Abstract
The zinc finger protein 217 (ZNF217) is an important oncogene based on the high frequency of amplification and overexpression in many cancer types, but its molecular mode of gene regulation is poorly understood. We purified a complex of nuclear ZNF217-binding proteins by affinity chromatography and identified its components by mass spectrometry as Jarid1b/Plu-1, G9a, LSD1, CoREST and CtBP1. Individual binding of these with ZNF217 was confirmed by co-immunoprecipiation (IP). Known activities of these proteins suggested a role of the ZNF217 complex in histone modification. Using in vitro assays the following activities were demonstrated: Histone H3 lysine 4 trimethyl (H3K4me3) demethylase activity, which co-fractionated with Jarid1b/Plu-1 in anion-exchange chromatography; H3K9 methylation, the known principal activity of G9a; and H3K27 methylation. The latter suggested EZH2 as another ZNF217 binding candidate, which could be confirmed by co-IP. Taken together, these findings suggest that ZNF217 assembles a distinct set of histone modifying proteins at target DNA sites that act synergistically in transcriptional repression.
Keywords: ZNF217, histone modifying enzymes, repressive complexes, oncogene, gene regulation
Introduction
Zinc finger protein 217 (ZNF217) has been recognized as a human oncogene based on rapidly accumulating genetic evidence for its role in carcinogenesis. We identified 30 independent studies reporting ZNF217 overexpression and/or genetic amplification in cancers of the ovary (seven reports), uterus (1), ovarian sarcoma (1), melanoma (1), colon (4), breast (5), prostate (1), stomach or esophagus (6), bladder (1), lung (1), pancreas (1), and head and neck (1), most of which are included in a recent review.1 While such genetic evidence for an involvement of ZNF217 in carcinogenesis is strong, little is known about its molecular mechanism(s).
ZNF217 binds to DNA through its zinc fingers thereby targeting potential protein binding partners to specific sites in the genome. Questions remain as to whether the ZNF217 oncoprotein operates solely as a transcription factor, as a co-repressor, or serves as a scaffold between other DNA-binding transcription factors that assemble into an array of complex chromatin modifying proteins.1 ZNF217 has been consistently identified as a polypeptide component of the CoREST/CtBP1 complex along with the first lysine demethylase identified as BHC11/LSD1/AOF2.2–7 More recent studies of ZNF217 have been driven by an interest in identifying gene targets in the human genome consistent with its role as an oncogenic transcriptional repressor and as a component of the CoREST/CtBP complex.8–10 However, questions remain about the potential of ZNF217 to align with alternate or complementary histone-modifying activities.8,11 Recent examples have evolved a model to include a function of transcriptional regulators to integrate and exchange multiple enzymatic activities that give rise to a comprehensive biochemical program to reorganize chromatin architecture, thereby sustaining appropriate gene expression programs (reviewed in Rosenfeld et al.12).
We were interested in identifying such ZNF217-binding proteins in order to understand, which activities ZNF217 might direct in vivo. A controversy surrounding ZNF217 is whether ZNF217 can adopt various configurations by integrating diverse histone modifying activities as an assembly of differing complexes. Here, we report that a nuclear ZNF217 protein complex contains several known histone-modifying enzymes and has biochemical activities in vitro that are strongly associated with gene repression.
Results
ZNF217 binding proteins identified by affinity chromatography
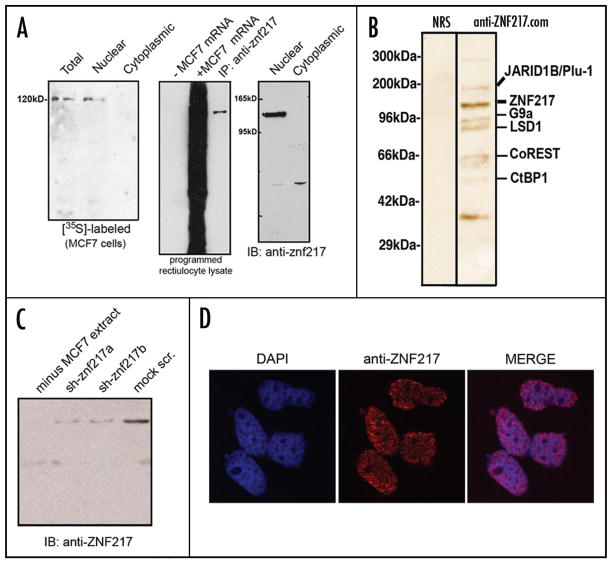
The polyclonal anti-ZNF217 antibody employed in the present study has high affinity and is highly specific identifying only its proper target (Fig. 1A). To assess the fidelity of the antibody to retain ZNF217 by affinity chromatography we tested the ZNF217 antibody by immunoprecipitation from MCF7 cell extracts pulse-labeled with [35S]. Results indicate that ZNF217 antibody can immune precipitate a homogenous [35S]-labeled ZNF217 by denaturing SDS-PAGE from nucleus but remains absent in the cytosol of MCF7 cells. In order to further characterize the ZNF217 antisera, we tested the antibody by immunoprecipitation of ZNF217 from rabbit reticulocyte lysate programmed with MCF7 cell polyA+ mRNA as shown Figure 1A. Immunocytochemical staining of ZNF217 (Fig. 1D) reveals that ZNF217 resides primarily in the nucleus of MCF7 cells. The high quality of this antibody enabled us to perform anti-ZNF217 affinity chromatography at physiological ZNF217 levels, i.e., without a need for overexpressing any protein or using any artificial tagged constructs. ZNF217 is localized exclusively in the nucleus (Fig. 1). We prepared nuclear extract from MCF7 breast carcinoma cells under non-denaturing conditions. An affinity chromatography column was prepared by covalently linking the anti-ZNF217 antibody to the cynanogen bromide (CnBr)-sepharose matrix. Unfractionated nuclear extract was applied to the column, unbound protein washed off, and then anti-ZNF217 bound protein complexes eluted with high salt.
