Abstract
Translational control plays a key role in late spermiogenesis. A number of mRNAs encoding proteins required for late spermiogenesis are expressed in early spermatids but are stored as translationally inactive messenger ribonucleoprotein particles (mRNPs). The translation of these mRNAs is associated with shortening of their poly(A) tail in late spermiogenesis. Poly(A)-binding protein (Pabp) plays an important role in mRNA stabilization and translation. Three Pabp-interacting proteins, Paip1, Paip2a, and Paip2b, have been described. Paip2a is expressed in late spermatids. To investigate the role of Paip2 in spermiogenesis, we generated mice with knockout of either Paip2a or Paip2b and double-KO (DKO) mice lacking both Paip2a and Paip2b. Paip2a-KO and Paip2a/Paip2b-DKO mice exhibited male infertility. Translation of several mRNAs encoding proteins essential to male germ cell development was inhibited in late spermiogenesis in Paip2a/Paip2b-DKO mice, resulting in defective elongated spermatids. Inhibition of translation in Paip2a/Paip2b-DKO mice was caused by aberrant increased expression of Pabp, which impaired the interaction between eukaryotic initiation factor 4E (eIF4E) and the cap structure at the 5′ end of the mRNA. We therefore propose a model whereby efficient mRNA translation in late spermiogenesis occurs at an optimal concentration of Pabp, a condition not fulfilled in Paip2a/Paip2b-DKO mice.
Introduction
Spermiogenesis is the process by which postmeiotic round spermatids (RSs) differentiate into elongated spermatids. The final step of spermiogenesis consists of dynamic morphological changes such as nuclear condensation, acrosome and sperm tail formation, and reorganization of organelles including the formation of the mitochondrial sheath along the midpiece of the flagellum (1). Regulated protein expression of components in each step is essential for proper germ cell differentiation.
While regulated gene expression is essential for germ cell development, transcription ceases at mid-spermiogenesis in RSs (2, 3). Gene expression in late spermiogenesis, therefore, is under translational control. mRNAs encoding proteins that are needed for late spermiogenesis are expressed in early spermatogenesis and stored as translationally repressed messenger ribonucleoprotein particles (mRNPs). These mRNAs are subsequently translated in elongating and elongated spermatids (ESs) (3–5), thus resulting in uncoupling between transcription and translation (2, 6–8). Defects in control of mRNA translation lead to abnormal sperm formation and male infertility (9). Since the regulation of protein expression at specific steps of male germ cell development is essential for proper germ cell differentiation and male fertility, understanding the molecular basis of this regulation is central to understanding how male germ cells are produced.
Polyadenylation and translational control via the poly(A) tail play important roles during late spermiogenesis. For instance, mRNAs encoding basic nuclear proteins such as protamines (Prms) and transition proteins (Tps) are produced in RSs and stored as mRNPs, which become translationally activated in elongated spermatids (10–16). Prm and Tp mRNAs that are translationally repressed in RSs contain poly(A) tails of approximately 180 nucleotides. They become activated in ESs, concomitant with the shortening of the poly(A) tail to approximately 30 nucleotides (17). The mechanism of translational activation and what triggers the shortening of the poly(A) tails are not understood (9).
RNA-binding proteins function in the stabilization and translation of mRNAs in differentiating male germ cells (18). For example, the germ cell Y-box protein Msy2 is a DNA/RNA-binding protein that acts as a stabilizer and a translational suppressor of mRNAs in RNP complexes (19, 20). Msy2-KO mice display spermatogenic arrest (20, 21). Another RNA-binding protein, polypyrimidine tract binding protein 2 (Ptbp2), stabilizes the testis-specific glycolytic enzyme Pgk2 mRNA and delays its translation for up to 2 weeks (22).
The poly(A)-binding protein (Pabp) contains 4 RNA recognition motifs (RRMs) and a proline-rich C-terminal domain and binds to the poly(A) tail of the mRNA with a periodicity of about 25–27 nucleotides (23, 24). Pabp functions in mRNA biogenesis, mRNA stabilization, and translation (25–27). In spermatogenesis, Pabp is associated with RNPs and polysomal mRNAs, suggesting that Pabp plays a role in both mRNA storage and translation (28). There are two isoforms of Pabp in mouse spermatogenic cells: Pabpc1 and the testis-specific Pabpc2 (29). Both isoforms are present in pachytene spermatocytes (PSs) and RSs, while only Pabpc1 is expressed in elongating spermatids (30). Interestingly, whereas Pabpc1 is associated with both monosomes and polysomes, Pabpc2 is present only in monosomes, suggesting that the isoforms serve distinct functions during spermatogenesis (30).
Pabp activity is modulated in metazoans by two Pabp-interacting proteins (Paips): Paip1 and Paip2 (of which there are two related gene products in mammals, Paip2a and Paip2b). Paip1 stimulates translation through its interaction with eukaryotic initiation factor 3 (eIF3) (31). Paip2 is a translational inhibitor, as it competes with eIF4G for Pabp binding and decreases the affinity of Pabp for the poly(A) tail (32, 33). Paip2 is a highly acidic protein that contains two separate Pabp-binding motifs (PAMs) (34). The two isoforms of Paip2 bind to Pabp but are expressed differentially in tissues, implying tissue-specific functions and responses to distinct stimuli (35).
Given Pabp’s important role in spermatogenesis (36–38), we reasoned that Paip2 may play a key role in this process. To investigate the physiological role of Paip2 in translational control during spermatogenesis, Paip2a-KO, Paip2b-KO, and Paip2a/Paip2b-double-KO (Paip2a/Paip2b-DKO) mice were generated. Here, we describe the phenotype of these mice with respect to fertility and propose a model for translational control in late spermiogenesis via the control of Pabpc1-Paip2a association. Our data implicate Paip2a as an important player in translational control of late spermiogenesis.
Results
Mouse tissue distribution of Paip2a and Paip2b.
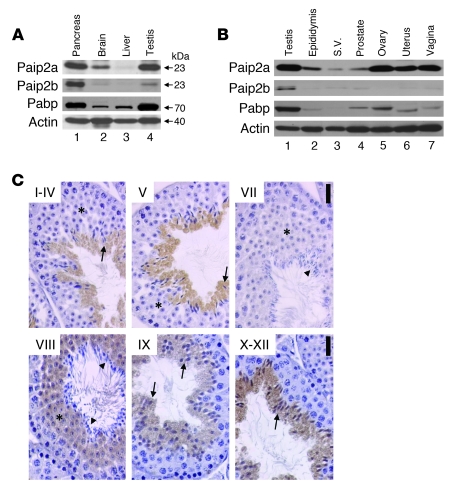
Paip2a and Paip2b expression was assessed in tissues by Western blot analysis. Paip2a protein was highly expressed in pancreas and testis, while Paip2b was highly expressed in pancreas, but barely in testis (Figure 1A). Paip2a was also expressed in brain, where the expression of Paip2b was minimal. Paip2a and Paip2b were barely detected in liver. Pabp was expressed at high levels in pancreas and testis and at lower levels in brain and liver. Thus, expression of the Paip2 isoforms and Pabp is tissue specific, with overlapping high expression of Paip2a and Pabp in testis. Paip2a and Paip2b expression in other reproductive tissues was assessed by Western blot analysis (Figure 1B). Paip2a was weakly expressed in male reproductive tissues such as epididymis, seminal vesicle, and prostate. However, Paip2a was highly expressed in female reproductive tissues such as ovary, uterus, and vagina. Paip2b was weakly expressed in testis but barely expressed in other male or female reproductive tissues. Pabp also showed high expression levels in testis, but not in other reproductive tissues.
Figure 1. Paip2a and Paip2b expression in mouse.
(A) Tissue distribution of Paip2a, Paip2b, and Pabp in pancreas, brain, liver, and testis of WT mouse by Western blot analysis. (B) Paip2a, Paip2b, and Pabp expression in reproductive tissues of WT mouse. S.V., seminal vesicle. (C) Paip2 expression in WT testis visualized by immunohistochemistry. Asterisks show RSs. Arrows indicate elongated spermatids, and arrowheads show mature sperm. Scale bars: 25 μm. Paip2a-specific and Paip2b-specific antibodies were used for Western blotting. Affinity-purified Paip2 antibody that mainly reacts with Paip2a and weakly cross-reacts with Paip2b was used for immunohistochemistry.
To determine which testicular cells express Paip2, we performed immunohistochemistry with an affinity-purified Paip2 antibody (Figure 1C). This antibody was generated against full-length human PAIP2A and weakly cross-reacts with PAIP2B (35). Paip2a was expressed only in germ cell cytoplasm and started being expressed in spermatids from steps 7 to 8 until the end of spermiogenesis, indicating that it may function during the last steps of this process.
