Abstract
Breast cancer is a highly heterogeneous disease, with several different subtypes being characterized by distinct histology, gene expression patterns, and genetic alterations. The tumor suppressor gene retinoblastoma 1 (RB1) is frequently lost in both luminal-B and triple-negative tumor (TNT; i.e., estrogen receptor–, progesterone receptor–, and human epidermal growth factor receptor 2–negative) breast cancer subtypes. However, a causal role for RB1 loss in different subtypes remains undefined. Here we report that deletion of Rb alone or together with its relative p107 in mouse mammary stem/bipotent progenitor cells induced focal acinar hyperplasia with squamous metaplasia. These lesions progressed into histologically diverse, transplantable mammary tumors with features of either luminal-B or TNT subtypes. The TNTs included basal-like tumors as well as tumors that exhibited epithelial-to-mesenchymal transition (EMT). The EMT-type tumors and a subset of the basal-like tumors, but not luminal-B–like tumors, expressed mutant forms of the tumor suppressor p53. Accordingly, targeted deletion of both Rb and p53 in stem/bipotent progenitors led to histologically uniform, aggressive, EMT-type tumors. Reintroduction of Rb into these tumor cells suppressed growth in vitro and tumor formation in vivo. These results establish a causal role for Rb loss in breast cancer in mice and demonstrate that cooperating oncogenic events, such as mutations in p53, dictate tumor subtype after Rb inactivation.
Introduction
Breast cancer is a highly heterogeneous disease representing tumors with distinct histology, gene expression patterns, and genetic alterations (1–4). Marker analysis revealed the existence of human epidermal growth factor receptor 2–positive (HER-2/NEU+) and estrogen receptor–positive (ER+) breast tumors as well as triple-negative tumors (TNTs), which do not express hormone receptors or HER-2/NEU (5). Likewise, microarray profiling identified luminal-A–like, luminal-B–like, HER2/NEU+, and basal-like breast carcinomas (2, 6–8). More recently, basal-like breast carcinoma were recognized as a subgroup of TNTs that also include metaplastic and claudin-low subtypes (9, 10, 11–14). The metaplastic and claudin-low TNTs exhibit epithelial-to-mesenchymal transition (EMT) and a cancer stem cell expression signature (9, 10, 14, 15). Although the basal-like subtype responds to chemotherapy, no effective treatment is available for metaplastic and claudin-low tumors; metastatic disease is virtually untreatable.
The tumor suppressors breast cancer 1 (Brca1), p53, phosphatase and tensin homolog (Pten), and retinoblastoma 1 (RB1) are commonly lost in familial and sporadic forms of TNTs (16–19). RB1 gene rearrangement was reported in approximately 10% of primary breast carcinomas of undefined subtypes and in approximately 20%–25% of breast cancer cell lines, including MDA-MB436, MDA-MB468, and BT549, which are derived from TNTs (20–27). Microarray analysis revealed that RB1 transcripts are downregulated in about 70% of basal-like breast tumors with concomitant induction of the CDK4/6 inhibitor p16Ink4a, a target for transcriptional repression by the RB1 protein, pRb (28). Low RB1 gene expression and loss of heterozygosity (LOH) at the RB1 locus were subsequently identified at high frequency in luminal-B–like and basal-like/TNTs (29). In accordance, a recent study has demonstrated loss of pRb expression coupled with high expression of p16Ink4a and p53, presumably a stabilized mutant form, in most basal-like/TNTs (30).
pRb regulates cell growth and differentiation by modulating the activity of transcription factors such as E2F family members (31, 32). Among E2F-responsive genes are factors required for cell cycle progression and apoptotic cell death (33). Apoptosis downstream of pRb is often, but not always, mediated by the tumor suppressor p53 (34, 35). Accordingly, pRb and p53 are commonly lost together in cancer; various DNA viruses harbor oncoproteins, such as SV40 large T antigen (SV40 Tag), that transform host cells by sequestering pRb and its relatives p107 and p130 as well as p53 (36, 37).
Much insight into Rb function was gained through analysis of mutant mice. Most Rb+/– heterozygote mice die at approximately 11 months of age with a wasting disease caused by pituitary tumors, whereas Rb–/– embryos die at midgestation (38). Rb loss in many tissues leads to ectopic cell proliferation, apoptosis, and incomplete differentiation (38). Mammary placodes from Rb–/– embryos develop normally when transplanted into recipient mammary glands (39). In contrast, mammary gland–specific transgenic expression of SV40 Tag, which binds the pRb protein family (pRb, p107, and p130), p53, and other factors, such as p300/CBP, or of a truncated form (T121), which binds the Rb family but not p53, induces mammary tumors (40–42). Thus, the long-term consequences of Rb inactivation alone on mammary gland tumorigenicity and breast cancer subtypes are yet to be determined.
Here we describe the effects of somatic inactivation of Rb in mammary epithelium using a floxed Rb allele and mammary-specific deleter Cre lines. We found that deletion of Rb in stem/early progenitor cells led to focal metaplastic lesions with squamous transdifferentiation and progression to form mammary tumors with features of luminal-B or basal-like/EMT breast carcinomas, with a subset of the latter exhibiting p53 mutations. Combined inactivation of Rb and p53 led to a uniform type of EMT tumor that was highly aggressive, yet readily inhibited upon reintroduction of pRb.
Results
Targeted inactivation of Rb in mammary epithelium via WAP-Cre induces lactation defects, but not mammary tumors.
Loss of Rb was previously modeled by targeted expression of SV40 Tag in the mammary gland under control of the whey acidic protein (WAP) promoter. This promoter directs gene expression to differentiating mammary epithelium during the estrous cycle and lobuloalveologenesis as well as to parity-identified stem cells during pregnancy (43, 44). The WAP-Cre deleter line was crossed with Rbfl mice, which harbor a floxed Rb exon 19 allele (45), yielding WAP-Cre:Rbfl/fl mice. In many tissues, concurrent inactivation of Rb with p107 or p130 is required to induce tumors (46, 47). We therefore also inactivated p107, which is coexpressed with Rb in the mammary gland, by crossing WAP-Cre:Rbfl/fl mice into a p107–/– background (48–50).
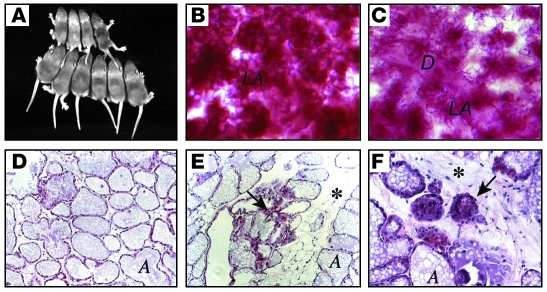
Mammary gland development and lactation proceeded apparently normally in young nulliparous and parous WAP-Cre:Rbfl/fl:p107–/– females (data not shown). However, by 6 months of age, about half of multiparous WAP-Cre:Rbfl/fl:p107–/– females exhibited lactation defects, leading to impaired development and death of their offspring (Figure 1A). Newborns from these WAP-Cre:Rbfl/fl:p107–/– females suckled, but contained very little milk in their stomachs, and their growth retardation and morbidity could be rescued by transferring to foster mothers (data not shown). Consistent with this defect, lobuloalveoli of lactating mammary glands from WAP-Cre:Rbfl/fl:p107–/– females were less dense compared with those from controls (Figure 1, B and C) and contained adipocytes, which are normally absent at late pregnancy and lactation, as well as many involuting lobuloalveoli (Figure 1, D–F). Whole-mount analysis of 10 mammary glands from 1- to 2-year-old nulliparous and multiparous WAP-Cre:Rbfl/fl:p107–/– mice failed to detect any microscopic tumors, and of more than 40 WAP-Cre:Rbfl/fl:p107–/– females that survived for up to 2 years, only 1 developed a tumor (solid adenocarcinoma; data not shown).
Figure 1. A lactation defect in WAP-Cre:Rbfl/fl:p107–/– mutant mice.
(A) Shown are 2 litters of 10-day old pups fed by a multiparous WAP-Cre:Rbfl/fl:p107–/– female (top, 4 pups) or control Rbfl/fl:p107–/– female (bottom, 6 pups). (B and C) Whole-mount staining of representative mammary glands from multiparous (third pregnancy) Rbfl/fl:p107–/– control (B) and WAP-Cre:Rbfl/fl:p107–/– (C) females at day 15 of lactation. Note more condensed lobuloalveoli in control relative to RbΔfl mammary gland. LA, lobuloalveoli; D, duct. (D–F) H&E staining of cross-sections through mammary glands from Rbfl/fl:p107–/– control (D) and WAP-Cre:Rbfl/fl:p107–/– (E and F) mice at day 15 of lactation. Arrows point to abnormal/involuting lobuloalveolar structures; asterisks mark abundant adipocyte tissue in RbΔfl lactating gland. A, alveolar lumen. Original magnification, ×50 (B and C); ×200 (D and E); ×400 (F).
Targeted inactivation of Rb in mammary epithelium via MMTV-Cre induces squamous metaplasia and acinar hyperplasia.
