Abstract
We introduce a system developed for the BioCreativeII.5 community evaluation of information extraction of proteins and protein interactions. The paper focuses primarily on the gene normalization task of recognizing protein mentions in text and mapping them to the appropriate database identifiers based on contextual clues. We outline a “fuzzy” dictionary lookup approach to protein mention detection that matches regularized text to similarly-regularized dictionary entries. We describe several different strategies for gene normalization that focus on species or organism mentions in the text, both globally throughout the document and locally in the immediate vicinity of a protein mention, and present the results of experimentation with a series of system variations that explore the effectiveness of the various normalization strategies, as well as the role of external knowledge sources. While our system was neither the best nor the worst performing system in the evaluation, the gene normalization strategies show promise and the system affords the opportunity to explore some of the variables affecting performance on the BCII.5 tasks.
Index Terms: biomedical natural language processing, information extraction, gene normalization, text mining
1 INTRODUCTION
BioCreative II.5 (BCII.5) addressed three text mining tasks in the molecular biology domain, evaluating systems performing article classification, interactor normalization, and interaction pair extraction. The evaluation thus pursued the same classes of tasks as BioCreative II [1], but increased the complexity of each task in various ways, in particular adding a focus on proteins that are specifically identified as participating in an experimentally validated interaction supported by the publication. This narrowing of the set of relevant entities while simultaneously increasing the ambiguity in the task through the use of (a) full text for all tasks, and (b) not providing the target species for gene normalization, resulted in a significantly more challenging evaluation than the previous incarnation of the tasks.
The Center for Computational Pharmacology approach to information extraction for the BioCreative II.5 challenge takes advantage of our open-source tools for Biomedical Natural Language processing1 (BioNLP), specifically the OpenDMAP system [2] and the BioNLP-UIMA framework [3], built on the Apache Unstructured Information Management Architecture2 [4]. We submitted responses to the gene normalization task (INT) and the interaction pair task (IPT).
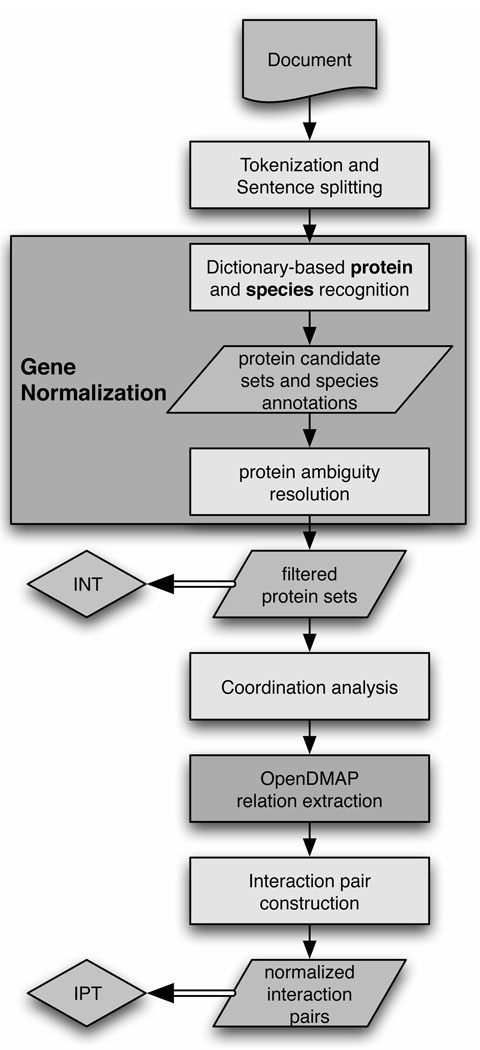
In this paper, we describe our system and introduce several different strategies for approaching gene normalization based on organism mentions in the text. We will explore the implications of various extensions to the original system. The architecture of our system can be seen in Figure 1; each component will be described in more detail below.
Fig 1.
The system architecture of our BioCreative II.5 submission.
2 GENE NORMALIZATION
The participants of BCII.5 tackled what has been called “inter-species” gene normalization, referring to the lack of a priori specification of the relevant target species for proteins of interest. In contrast, the gene normalization task in BioCreative II (BCII) [5] was a significantly easier task than the BCII.5 task in that the BCII normalization task was constrained to human genes and that it considered abstracts rather than full text publications. The best results on this harder task (by the Hakenberg et al. system; Raw F score of 0.429; mapped and filtered F score of 0.551) therefore are lower than the best results from the gene normalization task in the previous evaluation (F score of 0.81), even with the corrections used to generate the adjusted scores (see Section 4).
Our approach to gene normalization consists of a two-step process of (1) dictionary lookup of gene names using the Swiss-Prot subset of the UniProt database and (2) ambiguity resolution of competing candidates utilizing various document-internal clues including, most prominently, the species references identified in the document. This two-step process allows us to identify a set of candidates for the gene mentions in the text using simple string matching techniques, and then use other information to filter that set and ultimately select a small number (in most cases just one) of likely possibilities from among the candidates. The two steps are described in more detail below.
2.1 Dictionary-based Protein Name Matching
Our approach to recognizing gene mentions in text is dictionary based. We directly search the text for all matches to any name associated with a UniProt entry, without a separate gene mention detection step (see Section 6.1).
To incorporate the requirement for relaxed dictionary matching, since exact string matching has been shown to perform worse than “fuzzy” matching [6], we regularize both the name (or synonym) terms in the UniProt database and each sentence in the input text. We make the string lowercase, eliminate punctuation such as apostrophes, hyphens, and parentheses, convert Greek letters and Roman numerals to a standard form, and finally remove spaces, following previous regularization strategies. We load all UniProt names and synonyms into a trie data structure, and then attempt to match substrings of the normalized sentence to the strings stored in the trie. To reduce false positives, we begin matches into the trie only at token boundaries. We further require the matches to end on a token boundary, unless there is a plural in which case the right token boundary constraint is relaxed slightly.