Figure 1.
Nuclear ZNF217 binding proteins identified by affinity chromatography. (A) Fidelity of ZNF217 antisera. Western blotting and immunoprecipitation illustrates the specificity of the anti-ZNF217 antibody used in this study, which stains ZNF217 as a single band. Using [35S]-labeled cellular extract fractions pulse-labeled with [35S]-methionine/cysteine was used to detect ZNF217 by immunopreciptation in nuclear and cytosolic fractions. (B) Affinity chromatographic purification of ZNF217 binding proteins was performed capturing endogenous ZNF217 from nuclear extract with anti-ZNF217 antibodies (bound to the column) under non-denaturing conditions. Shown is an SDS gel that was silver-stained. Individual protein bands were extracted, subjected to tryptic digestion, and analyzed with MALDI-TOF mass spectrometry. The molecular weight of all proteins identified by this method was consistent with their known sizes. (C) RNA interference of ZNF217 by two specific siRNAs against ZNF217 in MCF7 cells was used to determine the presence of ZNF217 following transfections of siRNAs. (D) Immunocytochemical staining of MCF7 cells using ZNF217 antisera followed by secondary labeling with anti rabbit IgG conjugated with Texas Red (Thermo Scientific/Pierce). Fixed cells were counter-stained with DAPI and DAPI labeling was used to merge contrasting images to confirm co-localization.
SDS electrophoresis of the eluate revealed ten distinct silver-stainable protein bands. The most prominent had an apparent molecular weight of 140 kDa, the size for ZNF217 with this technique. The other nine bands were of various intensities and did not match protein candidates in any obvious way. Therefore, we excised individual bands, performed tryptic digests on each, and subjected the resulting peptide mixtures to MALDI-TOF mass spectrometry. This approach led to the identification of five proteins, namely Jarid1b/Plu-1, G9a, LSD1, CoREST and CtBP1 (Fig. 1B).
Confirmation of Jarid1b/Plu-1-, CtBP1-, CoREST-, G9a- and LSD1-binding to ZNF217 by co-immunoprecipitation
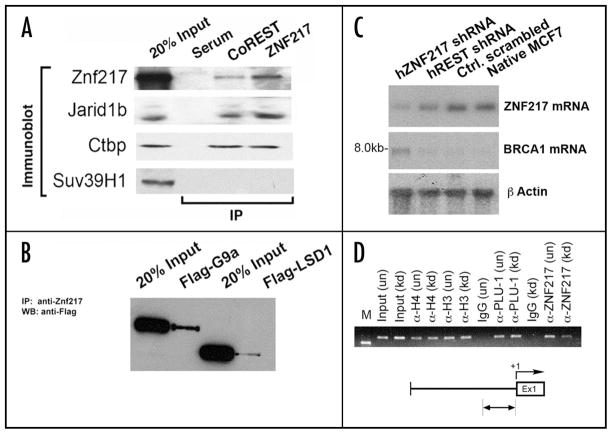
Affinity chromatography may in certain cases be affected by unspecific protein interactions (directly with the target or through intermediary proteins). MALDI-TOF may in certain cases not be satisfactory as a definitive proof for the identification of an unknown protein. For these reasons, we set out to confirm specific binding to ZNF217 for each of the identified proteins by direct co-immunoprecipitation. Figure 2A demonstrates Jarid1b and CtBP1 by western blotting in anti-ZNF217 immunoprecipitate. Performing anti-CoREST immunoprecipitation, we found ZNF217 detectable by western blotting. Thus, direct interactions for Jarid1b/Plu-1, CtBP1 and CoREST with ZNF217 were confirmed.
Figure 2.