Generation of Paip2a-KO, Paip2b-KO, and Paip2a/Paip2b-DKO mice.
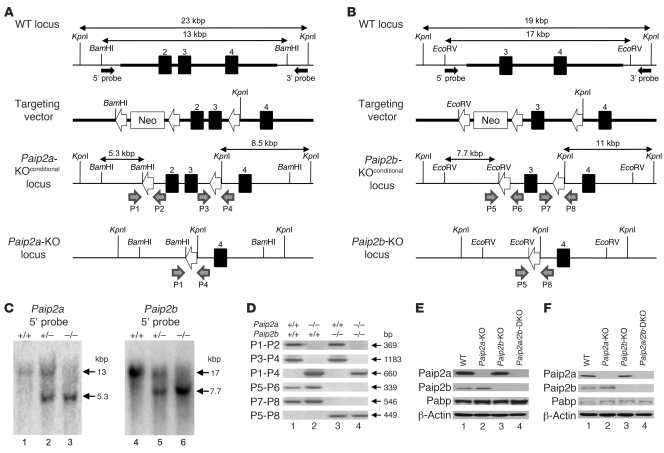
To study the role played by Paip2a in late spermiogenesis, we generated knockout mice. A Paip2a targeting vector was electroporated into R1 ES cells derived from male blastocysts of a hybrid of two 129/SvJ substrains. After G418 selection, the LoxP-flanked Neo cassette was removed by transient expression of Cre recombinase (Figure 2A). The targeted ES cell clones were injected into C57BL/6 blastocysts, and subsequent chimeric male mice were mated with C57BL/6J females to achieve Paip2a allele germline transmission. The second and third exons were removed to generate Paip2a-KO mice by crossing with the CMV-Cre transgenic mouse. Paip2a heterozygotes were crossed to generate Paip2a-KO mice as confirmed by Southern blot analysis (Figure 2C and Supplemental Figure 1, A and C; supplemental material available online with this article; doi: 10.1172/JCI43350DS1). Paip2b-KO mice were also generated (Figure 2, B and C, and Supplemental Figure 1, B and D) and crossed with Paip2a-KO mice to obtain Paip2a/Paip2b-DKO mice. The genotypes were confirmed by PCR targeting the loxP sites (Figure 2D). Western blot analysis confirmed the absence of Paip2a and Paip2b in testes (Figure 2E) and epididymides (Figure 2F).
Figure 2. Generation of Paip2a/Paip2b-KO mouse.
(A) Targeted disruption of Paip2a. Exons 2, 3, and 4 are depicted as filled boxes. The targeting regions are shown as bold lines. Filled arrows indicate 5′ probe and 3′ probe for Southern blot analysis. Open arrows indicate LoxP sites. Gray arrows indicate primers for genotyping (P1, P2, P3, and P4). (B) Targeted disruption of Paip2b. Exons 3 and 4 are shown as filled boxes. Gray arrows indicate primers for genotyping (P5, P6, P7, and P8). (C) Southern blot analysis of WT, Paip2a heterozygote, Paip2a-KO, Paip2b heterozygote, and Paip2b-KO mice probed by 5′ probes (see also Supplemental Figure 1). (D) Genotyping of WT, Paip2a-KO, Paip2b-KO, and Paip2a/Paip2b-DKO mice. Primer sets of P1 and P2, P3 and P4, and P1 and P4 were used to detect the first and second LoxP sites and the recombined loxP site of Paip2a, respectively. Primer sets of P5 and P6, P7 and P8, and P5 and P8 were used to detect the first LoxP site, the second LoxP site, and the recombined loxP site of Paip2b, respectively. (E) Western blot analysis of Paip2a, Paip2b, and Pabp in testes. (F) Western blot analysis of Paip2a, Paip2b, and Pabp in epididymides.
Paip2a-KO male mice are infertile.
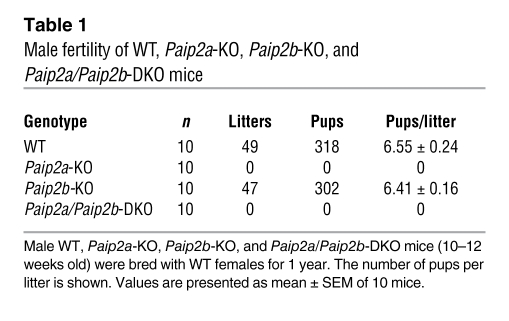
Male fertility was studied by mating 10 males of each strain with WT females for 1 year (Table 1). The lack of Paip2a resulted in male infertility, with no litters in Paip2a-KO or Paip2a/Paip2b-DKO males. Mating of transgenic females with WT males was also performed. No change in the litter per female or in the size of each litter was observed (data not shown), thus demonstrating that even though Paip2a is expressed in female reproductive organs, it is not essential for female fertility. Given that Paip2a-KO and Paip2a/Paip2b-DKO mice exhibit the same infertile phenotype, we pursued our studies with Paip2a/Paip2b-DKO mice because of the possibility, albeit remote, that the Paip2 antibody might cross-react with Paip2b in immunohistological assay at low intensity.
Table 1 .
Male fertility of WT, Paip2a-KO, Paip2b-KO, and Paip2a/Paip2b-DKO mice
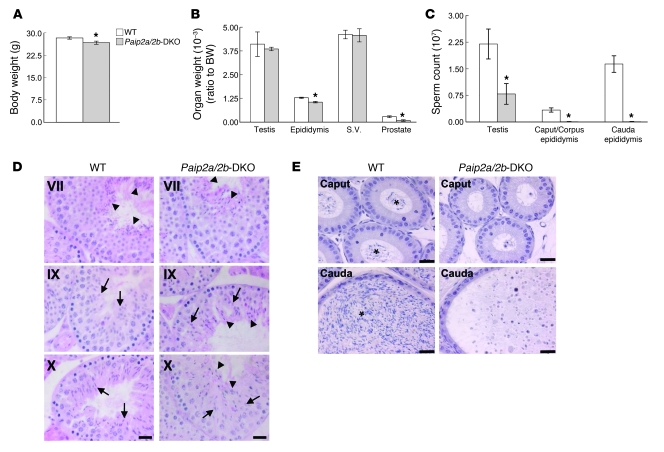
Body weight of Paip2a/Paip2b-DKO mice was lower than that of WT mice (5.6% less than WT, P < 0.05 using an unpaired t test) (Figure 3A). Statistically significant differences in weight between Paip2a/Paip2b-DKO and WT mice were observed for epididymis (19.8% lower than WT) and for prostate (71.2% lower than WT) (Figure 3B). There was no significant difference in weight of testis and seminal vesicle (non-normalized organ weights are shown in Supplemental Figure 2).
Figure 3. Phenotypes of Paip2a/Paip2b-KO mice.
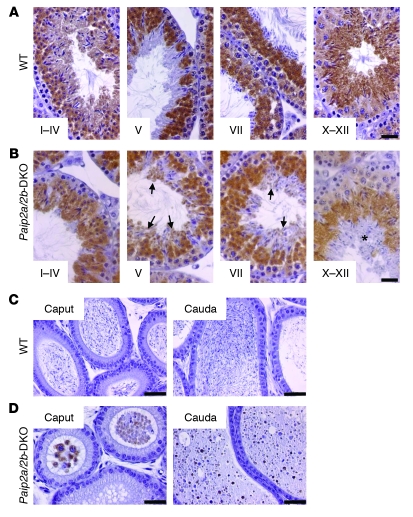
(A) Body weights of WT and Paip2a/Paip2b-DKO mice. Values are mean ± SEM of 20–23 mice at 10–12 weeks old. *P < 0.05 using an unpaired t test. (B) Reproductive organ weights of WT and Paip2a/Paip2b-DKO mice. Organ weights per body weight (BW) are shown. Values are mean ± SEM of 10–16 mice for testis and epididymis and 8–14 mice for seminal vesicle and prostate. *P < 0.05 using an unpaired t test. (C) Sperm count in testes, caput/corpus epididymides, and cauda epididymides of WT and Paip2a/Paip2b-DKO mice. Values are mean ± SEM of 5 mice. *P < 0.05 using an unpaired t test. (D and E) Histology of testes (D) and caput and cauda epididymides (E) from WT and Paip2a/Paip2b-DKO mice by H&E staining. Arrows indicate elongating spermatids; arrowheads in WT mice indicate mature sperm released from the epithelium at stage VII; and arrowheads in Paip2a/Paip2b-DKO mice show sperm that could not be released into the lumen in later stages (D). The asterisks indicate mature sperm in the lumen of WT epididymis (E). Scale bars: 25 μm.
