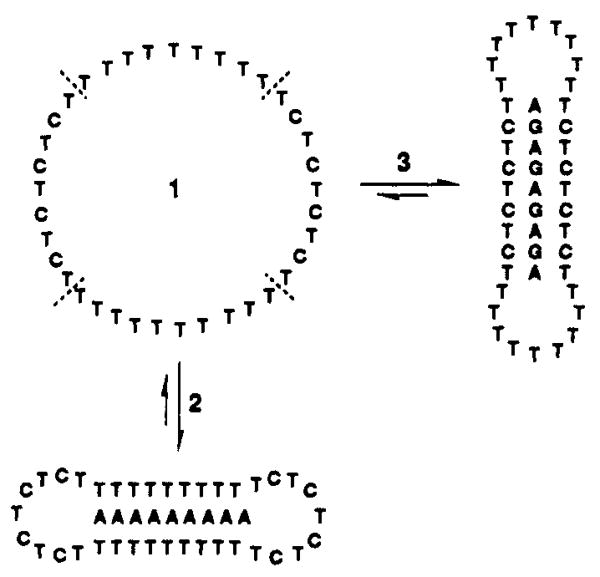
Proteins can recognize varied structures by making use of conformational flexibility. For example, DNA-binding proteins can have affinity for multiple sequences using a single binding pocket, by changing conformation in response to varied structures. To date, we know of no synthetic ligand which can recognize multiple sequences in this way. We now wish to report the design and synthesis of a macrocyclic ligand which can bind to two distinct DNA sequences in a mutually exclusive way by changing conformation on binding.
We constructed the 36-base oligonucleotide 1 by a template-directed cyclization of the linear precursor, 5′-dTCTCTTTTTTTTTTTCTCTCTCTTTTTTTTTTTCTCp.1,2 The template oligomer 5′-dAAAGAGAGAGAAA (1 equiv) was used to align the reactive ends in an aqueous esterification reaction, as described previously.3,4 The yield of purified circular 1 was 48%.5,6 Also prepared were two nine-base purine oligomers, dAAAAAAAAA (2) and dAGAGAGAGA (3) (Figure 1).
Figure 1.
Sequence of macrocycle 1 and the conformational changes involved in the binding of 1 to the sequences 2 and 3. Dotted lines demarcate the four domains in 1. When both sequences are present, the macrocycle binds to one of the two as shown, but cannot bind to both simultaneously.
As expected from its non-self-complementary sequence, compound 1 alone shows no significant temperature-dependent melting behavior when examined at 260 nm in a pH 7.0 buffer containing 100 mM NaCl and 10 mM MgCl2. In the presence of 1 equiv of the oligomer dA9 (2), however, there is a sharp, apparently two-state transition showing a hyperchromicity of 23% and a melting temperature (Tm) of 48.0 °C (Figure 2 (inset)).7 This is a considerably higher Tm than that observed for the duplex dA9·dT9 (Tm = 28.7 °C) under identical conditions. This high affinity is consistent with the bimolecular triple helical structure expected for this complex,4 with the purine oligomer held between opposing dT9 domains in the macrocycle.
Figure 2.