As an example, the sentence Affixin/beta-parvin is an integrin-linked kinase (ILK)-binding focal adhesion protein highly expressed in skeletal muscle and heart. is regularized to affixinbparvinisanintegrinlinkedkinase ilkbindingfocaladhesionproteinhighly expressedinskeletalmuscleandheart and then the substring Affixin is matched to the dictionary entry affixin (Uniprot:Q9HBI1), beta-parvin is matched to bparvin (Uniprot:Q9HBI1), and ILK is matched to the dictionary entry ilk (Uniprot:P57044), inter alia. Other substring matches are prevented due to the boundary constraints. The effect of the regularization is to enable dictionary matching that is tolerant to variations in case, punctuation, and spacing. Thus, beta-parvin, β-parvin, B parvin and similar variants will all match bparvin (Uniprot:Q9HBI1).
The dictionary that we utilize for the BCII.5 evaluation is comprised of names and synonyms extracted from the UniProt database files specified by the challenge organizers, specifically UniProt release 14.8 from February 10, 20093. We restrict our dictionary to only the Swiss-Prot subset of that release, due to memory issues in loading the dictionary when including the TrEMBL subset. We extract all names associated with each database entry: full names or short names, recommended names or alternative names, allergen names or the name as used in a biotechnological context, etc. We also extract and associate the organism, represented as an NCBI Taxonomy database identifier, for each UniProt ID.
The dictionary loading utilizes a stop word list to prevent the loading of (complete) terms in the dictionary that were found to contribute to false positive matches. These were derived from manual inspection of a list of frequency-sorted false positive matches over the training data. A biologist reviewed all false positive results that were matched more than once (a list of about 300 terms) and determined whether the term referred to a protein, didn’t refer to a protein, or was indeterminable. Terms that did not refer to proteins were added to the stop list. Terms that did refer to proteins or that were indeterminable were excluded from the stop word list, as well as any term with a frequency of one; this decision was made to promote recall rather than precision. Examples of terms that were added to the stop word list include general terms for molecules such as ”protein”, ”DNA”, and ”terminus”, variations of DNA sequences such as ”GGT”, short and highly polysemous acronyms such as ”ER”, ”B”, and ”E2”, and other non-protein terms, such as ”impact”, ”real time” and ”similar”.
Due to protein name ambiguity, after the protein name matching step it is common for there to exist multiple UniProt IDs associated with a single span of text. Each group of IDs corresponding to the same span of text is organized into a single set of protein candidates, or an ambiguity set, for downstream processing.
2.2 Dictionary-based Species Name Matching
The dictionary lookup of species names is done using a component from the UIMA framework repository sandbox [4] called the ConceptMapper4. The dictionary is derived from the NCBI Taxonomy OBO file5, as downloaded on October 27, 2008. The OBO file was mapped to the ConceptMapper dictionary format, with names and IDs preserved for all levels of the taxonomy. This module uses exact dictionary matching. We generalize from organisms at the sub-species level to the appropriate organism at the species level for subsequent species analysis, using the hierarchical structure of the taxonomy.
The dictionary was edited slightly to remove the taxonomy category names (“family”, “genus”, etc.). A few entries that led to numerous false positives were also removed, specifically NCBITaxon:37965 ”hybrid”, NCBITaxon:169495 ”This”, NCBITaxon:28384 ”other sequences”, NCBITaxon:32644 ”unidentified”, NCBITaxon:32630 ”synthetic construct”, and NCBITaxon:45202 ”unidentified plasmid”.
We observe that somewhat more sophisticated techniques for species name detection have been explored, and in future work we will consider integrating such methods. For instance, [7] start with a simple dictionary look-up for species mentions involving minimal string regularization (e.g. removing hyphens) but then augment it with statistical analysis based on analysis of a background data set. However, [8] argue that since the ambiguity of species words is low, a simple dictionary look-up method is adequate. That work includes additional species terminology resources; in the case of [7] they include cell line information, and [8] add species terms from UniProt. Unlike that work, we do include the full NCBI Taxonomy hierarchy, though the utility of matches to terms above the genus level is a question for future work.
2.3 Protein Ambiguity Resolution
Each set of protein candidates at a given span is processed to select a subset corresponding to the most likely document-relevant proteins based on contextual clues. That is, given a string such as ”Beta parvin” which has matched both UniProt:Q9HBI1, a human protein, and UniProt:Q9ES46, a mouse protein, the algorithm must select which of the two matches is more likely to be correct based on information in the document. The primary mechanism for ambiguity resolution used in our system considers explicit mentions of species names in the document, as detected by the dictionary-based species recognition introduced above.
Both global and local strategies for species detection are utilized, and a confidence score is attributed to specific gene normalizations based on what combination of evidence supports the normalization. By global, we refer to a strategy which selects a single relevant species for a document, and uses that to remove any protein candidates in ambiguity sets which do not correspond to that relevant species. A local strategy considers each protein mention in context (i.e. in its immediate linguistic environment, such as within the same sentence or paragraph) and determines the most relevant species for that individual mention. With a local strategy, it is possible for different ambiguous protein mentions in the document to be resolved to proteins associated with different organisms. With the global strategy, this can only occur if none of the protein candidates in an ambiguity set is known to be associated with the global species (as given by the database), in which case a “fallback” normalization strategy is used. This fallback strategy prefers human proteins if available, and otherwise selects randomly (with an appropriate confidence penalty).
Several existing works [9], [10], [11] perform local species disambiguation, assigning each gene mention in the document a mention-specific species tag, while Kappeler et al. [7] focuses on global species disambiguation for the document as a whole. In Wang and Matthews [11], species detection is tackled with a maximum entropy machine learning model based on document context features, such as the words (or more specifically the nouns or adjectives) to the left or right of an entity mention, and species words and IDs identified in the document. Interestingly, they show that adding filtering rules based on immediate local context increases the accuracy of their system. Kappeler et al. consider counts of species mentions within a document, combined with background species probabilities derived from the IntAct protein interaction database6. The algorithm they propose combines the frequency of species mentions within a document with the relative frequency of the species in the IntAct database to achieve a final ranking of document-relevant species.
We experimented with 5 basic strategies that incorporate insights from the previous work. Our global strategies explore different ways of counting species mentions in the document, and the local strategies also consider species mention counts, but are restricted to the local context of a protein mention. Where there are multiple possible protein mentions normalized to a given UniProt identifier, the maximum confidence associated with that normalization anywhere in the document is used in the final output.
Abstract: a global strategy that determines the species most frequently mentioned in the abstract of the article being processed. Protein candidates in ambiguity sets that match the abstract species are retained and given a confidence of 1.0; competing candidates are removed.
