Abstract
Nitric oxide (NO) is an important regulator of the catalytic activity of aldose reductase (AR). It reacts with the active site cysteines of AR and this reaction results in the formation of several kinetically distinct forms of the protein. The catalytic activity of AR is increased in the ischemic heart and this increase in activity is associated with NO-dependent modification of AR. During reperfusion, the enzyme reverts back to its un-activated form. Although, AR activation has been linked to thiol oxidation, the mechanisms of de-activation remain unclear. Here we report that treatment of recombinant human AR (AKR1B1) by a non-thiol based NO-donor (DEANO) results in activation and S-nitrosylation of the protein. The nitrosylated (ARSNO), but not the reduced (ARSH), protein reacted with reduced glutathione (GSH) and this reaction resulted in the formation of glutathiolated AR (ARSSG). The modification of AR by NO was site-specific at Cys-298 and was not affected by selective mutation of the neighboring residue, Cys303 to an alanine. Incubation of the glutathiolated AR (AR-SSG) with GSH resulted in the regeneration of the reduced form of the protein (ARSH). Treatment of nitrosylated AR (AR-SNO) with ascorbic acid also led to the conversion of the protein to its reduced form. These observations suggest that intracellular reductants such as GSH and ascorbate could convert the nitrosylated form of AR to its basal or reduced state. In general, such reductive reactions might represent a common mechanism for denitrosylating proteins or an “off” switch in NO-mediated signaling pathways involving protein S-nitrosylation reactions.
1. Introduction
Aldose reductase (AKR1B1) is a stress-responsive protein that catalyzes the reduction of a wide range of structurally diverse aldehydes. It is an efficient catalyst for the reduction of short- to medium-chain aldehydes including aldehydes derived from lipid peroxidation and the conjugates of aldehydes with glutathione [1]. The protein also catalyzes the reduction of glucose to sorbitol and has been assigned to the first transformative step in the polyol pathway for the generation of fructose from glucose. Excessive metabolism of glucose via the polyol pathway has been linked to the development of several secondary diabetic complications [1] and pharmacological inhibition of the enzyme has been shown to ameliorate diabetic neuropathy [2] and nephropathy [3] in animal studies and in phase II clinical trials [4].
In contrast to its deleterious effects in diabetes, our studies with non-diabetic hearts show that the enzyme plays a protective role during ischemia-reperfusion injury [5]. We have found that the activity of the enzyme is increased in the ischemic heart, and that the activated enzyme was resistant to inhibition by inhibitors such as sorbinil. The activated enzyme from the ischemic heart could be de-activated upon reduction by DTT, indicating that activation of the enzyme was due to the oxidation of its active site cysteine residues. The enzyme from the ischemic heart was also inhibited by dimedone (a sulfenic acid-specific reagent [6]) indicating that ischemia results in the oxidation of AR cysteines to sulfenic acid. Indeed, oxidation of recombinant AR by hydrogen peroxide results in the formation of cysteine oxy acids and the oxidized enzyme displays kinetic properties similar to those of the enzyme in the ischemic myocardium. MALDI-MS analysis of the enzyme from ischemic hearts shows that the active site cysteines of the enzyme (Cys-298 and Cys-303) are oxidized to sulfenic acids [5].
Our studies show that the activation of AR in the ischemic heart is mediated by NO, since inhibition of NO synthesis by L-NAME prevented ischemic activation of AR in isolated perfused rat hearts and in the hearts subjected to coronary ligation in situ [7]. This increase in AR activity was accompanied by an increase in eNOS phosphorylation and was prevented by inhibiting PI3Kinase (PI3K), suggesting that the activation of the PI3K/Akt/eNOS pathway is necessary for ischemia-induced activation of AR [7]. An increase in myocardial generation of NO by bradykinin or insulin in hearts perfused under aerobic conditions or those subjected to ischemia led to an increase in AR activity, indicating again that increased production of NO in the heart leads to AR activation. The increase in AR activity in hearts treated with bradykinin or insulin was reversed by DTT indicating that activation of AR by NO is due to the nitrosylation of its cysteine residues [7].
The mechanisms by which ischemia oxidizes AR cysteines are not clear, but may relate to the formation of a nitrosylated forms of the protein which are then oxidized to sulfenic acid. Our previous work shows that in several cells and tissues exogenous treatment with NO donors results in enzyme inactivation. Moreover, feeding diabetic mice with L-arginine [8] or increasing NO bioavailability by administration of nitroglycerine patches prevents tissue accumulation of sorbitol in diabetic rats (9), or in rat aortic vascular smooth muscle cells (VSMC) in culture [10]. These changes were accompanied by an increase in a glutathiolated form of the protein (AR-SSG) [10]. We interpret these finding to suggest that when NO is exogenously delivered, it reacts first with intracellular GSH to form GSNO. The GSNO formed in the cells then reacts with AR to form AR-SSG, which is a catalytically inactive form of the protein. However, when NO is generated intrinsically, as in the ischemic heart, it reacts directly with AR to form AR-NO which results in an increase in enzyme activity. Thus NO could lead to both activation and inactivation of the enzyme depending upon its site of generation and the tissue content of reduced glutathione.
In addition to AR, other proteins are also S-nitrosylated by NO. Several nitrosylated proteins have been detected both in vitro and in vivo [11] and because in many instances nitrosylation affects protein function, protein-nitrosylation and denitrosylation reactions have been suggested to be discrete modes of cell signaling, analogous to protein phosphorylation and dephosphorylation [12]. Nevertheless, the intracellular fate of nitrosylated proteins remains unknown. Specifically, it remains to be established whether the nitrosylated proteins are degraded as such, or are oxidized to sulfenic acids or they are de-nitrosylated by thiol-disulfide exchange reactions. Hence, using AR as a model protein, we tested the hypothesis that reduced glutathione (GSH) plays a central role in protein denitrosylation. Our data shows that S-nitrosylated AR is readily glutathiolated and that the reduced form of the protein is generated by thiol-disulfide exchange reaction involving GSH. AR was also de-nitrosylated by ascorbic acid. Based on these observations, we speculate that reductants such as glutathione and ascorbate play a pivotal role in mediating the denitrosylation of AR and other nitrosylated proteins and that these reactions (analogous to phosphatases) may represent a general mechanism for turning off NO-mediated signaling initiated by protein S-nitrosylation.
2. Materials and methods
2.1. Chemicals and reagents
GSNO, DEANO and N-ethylmaleimide (NEM) were obtained from Calbiochem. Reduced glutathione (GSH) was purchased from Sigma-Aldrich Chemicals and Sephadex G-25 columns were obtained from VWR. Recombinant human WT and C303A AR were purified from E.coli, as described earlier [13].
