA levels (Figure 3, E and F). This inverse correlation of RUNX3 versus PR3 and MPO expression is consistent with a silencing mechanism for PR3 and MPO potentially mediated by RUNX3. Consequently, we tested the hypothesis that RUNX3 regulates PR3 and MPO expression.
RUNX3 represses PR3 and MPO expression.
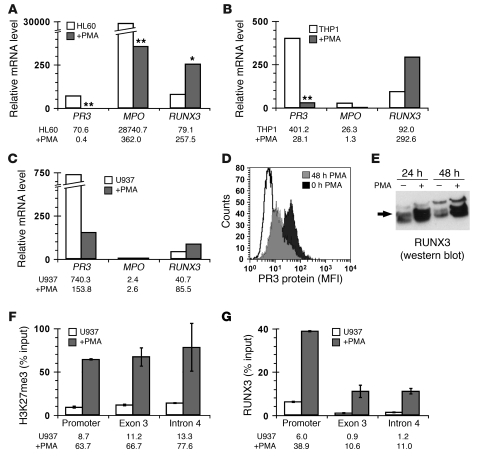
Human myelocytic cell lines, which undergo myeloid differentiation when treated with PMA, were used to test whether coordinated changes in gene expression for RUNX3 versus PR3 and MPO account for the inverse correlation observed in ANCA patients. PMA treatment resulted in significantly decreased PR3 mRNA in HL60, THP-1, and U937 cells and MPO mRNA in HL60 cells, as well as concomitantly increased RUNX3 expression in all 3 cell lines (Figure 4, A–C). We detected decreased PR3 protein and increased RUNX3 protein in differentiated U937 cells (Figure 4, D and E). These data confirm the inverse correlation in mRNA levels between RUNX3 and PR3 or MPO and are consistent with RUNX3-dependent silencing of PR3 and MPO.
Figure 4. Inverse correlation between RUNX3 expression and PR3 or MPO expression results from coordinated changes in gene expression.
(A–C) Relative mRNA levels for PR3, MPO, and RUNX3 from undifferentiated (white bars) or PMA-differentiated (gray bars) human acute promyelocytic leukemia cell line HL60 (A), acute monocytic leukemia cell line THP-1 (B), and histiocytic lymphoma (monocyte) cell line U937 (C) were determined by quantitative real-time RT-PCR with SYBR green (RUNX3) or Taqman (PR3 and MPO). Data represent median of 4–12 separate experiments, with exact values shown below graphs. *P < 0.05, **P = 0.001 versus untreated, paired t test. (D) PR3 protein levels were determined by flow cytometry on undifferentiated (black histogram) or PMA-differentiated (gray histogram) U937 cells. (E) RUNX3 protein levels (arrow) were detected by Western blot in U937 cells that were undifferentiated or differentiated with PMA for 24 and 48 hours. (F and G) H3K27me3 levels (F) and RUNX3 occupancy (G) at PR3 were determined by quantitative ChIP analysis in undifferentiated (white bars) and PMA-differentiated (black bars) U937 cells. Data (mean ± SD) represent percent DNA input from 2 experiments, with exact values shown below graphs.
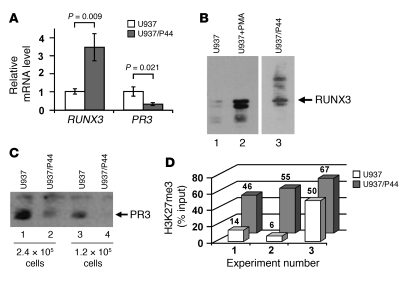
To demonstrate more directly a RUNX3-mediated mechanism for silencing PR3 and MPO, we used the U937-RUNX3/P44 cell line, which stably expresses RUNX3 (referred to herein as U937/P44 cells; ref. 36). We detected increased RUNX3 message and protein, as determined by Western blot, in U937/P44 cells compared with untransfected U937 cells (Figure 5, A and B). Because U937 cells express little or no MPO, our analysis was confined to PR3. In U937/P44 cells, reduced levels of PR3 message and total PR3 protein were detected (Figure 5, A and C). These data correlate RUNX3 overexpression with reduction of PR3 mRNA and PR3 protein levels. A reduction in PR3 mRNA with concomitant reduction in PR3 protein indicates that transcriptional changes in PR3 directly alter the amount of PR3. Importantly, mechanisms that alter transcriptional activity of PR3 may alter antigen availability.
Figure 5. Overexpression of RUNX3 in U937 myeloid cells silences PR3 expression.
(A) Relative RUNX3 and PR3 mRNA levels in undifferentiated U937 cells and undifferentiated U937/P44 cells were determined by quantitative real-time RT-PCR with SYBR green (RUNX3) and Taqman (PR3). (B and C) Western blot analysis was performed to detect RUNX3 (B) and PR3 (C) protein in undifferentiated U937 and U937/P44 cells and in PMA-differentiated U937 cells as indicated. Lanes in B were run on the same gel but were noncontiguous (white line). (D) H3K27me3 levels at PR3 were determined by quantitative ChIP analysis in undifferentiated U937 and U937/P44 cells. Data show percent DNA input for 3 separate experiments, with exact values shown above bars.
RUNX3 binds to PR3 and MPO genes.