First: a global strategy that determines the first species mentioned in the article as a whole. Protein candidates in ambiguity sets that match the first species are retained and given a confidence of 1.0; competing candidates are removed.
Majority: a global strategy that determines the species mentioned most frequently overall in the article. Protein candidates in ambiguity sets that match the majority species are retained and given a confidence of 1.0; competing candidates are removed.
Recency: a local strategy that for each protein determines the most recent previous species mention. Confidence is set based on the distance between the mention of the protein and the mention of the species.
Window: a local strategy that counts species mentions in a window of a given token length prior to the protein mention and uses the most frequent species in that window. Confidence for a mention is set based on the number of competing species in the window.
After experimenting with these strategies using the provided training data, we implemented a sixth Mixed global strategy, which was used in the official submission. This strategy essentially integrates each of the various strategies in a prioritization scheme.
The Mixed strategy sets the confidence of a given normalization in an ambiguity set differently depending on whether the species associated with that normalization has support in the document. Specifically, if a candidate normalization is associated with a species that matches the first species in the document (First strategy), that normalization is assigned a confidence of 1.0. If the candidate is associated with a species that matches the most recent species mention (Recency strategy), that normalization is assigned a confidence of 0.9. If the candidate normalization does not correspond to either the first or the most recent species mention, the strategy defaults to the Majority strategy.
In addition to species mentions, our algorithm also considers abbreviations explicitly identified from the text using the abbreviation detection system of Schwartz and Hearst [12], attempting to relate “short forms” of proteins to longer forms mentioned in the document that can be mapped to a specific UniProt identifier with higher certainty. For instance, if the symbol ”THG-1” has been directly associated with the longer form ”TSC-22 homologous gene-1” (Uniprot:Q9Y3Q8) in the document, as is done in PMID 18325344, then the competing mappings of ”THG-1” to ”tRNA(His) guanylyltransferase” (Uniprot:P53215, inter alia) can be ruled out.
3 PROTEIN INTERACTION EXTRACTION
For the interaction pair task, we employ the concept recognition mechanisms of our OpenDMAP [2] system to search for phrases in the input documents that match pre-defined patterns of expression for various interaction types. OpenDMAP is an ontology-driven concept analysis system that uses a context free pattern language combining text literals, syntactic consituents, and semantically-defined classes of entities. A pattern defines a class – in the case of BioCreative that class is ”protein-protein interaction” – and consists of pattern elements that define both the slots of the class – here, the interacting proteins – and the words that connect those slots in text. OpenDMAP searches for phrases in the input text that match the pre-defined patterns of expression for the defined classes. This pattern-based approach has been shown to result in high-precision extraction of interaction events [13].
We used the patterns from our BioCreative II submission7 [6], with only minor modifications. The pattern set was created manually by inspecting protein-protein interaction instances from the BioCreative II training set, and incorporating corpus frequency information for the interaction trigger words. Patterns consisted of text literals that defined the interaction trigger words (e.g. ”activate”, ”activated”, ”activation”), semantically-defined entities for the interacting protein slots (e.g. the slot ”interactor” was required to be a protein by the ontology), and basal syntactic patterns that characterized the linguistic interstitial material between the protein interactor mentions (e.g. determiners, auxiliary verbs and prepositions). There were about 70 patterns that described the ”protein-protein interaction” class.
We additionally took advantage of a coordination module that we utilized in our BioNLP09 system [13] to support handling of coordinated lists of interactors.
Confidence for the interaction pairs was derived by multiplying the confidence of the two component normalized proteins.
4 OFFICIAL RESULTS
Our system was run on the data selected by the evaluation organizers, the BioCreativeII.5 Elsevier corpus8. This corpus consists of 1190 articles from the journal FEBS Letters that were annotated with structured digital abstracts including formal protein interaction statements [14]. The corpus was split in two halves, for training and testing. The test data was unknown to our system until the official runs were executed. The training data was used to assess the effectiveness of our various gene normalization strategies during development.
Results for both the normalization and interaction pair tasks were evaluated according to the standard information extraction metrics of precision (P), recall (R), and F-score (F). In addition, the organizers calculated a value known as the AUC (area under the curve) of interpolated Precision/Recall curves (iP/R). See http://www.biocreative.org/tasks/biocreative-ii5/biocreative-ii5-evaluation for a detailed description of this measure. It measures the highest possible precision at each achievable recall given a ranked set of results, and therefore provides the opportunity to assess confidence rankings provided in the submitted results. Each of these measures was calculated twice for each result set: first, using ”micro” (”m-”) averaging, which calculates the scores across all documents and then divides by the number of documents, which essentially treats the entire corpus as one large document, and second, using ”macro” (”M-”) averaging, which is the arithmetic mean of the individual document scores. It is also important to explain that scores are calculated only over documents for which some prediction was generated, a maximum of 61 articles for the INT and IPT tasks. If no result was generated by the system for a given document, that document was ignored during scoring. This effectively inflates precision and recall for systems which generate results for less than the full set of articles. The ”Docs” column of each data table indicates how many articles were considered in scoring results.
The organizers additionally introduced two corrections to the scores, homonym ortholog mapping (”HO”) and organism filtering (”OF”). Homology ortholog mapping involves giving credit to protein identifiers that are orthologs of the correct normalization, potentially increasing true positive answers. Organism filtering takes advantage of provided information about which species are relevant to each document and removes protein identifiers associated with an organism that is not known to be relevant to the document, thereby decreasing false positive answers. Both corrections can be applied simultaneously (”HOOF”).
The official results for our submission can be found in Tables 1–2. We were known as team 32. The “server” version of our system was run via web services through the BioCreative meta-server [15], using the “Mixed” gene normalization strategy. The version of the system used for the official submission employed a less fine-grained scheme for setting confidence than described above, using 1.0 for an identifier selected through the primary normalization strategy employed in the run and 0.5 for normalizations selected through the fallback strategy. The “offline” system was nearly identical, but incorporated a slight modification to the confidence values. We see that this change had a very modest impact on the scores; the INT results are slightly better for the offline system and the IPT results are identical, apart from slightly lower iP/R AUC scores for the offline system, reflecting small changes in the confidence ranking that only affect the calculation of iP/R.
TABLE 1.
