Abstract
For more than a decade the Arp2/3 complex, a handful of nucleation-promoting factors, and formins were the only molecules known to directly nucleate actin filament formation de novo. However, the past several years have brought a surge in the discovery of mammalian proteins with roles in actin nucleation and dynamics. Newly recognized nucleation-promoting factors, such as WASH, WHAMM, and JMY stimulate Arp2/3 complex activity at distinct cellular locations. Formin nucleators with additional biochemical and cellular activities have also been uncovered. Finally, the Spire, Cordon-bleu, and Leiomodin nucleators have revealed new ways of overcoming the kinetic barriers to actin polymerization.
Actin is one of the most abundant and highly-conserved proteins in eukaryotic cells, and is structurally related to prokaryotic actin-like proteins, hinting at its ancient evolutionary origins (Box.1). A 42kDa monomeric ATP-binding protein, globular (G)-actin, can undergo cycles of self-assembly into filamentous (F)-actin, ATP hydrolysis, and depolymerization. Actin filaments contain dynamic barbed ends and less active pointed ends that are differentiated by their structural and biochemical characteristics. Filament turnover is controlled by many actin-binding proteins, including some that function in monomer sequestration or delivery, and others that promote filament nucleation, elongation, capping, severing, or depolymerization. The ability of this versatile cytoskeletal system to generate force, create structural scaffolds, and act as tracks for motor proteins makes it a critical participant in numerous cellular functions, including morphogenesis, migration, cytokinesis, and membrane transport (Fig.1).
Box 1 The Actin Cytoskeleton: Ancient evolutionary origins and functions.
Several prokaryotic proteins have structural similarities to eukaryotic actin and assemble into filaments, suggesting that they represent an ancestral actin cytoskeleton. These bacterial factors include MreB (murein formation cluster E B) and ParM (partitioning M) (reviewed in Ref.159; see the figure). MreB is expressed primarily in rod-shaped bacteria, and is an ATPase, similar to eukaryotic actin. Also like actin, MreB subunits are preferentially incorporated into filaments in their ATP-bound form, and depolymerize mainly in the ADP-bound form. However unlike F-actin, MreB subunits generally have a non-helical structure, and form short coils, spirals, and ribbons. These filaments help determine bacterial shape by forming a scaffold beneath the inner plasma membrane and positioning enzymes involved in cell wall biosynthesis. Another actin-like bacterial protein, ParM, is encoded on the E.coli R1 plasmid. ParM polymerizes in an ATP-dependent manner to form helical filaments like F-actin, but with the opposite handedness. Moreover, its assembly kinetics are drastically different from eukaryotic actin. ParM spontaneously nucleates more efficiently than actin and might not need nucleation factors. ParM filaments continuously cycle between phases of elongation and rapid catastrophic disassembly, a behavior that was first described for microtubules and is termed dynamic instability. Finally, ParM filaments are not polar, and can elongate from both ends. This has important functional consequences, because during bacterial division, bidirectionally elongating ParM filaments drive the physical partitioning of R1 plasmids into the two daughter cells. While a generic role for actin-like cytoskeletons in determining cell shape appears to be conserved from bacterial to mammalian cells, thus far a function for the actin-based movement of nucleic acids has only been well-characterized in prokaryotes.
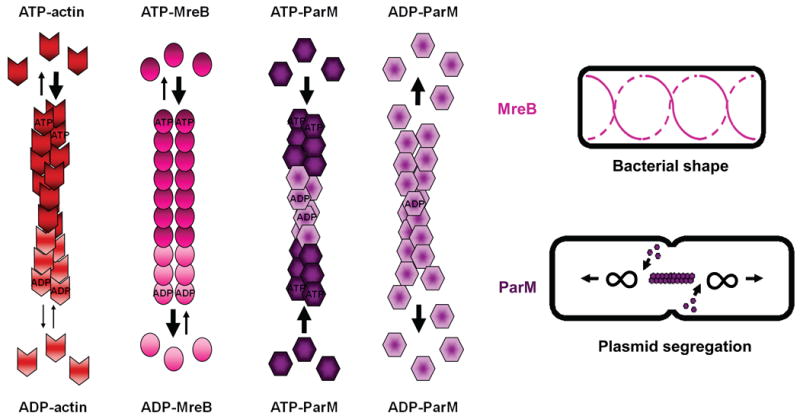
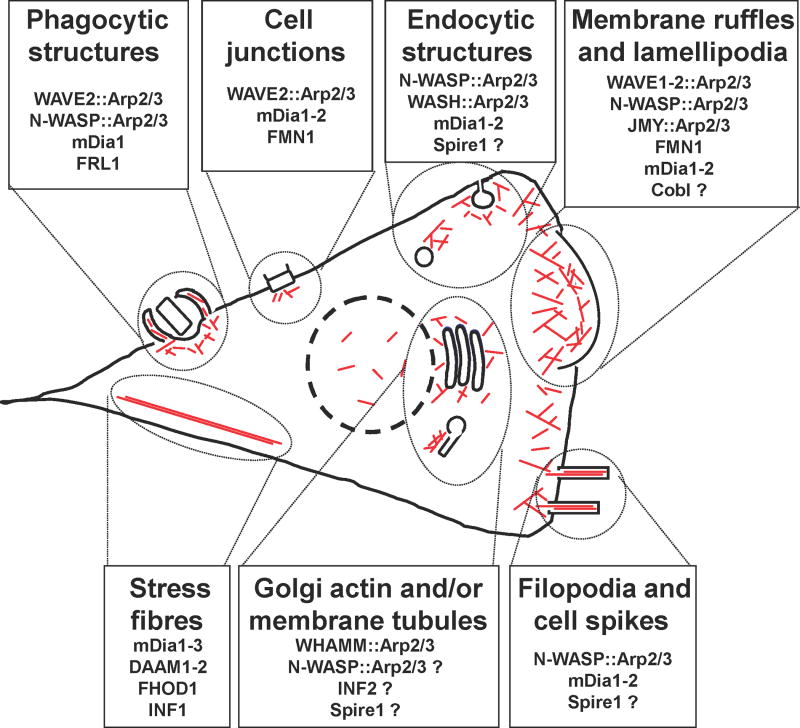
Figure 1. Localization and function of actin nucleation factors in mammalian cells.
Actin filaments (red lines) are nucleated and organized into branched networks by the Arp2/3 complex and its nucleation-promoting factors, or are generated in unbranched forms by formins and tandem WH2-domain nucleators. Functional roles for different nucleation factors are depicted during phagocytosis, cell junction assembly, endocytosis, membrane ruffling and lamellipodia dynamics, filopodia formation, Golgi and tubulo-vesicular membrane dynamics, and stress fiber formation in a generic mammalian cell. Question marks (?) indicate that the precise role for the depicted nucleation factor is unclear. Arp2/3, actin-related protein 2/3; Cobl, cordon-bleu; Daam, Dishevelled-associated activator of morphogenesis; FHOD, formin homology domain; FMN, formin; FRL, formin-related in leukocytes; INF, inverted formin; JMY, junction mediating regulatory; mDia, murine Diaphanous; N-WASP, neuronal-WASP; WASP, Wiskott-aldrich syndrome protein; WASH, WASP/Scar homolog; WAVE, WASP-verprolin homologous; WHAMM, WASP homologue associated with actin, membranes, and microtubules.
To initiate actin assembly during such processes, cells generate free barbed ends that act as templates for polymerization by uncapping or severing existing filaments or by nucleating from monomers de novo. Spontaneous actin assembly is inefficient, however, as the formation of actin dimers and trimeric “nuclei” is kinetically unfavorable. To overcome this obstacle, cells employ factors that directly nucleate actin. For many years, only the Arp2/3 (actin-related protein 2/3) complex, a handful of its activators, and formin proteins were known to directly participate in nucleation1-3. However, recent discoveries have revealed new factors with roles in nucleation. In this Review, we discuss the latest advances in our understanding of the cellular regulation and function of the Arp2/3 complex and formins, and examine how the characterization of new nucleation factors has provided fresh perspectives on actin dynamics in mammalian cells.
Arp2/3 complex: a Y-branching nucleator
The first major actin nucleator to be identified was the Arp2/3 complex, a 220kDa factor composed of seven stably-associated polypeptides that are highly conserved in virtually all eukaryotic organisms (for an in-depth review on the Arp2/3 complex, see Ref.1). These include Arp2 and Arp3 and five additional subunits, ARPC1-5 (Fig.2A). Among the known nucleators, the Arp2/3 complex is unique in its ability to both nucleate filaments and organize them into branched networks.
Figure 2. Structure of the Arp2/3 complex in Y-branches and model for nucleation and branching.
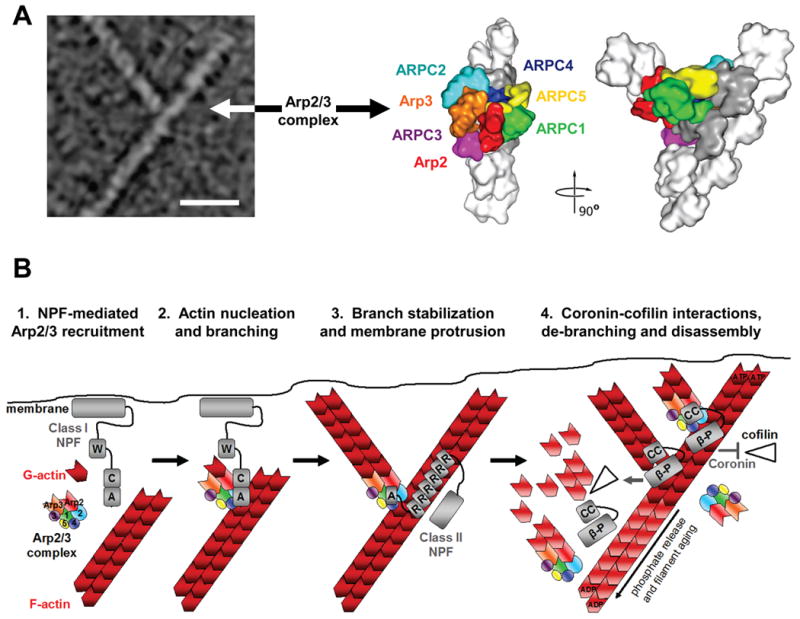
A) The morphology of a y-branched actin filament and the Arp2/3 complex is shown, both in an electron micrograph and in structural models based on electron tomography (modified with permission from Ref.13). The complex consists of the actin-related proteins Arp2 and Arp3 plus the additional Arp complex subunits ARPC1-5. In this model, all seven subunits participate in binding to the existing filament, while Arp2 and Arp3 act as the first two subunits of the nascent filament.
B) The Arp2/3 complex is recruited by the WCA domains of Class I NPFs in proximity to cellular membranes (1). The collective activities of WH2 and CA segments serve the basic purpose of bringing the Arp2/3 complex together with the first actin subunit in the new filament and generate a branch (2). Arp2/3 branchpoints can be stabilized by F-actin-binding Class II NPFs, like cortactin (3). Coronin-family proteins interact with the Arp2/3 complex and F-actin to prevent cofilin-mediated disassembly of newly-formed filaments (4, top). Coronin can also replace the Arp2/3 complex and synergize with cofilin to trigger debranching and disassembly of older ADP-actin filaments (4, middle). Disassembly of older branches can also occur spontaneously, following phosphate release from Arp2 and actin (4, bottom). A, acidic; β-P, beta-propellor; C, connector; CC, coiled-coil; R, repeat; W, WH2. Structural models in part a modified, with permission, from Ref.13 © (2008) Rockefeller University Press.