Co-immunoprecipitation confirming the interaction of ZNF217 with the proteins identified in the affinity purified complex. (A) Anti-ZNF217 antibody mediated immunoprecipitation of Jarid1b/Plu-1 and CtBP1, which were detected by western blotting. Anti-CoREST antibody mediated immunoprecipitation of the same proteins as well as of ZNF217. Anti-Suv39H1 western blotting served as a negative control. (B) Anti-ZNF217 antibody-mediated immunoprecipitation of Flag-tagged G9a and LSD1. (C) Northern hybridization analysis of ZNF217 and BRCA1 following RNA interference of ZNF217 and REST with siRNAs in MCF7 cells. (D) Chromatin immunopreciptation (ChIP) of ZNF217 and Jarid1b/Plu-1 within the human BRCA1 locus following RNA interference of ZNF217 with siRNAs.
To confirm ZNF217 interactions with G9a and LSD1, for which good antibodies were unavailable to us at the time of experimentation, we expressed tagged versions of these proteins. Immunoprecitation was again with anti-ZNF217 antibody under conditions of physiological ZNF217 levels (endogenous ZNF217 only). As shown in Figure 2B, specific interactions of G9a and LSD1 with ZNF217 could be confirmed.
Repression of BRCA1 by ZNF217 corresponds with occupation by the BRCA1 locus
To delineate whether BRCA1 is a target of ZNF217 we depleted ZNF217 by RNA interference using short interfering oligoribonucleotides (siRNAs) in MCF7 cells. As shown in Figure 2C, we successfully depleted MCF7 cells of ZNF217 mRNA by measuring poly A+ mRNA using northern hybridization methods. To assure that depletion of ZNF217 was specific, we compared ZNF217 mRNA depletion with oligoribonucleotides specific for knockdown of the REST transcriptional repressor as a control. As shown in Figure 2C, depletion of ZNF217 was commensurate with the elevation in BRCA1 mRNA within MCF7 cells. As suggested previously,9 Thillainadesan et al. revealed BRCA1 as a putative target gene for ZNF217. To assess that the relationship between ZNF217 and regulation of BRCA1 was the result of ZNF217 occupation of the BRCA1 locus we tested both ZNF217 and Jarid1b/Plu-1 to occupy the BRCA1 promoter immediately upstream of the first exon. Our results show that ZNF217 and Jarid1b/Plu-1 can co-occupy the BRCA1 promoter within MCF7 cells (Fig. 2D). Depletion of ZNF217 by siRNAs corresponded with a noticeable depletion in the occupation of BRCA1 by ZNF217, but did not appreciably alter the level of Jarid1b/Plu-1 occupation within the BRCA1 promoter (Fig. 2D). Our results suggest that Jarid1b/Plu-1 occupation of BRCA1 is likely independent of ZNF217 occupation, however, repression of BRCA1 by Jarid1b/Plu-1 is reinforced by ZNF217.
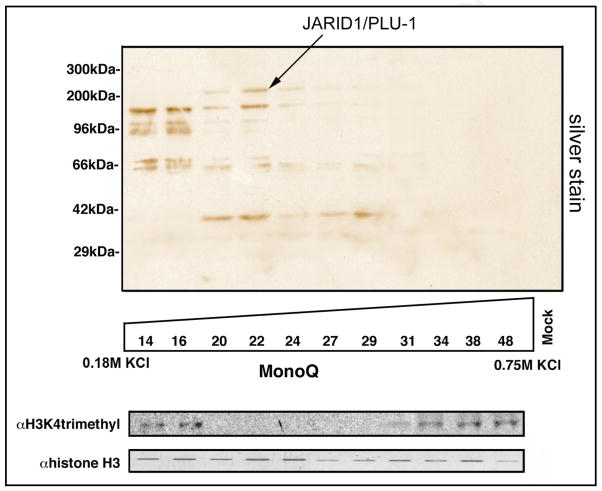
H3K4me3 demethylase activity
Jarid1b/Plu-1 has been shown to function as a histone H3 lysine 4 (H3K4) trimethyl demethylase.19 Therefore, we predicted that the affinity-purified ZNF217 complex containing Jarid1b/Plu-1 would have H3K4 trimethyl (H3K4me3) demethylase activity and that fractionation of the protein-components of the complex by anion-exchange chromatography should lead to co-elution of the protein identified by mass spectrometry as Jarid1/Plu-1 with this activity. Figure 3 demonstrates that this prediction was correct showing loss of H3K4me3 in a me3-specific Western blot, when H3K4me3 was exposed to proteins from the fractions 6–10, in which Jarid1/Plu-1 eluted.
Figure 3.