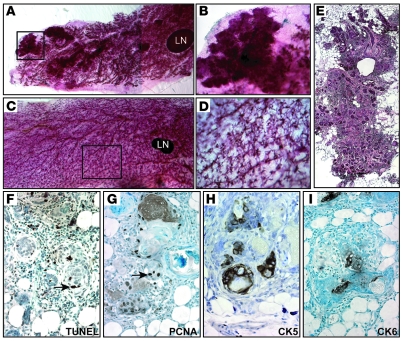
To delete Rb during ductal morphogenesis we used a mouse mammary tumor virus–Cre (MMTV-Cre) deleter, MMTV-CreNLST (44). As with WAP-Cre, ductal morphogenesis, lobuloalveologenesis, involution, and lactation proceeded apparently normally in MMTV-CreNLST:Rbfl/fl and MMTV-CreNLST:Rbfl/fl:p107–/– females (data not shown). However, at 10–14 months of age, mammary glands from 16 of 20 multiparous Rb mutant (RbΔfl) females were found to contain micronodules (Figure 2, A and B). None of 10 control mammary glands contained such lesions (Figure 2, C and D). Histopathology of nodules from 2 MMTV-Cre:Rbfl/fl:p107–/–, 4 MMTV-Cre:Rbfl/fl:p107+/–, and 2 MMTV-Cre:Rbfl/fl:p107+/+ mice revealed that each represented acinar hyperplasia with keratinizing squamous metaplasia (Figure 2E).
Figure 2. MMTV-Cre:Rbfl/fl mice develop focal hyperplastic lesions.
(A–D) Representative whole-mount carmine alum staining of MMTV-Cre:Rbfl/fl:p107+/– mutant (A and B) and control (C and D) mammary glands. Note lesions in the RbΔfl gland. Boxed regions in A and C are shown at higher magnification in B and D. A is a composite of 2 images. (E) H&E staining of a lesion from MMTV-Cre:Rbfl/fl:p107+/– mutant mammary gland, reproduced from 3 low-magnification micrographs. (F–I) Representative analysis of nodules from 8 independent mutant mice. Shown are TUNEL analysis for apoptosis (F) and IHC for PCNA (G), CK5 (H), and CK6 (I). Arrows point to representative stained cells. Original magnification, ×8 (A); ×10 (C); ×50 (B and D); ×100 (E); ×400 (F–I).
The hyperplastic lesions (n = 8) exhibited high levels of apoptosis and cell proliferation, as determined by in situ TUNEL and immunohistochemistry (IHC) for proliferating cell nuclear antigen (PCNA), respectively (Figure 2, F and G). The lesions expressed cytokeratin 5 (CK5), a marker of mammary myoepithelium and epidermis, and the epidermis-specific differentiation marker CK6 (Figure 2, H and I). Similar results were obtained regardless of p107 status. Thus, inactivation of Rb or Rb plus p107 via MMTV-CreNLST induced focal lesions characterized by hyperplastic growth and apoptosis with transdifferentiation into epidermal-like structures.
MMTV-CreNLST and WAP-Cre delete Rbfl/fl in different cells within the mammary stem cell hierarchy.
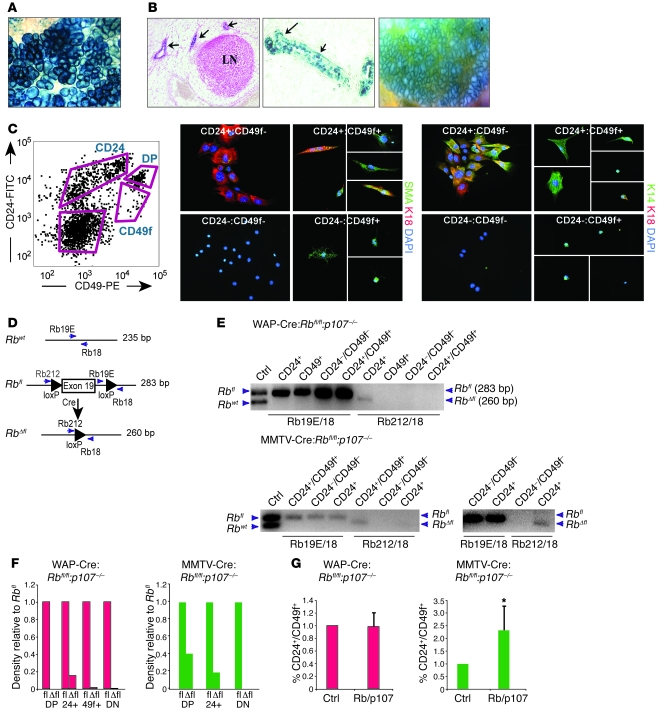
X-gal staining of mammary glands from WAP-Cre:ROSA26 females revealed high level of recombination during pregnancy and lactation (Figure 3A and ref. 51). In contrast, MMTV-CreNLST:ROSA26 mice exhibited high levels of recombination during pregnancy and lactation as well as during ductal morphogenesis and alveologenesis (Figure 3B).
Figure 3. Differential effects of MMTV-Cre and WAP-Cre on Rb deletion and stem/bipotent cell compartments.
(A) Whole-mount X-gal staining of a lactating WAP-Cre:ROSA26 gland. (B) X-gal staining of developing endbuds and ducts during ductal morphogenesis (left, cross section; center, transverse section), and lobuloalveoli during lactation (right, whole-mount) in MMTV-CreNLST:ROSA26 mice. (C) CD24-CD49f FACS profile of multiparous Lin– normal mammary epithelium and gating used for cell sorting, and immunofluorescent staining for SMA, CK14, and CK18 on the indicated cell populations. DP, CD24+CD49f+ double positive. (D) Schematic representation of floxed Rb exon 19 and primers used to detect WT Rb, Rbfl, and recombined RbΔfl alleles. (E) PCR analysis of DNA extracted from pooled, sorted Lin– mammary epithelium from multiparous WAP-Cre:Rbfl/fl:p107–/– (n = 5) or MMTV-Cre:Rbfl/fl:p107–/– (n = 4) mice. Cells were gated as in C. Blot at bottom right shows overexposure after loading twice as much DNA, revealing low-level recombination in CD24+ cells. (F) Densitometry analysis of RbΔfl and Rbfl PCR fragments from E. DN, double negative. (G) Percentage of Lin–CD24+CD49f+ cells in multiparous WAP-Cre:Rbfl/fl:p107–/– (n = 5) and MMTV-Cre:Rbfl/fl:p107–/– (n = 5) mice relative to control. *P = 0.034, 1-tailed paired Student’s t test. Original magnification, ×63 (A); ×100 (B, left); ×400 (B, center, and C); ×50 (B, right).
Next, we sought to identify the cells within the mammary stem cell (MSC) hierarchy in which Rb is deleted by WAP-Cre versus MMTV-Cre. MSCs, luminal progenitors, and myoepithelial progenitors were identified on the basis of expression of the cell surface markers CD24 (52) and either β1-integrin (CD29) or α6-integrin (CD49f; refs. 53, 54). A typical FACS profile of lineage-depleted (Lin–) mammary epithelial cells sorted for CD24 and CD49f expression is shown in Figure 3C. The presumptive MSCs or mammary repopulating units (MRU; CD24+CD49f+), myoepithelial progenitors (MYO; CD49f+), and colony-forming luminal progenitor cells (CFC; CD24+), as defined by Stingl et al. (54), are indicated. To test their differentiation potential, sorted cells were seeded onto collagen-1–coated plates, induced to differentiate for 3 days, and immunostained with the luminal marker CK18, the stem/myoepithelial marker CK14, and the myoepithelial marker SMA. The CD24–CD49f+ fraction differentiated into CK14+ and SMA+ cells; the CD24+CD49f– fraction differentiated into mostly CK14+CK18+ double-positive cells and was negative for SMA; and the CD24+CD49f+ fraction differentiated into SMA+, CK14+, and CK14+CK18+ cells (Figure 3C). Although the nature of the CK14+CK18+ cells is yet to be defined, these results are consistent with the assignment of CD24–CD49f+ cells as myoepithelial progenitors, CD24+CD49f– cells as luminal progenitors, and CD24+CD49f+ cells as MSCs or bipotent common progenitors.
To determine in which cells Rbfl/fl was excised, 6 mammary glands each (nos. 3, 4, and 5) from 4 multiparous MMTV-CreNLST:Rbfl/fl:p107–/– or multiparous WAP-Cre:Rbfl/fl:p107–/– mice approximately 6 months old were pooled (24 mammary glands), Lin– mammary epithelial cells were sorted as in Figure 3C, and DNA was extracted from each fraction. Cre-mediated recombination was then assessed by PCR with primers flanking exon 19, which can amplify 235-bp WT, 283-bp nonrecombined Rbfl, or 260-bp recombined RbΔfl alleles (Figure 3D and ref. 45). In WAP-Cre:Rbfl/fl:p107–/– mammary epithelium, the recombined RbΔfl allele was reproducibly observed only in the CD24+ luminal fraction, not in the CD24+CD49f+, CD49f+, or CD24–CD49f– fractions (Figure 3E). Densitometry analysis revealed that the percentage of RbΔfl relative to Rbfl was 15% in CD24+ cells, but undetectable in other cell fractions (Figure 3F). In contrast, in MMTV-CreNLST:Rbfl/fl:p107–/– mammary glands, recombination was observed primarily in the CD24+CD49f+ MSC/bipotent cell fraction, with 41% of the cells containing the recombined allele relative to Rbfl (Figure 3, E and F). Recombined RbΔfl was also observed in CD24+ and CD24–CD49f+ fractions upon loading more DNA and after a second round of PCR amplification, respectively (Figure 3, E and F, and Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI41490DS1). Together, these results indicated that MMTV-Cre and WAP-Cre delete Rbfl in different cells within the MSC hierarchy, and this may underlie the different phenotypes of MMTV-Cre:Rbfl/fl:p107–/– and WAP-Cre:Rbfl/fl:p107–/– mice.