Biochemical properties, structure and activation
In its purified form, the Arp2/3 complex binds to the side of an existing filament and initiates the assembly of a new filament, joining the two at a ∼70° Y-branch angle1. The nascent filament is capped by the complex at its pointed end, but free to elongate at its barbed end. Like actin, Arp2 and Arp3 bind ATP, and the conformation of the complex differs with and without nucleotide4,5. Importantly, ATP-binding is crucial for nucleation in vitro4,6,7. Arp2 has been shown to hydrolyze ATP, and although hydrolysis was initially implicated in either nucleation8 or debranching9, recent work suggests it primarily accelerates debranching and recycling of the complex10.
The crystal structure of the bovine Arp2/3 complex has been solved in various nucleotide-bound conditions6,11, but none of these represent the active version, because Arp2 and Arp3 are spaced too far apart to mimic a short-pitch actin dimer, the most likely configuration for promoting actin nucleation. Although structures of the active complex have yet to be solved, a combination of homology modeling and electron tomography has helped reveal its conformation at Y-branches (Fig.2A). These studies suggest that ARPC2 and ARPC4 form important contacts with the parental filament, while the barbed ends of Arp2 and Arp3 associate with the pointed end of the nascent filament12,13. Moreover, interactions between the entire complex and the existing filament involve conformational changes by both structures that might increase branchpoint stability13. Thus, upon binding to an existing filament, Arp2 and Arp3 likely reorient into a dimer that acts as the first two subunits of the new filament.
By itself, the Arp2/3 complex is an inefficient nucleator, but filament-binding increases its activity and is coupled with nucleation and branching. Potent nucleation also requires phosphorylation of threonine and tyrosine residues in Arp214. A third contributor to Arp2/3 activation, and the best characterized, involves engagement of the complex by nucleation-promoting factors (NPFs). Most mammalian NPFs activate Arp2/3 using a WCA domain, which is comprised of one or more WH2 (WASP-homology-2) motifs that bind actin monomers, plus an amphipathic connector region and acidic peptide that collectively bind the Arp2/3 complex (Fig.2B).
To nucleate actin, the CA portion of the WCA domain contacts multiple subunits of the complex and causes a substantial conformational change that primes the complex for nucleation4,5,15,16. The C region can interact with both the Arp2/3 complex and actin17, and structural studies suggest that this region binds to Arp218, while the WH2 domains, by virtue of their ability to bind G-actin19,20, deliver monomers to the barbed ends of Arp2 and/or Arp3 in the primed complex. Following Arp2/3 complex activation, the WCA domain is thought to dissociate, enabling it to promote additional rounds of nucleation. Further biochemical and structural study is needed to precisely define how WCA domains alter Arp2/3 complex conformation and promote activation during nucleation and branching.
In cells, the Arp2/3 complex is important for nucleating and organizing branched filament networks in lamellipodia and other structures (Fig.1), based on many localization and functional inhibition studies1. However, much of what we have subsequently learned about cellular Arp2/3 complex functions has come indirectly, by studying its activators. These include WCA domain-containing proteins encoded by microbial pathogens and mammalian NPFs that comprise the WASP superfamily (Fig.3A), originally named after the Wiskott-Aldrich Syndrome protein.
Figure 3. Different groups of NPFs possess distinct modes of regulation.
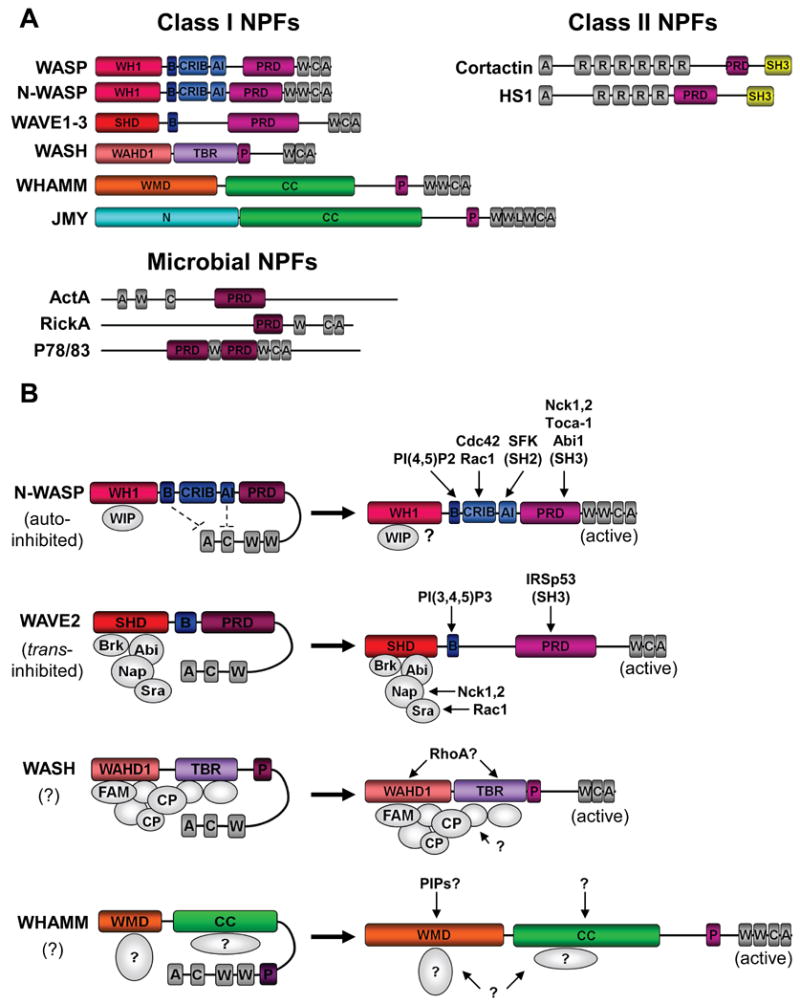
A) Mammalian Class I NPFs contain C-terminal WCA domains that bind G-actin and the Arp2/3 complex, plus diverse N-terminal regulatory regions. Microbial pathogens also express Class I NPFs. These include ActA from Listeria, RickA from Rickettsia, and P78/83 from baculoviruses. Class II NPFs contain N-terminal acidic domains that bind the Arp2/3 complex, central F-actin-binding repeats, and regulatory C-terminal domains. A, acidic; AI, autoinhibitory; B, basic; C, connector; CC, coiled-coil; CRIB, Cdc42-Rac-interactive-binding; L, linker; N, amino-terminal; P, polyproline; PRD, proline-rich-domain; R, repeat; SH3, Src-homology-3; SHD, Scar-homology-domain; TBR, tubulin-binding region; W, WASP-homology-2 (WH2) domain; WH1, WASP-homology-1; WAHD1, WASH-homology-domain-1; WMD, WHAMM-membrane interaction-domain.
B) N-WASP NPF activity is regulated both by autoinhibition and by interactions with proteins from the WIP family. It can be stimulated by direct binding of phosphoinositides, small GTPases, tyrosine phosphorylation, and SH2/SH3 domains. WAVE2 activity is controlled by a protein complex comprised of Brk1, Abi1, Nap1, and Sra1. It is stimulated by Nck-Nap1 and Rac-Sra1 interactions, or by binding of phosphoinositides or SH3 domains to WAVE2 itself. WASH activity is likely controlled by a multi-subunit complex that contains capping protein (CP) and FAM21. WHAMM NPF activity does not appear to be autoinhibited, and is likely to be controlled by factors that interact with its WMD and/or CC regions. Question marks (?) indicate that the depicted mechanism of activation is speculative.
NPFs: critical Arp2/3 collaborators
The largest group of mammalian NPFs (Class I) stimulate Arp2/3-mediated nucleation using C-terminal WCA domains, but contain diverse N-terminal sequences that enable different modes of regulation and functions in cells3 (Fig.3A-B). As a consequence, these NPFs are subcategorized into five groups: WASP and N-WASP, three WAVE (WASP-family verprolin homolog; also known as Scar or suppressor of cAR) isoforms, and the recently-identified factors WASH (WASP/Scar homolog), WHAMM (WASP homolog associated with actin, membranes and microtubules), and JMY (junction-mediating regulatory protein). Moreover, cortactin, a distinct type of NPF (Class II), binds to actin filaments and influences the stability of F-actin branchpoints in conjunction with proteins from the coronin family of Arp2/3 inhibitors.
WASP and N-WASP
WASP and N-WASP, the best-characterized NPFs, are found in animals, fungi, and protists. Mammalian WASP is expressed specifically in haematopoietic cells, and its mutation results in defective cell migration, phagocytosis, and T-cell signalling, leading to immunodeficiencies in mice and human Wiskott-Aldrich Syndrome patients21. In contrast, N-WASP is expressed in most cell types, and its deletion results in neurological and cardiac abnormalities and embryonic lethality in mice22,23. Our discussion will focus on this more ubiquitous NPF.
N-WASP possesses a modular domain organization consisting of an N-terminal WASP-homology-1 (WH1) domain, plus basic (B), Cdc42/Rac-interactive binding (CRIB), and auto-inhibitory (AI) motifs that are collectively called the GTPase-binding domain (GBD), and a proline-rich domain (PRD) adjacent to its WCA domain (Fig.3A). By itself, N-WASP has little NPF activity, because its CA region is inhibited by intramolecular interactions with the AI portion of the GBD (Fig.3B). This inactive conformation is stabilized by WH1-interacting proteins like WIP, a G- and F-actin-binding molecule that forms a complex with N-WASP24. WIP can inhibit N-WASP activity in vitro, but stimulates actin assembly in cells. Thus, it is unclear if WIP acts exclusively as an N-WASP inhibitor or if it can also enhance N-WASP activity. Notably, most studies of N-WASP have relied on derivatives purified in the absence of WIP. However, recent work has begun to explore the biochemical features of the N-WASP-WIP complex25-27. Continued analyses should reveal how WIP influences N-WASP activity in this complex.
To stimulate N-WASP, signal transduction pathways initiated at the plasma membrane often converge upon cellular factors that interact with the GBD-PRD. For example, binding of the small GTPase Cdc42 changes the conformation of the GBD to free the WCA domain, an allosteric activation mechanism that is enhanced by PI(4,5)P2-binding to the B region (Fig.3B). In addition, diverse PRD-binding proteins with Src-homology-3 (SH3) domains activate N-WASP. These include the adaptor proteins Nck1 and Nck2 (noncatalytic kinase-1-2)28, membrane-deforming factors like Toca-1 (transducer of Cdc42-dependent actin assembly-1)25,27, and the kinase-interacting protein Abi1 (Abl-interactor-1)29 (Fig.3B). Tyrosine kinases also phosphorylate the GBD to stabilize active N-WASP and enable further stimulation by SH2 domains30. Although each of these factors individually activate N-WASP in vitro, multiple inputs can be integrated to promote higher activity. Moreover, multimerization of N-WASP by proteins that oligomerize or contain multivalent N-WASP binding sites also increases N-WASP activity and its affinity for the Arp2/3 complex26,31,32.
Ultimately, in response to these diverse signaling molecules, N-WASP directs Arp2/3 to polymerize actin during a multitude of cellular functions (Fig.1). In the absence of proper N-WASP function, cells have multiple deficiencies in processes requiring actin dynamics at the plasma membrane. These include impaired filopodia formation22,33, dorsal membrane ruffling34, and membrane invagination35. N-WASP also contributes to endocytosis29,33,35,36, a process in which F-actin facilitates membrane fission and drives endosome movement. It seems likely that the essential function of N-WASP in mammals is linked to the abundance of actin-associated processes that it affects in cells.