Spermatogenesis was monitored by counting the number of sperm heads in testes and epididymides (Figure 3C). While no difference was observed in testis weight between Paip2a/Paip2b-DKO and WT strains, a significant decrease (64% ± 13%) in the number of sperm heads was detected in testes of Paip2a/Paip2b-DKO mice as compared with those of WT mice. This was consistent with a dramatic decrease in the number of sperm heads in caput/corpus epididymidis (99.2% ± 0.4%), as well as cauda epididymidis (99.8% ± 0.2%), in which some animals displayed a total absence of spermatozoa (Figure 3C).
To resolve the apparent differences between changes in weight and sperm counts, we undertook histological examination of testes and epididymides of WT and Paip2a/Paip2b-DKO mice. All spermatogenic stages could be observed in Paip2a/Paip2b-DKO testes, with normal gross morphological structures of Sertoli cells, spermatogonia, spermatocytes, and elongating spermatids (data not shown). However, aberrant spermatozoa were observed in the epithelia along the lumen of seminiferous tubules at stage VII in testes of Paip2a/Paip2b-DKO mice, which were not released into the lumen, as in WT mice (Figure 3D, arrowheads in Paip2a/Paip2b-DKO). The retained elongated spermatids were present even at stages IX–XII in testes of Paip2a/Paip2b-DKO mice (arrowheads in Figure 3D, Supplemental Figure 3B, and Supplemental Figure 4D). Strikingly, two populations of spermatids that are composed of a normal population of elongating spermatids (arrows in Figure 3D) and a population of aberrant condensed spermatids (arrowheads in Figure 3D) were observed through stages IX–XII in Paip2a/Paip2b-DKO mice. In contrast, spermatids in WT mice began elongating and formed a homogeneous population of elongating spermatids during stages IX–XII (arrows in Figure 3D), suggesting that the spermiation process, i.e., the release of completed spermatozoa into the lumen of seminiferous tubules, is impaired in Paip2a/Paip2b-DKO mice. Significantly, spermatids exhibiting aberrant morphology in Paip2a/Paip2b-DKO testis corresponded to those expressing Paip2a (Figure 1C) in late spermiogenesis. These observations demonstrate an important role of Paip2a in germ cell differentiation during late spermiogenesis.
Few sperm heads were observed in the lumen of caput and cauda epididymides of Paip2a/Paip2b-DKO mice as compared with WT mice (Figure 3E). Aberrant round structures were apparent in cauda epididymides of Paip2a/Paip2b-DKO mice (Figure 3E and Supplemental Figure 4D). The same phenotype was seen in testes and epididymides of Paip2a-KO mice (Supplemental Figure 4B) but not in Paip2b-KO mice (Supplemental Figure 4C). These data demonstrate that male infertility is caused, at least in part, by impaired release of spermatozoa from seminiferous tubules and migration from testis to epididymis.
Anomalous Pabp expression in late spermiogenesis in Paip2a/Paip2b-DKO mice.
To investigate the molecular basis of the Paip2a-KO phenotype, we performed immunohistochemistry to determine Pabp expression in testes of WT and Paip2a/Paip2b-DKO mice. In WT mice, Pabp was first expressed in the cytoplasm of PSs, reached its peak expression in step 7 and 8 of spermiogenesis, diminished as spermatids underwent nuclear condensation, and disappeared around step 15 of spermiogenesis (Figure 4A) (28). In sharp contrast, Pabp was still expressed in late spermiogenesis in Paip2a/Paip2b-DKO mice (Figure 4B): at stages X–XII, a layer of aberrant elongated spermatids was observed in which Pabp was still being expressed (asterisk in Figure 4B). Moreover, while Pabp was absent in the lumen of cauda epididymides in WT mice (Figure 4C), it was detected in Paip2a/Paip2b-DKO mice (Figure 4D). These data clearly indicate that Paip2a modulates Pabp expression and raise the possibility that translational control through Pabp-Paip2a interaction is important at the final steps of spermiogenesis.
Figure 4. Abnormal Pabp expression in late spermiogenesis in Paip2a/Paip2b-DKO mice.
(A and B) Immunohistochemistry of Pabp in testes of WT (A) and Paip2a/Paip2b-DKO (B) mice. Stages I–IV, V, VII, and X–XII are shown. Arrows show aberrant Pabp expression in elongated spermatids at stages V and VII of Paip2a/Paip2b-DKO mice. An asterisk indicates a layer of aberrant elongated spermatids in which Pabp is slightly expressed at stages X–XII in Paip2a/Paip2b-DKO testis. Scale bars: 25 μm. (C and D) Immunohistochemistry of Pabp in caput and cauda epididymides of WT (C) and Paip2a/Paip2b-DKO (D) mice. Scale bars: 50 μm.
Impairment of formation of mitochondrial sheath and chromatin condensation in Paip2a/Paip2b-DKO mice.
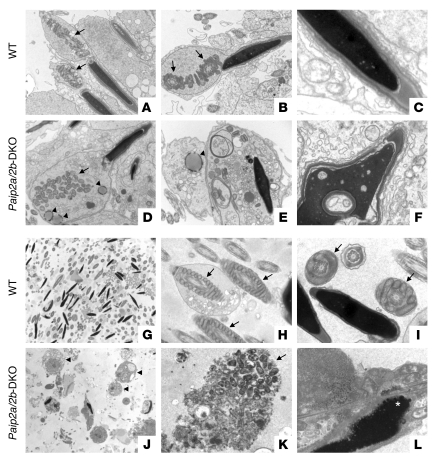
To determine how the defects in elongated spermatids of Paip2a/Paip2b-DKO mice affect ultrastructural aspects of sperm morphology, we examined testes at stage VII of spermatogenesis and epididymides by electron microscopy. While the mitochondrial sheath is formed around the middle piece of the spermatid flagellum in WT mice (arrows in Figure 5, A and B), the mitochondria cannot form the sheath in Paip2a/Paip2b-DKO mice (arrow in Figure 5D). This suggests that development is aberrant by step 15 of spermiogenesis. Moreover, abnormal spherical structures were also observed in the cytoplasm of Paip2a/Paip2b-DKO spermatids (arrowheads in Figure 5, D and E), but not in WT spermatids. The acrosome was detached from the nucleus, and abnormal chromatin condensation was evident in Paip2a/Paip2b-DKO mice (Figure 5F). Vacuoles of various size in the majority of nuclei were detected at stage VII of spermatogenesis in Paip2a/Paip2b-DKO mice, while these structures were rare in WT spermatids (Figure 5, C and F). The large number of cellular defects observed in Paip2a/Paip2b-DKO mice suggests that key components needed for cell differentiation in late spermiogenesis are not generated properly as a consequence of the loss of Paip2a, further supporting a model whereby Paip2a plays an important regulatory role in late spermiogenesis.
Figure 5. Abnormal chromatin condensation and mitochondrial alignment in Paip2a/Paip2b-DKO mice.
(A–F) Spermatids in testes from WT (A–C) and Paip2a/Paip2b-DKO (D–F) mice at stage VII of spermatogenesis. Mitochondria (arrows) and aberrant spherical structures in Paip2a/Paip2b-DKO mice (arrowheads) are shown. (G–L) Spermatozoa in cauda epididymides from WT (G–I) and Paip2a/Paip2b-DKO (J–L) mice. Mitochondria (arrows), ectopic residual bodies (arrowheads), and bubble-like structures in nucleus (asterisk) are shown. Original magnification, A and D: ×8100, B and E: ×10,200, C and F: ×24,400, G and J: ×2,120, H and K: ×13,800, I and L: ×24,400.
Consistent with the existence of abnormal spermatids in testis, abnormal spermatozoa were also present in cauda epididymides of Paip2a/Paip2b-DKO mice (Figure 5, J–L) as compared with WT mice (Figure 5, G–I). Residual bodies (arrowheads in Figure 5J) that are normally removed in testis remained in the lumen of cauda epididymidis of Paip2a/Paip2b-DKO mice. Proper tail structures in WT epididymis (Figure 5, G–I) were not evident in Paip2a/Paip2b-DKO epididymis (Figure 5, J–L). Disorganized clusters of mitochondria that failed to form the sheath around the flagellum were observed (arrow in Figure 5K). Some bubble-like structures in the nucleus (asterisk in Figure 5L) were also present in Paip2a/Paip2b-DKO epididymis. These data strongly suggest that the aberrant sperm and ectopic residual bodies in epididymides of Paip2a/Paip2b-DKO mice are likely the cause for male infertility.
Translation inhibition of mRNAs in late spermiogenesis.