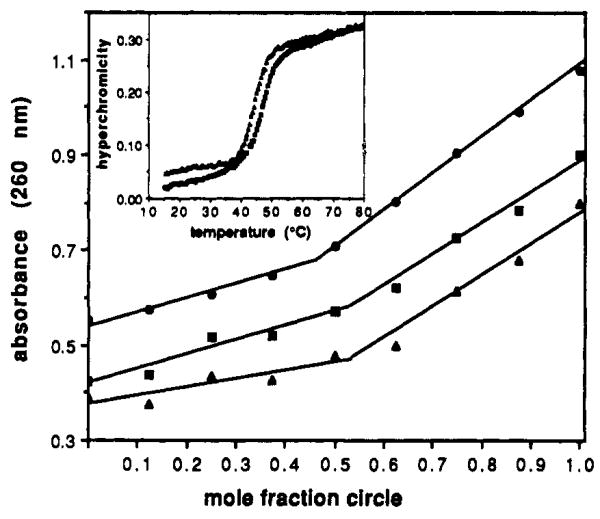
Mixing plots of mole fraction vs absorbance at 260 nm for circle 1 with purine complementary strands. Total DNA strand concentration was 3 μM, with molar ratios being varied as shown. Plots show 1 mixed with dA9 (▲), 1 with d(AG)4A (■, offset 0.1 AU), and 1 with a 1:1 mixture of dA9 and d(AG)4A (●, offset 0.3 AU). From the plots, mole fractions for full complexation are 0.52, 0.53, and 0.47, respectively. (Inset: Melting profiles of hyperchromicity vs temperature for 1:1 complexes of circle 1 with dA9 (Tm = 48.0 °C) and 1 with d(AG)4A (Tm = 44.6 °C) at pH 7.0. Experiments were carried out as described.7
Significantly, the macrocycle displays almost identical binding behavior with a second, distinctly different sequence. When added to the oligomer dAGAGAGAGA (3), a melting transition having a 22% hyperchromicity and a Tm of 44.6 °C is seen at pH 7 (Figure 2). Once again, this is significantly higher than the corresponding duplex d(AG)4A·d(TC)4T, which has a Tm of 32.7 °C under these conditions.
Several observations support the involvement of two opposing binding domains, resulting in triplexes, for the two complexes. Mixing plots carried out at 25 °C between the circle and 2 (Figure 2) show that the stoichiometry of the complex is 1:1; if both dT9 domains were involved in duplexes, the expected stoichiometry would be 2:1 dA9:circle. Similarly, a mixing plot for the circle and dAGAGAGAGA reveals a stoichiometry of 1:1 as well (Figure 2). In addition, the denaturation of complex 1·2 shows a significant hyperchromicity of 13% at 284 nm, consistent with reported properties of Tn·An·Tn triple helices and not duplexes.8 Finally, the melting transition for complex 1·3 is pH dependent, with a higher Tm of 56.2 °C being observed at pH 6.0; this is again consistent with a triplex protonated at cytosines in the Hoogsteen strand.9
The formation of these two different complexes requires that the macrocycle adopt two opposite conformations in the binding of 2 and 3 (Figure 1). In forming a complex, two opposed nine-base domains move inward and align themselves with the complementary purine strand on opposite sides, forming two or three hydrogen bonds on each side of a base. The unbound nine-base domains then act as loops for bridging the bound domains. In order to bind the other sequence, the pairs of domains reverse roles, with the former binding domains acting as bridging loops and vice versa.
Interestingly, a further mixing experiment reveals that this complexation is mutually exclusive: the act of binding to one sequence precludes the binding of the other, even though the necessary bases are unpaired in the loops. When the circle is mixed at varied molar ratios with a one-to-one mixture of the two sequences (Figure 2), the mixing plot shows a stoichiometry of 1:1. This indicates that a given complex cannot bind the second sequence with its loops at 25 °C. While this macrocycle at all times carries the bases necessary for forming complexes with the two sequences, the act of binding causes a conformational switch, holding the loops in an inaccessible or unproductive conformation.10
To confirm that the macrocycle can recognize these two sequences within a longer strand of DNA, we synthesized a 33-nucleotide oligomer having the sequence 5′-dCACAAGAGAGAGAATCCCTAAAAAAAAAAACAC (4). This oligomer contains the two recognition sequences separated by seven bases. When mixing experiments are performed (pH 7.0, 25 °C) with this oligomer and the macrocycle 1, a plot of absorbance versus mole fraction (not shown) clearly demonstrates a binding stoichiometry of two macrocycles to 1 equiv of 4; this confirms that the unusual binding property is also seen with two sites in one strand.
The results demonstrate a new strategy for the binding of two or more specific nucleic acid sequences with a single ligand. Such a strategy may be useful in the design of multifunctional biosensors or of therapeutics directed to multiple medicinally important targets.
