Abstract
shRNA overexpression from viral gene therapy vectors can trigger cytotoxicity leading to organ failure and lethality in mice and rats. This process likely involves saturation of endogenous cellular RNAi factors including exportin-5 (Xpo-5). Here, we have shown that Xpo-5 overexpression enhanced shRNA efficiency in the liver of adult mice but increased hepatotoxicity. We identified the 4 members of the human Argonaute (Ago) protein family as downstream factors involved in saturation of endogenous cellular RNAi, all of which were able to interact with shRNAs in cells and mice. In Ago/shRNA coexpression studies, Ago-2 (Slicer) was the primary rate-limiting determinant of both in vitro and in vivo RNAi efficacy, toxicity, and persistence. In adult mice, vector-based Ago-2/Xpo-5 coexpression enhanced U6-driven shRNA silencing of exogenous and endogenous hepatic targets, reduced hepatotoxicity, and extended RNAi stability by more than 3 months. Use of weaker RNA polymerase III promoters to minimize shRNA expression likewise alleviated in vivo toxicity and permitted greater than 95% persistent knockdown of hepatitis B virus and other transgenes in mouse liver for more than 1 year. Our studies substantiate that abundant small RNAs can overload the endogenous RNAi pathway and reveal possible strategies for reducing hepatotoxicity of short- and long-term clinical gene silencing in humans.
Introduction
RNAi has quickly moved from bench to bedside, as exemplified by the launch of phase I–III trials testing siRNAs or shRNAs against an extensive list of disorders within a decade from RNAi discovery (1). However, while emerging data suggest RNAi treatment to be effective and well tolerated and thus spur tremendous optimism (2), there are also escalating somber reports in animals of severe adverse side effects from in vivo RNAi. These include our early findings of substantial hepatotoxicities and/or fatalities in adult mice following hepatic high-level expression of more than 50 distinct shRNAs, delivered by a potent adeno-associated viral (AAV) vector (3, 4). Similarly, Davidson’s group noted striatal neurotoxicities and cerebellar Purkinje cell loss in mice treated with shRNAs against the huntingtin (5) and SCA1 (6) genes, respectively. Finally, others recently found considerable cell death in red nuclei of rats injected with AAV vectors encoding two distinct shRNAs (7).
Conspicuously, shRNA-induced cytotoxicity was usually neither sequence- nor target-specific, but rather correlated with the shRNA doses. Indeed, hepatotoxicity in mice was greatly reduced when we expressed shRNAs from a moderate liver-specific polymerase (pol) II promoter (8) as compared with the very strong U6 RNA pol III promoter used in our original study (3). A link between shRNA dose and cytotoxicity is further supported by findings in cell culture, where stable high-level U6 shRNA expression was frequently less well tolerated than that from weaker promoters, such as H1 (9, 10). Likewise, one group noted twice as many fetal and neonatal deaths in attempts to create mice transgenic for U6-driven shRNAs as compared with H1 shRNAs (11).
One explanation for all these observations we have raised before is that toxicities and fatalities were caused by inadvertent saturation of the endogenous RNAi machinery, resulting in dysregulation of cellular microRNAs (miRNAs) (3). We had indeed noted substantial changes in hepatic miRNA levels in livers of shRNA-treated ailing mice (3, 4), and miRNA activities were consistently disturbed in shRNA-transfected cells (3, 4, 6, 12). The fact that such adverse effects on miRNAs are seen with transfected siRNAs as well (12–16) implies that one or more key factors in the RNAi pathway are rate-limiting in cell culture. Indeed, a number of suspects have been controversially discussed in the past, such as Dicer, TRBP and components of RNA-induced silencing complex (RISC) (5, 12, 17–20), including Argonaute-2 (Ago-2) (13, 21–23). We and others also reported that siRNAs and shRNAs may overload the cellular karyopherins exportin-5 (Xpo-5) and CRM1, two crucial factors for nuclear-cytoplasmic shuttling of small RNAs (3, 12, 20, 24–27).
It has remained unclear whether the rate-limiting factors identified in vitro are equally determining the efficacy, toxicity, or persistence of therapeutic in vivo RNAi. This is partly because it is hard to faithfully recapitulate in vivo RNAi in cell culture, due to the inherent differences in cellular quiescence and gene expression, or in pharmacokinetics, strength, and delivery of RNAi expression (e.g., transfected plasmids versus infected viral vectors). Of note, we previously found that transient Xpo-5 overexpression increased shRNA efficacy and relieved competition between two shRNAs in murine livers and thus identified nuclear export as a central bottleneck in the hepatic RNAi pathway (3). Yet we did not resolve whether it was contributory to the observed hepatotoxicity. Clearly, a thorough dissection of these mechanisms and the identification of all rate-limiting components of RNAi are required for continued safe and successful clinical development of the technology. This need is further underscored by notions that even marginal RNAi toxicities can cause detrimental effects, such as increased emergence of viral resistance (12) or accelerated tumorigenesis in cancer-prone environments (4).
Results
Identification of Ago-2 as rate-limiting factor in vitro and in vivo.
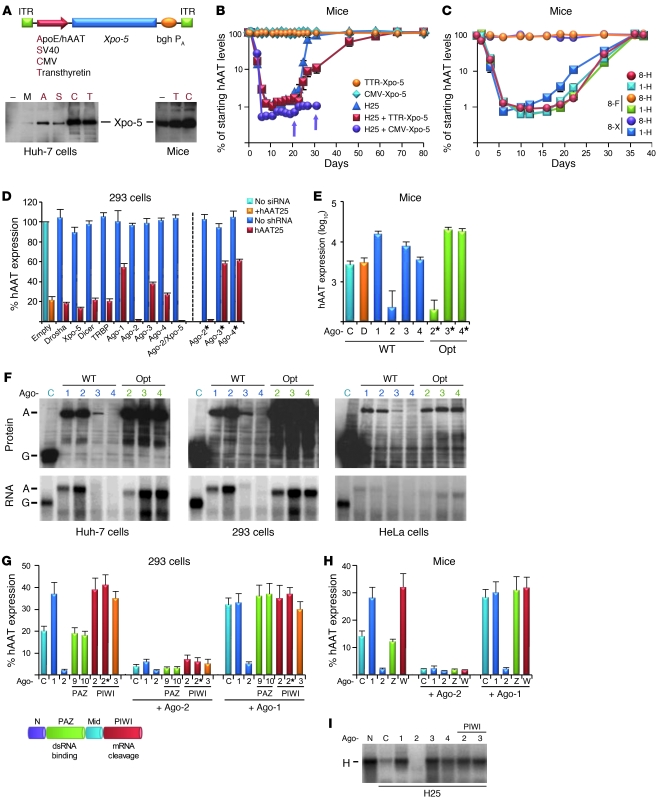
Knowing that Xpo-5 can limit shRNAs in mice (3), we now asked whether its stable expression in adult murine liver would suffice to alleviate shRNA hepatotoxicity and extend in vivo RNAi. We thus created AAV vectors expressing human Xpo-5 under various promoters (Figure 1A) and infused the two most potent into mice transgenic for human α1-antitrypsin (hAAT), together with a sublethal dose of a potent AAV-8 vector (3) encoding an anti-hAAT 25mer shRNA. Consistent with our prior data (3), controls receiving only shRNA showed robust yet transient hAAT knockdown (Figure 1B), due to hepatotoxicity, liver regeneration, and repopulation with parenchymal cells that had lost the episomal AAV vectors. Notably, Xpo-5 coexpression from a liver-specific transthyretin (TTR) promoter doubled the period of RNAi persistence. Curiously, Xpo-5 expression from the stronger CMV promoter not only further enhanced shRNA activity, but also promoted mortality (Figure 1B, arrows), implying that alleviating the Xpo-5 block had adversely saturated downstream rate-limiting factor(s). For validation, we infused TTR–Xpo-5 prior to AAV-shRNA (using distinct AAV serotypes for repeat injections) to overcome the lag in expression from single-stranded Xpo-5 AAV. The initial transient increase in shRNA efficacy, followed by accelerated RNAi loss (Figure 1C, dark blue line), a sign for hepatotoxicity (3), was again suggestive of in vivo saturation of factors downstream of Xpo-5.
Figure 1. Argonaute proteins are rate-limiting for shRNA in cells and mice.