H3K4me3 demethylase activity of the ZNF217 complex correlating with Jarid1b/Plu-1. The affinity-purified ZNF217 protein complex was fractionated on a MonoQ column. Individual fractions were tested for demethylase activity by incubation with H3K4me3 substrate. Western blotting with an H3K4 trimethyl-specific antibody demonstrates loss of methylation, i.e., presence of demethylase activity in fractions 6–10. This correlates with the presence of Jarid1/Plu-1 indicated in the corresponding silver stained SDS gel.
H3K4me3 is a histone mark that is strongly associated with transcription.20 Correspondingly, demethylation of H3K4 is a critical step in gene repression. Thus, the identification of Jarid1b/Plu-1 demonstrated that the ZNF217 protein complex includes at least one gene-repressive activity raising the question, whether it contains other histone modifying activities that could act synergistically.
H3K9 and H3K27 methylase activity
Gene repression is associated with the gain of certain histone methyl-marks, especially with methylation of H3K9 and H3K27. We tested, whether the affinity purified ZNF217 complex had methylating activity in vitro. The assay used histone tails as substrate. Histone tails consisted either of the normal sequence or were mutated at one or two lysine positions; specifically, the lysine H3K9 and/or H3K27 was/were mutated to arginine, i.e., to H3R9 or H3R27 respectively. Methylase activity was tested with the normal or mutated histone tails serving as substrate. As shown in Figure 4A, we detected marked methylase activity with normal histone tails (positive control). Methylation was abrogated if H3K9 and H3K27 were both mutated (negative control). Methylation was found with the H3R27 mutant demonstrating methylation of the intact H3K9. This is consistent with our finding that the affinity-purified ZNF217 complex contained G9a, the principal H3K9 methylase in euchromatin known to date.18
Figure 4.
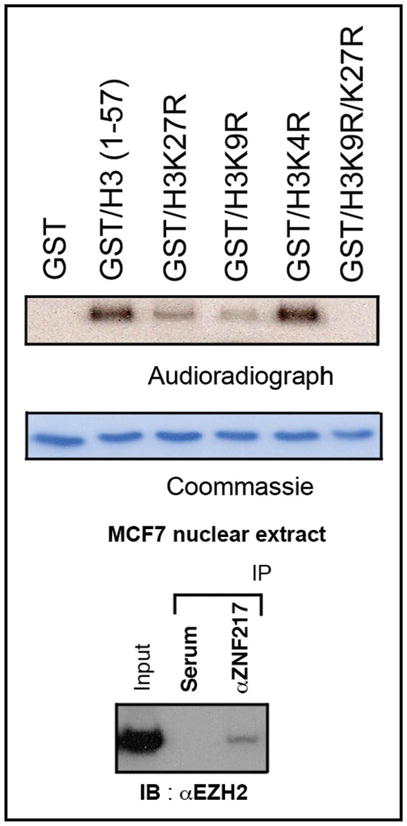
H3K9 and H3K27 methylase activity of the ZNF217 complex. (A) Histone H3 tails (1–57) were incubated with S-adenosyl-L- [methyl-14C]-methionine and aliquots of the affinity purified ZNF217 complex. Autoradiography demonstrated no modification of the GST tag by [14C] (negative control) and marked incorporation of [14C] into the H3 (1–57), i.e., into the native H3K9K27 tail sequence (positive control). Mutation to H3K27R or H3K9R decreased [14C] incorporation while double mutation H3K27R/H3K9R completely abrogated methylation with [14C]. (B) Anti-ZNF217 antibody mediated immunoprecipitation of EZH2, which was detected by western blot. EZH2 can account for H3K27 methylation, an activity that was not explained by any of the ZNF217 binding partners identified in Figure 1.
We also detected H3K27 methylase activity (using the H3R9 mutant; Fig. 4A). G9a has been reported to methylate H3K27,18 but this activity is relatively weak. In our experiment, H3K27 methylation was lower that dual methylation at H3K9 and H3K27 combined (positive control) but comparable in intensity with H3K9 methylation (H3R27 mutant) suggesting that it might not be fully accounted for by G9a. Therefore, we suspected that another protein might be present in the complex, which would be capable of the robust H3K27 methylase activity seen.
EZH2 binding to ZNF217
EZH2 has been reported as the principal H3K27 methylase21 making it a strong candidate. Figure 4B shows immunoprecipitation of ZNF217 with EZH2, confirming interaction and strongly suggesting that it was present in the original affinity purified ZNF217 complex among the unidentified components and is responsible for the H3K27 methylase activity demonstrated in Figure 4A.
Discussion
ZNF217 is an important oncogene in many cancer types (reviewed in the introduction). It impacts cell physiology markedly by shifting the apoptotic threshold of cancer cells causing resistance to the chemotherapeutical agent doxorubicin and contributing to telomere stability and immortalization under certain experimental conditions.22 While these observations are clearly relevant to the understanding of ZNF217’s role(s) in cancer, they represent an indirect effect through ZNF217’s aggregate activity on the large number of genes that it targets.