WAVE1-3
Like the WASPs, the WAVE-family NPFs are conserved in many animals and protists, but are also found in plants. The 3 mammalian isoforms are expressed in numerous cell types, with WAVE1 and WAVE2 distributed most broadly, although all are enriched in brain tissue. Likely as a consequence, WAVE1 knockout mice have sensorimotor defects, behavioral abnormalities, and reduced viability37,38, whereas WAVE2 ablation is lethal, and knockout embryos exhibit impaired angiogenesis, underdeveloped hearts, hemorrhaging, and brain malformation39,40.
In contrast to the WASPs, the WAVEs are fully active and not autoinhibited when purified as recombinant proteins. They lack a GBD, and their N-terminal Scar-homology domains (SHD) and proline-rich domains are distinct from the regulatory portions of N-WASP (Fig.3A). The SHD associates with a complex consisting of Brk1 (Brick1; also called HSPC300), Abi1, Nap1 (Nck-associated protein-1), and Sra1 (specifically Rac-associated-1)41-44. In the ubiquitous WAVE complex, Brk1 and Abi1 bind to WAVE2, while Nap1 interacts with Abi1 and Sra142 (Fig.3B). Unlike WAVE1 or WAVE2 by themselves, the activities of the reconstituted complexes are suppressed43,44. Removal of an individual subunit disrupts the stability and/or localization of the others, illuminating the fact that the complex behaves as a unit45,46.
Similar to N-WASP, the WAVE2 PRD can bind to SH3-domain-containing proteins, such as IRSp53 (insulin receptor substrate protein of 53kDa)47. WAVE2 also contains a basic peptide that binds phospholipid (PI(3,4,5)P3)47,48, and is tyrosine phosphorylated to modulate its activity49. However, in contrast to N-WASP, most signaling to WAVE2 occurs through interactions with the complex, rather than direct binding to WAVE2. Rac1, the best characterized WAVE activator, binds to Sra143,45, although it might also act via IRSp5347,50. Moreover, Nck adaptor proteins can activate the WAVE complex, probably by interacting with Nap141. Importantly, the cellular source of the WAVE complex and the manner in which it is purified can significantly influence its response to stimuli51. Rac1 and Nck were initially shown to dissociate the complex41, but more recent reports indicate that it remains intact after activation45,46. Moreover, physiological stimulation of the WAVE complex likely requires cooperativity among Rac1, PI(3,4,5)P3, and protein kinases51. A better understanding of how the WAVE complex subunits interact with one another and with the WAVE isoforms themselves is necessary to clarify how the complex is regulated and reorganized in response to upstream signaling.
In contrast to the functional versatility of N-WASP, the major function of WAVE NPFs is to activate the Arp2/3 complex during plasma membrane protrusion and cell motility (Fig.1). In these processes, WAVE1 and WAVE2 have partially-overlapping functions, as WAVE2-deficient cells exhibit severe defects in peripheral membrane ruffling, lamellipodia formation, and cell motility29,39,40,50,52, while dorsal ruffling and migration through extracellular matrix is impaired in the absence of WAVE150. WAVE2 might also help to organize and maintain cell-cell contacts53. It is interesting that there is not enough redundancy among the three WAVEs to prevent the major abnormalities that occur in mice lacking WAVE1 or WAVE2. Clearly, important differences in the regulation and function of the different WAVE isoforms remain to be uncovered.
WASH
For nearly a decade, the WASPs and WAVEs were the only known mammalian Class I NPFs. However, several new NPFs have gained recognition in recent years (Fig.3A). One such factor is WASH, a WCA-containing protein found in animals and protists, that was first identified as the product of a subtelomeric gene that has undergone extensive duplication in primates54. In Drosophila melanogaster the single WASH gene is essential54, suggesting that it has a role in early development and is not functionally redundant with WASP or WAVE.
In addition to its WCA domain, WASH possesses an adjacent polyproline region and distinct N-terminal sequence elements, termed WASH-homology-domain (WAHD1) and tubulin-binding region (TBR) (Fig.3A). As for other NPFs, these putative regulatory domains likely mediate the formation of a multiprotein complex (Fig.3B). In fact, WASH interacts with multiple proteins, including capping protein55, which caps filament barbed ends, and FAM21, a protein that links WASH to endosomes56. However, the mechanism by which the constituents of the native WASH complex control the activity and function of WASH are not well understood.
Mammalian WASH localizes to early and recycling endosomes55-57, where WASH-mediated Arp2/3 activity controls the shape of these membranes and also influences retromer-dependent trafficking to the trans-Golgi network, recycling to the plasma membrane, and trafficking to late endosomes. These functions might also be affected by interactions between WASH and tubulin subunits56. Interestingly, evidence from D.melanogaster suggests that WASH can bundle both F-actin and microtubules in a Rho GTPase-regulated manner58. Moreover, these studies suggest that WASH might act in concert with two other types of actin nucleators, formin and Spire (see below). Understanding how regulation by GTPases, bundling of actin and microtubules, and interactions with other nucleators contribute to WASH function in mammalian cells requires further investigation.
WHAMM and JMY
Unlike the WASP, WAVE, and WASH NPFs, two other recently-identified Arp2/3 activators, WHAMM and JMY, seem to be confined to vertebrate species59,60. Within mammals they are expressed in a variety of tissues and cell types. While WHAMM was identified based on its WCA sequence, JMY was discovered as a factor that interacts with the transcriptional regulators p300 and p53 before the effects of its WCA domain on actin polymerization were realized. WHAMM and JMY are nearly 35% identical, but can still be differentiated from one another and from other NPFs based on their N-terminal sequences.
In addition to their WCA domains and adjacent polyproline motifs, WHAMM and JMY possess central regions predicted to form coiled-coils (CC), but distinct N-termini that are only 25% identical to one another (Fig.3A). Like N-WASP, the WHAMM WCA segment includes two WH2 motifs, although WHAMM is a less potent NPF than N-WASP59. Notably, the JMY WCA domain contains three WH2 motifs. This third WH2 and an additional actin monomer-binding linker allow the JMY WCA domain to nucleate actin even in the absence of the Arp2/3 complex60. The filaments produced by JMY WCA without Arp2/3 are unbranched, unlike the networks that are generated in its presence. Future work will address whether the nucleating and NPF activities of JMY are coordinated, or whether one activity predominates in cells.
Whereas the NPF activity of full-length JMY has not been determined, full-length WHAMM is active in vitro, indicating that like WAVE2, it is not autoinhibited59. WHAMM might therefore be part of a multi-protein complex that modulates its NPF activity under native conditions, although the components of such a complex have not been identified (Fig.3B). Despite the relatedness of their domain organization, WHAMM and JMY likely interact with distinct cellular factors, as evidenced from differences in their localization and function.
Unlike other NPFs, WHAMM localizes to the cis-Golgi and to tubulovesicular membranes that move in an actin- and microtubule-dependent fashion59. These patterns are derived from the combined activities of the WHAMM membrane-interaction-domain (WMD) that mediates Golgi localization, and the coiled-coil region that directly binds microtubules (Fig.3A). Increasing or decreasing WHAMM protein levels perturbs Golgi structure and slows ER-Golgi transport, indicating that WHAMM influences Golgi morphology and anterograde trafficking. The ability of WHAMM to negotiate crosstalk between the actin and microtubule cytoskeletons seems central to its role in influencing membrane dynamics. A further characterization of how WHAMM activities are affected by its binding-partners should yield additional clues about how actin and microtubules cooperate during membrane transport in cells and during development.
In contrast to WHAMM, and in keeping with a role in transcriptional regulation, JMY is enriched in the nucleus of many cells60, and it appears to be recruited there in response to DNA damage61. In migrating cells, JMY relocates to lamellipodia, where its overexpression enhances motility and its depletion slows it60. The role of JMY in cell motility might involve its actin nucleating activity in conjunction with an ability to regulate the expression of cadherins61. However, the precise mechanisms controlling the localization and activity of JMY are unknown. It also remains to be determined if JMY and WHAMM share functional redundancy. Given its role in nuclear processes, advances in our understanding of JMY function might shed light on the role of actin in the nucleus.
Cortactin and coronin
In addition to WCA-containing NPFs, many animals express a second type of Arp2/3 complex activator, termed Class II62. In mammals, this category includes cortactin and its hematopoetic relative HS1 (hematopoetic-specific protein-1) (Fig.3A). These proteins possess Arp2/3-binding acidic peptides at their N-termini, but lack WH2 domains for binding G-actin. Instead, they harbor repetitive sequences that interact with F-actin. They also contain proline-rich regions with regulatory phosphorylation sites and C-terminal SH3 domains that interact with many proteins including N-WASP and WIP. Compared to Class I NPFs, Class II NPFs are considered weak Arp2/3 activators. Nevertheless, cortactin can enhance N-WASP-mediated activation of the Arp2/3 complex63-65. Cortactin also inhibits spontaneous dissociation of Arp2/3-bound filament branch junctions in vitro, thereby stabilizing Y-branches62 (Fig.2B).
The ability of cortactin to act as an NPF, cooperate with N-WASP, and stabilize Arp2/3-bound branches makes it an important participant in many cellular processes. Dominant negative and RNAi studies have revealed a role for cortactin in membrane fission during endocytosis and trans-Golgi export66-68, and in supporting cadherin adhesive zone formation and cell-cell contacts69. Similar manipulations of cortactin function also alter membrane dynamics and alter lamellipodial protrusion or persistence, resulting in impaired cell migration64,70-72. Deficiencies in membrane ruffling and motility have been recapitulated recently in cortactin knockout cells73, confirming that cortactin is a major regulator of cortical F-actin architecture.
Our understanding of how cells control the organization and turnover of lamellipodial actin networks has grown recently with the characterization of human coronin 1B. This protein is a member of the ubiquitous coronin family, and is the only known direct inhibitor of the Arp2/3 complex in vitro74,75. Coronin also binds F-actin and protects newly-formed ATP-actin filaments from the severing protein cofilin, yet can synergize with cofilin and AIP1 (actin-interacting protein-1) to promote severing or catastrophic bursts of disassembly of older ADP-actin filaments in vitro76-78 (Fig.2B). These disassembly processes may be coordinated with an ability of cofilin to dissociate Arp2/3 complex Y-branches79. In cells, the ability of coronin to inhibit Arp2/3-mediated actin nucleation, directly displace Arp2/3 from Y-branches just behind the leading edge, and interact with phosphatases that activate cofilin promotes recycling of actin and Arp2/3 to allow efficient lamellipodial protrusion75,80. This is one example of the highly orchestrated balance of nucleation, branching, and turnover that leads to the formation of dendritic actin networks following Arp2/3 activation.
Formins: nucleation and elongation
Unlike the Arp2/3 complex, all other known actin nucleators produce unbranched filaments. The best-characterized of these are the formins, which are present in virtually all eukaryotes (for an in-depth review of formins, see Ref.2). Their defining feature is the presence of the conserved formin-homology (FH) domains, FH1 and FH2 (Fig.4A-C). While much of what we know about formin structure and biochemical activity originated from studies of the yeast formins, nucleation activity has since been observed for many of the ∼15 mammalian formins. These proteins fall into 7 different subclasses based on FH2 sequence divergence: Dia (Diaphanous), FRL (formin-related proteins in leukocytes), DAAM (Dishevelled-associated activators of morphogenesis), FHOD (formin-homology domain proteins), FMN (Formin), Delphilin, and INF (inverted-formin)81.