(A) AAV vectors encoding human Xpo-5 cDNA under various promoters and resulting protein expression in transfected cells or murine livers. bghPA, bovine growth hormone polyadenylation signal; ITR, AAV replication/packaging signal; M, mock control. (B) In vivo Xpo-5 overexpression affects efficacy, toxicity and persistence of anti-hAAT shRNA (H25) in hAAT-transgenic mice (n = 3–6). Arrows indicate deaths of individual mice. (C) In vivo effects of preinfusion of Xpo-5–encoding vectors (n = 3). 1/8, AAV-1 or -8; F, human factor IX (control for anti–AAV-8 antibodies); H, H25 shRNA; X, Xpo-5. (D) In vitro screen to identify key RNAi factors limiting shRNA activity. 293 cells were cotransfected with hAAT plasmid, H25 shRNA, and the shown RNAi factors. *Codon-optimized Ago variants (see F). Data were normalized to a CMV-gfp control (Empty). (E) Effects of wild-type and optimized Ago overexpression in hydrodynamically transfected (hAAT, H25 shRNA, plus shown RNAi factors) mouse livers (n = 3, day 3). C, control; D, Dicer (representative example of an RNAi factor having no effect); Opt, codon-optimized; WT, wild-type Agos. For D and E, P < 0.01, Ago-2 versus controls. (F) Expression of different Ago cDNAs (top: Western blots; bottom: Northern blots) in transfected human cells. A, Ago; G, Gfp (expressed from plasmid control [C]). (G) Competition of wild-type and mutant Agos. They were cotransfected with hAAT and H25 shRNA plasmids, or additionally with 4-fold excess Ago-2 or -1. PIWI 3, Ago-3 Slicer mutant. N, N-terminal Ago domain; Mid, middle Ago domain. (H) Same in hydrodynamically transfected mice (n = 3–5, day 2). W, Ago-2 PIWI mutants (both gave similar results); Z, Ago-2 PAZ mutants. (I) Effects of Ago (codon-optimized Ago-3/4) overexpression on hAAT mRNA (H) cleavage with H25 shRNA in cotransfected cells (Northern blot).
To identify these factors, we first reverted to an in vitro system and transfected human 293 cells with plasmids encoding hAAT, anti-hAAT shRNA, or various RNAi proteins. Strikingly, Ago-2 overexpression enhanced shRNA potency by up to 10-fold (Figure 1D and Supplemental Figure 1 [P < 0.05, Ago-2 versus controls]; supplemental material available online with this article; doi: 10.1172/JCI43565DS1), corroborating the central role of Ago-2 for perfect RNAi duplexes in cell culture (13, 21–23). The only other factor yielding a similar, albeit milder effect was Xpo-5; accordingly, Xpo-5/Ago-2 coexpression gave the greatest boost in shRNA efficacy. Notably, in subsequent tests of the individual RNAi factors in mouse livers after hydrodynamic injection of plasmids, we consistently observed the most pronounced augmentation of shRNA activity by Ago-2 (Figure 1E). Our combined data from cells and mice thus strongly implied that Ago-2 was the main rate-limiting factor downstream of Xpo-5 predicted in our initial animal studies.
All 4 human Argonaute proteins interact with shRNAs in cells and mice.
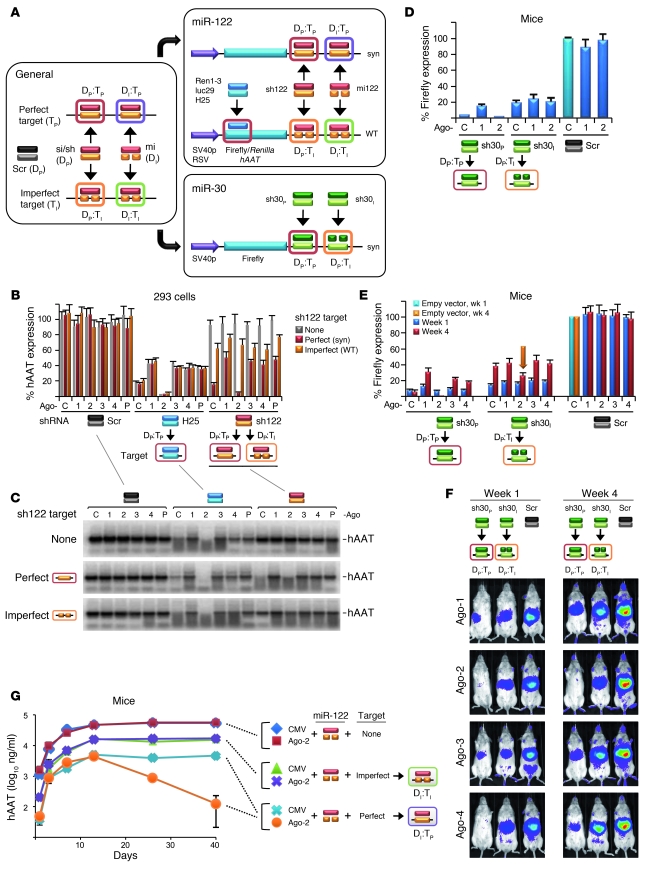
Curiously, unlike Ago-2, Ago-1 inhibited shRNA activity in vitro and in vivo (Figure 1, D and E). Ago-3 expression was also inhibitory, albeit less so, while Ago-4 had no effect. Protein and mRNA analyses showed that ectopic Ago-1 and -2 cDNA were expressed at much higher levels than Ago-3, while Ago-4 was undetectable (Figure 1F, WT). Analyses of all human Ago coding sequences showed that Ago-3/4 differed from Ago-1/2 in having lower GC content and higher numbers of RNA-destabilizing elements (Table 1). Codon adaptation index analyses (CAI) moreover implied that Ago-3 and -4 are translated suboptimally in mammals, including humans. We thus created synthetic Ago-3/4 cDNAs optimized for expression in human cells (see Methods and Supplemental Figures 2–4). Indeed, their mRNA and protein levels were drastically increased and even exceeded those of wild-type Ago-1 and -2 (Figure 1F). In vitro and in vivo, shRNA inhibition with the new Ago-3/4 clones was markedly elevated, to degrees at least matching Ago-1 (Figure 1, D and E, and Supplemental Figures 1 and 5). Two Ago-2 PIWI mutants (target cleavage–deficient) likewise competed with endogenous and exogenous wild-type Ago-2 in cells and mice, while two Ago-2 PAZ mutants (dsRNA binding–deficient) were inert (Figure 1, G and H, and Supplemental Figure 6). Together with recent in vitro data from our group and others (28–30), this implies that Ago proteins mainly compete for small RNA binding in vivo, although there may be additional effects on transcriptional and posttranslational levels (31–33). Paralleling protein expression, all inhibitory wild-type or mutant Ago variants blocked target mRNA degradation (Figure 1I).
Table 1 .
Comparison of wild-type and optimized Ago cDNA and RNA sequences
Ago-2 saturation depends on trigger-target sequence homologies.
Together, our data showed that shRNAs interact with all 4 human Ago proteins in vivo and are particularly prone to saturating Ago-2. Based on the critical role of Ago-2 in many cellular pathways (22, 23, 34, 35), we reasoned that its saturation might contribute to the observed toxicity and that relief will require RNAi strategies that avoid Ago-2 sequestration. Toward this goal, we studied the effects of relocating the shRNA target from coding regions to 3′UTRs and/or reducing its degree of homology (Figure 2A). Our rationale was that these alterations should allow the shRNA to engage the miRNA pathway, which in humans acts independently of specific Ago proteins, including Ago-2 (29, 34).
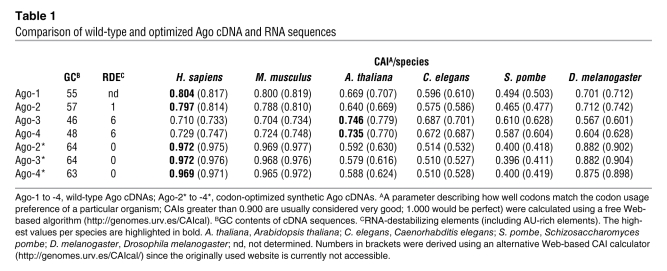
Figure 2. Influence of homology and target location on Ago-2 dependence.