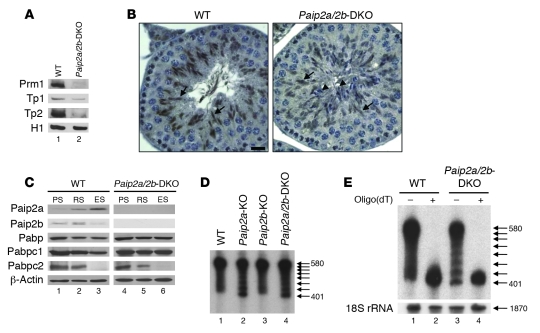
To understand the molecular basis of Paip2a’s control of late spermiogenesis, we investigated how Paip2a affects the translation of mRNAs encoding testicular basic nuclear proteins such as Prm1, Tp1, and Tp2, which are essential for chromatin condensation. These proteins are required for male fertility, and their mRNAs are translationally activated in late spermiogenesis. Strikingly, protein levels of all 3 basic nuclear proteins were reduced in Paip2a/Paip2b-DKO as compared with WT mice (Figure 6A). No difference was observed in Prm1, Tp1, and Tp2 mRNA levels of Paip2a/Paip2b-DKO testes as compared with WT testes as determined by real-time quantitative PCR (Supplemental Figure 5). These data indicate that Paip2a is required for efficient synthesis of Prm1, Tp1, and Tp2 proteins. Tp2 protein expression as assessed by immunohistochemistry was also reduced in testes of Paip2a/Paip2b-DKO mice (Figure 6B). Only Tp2-positive spermatids were detected at stages X–XII in WT testis (arrows), while two distinct populations that consisted of Tp2-positive cells and -negative cells (arrowheads) were observed in Paip2a/Paip2b-DKO testis. The Tp2-negative cells originated because of the impairment of spermiation, as elongated spermatids were retained in the epithelium of seminiferous tubule at stage VIII.
Figure 6. Translation inhibition in late spermiogenesis of Paip2a/Paip2b-DKO mice.
(A) Levels of basic nuclear proteins (Prm1, Tp1, Tp2, and histone H1) from testes of WT and Paip2a/Paip2b-DKO mice were analyzed by acid-urea polyacrylamide gel electrophoresis. (B) Tp2 expression in testes at stages X–XII from WT and Paip2a/Paip2b-DKO mice analyzed by immunohistochemistry. Arrows indicate Tp2-positive spermatids, and arrowheads indicate Tp2-negative spermatids. Scale bar: 10 μm. (C) Expression of Paip2a, Paip2b, Pabpc1, and Pabpc2 in PSs, RSs, and ESs of WT (lanes 1–3) and Paip2a/Paip2b-DKO (lanes 4–6) mice analyzed by Western blotting. (D) Prm1 mRNA in testes from WT, Paip2a-KO, Paip2b-KO, and Paip2a/Paip2b-DKO mice analyzed by Northern blot analysis. Numbers on the right denote nucleotides, and arrows indicate ladder RNA fragments. (E) Ladder RNA fragments were generated by cleavage of poly(A) tail. Total testis RNA of WT and Paip2a/Paip2b-DKO mice was incubated without (–) or with (+) oligo(dT) and then treated with RNase H, followed by Northern blot analysis. Ribosomal 18S RNA (1,870 nucleotides) was used as a control for the absence of nonspecific RNA degradation during this experiment.
To investigate in which germ cells Paip2a, Paip2b, Pabpc1, and the testis-specific Pabpc2 are expressed, we analyzed PSs, RSs, and ESs from WT and Paip2a/Paip2b-DKO mice by Western blot analysis. Both isoforms of Paip2 were expressed at low levels in PSs and RSs, but Paip2a was strongly expressed in ESs, whereas Paip2b was not (Figure 6C, lanes 1–3). Pabp was detected in all cell types of WT and Paip2a/Paip2b-DKO mice with a PABP antibody that cross-reacts with both Pabpc1 and Pabpc2 by immunohistochemistry. The testis-specific Pabpc2 isoform was predominantly expressed in PSs and RSs, while Pabpc1 was expressed in all cell types (Figure 6C, lanes 1–3). Whereas Pabpc1 was expressed at lower levels in ESs than in other cells in WT mice, its expression was equal in all cell types in Paip2a/Paip2b-DKO mice (Figure 6C and Supplemental Figure 6). Thus, Pabpc1 expression was increased in ESs of Paip2a/Paip2b-DKO mice. These data are in accordance with the histological findings, showing aberrant Pabp expression in elongated spermatids at stages V–XII of spermatogenesis in Paip2a/Paip2b-DKO mice (Figure 4B). No change was observed in Pabpc2 expression between WT and Paip2a/Paip2b-DKO mice. These results indicate that the absence of Paip2a causes a specific increase of Pabpc1 expression at late spermiogenesis.
The shortening of the poly(A) tail in elongated spermatids is a consequence of translation (17). It is thus predicted that the shortening of poly(A) tail would be impaired by the aberrant Pabp expression in late spermiogenesis in Paip2a/Paip2b-DKO mice. To investigate this, we isolated total RNA from testes and analyzed Prm1 mRNA by Northern blotting. Heterogeneous Prm1 mRNAs were observed as a diffused band in WT mice and also in Paip2b-KO mice reported earlier (Figure 6D, lanes 1 and 3) (28). In contrast, Paip2a-KO and Paip2a/Paip2b-DKO mice contained Prm1 mRNA species with ladder-like bands (lanes 2 and 4). The ladder bands arose from the limited spontaneous cleavage at the poly(A) tail at the sites not protected by bound Pabp. A repeating pattern of organization of poly(A) ribonucleoprotein, which is revealed after endonuclease digestion, has been documented earlier (23, 32). To make sure that the heterogeneity of mRNA was due to the different length of the poly(A) tail, the mRNA preparations were digested with RNase H in the presence of oligo(dT). As we expected, this treatment generated a single band (Figure 6E, lanes 2 and 4). The distance between the RNA-protected bands was approximately 27 nucleotides, the length required for binding of one molecule of Pabp (Figure 6, D and E). These data indicate that the mechanism for poly(A) shortening of mRNAs encoding proteins required for germ cell differentiation in late spermiogenesis is different in WT and Paip2a/Paip2b-DKO mice. The possible explanation for this difference is addressed in the Discussion.
Excess Pabp inhibits translation in late spermiogenesis.
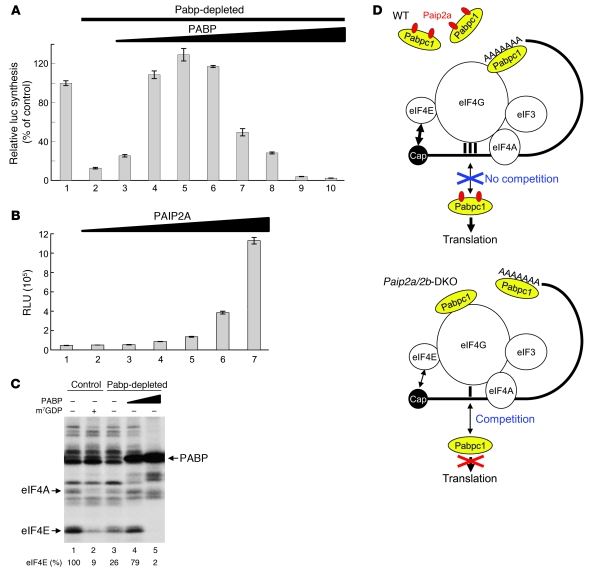
It is possible that translation is impaired by the increased Pabpc1 expression in late spermiogenesis of Paip2a/Paip2b-DKO mice. Pabpc1 stimulates translation, but the effect of excess Pabp on translation is not known. To address this question, we used established cell-free translation extracts because it is not possible to prepare a translationally competent cell-free extract from testis, primarily due to limited material. Pabp was depleted from a Krebs-2 ascites cell extract by using a GST-PAIP2 affinity matrix (39). Western blotting with PABP1 antibody confirmed the nearly complete depletion of Pabp (Supplemental Figure 7A). The ability of Krebs-2 extract to translate capped and polyadenylated (98 A’s) luciferase mRNA [Cap-Luc(A98)] decreased to 12.6% of control after Pabp depletion (Figure 7A, bar 2), and the addition of recombinant PABP restored and even stimulated translation relative to control (up to 129% at 4.5 μg/ml) (Figure 7A, bars 4–6). This is consistent with the well-established stimulatory role of PABP in translation initiation. However, optimal translation occurred at a narrow range of PABP concentrations, since the stimulation of translation was abolished as PABP concentrations reached 30 μg/ml (Figure 7A, bars 7–10). At this concentration of PABP, translation was even inhibited relative to the Pabp-depleted cell extract (compare bars 10 and 2). Thus, these results lead to a prediction that an increase in the abundance and activity of Pabp in elongated spermatids of Paip2a/Paip2b-DKO mice would cause translation inhibition.
Figure 7. Translational control of late spermiogenesis by Paip2a.