Official results for our submitted systems on the test data for the INT (normalization) task, offline runs refer to t32_INT_1_test in the formal results and server refers to t32_INT_s07_test. Raw results are the original calculations, “HO” refers to homology mapped results, “OF” refers to organism filtered results, and “HOOF” is both homology mapped and ontology filtered. Lowercase ”m” indicates micro-averaged calculations; uppercase ”M” indicates macro-averaged calculations.
| Run | Scored | Docs | TP | FP | FN | m-P | m-R | m-F | m-AUC | M-P | M-R | M-F | M-AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| offline | INT-raw | 61 | 105 | 1592 | 147 | 0.062 | 0.417 | 0.108 | 0.049 | 0.068 | 0.444 | 0.113 | 0.178 |
| server | INT-raw | 61 | 101 | 1575 | 151 | 0.060 | 0.401 | 0.105 | 0.044 | 0.067 | 0.420 | 0.110 | 0.166 |
| offline | INT-HO | 61 | 110 | 1577 | 142 | 0.065 | 0.437 | 0.113 | 0.053 | 0.071 | 0.461 | 0.118 | 0.183 |
| server | INT-HO | 61 | 106 | 1561 | 146 | 0.064 | 0.421 | 0.110 | 0.048 | 0.070 | 0.437 | 0.115 | 0.170 |
| offline | INT-OF | 58 | 105 | 458 | 140 | 0.187 | 0.429 | 0.260 | 0.180 | 0.232 | 0.467 | 0.268 | 0.337 |
| server | INT-OF | 58 | 101 | 453 | 144 | 0.182 | 0.412 | 0.253 | 0.167 | 0.229 | 0.442 | 0.262 | 0.320 |
| offline | INT-HOOF | 59 | 110 | 456 | 138 | 0.194 | 0.444 | 0.270 | 0.197 | 0.250 | 0.476 | 0.278 | 0.349 |
| server | INT-HOOF | 59 | 106 | 451 | 142 | 0.190 | 0.427 | 0.263 | 0.180 | 0.246 | 0.452 | 0.272 | 0.329 |
TABLE 2.
Official results for our submitted systems on the test data for the IPT (interaction pair) task, offline runs refer to t32_IPT_1_test and server refers to t32_IPT_s07_test.
| Run | Scored | Docs | TP | FP | FN | m-P | m-R | m-F | m-AUC | M-P | M-R | M-F | M-AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| offline | IPT-raw | 29 | 6 | 57 | 82 | 0.095 | 0.068 | 0.079 | 0.007 | 0.123 | 0.101 | 0.103 | 0.086 |
| server | IPT-raw | 29 | 6 | 57 | 82 | 0.095 | 0.068 | 0.079 | 0.011 | 0.123 | 0.101 | 0.103 | 0.091 |
| offline | IPT-HO | 29 | 6 | 57 | 82 | 0.095 | 0.068 | 0.079 | 0.007 | 0.123 | 0.101 | 0.103 | 0.086 |
| server | IPT-HO | 29 | 6 | 57 | 82 | 0.095 | 0.068 | 0.079 | 0.011 | 0.123 | 0.101 | 0.103 | 0.091 |
| offline | IPT-OF | 13 | 6 | 12 | 37 | 0.333 | 0.140 | 0.197 | 0.047 | 0.333 | 0.224 | 0.252 | 0.213 |
| server | IPT-OF | 13 | 6 | 12 | 37 | 0.333 | 0.140 | 0.197 | 0.064 | 0.333 | 0.224 | 0.252 | 0.224 |
| offline | IPT-HOOF | 13 | 6 | 12 | 37 | 0.333 | 0.140 | 0.197 | 0.047 | 0.333 | 0.224 | 0.252 | 0.213 |
| server | IPT-HOOF | 13 | 6 | 12 | 37 | 0.333 | 0.140 | 0.197 | 0.064 | 0.333 | 0.224 | 0.252 | 0.224 |
We note that in these results, the HO mapping correction had very little effect on our scores (and for the IPT task, no effect, though this is likely due to the limited number of results overall), suggesting that confusion between homologous proteins is not a large source of the false positives for our system. The OF adjustment, in contrast, did have a big effect in reducing false positives. Thus it is likely that most of the false positives recorded for our system derive from spurious dictionary matches that identify completely irrelevant proteins.
The system precision on both tasks is very low; on the normalization task recall is substantially higher but still not close to the performance of the best-performing systems. The top systems are able to achieve micro-averaged P/R/F/AUC of 0.30/0.26/0.28/0.10 (team 14) and 0.008/.63/0.015/.245 (team 10), with similar macro-averaged numbers, on the INT task. For the IPT task, the best system achieved macro-averaged performance of 0.128/0.146/0.116/0.128 (team 14), only surpassed by a team that made use of gold standard information and was able to achieve scores of 0.547/0.560/0.502/0.533, and with most teams far below team 14’s results. For the IPT task, our system shows a relatively large difference between the micro-averaging and macro-averaging calculations (with macro-averaged scores higher), indicating that there are likely several documents with a large number of results, on which we are not performing well. Our overall IPT performance was low and reflects the lack of specific development done for BCII.5. We note that organism filtering boosted the IPT scores substantially.
5 EXPERIMENTATION
5.1 System improvements post-submission
After the final submission for the official evaluation, a few changes were implemented in the system. First, the stop word list used by the dictionary matcher was expanded to include a broader range of English words (such as prepositions and other function words). Second, a bug in the initial Dictionary Matcher code that artificially capped the number of possible matches into the dictionary was fixed. This bug could have filtered out some potential True Positive gene candidates. Third, the algorithm for setting confidence of the gene normalizations was adjusted to completely conform with what is described in Section 2.3.
The impact of these changes can be seen by comparing the offline INT-raw run of Table 1 with the “Mixed” line in Table 3. We saw an important reduction in False Positives (1592 to 907), with only a slight loss in True Positives (105 to 95) and a slight increase in False Negatives (147 to 157). The net effect was an improvement in both the micro- and macro-averaged Precision, F, and iP/R AUC scores, with only a small drop in Recall. The comparison of the offline IPT-raw run results of Table 2 with the “Mixed” line in Table 4 also shows a substantial across the board improvement, though there were fewer results overall.
TABLE 3.