Figure 4. FH2 domain structure and elongation model of formin-mediated actin polymerization.
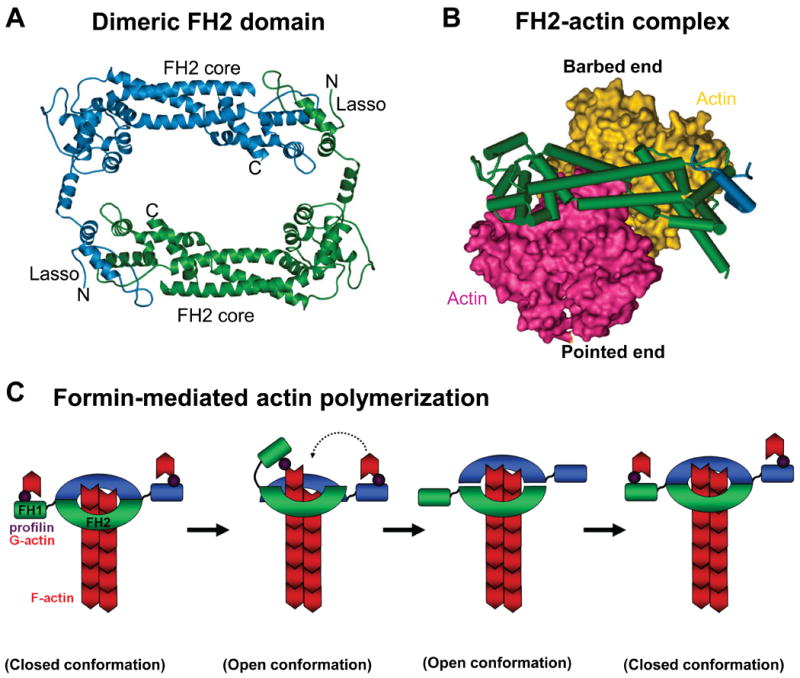
A) A ribbon diagram of the dimeric FH2 domain from S.cerevisiae Bni1 is shown (Ref.89). A “lasso” extends from the knob of one monomer and wraps around the “post” of the other monomer to stabilize this dimeric configuration. FH, formin homology.
B) The Bni1p FH2 domain wrapped around a space-filling model of an actin dimer is shown.
C) An FH2 dimer associates with the barbed end of an actin filament, while the FH1 domains recruit profilin-actin (1). The FH1 domain delivers profilin-actin to the barbed end, and this is either preceded by (Ref.92) or follows (Ref.93) the FH2 domain stepping towards the barbed end (2). The second FH2 repeats this process (3). The formin closed conformation prevents capping by other factors (4). Image in part a is modified, with permission, from Ref. 89 © (2004) Elsevier. Image in part b is modified, with permission, from Nature Ref. 92 © (2005) Macmillan publishers Ltd. All rights reserved.
Biochemical properties, structure and activation
FH2 domains are sufficient to trigger nucleation of purified actin2. In contrast to the Arp2/3 complex, which caps pointed ends, these domains bind to barbed ends and act as processive caps on elongating filaments (Fig.4C). As a result, they prevent other capping proteins from terminating elongation82-84, and compete with displacement factors at the barbed end to determine filament length85. FH2 domains are active as homodimers82,83,86,87, and mutations that disrupt dimerization abolish actin polymerization activity83,88,89. Crystal structures of the yeast Bni1 FH2 domain89 and mammalian formin FH2 domains90,91 further indicate that the two monomers are connected by flexible tethers to form a ring (Fig.4A). Moreover, a co-crystal structure of Bni1 with tetramethylrhodamine-actin revealed that each FH2 bridges two actin subunits in a configuration resembling the short-pitch actin dimer of a filament92 (Fig.4B), implying that FH2-mediated nucleation involves actin dimer stabilization.
The FH2-actin structure, together with the behavior of heterodimeric FH2 mutants, suggests a model in which the FH2 dimer exists in alternating morphologies at the barbed end (Fig.4C). In a closed conformation, both FH2 monomers bind the two terminal actin subunits, blocking further actin incorporation. In an open conformation, actin incorporation is enabled. Two potential open states have been proposed, one in which the FH2 steps toward the barbed end to allow addition of a new actin subunit92, and another in which actin addition occurs prior to FH2 stepping93. Further work will be needed to resolve how monomer addition and stepping are coordinated. Apart from the core FH2 nucleator, an adjacent proline-rich FH1 domain binds the actin monomer-delivery protein profilin. FH1-profilin interactions accelerate filament elongation84,93,94, most likely because binding of profilin-actin complexes to the FH1 domain increases the local concentration of G-actin that can be delivered directly to the barbed end95 (Fig.4C). A more refined view of formin-mediated nucleation, processive capping, and monomer addition will require solving the structures of active FH2 and FH1 domains in different conformations on an actual filament barbed end.
Importantly, the abilities of individual formins to nucleate actin, associate with barbed ends, and cooperate with profilin can vary by orders of magnitude. Sequence divergence in the FH1-FH2 module is likely a major contributor to these differences in vitro, although the molecular bases for such variation, and any resulting cellular consequences, are not well defined. Interestingly, apart from the FH1-FH2 module, mammalian formin domains are quite diverse (Fig.5A), and recent characterizations of these nucleators have expanded our understanding of their biochemical and cellular activities.
Figure 5. Different formins possess distinct domain organizations.
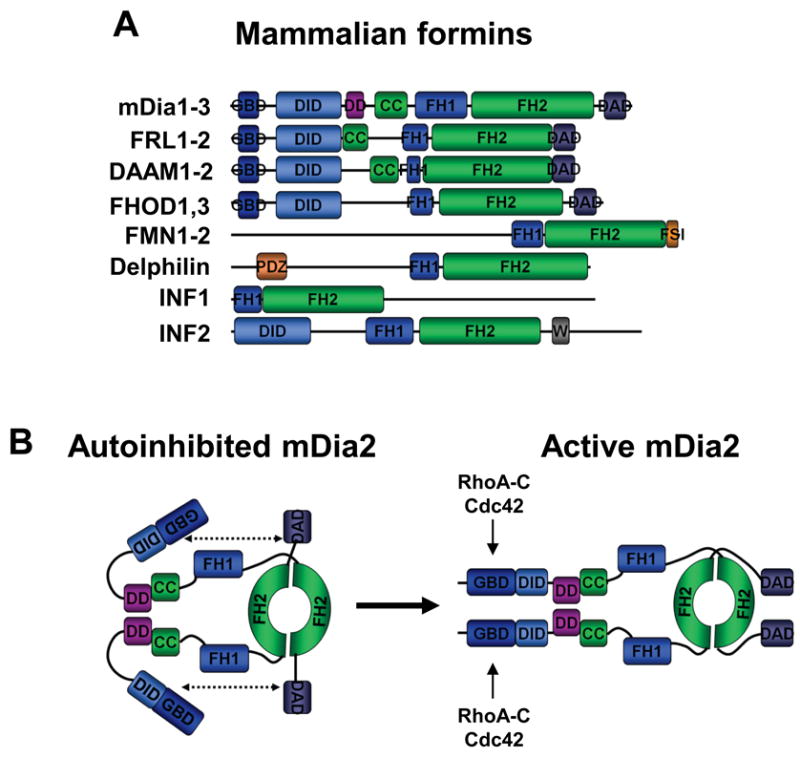
A) Mammalian formins share a conserved FH1-FH2 actin polymerizing module along with diverse regulatory motifs. The GBD-DID region of FHOD1-2 is structurally distinct from the analogous portions of other formins (non-bold typeface). CC, coiled-coil; DAD, diaphanous-autoinhibitory-domain; DID, diaphanous-inhibitory-domain; DD, dimerization-domain; FH, formin-homology; FSI, formin-spire-interaction; GBD, GTPase-binding-domain; PDZ, PSD95-DlgA-ZO1; W, WH2 domain.
B) Diaphanous-related formins like mDia2 are dimeric and regulated by autoinhibition. Their actin nucleation functions are stimulated by binding to Rho family GTPases, such as RhoA-C and Cdc42.
Diaphanous-related formins (DRFs) mDia1-3
The best characterized mammalian formins are called the DRFs based on their similarity to the diaphanous protein from D.melanogaster. In mice and other mammals, the DRFs are commonly named mDia1-3 and are thought to be widely-expressed. The gene encoding mDia1 has been knocked out, and T-cells and neutrophils from these mice exhibit impaired polarization, chemotaxis, and trafficking to secondary lymphoid organs96-98. The mice also develop age-dependent myeloproliferative defects99. Roles for other DRFs in organismal development and disease have not yet been elucidated.
The DRFs have a modular domain organization that can be divided into three functional regions (Fig.5A). Their N-termini contain regulatory sequences, including a GBD and a partially-overlapping diaphanous-inhibitory-domain (DID) that participates in autoinhibition87,100. These motifs are followed by central coiled-coil and dimerization domains (DD) that influence autoregulation. Lastly, their C-termini encompass the FH1-FH2 module and the diaphanous-autoregulatory domain (DAD) that binds the GBD-DID to inhibit the actin polymerizing activity of the FH2 domain101 (Fig.5B). This inhibitory interaction is disrupted by binding of RhoA to the GBD-DID102, and results in FH2 activation in vitro87,100. Structural comparisons of the GBD-DID bound to either a DAD87,102,103 or Rho GTPases104,105 indicate that binding of Rho and DAD is mutually exclusive, and their binding sites within the GBD-DID partially overlap. Importantly, DRF activation upon GTPase-binding in vitro is incomplete, raising the possibility that additional cellular factors are required for full activation87,100,106. The characterization of such binding-partners will be critical for fully understanding DRF regulation.
Although the aforementioned model suggests a simple linear DRF activation pathway, cellular signal transduction cascades are more complex. For example, different GTPases including RhoA-D, Cdc42, Rac, and Rif signal to multiple DRFs102,107-109. Moreover, aside from being a Rho effector, mDia1 has been proposed to act upstream of Rho, suggesting that positive feedback might control DRF and Rho activities110. Lastly, SH3 domain-containing proteins such as Src family kinases interact with DRFs, suggesting a connection with tyrosine kinase signaling, although how DRFs are integrated into these signaling pathways remains unresolved111. Despite the fact that few of these signaling pathways have been completely defined in vitro, the ultimate result of the dialogue between GTPases, kinases, and mDia1-3 in cells is actin polymerization.
DRF-mediated actin assembly participates in the formation of a remarkable variety of cellular structures (Fig.1). Key examples include stress fibers, dorsal filaments that emerge from focal adhesions at the leading edge of migrating cells, and the contractile ring that forms during cytokinesis112,113. DRFs also localize to filopodia, most prominently upon co-expression of full-length proteins with Rif or of truncated proteins missing the GBD, where their ability to nucleate actin correlates positively with the frequency of filopodia formation107,108,114,115. Multiple DRFs are also found in lamellipodia and contribute to cell motility114-117, or in phagocytic cups and participate in phagocytosis116,118. The DRFs additionally help organize cell adhesion junctions119,120, and coordinate actin assembly with endosome dynamics121. Lastly, actin polymerization triggered by DRF FH2 domains results in activation of serum response factor (SRF)-mediated transcription, an expression program that controls developmental processes which rely on cell migration, contractility, or morphogenesis111. FH2 domains from most formins share the ability to trigger the SRF response, suggesting that depletion of cytosolic G-actin is important for this process.