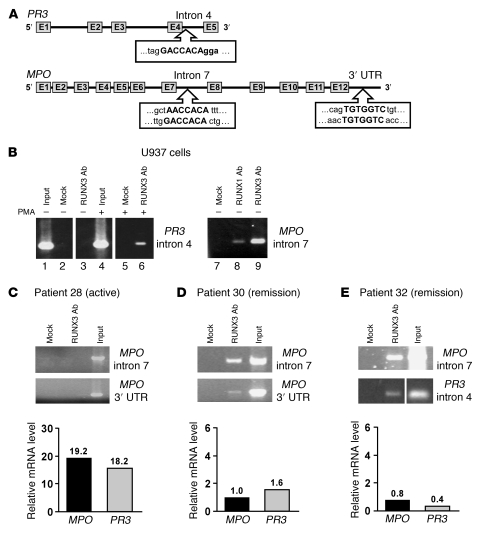
We addressed the possibility that RUNX3-mediated silencing of PR3 and MPO involve direct interaction with PR3 and MPO by searching for RUNX3 binding sites within the genes. Consensus RUNX3 binding sites (5′-PuACCPuCA-3′ and 5′-TGPyGGTPy-3′) were identified in intron 4 of PR3 and intron 7 and 3′ untranslated region (UTR) of MPO (Figure 6A). These sites were confirmed to be RUNX3 targets by ChIP. Chromatin from U937 cells was immunoprecipitated with a nonspecific antibody and a RUNX3 antibody. The RUNX3 site in intron 4 of PR3 was only immunoprecipitated with the RUNX3 antibody (Figure 6B, lanes 3 and 6). In addition, the interaction of RUNX3 with PR3 intron 4 appeared to improve upon PMA treatment, after which RUNX3 protein levels increased (Figure 6B, lane 6). RUNX3 was also detected at MPO intron 7 in U937 cells (Figure 6B, lane 9). RUNX3 binding to MPO intron 7 was robust in undifferentiated U937 cells, which may contribute to the undetectable levels of MPO message in this myeloid cell line.
Figure 6. RUNX3 binds PR3 and MPO genes in myeloid cell lines and in ANCA patients in remission.
(A) Consensus RUNX3 binding sites in PR3 intron 4 and MPO intron 7 and 3′ UTR. (B) ChIP analysis for RUNX3 binding to PR3 intron 4 and MPO intron 7 in undifferentiated or PMA-differentiated U937 cells under the indicated immunoprecipitation conditions. Normal rabbit serum was used in mock immunoprecipitation. Lanes were run on the same gel but were noncontiguous (white lines). (C–E) ChIP analysis for RUNX3 binding to MPO intron 7 and 3′ UTR (C and D) or to MPO intron 7 and PR3 intron 4 (E) in neutrophils from an active ANCA vasculitis patient (C) and from ANCA vasculitis patients in remission (D and E). Relative mRNA levels were determined by Taqman quantitative RT-PCR for MPO and PR3 from total leukocytes of ANCA vasculitis patients and calculated as fold change relative to healthy control, with exact values shown above bars. Lanes in E were run on the same gel but were noncontiguous (white line).
If RUNX3 binding to PR3 and MPO mediates gene silencing, it may be that RUNX3 binding inversely correlates with PR3 and MPO expression. RUNX3 binding to PR3 and MPO was not detected in patients with active disease, and generally high PR3 and MPO message, compared with patients in remission, with less PR3 and MPO message (Figure 6, C–E). Although recovering RUNX3 binding to PR3 or MPO varied in patients, surprisingly, in healthy controls, with the lowest PR3 and MPO message levels, RUNX3 binding was difficult to detect (data not shown). This latter observation is not incompatible with the concept of RUNX3-mediated gene silencing. Indeed, if RUNX3 ordinarily establishes silencing of PR3 and MPO in neutrophil progenitors during myelopoiesis by an epigenetic mechanism, rather than through active repression, persistent recruitment of RUNX3 to those genes would not be obligatory to maintain silencing in mature, circulating neutrophils. Thus, instead of differential RUNX3 binding at PR3 and MPO between healthy controls and ANCA patients, epigenetic differences may distinguish healthy controls from patients and signal the transcriptionally silent state of PR3 and MPO in neutrophils of healthy controls. The depletion of H3K27me3 in ANCA patients relative to healthy controls at PR3 and MPO (Figure 2, A and B) is consistent with this hypothesis. Accordingly, we next sought a mechanism that directly links RUNX3 regulation of myeloid gene expression to their epigenetic silencing via H3K27 trimethylation.
RUNX3 stimulates methylation of H3K27.
Methylation of H3K27 is catalyzed by the histone methyltransferase enhancer of zeste homolog 2 (EZH2), a Polycomb group (PcG) protein and subunit of Polycomb repressive complex 2 (PRC2; refs. 37–40). PcG proteins regulate gene expression in Drosophila through distinct genomic regions termed Polycomb response elements (PREs; ref. 41). In mammalian cells, similar PREs have not been characterized. However, interaction between site-specific DNA binding proteins and PRC2 subunits has been implicated in targeting PRC2 (42). We tested whether RUNX3 stimulates methylation of H3K27 by recruiting PRC2 to PR3 and MPO. ChIP for H3K27me3 was performed on cell lines with forced expression of RUNX3. Compared with undifferentiated U937 cells, PMA-treated U937 cells were enriched for H3K27me3 at PR3 promoter, exon 3, and intron 4 and showed increased RUNX3 binding to PR3 (Figure 4, F and G). The enrichment of RUNX3 at the PR3 promoter after PMA treatment may reflect RUNX3 recognition of DNA sequences similar to the RUNX3 consensus sequence. The element 5′-GGCCACA-3′, which differs from the consensus RUNX3 site (5′-PuACCPuCA-3′) at a single nucleotide, is present at PR3 at –292 and –223 of the transcription start site. We also observed enrichment of H3K27me3 at the PR3 promoter in U937/P44 cells (Figure 5D). These data suggest that RUNX3 is sufficient to increase H3K27me3 levels.
RUNX3 interacts with PRC2 subunits.