INT experiments with raw scores.
| Run | Docs | TP | FP | FN | m-P | m-R | m-F | m-AUC | M-P | M-R | M-F | M-AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mixed | 61 | 95 | 907 | 157 | 0.095 | 0.377 | 0.152 | 0.062 | 0.106 | 0.416 | 0.158 | 0.207 |
| Abstract | 61 | 74 | 952 | 178 | 0.072 | 0.294 | 0.116 | 0.082 | 0.079 | 0.316 | 0.118 | 0.231 |
| First | 61 | 85 | 930 | 167 | 0.084 | 0.337 | 0.134 | 0.095 | 0.093 | 0.361 | 0.139 | 0.248 |
| Recency | 61 | 98 | 971 | 154 | 0.092 | 0.389 | 0.148 | 0.058 | 0.095 | 0.424 | 0.146 | 0.173 |
| Majority | 61 | 95 | 907 | 157 | 0.095 | 0.377 | 0.152 | 0.132 | 0.106 | 0.416 | 0.158 | 0.297 |
| Window15 | 49 | 55 | 684 | 138 | 0.074 | 0.285 | 0.118 | 0.033 | 0.082 | 0.301 | 0.121 | 0.128 |
| Window30 | 49 | 59 | 684 | 134 | 0.079 | 0.306 | 0.126 | 0.045 | 0.087 | 0.320 | 0.129 | 0.144 |
| DefaultHuman | 61 | 74 | 952 | 178 | 0.072 | 0.294 | 0.116 | 0.082 | 0.079 | 0.316 | 0.118 | 0.231 |
| NoGN | 61 | 119 | 15550 | 133 | 0.008 | 0.472 | 0.015 | 0.044 | 0.017 | 0.530 | 0.031 | 0.139 |
| CellLine (OT) | 61 | 50 | 962 | 202 | 0.049 | 0.198 | 0.079 | 0.015 | 0.053 | 0.214 | 0.079 | 0.092 |
| CellLine (PBT) | 61 | 46 | 885 | 206 | 0.049 | 0.183 | 0.078 | 0.015 | 0.053 | 0.218 | 0.081 | 0.089 |
| EntrezGene | 61 | 101 | 3333 | 151 | 0.029 | 0.401 | 0.055 | 0.024 | 0.036 | 0.429 | 0.063 | 0.112 |
TABLE 4.
IPT experiments with raw scores.
| Run | Docs | TP | FP | FN | m-P | m-R | m-F | m-AUC | M-P | M-R | M-F | M-AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mixed | 15 | 5 | 19 | 54 | 0.208 | 0.085 | 0.120 | 0.019 | 0.194 | 0.139 | 0.141 | 0.101 |
| Abstract | 15 | 3 | 18 | 56 | 0.143 | 0.051 | 0.075 | 0.008 | 0.122 | 0.050 | 0.060 | 0.029 |
| First | 15 | 5 | 19 | 54 | 0.208 | 0.085 | 0.120 | 0.019 | 0.194 | 0.139 | 0.141 | 0.101 |
| Recency | 15 | 5 | 19 | 54 | 0.208 | 0.085 | 0.120 | 0.021 | 0.194 | 0.139 | 0.141 | 0.102 |
| Majority | 15 | 5 | 19 | 54 | 0.208 | 0.085 | 0.120 | 0.019 | 0.194 | 0.139 | 0.141 | 0.101 |
| Window15 | 3 | 1 | 2 | 16 | 0.333 | 0.059 | 0.100 | 0.020 | 0.333 | 0.042 | 0.074 | 0.042 |
| Window30 | 5 | 1 | 4 | 19 | 0.200 | 0.050 | 0.080 | 0.010 | 0.200 | 0.025 | 0.044 | 0.025 |
| DefaultHuman | 15 | 3 | 18 | 56 | 0.143 | 0.051 | 0.075 | 0.008 | 0.122 | 0.050 | 0.060 | 0.029 |
| NoGN | 15 | 7 | 4578 | 52 | 0.002 | 0.119 | 0.003 | 0.004 | 0.046 | 0.228 | 0.037 | 0.054 |
| CellLine (OT) | 15 | 4 | 18 | 55 | 0.182 | 0.068 | 0.099 | 0.013 | 0.106 | 0.131 | 0.109 | 0.060 |
| CellLine (PBT) | 13 | 3 | 17 | 51 | 0.150 | 0.056 | 0.081 | 0.009 | 0.083 | 0.112 | 0.087 | 0.050 |
| EntrezGene | 24 | 4 | 65 | 78 | 0.058 | 0.049 | 0.053 | 0.004 | 0.051 | 0.036 | 0.041 | 0.032 |
| BasePatterns | 16 | 5 | 20 | 57 | 0.200 | 0.081 | 0.115 | 0.018 | 0.182 | 0.130 | 0.132 | 0.095 |
| GenericPatterns | 31 | 6 | 82 | 123 | 0.068 | 0.047 | 0.055 | 0.003 | 0.072 | 0.078 | 0.068 | 0.050 |
| NoCoord | 13 | 5 | 14 | 49 | 0.263 | 0.093 | 0.137 | 0.026 | 0.237 | 0.160 | 0.168 | 0.118 |
We also introduced an additional global gene normalization called ”Default Human” for comparison purposes. This strategy always prefers normalizations corresponding to human proteins in the case of ambiguity.
5.2 Impact of normalization strategy
To better understand the impact of the various normalization schemes, we ran the updated system over the test data in turn with each normalization strategy. We see from the top sections of Tables 3 and 4 that choice of the normalization strategy does affect the overall system performance, with the Majority strategy and the Mixed strategy identical on the test data for every score apart from the INT task iP/R AUC scores. This latter difference is due to the differences in confidence ranking of these two strategies. The similar performance of these two strategies was determined to be the result of a programming error which prevented filtering in the Mixed prioritization scheme: when there was a match of first or recent species in the ambiguity set, other candidates were not removed; only confidence values were affected. Thus the filtering all happened via the Majority strategy.
The local strategies, Recency and Window, performed worse on this data set with the current confidence settings, though the differences for P/R/F-score are modest. These local strategies had performed a bit better than the global strategies on the training data.
The Default Human strategy and the Abstract strategy showed identically poor performance on both tasks, suggesting that there were few significant species mentions identified in the paper abstracts, and that the system had to use the fallback strategy of preferring human proteins. The poor performance of the Default Human strategy also indicates that the test corpus is probably quite diverse in its coverage of organisms, and certainly is not biased to human proteins.