In addition to their role in actin assembly, mDia1-3 also have the intriguing ability to bind both to microtubules122 and to proteins that interact with microtubule plus-ends123,124 to promote microtubule stability. These results suggest that DRF-mediated crosstalk between the actin and microtubule cytoskeletons can influence cell polarity and directional migration. Much like the study of WHAMM and WASH at the interface of the actin and microtubule cytoskeletons, future DRF characterizations will undoubtedly reveal more information about communication between these two cytoskeletal systems during membrane dynamics.
FRL and DAAM proteins
Several formins are structurally related to the DRFs and are regulated similarly. These include FRL1-3 (also known as FMNL1-3) and Daam1-281, which contain GBD-DID, CC/DD, and FH1-FH2-DAD elements106,125 (Fig.5A). In addition to polymerizing actin, the FH2 domains of FRL1-3 bundle F-actin, an activity they share with mDia286,126,127. The FH1-FH2-DAD region of FRL1 also severs filaments82. The cellular regulation and function of these activities await determination.
As with the DRFs, FRL1 can assume an autoinhibited conformation mediated by DID-DAD interactions. Its actin polymerization activity can be stimulated by Cdc42 in vitro, but only at high Cdc42 concentrations106, implying that activation in cells requires additional regulatory inputs. FMNL2 is also autoinhibited, but FMNL3 seems to be constitutively active in cells even though its DID and DAD bind one another in vitro127. This suggests that DID-DAD interactions are not uniformly inhibitory, and that other layers of autoregulation exist. In support of this possibility, the Daam1 FH2 domain crystallizes in a closed conformation that occludes its actin-binding surfaces90. Daam1 is also regulated by a DID-DAD interaction, and its inhibited state can be relieved by RhoA-C binding to the GBD128 and by the phosphoprotein Dishevelled binding to the DAD125,129, implying that multiple signaling inputs are integrated to promote Daam1 activation.
Compared with mDia1-3, FRL1-3 and Daam1-2 have not been extensively characterized in cells. Nevertheless, FRL1 has been implicated in Fc-receptor-mediated phagocytosis106. In contrast, Daam1 is critical for non-canonical Wnt signaling and RhoA activation during cell polarization and gastrulation in frog embryos129. In cells, Daam1 translocates to stress fibers in response to Wnt signaling130, and affects cell shape125,130. Further investigation into the location and regulation of FRL- and Daam-mediated actin nucleation should paint a more complete picture of the cellular and organismal functions of these formins.
FMN and FHOD proteins
The founding mammalian formins, FMN1-2, along with FHOD1 and FHOD3, share some characteristics with the Dia, FRL, and Daam groups, but do not have primary sequence similarity in their N-termini81 (Fig.5A). While the regulation of FMN1-2 has not been extensively characterized, FHOD1 is autoinhibited by interactions between its C-terminal DAD and divergent N-terminus131,132, and associates with Rac via a GBD132. Interestingly, the FHOD1 GBD has a different fold than other formins, although the remainder of its N-terminus resembles a DID133. Apparently, the modular organization of formins allows incorporation of variable structural elements.
In cells, an activated FHOD1 truncation lacking its DAD promotes stress fiber formation and elongated cell morphologies131,132. The effects of FHOD1 depend on the activities of RhoA, the Rho-kinase ROCK, and possibly Rac131,134. Interestingly, FHOD1 physically interacts with ROCK and is also a ROCK substrate134,135. ROCK phosphorylates the DAD, which relieves autoinhibition in vitro and promotes FHOD1-mediated stress fiber assembly in cells135. It remains to be seen if ROCK phosphorylation directly regulates other formins.
In contrast to FHOD1, FMN1 has been reported to localize to adherens junctions, and expression of truncations disrupts junction formation136. However, analysis of cells from mice expressing GFP-FMN1-isoform-IV (FMN1-IV) suggests that this formin is cytoplasmic137. A FMN1-IV knockout also failed to reveal junction abnormalities, but uncovered defects in focal adhesion formation, lamellipodial dynamics, and migration137. Thus, the role of FMN1 requires clarification. Interestingly, FMN2 might function distinctly from FMN1. Female mice lacking FMN2 have poor fertility as a result of the inability of FMN2-deficient oocytes to position the metaphase spindle at the cell cortex during meiosis and undergo cytokinesis to form a polar body138,139. Moreover, live imaging has revealed that FMN2 organizes an actin network connected to the oocyte cortex, and that actin assembly is critical for spindle and chromosome movement to subcortical regions140,141. Whether actin polymerization itself or myosin-II motor activity drives chromosome movement is debatable. These exciting results hold promise for illuminating the roles of the actin cytoskeleton during meiosis and mitosis.
Delphilin and INF1-2
The delphilin and INF1-2 proteins do not contain significant sequence similarity to other formins outside of their FH1-FH2 module, and are the most unique formins (Fig.5A). Delphilin, which is highly-expressed in mammalian brain tissue, lacks apparent GBD, DID, or DAD regulatory regions81, and its actin polymerization activity has not been investigated. Different delphilin splice variants contain a palmitoylation site and PDZ domains, sequences that likely influence its ability to associate with cellular membranes142,143. Consistent with a role in neuronal function, mice lacking delphilin have altered synaptic plasticity144.
The INFs are present in animals and differ from one another in their domain composition. INF1 lacks GBD-DID and DAD motifs, and its actin assembly activity has not been determined. When expressed in cells, INF1 induces stress fiber formation, but the protein localizes to microtubules145. INF2, on the other hand, both nucleates actin and causes actin severing and disassembly146. The latter activities require a DAD-like peptide that doubles as a WH2 domain (Fig.5A). Interestingly, DID-WH2 interactions inhibit the depolymerizing, but not the nucleating activity, of INF2147. Unlike other formins, INF2 localizes to the ER in some cell lines, and expression of WH2 mutants causes the ER to collapse around the nucleus147, suggesting that INF2 influences membrane organization. Overall, despite many recent advances, current characterizations have likely just scratched the surface of the numerous functions that the formins perform in mammalian cells.
WH2 domain nucleators
Recently, new classes of nucleating proteins have been identified that contain a tandem cluster of three or more G-actin-binding motifs, including WH2 domains, as their signature. Such factors include the mammalian Spire, Cordon-bleu (Cobl), and Leiomodin (Lmod) families, as well as several bacterial nucleators (Fig.6A). The WH2 element is shared with NPFs and these proteins may be evolutionarily related148, but the tandem WH2 nucleators do not appear to bind the Arp2/3 complex, and their WH2 domains are proposed to cause nucleation by tethering three or more actin monomers in either a single-stranded long-pitch multimer (Fig.6B) or a short-pitch trimer. However, without high-resolution structures of native WH2-actin complexes, precisely how the WH2 motifs promote intersubunit contacts to form an actin nucleus remains unclear.
Figure 6. WH2 domain-containing actin nucleators and models for polymerization.
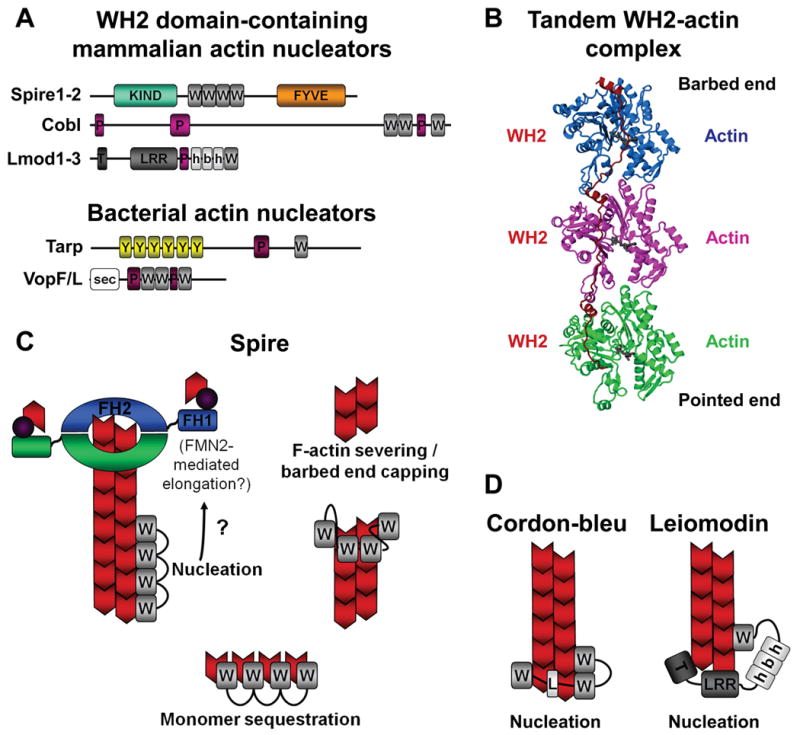
A) Members of the Spire, Cobl, and Lmod families use WH2 domains and additional actin monomer-binding sequences to nucleate and assemble unbranched actin filaments. They also contain other domains that likely regulate their nucleation activities. Bacterial pathogens also express WH2-based nucleators to induce plasma membrane remodeling. These include Tarp from Chlamydia and VopF/VopL from Vibrio. FYVE, Fab1-YOTB-Vac1-EEA1; h-b-h, helix-basic-helix; KIND, kinase noncatalytic domain; LRR, leucine-rich-repeats; P, polyproline; PRD, proline-rich-domain; sec, secretion signal; T, tropomyosin and actin-binding helices; W, WH2; Y, tyrosine-rich motifs.
B) A ribbon diagram of three tandem WH2 domains bound to actin monomers is shown (modified with permission from Ref.151). How bound actin monomers are reorganized into a conformation that favors polymerization is not yet known.
C) Spire is a versatile regulator of actin dynamics that can nucleate actin filaments (which might be further elongated by formins, as denoted by the question mark (?)), cap and sever existing filaments, and sequester monomers.
D) Cobl- and Lmod-family nucleators use multiple monomer-binding sequences to assemble trimeric actin nuclei and may remain associated with their pointed ends.
Image in part b is modified, with permission, from Ref.151© (2008) National Academy of Sciences.
Spire: a versatile regulator of actin dynamics
Spire was identified in D.melanogaster as a factor required for egg and embryo development, and was subsequently shown to nucleate actin149. While much of what is known about Spire-mediated actin dynamics comes from characterizations of the fly protein, orthologues have been identified in metazoans, including Spire1-2 in mammals. The Spire domain organization includes an N-terminal kinase-noncatalytic C-lobe domain (KIND), a central region with four WH2 motifs, and a C-terminal FYVE domain (Fig.6A).
The Spire sequence that polymerizes actin consists of the tandem WH2 domains together with an additional actin monomer-binding linker149 (Fig.6A). This region binds cooperatively to four G-actin molecules, and in an actin assembly assay it nucleates actin at low Spire:actin ratios and sequesters actin at high Spire:actin ratios149,150. Its nucleation activity is similar to that of the FH1-FH2 module of the fly formin cappuccino and less than that of activated Arp2/3149, but the activity of the full-length protein has not been defined. Maximal nucleation requires all four WH2 domains, although the third and fourth are most important. The monomer-binding linker is also critical for actin assembly, and this sequence can convert N-WASP into an Arp2/3-independent nucleator when it is introduced between the two N-WASP WH2 motifs60.