2.2. Reduction and enzyme assay of human AR
Before use, AR was reduced by incubating with 0.1 M DTT at 37 °C for 1 h. The reduced enzyme was passed through the Sephadex G-25 column equilibrated with N2-saturated potassium phosphate, pH 7.0 containing 1 mM EDTA. AR activity as well as the sensitivity to sorbinil was measured by the rate of disappearance of NADPH monitored at 340 nm at 25°C in a 1 ml system containing 10 mM DL-glyceraldehyde, 10 mM HEPES (pH 7.0) and 0.15 mM NADPH. One unit of enzyme is defined as the amount of the enzyme required to oxidize 1 μmol of NADPH/min. The control cuvette contained all the components of the reaction mixture except the enzyme.
2.3. Modification of AR by DEANO, GSNO and GSH
Reduced, DTT-free enzyme (0.5 mg/ml) was incubated with 0.5 mM DEANO, or 50 μM GSNO at 25°C for 1 h. Aliquots were withdrawn at different time intervals to measure enzyme activity. Modified AR was incubated further with 2 mM GSH for 1 h at 25°C. At the end of the treatment, excessive modifying reagents were removed by passing through a Sephadex G-25 column equilibrated with N2-saturated 0.1 M potassium phosphate (pH 7.0) or 10 mM HEPES (pH 7.0) and assayed for enzyme activity.
2.4. Electrospray ionization mass spectrometry (ESI+-MS)
Modified forms of AR were identified by ESI/MS using micromass LCZ, mass spectrometer. Before electrospray, the enzyme was desalted and separated from the modifying reagents by gel filtration on a Sephadex G-25 column equilibrated with 10 mM ammonium acetate. The desalted protein was diluted with the flow injection solvent (acetonitrile: H2O: formic acid: 50:50:1, v/v/v). The mixture was injected into a micro mass LCZ spectrometer at a rate of 10 μl/min. The operating parameters were as follows: capillary voltage, 3.1 kV: cone voltage, 27 V; extractor voltage, 4 V: source block temperature, 100°C: desolvation temperature: 200°C. Spectra were acquired at the rate of 10 atomic mass unit/s over the range 20–2000 atomic mass units. The instrument was calibrated with myoglobin.
2.5. Western blot analysis
Approximately 100–300 ng of protein in sample buffer containing 100 mM NEM was loaded on a 12 % non-reducing gel and analyzed by using anti-protein-glutathione monoclonal antibody, diluted 1:1000 in 5 % (v/v) blocking solution for 2 h at room temperature. AR protein was detected after stripping and re-probing the PVDF membrane with anti-AR antibody (dilution 1:5,000). Immunoreactive proteins were detected with the ECL Plus Western blot detection kit.
2.6. Statistical analysis
All data are expressed as mean ± SD and were analyzed by one way ANOVA for multiple comparisons, or by Student’s t-test for unpaired data. Statistical significance was accepted at P < 0.05.
3. Results
3.1. Modification and activation of AR by NO-donors
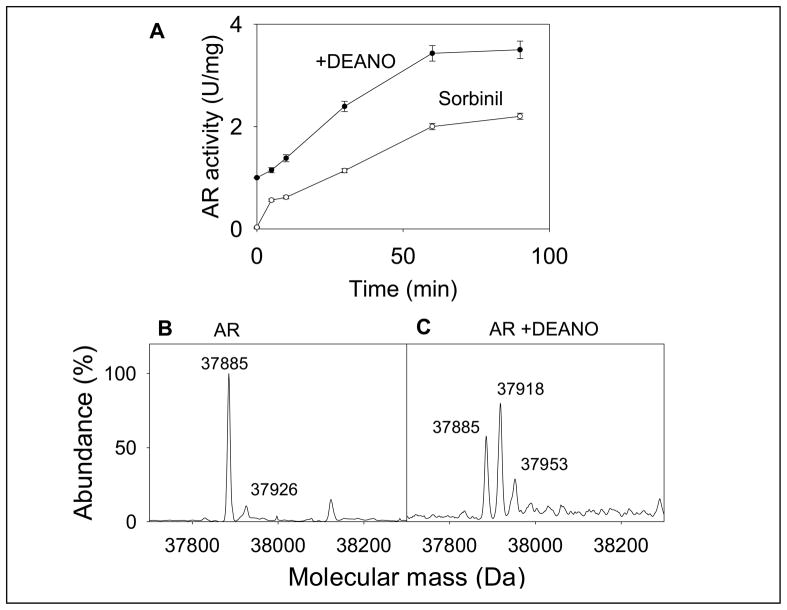
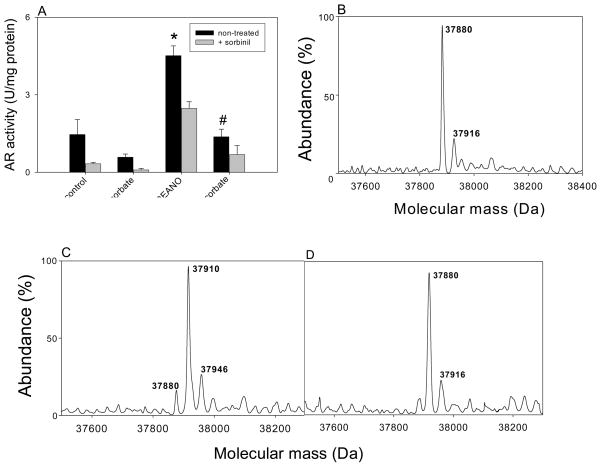
The active site of AR has two reactive cysteine residues (Cys-298 and Cys-303). Oxidation or site-directed substitution of Cys-298 results in an increase in enzyme activity [14]. Modification by NO-donors on the other hand, results in the formation of multiple species which have a higher or a lower enzyme activity than the native protein. To examine how S-nitrosylation by itself affects enzyme activity, the protein was completely reduced and then treated with 0.5 mM 2-(N,N-diethylamino)-diazenolate-2-oxide (DEANO), a NO donor that releases NO with a half-life of 16 min at 37°C. Aliquots of the reaction mixture were withdrawn and the activity of AR was determined with DL-glyceraldehyde as the substrate. As shown in Fig. 1A, treatment with DEANO resulted in 2–3 fold increase in enzyme activity (from 1.2 ± 0.09 to 3.8 ± 0.04 U/mg, n = 3; P < 0.05). The increase in enzyme activity achieved a steady-state within 60 min. Incubation of the enzyme with DEANO also altered the sorbinil sensitivity. Although the reduced enzyme (AR activity at time 0 in Fig. 1A) was completely inhibited by sorbinil, the modified enzyme (90 min after treatment with DEANO) was only partially (30–40 %) inhibited by sorbinil.