To establish a baseline for comparison, we also performed a run which attempted no gene normalization whatsoever – that is, it did no filtering of the ambiguous protein sets resulting from the dictionary lookup stage and simply allowed all protein candidates to persist to the output. This predictably resulted in a large increase in recall at the expensive of precision, with the system producing over 15 times the number of false positives of any of the other normalization strategies. These results appear as the “NoGN” run in the table. The fact that the large increase in recall was not even larger, and in fact still well below the highest recall produced by any submission in the evaluation (0.627 m-R and 0.683 M-R for t10_INT_RUN_1_test as compared with our NoGN scores of 0.472 m-R and 0.530 M-R) indicates that the dictionary matching was not successful in identifying a large quantity of protein mentions, effectively imposing a ceiling on the performance of the normalization algorithms.
Comparing the absolute numbers of true positives of the NoGN run with the basic normalization strategies (119 vs. ~95) also shows that our best normalization strategies are apparently only eliminating about 10% of the true positive mentions possible with our dictionary matching methodology, while significantly reducing false positives (though clearly there are many more to be removed).
It is interesting to observe that the performance of the run without gene normalization performs similarly to the top-ranked normalization system in BCII.5, in terms of the scores other than AUC. While we mentioned above that the NoGN run was still below the highest recall due to dictionary limitations, the m-F score is identical to the t10_INT_RUN_1_test run in the official results, and the NoGN M-F score is actually higher (0.024 vs. 0.031). Both systems exhibit relatively high recall and extremely low precision. It would appear that the team 10 submission is doing little more than dictionary lookup augmented with an effective ranking scheme, and it is not obvious that this is a desirable solution to the gene normalization problem in general.
5.3 Impact of additional knowledge modules
Having recognized that the dictionary matching was not recognizing sufficient protein mentions, and observing that the high-performing submissions to the INT task all seemed to augment the Swiss-Prot dictionary with EntrezGene9 terms, we augmented our initial dictionary with names extracted from EntrezGene and associated with UniProt identifiers via the UniProt idmapping file10. As shown in Table 3 for the run labeled “EntrezGene”, this only resulted in 6 additional true positive normalizations beyond the comparable Mixed run using the SwissProt only dictionary, while the number of false positives skyrocketed. Since other systems did appear to have positive benefit from adding this data, this could reflect a problematic interaction between the normalization strategy employed in our system and the data in EntrezGene, but at the very least it indicates that simply adding more names to a dictionary is not always beneficial.
Similarly, we noted that Kappeler et al. [7], as well as several other participants in the evaluation, utilized the Cell Line Knowledge Base11 to provide additional species clues. We therefore introduced a ConceptMapper-based module for recognizing cell line names to the dictionary lookup portion of our pipeline, and mapped each recognized cell line mention to the appropriate NCBI Taxonomy identifier for the organism that it is associated with. These mentions were then treated as species mentions within the gene normalization algorithms. For the reported experiments, we utilized the Mixed strategy.
As we were experimenting with the cell lines, we realized that many of them contained important punctuation that was ignored by the simple tokenizer that we were using for both dictionary and text processing, the basic OffsetTokenizer that comes with the ConceptMapper. We therefore substituted the tokenizer with the Penn Bio Tokenizer12, which does preserve punctuation, and ran again. The two results are runs “CellLine (OT)” for the OffsetTokenizer and “CellLine (PBT)” for the Penn Bio Tokenizer in Tables 3 and 4. In both cases, the number of true positive matches was reduced and the number of false negatives was increased. It seems clear that this simple, unfiltered and uncurated, addition of cell lines had a negative impact on system performance.
5.4 Impact of coordination resolution
In previous work [13], we introduced a constituent parser and associated modules for identification and handling of coordination structures in the text. For the BioNLP09 shared task described in that paper, we found that this coordination handling had a positive impact on performance, as it allowed us to produce distributed readings for interaction sentences involving coordinated proteins. To measure the effect of coordination on this task, we created a variant of the system removing coordination from the pipeline. This run is shown as “NoCoord” in the Table 4 (it is left out of table 3 as coordination has no effect on normalization). We see that in this particular case, removing the coordination handling module actually leads to a better precision and recall than the standard IPT run. This is likely because the patterns used by OpenDMAP for the BCII.5 IPT task were not written (originally for BCII) with a separate coordination module in mind, and so several of the patterns already accommodate coordinated structures. As an example, consider the pattern: c-interact := interact-noun preposition the? [interactor1] and the? [interactor2]; which would match a phrase such as the association of Protein-X and Protein-Y. The coordination module would in these cases be mirroring the effect of those patterns, therefore not providing positive benefit, with the added negative impact of errors in the coordination handling propagating through to the output.
5.5 Impact of information extraction patterns
In reviewing our submitted system, we identified some missing or misspelled interaction verbs in the original set of OpenDMAP patterns. We added these in and re-ran the system, with the results shown as run “BasePatterns”. The results indicate that these additions only resulted in false positives and false negatives, without any more true positive interaction pairs identified.
To compare our highly structured existing patterns with a looser, co-occurrence based approach, we created a variant of our system which augmented the linguistically specific patterns with a highly generic pattern that simply looks for two protein mentions with an interaction verb in between them. This run is shown as “GenericPatterns” in the table. While this run did find one additional true positive interaction pair, it also had quite a lot more false positives and false negatives, and overall performance was worse. This supports our strategy of more careful construction of patterns of expression.
The Hakenberg et al. system, which performed quite well on this task, utilizes a set of learned OpenDMAP patterns under the hood. This suggests that the relatively low number of patterns in our submission, rather than the general approach, is to blame for our low IPT performance.
6 DISCUSSION
6.1 The role of gene mention detection in gene normalization
We note that many systems approaching the gene normalization task in BCII.5, and the original BCII task, utilize a strategy of (1) performing named entity recognition on the text to detect gene/protein mentions, generally using a machine learning approach to recognize strings in the text that are very likely to correspond to gene mentions and (2) mapping those mentions to a UniProt identifier. In the context of the gene normalization task, where the system must eventually map gene mentions to some database entry via its known names or synonyms in any case, it does not superficially appear to provide much benefit to include a separate gene mention step. It introduces an extra processing step which can slow response time. More importantly, it limits the database search to the algorithmically identified gene mentions, meaning that the identification of genes in the document is subject to the false negatives of the gene mention system – any mention not picked out by the mention detection system will not be recognized.