(A) Excess amounts of PABP inhibit translation in vitro. Control (lane 1) or Pabp-depleted Krebs-2 cell extracts (lanes 2–10) were programmed with capped Luc(A98) mRNA (2 μg/ml). The translation in Pabp-depleted Krebs-2 extract was performed in the presence of increasing concentrations of recombinant PABP (lanes 2–10; 0, 1.5, 3.0, 4.5, 6.0, 12, 18, 24, and 30 μg/ml). Relative light units were measured, and the value in control (1,034,812 RLU) was set as 100%. Data are presented as mean ± SD. (B) Translation of capped Luc(A98) mRNA in Pabp-depleted Krebs-2 extract supplemented with excess PABP (30 μg/ml) was performed in the presence of increasing concentrations of recombinant GST-PAIP2A (lanes 1–7; 0, 2.5, 5.0, 10, 15, 20, and 30 μg/ml). Data are presented in RLU as mean ± SD. (C) Crosslinking of initiation factors to the cap structure. Control RRL (lanes 1 and 2) or Pabp-depleted RRL (lanes 3–5) was preincubated with control buffer (–) (lanes 1 and 3), 0.6 mM m7GDP (+) (lane 2), and PABP (15 or 30 μg/ml) (lanes 4 and 5, respectively). Oxidized [32P]cap-labeled Luc(A+) mRNA was then added. After reduction with NaIO4 and nuclease digestion, labeled proteins were analyzed by SDS-PAGE and autoradiography. Relative efficiencies of eIF4E crosslinking to the cap structure are indicated at the bottom. The value in control RRL (lane 1) was set as 100%. (D) Model of molecular mechanism of translational control in late spermiogenesis by Paip2a.
To investigate whether Paip2a can reverse the translation inhibition caused by excess Pabp, we added recombinant GST-tagged PAIP2A to a Pabp-depleted Krebs-2 ascites cell extract in which translation was inhibited by PABP (30 μg/ml) (Figure 7B). Increasing PAIP2A concentrations restored translation (Figure 7B, bars 2–7). Thus, Paip2a is expected to function in a stimulatory manner under conditions of Pabp abundance, such as those in late spermiogenesis.
At the optimal concentration, Pabp largely exerts its stimulatory activity on translation through binding to eIF4G, which is a subunit of the eIF4F heterotrimeric complex eIF4E/4G/4A, and stabilizing the association of eIF4E with the cap structure (40). To investigate whether excess Pabp inhibits the activity of eIF4F, we analyzed the interaction of eIF4E with the cap structure by chemical crosslinking in mock- and Pabp-depleted rabbit reticulocyte lysate (RRL) using Luc mRNA 32P-labeled at the 5′ cap. We previously established this assay as a reliable readout of eIF4F activity (41). eIF4E and eIF4A interacted specifically with the cap structure (Figure 7C, lane 1), as this interaction was inhibited by the cap analog m7GDP (9% of control; lane 2). In the Pabp-depleted RRL, eIF4E crosslinking was impaired (26% of control; lane 3). The addition of PABP (15 μg/ml) to Pabp-depleted RRL largely restored eIF4E crosslinking to the cap structure (79% of control; lane 4). Of significance, excess PABP (30 μg/ml) dramatically reduced eIF4E crosslinking (2% of control; lane 5). Thus, excess PABP impairs the interaction between eIF4E and the cap structure, resulting in translation inhibition. Nonspecific binding of PABP to the mRNA was observed (lanes 4 and 5), as this interaction was not inhibited by m7GDP (Supplemental Figure 7B). Although these results were obtained in a system different from testis, our findings are consistent with a model whereby Paip2a promotes efficient translation by inhibiting Pabp in late spermiogenesis (Figure 7D).
Discussion
In the present study, we demonstrated that Paip2a is required for late spermiogenesis through maintenance of Pabp homeostasis. In Paip2a/Paip2b-DKO mice, the translation of mRNAs encoding proteins such as basic nuclear proteins needed for late spermiogenesis was dramatically reduced, which was associated with persistent expression and elevated activity of Pabp. We propose a model whereby efficient mRNA translation in late spermiogenesis occurs at an optimal concentration of active Pabp, a condition not fulfilled in Paip2a/Paip2b-DKO mice. This model posits that in the absence of Paip2a buffering, excess Pabp competes with eIF4G for mRNA binding, resulting in reduced association of eIF4E with the cap structure and translational inhibition (42). While not prominently considered by current translation models (43), Pabp binding to internal mRNA sites has been documented in several studies (44–46). The 4 RRMs of Pabp have apparently different functions. While the combination of RRM domains 1 and 2 binds oligo(A) with the same affinity as intact Pabp, that of RRM domains 3 and 4 recognizes oligo(A) with reduced affinity and specificity (47, 48). Thus, Pabp interaction with the mRNA body is likely due to the promiscuous binding of RRM domains 3 and 4. In binding to the 5′ UTR, Pabp would compete with initiation factors for translation, i.e., display an activity characteristic of general RNA-binding proteins (41). In support of this idea, we showed that high concentrations of Pabp inhibit translation in vitro and also decrease the assembly of the eIF4F complex on the mRNA structure, as measured by eIF4E crosslinking. This is in sharp contrast to the prominent enhancement of translation that Pabp displays at subsaturating concentrations (40). In addition, free Pabp can potentially compete with poly(A)-bound Pabp to disrupt mRNA circular organization required for efficient translation. Further studies are needed to address this possibility.
When Pabp was limited, such as in late spermiogenesis, poly(A) was shortened in the 3′ to 5′ direction by an exonuclease, as evidenced by the heterogeneous appearance of Prm1 mRNA in WT mice. In contrast, when Pabp was in excess, in Paip2a/Paip2b-DKO mice, the poly(A) tail was protected against exonuclease digestion while being sensitive to a limited endonucleolytic cleavage at discrete sites. It is plausible that the poly(A) tail in Paip2a/Paip2b-DKO mice is more densely loaded with Pabp than WT mice and that this impairs mRNA degradation in the 3′ to 5′ direction by an exonuclease. It is not clear, however, whether these mechanistic differences in poly(A) shortening could affect mRNA translation.
The process of spermiation is not well understood (1), but it involves interactions between Sertoli and germ cells (49). Sertoli cells play a major role in spermatozoa release (50). Studying the mechanism of spermiation is essential for understanding how defects in spermiation cause idiopathic male infertility (51). Two knockout mouse models have been reported to exhibit a spermiation defect. Null mice for the transcription factor SRY-related HMG box 8 (Sox8) show age-dependant spermatogenesis disorganization and spermiation defect (52); and cleavage stimulation factor, 3′ pre-RNA subunit 2, tau–null (Cstf2t-null) mice have a failure of spermiation (53).
Spermatozoa in testes and cauda epididymides of Paip2a/Paip2b-DKO mice exhibit multiple morphological abnormalities, including impairment of flagellum formation associated with the absence of the mitochondrial sheath in the middle piece. We have also described a failure of Sertoli cells to remove residual bodies, a detachment of the acrosome from the nucleus, and aberrant vacuoles in the chromatin suggesting an impairment of the chromatin condensation process. These defects may be explained by Paip2a’s translational control of mRNAs encoding proteins needed for these processes.
Pabp expression is altered in late spermiogenesis of Paip2a/Paip2b-DKO mice. Paip2a protein levels are reduced in PABP-depleted HeLa cells due to its ubiquitination and degradation (54). Thus, Pabp and Paip2a affect each other’s cellular abundance. One possible explanation among many for the aberrant Pabp expression in Paip2a/Paip2b-DKO mice is that mRNAs encoding components responsible for Pabp degradation cannot be translated in late spermiogenesis because of the lack of Paip2a.
In summary, we have demonstrated a novel role for Paip2a in male germ cell development during late spermiogenesis. Considering the morphological abnormalities observed in elongated spermatids of Paip2a-KO and Paip2a/Paip2b-DKO mice, it is conceivable that translational control by Paip2a-Pabp interaction affects the synthesis of key proteins essential for male germ cell maturation during late spermiogenesis. Further studies on the Paip2a mouse model could provide important information about male germ cell development and fertility.
Methods
Targeted disruption of Paip2a.