Deletion of Rb in mammary epithelium via MMTV-Cre, but not WAP-Cre, expands MSCs/bipotent cells.
To assess the effect of Rb deletion on the CD24+CD49f+ MSC/bipotent cell population, we compared the percentage of CD24+CD49f+ cells in MMTV-Cre:Rbfl/fl:p107–/– (n = 5) or WAP-Cre:Rbfl/fl:p107–/– (n = 5) mice relative to control littermates. The proportion of CD24+CD49f+ cells in WAP-Cre:Rbfl/fl:p107–/– relative to control animals was nearly identical, at a ratio of 0.98 (SD, 0.22; P = 0.44; Figure 3G). In contrast, the percentage of CD24+CD49f+ cells in MMTV-Cre:Rbfl/fl:p107–/– mice was on average 2.32-fold higher than controls (SD, 0.94; P = 0.034), ranging from 1.44- to 3.5-fold (Figure 3G). In addition, we seeded Lin– epithelial cells from multiparous MMTV-Cre:Rbfl/fl:p107–/–, WAP-Cre:Rbfl/fl:p107–/–, and control mammary glands into ultra-low attachment plates and measured the number of primary mammospheres that formed 2 weeks later. Mammosphere frequency increased by 48% in MMTV-Cre:Rbfl/fl:p107–/– mammary glands and reduced by 30% in WAP-Cre:Rbfl/fl:p107–/– glands relative to controls (Supplemental Figure 1C). Thus, MMTV-CreNLST, but not WAP-Cre, induced excision of Rbfl/fl in CD24+CD49f+ stem/bipotent cells, leading to a moderate yet consistent increase in this cell population.
Deletion of Rb in mammary epithelium via MMTV-Cre deleter lines induces mammary adenocarcinomas and adenosquamous carcinomas.
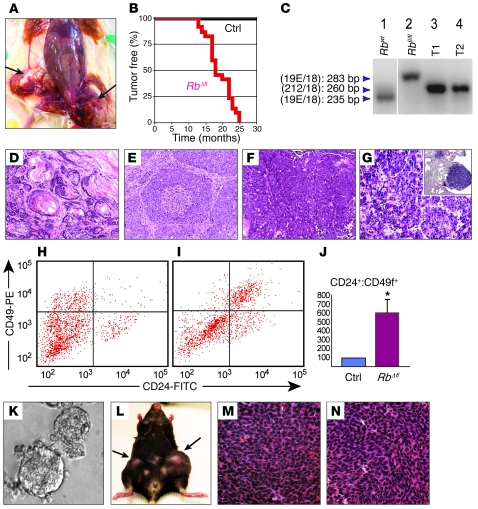
Virtually all multiparous MMTV-Cre:Rbfl/fl females developed frank mammary tumors, with a mean latency of 18.4 months (Figure 4, A and B). As with premalignant lesions, tumor latency did not appear to depend on p107, averaging 13, 18.7, and 19 months for MMTV-Cre:Rbfl/fl:p107+/+ (n = 2), MMTV-Cre:Rbfl/fl:p107–/– (n = 12), and MMTV-Cre:Rbfl/fl:p107+/– (n = 8) mice, respectively. Biallelic excision of exon 19 in these RbΔfl tumors was confirmed by complex PCR analysis (Figure 4C).
Figure 4. MMTV-Cre:Rbfl/fl and MMTV-Cre:Rbfl/fl:p107–/– mice develop transplantable mammary tumors.
(A) Dissected MMTV-Cre:Rbfl/fl:p107–/– female with 2 large tumors in mammary gland nos. 4 and 5 (arrows). (B) Tumor-free curve for a cohort of 22 RbΔfl mice (2 MMTV-Cre:Rbfl/fl:p107+/+, 12 MMTV-Cre:Rbfl/fl:p107–/– and 8 MMTV-Cre:Rbfl/fl:p107+/–) and 20 control (without MMTV-Cre) littermates. (C) Complex PCR analysis with the 3 primers. Lane 1, Rb+/+ mouse tail; lane 2, Rbfl/fl mouse tail; lane 3, MMTV-CreNLST:Rbfl/fl:p107–/– mammary tumor; lane 4, MMTV-CreD:Rbfl/+:p107+/– heterozygote mammary tumor, demonstrating LOH at the Rb locus. Lanes were run on the same gel but were noncontiguous (white line). (D–G) H&E staining of representative RbΔfl tumors and pulmonary metastasis. (D) Tumor 2, adenosquamous carcinoma. (E) Tumor 4, solid adenocarcinoma with comedo patterns. (F) Tumor 9, adenocarcinoma with cystic changes. (G) Pulmonary metastasis in MMTV-Cre:Rbfl/fl:p107–/– mutant shown in F. Note glandular tumors with histology similar to the original tumor. Inset: low-magnification image of the pulmonary metastasis (right; normal lung tissue on left). (H and I) CD24-CD49f flow cytometry profiles of Lin– control mammary gland (H) and RbΔfl tumor (I). (J) Mean Lin–CD24+CD49f+ cells in RbΔfl mammary tumors (n = 4) relative to Lin– mammary epithelial cells from control littermates (n = 6). P < 0.01. (K–N) RbΔfl mammary tumor cells were transplantable. (K) Secondary RbΔfl tumorspheres cultured for 2 weeks. (L) A Rag1–/– female mouse with secondary RbΔfl mammary tumors arising after transplantation of 10,000 and 1,000 tumorsphere cells into the right and left, respectively, of mammary gland no. 4. (M) Primary, poorly differentiated adenocarcinoma. (N) Secondary tumor after transplantation of tumorsphere cells derived from the tumor in M. Original magnification, ×200 (D–F); ×400 (G, M, and N); ×100 (G, inset); ×20 (K).
In contrast to the early focal lesions, the RbΔfl tumors were highly heterogeneous (Figure 4, D–F, and Supplemental Figure 2). About 40% of tumors were classified as adenosquamous carcinomas, and the rest represented various types of adenocarcinomas (Table 1 and Supplemental Figure 2). This tumor heterogeneity suggests that Rb inactivation may cooperate with different oncogenic pathways that transform the initially uniform hyperplastic lesions into distinct tumor subtypes. Macroscopic and microscopic examination of lungs from 14 RbΔfl tumor-bearing mice revealed pulmonary lesions with histology similar to that of the primary tumor in 6 mice (Figure 4, F and G).
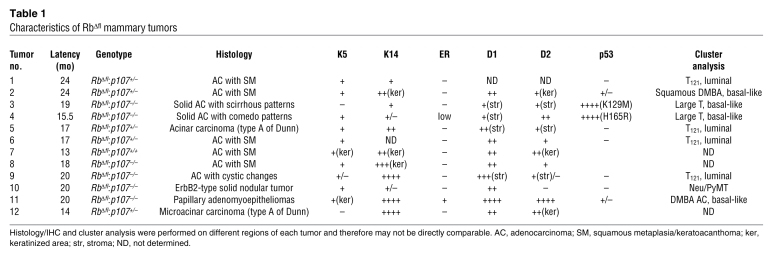
Table 1 .
Characteristics of RbΔfl mammary tumors
We also generated a colony of Rbfl/fl mice with a second deleter strain, MMTV-CreD (55). With this line, we obtained MMTV-CreD:Rbfl/fl mice on p107+/– heterozygous, but not p107–/– homozygous, background. Of approximately 50 mutant mice, 90% became morbid by 8–10 months of age. Large mammary tumors developed in 1 parous MMTV-CreD:Rbfl/fl:p107+/+ and 4 MMTV-CreD:Rbfl/fl:p107+/– females, with histology similar to those seen in MMTV-CreNLST:Rbfl/fl mice, yet with a shorter latency (average 10.5 months; data not shown). Moreover, 1 parous MMTV-CreD:Rbfl/+:p107+/– double-heterozygous female developed a mammary tumor. PCR analysis revealed LOH at the Rb locus (Figure 4C, lane 4). In contrast, PCR analysis of 4 tumors from MMTV-CreD:Rbfl/fl:p107+/– and MMTV-CreNLST:Rbfl/fl:p107+/– females (i.e., heterozygous for p107+/–) revealed that the remaining WT p107 allele was maintained (data not shown).
RbΔfl tumors are transplantable.
Flow cytometry analysis of Lin– cells from 4 different RbΔfl tumors revealed a 6.1-fold increase in the frequency of CD24+CD49f+ cells relative to control mammary epithelium (SD, 1.53; P < 0.05; Figure 4, H–J). To test for cell-autonomous growth, we assessed the ability of RbΔfl tumor cells to form spheres in nonadherent cultures and xenografts in vivo. After seeding Lin– tumor cells on nonadherent plates in defined medium containing EGF and FGF (56–58), 8 of 11 tumors gave rise to tumorsphere cultures that survived for 3 or more passages (Figure 4K). Upon transplantation into mammary glands of recipient Rag1–/– mice, 8 independent RbΔfl tumorspheres gave rise to secondary tumors (Figure 4L) that exhibited histology similar to that of the primary tumors from which they were derived (Figure 4, M and N). Thus, at least some RbΔfl tumor cells can disseminate new tumors independently of tumor-associated stroma.
RbΔfl tumors express basal cytokeratins, but not ERα.