On the other hand, a separate gene mention step does give the possibility of doing “lossier” mapping into the protein name space of the underlying database. That is, rather than doing only simple surface regularization as we have done, the dictionary lookup can involve more computationally complex algorithms, for instance utilizing a distance metric such as edit distance or the dice coefficient [5] to attempt to identify a close dictionary term. While it is not computationally feasible to do this for every substring of the document, if likely gene mentions have previously been picked out, this is viable and may ultimately give higher recall, effectively offsetting the loss of recall stemming from the false negatives inherent in the gene mention system.
Baumgartner et al. [3] explored the interactions between gene mention and gene normalization systems, and found that the performance of a gene normalization system is largely reflective of the recall, and less on the precision, of the underlying gene mention system. They argue that this suggests that the gene normalization system is itself filtering out many false positive gene mentions. This could be seen as an argument in favor of including a separate – high recall – gene mention system, in that the important function it is serving is to identify candidate substrings of the document to be mapped into the database. A thorough exploration of the benefits of the lossier dictionary matching that the gene mention step allows, as compared with the gain in positive matches resulting from avoiding missed mentions, is warranted to understand this choice better. This is particularly true in light of the good performance of systems such as ProMiner [16], which also does not utilize a separate gene mention step.
6.2 The importance of confidence ranking
The differences in performance among the variations of the system as measured by Precision, Recall and F-score were relatively modest in most cases. Where we see larger variation is in the iP/R AUC scores, and where two system variants may have identical performance on the absolute measures, the AUC measure can show a large difference. In Table 3, we observe the difference in the Mixed and Majority scores: micro AUCs of 0.062 and 0.132, respectively, and macro AUCs of 0.207 and 0.297, respectively, with the Majority AUC scores higher than any other system variant. In both cases, all other scores are identical and so the difference can only be attributed to the confidence ranking.
The specific confidence values associated with the various strategies, and the prioritization scheme developed for the Mixed strategy, were set through manual experimentation with the training data. The values were essentially selected in an ad hoc manner. Given the sensitivity of the scores to the confidence ranking, the obvious thing to attempt here would be to incorporate machine learning into the framework so that a model combining and weighting the various strategies can be constructed empirically from the data. This would also support determining the optimal window size for local clues. Experimentation with machine learning is planned for future work.
6.3 The role of knowledge resources
The performance of the experiments in which we supplied additional knowledge to the system is strikingly poor. Expanding the dictionary to include EntrezGene names and the introduction of extra species clues from the Cell Line Knowledge Base did not achieve the performance improvements that they were intended to produce, but rather resulted in inferior performance. While we have not yet performed separate analysis on these modules to understand why they introduced errors, we can guess that there are terms in each of those two sources that cause false positive matches. The stopword list used when processing the protein name dictionary would likely need to be expanded to include additional terms that are problematic for Entrez Gene. Similarly, there is likely a set of problematic terms in the cell line dictionary – either cell line names that overlap with common English words, or names that are inappropriately linked to particular organisms. This explanation is plausible given that other systems did benefit from inclusion of these resources, albeit with some filtering or with the use of ”black lists”.
What can be learned from these experiments is that simple augmentation of system knowledge resources without careful examination of those resources and assessment of their impact on system performance is not advisable because this could have undesirable side effects. Knowledge quality – and in particular the quality of the knowledge resources for the application context – must be considered. Taking knowledge developed for one purpose (e.g. database construction) and deploying it for another purpose (e.g. text mining) should not be done blindly. We even see that modifying knowledge resources that we are more familiar with can have unexpected consequences – as in the case of enlarging the list of interaction verbs for the information extraction patterns (“Base Patterns” experiment), which actually led to a slight performance decrease.
6.4 Experimentally validated interactions
Our system did not directly address the requirement to identify proteins involved in experimentally validated interactions as expressed in the document. It is likely that a number of the system’s false positive mentions result from this limitation – that is, the system picked up on valid gene mentions that are not involved in specific interactions. We considered limiting the submitted gene normalizations to only those genes that had been identified in the IPT task as interacting, but since our IPT system shows very low recall, and returns results for less than a quarter of the relevant documents in the corpus, this approach would overly restrict recall of the gene normalization system. Given improvements in the IPT system, however, this would be one approach to consider to implement the requirement.
7 CONCLUSION
We have introduced and experimented with a set of strategies for gene normalization that take advantage of document-internal clues for organisms relevant to mentioned proteins. The performance of these algorithms suggest that there is value in exploiting species references for the purpose of gene normalization, but that some references will be more significant than others. In this work, we found that a document-wide, global influence on gene normalization was more important than local clues, though we feel that experimentation with an empirically derived model that weights different clues will likely lead to a more accurate system, in particular due to the importance of confidence ranking for the primary iP/R AUC score. Finally, this work provides some important evidence that simple inclusion of knowledge resources without assessment of their quality in the context of use can be detrimental to overall system performance.
ACKNOWLEDGMENTS
This work was supported by NIH grants 5R01LM009254 and 2R01LM008111 to Lawrence Hunter and grant 1R01LM010120-01 to Karin Verspoor. The authors would also like to thank Florian Leitner at CNIO for his patience in responding to myriad questions about the evaluation. Thank you to Fabio Rinaldi who shared mappings from species names listed in the Cell Line Knowledge Base to NCBI Taxonomy identifiers. We also wish to thank our undergraduate research intern César Mejia Muñoz for his assistance in exploring the parameter space of the system, and our collaborator Philip Ogren for use of his coordination analysis module.
Biographies

Karin Verspoor has been a Research Assistant Professor in the Center for Computational Pharmacology at the University of Colorado Denver since March 2008. She is also a core faculty member of the Computational Bioscience graduate program at UCD. Prior to arriving in Colorado she was a Technical Staff Member at Los Alamos National Laboratory, and held positions as a computational linguist for two different startup companies. She has worked in biomedical natural language processing since 2003.

Christophe Roeder is a software engineer at the Center for Computational Pharmacology at the University of Colorado School of Medicine. He completed his M.S. in Computer Science at the University of Colorado, Denver in 2006. After two decades as a software developer in industry, he is involved in general software development, configuration management, and investigating ways to scale natural language processing.