Little is known about how ZNF217 operates at the level of individual genes, i.e., about its principal mode(s) of molecular action as transcription factor. The present study contributes to the understanding of this area. We demonstrated that ZNF217 forms a nuclear complex that can modify histones. Specifically, we documented H3K4me3 demethylation; H3K9 methylation; and H3K27 methylation. We identified five nuclear proteins contained in the ZNF217 complex, namely Jarid1b/Plu-1, a histone H3 lysine 4 (H3K4) trimethyl demethylase;19 G9a, a principal euchromatic H3K9 methylase;18 EZH2, a H3K27 methylase associated with the Polycomb Repressive Complex 2 (PRC2);21 LSD1, a H3K4 demethylase; CtBP1 and CoREST, which are both transcriptional co-repressors.
Detection of protein-protein interactions can be distorted by forced overexpression of proteins of interest increasing the risk for false-positive results. We avoided such approaches throughout our study. All experiments were performed at physiological protein levels assuring a stoichiometry reflective of the conditions, under which ZNF217 functions normally. Our findings are consistent with two reports about CtBP1 binding to ZNF217,7,23 and with scattered findings in the literature encountering ZNF217 in the context of studies focusing on other molecules (CoREST;24 LSD1,6).
The purification of ZNF217 complexes by an extensive fractionation scheme, to include anionic chromatography, reveals the presence of an alternate assembly of components associated with ZNF217 that include the H3K4me3 demethylase, Jarid1b/Plu-1. Because of the presence of H3K4me3 demethylase activity linked with ZNF217 (Fig. 3), it was difficult to reconcile the concept that ZNF217 remained static with LSD1 as the sole demethylase coupled with ZNF217. In light of novel H3K4me3 demethylase activity revealed from ZNF217 purification (Fig. 2A), several schemes for ZNF217 were tested to resolve H3K4me3 enzymatic activity only to find that anionic fractionation provided greater biochemical resolution of H3K4me3 demethylation from those using ZNF217 affinity chromatography. Our results suggest that the dynamic and promiscuous behavior of ZNF217 is to assemble with multiple histone modifying enzymes and co-regulatory proteins. Classic examples of how nuclear receptors and transcription factors, in general, adopt multiple configurations in the assembly of co-regulatory proteins has been widely accepted25,26 and has even been illustrated directly for LSD1 in adopting new partners during development and tissue specification,27 provide an example of how ZNF217 may operate to organize multiple repression complexes. Our studies to purify novel ZNF217-linked enzyme activities provides some insight as to how dynamic ZNF217 complexes may alternate between several co-regulatory proteins and histone modifying enzymes. Therefore, we envision that ZNF217 overexpression in several tumors types may allow a greater opportunity for ZNF217 to adopt alternate configurations in the assembly of transcriptional regulatory complexes. Such a configuration for ZNF217 to associate with Jarid1b/Plu-1 and EZH2 may further enforce cells to undergo cell transformation and to bypass several tumor-suppressing functions. It is of little surprise that ZNF217 could be linked with Jarid1b/Plu-1 and EZH2 as additional oncoproteins. Our experimental evidence reinforces a model that ZNF217, overall, is identified with additional protein interactions that are linked with specific carcinomas frequently associated with ZNF217 amplification.1,19,28–30
ZNF217 target genes were recently mapped in a ChIP-chip and ChIP-DSL studies.8,9 Quinlan et al. reviewing the study, noted that the “majority of the top-ranked target genes had low levels of RNA expression supporting the hypothesis that ZNF217 is a repressor of transcription and is silencing the promoters, to which it binds.”1 Our results indicate that among many genes associated with ZNF217 repression the conspicuous Jarid1b/Plu-1 target locus, BRCA1,19,31 was a rationale target for ZNF217, as indicated previously.9 Our result indicates that ZNF217 depletion by RNA interference corresponds with elevated BRCA1 mRNA from MCF7 cells. Despite the recent identification of BRCA1 as a putative target of ZNF217 in the one report,9 it was not the focus of further study. Reasoning that BRCA1 may lack the prototypic nucleotide consensus for ZNF217 binding, immediate to the transcription start site, may argue that ZNF217 may carry the capacity of a co-repressor as suggested.1 Thus, ZNF217 activity may interface with Jarid1b/Plu-1 as the primary DNA binding component of a higher-order complex further reinforcing repression of BRCA1 without requiring ZNF217 for Jarid1b/Plu-1 occupation of BRCA1, as indicated (Fig. 2D). Our result indicates that ZNF217 functions as a repressor of BRCA1 directly (Fig. 2C and D). Furthermore, ZNF217 occupies the promoter overlapping the Jarid1b/Plu-1 binding site within the BRCA1 locus. While it remains unclear precisely how ZNF217 may target BRCA1, our evidence indicates that ZNF217 repression may operate with Jarid1b/Plu-1. Although exceptions of ZNF217 repression were noted,1,8,9 these observations at the systems/genomics level appear to be highly consistent with the conclusions derived from the present study by biochemical means, i.e., ZNF217 as an organizer of repression.