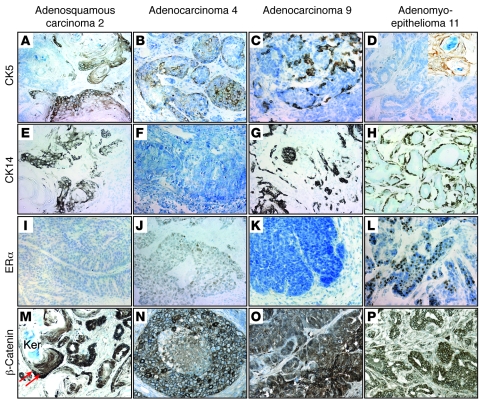
To begin to classify the RbΔfl tumors into subtypes, we stained histological sections for the myoepithelial/basal cell markers CK5 and CK14 as well as ERα. Of 12 RbΔfl tumors, 8 expressed CK5 in tumor epithelium or squamous areas (Figure 5, A–C, and Table 1). We also observed that 2 tumors were negative for CK5 in the epithelium compartment, but positive in keratinized areas (Figure 5D). CK14 was expressed at different levels in all tumors except for tumors 4 and 10 (Figure 5, E–H, and Table 1). Importantly, with the exception of the papillary adenomyoepithelioma (tumor 11), all RbΔfl tumors were negative for, or expressed very low levels of (tumor 4), ERα (Figure 5, I–L, and Table 1). Consistent with its role in epidermal transdifferentiation (59), we observed variable levels of β-catenin expression at the cell membrane in nodular areas of tumors (Figure 5, N–P), but almost exclusive nuclear staining in cells surrounding keratinized areas in adenosquamous tumors (Figure 5M, arrows).
Figure 5. Most RbΔfl mammary tumors express variable levels of basal cytokeratins, but not ERα.
Representative IHC of tumors 2, 4, 9, and 11 for CK5 (A–D), CK14 (E–H), ERα (I–L), and β-catenin (M–P). Inset in D shows CK5 expression in squamous area. Arrows in M mark cells surrounding keratinized area (Ker) with nuclear localized β-catenin. Original magnification, ×400 (A–P and D, inset).
RbΔfl mammary tumors cluster with T121- and Tag-induced tumors with features of luminal-B or basal-like breast cancer.
To further classify the RbΔfl mammary tumors, we subjected 9 tumor biopsies to microarray-based gene expression analysis. Previous gene expression profiling of 13 different mouse models of breast cancer, using 866 informative genes, revealed that tumors induced by SV40 Tag (C3-Tag and WAP-Tag) clustered with human basal-like breast cancer, whereas tumors induced by a truncated T antigen (WAP-T121), which binds Rb family but not p53, clustered primarily with luminal-B (9).
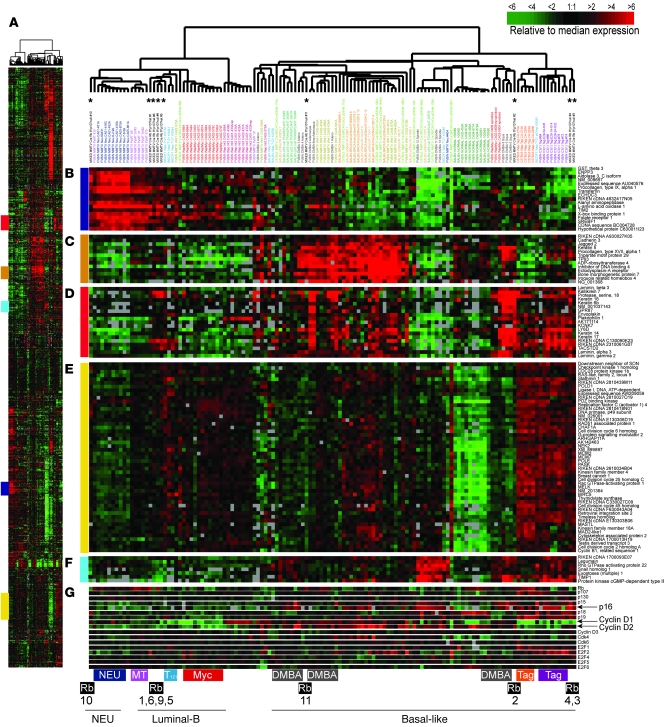
Figure 6A shows an overview of the complete 866-gene cluster diagram for the RbΔfl tumors and the 13 mouse models. Selected groups of genes are shown in detail: the luminal gene cluster (Figure 6B), the CK5 basal-like gene cluster in the mouse (Figure 6C), the CK14 basal-like gene cluster in the mouse (Figure 6D), proliferation-associated genes (Figure 6E), and genes associated with EMT (Figure 6F). In addition, Figure 6G highlights genes on the Rb pathway: E2Fs, G1-cyclins, and CDK inhibitors, including p16Ink4a.
Figure 6. RbΔfl mammary tumors cluster with DMBA and Tag-induced basal-like or T121-induced luminal-B–like breast tumors.
(A) Overview of expression of 866 reference genes in tumors from RbΔfl and 13 mouse models of breast cancer. Colored regions at left correspond to the regions shown in B–F. (B) Selected genes representing the luminal gene cluster. (C) CK5 basal-like gene cluster. (D) CK14 basal-like gene cluster. (E) Proliferation-associated gene cluster. (F) EMT gene cluster. (G) Subset of proliferation-associated genes. Asterisks at the top mark the 9 RbΔfl tumors; tumor models that clustered with RbΔfl tumors are highlighted at the bottom. DMBA, 7,12-dimethylbenz[a]-anthracene.
We found 6 RbΔfl tumors to cluster with either Tag (tumors 3 and 4) or T121 (tumors 1, 5, 6, and 9), which were previously classified as basal-like/TNT and luminal-B–like breast tumors, respectively (9). Tumors 2 and 11 clustered with squamous tumors induced by DMBA and also expressed markers associated with basal-like/TNTs. Tumor 10 clustered with NEU+ tumors. The basal-like tumors (nos. 2, 3, 4, and 11), but not luminal-B or NEU+ tumors (nos. 1, 5, 6, 9, and 10), expressed high levels of EMT associated genes including snail homolog 1, TIMP1, and Rho GTPase activator 22 (Figure 6F).
Among genes on the Rb pathway (Figure 6G), cyclin D1 was expressed at low levels in most tumors, with the exception of tumor 11, whereas cyclin D2 was expressed at levels comparable to or slightly higher than (tumors 2 and 4) those of normal mammary glands. Consistent with this, IHC analysis revealed high expression of nuclear cyclin D1 in glandular tumor cells only in tumor 11, whereas nuclear cyclin D2 was detected in epithelium, stroma, or keratinized areas in 10 of 11 RbΔfl mammary tumors (Supplemental Figure 3 and Table 1).
Array CGH analysis revealed a tendency toward more gains and/or losses in the Rb-deficient Tag/TNTs (tumors 3 and 4) compared with the 7 other Rb tumors (Supplemental Figure 4A and data not shown). Major losses and gains were found in chromosome 15 in tumors 3 and 4, but not in tumors 1, 5, 6, and 9 (Supplemental Figure 4B) or in tumors 2, 10, or 11 (data not shown). This region in chromosome 15 showed conservation of synteny with human chromosomes 5p13 and 8q22, which frequently undergo chromosomal gains in human breast cancer (60). Thus, these RbΔfl tumors could be divided into 2 major groups: T121/luminal-B–like (nos. 1, 5, 6, and 9) and basal-like (nos. 2, 3, 4, and 11). The basal-like group could be further divided into 2 subgroups: Tag-like (nos. 3 and 4) and DMBA-like (nos. 2 and 11).
Tag/basal-like, but not T121/luminal-B–like, RbΔfl mammary tumors express a human RB1-LOH proliferation gene signature.
Analysis of human breast tumors with LOH at the RB1 locus led to the generation of a RB1-specific proliferation signature (29). We created an average expression value for the human RB1-LOH signature and compared it with the levels of expression in RbΔfl mammary tumors. The top 2 samples were the Rb-deficient Tag/basal-like tumors 3 and 4, with values of 0.37 and 0.47, respectively. The average value for T121/luminal-B–like tumors (nos. 1, 5, 6, and 9) was 0.13 (0.04, 0.22, 0.09, and 0.17, respectively). Thus, the RbΔfl tumors that share the human RB1-LOH proliferation signature are tumors 3 and 4, which cluster with Tag/basal-like carcinoma.
Tag/basal-like RbΔfl tumors express high levels of p53.
To begin to understand the basis for tumor heterogeneity downstream of Rb, we investigated the status of p53. Remarkably, Tag/basal-like RbΔfl tumors exhibited high, near-uniform expression of nuclear p53, whereas DMBA-like RbΔfl tumors expressed nuclear p53 in a small fraction of the cells (Figure 7A, top, and Supplemental Figure 5). In contrast, RbΔfl luminal-B or NEU+ tumors did not express detectable levels of p53 (Figure 7A, bottom, and Supplemental Figure 5). Analysis of additional RbΔfl tumors identified 3 more with high expression of p53: a solid adenocarcinoma (tumor 48; Supplemental Figure 5), and 2 spindle-cell/EMT tumors (tumors 18 and 30; Figure 8, A and B, and see below).
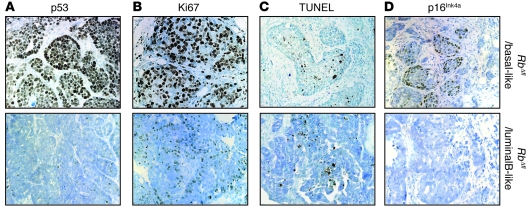
Figure 7. Tag/basal-like RbΔfl tumors express high levels of nuclear p53.