Although Spire triggers actin polymerization, its mechanism of nucleation is a matter of debate. Electron micrographs suggest that D.melanogaster Spire assembles an actin nucleus consisting of a long-pitch tetramer149, consistent with an x-ray light scattering study151, whereas kinetic analyses suggest that this might not be the case150. Moreover, initial work suggested that Spire caps pointed ends to inhibit disassembly149. However, a subsequent report indicates that human Spire interacts with barbed ends, inhibits profilin-actin assembly at these ends, and enhances pointed end disassembly by severing filaments150. Further structural and biochemical studies are needed to precisely define the configuration of the actin nucleus, the location of Spire on the filament, and which biochemical activity dominates under physiological conditions.
Importantly, Spire appears to cooperate with formins in actin assembly, as suggested by the initial observation that fly Spire and cappuccino physically interact152. Mammalian Spire1-2 and FMN1-2 also bind to one another, and the interaction is mediated by the Spire KIND153 and a formin/Spire-interaction sequence near the C-terminus of FMN1-2154. KIND binding to FMN2 or cappuccino inhibits formin-mediated actin nucleation while enhancing nucleation by the Spire WH2 cluster153. Spire also enhances actin-based motility of beads coated with mDia1-FH1-FH2150. It remains to be determined if Spire1-2 and FMN1-2 function sequentially such that Spire initiates nucleation and formins engage the barbed end to enable processive elongation using profilin-actin subunits153 (Fig.6C), or if Spire-mediated inhibition of profilin-actin assembly indirectly funnels actin to enhance formin-mediated elongation150.
Spire and FMN2 are also likely to function together in vivo. In the fly oocyte, Spire and cappuccino localize to the cortex152,153, while in mammalian cells tagged Spire variants colocalize with recycling endosome markers155, and Spire expression leads to recruitment of FMN2153. Spire might play a role in Golgi to plasma membrane transport, because this process can be inhibited by a C-terminal fragment of Spire lacking its WH2 sequences155. Lastly, Spire associates with and bundles microtubules, implying that it mediates actin-microtubule interactions152,153. Collectively, these results suggest that Spire plays important roles in membrane transport and cytoskeletal interactions, although the upstream signaling pathways that regulate these functions have yet to be fully explored.
Cordon-bleu (Cobl): Growing actin from trimeric seeds
Cobl was as identified in yeast two-hybrid screens for factors that interact with Abp1 and syndapin, proteins that promote actin polymerization in brain extracts156. Cobl appears to be a vertebrate-specific protein, and the human version does not have recognizable domains apart from three WH2 motifs near its C-terminus and multiple polyproline peptides that might bind profilin (Fig.6A).
As with Spire, the individual Cobl WH2 domains bind G-actin in vitro. Moreover, a WH2-encompassing fragment co-immunoprecipitates with actin after expression in mammalian cells156. This Cobl variant promotes nucleation of unbranched filaments at low Cobl:actin ratios, but sequesters actin at high Cobl:actin ratios. Cobl also binds to F-actin, and partially protects barbed ends from disassembly. Unlike Spire, all of the Cobl WH2s seem to be required for polymerization activity, even though the affinity of its third WH2 domain for actin is 10-fold lower than its first two. Interestingly, the length, but not the sequence, of the linker between the last two WH2 motifs is critical for actin assembly. These observations suggest that the closely-spaced WH2s create a long-pitch actin dimer, while the more distantly linked WH2 promotes lateral interactions with a third subunit, forming a trimeric configuration that favors polymerization (Fig.6D). However, it is not known how Cobl activity is modulated by other domains in the full-length protein, or if Cobl interacts with other nucleators.
The cellular functions of Cobl are just beginning to be explored. Cobl localizes throughout hippocampal neurons, where decreasing its expression suppresses neurite branching and overexpression increases the number of axonal branch points and the arborization of dendrites in an actin polymerization-dependent manner156. It will be interesting to determine if Cobl has functions in other cell types and whether its activities are influenced by Abp1 and syndapin, or by other signaling molecules.
Leiomodins: Muscle-specific F-actin makers
Lmod-2, the latest actin nucleator to be characterized157, is expressed in skeletal and cardiac muscle and has homologs in smooth muscle and perhaps other tissues (Lmod-1 and Lmod-3)158. The domain organization of Lmods is related to tropomodulins (Tmods), proteins that cap pointed ends. Similar to Tmods, the Lmod N-termini contain tropomyosin- and actin-binding helices (TMh) followed by a central leucine-rich-repeat (LRR) region that also binds actin (Fig.6A). Unlike Tmods, Lmod-2 contains an extended C-terminus featuring a polyproline peptide, two predicted helices, and a WH2 domain.
Lmod-2 potently nucleates actin filaments in vitro without affecting elongation rates at either end157. Although full nucleation activity requires elements within its N-terminus, LRR, and C-terminal extension, the combination of the LRR and C-terminus retains significant activity. Interestingly, the F-actin-binding protein tropomyosin enhances nucleation by full-length Lmod-2, but not a truncation lacking its N-terminus, implying that this portion of Lmod modulates nucleation efficiency. In the current model for Lmod-mediated nucleation, its three actin-binding motifs stabilize an actin trimer wherein the WH2 domain and C-terminal helices position the third monomer in a configuration that allows elongation from the barbed end (Fig.6D).
In cardiomyocytes, Lmod-2 localizes with tropomyosin in sarcomeres where F-actin pointed ends are positioned157. Depletion of Lmod-2 compromises sarcomere assembly and reduces cell adherence, indicating that Lmod-2 is important for sarcomere organization and cell morphology. Consistent with its nucleating ability, overexpression of Lmod-2 generates aberrant F-actin structures and disrupts sarcomere assembly. These interesting results provide a foundation for understanding the coordination of Leiomodin and tropomyosin activity during sarcomere assembly in skeletal muscle, and the function of Lmods in other cell types.
Concluding Remarks
The recent resurgence in discoveries related to actin nucleation has expanded and deepened our views of how the actin cytoskeleton influences cellular functions. We discussed the roles of nucleation factors in several genetic disorders and different aspects of mammalian development, but these proteins also influence numerous other pathogenic processes, including the invasiveness of cancer cells as well as the adhesion, invasion, and spread of microbes. Clearly, many critical questions regarding the structure and function of nucleation factors, and their potential roles in disease, have yet to be answered. For example, what do the structures of the active Arp2/3 complex, formins, and WH2 nucleators look like? How are the functions of the newly-identified proteins controlled by upstream regulators? How are actin- and microtubule-binding activities and the functions of the different nucleator classes orchestrated? It seems inevitable that continued investigations will reveal more nucleation factors and additional examples of cooperation and antagonism among the different classes of nucleators and cytoskeletal systems. Progress will yield a more comprehensive understanding of how the actin cytoskeleton operates in cells and how it contributes to human health and is altered during disease.
Acknowledgments
We thank Roberto Dominguez, Dorit Hanein, Mike Rosen, and Neils Volkmann for providing structural images and members of the Welch Lab for comments on this manuscript. We also thank investigators whose earlier work laid the foundation for the studies noted in this review, but who could not be cited due to space restrictions. KGC is supported by a Leukemia and Lymphoma Society special fellowship. MDW is supported by NIH/NIGMS RO1-GM059609 and NIH/NIAID RO1-AI074760.
Glossary
- WCA domain
The minimal sequence element required for potent activation of Arp2/3 complex-mediated actin nucleation and the formation of dendritic networks
- Lamellipodia
Sheet-like cellular protrusions that contain a dynamic Y-branched and cross-linked F-actin meshwork which elongates to drive membrane protrusion
- WASP homology 1 (WH1) domain
A regulatory domain found in WASP-like NPFs that bind proline-rich actin-binding proteins like WIP
- GTPase-binding domain (GBD)
A critical regulatory element found in some Arp2/3 complex activators and formin nucleators that promotes autoinhibitory interactions under resting conditions, but can bind to small GTPases like Cdc42, Rac, or Rho to relieve autoinhibition
- Proline-rich domain (PRD)
A domain commonly found in Arp2/3 complex activators that contains binding sites for SH3 domains and the actin monomer-associated protein profilin
- Filopodia
Finger-like cellular extensions that are composed of unbranched actin filaments which elongate to drive membrane protrusion
- Membrane ruffles
Dynamic cell surface protrusions containing a network of newly-assembled actin filaments. These can appear as dorsal circular waves or in peripheral cellular extensions
- Scar-homology-domains (SHD)
A regulatory element found in WAVE NPFs that bind multiple components of the WAVE complex
- WASH-homology-domain (WAHD)
A putative regulatory element found in WASH NPFs composed of two subdomains, WAHD1 and WAHD2 (also called tubulin-binding region (TBR))
- Retromer
A protein complex important for recycling transmembrane receptors from endosomes to the trans-Golgi
- Coiled-coil
A structural motif in proteins that consists of two or more helices that twist around each other to form a stable, rod-like structure
- Cadherin
Calcium-dependent adhesion transmembrane protein that plays important roles in cell-cell contacts
- Formin-homology (FH) domain
Domains found in formin proteins, including the conserved dimeric FH2 domain that nucleates actin, and the proline-rich FH1 domain that associates with the actin-binding protein profilin as well as SH3 and WW domains
- Diaphanous-inhibitory-domain (DID)
A regulatory region near the N-terminus of many formins that partially overlaps with a GBD and is involved in inhibiting actin nucleation activity by binding to a C-terminal DAD
- Diaphanous-autoregulatory-domain (DAD)
A short (∼20-residue) peptide located near the C-terminus of many formins that can bind to an N-terminal DID to inhibit actin nucleation activity
- Focal adhesion
A large transmembrane structure consisting of a cluster of integrins and associated proteins that binds to extracellular matrix molecules and intracellular actin stress fibers
- Wnt signaling
(Wingless and Int) A complex network of proteins with important roles in embryogenesis and signal transduction cascades involving intracellular responses to extracellular cues
- Adherens junction
A specialized intercellular junction of the plasma membrane in which cadherin molecules of adjacent cells interact with each other extracellularly in a calcium-dependent manner, and associate with actin filaments inside cells
- PDZ domain
(Postsynaptic-density protein, Discs large, and Zona occludens-1). A domain present in many scaffolding proteins that binds to specific short amino-acid sequences
- FYVE domain
A type of zinc-finger domain that binds phosphatidylinositol-3-phosphate and is found in many proteins involved in membrane trafficking
- Neurite
A projection (e.g., an axon or dendrite) from the cell body of a neuron
- Leucine-rich-repeat (LRR)
A structural motif that participates in protein-protein interactions and is comprised of repeating 20-30 residue peptides rich in leucine
- Sarcomere
A specialized structure in striated muscle composed of actin, myosin, and other proteins that serve as the smallest unit that generates contractile force
References
- 1.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nature Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 2.Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nature Rev Mol Cell Biol. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- 3.Stradal TE, Scita G. Protein complexes regulating Arp2/3-mediated actin assembly. Curr Opin Cell Biol. 2006;18:4–10. doi: 10.1016/j.ceb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 5.Zencheck WD, et al. Nucleotide- and activator-dependent structural and dynamic changes of arp2/3 complex monitored by hydrogen/deuterium exchange and mass spectrometry. J Mol Biol. 2009;390:414–427. doi: 10.1016/j.jmb.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolen BJ, Littlefield RS, Pollard TD. Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP. Proc Natl Acad Sci USA. 2004;101:15627–15632. doi: 10.1073/pnas.0407149101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin AC, et al. Effects of Arp2 and Arp3 nucleotide-binding pocket mutations on Arp2/3 complex function. J Cell Biol. 2005;168:315–328. doi: 10.1083/jcb.200408177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dayel MJ, Mullins RD. Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2004;2:E91. doi: 10.1371/journal.pbio.0020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Clainche C, Pantaloni D, Carlier MF. ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc Natl Acad Sci USA. 2003;100:6337–6342. doi: 10.1073/pnas.1130513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin AC, Welch MD, Drubin DG. Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nature Cell Biol. 2006;8:826–833. doi: 10.1038/ncb1443. [DOI] [PubMed] [Google Scholar]
- 11.Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltzner CC, Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J Mol Biol. 2004;336:551–565. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Rouiller I, et al. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper provides the highest resolution model of the Arp2/3 complex in filament branches.