Taken together, our findings suggest a model of ZNF217 as a transcription factor assembling a distinct set of histone modifying proteins at target DNA sites resulting in transcriptional repression. The model may be most applicable to malignancies, because all of our studies (as well as most reports on related proteins cited above) were performed in cancer cell lines. Additional studies will be required to understand, whether ZNF217 has similar activities in early development, when it appears to be most highly expressed.32 Overexpression of ZNF217 in the adult organism might contribute to carcinogenesis through reenacting gene repression programs characteristic of early development. Improved understanding of this process might ultimately help to define ZNF217-mediated gene repression as a target for transcription therapy.
Materials and Methods
Cell cultures, nuclear extraction preparation and chromatography
Human mammary adenocarcinoma MCF7 cells were obtained from ATCC (Rockville, MD) and cultured in DMEM (90%) and FBS (10%) with appropriate antibiotics. Antisera against ZNF217 was generated in rabbits using the highly-conserved C-terminal region of the mouse cDNA (nucleotides 2341–2693) expressed in bacteria. The ZNF217 antisera, described in this study, was provided as a generous gift (Gail Mandel, Ph.D., Vollum Institute, OHSU, Portland, OR). Anti-ZNF217 antibody was purified from the polyclonal antisera by affinity purification against a C-terminal mouse Zfp217 polypeptide fragment expressed as a recombinant poly-histidine tagged protein. MCF7 cells were cultured in 22 roller flasks and received as frozen cell pellets from the National Cell Culture Center under contract (Cellex Biosciences, Minneapolis, MN). Nuclear lysate was prepared and affinity chromatography performed using procedures described by this laboratory.13 For affinity chromatography 1 mg of purified antibody was covalently linked to CNBr-Sepharose (GE Life Sciences, Pistcataway, NJ) using the manufacturer’s instructions. Affinity chromatography and anion-exchange (MonoQ) chromatography were performed on an AKTA-purifier (GE Life Sciences, Pistcataway, NJ). Essentially, affinity chromatography was performed in column loading buffer (CLB) containing 20 mM HEPES (pH 7.9), 5% (v/v) glycerol, 5 mM MgCl2, 0.1 mM EDTA, protease inhibitor cocktail and 0.2 mM DTT. Soluble nuclear extracts were suspended and dialyzed in CLB and equilibrated in 40 mM KCl by dialysis. The total amount of nuclear protein loaded was separated by affinity chromatography using an anti-ZNF217 column. Dialyzed materials were immunoprecipitated directly from nuclear extracts with protein A Trisacryl (Thermo-Scientific/Pierce, Rockford, IL) conjugated with anti-ZNF217. After washing with 250–500 ml of CLB containing 0.15 M KCl on a column, the bound proteins were eluted from the column with 20 mM Hepes buffer (pH 7.9) containing 2% SDS. The bound proteins were eluted from the matrix by incubation at room temperature for 5 min. Anionic exchange MonoQ chromatography was conducted with an AKTA-FPLC (GE Life Sciences) using CLB. Typically, 100–200 ml of nuclear extracts were fractionated on a Mono Q HR 5/5 column at a flow rate of 1.0 ml/min and successively eluted through a gradient with 0.75 M KCl in CLB. Fractions were collected and dialyzed against CLB containing 0.15 M KCl for 3 hr. Eluted fractions were evaluated by high-resolution SDS-PAGE and silver stained with the visual bands excised for trypsin digestions, followed by analysis by matrix assisted laser desorption ionization-time-of flight (MALDI-TOF) mass spectrometry (Agilent Technologies, Foster City, CA) using MudPIT analysis.14,15
Metabolic labeling of MCF7 cells with [35S] in vivo
Mammary adenocarcinoma MCF7 cells were metabolically-labeled with [35S]-methionine/cysteine (Perkin-Elmer, Stamford, CT) using procedures described previously.16 Nuclear and cytosolic lysates of [35S]-labeled MCF7 cells were recovered using a nuclear isolation kit (ThemoScientific/Pierce) according to manufacturer’s instructions and immunoprecipitated with anti-ZNF217 rabbit serum and immunoprecipitated with protein A-Sepharose (GE, Piscataway, NJ). Protein immunoprecipitates were fractionated by SDS-PAGE, fluorographed and visualized by autoradiography.