(A) High nuclear p53 expression in RbΔfl basal-like, but not luminal-B–like, tumors. (B) Representative IHC for the proliferation marker Ki67. (C) Representative TUNEL analysis. (D) IHC for p16Ink4a showing a cluster of positive cells. Original magnification, ×400 (A–C); ×200 (D).
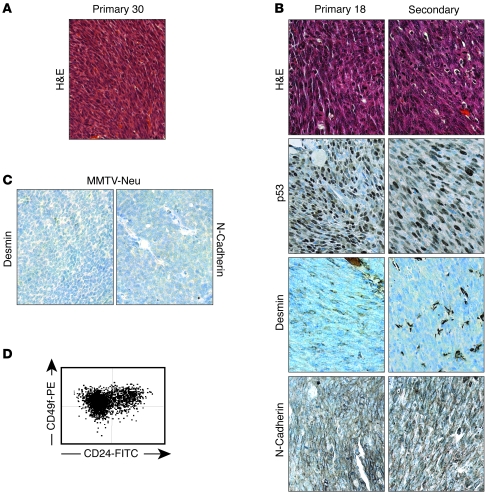
Figure 8. Spindle-cell RbΔfl tumors express nuclear p53 and markers of EMT.
(A) Histology staining of primary EMT tumor 30. (B) Histology staining and IHC for p53, desmin, and N-cadherin of primary EMT tumor 18 and a secondary tumor derivative. (C) Lack of desmin and N-cadherin expression in MMTV-neu tumor. (D) Representative flow cytometry profile of RbΔfl EMT tumors demonstrating a high CD49f to CD24 ratio. Original magnification, ×400.
The RbΔfl basal-like tumors exhibited higher rates of cell proliferation and similar levels of apoptosis compared with the luminal-like tumors (Figure 7, B and C), which suggested that they contained mutant p53 (see below). In addition, IHC analysis for p16Ink4a identified small pockets of positive cells in RbΔfl basal-like tumors, but not luminal-B–like tumors (Figure 7D). The relatively low IHC signal was likely related to the low sensitivity of anti-mouse p16Ink4a antibodies. Indeed, at the RNA level, p16Ink4a was highly expressed in the RbΔfl basal-like tumors, but not other tumors (Figure 6G). This elevated p16Ink4a expression is consistent with its high expression in RB1-negative human basal-like breast cancer (28–30).
RbΔfl spindle cell tumors express mutant p53 and markers of EMT.
As noted above, tumors 18 and 30 exhibited spindle-cell morphology (Figure 8, A and B). These tumors expressed high levels of p53 as well as the EMT-associated markers N-cadherin and desmin (Figure 8B, data not shown, and ref. 61). They readily grew as monolayer cultures and were highly aggressive; transplantation of 1,000 monolayer cells into the mammary glands of recipient mice gave rise to large tumors within 2–3 weeks that retained the same histology and marker expression as the parental tumors (Figure 8B). Consistent with their mesenchymal characteristics, flow cytometry analysis revealed that these tumors lost the CD24+ luminal marker, exhibiting a high CD49f+ to CD24+ ratio (Figure 8D). EMT tumors with spindle-cell morphology were previously described in several mouse models of breast cancer, including some rare mammary tumors with null mutations in p53 (62). In humans, EMT tumors in breast cancer have been recognized within the larger TNT group that includes claudin-low and metaplastic breast cancer (9–14). Thus, some RbΔfl mammary tumors express nuclear p53, are highly aggressive, and exhibit spindle-cell/EMT morphology.
Tag/basal-like and EMT RbΔfl tumors contain p53 mutations.
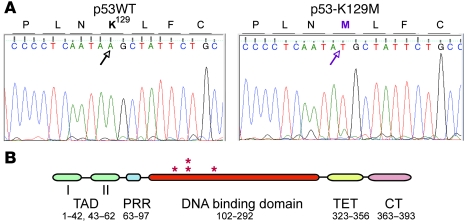
To determine whether high p53 expression in these tumors was caused by mutations in p53, which stabilizes the protein (63, 64), we sequenced exons 2–9 of p53 in tumors 3, 4, 18, and 30. We identified missense mutations K129M, H165R, C138R, and C138R, respectively, in the DNA-binding domain of these tumors (Figure 9). These amino acids were conserved in human p53 (K132, H168, C141, and C141, respectively) and are frequently mutated in human cancer (65, 66). Tumors 3 and 4 only harbored the mutant allele, whereas tumors 18 and 30 contained both the WT and mutant alleles, which suggests that C138R acts as dominant negative.
Figure 9. Identification of p53 mutations in Tag/basal-like and EMT RbΔfl tumors.
(A) DNA sequencing reaction around K129 in control DNA (left) and tumor 3 (right). (B) Schematic representation of human p53 and relative location (asterisks) of mutations identified in mouse RbΔfl tumors: Tag/basal-like tumor 3, K129M (human, K132M); Tag/basal-like tumor 4, H165R (human, H168R); EMT tumors 18 and 30, C138R (human, C141R). TAD, N-terminal transactivation domain; PRR, proline-rich region; TET, tetramerization domain, CT, C terminus.
Combined deletion of Rb and p53 in mammary epithelium via MMTV-Cre induces aggressive spindle cell/EMT tumors.
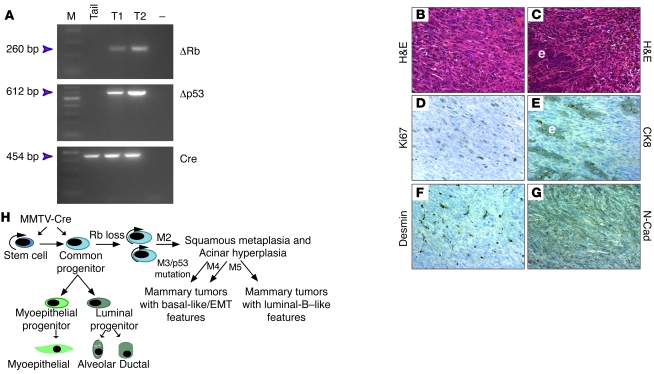
The above results suggest that p53 status dictates whether the focal hyperplastic lesions in MMTV-Cre:Rbfl/fl mice develop into luminal-B or basal-like/EMT tumors. We predicted that combined deletions of Rb and p53 would lead to uniform basal-like or EMT tumors at the expense of luminal-B. To test this notion, we crossed the MMTV-Cre:Rbfl/fl mice with floxed p53 (p53fl/fl) mice (67). Mammary-specific deletion of p53fl/fl leads to ERα-positive and -negative tumors with a latency of more than 11 months (68). MMTV-Cre:Rbfl/fl:p53fl/fl double-mutant mice were viable and fertile, yet nearly all developed lethal lymphomas within 2–6 months. To circumvent this, we purified Lin– mammary epithelial cells from 2 independent MMTV-Cre:Rbfl/fl:p53fl/fl mice and transplanted 10,000 cells each into the mammary glands of 4- to 5-week-old NOD/SCID mice. Tumors developed with an average latency of 126 days (6 tumors in 12 injections; 50%) and 58 days (10 tumors in 24 injections; 42%). Given that the mammary epithelium was derived from 76- and 119-day-old mice, the total latency for tumor formation was 202 and 177 days, an average of 190 days (i.e., about 6.5 months). PCR analysis confirmed deletion of Rb and p53 (Figure 10A). Analysis of 10 independent tumors revealed similar mesenchymal histology, characterized by spindle-shaped cells with abundant pink cytoplasm (Figure 10B). Some regions contained epithelial components surrounded by mesenchymal cells (Figure 10C), consistent with EMT. Indeed, expression of CK8 (Figure 10E) was restricted to epithelial cells within tumors, whereas mesenchymal areas expressed the EMT markers desmin and N-cadherin (Figure 10, F and G) and were indistinguishable from the Rb/p53 mesenchymal tumors derived from MMTV-Cre:Rbfl/fl mice (Figure 8).
Figure 10. MMTV-Cre:Rbfl/fl:p53fl/fl double mutant mice develop spindle cell/EMT tumors.
(A) PCR analysis of tail from MMTV-Cre:Rbfl/fl:p53fl/fl mice and tumor DNA for recombined Rb and p53 alleles or Cre. (B and C) Histology staining of Rb/p53 tumor showing spindle/mesenchymal and epithelial components (C). (D–G) Representative IHC staining for the indicated antibodies. e, epithelium. (H) Schematic model for tumor formation after Rb deletion in the mammary epithelium. See Discussion for details. Original magnification, ×400.
Reintroduction of Rb into aggressive RbΔfl/p53Δfl EMT tumor cells suppresses their growth.