Fig. 1. Modification and activation of AR by NO.
(A) Time-dependent changes in the catalytic activity and sorbinil sensitivity of AR. Recombinant human WT His-tag AR was purified from E.coli, reduced with 0.1 M DTT and desalted on a Sephadex G-25 column equilibrated with 0.1 M potassium phosphate (pH 7.0). The reduced enzyme was then incubated with 0.5 mM DEANO at 25°C. Aliquots were withdrawn at indicated times and the catalytic activity was measured with 10 mM DL-glyceraldehyde in the absence (●) and the presence (○) of 1μM sorbinil. Data are shown as discrete points (means ± SD; n=3). (B) Electrospray mass spectrum of WT AR and (C) AR after DEANO treatment. For modification studies the reduced enzyme was incubated with 0.5 mM DEANO in potassium phosphate (pH 7.0) for 60 min at 25 °C. Excess DEANO was removed by passing through Sephadex G-25 column equilibrated with N2-saturated 10 mM ammonium acetate. The deconvoluted electrospray-mass spectrum of unmodified protein corresponds to a single protein with an estimated mass of 37,885 Da, and unidentified minor peak of 37,926 Da. Modification by DEANO resulted in the appearance of protein forms with masses of 37,918 Da and 37,953 Da. The species corresponding to 37,885 Da was assigned to the unmodified protein.
To examine structural changes due to DEANO treatment, electrospray mass spectra of the reduced and modified proteins were compared. Upon ESI/MS the unmodified protein displayed a major peak corresponding to a molecular mass of 37,885 Da which is within an experimental error of the expected molecular mass of His-tagged AR (37,883 Da). A minor unidentified peak of molecular mass 37,926 Da was observed, (Fig. 1B). The identity of this peak is unknown. Mass analysis of AR incubated with DEANO (0.5 mM) for 30 min revealed two major peaks (Fig 1C) with a molecular mass of 37,885 Da that corresponds to the native form and a 37,918 Da form, which was ascribed to an adduct between AR and a single NO molecule. Taken together, this series of experiments suggest that AR is readily nitrosylated predominantly at a single susceptible site and that S-nitrosylation results in a 2–3 fold increase in the catalytic activity of the protein. We refer to the nitrosylated form (ARSNO) as the “activated” enzyme because it has significantly high catalytic activity than the reduced basal form of the protein (ARSH). The other main characteristic feature of the activated form of the enzyme is that it is insensitive to inhibition by sorbinil.
3.2. Inactivation and glutathiolation of S-nitrosylated AR by reduced glutathione
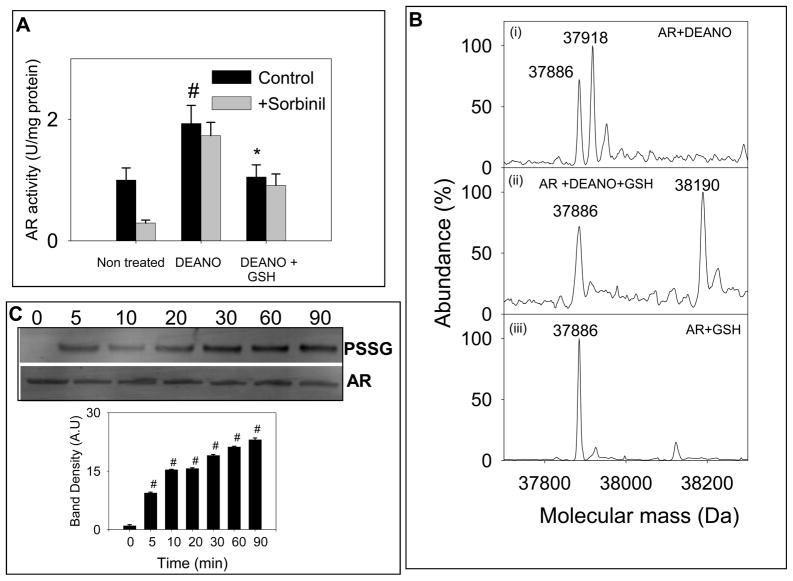
Previous studies show that AR and several other proteins are nitrosylated endogenously by NO or NO-donors [10–12]. However, further reactivity and metabolic fates of nitrosylated proteins remains unclear. To test whether addition of reduced glutathione may be one of the mechanisms by which the effects of S-nitrosylation are overcome, DEANO-treated protein was incubated with 2 mM GSH for 1 h at room temperature and the enzyme activity was determined using DL-glyceraldehyde as substrate. As before, treatment with DEANO resulted in an increase in enzyme activity and a decrease in its sensitivity to sorbinil (Fig. 2A). Treatment with GSH, however led to a decrease in enzyme activity. However, sorbinil sensitivity was not restored. These data suggest that treatment with GSH under these conditions results in deactivation of AR. Binding of glutathione to ARSNO results in enzyme inactivation and the residual activity remaining in the GSH-treated enzyme is due to unreacted protein (which still retains NO) because it was insensitive to sorbinil.
Fig. 2. Inactivation of S-nitrosylated AR by reduced glutathione.
(A) Reduced AR was incubated with 0.5 mM DEANO for 1h at 25°C, desalted and then incubated with 2 mM GSH in 0.1 M potassium phosphate (pH 7.0) for 1h at 25°C. Aliquots were withdrawn from the reaction mixture and the enzyme activity was measured as described under Materials and Methods in the presence and absence of 1 μM sorbinil, as indicated. Values are expressed as mean ± S.D, (n = 3) #P< 0.05 versus non-treated protein and *P <0.05 versus DEANO-treated protein. (B) Electrospray mass spectra of (i) reduced WT AR treated with DEANO showing molecular ions for unmodified AR (37,885 Da) and ARS-NO (37,918 Da); (ii) DEANO-modified AR incubated with 2 mM GSH for 1 h showing both, native (37,885 Da) and glutathiolated (+ 306 Da) AR (38,190 Da); (iii) reduced WT AR incubated with GSH showing only the native protein (37,885 Da). (C) Western blot analysis of DEANO-treated AR incubated with GSH. WT AR was treated with DEANO, desalted and treated with 2 mM GSH for the indicated time periods. The Western blots were developed using monoclonal anti-PSSG antibody. Lower panel shows group data for the intensity of the anti-PSSG immunopositive bands, normalized to AR. Data are presented as mean ± SD. #P < 0.005 versus DEANO-treated protein incubated with GSH for 0 min (n = 3).