Plasmids and cell transfection
The expression vector for FLAG-G9a was described previously.13 Flag-tagged human LSD1 was created by reverse transcription of poly A+ RNA from spleen and amplification of the open reading frame of LSD1 with primers corresponding to a N-terminal FLAG sequence followed by the ORF of human LSD1. The cDNA was then ligated into the TOPO vector and excised by endonuclease restriction and ligated into pcDNA3.1. MCF7 cells were transfected using lipofectamine (Invitrogen, Carlsbad, CA) according to manufacturer’s instruction.
RNA interference (RNAi)
Depletion of ZNF217 by RNAi was accomplished with siRNAs directed against ZNF217 (OnTargetplus; cat nos. J-004987-09 and J-004987-11) or REST (OnTargetplus; cat no. J-006466-11) and obtained from a commercial source (Thermo Scientific/Dharmacon, Lafayette, CO). Transfection of small interfering RNAs was performed with a prepared transfection reagent kit (DharmaFect; Thermo-Scientific/Dharmacon, Lafayette, CO) and was performed according to manufacturer’s instructions. Essentially, MCF7 cells (approximately 3 × 105 cells per 60 mm plate) were cultured in normal culture medium and then transfected with 100 ml of 2 mM siRNA mixture in to each. Two days following transfections, cells were processed as RNA recovery or cellular protein extracts for further experimentation.
Northern hybridizations
For northern hybridization of ZNF217, BRCA1 and b Actin cDNAs to mRNA, RNA was recovered from MCF7 cells and was isolated with the Oligotex direct polyA+ mRNA isolation kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. RNAs (0.5 mg per lane) were separated on 1% agarose gels containing 0.6% formaldehyde, transferred to Biodyne B nylon filters (Pall Corp., East Hills, NY) and hybridized with a random-primed, 32P-labeled 577 bp Eae1/Spe 1 cDNA fragment of BRCA1 isolated from plasmid pPD21, previously obtained as a generous gift (Peter Meldrum, Myriad Genetics, Salt Lake City, UT). Filters were exposed to X-Ray film and developed. Alternatively, images of filters were obtained digitally through a phosphoimager. Filters were stripped of cDNA probe and subsequently re-hybridized to ZNF217 and b-Actin cDNA fragments.
Immunoprecipitation and immunoblotting
To monitor interactions between ZNF217-associated proteins, MCF7 cells were cultured under normal conditions described above and nuclear lysates prepared. Immunoprecipitations were conducted with 50 μg nuclear lysate from MCF7 cells. Immunoprecipitation was performed in RIPA buffer. Precipitates were resolved by SDS-PAGE. Proteins from SDS-PAGE were then transferred onto PVDF membrane. The following antibodies were used for immunoprecipitation and immunoblots: anti-Suv39H1 (Cell Signalling Technology, Danvers, MA), anti-Plu-1 (cat no. ab50958, AbCam, UK), anti-CoREST (a gift G. Mandel), anti-CtBP1 (H-440) (catalogue no. sc-11390, Santa Cruz Biotechnology, Santa Cruz, CA), anti-EZH2 antibody (catalogue no. ab3748, AbCam, UK). Immunoblots conducted from samples taken from the ion exchange elution were subjected to immunoblot analysis with antibodies directed against either H3K4me3 (AbCam, UK) or total H3 (AbCam, UK).
Chromatin immunoprecipitation (ChIP)
ChIP was performed with the use of the MagnaEZ ChIP kit purchased commercially (Millipore/Upstate Biotech, Temecula, CA). Chromatin was immunoprecipitated by using antisera against ZNF217 and Jarid1b/Plu-1 (Bethyl Laboratories, Montgomery, TX; cat. no. A301-813A). Antisera against histone H3 and histone H4 (Millipore/Upstate Biotech, Temecula, CA) were used as controls to validate input signals for ChIP. Following the immunoprecipitation step, genomic DNA was extracted from immune complexes and amplified by standard end point PCR with primers designed to select for BRCA1 occupation (forward: 5′-CGC ACA GGT CTC CAA TCT ATC C-3′; reverse: 5′-TCG TAA GAA GAG GTC CCA ATC CCC-3′). PCR products were then resolved by gel electrophoresis in 1% agarose and images captured by CCD.
Immunofluorescence microscopy
For immunofluorescence microscopy, cells were fixed with methanol (5 min, 20°C) and acetone (30 sec, 20°C). Incubation with antibodies and DAPI staining was performed essentially as described.17 Dilutions in PBS of the primary antibodies raised during this study were 1:25 for anti-ZNF217.