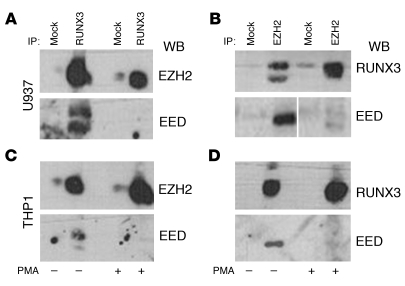
We evaluated whether the RUNX3-mediated increase in H3K27me3 involves direct interaction with PcG proteins. Western blot analysis detected EZH2 in U937 and THP-1 cell extracts immunoprecipitated with a RUNX3 antibody (Figure 7, A and C). RUNX3/EZH2 interaction was confirmed by Western blot analysis with a RUNX3 antibody on U937 and THP-1 cell extracts immunoprecipitated with an EZH2 antibody (Figure 7, B and D). Similarly, RUNX3 was found to interact with the PRC2 subunit embryonic ectoderm development (EED; Figure 7). The physical interaction between RUNX3 and the PRC2 components EZH2 and EED offers an explanation for recruitment of PRC2 and H3K27me3 to specific genes, including PR3 and MPO.
Figure 7. RUNX3 directly interacts with PRC2 subunits EZH2 and EED.
Western blot analysis on immunoprecipitated proteins from undifferentiated and PMA-differentiated U937 cells (A and B) and THP-1 cells (C and D). Cell extracts were immunoprecipitated with anti-RUNX3 (A and C) or anti-EZH2 (B and D) Ab, and Western blots were performed with anti-EZH2 (A and C), anti-RUNX3 (B and D), and anti-EED (A–D) Abs. Normal IgG was used in mock immunoprecipitations. Lanes in B were run on the same gel but were noncontiguous (white line).
The interaction between RUNX3 and EZH2 was detected in undifferentiated and differentiated (i.e., PMA-treated) U937 and THP-1 cells. However, in differentiated cells, the interaction between EED and RUNX3 and EZH2 was lost (Figure 7). This failure to detect EED in a complex with RUNX3 and EZH2 is surprising. Although EED does not catalyze histone methylation, it is thought to be necessary for methyltransferase activity by stabilizing the PRC2 components (43). In the absence of EED, H3K27me3 is lost and EZH2 protein is not detected (44). In contrast, the Drosophila ortholog of EZH2, enhancer of zeste [E(Z)], is found in a complex following embryogenesis that lacks the EED ortholog extra sex combs (ESC; ref. 45). Similarly, perhaps the RUNX3/EZH2 interaction stabilizes EZH2, and EED is no longer needed. Our observation warrants further investigation, as it has implications for regulating EZH2 methyltransferase activity.
Aberrant expression of neutrophil granule genes is not a consequence of ANCA disease.
An important question raised by our observation of increased PR3 and MPO expression and by the model we propose for the regulation of these genes is whether the increased expression and decreased H3K27me3 is a consequence of ANCA or more causally related to ANCA disease. To test the hypothesis that the changes we observed in PR3 and MPO are a consequence of ANCA disease, we turned to an animal model for MPO-ANCA. In this model, necrotizing and crescentic glomerulonephritis can be induced in otherwise normal and healthy mice by injecting anti-MPO IgG (5).
At 6 days after administering an intravenous dose of anti-MPO IgG into strain 129 mice, the animals were euthanized, kidneys were scored for glomerular lesions, and RNA was isolated from purified peripheral leukocytes to measure Mpo expression. Glomerular crescents were detected in all 5 mice that received anti-MPO IgG, with a range of 46%–80% affected glomeruli. As expected, this demonstrated that mice injected with anti-MPO IgG displayed symptoms of ANCA disease. However, the level of Mpo message in peripheral leukocytes of wild-type 129 strain mice 6 days after injection of anti-MPO IgG did not differ from that in peripheral neutrophils of wild-type 129 strain mice prior to injection (Figure 8). Interestingly, Mpo message levels in peripheral neutrophils of C57BL/6 mice were less than those observed in 129 mice (Figure 7). This is significant because 129 mice have a more severe response to anti-MPO IgG than do C57BL/6 mice: the average percent of glomeruli with crescents is 60%–70% in 129 mice and 5%–10% in C57BL/6 mice (H. Xiao, unpublished observations).
Figure 8. Mpo message level in peripheral neutrophils is insensitive to anti-MPO IgG.
Mouse Mpo mRNA levels from peripheral leukocytes were measured by quantitative real-time PCR, and ΔCt values were plotted against a standard curve of Mpo-positive cells serially diluted with Mpo-negative cells (see Methods and Supplemental Figure 5). Shown is Mpo message level for C57BL/6 and 129 mice not injected with anti-MPO IgG as well as for 129 mice 6 days after injection with anti-MPO IgG. Data represent average of 2 biological replicates of Mpo levels from leukocytes that were purified and pooled for each category prior to isolating RNA.
Our findings of ANCA disease symptoms in mice after anti-MPO IgG injection without an increase in Mpo message and increased response to antibody with higher baseline Mpo message levels are inconsistent with increased autoantigen gene expression occurring solely as a consequence of autoantibody exposure. Instead, they suggest that increased autoantigen gene expression is more causally related to disease. From this perspective, pathways that modulate autoantigen gene expression, such as the putative silencing mechanism described here, could be critical to the etiology of ANCA disease.
Discussion
Because patients with ANCA disease aberrantly express the ANCA autoantigen genes PR3 and MPO, we tested the hypothesis that aberrant expression results from disrupted epigenetic silencing. We demonstrate that aberrant expression in ANCA disease patients could result from active gene transcription, which suggests a failure in normal gene silencing. Importantly, in a mouse model of MPO-ANCA, gene silencing did not appear to be disrupted in response to autoantibody, because Mpo expression did not increase upon exposure to anti-Mpo antibodies. Instead, our evidence suggests that transcriptional regulation of PR3 and MPO may involve an epigenetic process in normal neutrophils that is perturbed in neutrophils of ANCA patients. Central to our case is the observation that, at PR3 and MPO loci, levels of H3K27me3, a mark of transcriptionally silent chromatin, were depleted in neutrophils of ANCA patients relative to those of healthy controls.