Helen L. Johnson has been a research assistant in the Center for Computational Pharmacology at the University of Colorado School of Medicine since 2005. Her research focuses on the computational linguistic aspects of biomedical text mining, in particular, named-entity recognition and normalization, and pattern-based information extraction systems. She received an M.A. in Linguistics with an emphasis in Human Language Technology from the University of Colorado in 2004.

K. Bretonnel Cohen leads the Biomedical Text Mining Group at the Center for Computational Pharmacology in the University of Colorado School of Medicine. He is a popular speaker at biomedical text mining and computational biology events and is the chairman of the Association for Computational Linguistics’ SIGBIOMED special interest group on biomedical natural language processing.

William A. Baumgartner Jr. is a member of the Center for Computational Pharmacology at the University of Colorado School of Medicine, where he currently serves as lead software engineer. He received his M.S.E. in Biomedical Engineering from Johns Hopkins University in 2001. His current research focuses on the application and evaluation of natural language processing and semantic integration technologies in the biomedical domain.

Lawrence E. Hunter is Professor in the Department of Pharmacology of the University of Colorado Denver, and Director of both the Center for Computational Pharmacology and the Computational Bioscience graduate program at UCD.
Footnotes
The Swiss-Prot database is available at http://www.uniprot.org/downloads.
The BioCreative II pattern set is available at http://sourceforge.net/projects/opendmap in the ”Supplemental Files” module.
The BCII.5 data is available at http://www.biocreative.org/resources/corpora/elsevier-corpus/.
Available from ftp://ftp.ncbi.nih.gov/gene/.
available from http://u-compare.org/components/components-syntactic_tools.html.
REFERENCES
- 1.Krallinger M, Morgan A, Smith L, Leitner F, Tanabe L, Wilbur J, Hirschman L, Valencia A. Evaluation of text-mining systems for biology: overview of the second BioCreative community challenge. Genome Biology. 2008;vol. 9 Suppl 2:S1. doi: 10.1186/gb-2008-9-s2-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hunter L, Lu Z, Firby J, B WA, Jr, Johnson HL, Ogren PV, Cohen KB. OpenDMAP: An open-source, ontology-driven concept analysis engine, with applications to capturing knowledge regarding protein transport, protein interactions and cell-specific gene expression. BMC Bioinformatics. 2008;vol. 9(no. 78) doi: 10.1186/1471-2105-9-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgartner WA, Jr, Cohen KB, Hunter L. An open-source framework for large-scale, flexible evaluation of biomedical text mining systems. Journal of Biomedical Discovery and Collaboration. 2008;vol. 3(no. 1) doi: 10.1186/1747-5333-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrucci D, Lally A. Building an example application with the unstructured information management architecture. IBM Systems Journal. 2004 July;vol. 43(no. 3):455–475. [Google Scholar]
- 5.Morgan AA, et al. Overview of BioCreative II gene normalization. Genome Biology. 2008;vol. 9 Suppl 2:S3. doi: 10.1186/gb-2008-9-s2-s3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumgartner WA, Jr, Lu Z, Johnson HL, Caporaso JG, Paquette J, Lindemann A, White EK, Medvedeva O, Cohen KB, Hunter L. Concept recognition for extracting protein interaction relations from biomedical text. Genome Biology. 2008;vol. 9 Suppl 2:S9. doi: 10.1186/gb-2008-9-s2-s9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kappeler T, Kaljurand K, Rinaldi F. TX task: Automatic detection of focus organisms in biomedical publications. Boulder, Colorado: Association for Computational Linguistics; Proceedings of the BioNLP 2009 Workshop. 2009 June;:80–88. [Online]. Available: http://www.aclweb.org/anthology/W09-1310.
- 8.Wang X, Tsujii J, Ananiadou S. Disambiguating the species of biomedical named entities using natural language parsers. Bioinformatics. 2010;vol. 26(no. 5):661–667. doi: 10.1093/bioinformatics/btq002. [Online]. Available: http://bioinformatics.oxfordjournals.org/cgi/content/abstract/26/5/661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Fan J-W, Friedman C. Biological, translational, and clinical language processing. Prague, Czech Republic: Association for Computational Linguistics; 2007. Jun, Combining multiple evidence for gene symbol disambiguation; pp. 41–48. [Online]. Available: http://www.aclweb.org/anthology/W/W07/W07-1006. [Google Scholar]
- 10.Xu H, Fan J-W, Hripcsa G, Mendon EA, Markatou M, Friedman C. Gene symbol disambiguation using knowledge-based profiles. Bioinformatics. 2007;vol. 23(no. 8):1015–1022. doi: 10.1093/bioinformatics/btm056. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Matthews M. Species disambiguation for biomedical term identification. Current trends in biomedical natural language processing: BioNLP 2008. 2008:71–79. [Google Scholar]
- 12.Schwartz A, Hearst M. A simple algorithm for identifying abbreviation definitions in biomedical text. Pacific Symposium on Biocomputing. 2003;vol. 8:451–462. [PubMed] [Google Scholar]
- 13.Cohen KB, Verspoor K, Johnson HL, Roeder C, Ogren PV, Baumgartner WA, Jr, White E, Tipney H, Hunter L. High-precision biological event extraction with a concept recognizer. BioNLP 2009 Companion Volume: Shared Task on Entity Extraction. 2009:50–58. [Google Scholar]
- 14.Ceol A, Chatr-Aryamontri A, Licata L, Cesareni G. Linking entries in protein interaction database to structured text: The febs letters experiment. FEBS Letters. 2008;vol. 582(no. 8):1171–1177. doi: 10.1016/j.febslet.2008.02.071. (1) The Digital, Democratic Age of Scientific Abstracts; (2) Targeting and Tinkering with Interaction Networks. [Online]. Available: http://www.sciencedirect.com/science/article/B6T36-4S0GY40-3/2/38c37d0da5f61f3e7945048281585ea3. [DOI] [PubMed] [Google Scholar]
- 15.Leitner F, et al. Introducing meta-services for biomedical information extractio. Genome Biology. 2008;vol. 9 Suppl 2:S6. doi: 10.1186/gb-2008-9-s2-s6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanisch D, Fundel K, Mevissen H-T, Zimmer R, Fluck J. ProMiner: rule-based protein and gene entity recognition. BMC Bioinformatics. 2005;vol. 6(no. Suppl. 1) doi: 10.1186/1471-2105-6-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]