(A) Combinations of perfect (P) or imperfect (I) RNAi duplexes (D) or targets (T), with corresponding constructs based on miR-122 or -30 (synthetic [syn] perfect or wild-type imperfect targets). Both sh30 duplexes are perfect but bind with perfect or imperfect homology. Standard shRNAs against ORFs served as positive and scrambled shRNAs as negative controls for Ago-2 dependence. RSV, Rous sarcoma virus; SV40p, SV40 promoter. (B) 293 cells were transfected with hAAT reporters from A and anti-ORF shRNA (H25), anti-3′UTR sh122, or a negative control. (C) Corresponding RNA expression (Northern blot). H25 and sh122 (perfect targets) were enhanced by Ago-2 and blocked by the other Agos. Target location thus does not affect Ago dependence of perfect duplexes. sh122 silenced imperfect 3′UTR targets irrespective of Ago and without mRNA degradation (C, orange), implying miRNA-like activity. (D) For in vivo validation, we transfected murine livers (n = 3–5, day 3) with miR-30–tagged luciferase and shRNA/Ago plasmids. Only the shRNA against the perfect target was activated by Ago-2 (red box). (E) When expressed long-term in mice (n = 3) via AAVs, the shRNA against the imperfect target also saturated Ago-2 (orange box/arrow). (F) Representative luciferase expression. (G) Coinfection of adult mice (n = 3) with AAV encoding hAAT fused with no target or with imperfect or perfect targets for hepatic miR-122, plus AAV expressing Ago-2 (or a CMV control). The drop in expression from the perfectly tagged hAAT construct in the presence of Ago-2 (orange line/purple box) shows that highly abundant miRNAs can also saturate Ago-2. The only combination yielding no Ago-2 saturation was imperfect miRNA duplexes against imperfect targets (purple line/green box).
Indeed, we found that shRNAs directed against imperfect 3′UTR targets retained partial activity in the absence of Ago-2, whereas conventional anti-ORF shRNAs were inactive (Supplemental Figure 7; S. Gu et al., unpublished observations). Yet we also noted that regardless of target location or homology, shRNA expression principally saturated Ago-2 in cells (Figure 2, B and C) and mice (Figure 2, D–F, and Supplemental Figure 7). For the shRNAs with imperfect 3′UTR targets, the effect was not obvious in transiently transfected cells or mice (Figure 2, B–D) but was clearly apparent after stable in vivo expression (Figure 2, E and F, compare 1- and 4-week time points). This underscores our conviction that in vitro data are not necessarily conclusive or predictive and that proper dissection of RNAi toxicities will require long-term animal studies. This complexity may also explain why some shRNAs we previously classified as safe in mice (e.g., luc29) could still saturate Ago-2 when rapidly overexpressed here in cultured cells.
Our finding that shRNAs can generally be enhanced by Ago-2 overexpression is conspicuously congruent with our prior data that mere shRNA expression is often toxic or lethal in mice, even without a target (3). A likely explanation for both observations is our recent notion that thermodynamically stable RNAi triggers (si/shRNAs) effectively and target-independently associate with Ago-2/RISC (28) (S. Gu et al., unpublished observations), thereby potentially causing saturation. Interestingly, we then also obtained evidence for in vitro and in vivo Ago-2/RISC saturation with miRNAs, i.e., imperfect duplexes, but only when they were directed against perfect targets (Figure 2G, Supplemental Figure 7). We thus concluded that any trigger-target pair comprising at least one layer of perfect homology, between the two duplex strands or to the target mRNA, can quantitatively sequester Ago-2/RISC in cells and adult liver, albeit at varying efficacies. One intriguing possible explanation is that Ago-2 saturation occurs on multiple subsequent levels, from shRNA association and ensuing activation via strand separation (together resulting in RISC loading) to target recognition and cleavage. While a more thorough investigation into and further speculation on this specific topic in RNAi biology were beyond the scope of this study, we clearly believe that unraveling such fundamental intricacies will ultimately be critical in order to further advance the clinical translation of RNAi.
In the present study, the only pairing yielding no evidence for Ago saturation, in either cells or mice, was imperfect triggers against imperfect targets within 3′UTRs (Figure 2G, Supplemental Figure 7). However, this is the least relevant option for human RNAi therapies, for numerous reasons: (a) the rules for designing imperfect triggers or for targeting imperfect sites are ill established, hampering rational clinical vector design; (b) miRNA-like silencing is usually less potent than shRNA-induced target cleavage; (c) shortening of 3′UTRs during cell proliferation may lead to a loss of potential target sites (36); (d) targeting of coding regions is more translatable for many clinically pertinent genes, whose 3′UTR sequences are often poorly characterized; and (e) proficient targeting of coding regions requires perfect binding, regardless of duplex structure, as ongoing translation impedes miRNA-RISC activity (37).
Strategies to alleviate potentially adverse Ago-2 saturation in adult mouse livers.
Together, the results indicated that the combination most prone to induce Ago-2 saturation is the same combination that is also most potent and specific, and thus most relevant for clinical RNAi therapies, i.e., perfect triggers against perfect sites (Figure 2 and Supplemental Figure 7). This prompted us to search for alternative strategies enabling effective shRNA-mediated in vivo RNAi without inherent toxicity. We specifically envisioned two options that we next wanted to explore in mice: (a) overexpression of all saturable RNAi factors, especially Xpo-5 and Ago-2; or (b) intracellular shRNA expression at minimal levels avoiding saturation altogether.
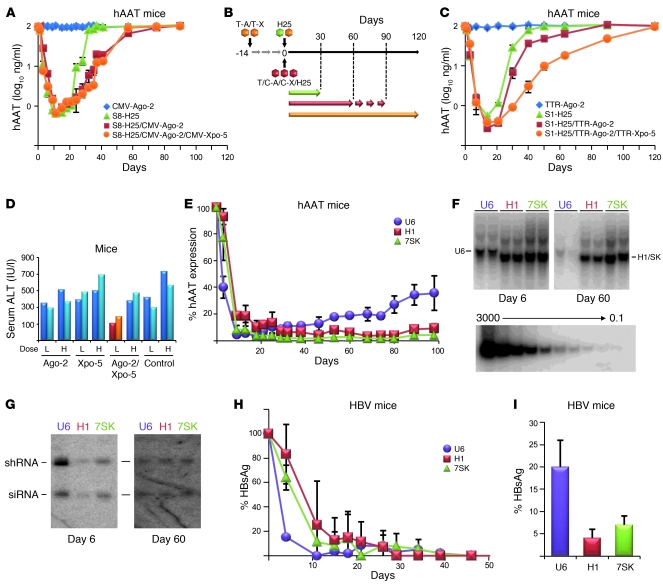
To validate our first idea with an endogenous gene, we infused hAAT-transgenic mice with a sublethal stabilized double-stranded AAV-8 (sdsAAV-8) dose encoding toxic H25 shRNA (3). As in our initial Xpo-5 studies (Figure 1B), we first coinjected two AAVs expressing shRNA or CMV-driven Ago-2 (Figure 3A). While an shRNA alone yielded typical transient RNAi (due to liver toxicity and regeneration; ref. 3), hAAT silencing persisted 2 months longer in the presence of excess Ago-2. Xpo-5 coinfusion had no effect, perhaps due to improper timing of Xpo, Ago, and shRNA expression in this complex scenario. We thus injected new mice using a staggered regime, whereby we preinfused the Ago/Xpo vectors (from AAV-8), followed 2 weeks later by anti-hAAT shRNA (from AAV-1) (Figure 3, B and C). Like CMV–Ago-2, the weaker yet preinjected TTR-driven Ago-2 vector extended in vivo RNAi persistence by 2 months and transiently increased shRNA efficacy (Figure 3C). Impressively, joint preinfusion of TTR-driven Xpo-5 and Ago-2 gave the strongest effect and extended RNAi by 3 months (Figure 3C). Xpo-5/Ago-2 upregulation also relieved hepatotoxicity, as evidenced by reduced serum ALT levels (Figure 3D).
Figure 3. Strategies to improve in vivo RNAi efficacy, toxicity, and persistence.
(A and C) Adult hAAT-transgenic mice (n = 3 to 5) were infused with the shown vectors, following the injection regime in B. Arrows in B indicate periods of measurable hAAT knockdown. C-X, CMV–Xpo-5; S1/8, serotypes 1/8; T-A, TTR–Ago-2; T-X, TTR–Xpo-5. (D) Measurements of serum ALT levels in mice (each bar represents 1 mouse) 12 days after injection of a low (L) or high (H) dose (8 × 1010 or 3 × 1011) of H25 shRNA-encoding sdsAAV-8, together with the indicated Ago-2/Xpo-5 AAV expression vectors (or none as control). (E) In vivo hAAT knockdown in hAAT-transgenic mice (n = 3–5) infused with sdsAAV-8 vectors expressing hAAT-19 shRNA (a shorter and less toxic form of hAAT-25; ref. 3) under the U6, H1, or 7SK pol III promoter. Note the much more persistent long-term hAAT silencing with the H1 and 7SK constructs, which correlated well with their prolonged persistence in the livers (Southern blots in F: each lane represents 1 mouse; bottom panel, DNA copy standard) and is most likely due to their inherently weaker shRNA expression (Northern blots in G). Also note the consistent drops in vector genomes and shRNA levels with the stronger and thus toxic U6 promoter constructs. (H and I) Validation in the HBV transgenic mouse model, showing delayed but ultimately safe and potent long-term in vivo shRNA expression and target suppression with the H1 and 7SK promoters, but not the more toxic U6 vectors. (I) Relative HBV surface antigen levels 13 months after infusion (P < 0.01, H1 versus U6; P < 0.05, 7SK versus U6).