To determine the chemical nature of modification, the GSH-treated nitrosylated protein was studied by ESI/MS. As before, the DEANO-treated AR displayed two major peaks with a molecular mass of 37,885 Da and 37,918 Da, corresponding to the native and the NO-bound forms of the protein. As shown in Fig 2B(ii), incubation of the S-nitrosylated form of protein (ARSNO) with GSH displayed two peaks with a molecular mass of 37,885 Da, the native form (ARSH), and 38,190 Da (an increased molecular mass of 305 Da due to AR-SSG) corresponding to the addition of a single GSH molecule (306 Da) to the protein. Glutathiolation of the protein was accompanied by a loss of NO. To determine whether the native (ARSH) form of the enzyme can directly bind to GSH, reduced AR was incubated with GSH, however, the spectrum of the protein was not affected by GSH (Fig 2B iii), indicating that native AR does not directly bind to GSH, and that only S-nitrosylated AR binds to GSH.
To examine the time-course of glutathiolation, the reduced enzyme incubated with DEANO and GSH was analyzed by Western blots using anti-PSSG antibody. Immunoreactivity with anti-PSSG antibody was normalized to the anti-AR antibody staining to correct for changes in protein levels. As shown in the Fig. 2C, no signal was detected with S-nitrosylated AR alone (t=0) indicating that the anti-PSSG antibody does not recognize the nitrosylated protein (ARSNO). Treatment of ARSNO with GSH led to glutathiolation. Within 5 min of incubation, a significant increase in glutathiolated protein was observed. The rate of glutathiolation was, however, slow and complete glutathiolation was achieved after 90 min of incubation with GSH. These results suggest that nitrosylated (ARSNO), but not reduced AR (ARSH), reacts directly with GSH and that addition of GSH to AR replaces NO from the active site as shown:
| [1] |
In this scheme, ARSNO is activated (with higher catalytic activity and reduced sorbinil sensitivity than ARSH), whereas ARSSG is catalytically inactive.
3.3. Identification of AR cysteine involved in S-nitrosylation and glutathiolation
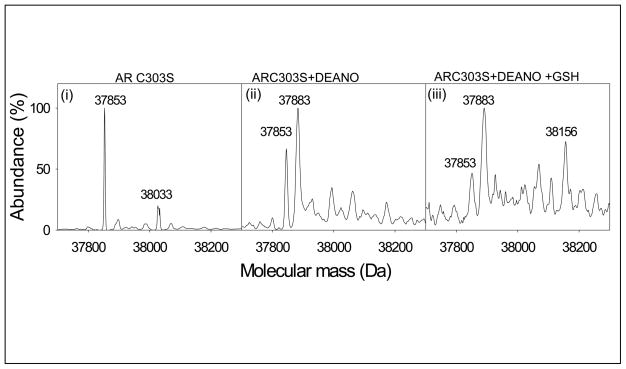
Our previous studies show that AR is nitrosylated at Cys298 [15]. The active site of AR, however, has two reactive cysteines – Cys298 and Cys303. To determine whether Cys-303 also plays a role in protein S-nitrosylation or glutathiolation, a site-directed mutant of AR in which cysteine 303 was replaced with alanine was generated. The AR:C303A mutant protein was purified and reduced and then treated with DEANO (0.5 mM). As shown in Fig. 3(i) the electrospray mass spectrum of the untreated AR:C303A displayed a major peak corresponding to a molecular mass of 37,853 Da and a minor peak (38,033 Da) of unknown chemical nature. The spectrum of AR treated with DEANO displayed two major species with a molecular mass of 37,853 Da representing the native protein, and 37,883 Da which is consistent with the addition of a single NO molecule (30 Da) to the native form of the protein (Fig. 3, ii). To examine whether GSH would bind to S-nitrosylated AR:C303A, the S-nitrosylated protein was incubated with GSH (2 mM). The electrospray mass spectrum of the S-nitrosylated protein treated with GSH, (Fig. 3, iii) displayed all three forms of the protein; 37,853 Da representing the native form, 37,883 Da and 38,156 Da species corresponding to the nitrosylated and glutathiolated forms of the protein, respectively. The observation that AR:C303A behaves like the WT protein, suggests that Cys303 does not participate in S-nitrosylation and glutathiolation reactions and that an intramolecular disulfide between Cys298 and Cys303 is not a significant intermediate between the nitrosylation or subsequent glutathiolation of AR.
Fig. 3. Identification of the glutathione binding site of AR.
Electrospray mass spectrum of (i) AR:C303A (37,853Da) and (ii) AR:C303A incubated with 0.5 mM DEANO in N2-saturated potassium phosphate (0.1 M, pH 7.0) at 25 °C for 60 min. Excess DEANO was removed by passing the reaction mixture through the Sephadex G-25 column and eluted with N2-saturated 10 mM ammonium acetate. Spectra show the native (37,853 Da) and S-nitrosylated protein (37,883 Da); and (iii) AR:C303A incubated with DEANO, desalted and treated with 2 mM GSH. The spectrum shows native (37,853 Da), S-nitrosylated (37,883 Da) and glutathiolated (38,156 Da) forms of the protein.
3.4. Dethiolation of glutathiolated AR by GSH
Our results so far showed that AR is readily nitrosylated and that nitrosylated protein reacts readily with GSH to form a glutathiolated protein. How then is glutathiolated form of AR (ARSSG) reverted to the reduced form (ARSH) of the protein? To address this issue, we studied the role of two major cellular reductants, GSH and ascorbic acid. To assess the role of GSH, a glutathiolated form of AR was generated. Because treatment of nitrosylated AR is an efficient way to generate glutathiolated AR (Fig. 2A) we treated AR with GSNO, which reacts with AR to generate, a mostly AR-SSG form of the protein by the following reaction:
| [2] |
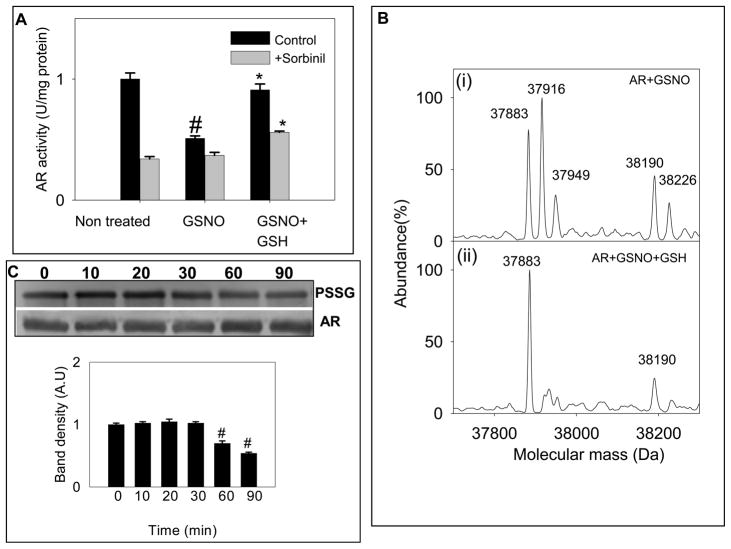
Consistent with the formation of ARSSG, treatment of reduced AR with GSNO resulted in a loss of enzyme activity (from 1.2 ± 0.2 to 0.55 ± 0.04 U/mg protein n=3; Fig. 4A) indicating the formation of catalytically-inactive ARSSG. Treatment with GSNO also reduced the sorbinil sensitivity of the enzyme, presumably due to the formation of a nitrosylated form of the enzyme generated by the following side reaction:
| [3] |
Fig. 4. Reduction and reactivation of GSNO modified AR by GSH.