A PCR fragment amplified with a primer set of Paip2a 5′ Fwd and Rev (Supplemental Table 1) was used as a probe to isolate genomic BAC DNA clone 103F10 from the 129/Sv mouse BAC genomic library RPCI-22. The targeting vector was constructed by recombination (55), and routine cloning methods were employed with a 10.5-kbp mouse Paip2a genomic fragment from clone 103F10, as illustrated in Figure 2A. The final targeting fragment was excised from its cloning vector backbone by NotI and electroporated into R1 ES cells. Southern blot analysis was performed with two probes corresponding to the 5′ and 3′ sequences outside the targeting region, as indicated in Figure 2A. To generate the 5′ and 3′ probes, the primer sets (Paip2a 5′ Fwd and Rev for the 5′ probe and Paip2a 3′ Fwd and Rev for the 3′ probe) listed in Supplemental Table 1 were used. After the LoxP-flanked Neo cassette was eliminated by subsequent transient transfection with a cre recombinase expression vector, these genomic engineering steps in ES cells resulted in two net insertions: the first 100-bp LoxP-containing sequence at the BstBI site is located 1 kb upstream of exon 2, and the second 107-bp LoxP-containing sequence at the BlnI site is located 302 bp downstream of exon 3 (Supplemental Table 2). Chimeric male mice were generated by the injection of the Paip2a-targeted ES cells into C57BL/6 blastocysts. The resultant chimeric mice were crossed with C57BL/6J mice for the Paip2a allele germline transmission to obtain Paip2afl/+ mice. After germline transmission, Paip2afl/+ mice were crossed with CMV-Cre transgenic mice constitutively expressing a cre gene under the control of the CMV promoter (56) to eliminate the second and third exons of Paip2a. Mice were then backcrossed with C57BL/6J mice from The Jackson Laboratory for 10 generations using JAX Speed Congenic Service. The mice were genotyped with the primer sets for amplification of the first LoxP site flanking region (Paip2a P1 and P2; 369 bp), the second LoxP site flanking region (Paip2a P3 and P4; 1,183 bp), and the recombined loxP site flanking region (Paip2a P1 and P4; 660 bp) (Figure 2, A and D, and Supplemental Table 1). Ten- to 12-week-old mice were used for experiments. All experiments were approved by the McGill University Animal Care Committee.
Targeted disruption of Paip2b.
To generate Paip2b-KO mice, PCR fragment amplified with a primer set of Paip2b 5′ Fwd and Rev (Supplemental Table 3) was used as a probe to isolate genomic BAC DNA clone 249C9 from the 129/Sv mouse BAC genomic library RPCI-22. The targeting vector was constructed using a 13-kbp mouse Paip2b genomic fragment from clone 249C9 as illustrated in Figure 2B. The final targeting fragment was excised from its cloning vector backbone by NotI and electroporated into R1 ES cells. The LoxP-flanked Neo cassette was eliminated. Southern blot analysis was performed with two probes corresponding to the sequences outside the targeting region, as indicated in Figure 2B, with the primer sets (Paip2b 5′ Fwd and Rev for the 5′ probe and Paip2b 3′ Fwd and Rev for the 3′ probe) listed in Supplemental Table 3. These genomic engineering steps in ES cells resulted in two insertions: a 115-bp LoxP-containing sequence at the first SpeI site located 1,053 bp upstream of exon 3, and a 112-bp LoxP-containing sequence at the second SpeI site located 529 bp downstream of exon 3 (Supplemental Table 4). Paip2b-KO mice in which the third exon was removed were generated as described for the targeted disruption of Paip2a. The mice were genotyped with the primer sets for amplification of the first LoxP site flanking region (Paip2b P5 and P6; 339 bp), the second LoxP site flanking region (Paip2b P7 and P8; 546 bp), and the recombined loxP site flanking region (Paip2b P5 and P8; 449 bp) (Figure 2, B and D, and Supplemental Table 3).
Generation of Paip2a/Paip2b-DKO mice.
Paip2a-KO and Paip2b-KO heterozygous mice were intercrossed to obtain Paip2a/Paip2b-DKO mice.
Antibodies.
Anti-PAIP2 antibody (P0087) was purchased from Sigma-Aldrich and used as Paip2a-specific antibody for Western blot analysis. Paip2b antibody (35) was used as Paip2b-specific antibody for Western blot analysis. Paip2 antibody (32) was purified with AminoLink Plus Immobilization Kit (Pierce) according to the manufacturer’s instructions and used for immunohistochemistry. This antibody mainly reacts with Paip2a and weakly cross-reacts with Paip2b. PABP1 antibody (no. 4992) was purchased from Cell Signaling Technology and used for Western blot analysis and immunohistochemistry. Antibodies against Pabpc1 and Pabpc2 were a gift from T. Baba (University of Tsukuba, Tsukuba, Ibaraki, Japan) (30) and used for Western blot analysis. Monoclonal mouse anti–human smooth muscle actin clone 1A4 (N0851) was purchased from Dako to detect α-actin, and monoclonal anti–β-actin antibody (A5441) was purchased from Sigma-Aldrich for Western blot analysis. Histone H1 (AE-4) antibody (sc-8030) was purchased from Santa Cruz Biotechnology Inc. Prm1 antibody (Hup1N monoclonal antibody for human protamine 1) was a gift from R. Balhorn (Lawrence Livermore National Laboratory, Livermore, California, USA) (57). Antibodies against Tp1 and Tp2 were gifts from S. Kistler (University of South Carolina, Columbia, South Carolina, USA) (58, 59).
Fertility.
Male fertility was assessed by mating 10 males of each strain with WT females for 1 year. Female fertility was assessed similarly by mating 10 females of each genotype with WT males.
Genotyping.
Mouse tails or toes were collected and boiled in solution 1 (25 mM NaOH, 0.2 mM EDTA) for 20 minutes at 100°C. After centrifugation, solution 2 (40 mM Tris-HCl [pH 7.5]) was added to the reaction, followed by vigorous vortexing. Genotyping was performed by PCR using TITANIUM Taq DNA polymerase (Clontech) according to the manufacturer’s instructions.
Western blot analysis.
Tissues were collected and lysed in RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, and 0.1% SDS) containing protease inhibitor cocktail (10 μl/ml) (Sigma-Aldrich, P8340). Cell lysates were clarified by centrifugation at 13,000 g for 14 minutes at 4°C. Protein concentrations were measured using Bio-Rad protein assay. Samples (100 μg) were resolved by SDS-12% PAGE and transferred onto a nitrocellulose membrane (Protein, PerkinElmer). Membranes were blocked in 5% nonfat milk in PBS containing 0.1% Tween 20 and probed with primary antibodies against Paip2a, Paip2b, Pabp, Pabpc1, and Pabpc2 at a dilution of 1:2,000 overnight at 4°C and subsequently with secondary antibody (ECL anti-rabbit IgG horseradish peroxidase-linked whole antibody [GE Healthcare]). Immunoreactive proteins were visualized using enhanced chemiluminescence (PerkinElmer).
Sperm count.
The testis, the caput/corpus epididymidis, and the cauda epididymidis were sonicated twice for 15 seconds separated by a 30-second interval in 0.9% saline, 0.1% merthiolate, and 0.05% Triton X-100 using a sonicator (VWR International). Heads of spermatozoa were counted using a hemocytometer.
Immunohistochemistry.
Testes were fixed in modified Davidson’s solution to detect Paip2 and Pabp (Electron Microscopy Sciences). Testes were fixed in Bouin solution for Tp2. Paraffin sections (5 μm) were rehydrated and incubated with 3% H2O2 for 10 minutes at room temperature. To detect Tp2, slides were boiled for 5 minutes with 0.01 M citrate buffer (pH 6.0) and incubated with blocking buffer (PBS containing 10% goat serum, 0.1% Triton X-100, and 5% BSA fraction V) for 30 minutes at room temperature. Slides were then incubated with blocking buffer containing affinity-purified Paip2 antibody (1:1,000), PABP1 antibody (1:1,000), or Tp2 antibody (1:1,000) overnight at 4°C. After washing in PBS, slides were incubated with biotinylated goat anti-rabbit IgG and subsequently with VECTASTAIN ABC reagents (Vector Laboratories) according to the manufacturer’s instructions. Paip2, Pabp, and Tp2 were detected with DAB Peroxidase Substrate Kit (Vector Laboratories) according to the manufacturer’s instructions and then counterstained with hematoxylin. Pictures were taken using a CoolSnap camera attached to a Leica DM LB2 microscope.
Electron microscopy.
To prepare samples for electron microscopy, testes and epididymides were fixed in fixation buffer (0.1 M sodium cacodylate [pH 7.4], 2.5% glutaraldehyde) overnight at 4°C and cut into pieces (less than 1-mm to 3-mm cubes). After washing with 0.1 M sodium cacodylate buffer (pH 7.4) containing 3% sucrose, samples were postfixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide and embedded in epoxy resin, and then analyzed using an electron microscope (Philips 410 Electron Microscope).
Cell separation.