To test the second strategy, we utilized a 19mer shRNA against hAAT that previously gave approximately 70% stable knockdown in hAAT-transgenic mice when expressed from the potent U6 RNA pol III promoter (3). Here, we repackaged this shRNA under the weaker H1 or 7SK pol III promoters and compared in vivo anti-hAAT RNAi with the original vector. As seen before (3), U6 RNAi peaked at greater than 99% at 2 weeks after infusion and then declined to approximately 65%, due to minor hepatotoxicity (Figure 3E). Conversely, despite a slower onset, H1 or 7SK RNAi was ultimately more effective and persisted at greater than 90% for the entire experiment (3.5 months), implying that the lower shRNA levels remained below the saturation threshold of the hepatic RNAi machinery and thus averted toxicity. This was paralleled by DNA copy number and shRNA expression analyses, both showing drastic gradual drops for the U6 but not the H1 or 7SK vectors (Figure 3, F and G). The latter also expressed less shRNA initially, explaining the slower RNAi kinetics and supporting that minimal shRNA doses can mitigate cytotoxicity and still yield potent RNAi.
Finally, we aimed for validation in the clinically relevant mouse model of chronic HBV infection from our initial report of in vivo RNAi toxicity (3). We thus recloned our original anti-HBV 19mer shRNA under the H1 or 7SK promoters and infused the resulting new AAV-8 vectors into HBV-transgenic mice (Figure 3, H and I). As in the experiments on hAAT-transgenic mice, HBV RNAi from the two weaker promoters was delayed compared with the more potent U6 vector, but then persisted much longer (>1 year) and up to 4-fold more efficiently (>90% or >95% for 7SK or H1, respectively), while U6-mediated knockdown gradually reverted to approximately 80%.
Discussion
Here, our aim was to further unravel the cellular determinants of in vivo RNAi efficacy, toxicity, and persistence, sparked by previous findings by us and others that high-level shRNA expression can cause cytotoxicities and fatalities in animals. As a whole, our new data verify and expand our prior conclusion that adverse in vivo shRNA effects are highly complex and at least partly due to saturation of cellular rate-limiting components (3). Still, we do not rule out additional previously indicated explanations that may also be consistent with our phenotypical observations and that require further consideration in the design of RNAi therapies, including innate immune responses and off-targeting (2, 38). Also, since our model was derived in the liver, future experiments must be performed to comprehensively determine the concentrations of key RNAi components, including the dsRNA trigger and target mRNAs, in further clinically relevant cell and tissue types.
Importantly, we now believe that in vivo shRNA overexpression potentially saturates not only Xpo-5, but also all 4 human Ago proteins (Figure 4). Among these, Ago-2 sequestration is probably most toxic due to its key role in numerous cellular pathways, from miRNA biogenesis, nuclear RNAi, embryogenesis, and oogenesis to hematopoiesis (33–35, 39–43). Strong additional support for this conclusion is provided by prior findings that expression of most miRNAs was reduced by more than 80% in Ago-2–knockout or –knockdown cells, causing substantial global dysregulation of thousands of genes (22, 32, 41). Likewise, in mouse oocytes, Ago-2 loss resulted in decreases in endogenous siRNAs (esiRNAs) and elevated expression of retrotransposons and other esiRNA-regulated transcripts (44). Thus, unsurprisingly, homozygous Ago-2–knockout mice display severe developmental abnormalities and are embryonic lethal (22, 35, 45). Besides, Ago-2 saturation may promote shRNA entry into non-Slicer RISCs and thus enforce off-targeting, in turn reducing the specificity of therapeutic RNAi and further increasing risks of adverse side effects (28, 30).
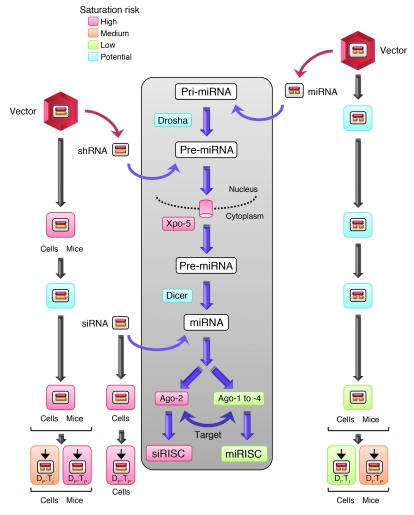
Figure 4. Comparison of the 3 major classes of therapeutic RNAi triggers (vector-encoded shRNA or miRNA, or delivered siRNA) and their potential risk for saturation (indicated by different colors; see key) of the cellular RNAi machinery (gray box).
Specific steps for which we have provided experimental evidence in cells and/or mice in the present study are indicated by the respective symbols. Blue boxes denote that saturation is theoretically possible at this step, but experimental proof is lacking.
Co-sequestration of the other 3 Ago proteins may even further potentiate shRNA cytotoxicity, a hypothesis supported by findings that in vitro knockout of all 4 Ago proteins triggers apoptosis (29). This would also explain the eventual RNAi loss in our mice despite Xpo-5/Ago-2 codelivery (Figure 3, A and C), as complete rescue from toxicity may require simultaneous overexpression of all 4 Ago proteins. Mechanistically, it is likely that Ago-1 to -4 cosaturation can further perturb miRNA function and/or cause global changes in the transcriptome. The latter was indeed observed in cells depleted of individual Ago proteins (32), and altered miRNA expression and activity were also among our most striking phenotypes in shRNA-expressing cells and mice (3, 4).
Interestingly, one earlier study found that selective Ago-1 or -3 depletion impaired only up to 50% of mRNAs compared with Ago-2, and the effect of Ago-4 depletion was even smaller (32). This adds to the ongoing controversy regarding the biological role of the 4 human Ago proteins, including the question as to whether at least Ago-1, -3, and -4 are redundant. Arguing for this might be our notion that wild-type human Ago-3 and -4 are expressed suboptimally. Together with their location on the same chromosome, this may suggest that they are Ago-1 pseudogenes. On the other hand, all 4 human Ago variants are expressed in a highly tissue- and developmental-specific manner (35, 46–48). Moreover, some groups reported association of Ago-2/3 with specific miRNAs (49), or of Ago-1/2 with unique mRNAs or proteins (31, 50, 51). We and others also found distinct roles of Ago proteins in human viral infection (52, 53). Furthermore, Ago-1/2 may preferably load perfect siRNAs/shRNAs (28, 29), while Ago-3/4 may more potently engage miRNAs (30). Finally, Ago-1 and -2 are involved in nuclear RNAi, while the role of Ago-3/4 is unclear (33, 42, 43). These data may indicate that all 4 human Ago proteins are indeed crucial and thus further support our belief that their cosaturation and the associated perturbation of cellular gene silencing pathways are detrimental and must be avoided in clinical RNAi therapies.
Particularly notable in this context are two recent studies that reported global loss of miRNA expression and function in livers of mice, caused by conditional Dicer knockouts (54, 55). Curiously, the resulting phenotypes were remarkably mild in the early postnatal phase, implying that loss or dysregulation of hepatic miRNA expression can be tolerated at least for a limited period; still, affected mice also eventually exhibited progressive hepatocyte death and severe liver damage. Moreover, expression of more than 1,600 miRNA-regulated genes was significantly altered, including those controlling lipid and glucose metabolism, and particularly fetal stage–specific and cell cycle–promoting genes (likely explaining why mice that escaped toxicity later developed hepatocellular carcinoma). This implies that hepatocytes lacking mature miRNAs function normally for a short period of time, before the loss of miRNA-mediated gene regulation causes cell death and tissue failure, strongly supporting our own previous (3) and present conclusions.