The depletion of H3K27me3 could result from a failure to methylate PR3 and MPO loci in ANCA patients. In the absence of a characterized PRE that would target PRC2 to PR3 and MPO, we hypothesize that RUNX3 directs PRC2 to PR3 and MPO. We propose this hypothesis based on the following data: (a) RUNX3 expression inversely correlated with PR3 and MPO message in leukocytes; (b) forced expression of RUNX3 in cell lines reduced PR3 and MPO expression; and (c) RUNX3 bound its cognate sites in PR3 and MPO in cell lines and in leukocytes of ANCA patients in remission. Furthermore, we detected a physical interaction between RUNX3 and EZH2, the PRC2 subunit with methyltransferase activity. This evidence is consistent with a model of epigenetic gene silencing in which reduced levels of H3K27me3 at PR3 and MPO in ANCA patients compared with healthy controls result from a failure of RUNX3 to recruit EZH2 and methylate H3K27 (Figure 9). An equally likely scenario is that both methylation and demethylation of H3K27 are involved in determining the epigenetic state of PR3 and MPO. In support of this, we measured expression of the H3K27me3 demethylase JMJD3 and found that it was elevated in leukocytes from ANCA patients compared with healthy controls. This suggests that a balance of these 2 processes determines the level of H3K27me3 at PR3 and MPO in mature neutrophils.
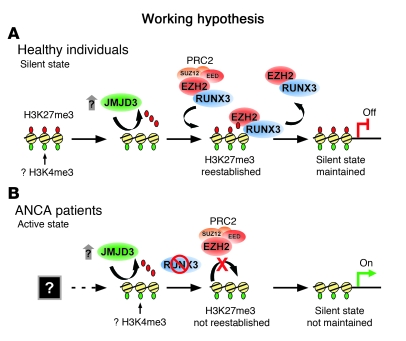
Figure 9. Model for PR3 and MPO gene silencing in normal neutrophils and disruption in ANCA vasculitis patients, highlighting the role of the H3K27me3 modification and enzymes that regulate this histone mark.
(A) Granulocyte genes are silenced in normal mature circulating neutrophils. We hypothesize that maintenance of a transcriptionally silent state is a dynamic process. Inflammatory cytokines can induce JMJD3 (26). Our present demonstration of an interaction between RUNX3 and PRC2 subunits suggests RUNX3 can recruit EZH2 and reestablish H3K27me3. This epigenetic modification would maintain the transcriptionally silent state. (B) In neutrophils of ANCA vasculitis patients, H3K27me3 is depleted at PR3 and MPO because of an unknown process, perhaps during neutrophil development. An alternative, but not mutually exclusive, possibility is that increased JMJD3 in ANCA patients reverses the silent state at PR3 and MPO by erasing the histone mark necessary for PcG-mediated silencing. Low levels of RUNX3 fail to recruit the EZH2 necessary to reestablish H3K27me3 and maintain transcriptional silence. Furthermore, DNA methylation at MPO may also contribute to maintaining the transcriptionally silent state (not shown). An intriguing possibility is that PR3 and MPO are in a transcriptionally poised state marked by both H3K27me3 and H3K4me3 (59) in neutrophils of healthy individuals. In neutrophils of patients with ANCA vasculitis, depletion of H3K27me3 allows transcription of PR3 and MPO.
Whereas the transcriptional activity of PR3 and MPO likely depends on their epigenetic status, the level of H3K27me3 may be only one predictor of transcriptional status. H3K27me3 levels at PR3 inversely correlated with PR3 mRNA levels, which suggests that H3K27me3 can predict PR3 expression. Only a slight inverse correlation was found between H3K27me3 levels and MPO mRNA levels, which suggests that transcriptional regulation of MPO appears to use additional mechanisms. A potential additional silencing mechanism may be DNA methylation. A CpG island in MPO was unmethylated in patients, and there was a significant increase in methylated DNA at this CpG island in healthy controls. The observation of another epigenetic difference between ANCA patients and healthy controls suggests that epigenetic dysregulation is an important hallmark of ANCA disease. While a causal link between these epigenetic changes and expression of PR3 and MPO can be inferred in ANCA patients, results from our in vitro experiments are consistent with our hypothesis that ANCA autoantigen genes are silenced by a RUNX3-dependent mechanism via H3K27me3.
Our findings are significant because they reveal a role for epigenetics in autoimmune vasculitis. Epigenetics has been implicated in autoimmunity, in particular DNA methylation in SLE (46). Our data highlight the role in ANCA of the H3K27me3 modification and enzymes that regulate this epigenetic histone mark. Failure to methylate histone H3K27 by EZH2 may result from impaired targeting of EZH2 to PR3 and MPO. This assertion is based on our evidence that RUNX3 recruited EZH2. Although this is not the first example of EZH2 interaction with a site-specific DNA binding protein (42), it is the first example to our knowledge of a RUNX3/EZH2 association. In addition, active demethylation would also explain the reduction in H3K27me3 at PR3 and MPO loci in ANCA patients. The H3K27me3 demethylase JMJD3 functions in cellular differentiation (47, 48) and is induced by inflammatory cytokines (26, 49). In ANCA patients, the increase in JMJD3 expression may reverse the silent state at PR3 and MPO by erasing the histone mark necessary for PcG-mediated silencing.
The implication that a decrease in RUNX3 is a predisposing factor for ANCA vasculitis is intriguing in light of previous studies investigating RUNX proteins in autoimmune diseases. RUNX proteins have been proposed to be susceptibility factors for SLE, rheumatoid arthritis, and psoriasis (50). Moreover, RUNX3 deficiency in the mouse leads to spontaneous inflammatory bowel disease (51). Our present observation that RUNX3 mRNA was reduced in ANCA vasculitis patients adds ANCA vasculitis to the list of autoimmune diseases in which RUNX3 plays a role, which may be to establish the epigenetic state of select target genes.