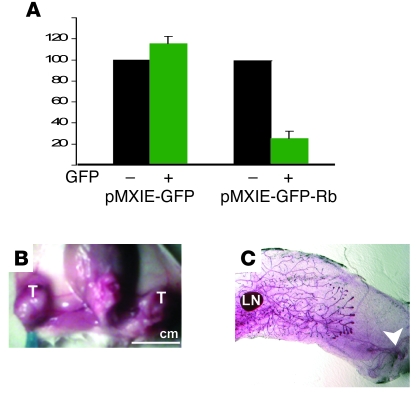
Many tumors are dependent on (i.e., addicted to) the continuous expression of oncogenes that drive them (reviewed in refs. 69, 70). Although introduction of constitutively activated, unphosphorylatable pRb alleles often suppress cell proliferation, WT pRb seems to inhibit only certain cells, likely with low levels of cyclins (71–75). To test whether growth of RbΔfl/p53Δfl tumor cells is dependent on the continuous absence of pRb, we transduced WT Rb via a retrovirus vector that also encodes GFP. Expression of pRb was confirmed by immunofluorescent staining (data not shown). GFP+ cells transduced with Rb (pMXIE-GFP-Rb) or vector alone (pMXIE-GFP) were sorted, and 500 GFP+ cells were seeded onto 96-well plates. After 1 week, MTT assays revealed approximately 5-fold inhibition of cell growth in Rb-transduced cultures relative to controls (Figure 11A). The GFP-sorted cells were also transplanted into mammary glands of 4- to 5-week-old NOD/SCID mice. Mice transplanted with 1,000 control tumor cells (empty vector) developed large tumors within 2–3 weeks (12 of 12; Figure 11B). In contrast, mice transplanted with Rb-GFP+ tumor cells did not develop tumors (0 of 12; Figure 11C). Thus, despite their aggressive nature, the RbΔfl/p53Δfl mammary tumors were addicted to continual absence of Rb and readily inhibited when reconstituted with this tumor suppressor.
Figure 11. Aggressive spindle cell/EMT RbΔfl tumors are addicted to Rb loss.
(A) Retrovirally mediated transduction of Rb into mesenchymal RbΔfl tumor cells inhibited cell proliferation in vitro. RbΔfl tumor cells were infected with the indicated virus, sorted for GFP+ and GFP– cells, seeded at 500 cells per well, and analyzed by MTT assay for cell proliferation 1 week later. Shown is mean ± SD from 3 independent experiments relative to GFP– cells. P = 0.019 for suppression by Rb. (B and C) Retrovirally mediated transduction of Rb into EMT RbΔfl tumor cells inhibited tumor formation. RbΔfl tumor cells were infected with GFP or Rb-GFP viruses, sorted, and 1,000 GFP+ cells were transplanted into no. 4 mammary glands of NOD/SCID mice. Tumors were detected 2 weeks later and scored after 3 weeks. (B) A dissected mouse carrying tumors (T) after transplantation of GFP+ cells. (C) Whole-mount staining revealing endogenous mammary epithelium, but no tumor, after transplanting Rb-GFP+ cells. Arrowhead points to site of injection. Original magnification, ×10. Scale bar: 1 cm (B).
Discussion
We describe the consequences of Rb deletion in the mammary epithelium and delineate several stages in tumor initiation and progression leading to distinct breast cancer subtypes (Figure 10H). First, loss of Rb in stem/bipotent progenitor cells expands this cell population. As these cells attempt to exit the cell cycle and differentiate into single-lineage (luminal or myoepithelial) progenitors, they undergo apoptotic death, which restricts their growth. Second, in cooperation with a limited number of oncogenic events (M2), some RbΔfl epithelial cells continue to proliferate to form focal acinar hyperplasia with propensity to squamous transdifferentiation. These lesions exhibit high levels of both cell proliferation and apoptosis. Third, additional mutations, which increase cell proliferation and/or inhibit apoptosis or differentiation, further transform these hyperplastic lesions into distinct tumor subtypes. We showed here that mutations in p53 induced basal-like or EMT tumors. We note that whereas the basal-like tumors were defined by microarray cluster analysis, the EMT tumors were identified by their spindle-cell morphology and expression of mesenchymal markers. Of interest, knockdown of RB1 in MCF7 breast cancer cells induces EMT (76). However, our results demonstrated that Rb loss also induced luminal-B–like as well as HER2+-like tumors that did not express EMT markers. Thus, other mutations, which are yet to be defined, cooperate with Rb loss to induce luminal-B–like tumors or other subtypes. Remarkably, combined mutations in Rb and p53 led to uniform types of EMT tumors at the expense of all others, which suggests that the timing of tumor suppressor inactivation may also dictate tumor subtype.
Why does deletion of Rb in mammary epithelium lead to mammary tumors, whereas Rb–/– mammary epithelial cells from embryonic placodes transplanted into recipient mice do not (39)? We suggest the discrepancy may be due to dependency on pregnancy for tumor formation in Rb-deficient mammary epithelium. Indeed, we found that mammary tumors developed only in parous MMTV-Cre:Rbfl/fl female mice. In contrast, the transplanted embryonic Rb–/– mammary epithelial cells were not taken through pregnancy (39). Notably, mammary tumors developed from transplanted MMTV-Cre:Rbfl/fl:p53fl/fl epithelium without pregnancy, indicating that loss of p53 bypassed this requirement.
The effect of Rb deletion was critically dependent on the cell within the mammary epithelial hierarchy in which it was disrupted: deletion of Rb via WAP-Cre led to minor lactation defects but not tumor formation, whereas deletion via 2 different MMTV-Cre lines led to mammary tumors. We showed that MMTV-Cre induced efficient Rb deletion primarily in CD24+CD49f+ stem/bipotent progenitors, whereas WAP-Cre induced deletion in CD24+ luminal progenitors. WAP-Cre is expressed in parity-identified multipotent alveolar stem cells (43, 77). Some low level of recombination within the stem cell/bipotent cell compartment may occur in WAP-Cre:Rbfl/fl mammary glands, but this is masked and greatly underrepresented relative to CD24+ cells, in which WAP-Cre is most abundantly expressed (78). The fact that tumors arose in MMTV-Cre:Rbfl/fl mice, but not WAP-Cre:Rbfl/fl mice, indicates that even if some low level of Rb deletion occurs in parity-identified multipotent epithelial cells, these cells are inherently different or exposed to a different microenvironment that diminishes their susceptibility to neoplastic transformation compared with ductal stem/bipotent progenitors.
Given the high frequency with which Rb and p53 are lost in TNT/basal-like breast cancer, and our results demonstrating that mutations in p53 cooperated with Rb loss to induce aggressive EMT tumors, these tumor suppressor pathways may be ideal therapeutic targets for TNTs. Importantly, we showed that transduction of WT pRb completely abolished growth of RbΔfl/p53Δfl TNTs. Thus, these tumors were addicted to Rb loss; drugs that mimic pRb function (79), perhaps in combination with drugs that activate mutant p53 (80), may therefore suppress the growth these tumors. The mouse models described here as well as 2 other models of TNTs — combined deletion of Brca1 and p53 (81) and overexpression of the Met receptor tyrosine kinase (82) — provide invaluable preclinical platforms to identify and assess potential therapeutics for this aggressive breast cancer subtype (83).
Methods
Mouse genotyping and genetic analysis.
All procedures were done in accordance with the current Canadian Animal Care Council guide for the care and use of laboratory animals and were approved by the Toronto General Research Institute Animal Research Committee. MMTV-Cre:Rbfl/fl, MMTV-Cre:Rbfl/fl:p107–/–, and WAP-Cre:Rbfl/fl:p107–/– mice were on mixed genetic backgrounds. Rbfl/fl:p107–/– mice were provided by R. Bremner (Toronto Western Hospital, Toronto, Ontario, Canada). MMTV-CreD was purchased from Jackson Laboratory. p53fl/fl mice were obtained from the NCI Mouse Repository. Genotyping was based on published primers and confirmed in tumor-bearing animals by second rounds of PCR reactions. Recombined Rb alleles were resolved on 3% NuSieve gel.
Whole mount and X-gal staining.
Mammary glands were fixed in 4% PFA, dehydrated in 100% ethanol, and stained with 0.2% carmine alum plus 0.5% aluminum potassium sulfate. Glands were destained in 70% ethanol plus 2% HCl, 90% ethanol, and 100% ethanol at 1 hour each, immersed in Toluene for 4 hours, and stored in methyl salicylate. Whole-mount X-gal staining was as described previously (74). Images were obtained via Leica stereomicroscope.
IHC and TUNEL analysis.
IHC on 4-μm sections was performed with a Vector ABC kit (PK6100). Antibodies were cyclin D1 (Neo Markers, MS-210-P1, catalog no. sc20044; Santa Cruz Biotechnology Inc.), cyclin D2 (catalog no. sc593; Santa Cruz Biotechnology Inc.) ER (catalog no. sc542; Santa Cruz Biotechnology Inc.), CK6 (catalog no. PRB 169P; Covance), CK14 (catalog no. E2624; Panomics), β-catenin (catalog no. 610153; BD Biosciences), p53 (FL-393; catalog no. sc-6243; Santa Cruz Biotechnology Inc.), Ki67 (catalog no. CRM325, clone SP6; Biocare Medical), Desmin (catalog no. D33; DAKO), and Rb (catalog no. 554136; BD) at 1:200 dilutions. PCNA monoclonal antibody (catalog no. P8825; Sigma-Aldrich) was used at 1:400 dilution, CK5 at 1:500 (catalog no. PRB-160P; Covance), and N-cadherin at 1:600 (catalog no. NB200-592; Novus Biologicals). Secondary biotinylated anti-mouse or anti-rabbit antibodies (Vector) were used at 1:200 dilution. Sections were counterstained with methyl green. TUNEL analysis was performed as described previously (74).
Mammary epithelial cells, flow cytometry analysis, and cell sorting.
Enrichment for mammary epithelial cells, flow cytometry, and cell sorting were performed as described previously (58) using PE-CY5-CD49f (catalog no. 555736; BD Biosciences — Pharmingen), CD24-FITC (catalog no. 553261; BD Biosciences — Pharmingen), and FACSCalibur flow cytometer or FACSAria-13 color Cell Sorter (BD) at 30 psi.