Of note, the fact that toxicity manifested later in the Dicer-knockout mice as compared with our shRNA overexpression studies (several weeks versus a few days, respectively) is fully consistent with an expected much more gradual relief of miRNA-dependent gene regulation after conditional Dicer ablation, which will mainly affect miRNA processing, a consequence that may initially go unnoticed due to the long half-life of many miRNAs. In contrast, shRNA-mediated overloading of the Ago/RISC complex will instantly block miRNA function and in the case of Ago-2, moreover, impact miRNA biogenesis and maturation (35, 39). Thus, although the phenotypes of Dicer or Ago loss are typically very similar (also in other cells and tissues, e.g., oocytes; ref. 41), the underlying molecular mechanisms may differ. This may also include specific classes of small regulatory RNAs that require Ago proteins yet are generated Dicer-independently, such as those derived from tRNAs that we discovered recently (28). This could help to explain why only up to one-third of genes were similarly dysregulated in Dicer- or Ago-2–knockdown cells (32) or in knockout oocytes (41). Another particularly striking example is spermatogenesis, which was retarded in Dicer- yet not in Ago-2–knockout mice (56). Last but not least, our own findings that Dicer, unlike Xpo and the 4 Ago proteins, does not appear to rate-limit shRNA efficacy in cells or in mice (Figure 1, D and E) add further evidence for marked principal differences between the various key RNAi proteins. Notably, our data on Dicer are fully congruent with previous reports that it restricts the potency of neither transfected pri-miRNA constructs in cultured cells (21) nor of siRNAs (13). Still, Dicer is of course essentially required for principal processing and activity of longer RNAi duplexes (shRNAs and miRNAs), as confirmed in Dicer-knockout studies (13, 57, 58).
Importantly, our saturation model also suggests that caution must be exercised in using siRNAs and vectors based on miRNAs (Figure 4), since we found that both RNAi triggers effectively compromise Ago-2 when delivered or stably expressed at high doses, as may be required for certain clinical applications. Evidence (Figures 2 and 5, and Supplemental Figure 7) was that (a) transient or stable Ago-2 overexpression increased siRNA (and shRNA) activity in cells and relieved si-siRNA or si-shRNA competition; (b) miRNAs were also enhanced by Ago-2 (at least when directed against a perfect target); and (c) siRNAs and shRNAs were inactive in Ago-2–knockout cells and rescued only by Ago-2, but not any other Ago protein. Together, these data not only confirm, extend, and help to explain prior notions of, for example, si-si/shRNA competition in cells (12–14), but also validate that Ago-2 is vital for and saturable by all classes of RNAi triggers. Hence, as with shRNAs, care must likewise be taken especially with the latest proficient siRNA formulations (59, 60) to avoid in vivo Ago-2 saturation and associated adverse effects in clinical settings.
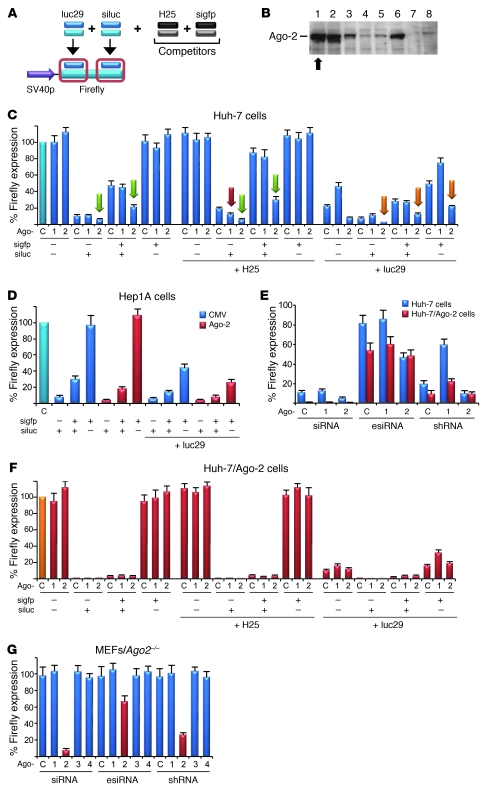
Figure 5. Ago-2 is critical for basal and competitive activity of perfect RNAi duplexes.
(A) Scheme depicting shRNA (luc29) or siRNA (siluc) against Firefly luciferase, plus competitors (H25 shRNA and sigfp siRNA). (B) New Huh-7 cell lines stably overexpressing human Ago-2 (8 representative examples; all studies were performed in clone 1 [arrow] and validated in clone 2). Arrow indicates the best-expressing cell line, in which all studies were performed. (C) Huh-7 cells were cotransfected with: (a) Firefly (target) and Renilla (normalization) luciferase, (b) shRNA expression plasmid (encoding H25 or luc29 shRNA, or only U6 promoter), (c) plasmids encoding Ago-1 or -2 (or CMV promoter as control), (d) anti-gfp competitor siRNA (sigfp), and (e) anti-luciferase siRNA (siluc). Ago-2 overexpression enhanced basal luc siRNA activity and diminished competition with gfp siRNA or H25 shRNA (green arrows). Ago-2 likewise enhanced luciferase knockdown when cotransfected with luc29 shRNA (plus or minus luc siRNA; orange arrows). Ago-1 overexpression had no significant effect on the siRNA, but also reduced si/shRNA competition (red arrow), perhaps by sequestering the shRNA and thus freeing Ago-2/RISC for the siRNA. P < 0.05, all Ago-2 bars labeled with green/orange arrows versus controls. (D) Confirmation of the Ago-2 effects in a different cell line. (E) Validation in stable Ago-2 Huh-7 cells (see B), transfected with Firefly/Renilla, various RNAi triggers (siRNA, esiRNA, or shRNA, as indicated) against Firefly, and Ago expression plasmids (or a control). Basal siRNA, esiRNA (enzymatically created siRNA), and shRNA activities were all elevated (red) compared with parental Huh-7 (blue), confirming that Ago-2 restricts all classes of perfect RNAi duplexes. (F) Repeat of the transfections from C in stable Ago-2 cells. Competition between 2 siRNAs, or between luc siRNA and H25 shRNA, was greatly diminished compared with that in the parental cells (C). (G) Transfection of all perfect RNAi duplexes into Ago-2–deficient MEFs confirmed their strict Ago-2 dependence.
Concerning miRNAs as vector templates, intriguing data from the Rossi and Davidson laboratories imply that their in vitro and in vivo safety may in fact be relatively high as compared with that of shRNAs (5, 6, 12, 20). While counterintuitive in view of their potential for Ago-2 sequestration noted here, the expressed steady-state levels of active RNA strands are reportedly far below those achieved with conventional shRNAs (6, 20). In addition, as miRNA vectors are being shunted through the cellular miRNA pathway, it was speculated that this may slow their loading into RISC and thereby also help to reduce their competition with other small RNAs or RNAi factors (12). Finally, as mentioned, it is possible that Ago-2 competition occurs on multiple levels and that miRNA vectors specifically overload the final step (binding to, and cleavage of, a perfect target). All explanations (which are not exclusive) thus further support our saturation model and underscore the necessity of avoiding obstruction of RISC and other RNAi factors at any level in therapeutic settings.
In this regard, our results and models allow us to envision multiple new avenues to concurrently improve the efficacy, safety, and persistence of future RNAi gene therapies in humans, especially those involving shRNA vectors (Figure 6). For their routine applications, stably coincreasing all limiting factors is impractical, as it would require staggered high-dose multiple vector delivery to patients and may also yield complex results, involving the opposing roles of Ago proteins for shRNAs and potentially for miRNAs. Still, deliberate upregulation of Xpo-5 and Ago-2 (e.g., using our AAV vectors) may benefit short-term RNAi applications, such as tumor therapies, where transient rapid and potent knockdown are desirable and where limiting toxicity is typically not the highest priority. Of note, we have found no evidence thus far for general adverse effects from Ago/Xpo expression in mice, and our stable Ago-2 cell lines have not displayed abnormalities after more than 1 year in culture (data not shown).
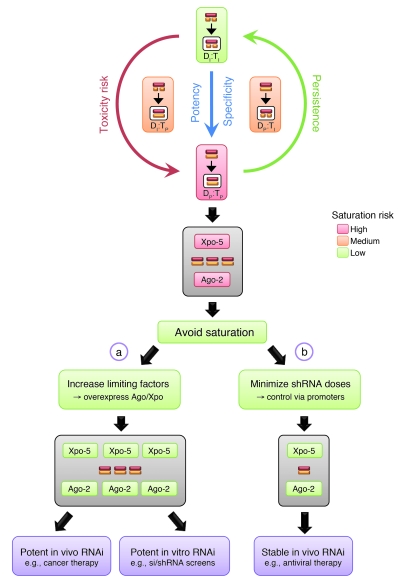
Figure 6. Options to improve human RNAi gene therapies and other RNAi applications.