Our data suggest a mechanism for determining the epigenetic status of PR3 and MPO, but it remains to be determined when the epigenetic state is established in normal neutrophils and when it is disrupted in neutrophils of patients with ANCA disease. Additionally, although the present work highlights a gene-silencing mechanism that has gone awry in ANCA disease, further studies will be needed to determine what triggers the gene-silencing mechanism to fail. One possibility is that decreased RUNX3 expression, perhaps via preferential promoter DNA methylation in ANCA disease patients, is the upstream event that disrupts PcG mediated silencing of PR3 and MPO. Another equally likely possibility is that RUNX3 is a PRC2 target (52) and the upstream event is aberrant PcG-mediated silencing, which triggers misregulation of a class of neutrophil genes in ANCA vasculitis patients. Investigating these limitations and experimentally testing predictions of our model may demonstrate a causal link between epigenetic status and transcriptional status of PR3 and MPO, and potentially other genes aberrantly expressed in ANCA vasculitis patients. Finally, understanding the establishment and maintenance of the transcriptionally silent state of ANCA autoantigen genes could unveil new therapeutic approaches.
Methods
Patients.
ANCA patients were enrolled at UNC–Chapel Hill clinics, and healthy volunteers from UNC were recruited, according to the guidelines of the UNC Institutional Review Board (IRB protocol no. 97-MED-44). Informed consent was obtained from all participants prior to enrollment. The demographics of the entire study population are summarized in Supplemental Table 2. ANCA patients were categorized as microscopic polyangiitis (MPA), Wegener granulomatosis (WG), Churg-Strauss syndrome (CSS), or renal-limited pauci-immune necrotizing and crescentic glomerulonephritis (NCGN), as defined by the Chapel Hill Consensus Conference (53). The severity of disease activity was ranked on the basis of the Birmingham Vasculitis Activity Score (BVAS) 2003 version (18).
Cell culture.
Human cell lines Jurkat (lymphoid leukemia), THP1 (monocytic leukemia), HL60 (promyelocytic leukemia), and U937 (histiocytic lymphoma) were obtained from Tissue Culture Facility of UNC and cultured in RPMI1640 supplemented with 10% FCS, glutamine, and antibiotics. Induction of cell differentiation was accomplished in the presence of PMA (Sigma-Aldrich) at concentrations of 25 nM (THP1) or 100 nM (HL60 and U937) for 24 and 48 hours. The U937/P44 cell line (36), a G418-resistant clone of U937 stably transfected with RUNX3 expression construct, was provided by A.L. Corbí López (Consejo Superior de Investigaciones Cientificas, Madrid, Spain) and maintained in complete medium with 1.0 mg/ml G418 (Gibco, Invitrogen) and 25 mM HEPES (Gibco, Invitrogen).
Microarray and quantitative PCR.
Total leukocytes were obtained from EDTA-treated human blood. Erythrocytes were lysed using hypotonic NH4Cl buffer. RNA was extracted using RNA STAT-60 (Tel-Test), and all samples were treated with DNase. Leukocyte gene expression was examined on Affymetrix HU133A and B microarrays at Gene Logic Inc. as recommended by the manufacturer (Affymetrix) and previously described (12, 54).
Quantitative PCR assays were performed on an ABI PRISM 7900HT sequence detection system (Applied Biosystems) using the TaqMan EZ RT-PCR kit (Applied Biosystems). Primers and probes were designed using Primer Express software (Applied Biosystems). Primers used with SYBR green fluorescence assays were validated for linear amplification and verified for only a single product. Primer sequences are listed in Supplemental Table 3. Relative level of a specific mRNA was determined by 2(–ΔΔCt) calculations and expressed as fold change of reference control samples (55). Cytochrome c oxidase COX5b was used as RNA standard.
For the analysis of mouse Mpo mRNA, EDTA-treated whole blood was collected and pooled from 3 mice, and 2 mice in a biological replicate, with or without injection of anti-MPO antibodies. From the pooled samples, total leukocytes were isolated after lysing erythrocytes with hypotonic NH4Cl buffer. RNA was purified using RNA STAT-60 (Tel-Test) and treated with DNase. To quantify the relative level of mouse Mpo mRNA in peripheral leukocytes, ΔCt values were compared with a standard curve for mouse Mpo mRNA (Supplemental Figure 4). The standard curve was made by measuring mouse Mpo mRNA from Mpo-positive cells (WEHI cells) that were serially diluted into Mpo-negative cells (mouse ES cells). The final value of mouse Mpo mRNA from the isolated leukocytes was reported as the number of Mpo-positive cells per 1,000 cells.
Anti-MPO antibody injection.
The protocol for injecting mice with anti-MPO antibodies was similar to that previously reported (5). All mouse experiments were performed using protocols approved by the UNC–Chapel Hill Institutional Animal Care and Use Committee. Female strain 129 mice 6–8 weeks of age received anti-MPO antibodies by tail vein injection at a dose of 50 μg protein/g body weight. At 6 days after injection, mice were euthanized, RNA was extracted from peripheral leukocytes, and kidneys were dissected and prepared for histological examination. Kidneys were processed as described previously (5) and examined with a light microscope to determine the percent of glomeruli with crescents. Glomerular crescents were detected in all 129 mice, with 46%, 64%, 78%, 79%, and 80% of glomeruli affected.
ChIP.