Spermatogenic cells were obtained by cell separation as described previously (60). Briefly, the testes of 6 mice were decapsulated and digested by enzymatic treatment at 34°C with 0.5 mg/ml collagenase (Sigma-Aldrich, C9891) for 12 minutes, followed by 0.5 mg/ml trypsin (Sigma-Aldrich, T8003) for 16 minutes. Cells were then suspended by gentle pipetting in the presence of 1 μg/ml DNase I (Sigma-Aldrich, DN-25), filtered through a nylon mesh, and washed with RPMI medium containing 0.5% BSA fraction V. Cells (6 × 108) were loaded in the velocity sedimentation apparatus (STA-PUT; Proscience), followed by 2%–4% BSA gradient in RPMI. Fractions containing PSs, RSs from spermiogenesis steps 1–9, and ESs from spermiogenesis steps 10–16 were identified by phase contrast microscopy. Cell morphology was compared in WT and Paip2a/Paip2b-DKO mice. Fractions with greater than 85% purity were pooled and stored at –80°C for protein extraction.
Analysis of basic nuclear proteins.
Total basic nuclear protein extracts were prepared as described previously (61, 62). Briefly, testes were homogenized in cold water containing protease inhibitor cocktail (10 μl/ml) (Sigma-Aldrich) and 0.1 mM PMSF. The cell suspension was filtered through 4 layers of gauze, and the filtrate was centrifuged at 600 g for 10 minutes. The pellet was suspended in cold water containing protease inhibitor and PMSF and sonicated for 1 minute, followed by incubation in water containing protease inhibitor and 10 mM DTT for 30 minutes. Basic nuclear proteins were extracted with 0.5 M HCl and precipitated overnight with 25% TCA. Resultant pellets were washed with acidified acetone, followed by acetone. Proteins (50 μg) were separated by electrophoresis in acid-urea 15% polyacrylamide gels. The proteins were blotted onto a Hybond polyvinylidene difluoride membrane in 0.7% acetic acid. Membranes were blocked in 5% nonfat milk in PBS containing 0.1% Tween 20 and incubated with primary antibodies (histone H1 antibody [1:1,000], Prm1 antibody [1:1,000], Tp1 antibody [1:10,000], and Tp2 antibody [1:10,000]) and secondary antibodies (ECL anti-rabbit IgG horseradish peroxidase–linked whole antibody [GE Healthcare]). Immunoreactive proteins were visualized using enhanced chemiluminescence (PerkinElmer).
Northern blot analysis.
Total RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol. RNA samples were separated on 4.5% acrylamide–50% (w/v) urea gel using TBE (89 mM Tris-HCl, 89 mM boric acid, 2 mM EDTA [pH 8.9]) as a running buffer. RNAs were electroblotted onto an Amersham Hybond-N (GE Healthcare, RPN303B). The probes were generated by RT-RCR using total testis RNA. The blots were incubated with the probes labeled with [32P]dCTP using Amersham Ready-To-Go DNA Labeling Beads (–dCTP) (GE Healthcare) according to manufacturer’s instructions. The signal was detected by autoradiography.
RNase H treatment.
Total testis RNA (5 μg) was incubated with RNase H (New England BioLabs Inc.) in the presence or absence of Oligo(dT)12–18 Primer (Invitrogen) according to the manufacturer’s instructions. After phenol-chloroform extraction, RNA samples were subjected to Northern blot analysis.
In vitro translation.
Translation of capped Luc(A98) mRNA in Krebs-2 extract or Pabp-depleted Krebs-2 extract was performed as previously described (39). Luc activity was measured by a Lumat LB 9507 bioluminometer (EG&G Bertold).
Chemical crosslinking assay.
The 3′ poly(A) tail of uncapped Luc mRNA (Promega) was extended using a Poly(A) Tailing Kit (Ambion). Luc(A+) mRNA was radioactively labeled at the m7G cap using vaccinia virus guanylyltransferase (Ambion) with [α-32P]GTP (PerkinElmer) and S-adenosylmethionine according to the manufacturer’s instructions. After oxidation with NaIO4, the cap-labeled Luc(A+) mRNA was used for chemical crosslinking studies in RRL (Promega) as described previously (41, 63). Pabp-depleted RRL was prepared as described (39).
Statistics.
The statistical significance of the difference between the mean values for the different genotypes was evaluated using 2-tailed, unpaired Student’s t test. The data were considered significant when the P value was less than 0.05.
Supplementary Material
Acknowledgments
We thank Zhu Qinzhang for technical assistance in generating Paip2a/Paip2b-KO mice; Johanne Ouellette for assistance with electron microscopy; Noriko Uetani, Annie Sylvestre, Sandra Perreault, and Colin Lister for technical assistance; Tadashi Baba for antibodies against Pabpc1 and Pabpc2; Rod Balhorn for providing monoclonal antibody Hup1N; Stephen Kistler for antibodies against Tp1 and Tp2; and Trang Luu for technical assistance in designing the probes for Northern blot analysis. A. Yanagiya was the recipient of a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad. This work is supported by the NIH grant 2R01GM066157 to N. Sonenberg and a Canadian Institutes of Health Research (CIHR) grant to B. Robaire.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(9):3389–3400. doi:10.1172/JCI43350.
See the related Commentary beginning on page 3090.
References
- 1. Kerr JB, Loveland KL, O’Bryan MK, de Kretser DM. Cytology of the testis and intrinsic control mechanisms. In: Neill JD, ed.Physiology of Reproduction . 3rd ed. Maryland Heights, Missouri, USA: Elsevier Press; 2006:1. [Google Scholar]
- 2.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20(7):555–561. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 3.Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat Embryol (Berl). 1999;199(6):471–487. doi: 10.1007/s004290050245. [DOI] [PubMed] [Google Scholar]
- 4.Kleene KC. Multiple controls over the efficiency of translation of the mRNAs encoding transition proteins, protamines, and the mitochondrial capsule selenoprotein in late spermatids in mice. Dev Biol. 1993;159(2):720–731. doi: 10.1006/dbio.1993.1277. [DOI] [PubMed] [Google Scholar]
- 5.Penttila TL, Yuan L, Mali P, Hoog C, Parvinen M. Haploid gene expression: temporal onset and storage patterns of 13 novel transcripts during rat and mouse spermiogenesis. Biol Reprod. 1995;53(3):499–510. doi: 10.1095/biolreprod53.3.499. [DOI] [PubMed] [Google Scholar]
- 6.Braun RE. Post-transcriptional control of gene expression during spermatogenesis. Semin Cell Dev Biol. 1998;9(4):483–489. doi: 10.1006/scdb.1998.0226. [DOI] [PubMed] [Google Scholar]
- 7.Eddy EM. Regulation of gene expression during spermatogenesis. Semin Cell Dev Biol. 1998;9(4):451–457. doi: 10.1006/scdb.1998.0201. [DOI] [PubMed] [Google Scholar]
- 8.Braun RE. Temporal control of protein synthesis during spermatogenesis. Int J Androl. 2000;23(suppl 2):92–94. doi: 10.1046/j.1365-2605.2000.00027.x. [DOI] [PubMed] [Google Scholar]
- 9.Yan W. Male infertility caused by spermiogenic defects: lessons from gene knockouts. Mol Cell Endocrinol. 2009;306(1–2):24–32. doi: 10.1016/j.mce.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alfonso PJ, Kistler WS. Immunohistochemical localization of spermatid nuclear transition protein 2 in the testes of rats and mice. Biol Reprod. 1993;48(3):522–529. doi: 10.1095/biolreprod48.3.522. [DOI] [PubMed] [Google Scholar]
- 11.Balhorn R, Weston S, Thomas C, Wyrobek AJ. DNA packaging in mouse spermatids. Synthesis of protamine variants and four transition proteins. Exp Cell Res. 1984;150(2):298–308. doi: 10.1016/0014-4827(84)90572-X. [DOI] [PubMed] [Google Scholar]
- 12.Heidaran MA, Showman RM, Kistler WS. A cytochemical study of the transcriptional and translational regulation of nuclear transition protein 1 (TP1), a major chromosomal protein of mammalian spermatids. J Cell Biol. 1988;106(5):1427–1433. doi: 10.1083/jcb.106.5.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kleene KC. Patterns of translational regulation in the mammalian testis. Mol Reprod Dev. 1996;43(2):268–281. doi: 10.1002/(SICI)1098-2795(199602)43:2<268::AID-MRD17>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 14.Mali P, et al. Stage-specific expression of nucleoprotein mRNAs during rat and mouse spermiogenesis. Reprod Fertil Dev. 1989;1(4):369–382. doi: 10.1071/RD9890369. [DOI] [PubMed] [Google Scholar]
- 15.Saunders PT, Millar MR, Maguire SM, Sharpe RM. Stage-specific expression of rat transition protein 2 mRNA and possible localization to the chromatoid body of step 7 spermatids by in situ hybridization using a nonradioactive riboprobe. Mol Reprod Dev. 1992;33(4):385–391. doi: 10.1002/mrd.1080330404. [DOI] [PubMed] [Google Scholar]
- 16.Shih DM, Kleene KC. A study by in situ hybridization of the stage of appearance and disappearance of the transition protein 2 and the mitochondrial capsule seleno-protein mRNAs during spermatogenesis in the mouse. Mol Reprod Dev. 1992;33(2):222–227. doi: 10.1002/mrd.1080330216. [DOI] [PubMed] [Google Scholar]