(A) Reversal of GSNO-induced inhibition of the enzyme by GSH. The reduced enzyme was incubated with 50 μM GSNO in 0.1 M potassium phosphate (pH 7.0) for 1 h at 25°C. The treated protein was desalted and incubated with 2 mM GSH for 1 h at 25°C. At the end of the treatment aliquots were withdrawn and assayed for enzyme activity without and with 1 M sorbinil, as indicated. Values are expressed as mean ± S.D, (n = 3) # P< 0.05 versus non-treated protein and * P < 0.05 versus GSNO-treated protein. Electrospray mass spectra of AR (i) incubated with 50 μM GSNO or 1 h at room temperature showing major species corresponding to native (37,883 Da), S-nitrosylated (37,916 Da) and glutathiolated (38,190 Da) forms of the protein and (ii) treated with GSNO, desalted and incubated with 2 mM GSH for 1 h at room temperature. The spectrum shows a major species corresponding to the native (37,883 Da) and a minor peak corresponding to a glutathiolated (38,190 Da) form of the protein. (C) Western blot analysis of time-dependent changes in glutathiolated AR incubated with 2 mM GSH. AR was treated with GSNO and incubated with GSH for the indicated time periods. PVDF membrane was immunoblotted by using anti-PSSG antibody. Lower panel shows group data for the intensity of the anti-PSSG immunopositive bands normalized to AR. Data are expressed as Mean ± SD. #P < 0.005 versus GSNO-treated protein incubated with GSH for 0 min (n = 3).
Incubation of the GSNO-treated protein with GSH restored both, the enzyme activity and the sorbinil sensitivity of the enzyme to levels similar to those of the native (reduced) form of the protein. To determine the structural changes that accompany the kinetic alterations, changes in the protein were examined by ESI/MS. Electrospray mass spectra showed three major peaks with molecular masses of 37,883 Da, 37,916 Da and 38,190 Da corresponding to the native (ARSH), S-nitrosylated (ARSNO) and the glutathiolated (ARSSG) forms of the protein, respectively (Fig. 4B, i). Incubation of the glutathiolated form with GSH resulted in the removal of glutathione bound to the protein and the appearance of native protein with a molecular mass of 37,883 Da and a minor peak of 38,190 Da corresponding to the glutathiolated form of AR (Fig. 4B, ii). The reaction could be written as:
| [4] |
The time course of deglutathiolation was also assessed by Western blot analysis. Aliquots of the GSNO-modified protein were incubated with GSH (1 mM) for different time intervals and the protein glutathiolation was analyzed by Western blotting using the anti-PSSG antibody. To correct for loading differences, the intensity of the band was normalized to the band obtained with the anti-AR antibody. The rate of dethiolation by GSH is summarized in Fig. 4C. After 30 min of incubation with GSH, the intensity of the thiolated protein was reduced by 20–30%, which was further decreased to 40–50% after 90 min of incubation. These results provide clear evidence that both, the S-nitrosylated and the glutathiolated forms of the protein are converted back to the reduced form by thiol exchange reactions.
3.5. Denitrosylation of AR by ascorbate
In addition to glutathione, ascorbic acid is another potent intracellular reducing agent that could reduce S-nitrosylated proteins and restore them to their native, reduced form. To test the role of ascorbate, untreated or DEANO-treated WT AR was incubated with ascorbate (10 mM). Treatment with ascorbate decreased the activity of the native form of the enzyme slightly, indicating basal oxidation of the protein (Fig. 5A). As observed before, treatment with DEANO increased AR activity, but decreased sorbinil sensitivity. Treatment of the S-nitrosylated protein with ascorbate decreased the activity of the protein and increased sorbinil sensitivity (Fig. 5A), indicating that ascorbate could abolish the kinetic effects of AR S-nitrosylation.
Fig. 5. De-activation and de-nitrosylation of AR by ascorbic acid.
(A) Catalytic activity of reduced and S-nitrosylated WT AR treated with ascorbic acid. Aliquots of reduced AR were treated with 0.5 mM DEANO alone for 1 h, or treated with DEANO followed by the incubation with 10 mM sodium ascorbate for 1 h, as indicated. Prior to enzyme activity measurements, salts were removed by the gel filtration and AR activity was measured as described under Materials and Methods. Sensitivity of the modified enzyme to 1 μM sorbinil was measured in parallel. Data are reported as mean ± SD (n=3). *P < 0.01 versus control; #P < 0.02 versus DEANO. Electrospray mass spectra of (B) native AR (37,880) (C) native AR treated with 0.5 mM DEANO showing the addition of one (37,910 Da) NO molecule bound to the protein; and (D) AR treated with 0.5 mM DEANO followed by the incubation with 10 mM sodium ascorbate, showing only the reduced native form of the AR protein (37,880 Da).
To examine structural changes, ESI/MS analysis was performed. As expected, the native protein conformed to a single molecular ion with an estimated mass of 37,880 Da (Fig. 5B). Treatment with DEANO increased the molecular mass of the protein by 30 Da, consistent with the addition of a single NO molecule to the protein (Fig. 5C). Treatment with ascorbate restored the native form of the protein such that the mass spectrum of the ascorbate-treated S-nitrosylated protein displayed a single ion corresponding to the native form of the protein (Fig 5D). These data suggest that ascorbate is able to de-nitrosylate AR and to restore the enzyme to its native form.