Isolated polymorphonuclear leukocytes, leukocytes from NH4Cl lysis buffer, or cell lines (2 × 106 cells/ml/sample) were incubated with formaldehyde at a final concentration of 0.4% to crosslink DNA and proteins. Glycine was added to a final concentration of 0.125 M to stop crosslink reaction. Cells were lysed in 0.5 ml SDS lysis buffer (1% SDS; 10 mM EDTA, pH 8.0; and 50 mM Tris-HCl, pH 8.1), and chromatin was sonicated with Branson Digital Sonifier 450 (Branson Ultrasonics Corp.). Samples were diluted 3-fold with ChIP dilution buffer (0.1% SDS; 1.1% Triton X-100; 1.2 mM EDTA, pH 8.0; 16.7 mM Tris-HCl, pH 8.1; and 167 mM NaCl), and 150 μl, or 1:10, of this solution was removed to serve as input. Samples were precleared with Protein A/G Plus-Agarose beads (Santa Cruz Biotechnology). Lysates were immunoprecipitated overnight at 4°C with 5–10 μg of the following antibodies, all from Abcam: anti-H3K9me2 (catalog no. ab1220), anti-H3K27me3 (catalog no. ab6002), and anti-RUNX3 (catalog no. ab11905). Immune complexes were collected with Protein A/G Plus-Agarose beads. Bound immune complexes were eluted, incubated at 65°C overnight with NaCl (0.2 M final) to reverse crosslinks, and treated with proteinase K (Invitrogen) to a final concentration of 400 μg/ml. DNA was extracted with phenol/chloroform and analyzed by real-time PCR. PCR products were measured by SYBR green fluorescence, and the relative level of a specific PCR product was expressed as percent of input chromatin using raw Ct values, not adjusted for dilution. Primers listed in Supplemental Table 4 were designed using Primer Express software (Applied Biosystems) and validated for linear amplification against input control template.
RNA immunoprecipitation.
RNA immunoprecipitation was performed using a procedure similar to the one described above for ChIP and previously reported for RNA immunoprecipitation by Gilbert et al. (56). Antibody 4H8 (catalog no. ab5408; Abcam), which recognizes the phosphorylated and unphosphorylated carboxyterminal tail domain of RNA polymerase II, was used for immunoprecipitations. The primers used to detect PR3 and MPO mRNA are listed in Supplemental Table 4.
DNA methylation.
DNA methylation at the PR3 (CpG island at chromosome 19; 843,469–844,085), MPO (CpG island at chromosome 17; 56,356,375–56,357,013), and RUNX3 (CpG island at chromosome 1; 25,255,528–25,259,005) was determined using the SA Biosciences Methyl-Profiler DNA Methylation assay (MEA-03). All coordinates correspond to UCSC Genome Browser annotation of Human GRCh37 Assembly (hg19). Primers for quantitative real-time PCR for PR3 (catalog no. MePH23428-1A), MPO (catalog no. MePH22382-1A), and RUNX3 (catalog no. MePH28535-1A) were from SABiosciences. Briefly, 50–100 ng of purified DNA from total leukocytes was used in a mock restriction enzyme digest, DNA methylation–sensitive restriction enzyme digest, DNA methylation–dependent restriction enzyme digest, and double digest. After an overnight incubation at 37°C, the DNA was analyzed by real-time PCR. Ct values were used to determine the relative DNA methylation levels. A detailed explanation of the calculations used to determine DNA methylation states is described in Supplemental Methods and available from SABiosciences (57).
Flow cytometry.
U937 cells (1 × 106) were fixed and permeabilized using FACS Lysing Solution and FACS Permeabilizing Solution 2 (BD Biosciences) as prescribed by the manufacturer. Cells were labeled with primary antibodies rabbit anti-PR3 or mouse anti-PR3 (gift of J. Wieslander, Lund University, Lund, Sweden), followed by secondary antibodies FITC-conjugated affinpure F(ab′)2 fragment donkey anti-rabbit or anti-mouse IgG (H+L) (Jackson ImmunoResearch Laboratories). Cells were gated and analyzed by a FACSCalibur (BD) with Summit software (DakoCytomation).
Co-IP and Western blot.
U937 and THP1 cells were cultured with or without PMA (100 nM for 24 or 48 hours). Approximately 2.0 × 108 cells were rinsed in cold PBS and placed in nuclear lysis buffer (60 mM KCl; 2 mM EDTA; 0.5 mM EGTA; 15 mM Tris-HCl, pH 7.4; 0.5M sucrose; and 1.0 mM PMSF) for 10 minutes on ice. Pelleted nuclei (2,000 g for 10 minutes at 4°C) were lysed (25 mM HEPES, pH 7.5; 12.5 mM MgCl2; 150 mM KCl; 0.5% NP-40; 1.0 mM dithiothreitol plus 10 μg/ml leupeptin; 1% aprotinin; and 0.5 mM phenylmethylsulfonyl fluoride). Glycerol was added to a final concentration of 10%. After incubation (1 hour at 4°C), extracts were centrifuged at 16,000 g for 30 minutes, and supernatants were collected. Extracts were precleared with an excess of Protein A/G Plus-Agarose beads (Santa Cruz Biotechnology). One-fourth of each precleared extract was immunoprecipitated with rabbit polyclonal anti-RUNX3 (catalog no. ab11905; Abcam), one-fourth was immunoprecipitated with rabbit anti-EZH2 (catalog no. ab3748; Abcam), and the remaining was used for mock controls with normal IgG (Santa Cruz Biotechnology). Immunoprecipitations were incubated overnight at 4°C. Immune complexes were retrieved with Protein A/G, washed 3–5 times in lysis buffer, eluted in 2× Laemmli sample buffer, and analyzed by Western blot. RUNX3 Western blots were hybridized with monoclonal anti-RUNX3 (MBL). EZH2 Western blots were hybridized with monoclonal anti-EZH2 (M5; ref. 58). Blots were reprobed for EED using a chicken polyclonal antibody (catalog no. ab14294; Abcam). To assess PR3 and RUNX3 protein levels in U937 and U937/p44 cells, Western blots were performed on cells lysed in 1.5× Laemmli sample buffer at a concentration of 6 × 106 cells/ml. Blots were hybridized with rabbit anti-PR3 or with rabbit polyclonal anti-RUNX3 (Abcam).
Statistics.