- 17.Kleene KC. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development. 1989;106(2):367–373. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- 18.Xu M, Hecht NB. MSY2 and polypyrimidine tract binding protein 2 stabilize mRNAs in the mammalian testis. Int J Androl. 2008;31(5):457–461. doi: 10.1111/j.1365-2605.2008.00885.x. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Medvedev S, Reddi PP, Schultz RM, Hecht NB. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc Natl Acad Sci U S A. 2005;102(5):1513–1518. doi: 10.1073/pnas.0404685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J, et al. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci U S A. 2005;102(16):5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang J, Morales CR, Medvedev S, Schultz RM, Hecht NB. In the absence of the mouse DNA/RNA-binding protein MSY2, messenger RNA instability leads to spermatogenic arrest. Biol Reprod. 2007;76(1):48–54. doi: 10.1095/biolreprod.106.055095. [DOI] [PubMed] [Google Scholar]
- 22.Xu M, Hecht NB. Polypyrimidine tract binding protein 2 stabilizes phosphoglycerate kinase 2 mRNA in murine male germ cells by binding to its 3′UTR. Biol Reprod. 2007;76(6):1025–1033. doi: 10.1095/biolreprod.107.060079. [DOI] [PubMed] [Google Scholar]
- 23.Baer BW, Kornberg RD. The protein responsible for the repeating structure of cytoplasmic poly(A)-ribonucleoprotein. J Cell Biol. 1983;96(3):717–721. doi: 10.1083/jcb.96.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khanam T, Muddashetty RS, Kahvejian A, Sonenberg N, Brosius J. Poly(A)-binding protein binds to A-rich sequences via RNA-binding domains 1+2 and 3+4. RNA Biol. 2006;3(4):170–177. doi: 10.4161/rna.3.4.4075. [DOI] [PubMed] [Google Scholar]
- 25.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19(17):4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103(1):29–40. doi: 10.1016/S0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 27.Copeland PR, Wormington M. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA. 2001;7(6):875–886. doi: 10.1017/S1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu W, Kwon Y, Oko R, Hermo L, Hecht NB. Poly (A) binding protein is bound to both stored and polysomal mRNAs in the mammalian testis. Mol Reprod Dev. 1995;40(3):273–285. doi: 10.1002/mrd.1080400303. [DOI] [PubMed] [Google Scholar]
- 29.Kleene KC, Wang MY, Cutler M, Hall C, Shih D. Developmental expression of poly(A) binding protein mRNAs during spermatogenesis in the mouse. Mol Reprod Dev. 1994;39(4):355–364. doi: 10.1002/mrd.1080390403. [DOI] [PubMed] [Google Scholar]
- 30.Kimura M, Ishida K, Kashiwabara S, Baba T. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol Reprod. 2009;80(3):545–554. doi: 10.1095/biolreprod.108.072553. [DOI] [PubMed] [Google Scholar]
- 31.Martineau Y, et al. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol. 2008;28(21):6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khaleghpour K, et al. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell. 2001;7(1):205–216. doi: 10.1016/S1097-2765(01)00168-X. [DOI] [PubMed] [Google Scholar]
- 33.Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci U S A. 2006;103(25):9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khaleghpour K, et al. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol. 2001;21(15):5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berlanga JJ, Baass A, Sonenberg N. Regulation of poly(A) binding protein function in translation: characterization of the Paip2 homolog, Paip2B. RNA. 2006;12(8):1556–1568. doi: 10.1261/rna.106506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gray NK, Wickens M. Control of translation initiation in animals. Annu Rev Cell Dev Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- 37.Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14(20):2596–2609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorgoni B, Gray NK. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct Genomic Proteomic. 2004;3(2):125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 39.Svitkin YV, Sonenberg N. An efficient system for cap- and poly(A)-dependent translation in vitro. Methods Mol Biol. 2004;257:155–170. doi: 10.1385/1-59259-750-5:155. [DOI] [PubMed] [Google Scholar]
- 40.Kahvejian A, Svitkin YV, Sukarieh R, M’Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19(1):104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svitkin YV, et al. General RNA-binding proteins have a function in poly(A)-binding protein-dependent translation. EMBO J. 2009;28(1):58–68. doi: 10.1038/emboj.2008.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol. 2009;29(6):1661–1669. doi: 10.1128/MCB.01187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jacobson A. Poly(A) Metabolism and translation: the closed-loop model. In: Hershey JWB, Mathews MB, Sonenber N, eds.Translational Control . Woodbury, New York, USA: Cold Spring Harbor Laboratory Press; 1996:451–480. [Google Scholar]
- 44.Burd CG, Matunis EL, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11(7):3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gorlach M, Burd CG, Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp Cell Res. 1994;211(2):400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 46.Afonina E, Neumann M, Pavlakis GN. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J Biol Chem. 1997;272(4):2307–2311. doi: 10.1074/jbc.272.4.2307. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol. 1996;256(1):20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 48.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98(6):835–845. doi: 10.1016/S0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 49.Mruk DD, Cheng CY. Sertoli-Sertoli and Sertoli-germ cell interactions and their significance in germ cell movement in the seminiferous epithelium during spermatogenesis. Endocr Rev. 2004;25(5):747–806. doi: 10.1210/er.2003-0022. [DOI] [PubMed] [Google Scholar]
- 50.Mruk DD, Cheng CY. Cell-cell interactions at the ectoplasmic specialization in the testis. Trends Endocrinol Metab. 2004;15(9):439–447. doi: 10.1016/j.tem.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 51.Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14(11):1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O’Bryan MK, et al. Sox8 is a critical regulator of adult Sertoli cell function and male fertility. Dev Biol. 2008;316(2):359–370. doi: 10.1016/j.ydbio.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dass B, et al. Loss of polyadenylation protein tauCstF-64 causes spermatogenic defects and male infertility. Proc Natl Acad Sci U S A. 2007;104(51):20374–20379. doi: 10.1073/pnas.0707589104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida M, et al. Poly(A) binding protein (PABP) homeostasis is mediated by the stability of its inhibitor, Paip2. EMBO J. 2006;25(9):1934–1944. doi: 10.1038/sj.emboj.7601079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13(3):476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dupe V, et al. In vivo functional analysis of the Hoxa-1 3′ retinoic acid response element (3′RARE). Development. 1997;124(2):399–410. doi: 10.1242/dev.124.2.399. [DOI] [PubMed] [Google Scholar]
- 57.Stanker LH, et al. Immunological evidence for a P2 protamine precursor in mature rat sperm. Mol Reprod Dev. 1992;33(4):481–488. doi: 10.1002/mrd.1080330415. [DOI] [PubMed] [Google Scholar]
- 58.Heidaran MA, Kozak CA, Kistler WS. Nucleotide sequence of the Stp-1 gene coding for rat spermatid nuclear transition protein 1 (TP1): homology with protamine P1 and assignment of the mouse Stp-1 gene to chromosome 1. Gene. 1989;75(1):39–46. doi: 10.1016/0378-1119(89)90381-8. [DOI] [PubMed] [Google Scholar]
- 59.Green GR, Balhorn R, Poccia DL, Hecht NB. Synthesis and processing of mammalian protamines and transition proteins. Mol Reprod Dev. 1994;37(3):255–263. doi: 10.1002/mrd.1080370303. [DOI] [PubMed] [Google Scholar]
- 60.Bellve AR, Millette CF, Bhatnagar YM, O’Brien DA. Dissociation of the mouse testis and characterization of isolated spermatogenic cells. J Histochem Cytochem. 1977;25(7):480–494. doi: 10.1177/25.7.893996. [DOI] [PubMed] [Google Scholar]
- 61.Platz RD, Meistrich ML, Grimes SR., Jr Low-molecular-weight basic proteins in spermatids. Methods Cell Biol. 1977;16:297–316. doi: 10.1016/s0091-679x(08)60107-7. [DOI] [PubMed] [Google Scholar]
- 62.Zhao M, et al. Targeted disruption of the transition protein 2 gene affects sperm chromatin structure and reduces fertility in mice. Mol Cell Biol. 2001;21(21):7243–7255. doi: 10.1128/MCB.21.21.7243-7255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonenberg N. ATP/Mg++-dependent cross-linking of cap binding proteins to the 5′ end of eukaryotic mRNA. Nucleic Acids Res. 1981;9(7):1643–1656. doi: 10.1093/nar/9.7.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.