Red box: Preferred setting (due to highest specificity and potency) for therapeutic RNAi, i.e., perfect RNAi duplexes against perfect targets. As shown, this is also the trigger-target combination that is most prone to saturating Ago-2 and thus causing cytotoxicity. As further implied by our data, this risk may be lowered by reducing the homology either between the two dsRNA strands or to the target. Hence, imperfect triggers against imperfect targets were least dependent on Ago-2. Yet they may also be least useful for human RNAi therapies. We thus favor two alternative strategies that both aim at avoiding saturation and thus alleviating shRNA toxicity and for which we have provided proof-of-concept in this article. One (a) is to transiently overexpress (e.g., using AAV vectors) limiting cellular key RNAi components, especially Xpo-5 and Ago-2. This should for instance benefit cancer-directed RNAi gene therapies, where instant and maximally potent target knockdown might be critical. It should likewise improve functional in vitro RNAi screens, where short-term and robust siRNA or shRNA activity is typically desired. An alternative (but not exclusive) second strategy (b) is to utilize minimal yet effective trigger doses in humans, by employing weak and/or tissue-specific promoters for shRNA expression. Based on our mouse data, we predict that this can also mitigate saturation-induced toxicity and prevent vector loss and thus enhance both efficacy and persistence of RNAi. Accordingly, this specific strategy should be particularly useful for long-term RNAi gene therapies against persisting exogenous targets, such as HIV or hepatitis viruses.
In contrast, persisting human viral pathogens as another clinically relevant target may instead require long-term strategies aimed at stabilizing RNAi. Avoiding global saturation is then particularly critical to avert toxicity and to preserve combinatorial RNAi therapies designed to thwart viral escape (12, 61). One strategy outlined here is the use of weak pol III promoters for shRNA expression, which yielded more than 1 year of potent HBV silencing in adult mice in this study. Beyond viral infections, we consider such promoters highly useful for any RNAi therapies requiring stable and safe in vivo gene silencing. Further assets are their small sizes (~95/239 for H1/7SK versus ~500 bp for U6), making them ideal for combined gene silencing/addition strategies employing viral gene therapy vectors (61). An alternative strategy already indicated above is expression within the context of a miRNA vector, which will likewise help to avoid RNAi saturation and thus attenuate in vivo toxicities, as recently demonstrated in mouse brain (5). Yet in contrast to straightforward shRNA expression from pol III promoters, many aspects of artificial miRNA vectors, including designing robust vectors and elucidating their intracellular processing, remain uncertain, and substantial further optimization is warranted to improve this particular strategy. Also, the limited silencing efficacy that characterizes the current generation of these vectors inherently precludes their use in clinical RNAi applications requiring maximum potency. Moreover, an additional looming concern is potential in vivo competition for Drosha or other nuclear factors that are specific to miRNA processing.
Finally, we and others recently documented that in vivo RNAi safety can be further enhanced by judicious shRNA selection, use of minimal effective vector doses, or shRNA expression from tissue-specific pol II promoters (3, 8, 62). It will be exciting to juxtapose these avenues with other promising advances, such as molecularly evolved viral vectors with inherent assets for safe and long-term therapeutic RNAi in humans (63). Such combinations of experimental strategies will then provide an additional wealth of options that researchers and clinicians can empirically test for their given RNAi application before selecting the most suitable system based on careful consideration of the balance of efficacy, specificity, delivery, persistence, and, last but not least, toxicity.
Methods
Plasmids and vectors.
Constructs expressing human Drosha, Dicer, or exportin-5 from a CMV promoter were gifts from Ian G. Macara (University of Virginia, Charlottesville, Virginia, USA) and/or were reported before (3). Plasmids expressing human wild-type Ago-1 to -4 or TRBP from a CMV promoter were purchased from Addgene (Ago-1: plasmid number 10820, Ago-2: 10822, Ago-3: 10823, Ago-4: 10824, TRBP: 15666). A plasmid expressing the gfp gene from a CMV promoter (Addgene, plasmid number 10825) served as negative control for all above expression constructs.
To generate AAV vector plasmids utilizing various promoters to drive expression of human Xpo-5 cDNA, the latter was first PCR amplified using as primers EXPFOR 5′-GAGTCCAATTGACGCGTCCTGAGTTCGTCTAGAGGATCTATGGCGATGGATC-3′ and EXPREV 5′-CCTGAGTCGACGATCTTCAGGGTTCAAAGATGGTG-3′. These primers contained sites for the restriction enzymes MfeI (bold), MluI (italics), or XbaI (part of the Xpo-5 cDNA; bold/italics) (all in EXPFOR primer), or SalI (underlined, in EXPREV), to allow subsequent cloning into appropriately digested AAV vector plasmid pTRUF3 (64) (containing two identical AAV-2 inverted terminal repeats and serving to produce conventional single-stranded DNA AAV vectors; see below). To clone a CMV–Xpo-5 plasmid, the PCR product was directly ligated into pTRUF3 (already containing a CMV promoter and a bovine growth hormone polyadenylation signal) digested with XbaI and SalI. To bring the Xpo-5 cDNA under the SV40 promoter, the latter was PCR amplified from plasmid pGL3 (Promega), using as primers SVFOR 5′-CATTTCTCTATCGATAGGTACCGAGCTCTTAC-3′ and SVREV 5′-GACCGTCTAGAGCTTTACCAACAGTACCGGAATGCCAAG-3′, and cloned as KpnI (bold)/XbaI (bold/italics) fragment into the KpnI/XbaI-cut CMV–Xpo-5 vector plasmid (resulting in replacement of the CMV with the SV40 promoter). For the two other promoters shown in Figure 1A, the original Xpo-5 PCR product was cut with MfeI and SalI and cloned into appropriately digested pTRUF3, eliminating the genuine CMV promoter and introducing an additional MluI site upstream of the Xpo-5 cDNA. The apoE/hAAT promoter/enhancer (gift from Jeff Giering, Stanford University, Stanford, California, USA) was then PCR amplified using as primers APOFOR 5′-GAGTCACGCGTGTTTCTGGGCTCACCCTGCCCCCTTC-3′ and APOREV 5′-CCTGGTCTAGAACTGTCCCAGGTCAGTGGTGGTGC-3′, and cloned as a MluI (bold)/XbaI (bold/italics) fragment into the MluI/XbaI-cut Xpo-5 vector plasmid. Similarly, the TTR promoter (gift from Michael Hebert, Stanford University, Stanford, California, USA) was PCR amplified using as primers TTRFOR 5′-GAGTCCACGCGTGGATCTGTCAATTCACGCGAG-3′ and TTRREV 5′-CCTGGAACTAGTCAGCTGGGCTTCTCCTGGTGAAG-3′, and cloned as MluI (bold)/SpeI (bold/italics; compatible with XbaI) fragment into the MluI/XbaI-cut Xpo-5 vector plasmid.
To create AAV vector plasmids utilizing a CMV promoter to express wild-type or codon-optimized (see below) human Ago cDNAs, the latter were PCR amplified and then cloned as XbaI/XhoI (Ago-1, XhoI is compatible with SalI) or XbaI/SalI (Ago-2 to -4, and gfp control) fragments into XbaI/SalI-digested pTRUF3. Primers were AGOF1234G 5′-GACTCCTCTAGACAAGCTTGGTACCGAGCTCGGATCG-3′ (forward primer for all PCRs; XbaI in bold), AGOR1 5′-CTGGACCTCGAGCTAGATGCATGCTCGACCTGCAGTTG-3′ (reverse primer for Ago-1; XhoI in bold/italics), AGOR234 5′-CTGGACGTCGACCTAGATGCATGCTCGACCTGCAGTTG-3′ (reverse primer for Ago-2 to -4; SalI in bold/italics), or GFPR 5′-CTGGACGTCGACCTCAGAAGAACTCGTCAAGAAGGCGATAG-3′ (reverse primer for gfp control; SalI in bold/italics).
To express Ago cDNA under the TTR or apoE/hAAT promoters (in an AAV vector context), a strategy similar to that described for Xpo5 above was used. Briefly, based on the construct containing Xpo-5 cDNA flanked by MluI and XbaI sites (5′ of Xpo-5), the Xpo-5 cDNA was replaced by the different Ago cDNAs (see above, amplified with XbaI/XhoI- or SalI-containing primers). The TTR or apoE/hAAT promoters were next inserted as an MluI/SpeI or XbaI PCR fragment, as also described above.
Mutations in the Ago-2 PIWI domain were generated by D597A (aspartate to alanine at nucleotide position 597) or D669A site-directed mutagenesis (Quikchange II Kit, Stratagene), based on previous reports (22). Corresponding mutations in the codon-optimized (see below) Ago-3 cDNA were generated similarly, using as forward primers A3M1 5′-CTGTGATCTTCCTGGGCGCCGCCGTGACCCACCCC-3′ or A3M2 5′-GGATCATCTTCTACCGGGCCGGCGTGAGCGAGGGCC-3′ (point mutations are underlined). Ago-2 PAZ mutants containing 9 or 10 dispersed point mutations throughout the PAZ domain were reported before and obtained as gifts from Greg Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA) and Roy Parker (University of Arizona, Tucson, Arizona, USA) (65, 66).