Comparisons between 2 groups were done by Wilcoxon rank-sum test. The overall P value for each subset of genes was 0.05, with adjustment using a Bonferroni correction (α/n comparisons per group). Spearman rank analysis was used to calculate r2 and P values between continuous measures.
Supplementary Material
Acknowledgments
We thank Julian Preston for advice early in the project, our colleagues in the Magnuson laboratory for continued helpful discussions, and Susan L. Hogan and Yichun Hu for statistical assistance. We are grateful to Angel L. Corbí López for providing the U937/P44 cell line. This work was supported by NIH grant PO1 DK058335 to R.J. Falk and J.C. Jennette.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(9):3209–3219. doi:10.1172/JCI40034.
Peter Hewins’s present address is: Department of Renal Medicine, University Hospital Birmingham, Birmingham, United Kingdom.
Carla M. Nester’s present address is: Adult and Pediatric Nephrology, University of Iowa Hospitals and Clinics, Iowa City, Iowa, USA.
William F. Pendergraft III’s present address is: Department of Medicine, University of California at San Francisco Medical Center, San Francisco, California, USA.
References
- 1.Bansal PJ, Tobin MC. Neonatal microscopic polyangiitis secondary to transfer of maternal myeloperoxidase-antineutrophil cytoplasmic antibody resulting in neonatal pulmonary hemorrhage and renal involvement. Ann Allergy Asthma Immunol. 2004;93(4):398–401. doi: 10.1016/S1081-1206(10)61400-7. [DOI] [PubMed] [Google Scholar]
- 2.Falk RJ, Terrell RS, Charles LA, Jennette JC. Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc Natl Acad Sci U S A. 1990;87(11):4115–4119. doi: 10.1073/pnas.87.11.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Little MA, et al. Antineutrophil cytoplasm antibodies directed against myeloperoxidase augment leukocyte-microvascular interactions in vivo. Blood. 2005;106(6):2050–2058. doi: 10.1182/blood-2005-03-0921. [DOI] [PubMed] [Google Scholar]
- 4.Schlieben DJ, Korbet SM, Kimura RE, Schwartz MM, Lewis EJ. Pulmonary-renal syndrome in a newborn with placental transmission of ANCAs. Am J Kidney Dis. 2005;45(4):758–761. doi: 10.1053/j.ajkd.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Xiao H, et al. Antineutrophil cytoplasmic autoantibodies specific for myeloperoxidase cause glomerulonephritis and vasculitis in mice. J Clin Invest. 2002;110(7):955–963. doi: 10.1172/JCI15918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao H, et al. The role of neutrophils in the induction of glomerulonephritis by anti-myeloperoxidase antibodies. Am J Pathol. 2005;167(1):39–45. doi: 10.1016/S0002-9440(10)62951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kallenberg CG, Heeringa P, Stegeman CA. Mechanisms of Disease: pathogenesis and treatment of ANCA-associated vasculitides. Nat Clin Pract Rheumatol. 2006;2(12):661–670. doi: 10.1038/ncprheum0355. [DOI] [PubMed] [Google Scholar]
- 8.Xiao H, Schreiber A, Heeringa P, Falk RJ, Jennette JC. Alternative complement pathway in the pathogenesis of disease mediated by anti-neutrophil cytoplasmic autoantibodies. Am J Pathol. 2007;170(1):52–64. doi: 10.2353/ajpath.2007.060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jennette JC, Falk RJ. Small-vessel vasculitis. N Engl J Med. 1997;337(21):1512–1523. doi: 10.1056/NEJM199711203372106. [DOI] [PubMed] [Google Scholar]
- 10.Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997;89(10):3503–3521. [PubMed] [Google Scholar]
- 11.Cowland JB, Borregaard N. The individual regulation of granule protein mRNA levels during neutrophil maturation explains the heterogeneity of neutrophil granules. J Leukoc Biol. 1999;66(6):989–995. doi: 10.1002/jlb.66.6.989. [DOI] [PubMed] [Google Scholar]
- 12.Yang JJ, et al. Circumvention of normal constraints on granule protein gene expression in peripheral blood neutrophils and monocytes of patients with antineutrophil cytoplasmic autoantibody-associated glomerulonephritis. J Am Soc Nephrol. 2004;15(8):2103–2114. doi: 10.1097/01.ASN.0000135058.46193.72. [DOI] [PubMed] [Google Scholar]
- 13.Ohlsson S, Hellmark T, Pieters K, Sturfelt G, Wieslander J, Segelmark M. Increased monocyte transcription of the proteinase 3 gene in small vessel vasculitis. Clin Exp Immunol. 2005;141(1):174–182. doi: 10.1111/j.1365-2249.2005.02819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newburger PE, Subrahmanyam YV, Weissman SM. Global analysis of neutrophil gene expression. Curr Opin Hematol. 2000;7(1):16–20. doi: 10.1097/00062752-200001000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Tsukahara Y, et al. Gene expression in human neutrophils during activation and priming by bacterial lipopolysaccharide. J Cell Biochem. 2003;89(4):848–861. doi: 10.1002/jcb.10526. [DOI] [PubMed] [Google Scholar]
- 16.Yang JJ, et al. Expression profile of leukocyte genes activated by anti-neutrophil cytoplasmic autoantibodies (ANCA). Kidney Int. 2002;62(5):1638–1649. doi: 10.1046/j.1523-1755.2002.00619.x. [DOI] [PubMed] [Google Scholar]
- 17.Coldren CD, et al. Functional and genomic changes induced by alveolar transmigration in human neutrophils. Am J Physiol Lung Cell Mol Physiol. 2006;291(6):L1267–L1276. doi: 10.1152/ajplung.00097.2006. [DOI] [PubMed] [Google Scholar]
- 18.Luqmani RA, et al. Birmingham Vasculitis Activity Score (BVAS) in systemic necrotizing vasculitis. QJM. 1994;87(11):671–678. [PubMed] [Google Scholar]
- 19.Kalantry S, Mills KC, Yee D, Otte AP, Panning B, Magnuson T. The Polycomb group protein Eed protects the inactive X-chromosome from differentiation-induced reactivation. Nat Cell Biol. 2006;8(2):195–202. doi: 10.1038/ncb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 21.Lubbert M, Miller CW, Koeffler HP. Changes of DNA methylation and chromatin structure in the human myeloperoxidase gene during myeloid differentiation. Blood. 1991;78(2):345–356. [PubMed] [Google Scholar]