In vitro differentiation and immunocytostaining.
Sorted cells were seeded onto collagen-1–coated coverslips (catalog no. 354089; VWR, BD Biosciences) in growth media (DMEM/F-12, 10% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 5 ng/ml EGF, and penicillin/streptomycin) for 3 days. Differentiation was induced for 3 additional days in DMEM/F-12 containing 5 μg/ml insulin, 1 μg/ml hydrocortisone, 3 μg/ml prolactin, and penicillin/streptomycin. Cells were stained as described previously (58) using rabbit anti-SMA (1:200 dilution; catalog no. 600-531; Novus Biologics), mouse anti-K18 (1:200 dilution; catalog no. RDI-PR061028; Fitzgerald), and rabbit anti-CK14 (1:200 dilution; catalog no. E2624; Panomics). Secondary antibodies were goat anti-rabbit Alexa Fluor 488 (green, 1:200 dilution; catalog no. A11008; Invitrogen) or goat anti-mouse Alexa Fluor 568 (red, 1:200 dilution; catalog no. A11004; Invitrogen). Nuclei were visualized with DAPI (catalog no. D9542; Sigma-Aldrich) and viewed under Zeiss Axioskop 2 fluorescent microscope.
Tumorsphere and mammosphere culture and transplantation.
Lin– tumor cell suspensions were seeded onto ultra-low attachment plates (catalog no. 3471; Corning Costar) as described previously (58). For transplantation, primary tumorspheres were dissociated, resuspended in 10 μl media, mixed at a 1:1 ratio with 10 μl Matrigel (BD Bioscience) on ice, and injected into no. 4 mammary glands of immunocompromised Rag1–/– or NOD/SCID mice. Lin– mammary epithelial cells were seeded at 5,000 cells/well onto 96-well plates, and mammospheres were counted 2 weeks later.
Retrovirus infection.
Plasmids pMXIE, pMXIE-WTRb, and pMXIE-RbΔK11 (provided by R. Bremner) as well as p-Helper and p-VSVG were purified by endotoxin-free QIAGEN Maxi kit, transfected into Phoenix Eco packaging cells using TransIT-LT1 transfection reagent (product nos. MIR 2300, 2305, and 2306; Mirus Bio), and harvested after 40–68 hours. Rb/p53 tumor cells maintained in DMEM with 10% FBS were infected with viral supernatants at 1:10 dilution with 8 μg/ml polybrene. After 48 hours, cells were immunostained for Rb or sorted for GFP, and either replated in 96-well plates (500 cells/well) for MTT analysis or transplanted into mammary glands of NOD/SCID mice.
Microarray analysis and array CGH analysis.
Tumors were snap-frozen in liquid nitrogen. RNA and DNA were purified from the same biopsies using RNeasy and DNAeasy kits from Qiagen. RNA was labeled with Cy5 and used for microarray analysis as described previously (9). The dataset was deposited in the GEO database ( http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE14457). DNA was labeled according to the direct incorporation method of Agilent, hybridized to Agilent 244k CGH arrays (model G4415A), and scanned (model G2505B) at a resolution of 5 μm. Tail DNA from 4 Rb mutant female mice was used as reference.
p53 sequencing.
Tumors 3 and 4 were sequenced from biopsies, tumor 18 from a derived cell line, and tumor 30 from both biopsy and a derived cell line. Primers were as follows: exons 2–4 forward, 5′-GCAGGATCCACAATCTCTTCTCTCTACAG-3′; exons 2–4 reverse, 5′-CCGGAATTCAAAGGTCACACGAAAGACAAC-3′; exons 5–6 forward, 5′-TAAGGATCCTTAGTTCCCCACCTTGACACC-3′; exons 5–6 reverse, 5′-TCCGAATTCTCTAGGCTGGAGTCAACTGTC-3′; exons 7–9 forward, 5′-TAAGGATCCTTCACCTGGATCCTGTGTCTTC-3′; exons 7–9 reverse, 5′-TCCGAATTCAATGGGTACAGCTTGTCTCTGG-3′. PCR amplification reactions were purified on Qiagen PCR purification columns and subjected to high-quality capillary-based fluorescent sequencing on dual ABI 3730XL instruments at the Hospital for Sick Children TCAG Sequencing Facility. Identified mutations were verified by 2 additional PCR reactions/sequencing.
Statistics.
Average across independent experiments is presented as mean of each experiment relative to control, which was set as 1 or 100%, and analyzed by 2-tailed Student’s t test (except 1-tailed analysis in Figure 3G). A P value of 0.05 or less was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Robert Cardiff for comments on histopathology, George Murrow for help with array CGH analysis, Tara Paton for sequencing data acquisition, Anton Berns and Rod Bremner for Rbfl/fl mice, and Sean Egan for comments on the manuscript. This work was supported by funds from the V Foundation for Cancer Research and the Breast Cancer Research Foundation to C.M. Perou, and from the Canadian Breast Cancer Foundation (Ontario Chapter), Ontario Institute for Cancer Research (OICR) — Cancer Stem Cell Program, OICR/Terry Fox Research Institute — Selective Therapeutics Program, and Canadian Breast Cancer Research Alliance grant 020456 to E. Zacksenhaus.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(9):3296–3309. doi:10.1172/JCI41490.
Jason I. Herschkowitz’s present address is: Department of Molecular and Cellular Biology, Baylor College of Medicine, Houston, Texas, USA.
References
- 1.Cardiff RD, Wellings SR. The comparative pathology of human and mouse mammary glands. J Mammary Gland Biol Neoplasia. 1999;4(1):105–122. doi: 10.1023/A:1018712905244. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 3.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318(5853):1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 4.Chan TA, et al. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med. 2008;5(5):e114. doi: 10.1371/journal.pmed.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneider BP, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14(24):8010–8018. doi: 10.1158/1078-0432.CCR-08-1208. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sorlie T. Molecular portraits of breast cancer: tumour subtypes as distinct disease entities. Eur J Cancer. 2004;40(18):2667–2675. doi: 10.1016/j.ejca.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Fan C, et al. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355(6):560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 9.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69(10):4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fulford LG, et al. Specific morphological features predictive for the basal phenotype in grade 3 invasive ductal carcinoma of breast. Histopathology. 2006;49(1):22–34. doi: 10.1111/j.1365-2559.2006.02453.x. [DOI] [PubMed] [Google Scholar]
- 12.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52(1):108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]
- 13.Yehiely F, Moyano JV, Evans JR, Nielsen TO, Cryns VL. Deconstructing the molecular portrait of basal-like breast cancer. Trends Mol Med. 2006;12(11):537–544. doi: 10.1016/j.molmed.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 15.Weigelt B, Kreike B, Reis-Filho JS. Metaplastic breast carcinomas are basal-like breast cancers: a genomic profiling analysis. Breast Cancer Res Treat. 2009;117(2):273–280. doi: 10.1007/s10549-008-0197-9. [DOI] [PubMed] [Google Scholar]
- 16.Foulkes WD, et al. Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst. 2003;95(19):1482–1485. doi: 10.1093/jnci/djg050. [DOI] [PubMed] [Google Scholar]
- 17.Turner NC, Reis-Filho JS. Basal-like breast cancer and the BRCA1 phenotype. Oncogene. 2006;25(43):5846–5853. doi: 10.1038/sj.onc.1209876. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saal LH, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40(1):102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee EY, To H, Shew JY, Bookstein R, Scully P, Lee WH. Inactivation of the retinoblastoma susceptibility gene in human breast cancers. Science. 1988;241(4862):218–221. doi: 10.1126/science.3388033. [DOI] [PubMed] [Google Scholar]
- 21.T’Ang A, Varley JM, Chakraborty S, Murphree AL, Fung YK. Structural rearrangement of the retinoblastoma gene in human breast carcinoma. Science. 1988;242(4876):263–266. doi: 10.1126/science.3175651. [DOI] [PubMed] [Google Scholar]
- 22.Bookstein R, Lee EY, Peccei A, Lee WH. Human retinoblastoma gene: long-range mapping and analysis of its deletion in a breast cancer cell line. Mol Cell Biol. 1989;9(4):1628–1634. doi: 10.1128/mcb.9.4.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varley JM, Brammar WJ, Walker RA. Oncogene organisation and expression: prediction in breast cancer. Horm Res. 1989;32(suppl 1):250–253. doi: 10.1159/000181357. [DOI] [PubMed] [Google Scholar]
- 24.Varley JM, et al. The retinoblastoma gene is frequently altered leading to loss of expression in primary breast tumours. Oncogene. 1989;4(6):725–729. [PubMed] [Google Scholar]
- 25.Wang NP, To H, Lee WH, Lee EY. Tumor suppressor activity of RB and p53 genes in human breast carcinoma cells. Oncogene. 1993;8(2):279–288. [PubMed] [Google Scholar]
- 26.Trudel M, Mulligan L, Cavenee W, Margolese R, Cote J, Gariepy G. Retinoblastoma and p53 gene product expression in breast carcinoma: immunohistochemical analysis and clinicopathologic correlation. Hum Pathol. 1992;23(12):1388–1394. doi: 10.1016/0046-8177(92)90059-C. [DOI] [PubMed] [Google Scholar]
- 27.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gauthier ML, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12(5):479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herschkowitz JI, He X, Fan C, Perou CM. The functional loss of the retinoblastoma tumour suppressor is a common event in basal-like and luminal B breast carcinomas. Breast Cancer Res. 2008;10(5):R75. doi: 10.1186/bcr2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subhawong AP, et al. Most basal-like breast carcinomas demonstrate the same Rb–/p16+ immunophenotype as the HPV-related poorly differentiated squamous cell carcinomas which they resemble morphologically. Am J Surg Pathol. 2009;33(2):163–175. doi: 10.1097/PAS.0b013e31817f9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burkhart DL, Sage J. Cellular mechanisms of tumor suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8(9):671–682. doi: 10.1038/nrc2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bremner R, Zacksenhaus E. Cyclins, Cdks, E2f, Skp2, and more at the first international RB tumor suppressor meeting. Cancer Res. doi: 10.1158/0008-5472.CAN-10-0358. [published online ahead of print July 7, 2010]. doi:10.1158/0008-5472.CAN-10-0358. [DOI] [PubMed] [Google Scholar]