Acknowledgments
We thank the National Institutes of Health (R01-GM46625) for partial funding of this work. E.T.K. is an Office of Naval Research Young Investigator (1992-1995) and an Arnold and Mabel Beckman Foundation Young Investigator (1992-1994).
References
- 1.The phosphorylated 36-mer was purchased from Midland Certified Reagents (Midland, TX) and was prepared using the standard β-cyanoethyl phosphoramidite method.2 Other oligomers were synthesized on a Pharmacia-LKB instrument, using this same chemistry.
- 2.Beaucage SL, Caruthers MH. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 3.The BrCN/imidazole/Ni2+ ligation method has been shown to give the natural DNA phosphodiester linkage: Kanaya E, Yanagawa H. Biochemistry. 1986;25:7423–7430. doi: 10.1021/bi00371a026.Ferris JP, Huang CH, Hagan WJ. Nucleosides Nucleotides. 1989;8:407–414. doi: 10.1080/07328318908054184.Luebke KJ, Dervan PB. J Am Chem Soc. 1989;111:8733–8735.
- 4.(a) Kool ET. J Am Chem Soc. 1991;113:6265–6266. [Google Scholar]; (b) Prakash G, Kool ET. J Chem Soc, Chem Commun. 1991:1161M–1163. doi: 10.1039/C39910001161. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Prakash G, Kool ET. J Am Chem Soc. 1992;114:3523–3527. doi: 10.1021/ja00035a056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oligonucleotide concentrations were measured by absorbance at 260 nm. Extinction coefficients were calculated by the nearest-neighbor method.6 The reaction was allowed to proceed for 12 h at 23 °C, and the product was isolated by preparative denaturing gel electrophoresis. The mobility of the circular product on a 20% denaturing polyacrylamide gel was 0.90 that of the 36-nucleotide linear precursor. Conversion to circular product was ≥95% in 12 h, as judged by UV shadowing. The circularity of the product was confirmed by complete resistance to cleavage by exonuclease activity (T4 polymerase, Promega) under conditions that completely cleave a linear sequence to mononucleotides.
- 6.Borer PN. In: Handbook of Biochemistry and Molecular Biology. Fasman GD, editor. CRC Press; Cleveland: 1975. p. 589. [Google Scholar]
- 7.Thermal denaturations were performed on a Cary 1 spectrophotometer in 1 cm path length stoppered quartz microcells under an N2 atmosphere. The cells were monitored at 260 nm, with a temperature rise of 0.5 °C/min. Mixtures were 3 μM in each DNA strand and contained 10 mM Na·PIPES buffer (pH 7.0), 100 mM NaCl, and 10 mM MgCl2. Tm's were taken as the inflection point in the denaturation curve and are estimated to be accurate to within ±0.5 °C.
- 8.(a) Riley M, Maling B, Chamberlin MJ. J Mol Biol. 1966;20:359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]; (b) Pilch DS, Levenson C, Shafer RH. Proc Natl Acad Sci USA. 1990;87:1942–1946. doi: 10.1073/pnas.87.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Pilch DS, Brousseau R, Shafer RH. Nucleic Acids Res. 1990;18:5743–5750. doi: 10.1093/nar/18.19.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Lipsett MN. Biochem Biophys Res Commun. 1963;11:224–228. doi: 10.1016/0006-291x(65)90350-5. [DOI] [PubMed] [Google Scholar]; (b) Morgan AR, Wells RD. J Mol Biol. 1968;37:63–80. doi: 10.1016/0022-2836(68)90073-9. [DOI] [PubMed] [Google Scholar]; (c) Moser HE, Dervan PB. Science. 1987;238:645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]; (d) Rajagopal P, Feigon J. Nature. 1989;339:637–640. doi: 10.1038/339637a0. [DOI] [PubMed] [Google Scholar]
- 10.For the related hairpin loops in duplex DNA it has been shown that full-length hybridization is weak because of the curvature in the loop: Ecker DJ, Vickers TA, Bruice TW, Freier SM, Jenison RD, Manoharan M, Zounes M. Science. 1992;257:958–961. doi: 10.1126/science.1502560.