- 22.Lubbert M, Tobler A, Daskalakis M. Cytosine demethylation of the proteinase-3/myeloblastin primary granule protease gene during phagocyte development. Leukemia. 1999;13(9):1420–1427. doi: 10.1038/sj/leu/2401486. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto K, Kokubun S, Itoi E, Roach HI. Improved quantification of DNA methylation using methylation-sensitive restriction enzymes and real-time PCR. Epigenetics. 2007;2(2):86–91. doi: 10.4161/epi.2.2.4203. [DOI] [PubMed] [Google Scholar]
- 24.Oakes CC, La Salle S, Robaire B, Trasler JM. Evaluation of a quantitative DNA methylation analysis technique using methylation-sensitive/dependent restriction enzymes and real-time PCR. Epigenetics. 2006;1(3):146–152. doi: 10.4161/epi.1.3.3392. [DOI] [PubMed] [Google Scholar]
- 25.Schmelz K, Sattler N, Wagner M, Lübbert M, Dörken B, Tamm I. Induction of gene expression by 5-Aza-2′-deoxycytidine in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) but not epithelial cells by DNA-methylation-dependent and -independent mechanisms. Leukemia. 2005;19(1):103–111. doi: 10.1038/sj.leu.2403552. [DOI] [PubMed] [Google Scholar]
- 26.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 27.Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 28.Li QL, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109(1):113–124. doi: 10.1016/s0092-8674(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 29.Inoue K, et al. Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci. 2002;5(10):946–954. doi: 10.1038/nn925. [DOI] [PubMed] [Google Scholar]
- 30.Levanon D, et al. The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J. 2002;21(13):3454–3463. doi: 10.1093/emboj/cdf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taniuchi I, Littman DR. Epigenetic gene silencing by Runx proteins. Oncogene. 2004;23(24):4341–4345. doi: 10.1038/sj.onc.1207671. [DOI] [PubMed] [Google Scholar]
- 32.Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111(5):621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 33.Fainaru O, et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23(4):969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puig-Kroger A, et al. RUNX3 negatively regulates CD36 expression in myeloid cell lines. J Immunol. 2006;177(4):2107–2114. doi: 10.4049/jimmunol.177.4.2107. [DOI] [PubMed] [Google Scholar]
- 35.Ku JL, et al. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004;23(40):6736–6742. doi: 10.1038/sj.onc.1207731. [DOI] [PubMed] [Google Scholar]
- 36.Puig-Kroger A, et al. RUNX/AML and C/EBP factors regulate CD11a integrin expression in myeloid cells through overlapping regulatory elements. Blood. 2003;102(9):3252–3261. doi: 10.1182/blood-2003-02-0618. [DOI] [PubMed] [Google Scholar]
- 37.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 38.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111(2):185–196. doi: 10.1016/S0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 39.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111(2):197–208. doi: 10.1016/S0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 41.Ringrose L, Paro R. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development. 2007;134(2):223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 42.Villa R, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11(6):513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 43.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15(1):57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 44.Montgomery ND, et al. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15(10):942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 45.Furuyama T, Tie F, Harte PJ. Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis. 2003;35(2):114–124. doi: 10.1002/gene.10173. [DOI] [PubMed] [Google Scholar]
- 46.Strickland FM, Richardson BC. Epigenetics in human autoimmunity. Epigenetics in autoimmunity – DNA methylation in systemic lupus erythematosus and beyond. Autoimmunity. 2008;41(4):278–286. doi: 10.1080/08916930802024616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jepsen K, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 48.Sen GL, Webster DE, Barragan DI, Chang HY, Khavari PA. Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3. Genes Dev. 2008;22(14):1865–1870. doi: 10.1101/gad.1673508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishii M, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alarcon-Riquelme ME. Role of RUNX in autoimmune diseases linking rheumatoid arthritis, psoriasis and lupus. Arthritis Res Ther. 2004;6(4):169–173. doi: 10.1186/ar1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brenner O, et al. Loss of Runx3 function in leukocytes is associated with spontaneously developed colitis and gastric mucosal hyperplasia. Proc Natl Acad Sci U S A. 2004;101(45):16016–16021. doi: 10.1073/pnas.0407180101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283(25):17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jennette JC, et al. Nomenclature of systemic vasculitides. Proposal of an international consensus conference. Arthritis Rheum. 1994;37(2):187–192. doi: 10.1002/art.1780370206. [DOI] [PubMed] [Google Scholar]
- 54.Alcorta DA, et al. Leukocyte gene expression signatures in antineutrophil cytoplasmic autoantibody and lupus glomerulonephritis. Kidney Int. 2007;72(7):853–864. doi: 10.1038/sj.ki.5002371. [DOI] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol Cell. 2004;14(4):457–464. doi: 10.1016/S1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- 57.Methyl-Profiler DNA Methylation PCR Assays . Frederick, Maryland, USA: SABiosciences; 2010. [Google Scholar]
- 58.Hamer KM, Sewalt RG, den Blaauwen JL, Hendrix T, Satijn DP, Otte AP. A panel of monoclonal antibodies against human polycomb group proteins. Hybrid Hybridomics. 2002;21(4):245–252. doi: 10.1089/153685902760213859. [DOI] [PubMed] [Google Scholar]
- 59.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.