- 14.LeClaire LL, 3rd, Baumgartner M, Iwasa JH, Mullins RD, Barber DL. Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments. J Cell Biol. 2008;182:647–654. doi: 10.1083/jcb.200802145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodal AA, et al. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nature Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- 16.Kreishman-Deitrick M, et al. NMR analyses of the activation of the Arp2/3 complex by neuronal Wiskott-Aldrich syndrome protein. Biochemistry. 2005;44:15247–15256. doi: 10.1021/bi051065n. [DOI] [PubMed] [Google Scholar]
- 17.Kelly AE, Kranitz H, Dotsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boczkowska M, et al. X-Ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hertzog M, et al. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- 20.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bosticardo M, Marangoni F, Aiuti A, Villa A, Grazia Roncarolo M. Recent advances in understanding the pathophysiology of Wiskott-Aldrich syndrome. Blood. 2009;113:6288–6295. doi: 10.1182/blood-2008-12-115253. [DOI] [PubMed] [Google Scholar]
- 22.Snapper SB, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nature Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- 23.Lommel S, et al. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2001;2:850–857. doi: 10.1093/embo-reports/kve197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anton IM, Jones GE, Wandosell F, Geha R, Ramesh N. WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol. 2007;17:555–562. doi: 10.1016/j.tcb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Ho HY, et al. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 26.Campellone KG, et al. Repetitive N-WASP-binding elements of the enterohemorrhagic Escherichia coli effector EspF(U) synergistically activate actin assembly. PLoS Pathog. 2008;4:e1000191. doi: 10.1371/journal.ppat.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J. 2008;27:2817–28. doi: 10.1038/emboj.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomasevic N, et al. Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry. 2007;46:3494–3502. doi: 10.1021/bi062152y. [DOI] [PubMed] [Google Scholar]
- 29.Innocenti M, et al. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nature Cell Biol. 2005;7:969–976. doi: 10.1038/ncb1304. [DOI] [PubMed] [Google Scholar]
- 30.Torres E, Rosen MK. Protein-tyrosine kinase and GTPase signals cooperate to phosphorylate and activate Wiskott-Aldrich syndrome protein (WASP)/neuronal WASP. J Biol Chem. 2006;281:3513–3520. doi: 10.1074/jbc.M509416200. [DOI] [PubMed] [Google Scholar]
- 31.Padrick SB, et al. Hierarchical regulation of WASP/WAVE proteins. Mol Cell. 2008;32:426–438. doi: 10.1016/j.molcel.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes allosteric and new multimerization-based mechanisms for activating NPFs.
- 32.Sallee NA, et al. The pathogen protein EspF(U) hijacks actin polymerization using mimicry and multivalency. Nature. 2008;454:1005–1008. doi: 10.1038/nature07170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bu W, Chou AM, Lim KB, Sudhaharan T, Ahmed S. The Toca-1-N-WASP complex links filopodial formation to endocytosis. J Biol Chem. 2009;284:11622–11636. doi: 10.1074/jbc.M805940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Legg JA, et al. N-WASP involvement in dorsal ruffle formation in mouse embryonic fibroblasts. Mol Biol Cell. 2007;18:678–687. doi: 10.1091/mbc.E06-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsujita K, et al. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benesch S, et al. N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. J Cell Sci. 2005;118:3103–3115. doi: 10.1242/jcs.02444. [DOI] [PubMed] [Google Scholar]
- 37.Dahl JP, et al. Characterization of the WAVE1 knock-out mouse: implications for CNS development. J Neurosci. 2003;23:3343–3352. doi: 10.1523/JNEUROSCI.23-08-03343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soderling SH, et al. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamazaki D, et al. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- 40.Yan C, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- 42.Gautreau A, et al. Purification and architecture of the ubiquitous Wave complex. Proc Natl Acad Sci USA. 2004;101:4379–4383. doi: 10.1073/pnas.0400628101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nature Struct Mol Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derivery E, Lombard B, Loew D, Gautreau A. The Wave complex is intrinsically inactive. Cell Motil Cytoskeleton. 2009;66:777–790. doi: 10.1002/cm.20342. [DOI] [PubMed] [Google Scholar]
- 45.Innocenti M, et al. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nature Cell Biol. 2004;6:319–327. doi: 10.1038/ncb1105. [DOI] [PubMed] [Google Scholar]
- 46.Steffen A, et al. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 2004;23:749–759. doi: 10.1038/sj.emboj.7600084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suetsugu S, et al. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oikawa T, et al. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nature Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- 49.Leng Y, et al. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc Natl Acad Sci USA. 2005;102:1098–1103. doi: 10.1073/pnas.0409120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell. 2003;5:595–609. doi: 10.1016/s1534-5807(03)00297-1. [DOI] [PubMed] [Google Scholar]
- 51.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that Rac, PI(3,4,5)P3, and phosphorylation cooperate during activation of the WAVE complex.
- 52.Steffen A, et al. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamazaki D, Oikawa T, Takenawa T. Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J Cell Sci. 2007;120:86–100. doi: 10.1242/jcs.03311. [DOI] [PubMed] [Google Scholar]
- 54.Linardopoulou EV, et al. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Derivery E, et al. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]; This paper suggests that WASH plays a role in endosome recycling to the plasma membrane.
- 56.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell. 2009;17:699–711. doi: 10.1016/j.devcel.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper indicates that WASH functions in endosome trafficking to the trans-Golgi network.
- 57.Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton. 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu R, et al. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development. 2009;136:2849–2860. doi: 10.1242/dev.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper begins to characterize the biochemical activities and cellular roles of the Drosophila WASH NPF.
- 59.Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies WHAMM as an NPF that interacts with actin and microtubules to modulate membrane dynamics and transport.
- 60.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nature Cell Biol. 2009;11:451–459. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies Jmy as both an NPF and an actin nucleator that shuttles between the nucleus and plasma membrane.
- 61.Coutts AS, Weston L, La Thangue NB. A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc Natl Acad Sci USA. 2009;106:19872–19877. doi: 10.1073/pnas.0906785106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil Cytoskeleton. 2008;65:687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol Cell Biol. 2004;24:5269–5280. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kowalski JR, et al. Cortactin regulates cell migration through activation of N-WASP. J Cell Sci. 2005;118:79–87. doi: 10.1242/jcs.01586. [DOI] [PubMed] [Google Scholar]
- 65.Tehrani S, Tomasevic N, Weed S, Sakowicz R, Cooper JA. Src phosphorylation of cortactin enhances actin assembly. Proc Natl Acad Sci USA. 2007;104:11933–11938. doi: 10.1073/pnas.0701077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao H, et al. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nature Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- 67.Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 68.Zhu J, et al. Regulation of cortactin/dynamin interaction by actin polymerization during the fission of clathrin-coated pits. J Cell Sci. 2005;118:807–817. doi: 10.1242/jcs.01668. [DOI] [PubMed] [Google Scholar]
- 69.Helwani FM, et al. Cortactin is necessary for E-cadherin-mediated contact formation and actin reorganization. J Cell Biol. 2004;164:899–910. doi: 10.1083/jcb.200309034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boguslavsky S, et al. p120 catenin regulates lamellipodial dynamics and cell adhesion in cooperation with cortactin. Proc Natl Acad Sci USA. 2007;104:10882–10887. doi: 10.1073/pnas.0702731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bryce NS, et al. Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr Biol. 2005;15:1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 72.Kempiak SJ, et al. A neural Wiskott-Aldrich Syndrome protein-mediated pathway for localized activation of actin polymerization that is regulated by cortactin. J Biol Chem. 2005;280:5836–5842. doi: 10.1074/jbc.M410713200. [DOI] [PubMed] [Google Scholar]
- 73.Lai FP, et al. Cortactin promotes migration and platelet-derived growth factor-induced actin reorganization by signaling to Rho-GTPases. Mol Biol Cell. 2009;20:3209–3223. doi: 10.1091/mbc.E08-12-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Humphries CL, et al. Direct regulation of Arp2/3 complex activity and function by the actin binding protein coronin. J Cell Biol. 2002;159:993–1004. doi: 10.1083/jcb.200206113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai L, Marshall TW, Uetrecht AC, Schafer DA, Bear JE. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 2007;128:915–929. doi: 10.1016/j.cell.2007.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brieher WM, Kueh HY, Ballif BA, Mitchison TJ. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J Cell Biol. 2006;175:315–324. doi: 10.1083/jcb.200603149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kueh HY, Charras GT, Mitchison TJ, Brieher WM. Actin disassembly by cofilin, coronin, and Aip1 occurs in bursts and is inhibited by barbed-end cappers. J Cell Biol. 2008;182:341–353. doi: 10.1083/jcb.200801027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gandhi M, Achard V, Blanchoin L, Goode BL. Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol Cell. 2009;34:364–374. doi: 10.1016/j.molcel.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper explains the paradoxical ability of coronin to both antagonize and synergize with cofilin.
- 79.Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cai L, Makhov AM, Schafer DA, Bear JE. Coronin 1B antagonizes cortactin and remodels Arp2/3-containing actin branches in lamellipodia. Cell. 2008;134:828–842. doi: 10.1016/j.cell.2008.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the competing activities of cortactin and coronin during the dynamics of Arp2/3-containing branches.