The majority of plasmids used in the various knockdown experiments and expressing either targets or shRNAs (under a U6 promoter) were previously generated in and reported by our laboratory (3). This also includes the control AAV vector expressing human factor IX from a liver-specific promoter (67). New constructs comprised a series of AAV/U6-shRNA expression plasmids encoding shRNAs against Renilla luciferase (present in the psiCheck2 plasmid [Promega]), as well as a construct expressing an shRNA encoding an artificial perfect miR-122 duplex. All these plasmids were assembled in the context of an sdsAAV vector backbone, basically as reported before (see also below, section on H1/7SK promoters) (3). Briefly, sequences of the sense strands were: REN1 5′-GCAACGCAAACGCATGATCAC-3′, REN2 5′-GCTGGACTCCTTCATCAAC-3′, REN3 5′-GGCCTTTCACTACTCCTACGA-3′, and miR-122 5′-TGGAGTGTGACAATGGTGTTTGT-3′. We also created a plasmid expressing wild-type human miR-122 (160-bp fragment), by PCR-amplifying the latter from genomic DNA from human 293 cells, using as primers HCRFOR 5′-GAGTCAAGCTTTGGAGGTGAAGTTAACACCTTCGTG-3′ and HCRREV 5′-CTCGAGGGCCCAAGCAAACGATGCCAAGACATTTATCG-3′. This fragment was digested with HindIII (bold) and ApaI (bold/underlined) and cloned under the control of a CMV promoter in plasmid pcDNA5/FT/TO/CAT (Invitrogen).
In addition, we created reporter constructs (in an AAV vector backbone) carrying perfect or imperfect binding sites for miR-122 in the 3′UTRs of the hAAT or luciferase (Firefly or Renilla) genes. The respective expression cassettes were reported before by our laboratory (RSV-hAAT; ref. 68) or isolated from commercial plasmids (pGL3 and psiCheck2, both Promega). Sequence 5′-ACAAACACCATTGTCACACTCCA-3′ was used as perfect and 5′-GCCAGCACCATTTCACACACACTCCT-3′ or 5′-ACTAAGGCTGCTCCATCAACACTCCA-3′ as imperfect sites (the miR-122 seed region is shown in bold/underlined).
Reporters and shRNA expression vectors based on miR-30 (including scrambled control) were reported before by our laboratory (37). To express these sequences from an AAV vector, the sh30 (here called sh30P) and mi30 (here called sh30I) shRNAs (plus SCR control) were recloned into our previously reported AAV/U6 vector (3), using the same cloning strategy as for all other shRNAs. The entire SV40 promoter-driven, miR-30 site-tagged luciferase expression cassette was cloned as a BglII/BamHI fragment into our double-stranded AAV vector backbone linearized with BglII (compatible with BamHI).
To generate “user-friendly,” reporter-tagged double-stranded AAV vectors for the expression of shRNAs under the H1 or 7SK promoters, 95-bp (H1) or 239-bp (7SK) promoter fragments were PCR isolated from commercial plasmids (pSilencer 3.1-H1 puro for H1 [Ambion] and psiRNA-h7SK for 7SK [Invivogen]) and cloned as AscI/NotI fragments into our original AAV/U6 vector plasmid (3), to replace the U6 promoter. Subsequently, shRNAs were cloned as annealed oligonucleotides with overhangs CACC (forward) or AAAA (reverse) into BbsI-cut vector plasmids (containing tandem BbsI sites leaving compatible overhangs for the annealed oligonucleotides). The sequences of the shRNAs and loops were reported before (3).
Ago cDNA codon optimization.
Optimization of the cDNAs for human Ago-2, -3, and -4 was performed by GeneArt, using proprietary algorithms and methods. Ago-2 was included as control, while the actual focus in this work was on improving the expression of Ago-3 and -4 in human cells. Optimized cDNA features comprised the codon usage (increased), GC content (increased), and the number of RNA-destabilizing elements (decreased) (see Table 1 and Supplemental Figures 2–4 for details). All optimized cDNAs were verified by sequencing and recloned into the original plasmids (Addgene) as NotI/EcoRI fragments (sites for these two enzymes were avoided during the optimization process).
AAV vector production.
Viral particles were generated via a standard triple-transfection method, purified via two rounds of cesium chloride density gradient centrifugation, and titered by dot blot, all as described previously (3, 67). AAV helper plasmids expressing capsid genes from serotypes 1 or 8 were gifts from James Wilson (University of Pennsylvania, Philadelphia, Pennsylvania, USA). Typical particle yields from approximately 2 × 109 transfected 293 cells were at least 5 × 1012 vector genomes per milliliter (in a total volume of ~4 ml).
Cell culture studies.
All cell lines in this study were maintained in DMEM (Invitrogen) containing 10% fetal calf serum, 2 mM l-glutamine, and 50 IU/ml each of penicillin and streptomycin at 37°C in 5% CO2. Human 293 hcr/mut cells conditionally expressing wild-type or mutant miR-122, respectively, were a gift from Jinhong Chang (Drexel University College of Medicine, Doylestown, Pennsylvania, USA). They were maintained in tetracycline-free medium and induced with 2 μg/ml doxycycline. Mouse embryonic Ago-2–knockout fibroblasts (MEFs) were a gift from Greg Hannon (Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA). To generate stably Ago-2–expressing Huh-7 cells, they were initially transfected (10-cm dish, Lipofectamine 2000 [Invitrogen]) with 15 μg of the wild-type Ago-2 expression plasmid, containing a neomycin resistance gene. The cells were subsequently passaged and then put and maintained under selection pressure (250 μg/ml G418) starting at day 3.
The siRNAs against Firefly luciferase or gfp were purchased in a ready-to-use format (20 μM stocks) from Invitrogen (Stealth RNAi reporter control duplexes) and transfected at 200-nM concentrations. Enzymatically prepared siRNAs (esiRNAs) were generated and purified using the siRNA Silencer Cocktail Kit (RNase III) (Ambion) and following the manufacturer’s instructions. Firefly luciferase cDNA was used as template, and primers were FIRFOR 5′-TAATACGACTCACTATAGGGCTCCCGCTGAATTGGAATCC-3′ and FIRREV 5′-TAATACGACTCACTATAGGGGGATCTCTCTGATTTTTCTTGCGTCG-3′. Final esiRNA concentrations were in a range of 10 to 26 μM, and 100 nM per well was used for transfections in 24-well plates.
All other transfections were also carried out in 24-well plates (with at least 3 independent replicates per sample), using Lipofectamine 2000 (Invitrogen) as transfection reagent and following the manufacturer’s recommendations for this format, including a total amount of DNA per well of 800 ng. This typically comprised 100 ng si/esi/shRNA target plasmid, 200 ng shRNA plasmid, and 500 ng Ago plasmid. In cases where Firefly luciferase (pGL3, Promega) was used as target, 10 ng of a separate Renilla luciferase plasmid was cotransfected for normalization. In control samples, empty expression plasmids were added to maintain identical total amounts (e.g., CMV-gfp for the CMV-Ago constructs, or the empty U6 plasmid for the U6 shRNA constructs). In competition experiments where several Ago plasmids and/or si/shRNAs were cotransfected in various combinations, total amounts were split up accordingly. Expression levels of hAAT in cell supernatants were measured via ELISA as described (3), and luciferase coexpression (target versus normalization control) was quantitated using the Dual Luciferase Reporter Assay System (Promega), following the manufacturer’s instructions.
Mouse studies.
Wild-type female FVB/NJ mice (6 to 8 weeks old) were purchased from The Jackson Laboratory. Mice of the FVB strain transgenic for the hAAT gene (under a hepatocyte-specific promoter), or carrying an integrated copy of the HBV genome (STC lineage), were previously reported (3). Animals were selected such that all groups had comparable average initial hAAT or HBsAg levels.
Hydrodynamic plasmid injections (in 1.8 ml 0.9% NaCl) and AAV vector infusions (via tail vein injection, in a total volume of 300 μl of 1× PBS) were performed as described before (3). Stabilized double-stranded AAV-8 vectors expressing shRNAs were typically infused at doses of 8 × 1010 to 3 × 1011 particles per mouse. The lowest dose (8 × 1010) was used particularly for the anti-hAAT 25mer (e.g., Figure 1, B and C), which we previously found to be severely toxic or even lethal at higher doses (3). To express the same shRNA from an AAV-1 capsid, 2 × 1011 particles were infused, to account for this serotype’s lower efficiency in liver. To compare shRNA expression and efficacy from the 3 different pol III promoters (Figure 3, E and F), intermediate (1 × 1011, hAAT 19mer) or higher (3 × 1011, HBV 19mer) doses were used. The luciferase expression vector in Figure 2F was infused at 2 × 1011 particles per mouse, together with 8 × 1010 particles of the sh30P/I expression vectors. Single-stranded AAV-8 vectors expressing exportin-5 or the different Ago proteins (or control human factor IX, also from AAV-1; Figure 1C) were infused at 5 × 1012 to 7 × 1012 particles per mouse, to ensure complete liver transduction.