- 33.Nahle Z, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4(11):859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 34.Macleod KF, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15(22):6178–6188. [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Z, et al. E2F1 and p53 are dispensable whereas p21Waf1/Cip1 cooperates with Rb to restrict endoreduplication and apoptosis during skeletal myogenesis. Dev Biol. 2000;227(1):28–41. doi: 10.1006/dbio.2000.9892. [DOI] [PubMed] [Google Scholar]
- 36.Whyte P, et al. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334(6178):124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 37.Ewen ME, et al. An N-terminal transformation-governing sequence of SV40 large T antigen contributes to the binding of both p110Rb and a second cellular protein, p120. . Cell. 1989;58(2):257–267. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 38.Vooijs M, Berns A. Developmental defects and tumor predisposition in Rb mutant mice. Oncogene. 1999;18(38):5293–5303. doi: 10.1038/sj.onc.1202999. [DOI] [PubMed] [Google Scholar]
- 39.Robinson GW, Wagner KU, Hennighausen L. Functional mammary gland development and oncogene-induced tumor formation are not affected by the absence of the retinoblastoma gene. Oncogene. 2001;20(48):7115–7119. doi: 10.1038/sj.onc.1204888. [DOI] [PubMed] [Google Scholar]
- 40.Husler MR, Kotopoulis KA, Sundberg JP, Tennent BJ, Kunig SV, Knowles BB. Lactation-induced WAP-SV40 Tag transgene expression in C57BL/6J mice leads to mammary carcinoma. Transgenic Res. 1998;7(4):253–263. doi: 10.1023/A:1008865911660. [DOI] [PubMed] [Google Scholar]
- 41.Maroulakou IG, Anver M, Garrett L, Green JE. Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci U S A. 1994;91(23):11236–11240. doi: 10.1073/pnas.91.23.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simin K, et al. pRb inactivation in mammary cells reveals common mechanisms for tumor initiation and progression in divergent epithelia. PLoS Biol. 2004;2(2):E22. doi: 10.1371/journal.pbio.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner KU, Boulanger CA, Henry MD, Sgagias M, Hennighausen L, Smith GH. An adjunct mammary epithelial cell population in parous females: its role in functional adaptation and tissue renewal. Development. 2002;129(6):1377–1386. doi: 10.1242/dev.129.6.1377. [DOI] [PubMed] [Google Scholar]
- 44.Li G, et al. Conditional loss of PTEN leads to precocious development and neoplasia in the mammary gland. Development. 2002;129(17):4159–4170. doi: 10.1242/dev.129.17.4159. [DOI] [PubMed] [Google Scholar]
- 45.Vooijs M, te Riele H, van der Valk M, Berns A. Tumor formation in mice with somatic inactivation of the retinoblastoma gene in interphotoreceptor retinol binding protein-expressing cells. Oncogene. 2002;21(30):4635–4645. doi: 10.1038/sj.onc.1205575. [DOI] [PubMed] [Google Scholar]
- 46.Chen D, Livne-bar I, Vanderluit JL, Slack RS, Agochiya M, Bremner R. Cell-specific effects of RB or RB/p107 loss on retinal development implicate an intrinsically death-resistant cell-of-origin in retinoblastoma. Cancer Cell. 2004;5(6):539–551. doi: 10.1016/j.ccr.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 47.MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18(14):1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MH, et al. Targeted disruption of p107: functional overlap between p107 and Rb. . Genes Dev. 1996;10(13):1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 49.LeCouter JE, et al. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol Cell Biol. 1998;18(12):7455–7465. doi: 10.1128/mcb.18.12.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang Z, Zacksenhaus E. Coordinated expression of Rb gene family in the mammary gland. Mech Dev. 2010;119(suppl 1):S39–S42. doi: 10.1016/S0925-4773(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 51.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 52.Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8(1):R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 54.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 55.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25(21):4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 57.Ponti D, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 58.Liu JC, Deng T, Lehal RS, Kim J, Zacksenhaus E. Identification of tumorsphere- and tumor-initiating cells in HER2/neu-induced mammary tumors. Cancer Res. 2007;67(18):8671–8681. doi: 10.1158/0008-5472.CAN-07-1486. [DOI] [PubMed] [Google Scholar]
- 59.Miyoshi K, et al. Activation of beta -catenin signaling in differentiated mammary secretory cells induces transdifferentiation into epidermis and squamous metaplasias. Proc Natl Acad Sci U S A. 2002;99(1):219–224. doi: 10.1073/pnas.012414099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walker LC, Harris GC, Wells JE, Robinson BA, Morris CM. Association of chromosome band 8q22 copy number gain with high grade invasive breast carcinomas by assessment of core needle biopsies. Genes Chromosomes Cancer. 2008;47(5):405–417. doi: 10.1002/gcc.20545. [DOI] [PubMed] [Google Scholar]
- 61.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damonte P, Gregg JP, Borowsky AD, Keister BA, Cardiff RD. EMT tumorigenesis in the mouse mammary gland. Lab Invest. 2007;87(12):1218–1226. doi: 10.1038/labinvest.3700683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller PA, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139(7):1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 64.Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130(4):597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 65. TP53 mutation distribution. http://p53.free.fr/Database/p53_database_dist.html . Accessed July 19, 2010. [Google Scholar]
- 66.Walker DR, et al. Evolutionary conservation and somatic mutation hotspot maps of p53: correlation with p53 protein structural and functional features. Oncogene. 1999;18(1):211–218. doi: 10.1038/sj.onc.1202298. [DOI] [PubMed] [Google Scholar]
- 67.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 68.Lin SC, et al. Somatic mutation of p53 leads to estrogen receptor alpha-positive and -negative mouse mammary tumors with high frequency of metastasis. Cancer Res. 2004;64(10):3525–3532. doi: 10.1158/0008-5472.CAN-03-3524. [DOI] [PubMed] [Google Scholar]
- 69.Weinstein IB, Joe A. Oncogene addiction. Cancer Res. 2008;68(9):3077–3080. doi: 10.1158/0008-5472.CAN-07-3293. [DOI] [PubMed] [Google Scholar]
- 70.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136(5):823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang MW, et al. Cytostatic gene therapy for vascular proliferative disorders with a constitutively active form of the retinoblastoma gene product. Science. 1995;267(5197):518–522. doi: 10.1126/science.7824950. [DOI] [PubMed] [Google Scholar]
- 72.Zhu L, et al. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7(7A):1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]
- 73.Muncaster MM, Cohen BL, Phillips RA, Gallie BL. Failure of RB1 to reverse the malignant phenotype of human tumor cell lines. . Cancer Res. 1992;52(3):654–661. [PubMed] [Google Scholar]
- 74.Jiang Z, Zacksenhaus E. Activation of retinoblastoma protein in mammary gland leads to ductal growth suppression, precocious differentiation, and adenocarcinoma. J Cell Biol. 2002;156(1):185–198. doi: 10.1083/jcb.200106084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chew YP, Ellis M, Wilkie S, Mittnacht S. pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene. 1998;17(17):2177–2186. doi: 10.1038/sj.onc.1202443. [DOI] [PubMed] [Google Scholar]
- 76.Arima Y, et al. Rb depletion results in deregulation of E-cadherin and induction of cellular phenotypic changes that are characteristic of the epithelial-to-mesenchymal transition. Cancer Res. 2008;68(13):5104–5112. doi: 10.1158/0008-5472.CAN-07-5680. [DOI] [PubMed] [Google Scholar]
- 77.Smith GH, Medina D. Re-evaluation of mammary stem cell biology based on in vivo transplantation. Breast Cancer Res. 2008;10(1):203. doi: 10.1186/bcr1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matulka LA, Triplett AA, Wagner KU. Parity-induced mammary epithelial cells are multipotent and express cell surface markers associated with stem cells. Dev Biol. 2007;303(1):29–44. doi: 10.1016/j.ydbio.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 79.Knudsen ES, Wang JY. Targeting the RB-pathway in cancer therapy. Clin Cancer Res. 2010;16(4):1094–1099. doi: 10.1158/1078-0432.CCR-09-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bassett EA, Wang W, Rastinejad F, El-Deiry WS. Structural and functional basis for therapeutic modulation of p53 signaling. Clin Cancer Res. 2008;14(20):6376–6386. doi: 10.1158/1078-0432.CCR-08-1526. [DOI] [PubMed] [Google Scholar]
- 81.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ponzo MG, et al. Met induces mammary tumors with diverse histologies and is associated with poor outcome and human basal breast cancer. Proc Natl Acad Sci U S A. 2009;106(31):12903–12908. doi: 10.1073/pnas.0810402106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rottenberg S, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.