4. Discussion
Results from this study demonstrate that the activity of AR (AKR1B1) is regulated by NO and that the de-nitrosylation of AR is mediated or facilitated by intracellular reductants such as glutathione and ascorbate. These findings suggest that like other forms of post-translational modification, S-nitrosylation is a reversible process and that intracellular reductants play a critical role in regulating both the extent and the duration of S-nitrosylation of proteins such as AR.
The sensitivity of AR to NO and other oxidants such as hydrogen peroxide or peroxynitrite stems from the high reactivity of Cys298 located at the active site of the enzyme. While this residue does not directly participate in substrate or cofactor binding or in catalysis, it is located near the NADPH binding domain of the protein and regulates cofactor binding [14]. Our previous studies have shown that oxidation of Cys298 increases kcat, which appears to be due to an increase in the off-rate of NADP+ release. Because NADP+ release from the protein at the end of the catalytic cycle is rate-limiting, an increase in NADP+ release increases the rate of the overall catalytic cycle. The kinetics of AR activation by thiol modification has been described in detail [16]. However, the physiological significance of changes in AR kinetics due to thiol oxidation is less clear. It has been known from some time that the thiol residues of AR are oxidized in tissues or during purification and that oxidation decreases the inhibitor-sensitivity of the enzyme [17]. We have reported recently that the enzyme is readily nitrosylated in vitro and that S-nitrosylation affects the kinetics and the inhibitor binding to the enzyme [18;19].
Our experiments show that treatment with the SIN-1, which generated both NO and superoxide decreases the activity of the enzyme [15], treatment with NO donors such as DEANO leads to an increase in the enzyme activity [20]. On the other hand treatment with GSNO results in enzyme inactivation and the loss of catalysis because of the formation of a single glutathione molecule to Cys-298 at the active site of the enzyme. Thus, depending upon the NO donor and the experimental conditions, NO donor could either increase or decrease enzyme activity.
In addition to modification in vitro, NO is also an in vivo regulator of AR. It has been shown, that treatment of SMC with S-nitroso-N-acetylpenicillamine (SNAP) increases AR mRNA expression and AR activity [21] and in diabetic animals increasing NO availability (by nitroglycerin patches or feeding L-arginine) prevents AR activation and tissue sorbitol accumulation [9]. The decrease in AR activity in VSMC was accompanied by an increase in glutathiolation of AR, indicating that in GSH replete cells, exogenous NO reacts first with GSH and that the product of this reaction (GSNO) then modifies AR by glutathiolating the active site cysteine [20]. Because glutathione is a large tripeptide, its binding prevents substrate access to active site which is located at the bottom of a β-barrel structure; thereby inhibiting catalysis. In contrast, binding of small molecules such as NO, which would happen if NO is generated endogenously in close proximity to the enzyme, merely opens up the active site and increases NADP+ release resulting in an increase in the catalytic rate. Hence, the kinetic change induced in the enzyme depends critically on the nature of the modifying group bound to Cys298.
We have recently reported that during ischemia AR is activated by NO derived from eNOS and that this is associated with the formation of protein-sulfenic acid formation [7]. Given that in the ischemic heart, Cys298 was oxidized to a sulfenic acid and that this oxidation was dependent upon NO derived from eNOS, we hypothesized that sulfenic acid formation may be consequence of AR S-nitrosylation, and that proteins that are nitrosylated are more likely to be oxidized to sulfenic acids than those which remain as free thiols. Hence, it appears likely that despite the appearance of oxy-acid forms of AR, the protein might be regulated in the heart by S-nitrosylation and denitrosylation.
Modification of the proteins by NO and subsequent change in their function may represent an important component of NO signaling and regulation of intracellular processes. Many myocardial proteins have been shown to be susceptible to S-nitrosylation in vitro [22]. Protein S-nitrosylation has been linked to the regulation of several physiological processes by NO, such as changes in myocardial contraction [23], apoptosis [24], mitochondrial energetics and calcium homeostasis [25]. Although NO does not directly react with cysteines, it can undergo oxidation to N2O3, which is an efficient nitrosylating agent. Because S-nitrosylation affects thiol reactivity, charge and ionization, it could induce changes in protein conformation or catalytic activity. In case of AR, S-nitrosylation increases catalysis and decreases the inhibitor sensitivity of the protein (Fig. 1). Similar changes have been reported for other enzymes or myocardial proteins, e.g., L-type Ca2+- channels [23] and mitochondrial complex I [26]. Once nitrosylated, these proteins have been implicated in mediating cardioprotection against ischemia-reperfusion injury [27].
S-nitrosylation plays a critical role not only in maintaining certain cellular function, but also in regulating, or mediating apoptosis [28]. S-nitrosylation of albumin is believed to serve as a NO-pool, whereas S-nitrosylation of creatine kinase was found to inhibit its activity [29]. Modification of actin by S-nitrosylation decreases its capacity of polymerization and might be a means to conserve NO bioavailability [30]. S-nitrosylation of Hsp90 has been shown to inhibit its ATPase activity and diminishes its positive control on endothelial nitric oxide synthase (eNOS) activity [31]. Among the new proteins identified as targets of GSNO-mediated S-nitrosylation are: destrin, myosin, Hsp27, triosephosphate isomerase, enolase and adenylate kinase 1 [32].
Even though overwhelming evidence supports the notion that S-nitrosylation is a significant oxidative post-translational mechanism, it is not clear how the effects of S-nitrosylation are terminated or “switched off”. Are nitrosylated proteins degraded? Do they inevitably or eventually get oxidized to sulfur-oxy acids? Are nitrosylated proteins de-nitrosylated and are these processes of de-nitrosylation regulated? To address some of these issues, we examined the case of AR and tested whether the protein could be de-nitrosylated by intracellular reductants.
Our results show that S-nitrosylated AR reacted readily with reduced glutathione. No GSH reactivity was observed with the native protein. This observation suggests that NO provides a susceptible target for the addition of glutathione and therefore glutathione removes NO bound to the proteins. Thus, S-nitrosylation of proteins in most eukaryotic cells, which contain 1–10 mM glutathione is likely to be transient and readily terminated by glutathione. As seen with AR, glutathione could directly add to the site of S-nitrosylation or when present in excess could remove NO and restore the cysteine partner to its original reduced form. The hypothesized sequence of events is illustrated in the Scheme 1 showing that S-nitrosylated protein could be either oxidized to sulfenic, sulfinic or sulfonic acids or reduced back to its native form by glutathione or ascorbate. Both, sulfinic and sulfonic acids are considered to be irreversible oxidations and hence glutathiolation may be a protective mechanism for preventing irreparable damage to nitrosylated proteins. In addition to nitrosylated proteins, the sulfenic acid forms of proteins could also be rescued, however, in this case the addition of glutathione to the oxidized protein may be catalyzed by enzymes such as glutathione-S-transferases [33].