- 81.Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Harris ES, Li F, Higgs HN. The mouse formin, FRLalpha, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem. 2004;279:20076–20087. doi: 10.1074/jbc.M312718200. [DOI] [PubMed] [Google Scholar]
- 83.Moseley JB, et al. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Romero S, et al. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 85.Chesarone M, Gould CJ, Moseley JB, Goode BL. Displacement of formins from growing barbed ends by bud14 is critical for actin cable architecture and function. Dev Cell. 2009;16:292–302. doi: 10.1016/j.devcel.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harris ES, Rouiller I, Hanein D, Higgs HN. Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J Biol Chem. 2006;281:14383–14392. doi: 10.1074/jbc.M510923200. [DOI] [PubMed] [Google Scholar]
- 87.Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- 88.Copeland JW, Copeland SJ, Treisman R. Homo-oligomerization is essential for F-actin assembly by the formin family FH2 domain. J Biol Chem. 2004;279:50250–50256. doi: 10.1074/jbc.M404429200. [DOI] [PubMed] [Google Scholar]
- 89.Xu Y, et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 90.Lu J, et al. Structure of the FH2 domain of Daam1: implications for formin regulation of actin assembly. J Mol Biol. 2007;369:1258–69. doi: 10.1016/j.jmb.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shimada A, et al. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell. 2004;13:511–522. doi: 10.1016/s1097-2765(04)00059-0. [DOI] [PubMed] [Google Scholar]
- 92.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 93.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 95.Vavylonis D, Kovar DR, O'Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eisenmann KM, et al. T cell responses in mammalian diaphanous-related formin mDia1 knock-out mice. J Biol Chem. 2007;282:25152–25158. doi: 10.1074/jbc.M703243200. [DOI] [PubMed] [Google Scholar]
- 97.Sakata D, et al. Impaired T lymphocyte trafficking in mice deficient in an actin-nucleating protein, mDia1. J Exp Med. 2007;204:2031–2038. doi: 10.1084/jem.20062647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shi Y, et al. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol. 2009;182:3837–3845. doi: 10.4049/jimmunol.0803838. [DOI] [PubMed] [Google Scholar]
- 99.Peng J, et al. Myeloproliferative defects following targeting of the Drf1 gene encoding the mammalian diaphanous related formin mDia1. Cancer Res. 2007;67:7565–7571. doi: 10.1158/0008-5472.CAN-07-1467. [DOI] [PubMed] [Google Scholar]
- 100.Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- 101.Wallar BJ, et al. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J Biol Chem. 2006;281:4300–4307. doi: 10.1074/jbc.M510277200. [DOI] [PubMed] [Google Scholar]
- 102.Lammers M, Rose R, Scrima A, Wittinghofer A. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 2005;24:4176–4187. doi: 10.1038/sj.emboj.7600879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nezami AG, Poy F, Eck MJ. Structure of the autoinhibitory switch in formin mDia1. Structure. 2006;14:257–263. doi: 10.1016/j.str.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 104.Otomo T, Otomo C, Tomchick DR, Machius M, Rosen MK. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell. 2005;18:273–281. doi: 10.1016/j.molcel.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 105.Rose R, et al. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature. 2005;435:513–518. doi: 10.1038/nature03604. [DOI] [PubMed] [Google Scholar]
- 106.Seth A, Otomo C, Rosen MK. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol. 2006;174:701–713. doi: 10.1083/jcb.200605006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Peng J, Wallar BJ, Flanders A, Swiatek PJ, Alberts AS. Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr Biol. 2003;13:534–545. doi: 10.1016/s0960-9822(03)00170-2. [DOI] [PubMed] [Google Scholar]
- 108.Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 109.Lammers M, Meyer S, Kuhlmann D, Wittinghofer A. Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem. 2008;283:35236–35246. doi: 10.1074/jbc.M805634200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kitzing TM, et al. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21:1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Young KG, Copeland JW. Formins in cell signaling. Biochim Biophys Acta. 2008 doi: 10.1016/j.bbamcr.2008.09.017. In press. [DOI] [PubMed] [Google Scholar]
- 112.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Watanabe S, et al. mDia2 induces the actin scaffold for the contractile ring and stabilizes its position during cytokinesis in NIH 3T3 cells. Mol Biol Cell. 2008;19:2328–2338. doi: 10.1091/mbc.E07-10-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sarmiento C, et al. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol. 2008;180:1245–1260. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang C, et al. Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 2007;5:e317. doi: 10.1371/journal.pbio.0050317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brandt DT, et al. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gupton SL, Eisenmann K, Alberts AS, Waterman-Storer CM. mDia2 regulates actin and focal adhesion dynamics and organization in the lamella for efficient epithelial cell migration. J Cell Sci. 2007;120:3475–3487. doi: 10.1242/jcs.006049. [DOI] [PubMed] [Google Scholar]
- 118.Colucci-Guyon E, et al. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr Biol. 2005;15:2007–2012. doi: 10.1016/j.cub.2005.09.051. [DOI] [PubMed] [Google Scholar]
- 119.Carramusa L, Ballestrem C, Zilberman Y, Bershadsky AD. Mammalian diaphanous-related formin Dia1 controls the organization of E-cadherin-mediated cell-cell junctions. J Cell Sci. 2007;120:3870–3882. doi: 10.1242/jcs.014365. [DOI] [PubMed] [Google Scholar]
- 120.Ryu JR, Echarri A, Li R, Pendergast AM. Regulation of cell-cell adhesion by Abi/Diaphanous complexes. Mol Cell Biol. 2009;29:1735–1748. doi: 10.1128/MCB.01483-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gasman S, Kalaidzidis Y, Zerial M. RhoD regulates endosome dynamics through Diaphanous-related formin and Src tyrosine kinase. Nature Cell Biol. 2003;5:195–204. doi: 10.1038/ncb935. [DOI] [PubMed] [Google Scholar]
- 122.Bartolini F, et al. The formin mDia2 stabilizes microtubules independently of its actin nucleation activity. J Cell Biol. 2008;181:523–536. doi: 10.1083/jcb.200709029. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that mDia2 formin can directly stabilize microtubules.
- 123.Wen Y, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nature Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 124.Lewkowicz E, et al. The microtubule-binding protein CLIP-170 coordinates mDia1 and actin reorganization during CR3-mediated phagocytosis. J Cell Biol. 2008;183:1287–1298. doi: 10.1083/jcb.200807023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu W, et al. Mechanism of activation of the Formin protein Daam1. Proc Natl Acad Sci USA. 2008;105:210–215. doi: 10.1073/pnas.0707277105. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the importance of the Dishevelled protein in regulating Daam1 activity.
- 126.Esue O, Harris ES, Higgs HN, Wirtz D. The filamentous actin cross-linking/bundling activity of mammalian formins. J Mol Biol. 2008;384:324–334. doi: 10.1016/j.jmb.2008.09.043. [DOI] [PubMed] [Google Scholar]
- 127.Vaillant DC, et al. Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity. J Biol Chem. 2008;283:33750–33762. doi: 10.1074/jbc.M803156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Higashi T, et al. Biochemical characterization of the Rho GTPase-regulated actin assembly by diaphanous-related formins, mDia1 and Daam1, in platelets. J Biol Chem. 2008;283:8746–8755. doi: 10.1074/jbc.M707839200. [DOI] [PubMed] [Google Scholar]
- 129.Habas R, Kato Y, He X. Wnt/Frizzled activation of Rho regulates vertebrate gastrulation and requires a novel Formin homology protein Daam1. Cell. 2001;107:843–854. doi: 10.1016/s0092-8674(01)00614-6. [DOI] [PubMed] [Google Scholar]
- 130.Sato A, et al. Profilin is an effector for Daam1 in non-canonical Wnt signaling and is required for vertebrate gastrulation. Development. 2006;133:4219–4231. doi: 10.1242/dev.02590. [DOI] [PubMed] [Google Scholar]
- 131.Koka S, et al. The formin-homology-domain-containing protein FHOD1 enhances cell migration. J Cell Sci. 2003;116:1745–1755. doi: 10.1242/jcs.00386. [DOI] [PubMed] [Google Scholar]
- 132.Schonichen A, et al. Biochemical characterization of the diaphanous autoregulatory interaction in the formin homology protein FHOD1. J Biol Chem. 2006;281:5084–5093. doi: 10.1074/jbc.M509226200. [DOI] [PubMed] [Google Scholar]
- 133.Schulte A, et al. The human formin FHOD1 contains a bipartite structure of FH3 and GTPase-binding domains required for activation. Structure. 2008;16:1313–1323. doi: 10.1016/j.str.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 134.Hannemann S, et al. The Diaphanous-related Formin FHOD1 associates with ROCK1 and promotes Src-dependent plasma membrane blebbing. J Biol Chem. 2008;283:27891–27903. doi: 10.1074/jbc.M801800200. [DOI] [PubMed] [Google Scholar]
- 135.Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 2008;27:618–628. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper defines a ROCK-mediated mechanism of FHOD1 regulation.
- 136.Kobielak A, Pasolli HA, Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nature Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dettenhofer M, Zhou F, Leder P. Formin 1-isoform IV deficient cells exhibit defects in cell spreading and focal adhesion formation. PLoS One. 2008;3:e2497. doi: 10.1371/journal.pone.0002497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Leader B, et al. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nature Cell Biol. 2002;4:921–928. doi: 10.1038/ncb880. [DOI] [PubMed] [Google Scholar]
- 139.Dumont J, et al. Formin-2 is required for spindle migration and for the late steps of cytokinesis in mouse oocytes. Dev Biol. 2007;301:254–265. doi: 10.1016/j.ydbio.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 140.Li H, Guo F, Rubinstein B, Li R. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nature Cell Biol. 2008;10:1301–1308. doi: 10.1038/ncb1788. [DOI] [PubMed] [Google Scholar]; This paper, along with Ref.141 implicates FMN2 in controlling spindle movements in mammalian oocytes.
- 141.Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18:1986–1992. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]; This paper, along with Ref.140 implicates FMN2 in controlling spindle movements in mammalian oocytes.
- 142.Miyagi Y, et al. Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J Neurosci. 2002;22:803–814. doi: 10.1523/JNEUROSCI.22-03-00803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Matsuda K, Matsuda S, Gladding CM, Yuzaki M. Characterization of the delta2 glutamate receptor-binding protein delphilin: Splicing variants with differential palmitoylation and an additional PDZ domain. J Biol Chem. 2006;281:25577–25587. doi: 10.1074/jbc.M602044200. [DOI] [PubMed] [Google Scholar]
- 144.Takeuchi T, et al. Enhancement of both long-term depression induction and optokinetic response adaptation in mice lacking delphilin. PLoS One. 2008;3:e2297. doi: 10.1371/journal.pone.0002297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Young KG, Thurston SF, Copeland S, Smallwood C, Copeland JW. INF1 is a novel microtubule-associated formin. Mol Biol Cell. 2008;19:5168–5180. doi: 10.1091/mbc.E08-05-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chhabra ES, Higgs HN. INF2 Is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem. 2006;281:26754–26767. doi: 10.1074/jbc.M604666200. [DOI] [PubMed] [Google Scholar]
- 147.Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN. INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci. 2009;122:1430–1440. doi: 10.1242/jcs.040691. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper suggests that the formin INF2 can associate with the ER and influences its morphology.
- 148.Dominguez R. Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol. 2009;44:351–366. doi: 10.3109/10409230903277340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 150.Bosch M, et al. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]; This paper provides a detailed biochemical analysis of the numerous effects of Spire on actin dynamics.
- 151.Rebowski G, et al. X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc Natl Acad Sci USA. 2008;105:10785–10790. doi: 10.1073/pnas.0801650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rosales-Nieves AE, et al. Coordination of microtubule and microfilament dynamics by Drosophila Rho1, Spire and Cappuccino. Nature Cell Biol. 2006;8:367–76. doi: 10.1038/ncb1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–128. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrates that Spire binds directly to and influences the actin nucleating activity of the formin Cappuccino.
- 154.Pechlivanis M, Samol A, Kerkhoff E. Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J Biol Chem. 2009;284:25324–25333. doi: 10.1074/jbc.M109.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kerkhoff E, et al. The Spir actin organizers are involved in vesicle transport processes. Curr Biol. 2001;11:1963–1968. doi: 10.1016/s0960-9822(01)00602-9. [DOI] [PubMed] [Google Scholar]
- 156.Ahuja R, et al. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies Cobl as a nucleator that regulates actin dynamics and dendrite branching in neurons.
- 157.Chereau D, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper identifies Lmod-2 as a potent actin nucleator that influences sarcomere assembly in muscle cells.
- 158.Conley CA, Fritz-Six KL, Almenar-Queralt A, Fowler VM. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73:127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 159.Thanbichler M, Shapiro L. Getting organized--how bacterial cells move proteins and DNA. Nature Rev Microbiol. 2008;6:28–40. doi: 10.1038/nrmicro1795. [DOI] [PubMed] [Google Scholar]