Blood was collected at the indicated time points via retro-orbital bleeding, and plasma hAAT as well as serum alanine aminotransferase (ALT) or HBV surface antigen levels were determined via specific ELISAs as previously described (3). Luciferase expression was monitored in live animals also as reported (3).
All procedures were approved by the Animal Care Committee of Stanford University.
Western, Southern, and Northern blotting.
Human or mouse exportin-5 was detected by Western blotting using a polyclonal rabbit anti-serum (gift from Dirk Görlich, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). Wild-type or codon-optimized Ago proteins were visualized using the anti-FLAG M2 monoclonal antibody (Sigma-Aldrich) at a 1:1,000 dilution. Southern blots to detect and quantify AAV vector copy numbers in treated mice were performed as described (3). Ago, hAAT, and gfp transcripts were detected by standard Northern blotting, using total RNA isolated with TRIzol (Invitrogen) and specific 32P-labeled probes. Small RNA Northern blot analyses were conducted essentially as reported previously (3).
Statistics.
Where appropriate, statistical significance for comparisons between two or multiple groups was calculated using 2-tailed Student’s t test or ANOVA, respectively. A P value of less than 0.05 was considered significant. All experiments were typically performed at least in triplicate, and all data are presented as mean ± SD.
Supplementary Material
Acknowledgments
We are grateful to Oliver Wicht for initial help with the generation of esiRNAs and to Hui Xu for help with hydrodynamic mouse injections. This work was supported by NIH grant AI 071068. K. Börner was supported by ViroQuant. D. Grimm and N. Schürmann were supported by the Cluster of Excellence CellNetworks and the Chica and Heinz Schaller Foundation.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(9):3106–3119. doi:10.1172/JCI43565.
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimm D, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 4.Beer S, et al. Low-level shRNA cytotoxicity can contribute to MYC-induced hepatocellular carcinoma in adult mice. Mol Ther. 2010;18(1):161–170. doi: 10.1038/mt.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride JL, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci U S A. 2008;105(15):5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther. 2009;17(1):169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehlert EM, Eggers R, Niclou SP, Verhaagen J. Cellular toxicity following application of adeno-associated viral vector-mediated RNA interference in the nervous system. BMC Neurosci. 2010;11:20. doi: 10.1186/1471-2202-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giering JC, Grimm D, Storm TA, Kay MA. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol Ther. 2008;16(9):1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 9.An DS, et al. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14(4):494–504. doi: 10.1016/j.ymthe.2006.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasior SL, Palmisano M, Deininger PL. Alu-linked hairpins efficiently mediate RNA interference with less toxicity than do H1-expressed short hairpin RNAs. Anal Biochem. 2006;349(1):41–48. doi: 10.1016/j.ab.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Cao W, Hunter R, Strnatka D, McQueen CA, Erickson RP. DNA constructs designed to produce short hairpin, interfering RNAs in transgenic mice sometimes show early lethality and an interferon response. J Appl Genet. 2005;46(2):217–225. [PubMed] [Google Scholar]
- 12.Castanotto D, et al. Combinatorial delivery of small interfering RNAs reduces RNAi efficacy by selective incorporation into RISC. Nucleic Acids Res. 2007;35(15):5154–5164. doi: 10.1093/nar/gkm543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickers TA, Lima WF, Nichols JG, Crooke ST. Reduced levels of Ago2 expression result in increased siRNA competition in mammalian cells. Nucleic Acids Res. 2007;35(19):6598–6610. doi: 10.1093/nar/gkm663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koller E, et al. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34(16):4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McManus MT, et al. Small interfering RNA-mediated gene silencing in T lymphocytes. J Immunol. 2002;169(10):5754–5760. doi: 10.4049/jimmunol.169.10.5754. [DOI] [PubMed] [Google Scholar]
- 16.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27(6):549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436(7051):740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr Biol. 2003;13(1):41–46. doi: 10.1016/S0960-9822(02)01394-5. [DOI] [PubMed] [Google Scholar]
- 19.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30(8):1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14(9):1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diederichs S, Jung S, Rothenberg SM, Smolen GA, Mlody BG, Haber DA. Coexpression of Argonaute-2 enhances RNA interference toward perfect match binding sites. Proc Natl Acad Sci U S A. 2008;105(27):9284–9289. doi: 10.1073/pnas.0800803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305(5689):1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 23.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR. Overexpression of exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA. 2005;11(2):220–226. doi: 10.1261/rna.7233305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci U S A. 2009;106(51):21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohrt T, Merkle D, Birkenfeld K, Echeverri CJ, Schwille P. In situ fluorescence analysis demonstrates active siRNA exclusion from the nucleus by Exportin 5. Nucleic Acids Res. 2006;34(5):1369–1380. doi: 10.1093/nar/gkl001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16(4):673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23(3):304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic ago proteins for on-target RNA interference. Curr Biol. 2008;18(17):1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hock J, et al. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8(11):1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitter D, et al. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34(17):4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4(11):e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26(5):611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465(7298):584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat Struct Mol Biol. 2009;16(2):144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8(3):173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 39.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131(6):1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 40.O’Carroll D, et al. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21(16):1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaneda M, Tang F, O’Carroll D, Lao K, Surani MA. Essential role for Argonaute2 protein in mouse oogenesis. Epigenetics Chromatin. 2009;2(1):9. doi: 10.1186/1756-8935-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Janowski BA, et al. Involvement of AGO1 and AGO2 in mammalian transcriptional silencing. Nat Struct Mol Biol. 2006;13(9):787–792. doi: 10.1038/nsmb1140. [DOI] [PubMed] [Google Scholar]
- 43.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci U S A. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453(7194):539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 45.Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89(6):687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez-Gonzalez E, Lopez-Casas PP, del Mazo J. The expression patterns of genes involved in the RNAi pathways are tissue-dependent and differ in the germ and somatic cells of mouse testis. Biochim Biophys Acta. 2008;1779(5):306–311. doi: 10.1016/j.bbagrm.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82(3):323–330. doi: 10.1016/S0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 48.Lu J, Qian J, Chen F, Tang X, Li C, Cardoso WV. Differential expression of components of the microRNA machinery during mouse organogenesis. Biochem Biophys Res Commun. 2005;334(2):319–323. doi: 10.1016/j.bbrc.2005.05.206. [DOI] [PubMed] [Google Scholar]
- 49.Azuma-Mukai A, et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A. 2008;105(23):7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beitzinger M, Peters L, Zhu JY, Kremmer E, Meister G. Identification of human microRNA targets from isolated argonaute protein complexes. RNA Biol. 2007;4(2):76–84. doi: 10.4161/rna.4.2.4640. [DOI] [PubMed] [Google Scholar]
- 51.Landthaler M, et al. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14(12):2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haussecker D, Cao D, Huang Y, Parameswaran P, Fire AZ, Kay MA. Capped small RNAs and MOV10 in human hepatitis delta virus replication. Nat Struct Mol Biol. 2008;15(7):714–721. doi: 10.1038/nsmb.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Randall G, et al. Cellular cofactors affecting hepatitis C virus infection and replication. Proc Natl Acad Sci U S A. 2007;104(31):12884–12889. doi: 10.1073/pnas.0704894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009;49(2):618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sekine S, et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136(7):2304–2315. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayashi K, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS One. 2008;3(3):e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis ME, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8(2):129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grimm D, Kay MA. Combinatorial RNAi: a winning strategy for the race against evolving targets? Mol Ther. 2007;15(5):878–888. doi: 10.1038/sj.mt.6300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.An DS, et al. Stable reduction of CCR5 by RNAi through hematopoietic stem cell transplant in non-human primates. Proc Natl Acad Sci U S A. 2007;104(32):13110–13115. doi: 10.1073/pnas.0705474104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grimm D, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol. 2008;82(12):5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zolotukhin S, Potter M, Hauswirth WW, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70(7):4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Rivas FV, Wohlschlegel J, Yates JR 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7(12):1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7(7):719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grimm D, Pandey K, Nakai H, Storm TA, Kay MA. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J Virol. 2006;80(1):426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Riu E, Grimm D, Huang Z, Kay MA. Increased maintenance and persistence of transgenes by excision of expression cassettes from plasmid sequences in vivo. Hum Gene Ther. 2005;16(5):558–570. doi: 10.1089/hum.2005.16.558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.