In vitro histone methylation studies
Methylation analysis using GST-tagged histone H3 tails were performed as described previously.13 Briefly, Immunoprecipitates were collected by centrifugation in microtubes washed four times in phisphate-buffered saline and finally resuspended in MTS buffer (containing 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 0.4 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride). Methyltransferase assays were conducted on GST fusions with wild-type and with mutated histone H3 in which specific residues were mutated as indicated in the figure legend. Some of these reagents were supplied as generous gifts (Y. Shinkai, Kyoto University, Kyoto, Japan) and are described in detail.18 Glutathione agarose-purified histone H3 proteins were incubated with individual immunoprecipitates with S-adenosyl-L-[methyl-14C]-methionine (Perkin-Elmer) in 30 ml of MTS buffer for 45 min at 30°C. Reactions were terminated by addition of a protein gel loading buffer containing 6 M urea and SDS and analyzed by 15% SDS-PAGE and visualized with film.
Acknowledgments
PC041225 (DOD) and K08DK068026 (to M.S.B.); R01HL 067099 (to M.J.W.).
References
- 1.Quinlan KG, Verger A, Yaswen P, Crossley M. Amplification of zinc finger gene 217 (ZNF217) and cancer: when good fingers go bad. Biochim Biophys Acta. 2007;1775:333–40. doi: 10.1016/j.bbcan.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26:6395–402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci USA. 2002;99:7420–5. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–5. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 5.Hakimi MA, Dong Y, Lane WS, Speicher DW, Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J Biol Chem. 2003;278:7234–9. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 6.Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–64. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 7.Quinlan KG, Nardini M, Verger A, Francescato P, Yaswen P, Corda D, et al. Specific recognition of ZNF217 and other zinc finger proteins at a surface groove of C-terminal binding proteins. Mol Cell Biol. 2006;26:8159–72. doi: 10.1128/MCB.00680-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krig SR, Jin VX, Bieda MC, O’Geen H, Yaswen P, Green R, et al. Identification of genes directly regulated by the oncogene ZNF217 using chromatin immunoprecipitation (ChIP)-chip assays. J Biol Chem. 2007;282:9703–12. doi: 10.1074/jbc.M611752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thillainadesan G, Isovic M, Loney E, Andrews J, Tini M, Torchia J. Genome analysis identifies the p15ink4b tumor suppressor as a direct target of the ZNF217/CoREST complex. Mol Cell Biol. 2008;28:6066–77. doi: 10.1128/MCB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowger JJ, Zhao Q, Isovic M, Torchia J. Biochemical characterization of the zinc-finger protein 217 transcriptional repressor complex: identification of a ZNF217 consensus recognition sequence. Oncogene. 2007;26:3378–86. doi: 10.1038/sj.onc.1210126. [DOI] [PubMed] [Google Scholar]
- 11.Sun Y, Wong N, Guan Y, Salamanca CM, Cheng JC, Lee JM, et al. The eukaryotic translation elongation factor eEF1A2 induces neoplastic properties and mediates tumorigenic effects of ZNF217 in precursor cells of human ovarian carcinomas. Int J Cancer. 2008;123:1761–9. doi: 10.1002/ijc.23708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epi-genetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–28. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 13.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci USA. 2004;101:11257–62. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graumann J, Dunipace LA, Seol JH, McDonald WH, Yates JR, 3rd, Wold BJ, et al. Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol Cell Proteomics. 2004;3:226–37. doi: 10.1074/mcp.M300099-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, et al. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–11. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, et al. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274:7803–15. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 17.Qian C, Zhang Q, Li S, Zeng L, Walsh MJ, Zhou MM. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol. 2005;12:1078–85. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
- 18.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–17. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 19.Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–12. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 21.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Huang G, Krig S, Kowbel D, Xu H, Hyun B, Volik S, et al. ZNF217 suppresses cell death associated with chemotherapy and telomere dysfunction. Hum Mol Genet. 2005;14:3219–25. doi: 10.1093/hmg/ddi352. [DOI] [PubMed] [Google Scholar]
- 23.Kuppuswamy M, Vijayalingam S, Zhao LJ, Zhou Y, Subramanian T, Ryerse J, et al. Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol Cell Biol. 2008;28:269–81. doi: 10.1128/MCB.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.You A, Tong JK, Grozinger CM, Schreiber SL. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc Natl Acad Sci USA. 2001;98:1454–8. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- 26.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–18. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–7. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- 28.Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sellers WR, Loda M. The EZH2 polycomb transcriptional repressor—a marker or mover of metastatic prostate cancer? Cancer Cell. 2002;2:349–50. doi: 10.1016/s1535-6108(02)00187-3. [DOI] [PubMed] [Google Scholar]
- 30.Barrett A, Madsen B, Copier J, Lu PJ, Cooper L, Scibetta AG, et al. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: a new cancer/testis antigen? Int J Cancer. 2002;101:581–8. doi: 10.1002/ijc.10644. [DOI] [PubMed] [Google Scholar]
- 31.Scibetta AG, Santangelo S, Coleman J, Hall D, Chaplin T, Copier J, et al. Functional analysis of the transcription repressor PLU-1/JARID1B. Mol Cell Biol. 2007;27:7220–35. doi: 10.1128/MCB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]