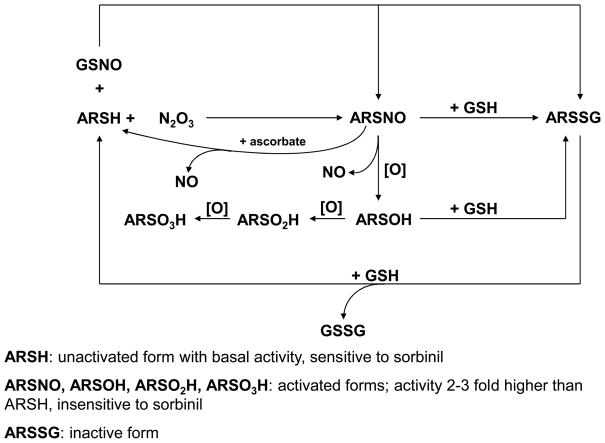
Scheme I. Modification of AR by NO and nitrosoglutathione (GSNO).
Reduced AR (ARSH) is nitrosylated by N2O3 (generated from the reaction of NO with molecular oxygen) to the activated (ARSNO) form. Reaction of AR with GSNO results mainly in the formation of glutathiolated AR (ARSSG). Nitrosylated AR (ASNO) is generated as a minor side product. The ARSSG form is catalytically inactive. The nitrosylated form of AR (ARSNO) could be oxidized and de-nitrosylated to generate oxy-sulfur acid forms of AR (ARSOH, ARSO2H, and ARSO3H). Like ARSNO, these forms of AR display 2–3 fold higher catalytic activity and are relatively insensitive to inhibition by sorbinil. In addition to oxidation, ARSNO could react with GSH to generate ARSSG which is catalytically inactive. Although not shown in the current study, ARSOH could also be converted to ARSSG by GSH. The ARSSG form is subsequently reduced to the basal ARSH form by another GSH molecule. ARSNO could also be reduced and converted back to ARSH by ascorbic acid. The catalytic activity of each of these forms is listed. Several forms of AR could, therefore, coexist in a cell depending upon the concentration of GSH, NO, and oxygen.
The observation that S-nitrosylated AR readily undergoes glutathiolation provides a new explanation for the presence of glutathiolated proteins in the cells. In most tissues, 15–20 % of the total glutathione pool is bound to the proteins. The intracellular concentration of oxidized glutathione (GSSG) is very low (< 1 μM), because GSSG once formed is either rapidly extruded (by GS-X transporters) or reduced by glutathione reductase [34]; hence the observation that a large fraction of glutathione remains bound to the proteins lacked an adequate explanation. Therefore, our observation that S-nitrosylation precedes glutathiolation suggests that the glutathiolated protein pool in cells may be due to a steady-state accumulation of protein between the cycles of S-nitrosylation and de-nitrosylation.
In addition to glutathione, ascorbic acid is another reductant that might facilitate protein de-nitrosylation. Most cells contain high levels of ascorbic acid, which like glutathione can also be recycled. Ascorbate is considered to be a weak reductant and has been used to selectively de-nitrosylate proteins in vitro in protein S-nitrosylation assays [35]. However, as demonstrated here for AR, reduction by ascorbic acid could be one of the mechanisms by which the S-nitrosylation is turned off.
The results reported here provide further understanding of redox mechanisms that regulate the intracellular activity of AR. We have found that modification of AR by NO could be selective (restricted to Cys-298) and reversible. The modification of Cys-298 was not affected by a site-directed replacement of Cys-303 with alanine, indicating that the intramolecular disulfide formation is unlikely to significantly affect protein S-nitrosylation or glutathiolation. Our findings support the possibility of glutathione-assisted denitrosylation of AR in vivo. Although intracellular glutathiolation and de-glutathiolation reactions have not been studied in detail, it is likely that proteins such as glutaredoxin could facilitate such reactions. Further studies are required to fully assess how S-nitrosylation and glutathiolation reactions regulate the role of AR in secondary diabetic complications or during myocardial ischemia-reperfusion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Srivastava SK, Ramana KV, Bhatnagar A. Role of aldose reductase and oxidative damage in diabetes and the consequent potential for therapeutic options. Endocr Rev. 2005;26:380–392. doi: 10.1210/er.2004-0028. [DOI] [PubMed] [Google Scholar]
- 2.Nakayama M, Nakamura J, Hamada J, Chaya S, Mizubayashi R, Yasuda Y, Kamiya K, Koh N, Hotta N. Aldose reductase inhibition ameliorates pupillary light reflex and F-wave latency in patients with mild diabetic neuropathy. Diabetes Care. 2001;24:1093–1098. doi: 10.2337/diacare.24.6.1093. [DOI] [PubMed] [Google Scholar]
- 3.Hodgkinson AD, Sondergaard KL, Yang BM, Cross DF, Millward BA, Demaine AG. Aldose reductase expression is induced by hyperglycemia in diabetic nephropathy. Kidney Int. 2001;60:211–218. doi: 10.1046/j.1523-1755.2001.00788.x. [DOI] [PubMed] [Google Scholar]
- 4.Chung SSM, Chung SK. Aldose reductase in diabetic microvascular complications. Curr Drug Targets. 2005;6:475–486. doi: 10.2174/1389450054021891. [DOI] [PubMed] [Google Scholar]
- 5.Kaiserova K, Srivastava S, Hoetker JD, Awe SO, Tang XL, Cai J, Bhatnagar A. Redox activation of aldose reductase in the ischemic heart. J Biol Chem. 2006;281:15110–15120. doi: 10.1074/jbc.M600837200. [DOI] [PubMed] [Google Scholar]
- 6.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proceedings of the Natl Acad Sci U S A. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaiserova K, Tang XL, Srivastava S, Bhatnagar A. Role of nitric oxide in regulating aldose reductase activation in the ischemic heart. J Biol Chem. 2008;283:9101–9112. doi: 10.1074/jbc.M709671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West MB, Ramana KV, Kaiserova K, Srivastava SK, Bhatnagar A. L-arginine prevents metabolic effects of high glucose in diabetic mice. FEBS Lett. 2008;582:2609–2614. doi: 10.1016/j.febslet.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra D, Jackson EB, Ramana KV, Kelley R, Srivastava SK, Bhatnagar A. Nitric oxide prevents aldose reductase activation and sorbitol accumulation during diabetes. Diabetes. 2002;51:3095–3101. doi: 10.2337/diabetes.51.10.3095. [DOI] [PubMed] [Google Scholar]
- 10.Ramana KV, Chandra D, Srivastava S, Bhatnagar A, Srivastava SK. Nitric oxide regulates the polyol pathway of glucose metabolism in vascular smooth muscle cells. FASEB J. 2003;17:417–425. doi: 10.1096/fj.02-0722com. [DOI] [PubMed] [Google Scholar]
- 11.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitro-sylation: purview and parameters. Nat Rev Mol Cell Biol. 2005:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 12.Stamler JS, Sun QA, Hess DT. A SNO storm in skeletal muscle. Cell. 2008;133:33–35. doi: 10.1016/j.cell.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Srivastava S, Spite M, Trent JO, West MB, Ahmed Y, Bhatnagar A. Aldose re-ductase- catalyzed reduction of aldehyde phospholipids. J Biol Chem. 2004;279:53395–53406. doi: 10.1074/jbc.M403416200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petrash JM, Harter TM, Devine CS, Olins PO, Bhatnagar A, Liu S, Srivastava SK. Involvement of cysteine residues in catalysis and inhibition of human aldose reductase. Site-directed mutagenesis of Cys-80, -298, and -303. J Biol Chem. 1992;267:24833–24840. [PubMed] [Google Scholar]
- 15.Chandra A, Srivastava S, Petrash JM, Bhatnagar A, Srivastava SK. Active site modification of aldose reductase by nitric oxide donors. Biochim Biophys Acta. 1997;1341:217–222. doi: 10.1016/s0167-4838(97)00084-8. [DOI] [PubMed] [Google Scholar]
- 16.Bhatnagar A, Liu SQ, Ueno N, Chakrabarti B, Srivastava SK. Human placental aldose reductase: role of Cys-298 in substrate and inhibitor binding. Biochim Biophys Acta. 1994;1205:207–214. doi: 10.1016/0167-4838(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 17.Bhatnagar A, Liu S, Das B, Srivastava SK. Involvement of sulfhydryl residues in aldose reductase-inhibitor interaction. Mol Pharmacol. 1989;36:825–30. [PubMed] [Google Scholar]
- 18.Srivastava S, Dixit BL, Ramana KV, Chandra A, Chandra D, Zacarias A, Petrash JM, Bhatnagar A, Srivastava SK. Structural and kinetic modifications of aldose reductase by S-nitrosothiols. Biochem J. 2001;358:111–118. doi: 10.1042/0264-6021:3580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixit BL, Ramana KV, Chandra D, Jackson EB, Srivastava S, Bhatnagar A, Srivastava SK. Metabolic regulation of aldose reductase activity by nitric oxide donors. Chem Biol Interact. 2001;130:573–581. doi: 10.1016/s0009-2797(00)00300-8. [DOI] [PubMed] [Google Scholar]
- 20.Chandra A, Srivastava S, Petrash JM, Bhatnagar A, Srivastava SK. Modification of aldose reductase by S-nitrosoglutathione. Biochemistry. 1997;36:15801–15809. doi: 10.1021/bi9714722. [DOI] [PubMed] [Google Scholar]
- 21.Seo HG, Nishinaka T, Yabe-Nishimura C. Nitric oxide up-regulates aldose reductase expression in rat vascular smooth muscle cells: A potential role for aldose reductase in vascular remodeling. Mol Pharmacol. 2000;57:709–717. doi: 10.1124/mol.57.4.709. [DOI] [PubMed] [Google Scholar]
- 22.Hare JM, Beigi F, Tziomalos K. Nitric oxide and cardiobiology-methods for intact hearts and isolated myocytes. Nitric Oxide, Pt G: Oxidative and Nitrosative Stress in Redox Regulation of Cell Signaling. 2008;441:369–392. doi: 10.1016/S0076-6879(08)01221-4. [DOI] [PubMed] [Google Scholar]
- 23.Sun JH, Picht E, Ginsburg KS, Bers DM, Steenbergen C, Murphy E. Hypercontractile female hearts exhibit increased S-nitrosylation of the L-type Ca2+ channel alpha 1 subunit and reduced ischemia/reperfusion injury. Circ Res. 2006;98:403–411. doi: 10.1161/01.RES.0000202707.79018.0a. [DOI] [PubMed] [Google Scholar]
- 24.Benhar M, Stamler JS. A central role for S-nitrosylation in apoptosis. Nat Cell Biol. 2005;7:645–646. doi: 10.1038/ncb0705-645. [DOI] [PubMed] [Google Scholar]
- 25.Sun JH, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 26.Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosylation of mitochondrial complex I. Biochem J. 2006;394:627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nadtochiy SM, Burwell LS, Brookes PS. Cardioprotection and mitochondrial S-nitrosylation: Effects of S-nitroso-2-mercaptopropionyl glycine (SNO-MPG) in cardiac ischemia-reperfusion injury. J Mol Cell Cardiol. 2007;42:812–825. doi: 10.1016/j.yjmcc.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol. 2005;7:665–U40. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 29.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 30.Quinn MT, Siemsen DW, Swain SD, Hanson AJ, Nelson-Overton LK, Rohn TT, Clements MK. Modulation of actin polymerization by peroxynitrite: Effect on neutrophil functional responses. FASEB J. 2003;17:C71. doi: 10.1189/jlb.0802401. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Ruiz A, Villanueva L, de Orduna CG, Lopez-Ferrer D, Higueras MA, Tarin C, Rodriguez-Crespo I, Vazquez J, Lamas S. S-nitrosylation of Hsp90 promotes the inhibition of its ATPase and endothelial nitric oxide synthase regulatory activities. Proc Natl Acad Sci U S A. 2005;102:8525–8530. doi: 10.1073/pnas.0407294102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saraiva RM, Hare J. Nitric oxide signaling in the cardiovascular system: implications for heart failure. Curr Opin Cardiol. 2006;21:221–228. doi: 10.1097/01.hco.0000221584.56372.dc. [DOI] [PubMed] [Google Scholar]
- 33.Shackelford RE, Heinloth AN, Heard SC, Paules RS. Cellular and molecular targets of protein S-glutathiolation. Antioxid Redox Signal. 2005;7:940–950. doi: 10.1089/ars.2005.7.940. [DOI] [PubMed] [Google Scholar]
- 34.Sanz-Alfayate G, Obeso A, Agapito MT, Gonzalez C. Reduced to oxidized glutathione ratios and oxygen sensing in calf and rabbit carotid body chemoreceptor cells. J Physiol (Lond) 2001;537:209–220. doi: 10.1111/j.1469-7793.2001.0209k.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]